Abstract
Objective
To develop a clinico‐genetic predictor of impulse control disorder (ICD) risk in Parkinson's disease (PD).
Methods
In 5770 individuals from three PD cohorts (the 23andMe, Inc.; the University of Pennsylvania [UPenn]; and the Parkinson's Progression Markers Initiative [PPMI]), we used a discovery‐replication strategy to develop a clinico‐genetic predictor for ICD risk. We first performed a Genomewide Association Study (GWAS) for ICDs anytime during PD in 5262 PD individuals from the 23andMe cohort. We then combined newly discovered ICD risk loci with 13 ICD risk loci previously reported in the literature to develop a model predicting ICD in a Training dataset (n = 339, from UPenn and PPMI cohorts). The model was tested in a non‐overlapping Test dataset (n = 169, from UPenn and PPMI cohorts) and used to derive a continuous measure, the ICD‐risk score (ICD‐RS), enriching for PD individuals with ICD (ICD+ PD).
Results
By GWAS, we discovered four new loci associated with ICD at p‐values of 4.9e‐07 to 1.3e‐06. Our best logistic regression model included seven clinical and two genetic variables, achieving an area under the receiver operating curve for ICD prediction of 0.75 in the Training and 0.72 in the Test dataset. The ICD‐RS separated groups of PD individuals with ICD prevalence of nearly 40% (highest risk quartile) versus 7% (lowest risk quartile).
Interpretation
In this multi‐cohort, international study, we developed an easily computed clinico‐genetic tool, the ICD‐RS, that substantially enriches for subgroups of PD at very high versus very low risk for ICD, enabling pharmacogenetic approaches to PD medication selection.
Introduction
Impulse control disorders and related behaviors (ICDs) have risen in importance in Parkinson's disease (PD) over the past few decades, following the introduction of D2 receptor‐selective dopamine agonists (DAs) for management of PD symptoms in the 1980s. Initial systematic studies showed that ICDs (most commonly compulsive gambling, buying, sexual behavior, and eating behaviors) occur frequently in people with PD treated with dopaminergic medications. 1 , 2 More recent studies confirmed that ICD rates are not elevated in people with de novo, untreated PD. 3
In the largest cross‐sectional epidemiological study done to date, encompassing 3090 PD patients, an ICD was identified in 14%. 4 A recent national multi‐site study reported a 5‐year cumulative ICD incidence rate of 46%, 5 and another study found clinically significant ICD symptoms in 36% of people with PD who also experienced dyskinesias. 6 DA treatment is by far the strongest PD medication correlate. 6 , 7 In addition, a personal or family history of alcoholism or gambling; impulsive or novelty‐seeking characteristics; younger age or early PD onset; male sex; and psychiatric comorbidity (depression, anxiety and REM sleep behavior disorder) are additional correlates of ICDs in PD. 1 , 6 , 8 , 9 , 10 In candidate gene studies, common variants in a number of neurotransmitter genes have also been linked to the development of ICDs in individuals with PD, but to date, evidence from these relatively small‐scale genetic studies is mixed. 11 , 12
Often problematic and at times disabling, ICDs remain under‐recognized in clinical practice. 2 , 13 , 14 , 15 To assist in diagnosis and management, several questionnaires and rating scales have been developed for detecting and monitoring ICDs and related behaviors in PD, including the Questionnaire for Impulsive‐Compulsive Disorders in Parkinson Disease (QUIP) 16 and the Questionnaire for Impulsive‐Compulsive Disorders in Parkinson Disease‐Rating Scale (QUIP‐RS), 17 both of which are recommended instruments for assessing ICD symptoms in PD. 18
Management strategies for ICDs are suboptimal. 19 Current clinical practice is to withdraw DA medications after ICD development, substituting with levodopa for symptom control. While ICD behaviors often resolve after discontinuing DA treatment, 20 considerable morbidity is still incurred in the process, and complicated withdrawal symptoms can occur (i.e., DA withdrawal syndrome). 21
Given the limited treatment strategies available to manage ICDs in PD, preventive strategies would be ideal. We reasoned that the evidence supporting a genetic basis for impulsivity, together with the high prevalence of DA use in PD, the high cumulative prevalence of ICDs in PD individuals, and the substantial morbidity incurred by the development of an ICD, represented an opportunity to develop pharmacogenetics in PD. Specifically, here we sought to develop a clinico‐genetic predictor of PD individuals at high versus low risk for the development of ICDs. Such a predictor could identify which individuals might be safest to receive DAs, an effective therapy for motor control but the PD symptomatic therapy class most likely to precipitate an ICD. To develop and then test this predictor, we used three large PD cohorts comprising 5770 individuals with PD: the Michael J. Fox Foundation Parkinson's Progression Markers Initiative (PPMI) cohort, the 23andMe PD cohort, and the University of Pennsylvania NIA U19 PD (UPenn) Cohort (Fig. 1).
Figure 1.
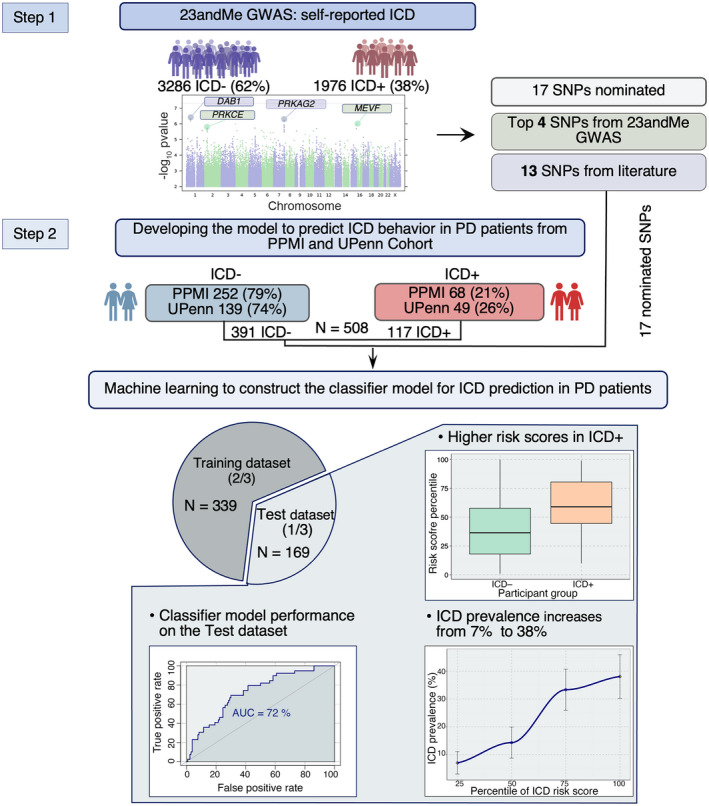
Overview of study. The study consisted of two major steps: (1) GWAS in the 23andMe Cohort for nomination of novel variants associated with ICD in PD and (2) development of a model to predict ICD behavior in PD subjects. The GWAS in the 23andMe Cohort (3286 ICD negative (ICD−) and 1976 ICD positive (ICD+) participants) uncovered four SNPs associated with ICD behavior in PD subjects at p < 1.3e‐06. These four and the additional 13 SNPs that were previously reported in the literature to associate with ICD were tested for association with ICD behavior and used to develop an ICD risk score in PD subjects. In particular, we obtained genotypes of 17 nominated SNPs for 320 (252 ICD− and 68 ICD+ participants) PPMI and 188 (139 ICD− and 49 ICD+ PD participants) UPenn Cohort PD subjects. We applied model selