Highlights
-
•
Complex I from E. coli is a membrane-bound enzyme comprising 13 subunits.
-
•
Subcomplexes were identified by expressing subsets of the 13 genes.
-
•
Assembly was negatively impacted by dividing the genes between 2 plasmids.
-
•
Assembly was most efficient when subunit L (ND5) was expressed alone, after a time-delay.
Keywords: Complex I; Protein assembly; Membrane protein; Bioenergetics; NADH dehydrogenase; Native gel electrophoresis; Abbreviations: ACMA, 9-amino 3-chloro 2-methoxy acridine; BA14, nuoA-N deletion strain in E. coli; Dodecyl maltoside, n-dodecyl-(-D-maltopyranoside; FCCP, carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone; dNADH, deamino-NADH
Abstract
Respiratory Complex I, a multi-subunit, membrane-bound enzyme, oxidizes NADH in the electron transport chains of mammalian mitochondria, and many bacterial species. We have examined in vivo assembly of the membrane subunits of Complex I from E. coli. Complexes of J-K, L-M, M-N, and J-K-L-M-N were observed by both native gel electrophoresis and co-immunoprecipitation, when subsets of the genes were expressed. Subunit L (ND5 in humans), the most distal membrane subunit, with an unusual extended C-terminal segment, did not join with M-N, and but could join with J-K-M-N. When the genes were split between two plasmids, with L, M, and N subunits expressed in various combinations from one plasmid, the resulting enzyme activity in membrane vesicles dropped to 19–60% relative to expression from the whole operon encoded on one plasmid. When L was expressed after a time-delay, rather than simultaneously, the activity increased from 28% to 100%, indicating that it can efficiently join a pre-formed complex lacking L. In contrast, when larger groups of membrane subunits were expressed last, LMN or JKLMN, assembly was much less efficient. The two-plasmid expression system was used to re-analyze C-terminal mutations in subunit K (ND4L), which occur near the overlapping nuoK and L genes. These mutations were found to disrupt assembly, indicating the importance of the junction of L, N and K subunits. The results highlight the temporal and spatial aspects of gene expression that allow efficient assembly of the membrane subunits of Complex I.
Introduction
Complex I, or NADH:ubiquinone oxidoreductase, is the first member of the respiratory chain in mammalian mitochondria, and in many bacteria (for recent reviews see [1], [2], [3], [4], [5], [6]). It is a multi-subunit enzyme with an overall L-shape, in which one arm is membrane-bound, and one arm is peripheral. The peripheral arm contains a flavin mononucleotide (FMN) that is directly reduced by NADH. The ubiquinone binding region is near the junction of the two arms, and is connected to the flavin by a series of seven FeS clusters that transmit electrons to the ubiquinone. The membrane arm translocates four hydrogen ions across the membrane per NADH by mechanisms not totally clear [7], [8], [9], [10], [11], [12], [13], [14]. One site of proton translocation appears to be near the quinone binding region, while the remaining three protons are likely translocated by three similar subunits, arranged linearly, that are related to bacterial antiporter proteins [15]. The reduced quinol is then available for downstream respiratory chain reactions, while the oxidized NAD+ is reduced by TCA cycle enzymes, or consumed in other reactions.
The subunit composition of Complex I varies according to species, with the mammalian enzyme containing 45 subunits, 43 unique and one in two copies [16, 17]. All species appear to contain a core group of subunits homologous to fourteen of the mammalian subunits. Subunits beyond the core are called supernumerary, or accessory, subunits [18], [19], [20]. In E. coli [21, 22], Complex I contains thirteen subunits nuoA-N, because one subunit is the result of a gene fusion of two core subunits, nuoCD. The peripheral arm is composed of two modules: The N-module is formed by subunits E, F, and G, in which subunit F binds the flavin and one FeS cluster, and subunit G binds 4 FeS clusters, one of which is not part of the electron transport chain. The Q-module comprises subunits B, CD, and I, and these subunits interact with the membrane arm subunits H and A. The remaining membrane subunits include L, M and N, the three antiporter-like subunits, and two smaller subunits, J and K, which form the interface between H and A and the other three. The subunits appear to come from three different evolutionary paths, which correspond to modules: an N-module (E, F, G), a Q-module (B, CD, I, A, H) and a P-module (J, K, L, M, N)
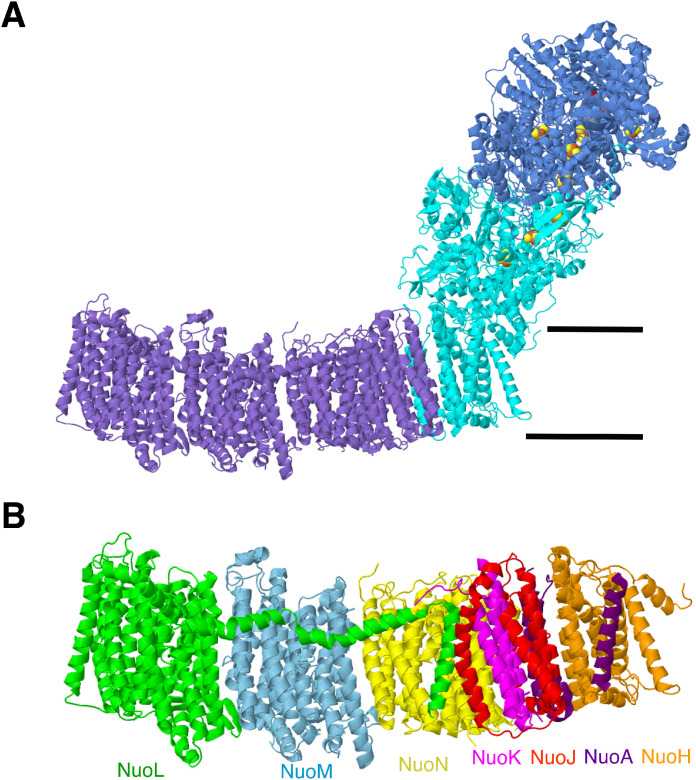
The assembly process of Complex I, as a large, multi-subunit, membrane-bound enzyme, is a question of fundamental interest, and one that can be addressed because the structure is known for various bacterial [23, 24], fungal [14, 25, 26] and mammalian species [27], [28], [29], [30], [31]. The bacterial enzyme provides a special opportunity for analysis of the assembly of a multi-subunit enzyme because of the relative simplicity of its gene expression from a single operon. The structure of a complete bacterial Complex I from Escherichia coli is shown in Fig. 1A, and the membrane arm of Complex I from E. coli is shown in Fig. 1B. Assembly of mammalian and plant Complex I has been addressed by studies in which individual genes were knocked out, and the resulting complexes analyzed [18, [32], [33], [34]]. Complexome analysis has also been applied to studies in which mitochondrial translation is shut down, and then reactivated [35, 36]. After the reactivation, complexes are isolated and analyzed by mass spectrometry to determine which subunits are present. The mammalian enzymes differ from the bacterial ones, by having many supernumerary subunits, and also, many assembly factors which bind the core subunits at different stages. The bacterial enzyme allows the possibility of expressing groups of genes in order to determine if various combinations of subunits can form subcomplexes, and how that relates to complete assembly of Complex I.
Fig. 1.
Structures of bacterial Complex I. A. Complex I from Escherichia coli. (pdb id=7nyr) [40]. The N-module, including E, F, and G subunits, is colored dark blue. The Q-module including subunits B, CD, I, H and A, is colored cyan. The P-module is colored purple, and includes subunit J, K, L, M, and N. Iron sulfur clusters and FMN are shown in spacefilling view. B. The membrane arm from E. coli. (pdb id=7nyh) [40]. Each subunit is colored differently and labeled with corresponding color.
In a previous analysis of Complex I from E. coli, the functional assembly of E-F-G has been detected by plasmid-derived over-expression, but only if subunits B and CD were also expressed [37]. Similar studies have not addressed assembly of any of the membrane subunits. Alternatively, chromosomal gene knockouts have been analyzed, but in each case, little or no membrane-bound Complex I was detected, indicating a low yield of assembly [38]. In the case of the nuoL deletion, a small amount of membrane-extracted material with activity was detected after sucrose gradient centrifugation, suggesting formation of a complex that lacked only subunit L. When the nuo operon was over-expressed from a pBAD plasmid, it was possible to isolate an assembled Complex I lacking subunit L or containing a truncated subunit L [39]. These forms of the enzyme retained considerable NADH:decylubiquinone activity, and about 50% of the proton translocation activity. That indicates that Complex I can assemble without subunit L, in a functional state, but that over-expression is necessary to drive the assembly.
Here, we have systematically examined whether groups of subunits from the membrane arm can form complexes, in the absence of the rest of Complex I subunits, using over-expression vectors. A time-delayed expression [41] of membrane subunits was used to gain insight into the unique role of subunit L in assembly.
Materials and methods
Materials
Restriction endonucleases, T4 DNA ligase, Q5 Site-Directed Mutagenesis Kit, Q5 High-Fidelity PCR Kit, and Monarch PCR Cleanup Kit were from New England BioLabs (Beverly, MA). DNA Miniprep kits were purchased from Qiagen (Valencia, CA) and ThermoFisher Scientific (Pittsburgh, PA). The QIAquick and Qia IIX Gel Extraction Kits were from Qiagen (Germantown, MD). Immunoblot PVDF (polyvinylidene difluoride), Mini-PROTEAN TGX gels (12%; 4–15%), Precision Plus Protein ™ Dual Color standards, Precision Plus Protein ™ unstained standards and the DC protein assay kit were from Bio-Rad (Hercules, CA). 9-Amino-6-chloro-2-methoxyacridine (ACMA) and NativeMark™ Unstained Protein Standards were from Invitrogen. The polyclonal antibodies against E. coli Complex I subunits L and M were prepared by Affinity BioReagents (Golden, CO). These antibodies were raised in rabbits against peptide QTYSQPLWTWMSVGD (corresponding to residues 58–72) in subunit L and peptide GKAKSQIASQELPGM (corresponding to residues 446–460) in subunit M. Polyclonal antibodies raised in rabbits against C-terminal oligopeptides from subunits J (residues DSAKRKTEEHA)(Kao et al. 2005a) and K (residues LNIDSVSEMRG) (Kao et al. 2005b), and from purified subunit CD (Kao et al. 2004) were a generous gift from T. Yagi and A. Matsuno-Yagi (Scripps Research Institute, La Jolla, CA USA). Subunit N was detected by the monoclonal mouse HA-probe (F7), from Santa Cruz Biotechnology (Dallas, TX), as were Goat anti-rabbit and Goat anti-mouse IgG-HRP, IgG (mouse monoclonal IgG1), and Protein A/G PLUS-Agarose. SuperSignal West Dura Extended Duration Substrate and p-nitro blue tetrazolium chloride (NBT) were from ThermoFisher Scientific (Pittsburgh, PA). n-dodecyl-β-D-maltopyranoside was from Anatrace (Maumee, OH). Other chemicals, including NADH, 6-aminocaproic acid, L-arabinose, D-fucose, glucose, Coomassie Brilliant Blue G, Protease Inhibitor Cocktail (P8465), Bis-Tris, deamino-NADH, FCCP, and IPTG were from MilliporeSigma (Burlington, MA). Oligonucleotides for plasmid construction, mutagenesis, and sequencing were synthesized by Eurofins Genomics (Louisville, KY). DNA sequencing was performed by Lone Star Labs. (Houston, TX).
Plasmids and strains
Plasmid pBA400 (18.2 kb), from pACYC184, CmR, contains a full length nuo operon and was used as the PCR template for plasmid construction [42]. pBAD33 (5.35 kb) and pET22b (5.49 kb) were used for construction of plasmids containing the nuo genes. Plasmid pT7Pol26(12.3 kb) was used to control the expression of the genes in pET22b. These 3 plasmids were generously provided by Dr. Gabriele Deckers-Hebestreit, Universität Osnabrück, Germany. For plasmid construction and propagation, XL1-Blue (recA, endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac {F′ proAB lacIq ZΔM15 Tn10 (TetR)}) was used. For transformation of ligation products from large fragments, forming plasmids greater than 15 kb, 10-Beta cells, a derivative of DH10B from New England Biolabs was used (Δ(ara-leu) 7697 araD139 fhuA ΔlacX74 galK16 galE15 e14-ϕ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL(StrR) rph spoT1 Δ(mrr-hsdRMS-mcrBC)). For gene expression, BA14a (bglR, thi-1, relA1, HfrPO1, ΔnuoA-N) with deletion of the entire nuo operon was used as the host. The construction of other plasmids, with one exception, is described in Supplementary Tables 2 and 3. pBAD33(LMN) -final version, was constructed by copying a 2 kb fragment of pET22b(L), including the ribosome binding site region and nuoL, and introducing a new Sac I site upstream of the ribosome binding site. Using the internal Apa I site in nuoL and the Sac I site which is found upstream of nuoL in pBAD33(LMN), a 0.8 kb fragment was excised, and ligated to pBAD33(LMN) digested with the same enzymes, Sac I and Apa I. This had the effect of increasing the expression of L from pBAD33(LMN). Low expression of M in pBAD33(A-K+MN) was solved in a different way, as described in Supplementary Table 2. Briefly, the K-L junction was preserved, including a few N-terminal amino acids of L before the sequence was switched to the N-terminus of M.
Growth and membrane preparation
For expression all of the E. coli Complex I subunits, the cells were grown in a rich chemical medium (0.5% yeast extract, 1.0% peptone, 15 mM Na3PO4, 10 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 50 mg/L riboflavin, 30 mg/L ferric ammonium citrate, and 0.5 mM L-cysteine) at 37 °C [43]. For expression of a subset of Complex I genes, cells were grown in a rich medium (3% tryptone, 1.5% yeast extract and 0.15% NaCl and 1% (v/v) glycerol) at 37 °C [44]. Harvested cells were suspended in 9 ml of 50 mM MES, 25% glycerol, and 10 mM MgSO4 (pH 6.0), and passed through the French press at 8000 psi. Cell debris and unbroken cells were removed by a low speed spin at 8000 rpm for 15 min in a JA-20 rotor. The supernatant was then centrifuged at 45,000 rpm for 1 h at 4 °C in a Beckman Ti-50 rotor. The pellet was resuspended in 1 ml of the same buffer, and centrifuged at 100,000 rpm for 1 h at 4 °C in a TLA100.2 rotor. The pellet was then resuspended in 1 ml of the same buffer, and used in the experiments or stored at −80 °C.
Time-delayed and simultaneous expression systems
The time-delayed, in vivo assembly procedures developed by Brockmann et al. [41] were used. Three plasmids were used in a single cell. A subset of genes was cloned into one plasmid, pBAD33, and the remaining genes are cloned into a second plasmid, pET22b. A third plasmid, pT7Pol26 is required to control expression of the genes cloned into pET22b. A subset of genes from pBAD33 and the remaining genes from pET22b were simultaneously transformed into pT7Pol26/BA14 competent cells and grown in 500 ml of rich chemical medium at 37 °C with 100 mg/l ampicillin, 40 mg/l chloramphenicol, and 50 mg/l kanamycin as selection markers. The cultures were induced with 0.03% (w/v) arabinose when A600 = 0.05, to express the genes cloned into pBAD33, and grown to A600 = 0.3. At this time, 0.5% (w/v) glucose and 0.045% (w/v) D-fucose were added to the medium, to repress further expression of the pBAD-controlled genes. Thirty minutes was shown to be sufficient for remaining mRNA to be degraded [41]. Induction of the second set of genes, cloned into pET22b, was started with addition of 0.1 mM IPTG, and the culture was grown for about one additional hour before harvesting the cells at a cell density of A600 = 1.0. For simultaneous expression from both plasmids, the cells were induced with 0.03% (w/v) arabinose and 0.1 mM IPTG at A600 = 0.2, and harvested at A600 = 0.5.
SDS electrophoresis and immunoblotting
50 μg of membrane protein (about 5 µl) were mixed with 10 μl 4% SDS and 5 μl of loading dye (60 mM Tris–HCl (pH 6.8), 25% glycerol, 14.4 mM 2-mercaptoethanol, 0.1% bromophenol blue) and then incubated at 37 °C for 15 minutes. Samples were run on 12% acrylamide gels at 150 V for 1 h and then transferred to PVDF membranes by applying 100 V for 1 h in transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol, pH 8.3). The PVDF membrane was blocked with 5% milk in TBS/Tween 20 (20 mM Tris, 500 mM NaCl, 0.05% Tween 20, pH 7.5) for overnight and then washed 3 times with TBS/ Tween 20. The PVDF membrane was incubated with rabbit custom antibodies diluted 1:5000 for subunit L and M, 1:10,000 for subunits J and K, 1:20,000 for subunit CD or mouse anti-HA probe, diluted 1:170 for subunit N at room temperature for 2 h. After washing 3 times with TBS/Tween 20, the blot was incubated with anti-rabbit IgG-HRP or anti-mouse IgG-HRP (diluted 1:10,000) for 1 h. After three more washings with TBS/Tween 20, the blot was detected by horseradish peroxidase (HRP) chemiluminescent substrates in a ChemiDoc Imaging System (Bio-Rad, Hercules CA). Images were exported by Image Lab software. Blots shown are representative of blots from 2–3 preparations, in total. The antibodies used in this report were validated by probing membrane samples from the deletion strain BA14 (negative control) and BA14 transformed with pBAD33(A-N) which expresses all subunits of Complex I. The blots are shown in Supplementary Figure 1.
Blue-native gel electrophoresis and NADH dehydrogenase activity assay
To analyze the assembly of the Complex I subunits blue-native gel electrophoresis was performed according to previous methods [45, 46]. In brief, E. coli membranes equivalent to 800 μg of protein were resuspended in 750 mM aminocaproic acid, 50 mM Bis-Tris–HCl (pH 7.0), 0.1 mg/ml DNase, and 0.8% (w/v) dodecyl-β maltoside to a total volume of 240 μl. After incubation on ice for 30 min, the samples were centrifuged at 149,000 g for 10 min. The supernatants were recovered, and 2% (w/v) of Coomassie Blue G in 1 M aminocaproic acid was added at a final concentration of Coomassie Blue of 0.08% (v/v). 50 μg protein from the samples were loaded on a 4–15% gradient gel, and electrophoresis was performed in a 4˚ C chamber at 100 V until entry of the protein sample into the stacking gel. After that, the cathode buffer, containing 0.02% of Coomassie Brilliant Blue G, was replaced by the cathode buffer without Coomassie Blue G, and the electrophoresis was continued at 150 V until the tracking dye ran out. After the electrophoresis, the gel was washed several times in 2 mM Tris–HCl (pH 7.5) before transfer to PVDF. For the NADH dehydrogenase activity assay, the gel was incubated in 2 mM Tris–HCl (pH 7.5) containing 2.5 mg/ml of p-nitroblue tetrazolium and 150 μM NADH for 40 min at room temperature. The reaction was terminated by 10% acetic acid and 50% methanol.
Immunoprecipitation
450 μl of protease inhibitor cocktail solution (prepared by adding the lyophilized powder to 1 ml of DMSO and 4 ml of deionized water) was added to 9 ml cell suspension before the French press procedure. 30 μl of Protein A/G Plus-Agarose beads were incubated with 1 μg of the pull-down antibody (anti-HA) in 300 μl of resuspension buffer (750 mM aminocaproic acid, 50 mM Bis Tris–HCl (pH 7.0), 0.1 mg/ml DNase, and 0.8% (w/v) dodecyl maltoside) for 1 h at 4 °C with constant rocking. Beads bound with pull-down antibody were centrifuged at 1470 g for 2 min, and the supernatant was discarded. Membranes equivalent to 500 μg of protein were resuspended in 200 μl of the same buffer. After incubation on ice for 30 mins, samples were centrifuged at 149,000 g for 10 min at 4 °C. To reduce non-specific binding, the supernatants were incubated with 25 μl of untreated A/G beads with constant rocking. After 1 h, the samples were centrifuged at 1470 g for 2 min. Supernatants were recovered and incubated overnight with the beads which had bound anti-HA antibodies, at 4 °C with constant rocking, followed by washes with 300 μl of the same resuspension buffer and centrifugation at 1470 g for 2 min. The flow-through was discarded, and washes steps were repeated for a total of three times. The immunoprecipitated material was dissolved in 30 µl of SDS sample buffer and incubated at 37 °C for 10 min. After centrifugation at 1470 g for 2 min, the beads were discarded and the supernatant was subjected to gel electrophoresis for immunoblot analysis. 50 μg of protein was loaded in “input” lanes.
Enzyme assay
The activity assays were modified for the Epoch 2 plate reader (BioTek, Burlington, VT) from methods described previously [42, 47]. Deamino-NADH (dNADH) was used to avoid oxidation by the alternative NADH oxidase in E. coli membranes [48]. In brief, NADH oxidase activity assays were started with 0.25 mM dNADH (extinction coefficient 6.22 mM−1 cm−1) and the absorbance was monitored at 340 nm for 2 minutes. dNADH oxidase activity assays were performed in 50 mM MOPS, 10 mM MgCl2, pH 7.3 at room temperature. The rate of dNADH oxidase from membranes of BA14/pBAD33(A-N) was about 1.0 µmoles dNADH per min per mg protein. Proton translocation assays were conducted by measuring the fluorescence quenching of the acridine dye ACMA (9-amino 3- chloro 2-methoxy acridine) as a ΔpH indicator using excitation and emission wave lengths of 410 and 490 nm, respectively. The uncoupler FCCP was added to 1 μM final concentration from a 1 mM ethanol stock to eliminate the buildup of a proton gradient during NADH oxidase assays. Proton translocation assays were conducted in 50 mM MOPS, 5 mM MgCl2, 50 mM KCl, at pH 7.3 at 25˚C, with additions of 1 μM valinomycin and 1 μM ACMA. The reactions were initiated with dNADH (0.25 mM) and after about 2 min the proton gradient was collapsed by the addition of FCCP (1 µM).
Results
Assembly of Complex I and detection of the L, M, and N subunits
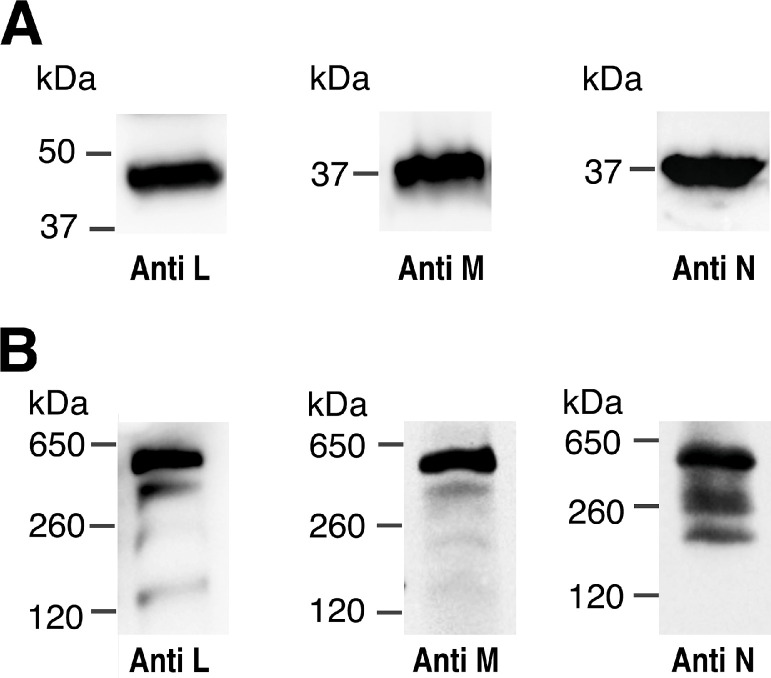
The L, M, and N subunits form the distal end of the membrane arm of bacterial Complex I (see Fig. 1). Assembly of Complex I was examined initially by expressing all subunits of the nuo operon (A-B-CD-E-F-G-H-I-J-K-L-M-N) from a pBAD plasmid, called pBAD33(A-N). Cultures were induced with arabinose, membrane vesicles were prepared from cells as described in “Materials and Methods”, and analyzed by both native and denaturing gel electrophoresis. In Fig. 2A it can be seen that in an SDS denaturing gel, both subunits M and N migrate approximately as 37 kDa proteins, while subunit L migrates as a protein of about 45 kDa. These sizes are much less than their actual masses of 66 (L), 56 (M) and 52 (N) kDa, respectively, but they are consistent with the behavior of highly hydrophobic membrane proteins. The bands were detected by immunoblotting with antibodies to subunits L and M, or to the HA tag of subunit N.
Fig. 2.
Immunoblots of L, M and N subunits by native and SDS gel electrophoresis. The entire nuo operon was expressed from pBAD33(A-N) in BA14 (∆nuo strain). The cells were grown in a chemical medium, induced with 0.03% (w/v) arabinose when A600 = 0.5 and harvested at A600 = 1.0. Membrane vesicles were prepared by French Press disruption. A. Samples were resolved by SDS electrophoresis, blotted, and probed by antibodies to subunits L, M and N (HA). B. Samples were solubilized with 0.8% (w/v) dodecyl maltoside and were run on a 4–15% gradient blue native gel. After electrophoresis, the immunoblots used antibodies against subunits L, M, N (HA). Blots are representative of at least 2 preparations with 2–3 replicates each.
The assembly of Complex I expressed from pBAD33(A-N) was analyzed by native gel electrophoresis, and the results are shown in Fig. 2B. Complex I from E. coli has a mass of about 540 kDa. The same three antibodies as in Fig. 2A were used in immunodetection of native gels. In each case, a strong band was seen between the two protein standards of 650 and 260 kDa, consistent with the expected size of 540 kDa. Also, minor bands were seen, particularly for subunit N, which migrated faster than the main band. The minor bands probably represent complexes that have lost subunits or failed to assemble them, most likely from distal positions such as E, F, G, L, or M. The conclusion is that native Complex I can assemble after expression from pBAD33(A-N), and that it can be detected in native gels.
Assembly of subunits L, M, and N
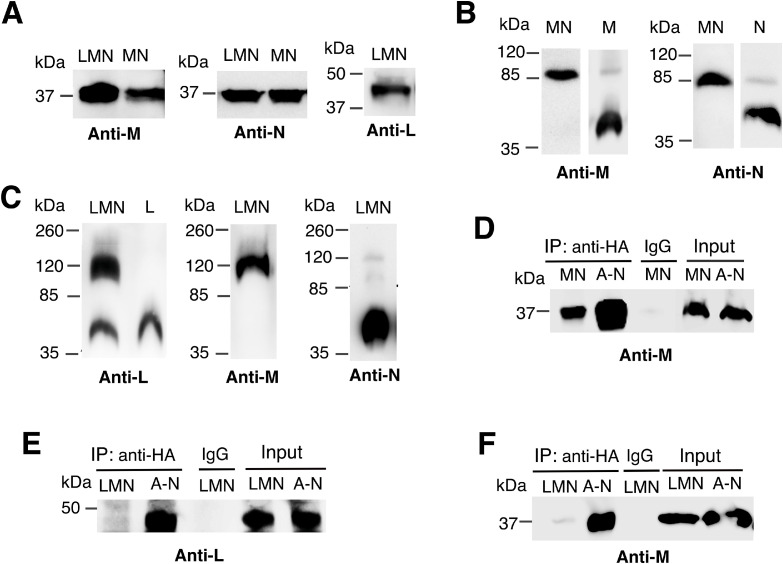
The question of whether subunits L, M and N could form a subcomplex during assembly of Complex I was addressed by expressing them in the absence of other subunits. The nuoL, M, and N genes were cloned as a unit in the pBAD33 plasmid, which allows induction by the addition of arabinose to the growth medium, or in the pET22b plasmid, which allows induction by IPTG. The preparation of membrane vesicles is described in “Materials and Methods”. Immunodetection of these subunits in membrane preparations after SDS polyacrylamide gel electrophoresis showed that all three subunits are expressed at high levels, as shown in Fig. 3A. Results are also shown for plasmids expressing nuoM and nuoN together, pBAD33(MN). In all cases, the subunits migrate at the same position as when expressed from the plasmid expressing the nuo operon (Fig. 2A).
Fig. 3.
Analysis of the interactions between subunits L, M and N. A. Membranes from cells expressing LMN or MN subunits from pBAD33(LMN) or pBAD33(MN) were resolved by SDS gel electrophoresis, and detected in immunoblots by antibodies to L, M or N (HA). B. Samples from cells expressing MN, M or N from pBAD33(MN), pBAD33(M), or pBAD33(N) were solubilized with dodecyl maltoside, resolved by blue native gel electrophoresis, and immunoblotted with antibodies against subunits M, and N (HA). C. Samples from pET22b(LMN) were solubilized with dodecyl maltoside, resolved by blue native gel electrophoresis, and immunoblotted with antibodies against subunits L, M, and N (HA). D. Membranes from cells expressing MN subunits, pBAD33(MN) or all nuo subunits pBAD33(A-N) were solubilized with dodecyl maltoside, and immunoprecipitated with HA antibody, using an anti-HA agarose slurry. Co-IP reactions were performed at 4 °C overnight. pBAD33(A-N) was used as a positive control, and IgG as a negative control. Blots were probed with antibody to subunit M. E. Membranes from cells expressing LMN subunits, pET22b(LMN) or all nuo subunits pBAD33(A-N) were solubilized with dodecyl maltoside, and immunoprecipitated with HA antibody, as before. Blots were probed with antibody to subunit L. F. The samples as described in (E) were probed with antibody to subunit M. Blots are representative of at least 2 preparations with 2–3 replicates each.
Next, samples were analyzed by native gel electrophoresis to look for evidence of subcomplex formation, and the results are shown in Fig. 3B and C. Membrane vesicle preparations from cells expressing M-N or L-M-N subunits were solubilized with dodecyl maltoside and loaded on native gels. When pBAD33(MN) was used subunits M and N were seen to co-migrate near the 85 kDa standard by immunodetection, as shown in Fig. 3B. When expressed individually, M and N subunits migrated in native gels faster than did the M-N subcomplex, consistent with a monomeric state. A minor band was seen at a higher molecular mass, possibly indicative of a low level of a dimeric form, when expressed alone. In contrast, when using pET22b(LMN), immunodetection revealed co-migration of subunits L and M near the standard protein of 120 kDa, as shown in Fig. 3C, with a small amount of monomeric subunit L. Subunit N was detected primarily as a monomer. The conclusions from these experiments are that subunits M and N can form a subcomplex when co-expressed. When subunit L is co-expressed with subunits M and N, it forms a complex with M, but N is excluded. Under no circumstances was a complex of L-M-N observed at a significant level.
Co-immunoprecipitation was used as a second method to determine the association of L, M and N subunits. Cells were grown expressing M and N subunits from pBAD33(MN), and membrane vesicles were prepared, as before, and solubilized with dodecyl maltoside. Cells with the whole operon plasmid, pBAD33(A-N), were used as a positive control. An HA antibody was used to precipitate subunit N, which had been cloned with an HA epitope at its C-terminus. After SDS gel electrophoresis the blots were probed with antibodies against subunit M, as shown in Fig. 3D. The results show that M is co-immunoprecipitated with N, consistent with an MN complex. Next, cells were grown expressing all 3 subunits, pET22b(LMN). After solubilization from membranes using dodecyl maltoside, samples were immunoprecipitated with the HA antibody (N). Cells with the whole operon plasmid, pBAD33(A-N), were again used as a positive control. In Fig. 3E, the samples were probed with antibodies against subunit L, and the results were negative. Similarly, in Fig. 3F, when probed with antibodies against subunit M, the results were negative. These results from co-immunoprecipitation agree with the analysis using native gel electrophoresis that L forms a complex with M, that M and N can form a complex in the absence of L, and that an LMN complex is not seen.
Assembly of subunits L, M, N, with J and K
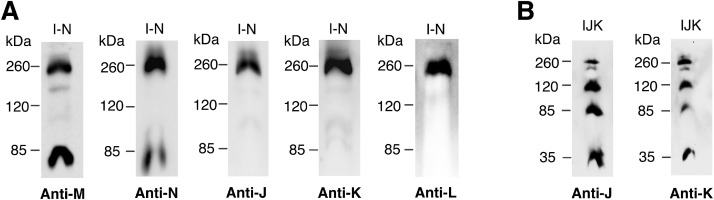
Since subunit L appeared unable to associate with M and N to form a subcomplex, we asked whether the addition of subunits J and K would facilitate the assembly of a larger subcomplex that included L. Subunit L has an interface with both J and K through its C-terminal transmembrane helix, in addition to its larger interfaces with subunits M and N (see Supplementary Table 1). Our initial construct, pBAD33(J-N), showed poor expression of J and K, as indicated by immunoblot. Therefore, the upstream gene nuoI was included, to make pBAD33(I-N) or pET22b(I-N), for these experiments, which produced a higher expression level of J and K. Subunit I does not interact with subunits J-N in the assembled Complex I (see Supplementary Table 1), and therefore should not complicate the analysis. Membrane vesicles were prepared from arabinose- or IPTG-induced cell cultures, as previously described, solubilized with dodecyl maltoside, and analyzed by native gel electrophoresis. A subcomplex was identified by immunodetection using antibodies against individual subunits, as shown in Fig. 4A. It ran near the 260 kDa standard, and subunits J, K, L, M, and N were all detected. In the case of subunits M and N, unincorporated subunits migrated near the 85 kDa standard, perhaps as a heterodimer. Subunits J, K, and L appeared only in the band near the 260 kDa standard. These results indicate that the presence of subunits J and K facilitates the joining of subunit L to a subcomplex with M and N. When subunits J and K were co-expressed in the absence of other membrane subunits from pBAD33(IJK), a subcomplex of J-K was detected in native gels, which had a tendency to oligomerize, with at least 4 distinct bands, as shown in Fig. 4B. Each band showed both J and K subunits. The band near the 35 kDa standard is a likely heterodimer of J (20 kDa) and K (10.8 kDa).
Fig. 4.
Analysis of the assembly of subunits J-N by blue native gel electrophoresis. Subunits I-N and IJK were expressed from pBAD33. Cells were grown in rich medium, induced with 0.2% (wt/vol) arabinose after 1 h and harvested at A600=0.8. A. Immunoblots of native gels of subunits I-N. Blue native gel electrophoresis was performed, followed by immunoblotting with antibody against M, N, J, K or L. B. Immunoblots of native gels of subunits IJK. Blots were probed with antibodies to subunits J or K. Blots are representative of at least 2 preparations with 2–3 replicates each.
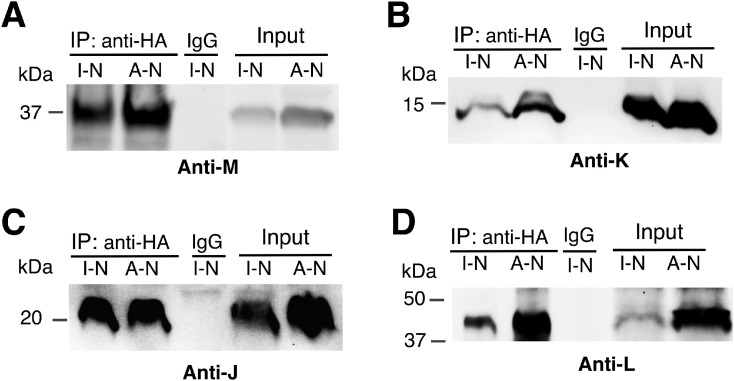
The formation of a J-K-L-M-N subcomplex was confirmed by co-immunoprecipitation, as performed previously, for Fig. 3. Cell cultures expressing either subunits I-N or A-N were grown, membrane vesicles were prepared, and solubilized with dodecyl maltoside. Immunoprecipitation was carried out using an HA antibody, targeting the C-terminal HA tag of subunit N. Immunodetection by an M antibody showed the presence of subunit M when either I-N or A-N were expressed, as shown in Fig. 5A. In Figs. 5B-D, similar results were obtained for subunits K, J, and L, respectively. In conclusion, these results support the findings by native gels in Fig. 4A, and indicate that the presence of subunits J and K enables subunit L to co-assemble with both subunits N and M.
Fig. 5.
Analysis of the assembly of subunits J-N by co-immunoprecipitation. Subunits I-N or all nuo subunits (A-N) were expressed from pBAD33. Subunits I-N were grown under the same conditions as in Fig. 4. Subunits A-N were grown under the same conditions as in Fig. 2. Samples were solubilized with dodecyl maltoside, and immunoprecipitated with HA antibody, using an anti-HA agarose slurry. Co-IP reactions were performed at 4 °C overnight. pBAD33(A-N) was used as a positive control, and IgG as a negative control. Blots were probed with antibodies to subunit M, K, J and L, and shown in panels A, B, C, and D, respectively. Blots are representative of at least 2 preparations with 2–3 replicates each.
Assembly of complex I by subunits expressed from two plasmids
The subunits of Complex I in E. coli are ordinarily expressed from a single operon, which allows all proteins to be made at the same time, and at the same location, due to the single transcript. The use of two plasmids causes the genes to be expressed at different locations, and also allows the genes to be expressed sequentially, at two different times. Pairs of plasmids were constructed with some genes on one plasmid, and the remaining genes on the other plasmid. Nine pairs of plasmids were constructed, as shown in Table 1. Procedures were followed that had been developed by Brockmann et al. [41], using pBAD33, which is induced by arabinose, and pET22b which is induced by IPTG, and are described in more detail in “Materials and Methods”. For sequential expression, induction by arabinose is first, followed by repression using D-fucose and glucose, and then induction by IPTG.
Table 1.
Pairs of plasmids for simultaneous and for time-delayed expression.
A-N refers to a native construct in which all genes from nuoA to nuoN are cloned.
A-K+N refers to a construct in which an artificial junction was constructed to add gene nuoN to follow a native nuoA-K construct.
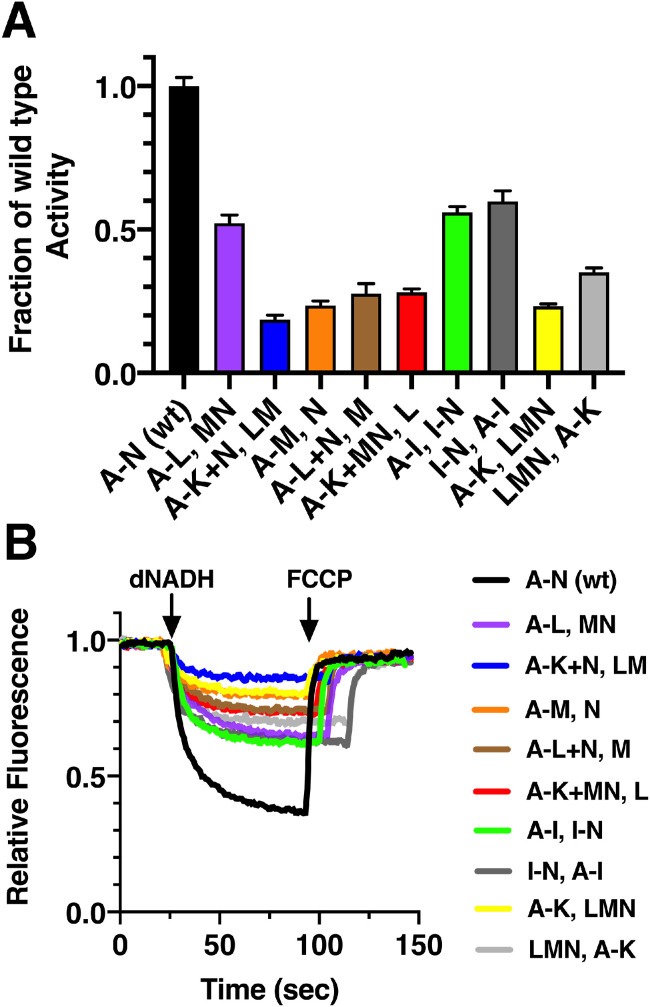
As a first step, the genes from both plasmids were induced simultaneously by the addition of both arabinose and IPTG. This allows the proteins to be made at the same time, but in different locations. It establishes a baseline for assembly and activity when compared to a time-delayed expression of a subset of genes. After simultaneous induction at A600 = 0.2, the cells were harvested at A600 = 0.5. Membrane vesicles were prepared, and assayed the same day. A summary of enzyme activities and proton translocation rates are shown in Fig. 6. A comparison of rates of dNADH oxidase activities is shown in Fig. 6A. Only three pairs achieved over 50% of the activity of the whole operon construct (A-N): pBAD33(A-I); pET22b(I-N), the reverse pBAD33(I-N); pET22b(A-I), and pBAD33(A-L); pET22b(MN). All of the others fell in the 19–35% range. Measurements of dNADH-driven proton translocation showed corresponding rates, as shown in Fig. 6B. Addition of dNADH to membrane vesicles caused rapid quenching of the fluorescence of ACMA, and reached a plateau after about one minute. This quenching corresponds to the buildup of a proton gradient across the membrane, as Complex I oxidizes NADH and translocates protons. The fluorescence was recovered rapidly upon the addition of the protonophore FCCP. The extent of fluorescence quenching follows the same order as the rate of dNADH oxidase activity. One common feature of the three highest activity combinations is the presence of the normal J-K-L gene sequence in one of the plasmids. The rather high levels of expression from these plasmids, in general, might tend to overcome the fact that some of the subunits are synthesized at different locations and will need to find each other to assemble Complex I.
Fig. 6.
Same-time expression of the nuo subunits split between two plasmids. Nine pairs of plasmids (Table 1) are used to transform strain BA14/ pT7Pol26 and grown in a chemical medium. The cells were induced with 0.03% (w/v) arabinose and 0.1 mM IPTG at A600 = 0.2, and harvested at A600 = 0.5. A. The level of dNADH oxidase activity in membrane vesicles is shown relative to that from cells expressing nuoA-N from one plasmid. Labeling of the cultures, e.g., A-I, I-N, means that genes A-I are expressed from pBAD33, and genes I-N are expressed from pET22b. Reactions included 100 μg/ml of membrane protein, 250 μM dNADH, and 1 μM FCCP. Results are the means and standard deviations from at least two membrane preparations with four replicates each. B. Proton translocation rates from the same samples as in panel A. The reactions included about 50 µg/ml membrane protein, and were initiated with dNADH at 250 μM final concentration. The fluorescence of ACMA (1 μM) was followed for several minutes, and then the uncoupler FCCP was added (1 μM) to collapse the generated proton gradient. The traces shown are representative of 2–3 preparations with 2–3 replicates for each.
Assembly of Complex I by subunits expressed from two plasmids in a time-delayed system
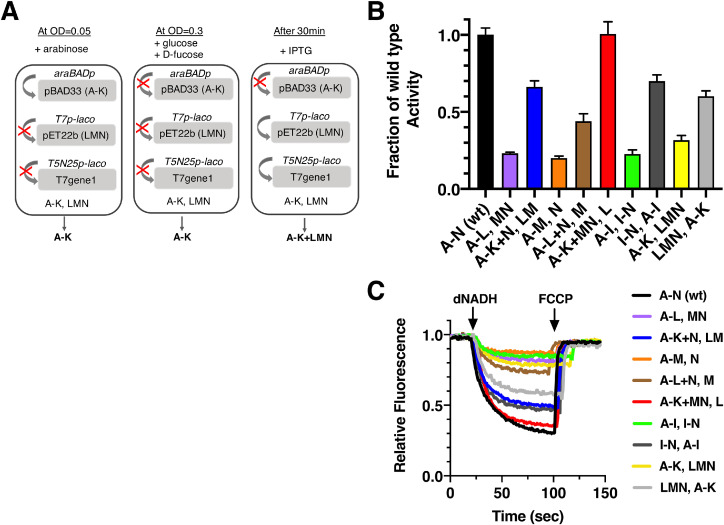
Next, cells with the same pairs of plasmids were grown with a time-delayed expression of genes on the pET22b plasmid. Fig. 7A presents the induction or repression states of the different promoters present. Cells were initially induced with arabinose at A600 = 0.05, araBADp was induced and allowed expression of nuo A-K. Followed by addition of 0.5% (w/v) glucose and 0.045% (w/v) D-fucose when A600 = 0.3, araBADp was repressed and therefore the transcription of A-K was stopped. After 30 minutes, the addition of 0.1 mM IPTG was added, which induced the lac operator-controlled T7 and T5N25 promoters then enabled the individual synthesis of nuo LMN. Cells were harvested after about one hour when the cultures reached A600 = 1.0. The results are shown in Fig. 7BC. Using this time-delayed expression of some genes, the activities for the same set of plasmids varied over the entire range of 20%-100%. Once again the levels of fluorescence quenching in the proton translocation assay closely followed the rates of dNADH oxidase activity, as shown in Fig. 7C. The highest rates were seen in a time-delayed expression of L alone, using pBAD33(A-K+MN); pET22b(L). In this case, the activity was essentially the same as the dNADH oxidase activity of whole operon construct (A-N). The time-delayed expression of LM, using pBAD33(A-K+N); pET22b(LM), also achieved a high level of activity, about 65% of the whole operon construct (A-N). The order for activity according to the time-delayed genes is L > LM > M > LMN > MN = N. Reaching about 70% of the activity from the whole operon construct was pBAD33(I-N); pET22b(A-I). In that case, interestingly, the entire sub-complex JKLMN would be allowed to form in the first stage of expression. One trend stood out: the three highest activities were found when J, K and N were expressed together in the first stage. In the fully assembled Complex I, subunit K sits between J and N, and has extensive contacts with each (see Supplementary Table 1). Two pairs were also analyzed in the reverse direction, i.e., LMN first and LMN last, and I-N first and I-N last. The results were striking, in that both LMN and I-N expressed first gave much higher activity than when they were expressed after a time delay. For LMN it was 60% (first) and 32% (last), while for I-N it was 70% (first) and 23% (last).
Fig. 7.
Time-delayed expression of the nuo subunits split between two plasmids. Nine pairs of plasmids (Table 1) were used to transform strain BA14/ pT7Pol26 and grown in chemical medium. Cells were initially induced with 0.03% (w/v) arabinose when A600 = 0.05, followed by addition of 0.5% (w/v) glucose and 0.045% (w/v) D-fucose when A600 = 0.3. After 30 minutes, 0.1 mM IPTG was added, and the cells grown to A600 = 1.0, and harvested. Labeling is the same as in Fig. 6. A. In vivo system for time-delayed assembly of subunits LMN into subunits A-K. A detailed description is given in the text. This panel is adapted from [41]. B. The level of dNADH oxidase activity in membrane vesicles is shown relative to that from cells expressing nuoA-N from one plasmid. Reactions included 300 μg/ml of membrane protein, 250 μM dNADH, and 1 μM FCCP. Results are the means and standard deviations from at least two membrane preparations with four replicates each. C. Proton translocation rates from the same samples as in panel A. The reactions were initiated with dNADH to 250 μM final concentration. The fluorescence of ACMA (1 μM) was followed for several minutes, and then the uncoupler FCCP was added (1 μM) to collapse the generated proton gradient. The traces shown are representative of 2–3 preparations with 2–3 replicates for each.
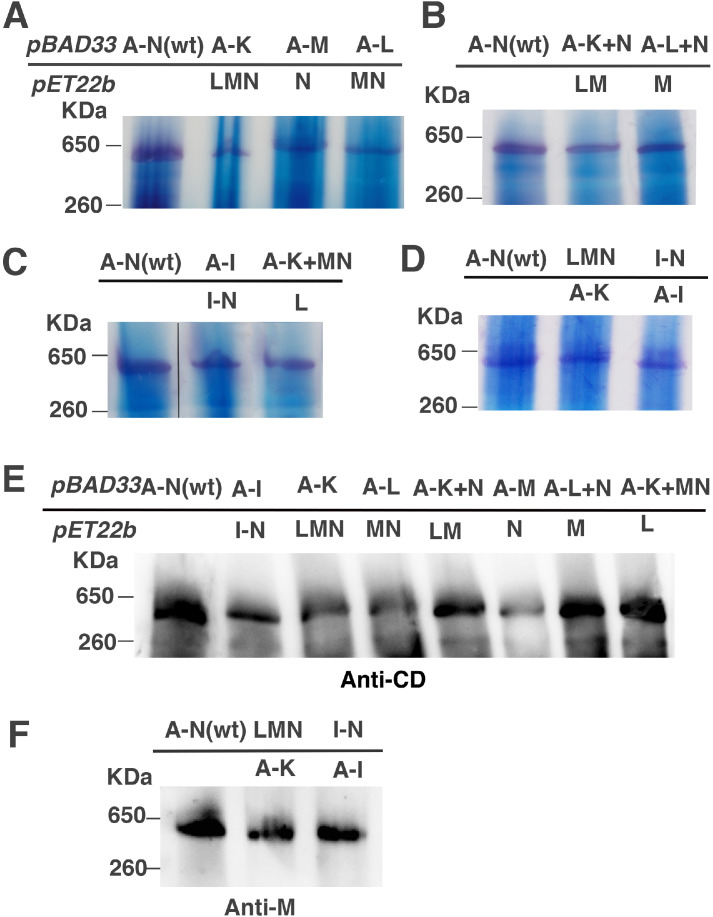
Membrane samples from the two-plasmid, time-delayed expression system were solubilized with dodecyl maltoside and analyzed by native gel electrophoresis. Membranes from cells expressing each of the nine pairs of plasmids showed a discrete band of activity indicative of fully assembled Complex I using an in-gel assay for NADH dehydrogenase activity, as shown in Fig. 8ABCD. Activity is detected by the color change of the electron acceptor NBT (nitro-blue tetrazolium chloride), and this requires the presence of the N-module, specifically the flavin of subunit F, but not the complete chain of FeS clusters. This was also shown by immunodetection using antibody against the peripheral arm subunit, CD (Fig. 8E) or subunit M (Fig. 8F). In this case the different yields of full-size Complex I roughly correlate with the differences in measured dNADH oxidase activities (Fig. 7B).
Fig. 8.
Native gel electrophoresis demonstrates assembly of Complex I expressed from two plasmids in a time-delayed system. Genes expressed from pBAD33 are shown above the line, and those from pET22b are shown below the line. Cells were grown under the same conditions as in Fig. 7. In panels A, B, C, D, the native gels are assayed for NADH dehydrogenase activity assay by incubating with 2.5 mg/ml p-nitroblue tetrazolium (NBT) and 150 μM NADH for 40 min at room temperature. A purple band appears near the 650 kDa standard, consistent with fully-assembled Complex I. E, F. Immunoblots of native gels of nine membrane samples from a time-delayed expression system. Blue native gel electrophoresis was performed, immunoblotting used antibody against subunit CD (E) or subunit M (F). Blots are representative of at least 2 preparations.
Assembly of Complex I in the absence of subunit L
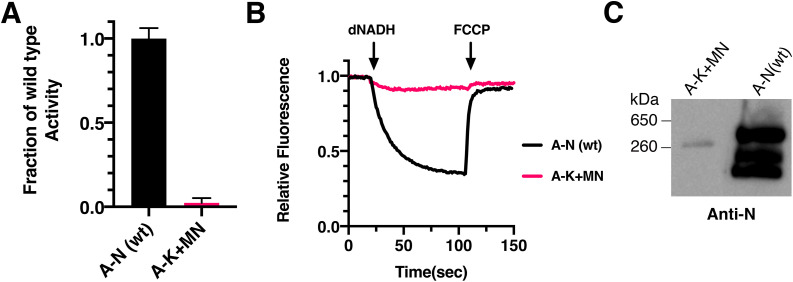
It has been reported that Complex I from E. coli lacking subunit L can assemble with significant activity [39]. If this occurred in our system, when subunit L is expressed after a time delay, the possibility would exist that some of the enzyme activity is due to complexes lacking subunit L. While we found no evidence for such complexes when all subunits were expressed in a time-delayed system (Fig. 8ABCD), we examined enzyme activity and assembly when subunit L was not expressed, shown in Fig. 9. In this case, cultures containing plasmid pBAD33(A-K+MN) were induced with arabinose and membrane vesicles were prepared. As shown in Fig. 9A, only very low levels of dNADH oxidase activity were measured (2%), and similarly low levels of dNADH-driven proton translocation (Fig. 9B). In analysis by native gel electrophoresis, a weak band was detected by the HA antibody (against HA-tagged subunit N) indicating a complex somewhat smaller than fully-assembled Complex I (Fig. 9C). Therefore, in the time-delayed expression of subunit L (Fig. 7B), essentially all of the activity measured must be due to the assembly of subunit L, and not due to Complex I that lacks subunit L.
Fig. 9.
Assembly of Complex I in the absence of subunit L. Cells expressing subunits A-K+MN from pBAD33 were grown in a rich chemical medium, induced with 0.03% (w/v) arabinose when A600 = 0.3, harvested at A600 = 0.8, and membrane vesicles were prepared. In each panel pBAD33(A-N) is shown as a positive control. A. dNADH oxidase activity of Complex I lacking subunit L. 50 μg/ml of membrane protein was used in each assay. The means and standard deviations are shown for 2 independent preparations with 4 replicates each. B. Proton translocation rates of Complex I lacking subunit L. Each assay included 25 μg/ml of membrane protein. The traces shown are representative of 2 preparations with 2–3 replicates each. C. Immunoblots of native gels of Complex I without subunit L. Immunoblotting used antibody against subunit N (HA). Blot is representative of 2 preparations.
Analysis of two C-terminal mutants in subunit K using a two-plasmid expression system
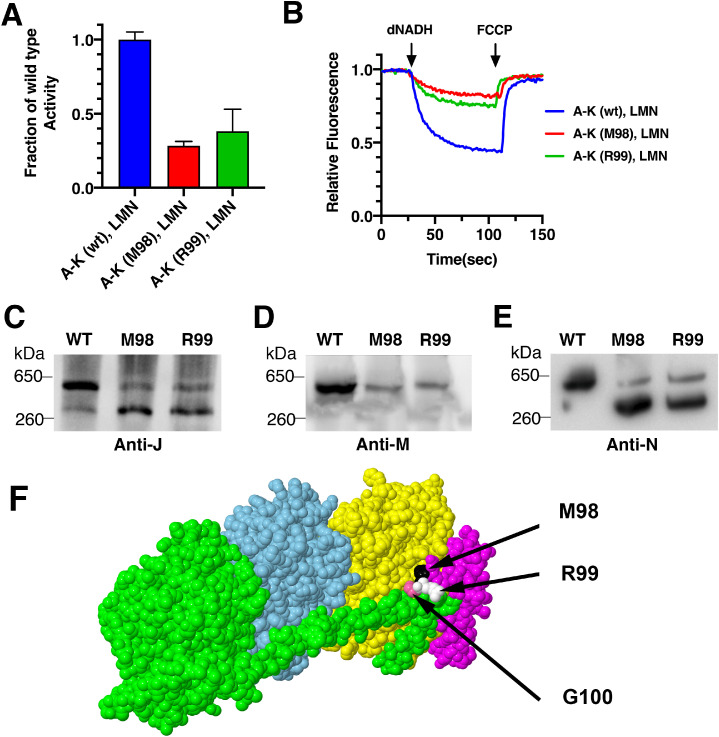
The two-plasmid, same-time expression system was used to analyze two intriguing mutations in subunit K, previously described by this lab [49]. This subunit has 100 residues, and the C-terminus rests along the junction of subunits N and L, on the cytoplasmic face (See Fig. 10F). If a stop codon replaces residue Gly100, there is little effect on Complex I activity [49]. When residue Arg99 or Met98 was replaced with a stop codon, there was essentially no activity in membrane vesicles, and subunits L, M and N could not be detected by immunoblotting. Since the genes for subunits K and L overlap (the same TG is part of the TGA stop codon for nuoK and the ATG start codon for nuoL) it seemed possible that these mutations in nuoK affected translation of nuoL. These mutations were now re-examined using the two plasmid system: pBAD33(A-K); pET22b(LMN), with same-time induction. As was shown in Fig. 6, this two-plasmid system is expected to generate moderate levels of Complex I activity. The results with the two mutations in subunit K are shown in Fig. 10AB. When compared to the wild type constructs, Arg99-stop shows 38% of the dNADH oxidase activity, while Met98-stop shows 28%. This is substantially higher than the background-level seen when the mutations were constructed in the whole operon plasmid pBA400 in the previous study [49], suggesting that the translation of L was affected when the mutations occurred in the whole operon. However, the level is still lower than the wild type construct, suggesting that the C-terminus of subunit K is critical for full activity of Complex I. Assays of proton translocation are also lower than the wild type construct. In these assays a larger amount of membrane protein was used, compared to Fig. 6B, resulting in greater quenching.
Fig. 10.
Analysis of C-terminal mutations in nuoK using the two-plasmid, same-time expression system. BA14 cells containing pET22b(LMN) and pBAD33(A-K) were grown in chemical medium and induced with 0.03% (w/v) arabinose and 0.1 mM IPTG at the same time (A600 = 0.2), and harvested at A600 = 0.5. The pBAD33(A-K) plasmid was wild type (WT), or had stop codons at residue Arg99 (R99) or Met98 (M98) in nuoK. A. The effect of the stop codon substitutions in nuoK on the dNADH oxidase activity. In each assay, 300 μg/ml of membrane protein was used. The mean and standard deviation are shown from 2 independent preparations with 4 replicates each. B. Proton translocation rates of the stop codon substitutions in K. Each assay included 150 μg/ml of membrane protein. The traces shown are representative of 2 preparations with 2–3 replicates each. C-E. Immunoblots of BN-PAGE gels of the membrane preparations from two stop codon substitutions in nuoK. Immunoblotting used antibody against subunits J (C), M (D) and N (E). Blots are representative of at least 2 preparations. F. Space filling views of the sites of stop codon substitutions in nuo K. Subunit L (green), subunit M (cyan), subunit N (yellow), subunit K(magenta) are shown. The three stop codon substitutions are indicated in black arrow.
Finally, native gel electrophoresis was used to analyze the assembly of Complex I by these mutants, as shown in Fig. 10CDE. In all three panels, the wild type construct is compared to the two mutants: Arg99-stop and Met98-stop. When antibodies against subunit J, M or N were used, it was seen that the wild type construct showed a strong band at the expected position for assembled Complex I. In the two mutants the detection of bands containing the subunits J, N and M varied according to the antibody used. The J antibody revealed that most of the J subunit was found in a faster moving band (Fig. 10C). The M antibody revealed only the full-size Complex I (Fig. 10D), indicating that when M is present, L is also present. The N antibody (HA tag) showed a distribution of subunit N between the two sizes, with the faster migrating band predominant, similar to J (Fig. 10E). The interpretation is that assembly is disrupted by these two mutations, resulting in a small amount of full sized Complex I, and a larger amount of assembled A-K+N. This appears to account for most or all of the loss of activity. We presume that the smaller complex must contain subunit K, which sits between J and N, but could not test it directly with the available K antibody, since our antibody does not recognize C-terminal mutants. The conclusion is that in expression of the complete nuo operon, C-terminal mutations in subunit K affect translation of subunit L as well as disrupting subunit interactions, leading to a nearly total loss of enzyme activity. The studies using two plasmids indicate that the C-terminus of subunit K is important for the incorporation of subunit M and L into Complex I.
Discussion
This was a systematic investigation of in vivo assembly of some of the membrane subunits of bacterial Complex I by expressing a subset of the subunits from a plasmid. With respect to the membrane subunits, chromosomal deletions of genes encoding individual membrane subunits have been analyzed in E. coli [44, 46, 50, 51] and Rhodobacter capsulatus [52]. In those cases, deletions of L, M or N have led to essentially a total loss of Complex I activity in membrane vesicles. Different results have been obtained when the operon containing the deletion was over-produced by expression from a plasmid [38, 39]. In that case, a smaller form of Complex I was found when subunit L was deleted, and it retained considerable activity [39]. From a structural viewpoint that seems reasonable, in that subunit L is found at the most distal region of the membrane arm. In this study, results support the view that subunit L can be efficiently added last to the assembling Complex I.
Subunits L, M and N compose the bulk of the distal end of the membrane arm, and in bacteria these subunits appear to have little or no contact with subunits of the peripheral arm. Interactions among membrane arm subunits for the E. coli enzyme are summarized in Supplementary Table 1, showing large contact surfaces between these three subunits. They each have a conserved, homologous core of fourteen transmembrane helices. Subunit L differs in that it has two additional transmembrane helices bridged by a lateral helix that runs along the membrane surface and serves to stabilize these three subunits [53], [54], [55]. Because of the extensive surface interactions among these three subunits, it could be expected that they would form a complex, perhaps as an assembly intermediate. However, we found no evidence for the association of these three subunits when they were co-expressed, by native gel electrophoresis or by co-immunoprecipitation. While M and N formed a subcomplex in the absence of subunit L, when all three subunits were co-expressed, only a subcomplex of L-M was seen in native gels, and L was not co-immunoprecipitated with N. Only with the additional subunits J and K, could a complex including L be seen. We propose that in the L-M subcomplex, L and M interact through transmembrane helices. And, that in the absence of subunits J and K, the C-terminus of L remains disordered, and prevents the binding of subunit N. The extra functionality of subunit L introduced by its C-terminal lateral helix likely complicates its folding and assembly, and that this is an evolutionary trade-off. This lateral helix appears necessary to bind the three subunits together for optimal proton translocation. But in the absence of some of its binding partners (J-K-M-N), the C-terminal region of L cannot fold to its native state. In eukaryotes, this problem appears to be solved by the binding of chaperones and supernumerary subunits to the ND5 protein [35].
When expressed alone the J and K subunits demonstrated a series of 4–5 oligomers. It is not surprising that these two subunits would associate, since they have the largest interface of any two subunits in bacterial Complex I, as shown in Supplementary Table 1. Oligomerization represents another potential problem for multi-subunit assembly, but that possibility is minimized by the simultaneous synthesis of the other membrane subunits. It is likely that subunits J and K must be expressed together, because they cannot form a native conformation without each other. Subunit K has three transmembrane helices, and lacks a core. Subunit J has five transmembrane helices, but the first and fifth ones do not interact with others, and it also lacks a core. These two subunits become interdigitated in the assembled complex, and form a core of six interacting transmembrane helices. The large interaction surface between J, K and N suggests that the co-expression of N might serve to prevent oligomerization of J-K.
Using two plasmids, in which all thirteen subunits are encoded, but split in different ways between the plasmids, allowed us to address the question of whether distal membrane subunits can add last to assembling Complex I. One feature of an operon is that if a single mRNA is transcribed, then all proteins will be made at the same time and in the same place. This eliminates the need for the subunits to find each other by long-range diffusion, and reduces the opportunities for degradation or misfolding. Furthermore, subunits encoded by adjacent genes in an operon commonly have interactions, which can facilitate assembly [56, 57]. In the nuo operon, A and B have interactions, also B and CD, E and F, F and G, H and I, J and K, K and L, L and M, and M and N. That system breaks down when subunits are expressed from two different plasmids, since some neighboring subunits will be translated by different ribosomes, and they will need to find each other. This is especially problematic when single-copy chromosomal genes in an operon are moved to separate chromosomal loci, as illustrated by the work of Shieh et al. [57]. When medium or high-copy number plasmids are used, the subunits might have a better chance to find each other, due to higher expression levels. There might be differences for soluble subunits that diffuse in the cytoplasm, and membrane subunits that diffuse in the lipid bilayer. In our studies, in which the L, M, and N subunits are lacking in the pBAD33(A-K) plasmid, and are placed in the pET22b plasmid for same-time expression, the Complex I activity typically decreased to about 20% of that from the whole operon plasmid. When the genes were switched between the two plasmids: pBAD33(A-K) and pET22b(LMN) the Complex I activity was about 35%, probably reflecting differences in copy number, and rates of induction. The highest activities for same-time expression were obtained when M-N were expressed from pET22b (50%), or in the two A-I, I-N splits, which had about 55% or 60% of the whole operon activity. These results were similar to those of Shieh et al [57], which examined the splitting of a two-gene operon. In the multi-gene nuo operon, the J-K-L sequence appeared important for highest activity, and it occurred in the context of the N and Q modules (A-L), or the complete P module (I-N). This situation would reduce the time of diffusion needed for L to arrive at its binding site in the assembling complex. In contrast, subunits M and N appear to diffuse more efficiently.
The possibility that some subunits can be efficiently added last to the growing Complex I was addressed by expressing some subunits in a time-delayed manner. We assume that successful assembly of Complex I using time-delayed expression requires that the first group of proteins must be stable over a period of 30 minutes, and likely forms a complex. The time-delayed expression of L alone was essentially as effective as when all genes were expressed from the same operon, and was more effective in assembling a functional Complex I than any pair of complementary plasmids induced at the same time. This suggests that allowing the rest of Complex I to assemble, i.e., subunits A-K + M-N, before L is expressed, ensures that L joins efficiently. Assembly when both L and M are expressed after a time delay, is not quite as efficient. The activity improves from the same-time level of 20% activity to the time-delayed level of 65%, but still less than the nearly 100% seen when L alone is expressed last. These results are complementary to earlier findings that showed that subunits L, or L and M, can dissociate from isolated Complex I, depending upon the detergent present [58]. Our results also support the interpretation that the L-M complex is not a productive intermediate, and that L and M must join the developing complex sequentially. When L-M-N are all expressed after a time-delay, the improvement in activity over same time expression is much less (30% vs. 20%). It can be seen that the key distinction is whether N is co-expressed with A-K in the first stage. That suggests that the complex of A-K is more stable when N can bind to J-K, resulting in more efficient assembly of the entire complex. Subunit N might also prevent oligomerization of the J-K complex.
When the nuo operon was split pBAD33(A-I)/pET22b(I-N) or pBAD33(I-N)/pET22b(A-I), assembly was moderately efficient during same time expression, at 55% or 60%, respectively. This might reflect the extensive subunit interactions between subunits A-I and J-N. When the pET22b genes were expressed after a time delay, efficiency of assembly diverged: 20% for the former and 70% for the latter, where the A-I genes were expressed last. Similar results were seen when comparing A-K expressed last (60%) compared to LMN last (30%). In the case of the A-I, I-N pairing, the results suggest that the stability of the JKLMN complex is key, when those genes are expressed first. In contrast, while the A-I subunits can likely form a complex, that complex might be less stable when expressed first, in particular regarding the membrane subunits H and A, in the absence of the binding partners J and K. That could explain the much lower assembly achieved when subunits A-I are expressed first. The results when J and K are joined with A-I support that interpretation. When LMN are expressed first, rather than I-N, the activity decreases from 70% to 60%, while when expressed last, the activity increases from 20% to 30%. Possible oligomerization of J-K subunits in subcomplexes could contribute to lower assembly, especially when expressed first.
Use of the two plasmid system allowed re-evaluation of C-terminal mutations in subunit K. Previously we had shown that genetic truncations of 2–4 amino acids at the C-terminus of subunit K resulted in a total loss of Complex I activity [49]. Several subunits could not be detected by immunoblotting of membrane fractions, suggesting that assembly of Complex I did not occur. However, due to overlapping of the nuoK and nuoL genes, it remained possible that downstream translation was negatively impacted, leading to limited assembly. This problem was averted by splitting the operon between nuoK and nuoL. Translational coupling regularly occurs in closely spaced genes within an operon [59, 60]. Recent results support the view that translation of the upstream gene opens up the ribosome binding site for the downstream gene [61]. Mutations at the 3′ end of the upstream reading frame might impair the binding of the ribosome for the downstream reading frame. Our results demonstrate that these truncations of only 2 or 3 amino acids, reduce the activity of Complex I relative to that from the wild type genes, and that the activity levels roughly correlate with the level of assembled Complex I. When translational coupling exists, it should be considered when genes within operons are mutated near the gene junctions.
In summary we have shown that subunit L can join last to a pre-assembled complex lacking only subunit L. Similar findings were reported for the ATP synthase by the originators of this time-delayed expression system [41, 62]. What was unexpected is that time-delayed expression of larger groups of membrane subunits is much less effective. That probably reflects the increased time necessary for several subunits to diffuse individually to the assembling enzyme, since the LMN complex is not stable. In contrast, the JKLMN complex appears to be stable, since if assembled first, a time-delayed expression of the remaining genes is relatively efficient (70%). In particular the group of J-K-N appears to be important for stabilizing a developing membrane arm. The unusual C-terminal structure of subunit L, along with its many interaction partners, appears to limit the ability of this protein to form productive subcomplexes with other subunits, outside of assembled Complex I or its complete complement of partners.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Hind Alkhaldi for construction of plasmids, including pBAD33(A-I) and pBAD33(A-N), Cambley Sassman for constructing the nuoK mutants in pBAD33(A-K), and Zainab Fatima for constructing pBAD33(N) and starting this project. We thank Bilal Amarneh for various contributions to the project. We also thank Gabriele Deckers-Hebestreit and her group at Universität Osnabrück, Germany for their hospitality during a sabbatical (SBV), and for supplying plasmids for this project.
Funding
This research was funded by the National Institutes of Health, USA, grant number 1R15GM126507. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding came from the American Heart Association, grant number 17AIREA33661165.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2021.100027.
Appendix. Supplementary materials
References
- 1.Parey K., Wirth C., Vonck J., Zickermann V. Respiratory complex I - structure, mechanism and evolution. Curr. Opin. Struct. Biol. 2020;63:1–9. doi: 10.1016/j.sbi.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Parey K., Haapanen O., Sharma V., Kofeler H., Zullig T., Prinz S., Siegmund K., Wittig I., Mills D.J., Vonck J., Kuhlbrandt W., Zickermann V. High-resolution cryo-EM structures of respiratory complex I: mechanism, assembly, and disease. Sci. Adv. 2019;5:eaax9484. doi: 10.1126/sciadv.aax9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agip A.A., Blaza J.N., Fedor J.G., Hirst J. Mammalian respiratory Complex I through the lens of cryo-EM. Annu. Rev. Biophys. 2019;48:165–184. doi: 10.1146/annurev-biophys-052118-115704. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi T., Ohnishi S.T., Salerno J.C. Five decades of research on mitochondrial NADH-quinone oxidoreductase (complex I) Biol. Chem. 2018;399:1249–1264. doi: 10.1515/hsz-2018-0164. [DOI] [PubMed] [Google Scholar]
- 5.Formosa L.E., Dibley M.G., Stroud D.A., Ryan M.T. Building a complex complex: assembly of mitochondrial respiratory chain complex I. Semin. Cell Dev. Biol. 2018;76:154–162. doi: 10.1016/j.semcdb.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Fiedorczuk K., Sazanov L.A. Mammalian mitochondrial Complex I structure and disease-causing mutations. Trends Cell Biol. 2018;28:835–867. doi: 10.1016/j.tcb.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Wang P., Dhananjayan N., Hagras M.A., Stuchebrukhov A.A. Respiratory complex I: bottleneck at the entrance of quinone site requires conformational change for its opening. Biochim. Biophys. Acta Bioenerg. 2021;1862 doi: 10.1016/j.bbabio.2020.148326. [DOI] [PubMed] [Google Scholar]
- 8.Ropke M., Saura P., Riepl D., Poverlein M.C., Kaila V.R.I. Functional water wires catalyze long-range proton pumping in the mammalian respiratory Complex I. J. Am. Chem. Soc. 2020;142:21758–21766. doi: 10.1021/jacs.0c09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mühlbauer M.E., Saura P., Nuber F., Di Luca A., Friedrich T., Kaila V.R.I. Water-gated proton transfer dynamics in respiratory Complex I. J. Am. Chem. Soc. 2020;142:13718–13728. doi: 10.1021/jacs.0c02789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaniya U., Gupta C., Cai X., Mao J., Kaur D., Zhang Y., Singharoy A., Gunner M.R. Hydrogen bond network analysis reveals the pathway for the proton transfer in the E-channel of T. thermophilus Complex I. Biochim. Biophys. Acta Bioenerg. 2020;1861 doi: 10.1016/j.bbabio.2020.148240. [DOI] [PubMed] [Google Scholar]
- 11.Kampjut D., Sazanov L.A. The coupling mechanism of mammalian respiratory complex I. Science. 2020;370:eabc4209. doi: 10.1126/science.abc4209. [DOI] [PubMed] [Google Scholar]
- 12.Haapanen O., Reidelbach M., Sharma V. Coupling of quinone dynamics to proton pumping in respiratory complex I. Biochim. Biophys. Acta Bioenerg. 2020;1861 doi: 10.1016/j.bbabio.2020.148287. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Fernandez J., Kaszuba K., Minhas G.S., Baradaran R., Tambalo M., Gallagher D.T., Sazanov L.A. Key role of quinone in the mechanism of respiratory complex I. Nat. Commun. 2020;11:4135. doi: 10.1038/s41467-020-17957-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grba D.N., Hirst J. Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation. Nat. Struct. Mol. Biol. 2020;27:892–900. doi: 10.1038/s41594-020-0473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swartz T., Ikewada S., Ishikawa O., Ito M., Krulwich T. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005;9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- 16.Carroll J., Fearnley I.M., Skehel J.M., Shannon R.J., Hirst J., Walker J.E. Bovine Complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 17.Pilkington S.J., Skehel J.M., Walker J.E. The 30-kilodalton subunit of bovine mitochondrial Complex I is homologous to a protein coded in chloroplast DNA. Biochemistry. 1991;30:1901–1908. doi: 10.1021/bi00221a024. [DOI] [PubMed] [Google Scholar]
- 18.Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., Salim A., Ryan M.T. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 19.Kmita K., Zickermann V. Accessory subunits of mitochondrial Complex I. Biochem. Soc. Trans. 2013;41:1272–1279. doi: 10.1042/BST20130091. [DOI] [PubMed] [Google Scholar]
- 20.Elurbe D.M., Huynen M.A. The origin of the supernumerary subunits and assembly factors of Complex I: a treasure trove of pathway evolution. Biochim. Biophys. Acta. 2016:971–979. doi: 10.1016/j.bbabio.2016.03.027. 1857. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich T. The NADH:ubiquinone oxidoreductase (Complex I) from Escherichia coli. Biochim. Biophys. Acta. 1998;1364:134–146. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich T., Dekovic D.K., Burschel S. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (respiratory Complex I) Biochim. Biophys. Acta. 2016;1857:214–223. doi: 10.1016/j.bbabio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Baradaran R., Berrisford J.M., Minhas G.S., Sazanov L.A. Crystal structure of the entire respiratory Complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efremov R.G., Sazanov L.A. Structure of the membrane domain of respiratory Complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 25.Parey K., Brandt U., Xie H., Mills D.J., Siegmund K., Vonck J., Kuhlbrandt W., Zickermann V. Cryo-EM structure of respiratory Complex I at work. eLife. 2018;7:e39213. doi: 10.7554/eLife.39213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zickermann V., Wirth C., Nasiri H., Siegmund K., Schwalbe H., Hunte C., Brandt U. Structural biology. Mechanistic insight from the crystal structure of mitochondrial Complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 27.Blaza J.N., Vinothkumar K.R., Hirst J. Structure of the deactive state of mammalian respiratory Complex I. Structure. 2018;26 doi: 10.1016/j.str.2017.12.014. 312–319 e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agip A.A., Blaza J.N., Bridges H.R., Viscomi C., Rawson S., Muench S.P., Hirst J. Cryo-EM structures of Complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 2018;25:548–556. doi: 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiedorczuk K., Letts J.A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L.A. Atomic structure of the entire mammalian mitochondrial Complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu J., Wu M., Guo R., Yan K., Lei J., Gao N., Yang M. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J., Vinothkumar K.R., Hirst J. Structure of mammalian respiratory Complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer E.H., Solheim C., Tanz S.K., Bonnard G., Millar A.H. Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 2011;286:26081–26092. doi: 10.1074/jbc.M110.209601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perales-Clemente E., Fernandez-Vizarra E., Acin-Perez R., Movilla N., Bayona-Bafaluy M.P., Moreno-Loshuertos R., Perez-Martos A., Fernandez-Silva P., Enriquez J.A. Five entry points of the mitochondrially encoded subunits in mammalian Complex I assembly. Mol. Cell. Biol. 2010;30:3038–3047. doi: 10.1128/MCB.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim S.C., Hroudova J., Van Bergen N.J., Lopez Sanchez M.I., Trounce I.A., McKenzie M. Loss of mitochondrial DNA-encoded protein ND1 results in disruption of Complex I biogenesis during early stages of assembly. FASEB J. 2016;30:2236–2248. doi: 10.1096/fj.201500137R. [DOI] [PubMed] [Google Scholar]
- 35.Guerrero-Castillo S., Baertling F., Kownatzki D., Wessels H.J., Arnold S., Brandt U., Nijtmans L. The assembly pathway of mitochondrial respiratory chain Complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Ligas J., Pineau E., Bock R., Huynen M.A., Meyer E.H. The assembly pathway of Complex I in Arabidopsis thaliana. Plant J. 2019;97:447–459. doi: 10.1111/tpj.14133. [DOI] [PubMed] [Google Scholar]
- 37.Braun M., Bungert S., Friedrich T. Characterization of the overproduced NADH dehydrogenase fragment of the NADH:ubiquinone oxidoreductase (Complex I) from Escherichia coli. Biochemistry. 1998;37:1861–1867. doi: 10.1021/bi971176p. [DOI] [PubMed] [Google Scholar]
- 38.Erhardt H., Steimle S., Muders V., Pohl T., Walter J., Friedrich T. Disruption of individual nuo-genes leads to the formation of partially assembled NADH:ubiquinone oxidoreductase (Complex I) in Escherichia coli. Biochim. Biophys. Acta. 2012;1817:863–871. doi: 10.1016/j.bbabio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Steimle S., Bajzath C., Dorner K., Schulte M., Bothe V., Friedrich T. Role of subunit NuoL for proton translocation by respiratory Complex I. Biochemistry. 2011;50:3386–3393. doi: 10.1021/bi200264q. [DOI] [PubMed] [Google Scholar]
- 40.Kolata P., Efremov R.G. Structure of Escherichia coli respiratory complex I reconstituted into lipid nanodiscs reveals an uncoupled conformation. eLife. 2021;10 doi: 10.7554/eLife.68710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockmann B., Koop Genannt Hoppmann K.D., Strahl H., Deckers-Hebestreit G. Time-delayed in vivo assembly of subunit a into preformed Escherichia coli FoF1 ATP synthase. J. Bacteriol. 2013;195:4074–4084. doi: 10.1128/JB.00468-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amarneh B., De Leon-Rangel J., Vik S.B. Construction of a deletion strain and expression vector for the Escherichia coli NADH:ubiquinone oxidoreductase (Complex I) Biochim. Biophys. Acta. 2006;1757:1557–1560. doi: 10.1016/j.bbabio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Bungert S., Krafft B., Schlesinger R., Friedrich T. One-step purification of the NADH dehydrogenase fragment of the Escherichia coli Complex I by means of Strep-tag affinity chromatography. FEBS Lett. 1999;460:207–211. doi: 10.1016/s0014-5793(99)01341-1. [DOI] [PubMed] [Google Scholar]
- 44.Amarneh B., Vik S.B. Mutagenesis of subunit N of the Escherichia coli Complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry. 2003;42:4800–4808. doi: 10.1021/bi0340346. [DOI] [PubMed] [Google Scholar]
- 45.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 46.Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 2007;282:36914–36922. doi: 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- 47.Michel J., DeLeon-Rangel J., Zhu S., Van Ree K., Vik S.B. Mutagenesis of the L, M, and N subunits of Complex I from Escherichia coli indicates a common role in function. PLoS One. 2011;6:e17420. doi: 10.1371/journal.pone.0017420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita K., Ohnishi T., Kaback H.R. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 49.Zhu S., Canales A., Bedair M., Vik S.B. Loss of Complex I activity in the Escherichia coli enzyme results from truncating the C-terminus of subunit K, but not from cross-linking it to subunits N or L. J. Bioenerg. Biomembr. 2016;48:325–333. doi: 10.1007/s10863-016-9655-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamaru-Ogiso E., Kao M.C., Chen H., Sinha S.C., Yagi T., Ohnishi T. The membrane subunit NuoL (ND5) is involved in the indirect proton pumping mechanism of Escherichia coli Complex I. J. Biol. Chem. 2010;285:39070–39078. doi: 10.1074/jbc.M110.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres-Bacete J., Sinha P.K., Matsuno-Yagi A., Yagi T. Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of Complex I from Escherichia coli. J. Biol. Chem. 2011;286:34007–34014. doi: 10.1074/jbc.M111.260968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupuis A., Darrouzet E., Duborjal H., Pierrard B., Chevallet M., van Belzen R., Albracht S.P., Lunardi J. Distal genes of the nuo operon of Rhodobacter capsulatus equivalent to the mitochondrial ND subunits are all essential for the biogenesis of the respiratory NADH-ubiquinone oxidoreductase. Mol. Microbiol. 1998;28:531–541. doi: 10.1046/j.1365-2958.1998.00814.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhu S., Vik S.B. Constraining the lateral helix of respiratory Complex I by cross-linking does not impair enzyme activity or proton translocation. J. Biol. Chem. 2015;290:20761–20773. doi: 10.1074/jbc.M115.660381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steimle S., Schnick C., Burger E.M., Nuber F., Kramer D., Dawitz H., Brander S., Matlosz B., Schafer J., Maurer K., Glessner U., Friedrich T. Cysteine scanning reveals minor local rearrangements of the horizontal helix of respiratory Complex I. Mol. Microbiol. 2015;98:151–161. doi: 10.1111/mmi.13112. [DOI] [PubMed] [Google Scholar]
- 55.Belevich G., Knuuti J., Verkhovsky M.I., Wikstrom M., Verkhovskaya M. Probing the mechanistic role of the long alpha-helix in subunit L of respiratory Complex I from Escherichia coli by site-directed mutagenesis. Mol. Microbiol. 2011;82:1086–1095. doi: 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells J.N., Bergendahl L.T., Marsh J.A. Operon gene order is optimized for ordered protein complex assembly. Cell Rep. 2016;14:679–685. doi: 10.1016/j.celrep.2015.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shieh Y.W., Minguez P., Bork P., Auburger J.J., Guilbride D.L., Kramer G., Bukau B. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 58.Holt P.J., Morgan D.J., Sazanov L.A. The location of NuoL and NuoM subunits in the membrane domain of the Escherichia coli complex I: implications for the mechanism of proton pumping. J. Biol. Chem. 2003;278:43114–43120. doi: 10.1074/jbc.M308247200. [DOI] [PubMed] [Google Scholar]
- 59.Pradhan P., Li W., Kaur P. Translational coupling controls expression and function of the DrrAB drug efflux pump. J. Mol. Biol. 2009;385:831–842. doi: 10.1016/j.jmb.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 60.Oppenheim D.S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saito K., Green R., Buskirk A.R. Ribosome recycling is not critical for translational coupling in Escherichia coli. eLife. 2020;9:e59974. doi: 10.7554/eLife.59974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilbers F., Eggers R., Pradela K., Friedrich K., Herkenhoff-Hesselmann B., Becker E., Deckers-Hebestreit G. Subunit delta is the key player for assembly of the H+-translocating unit of Escherichia coli FoF1 ATP synthase. J. Biol. Chem. 2013;288:25880–25894. doi: 10.1074/jbc.M113.484675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.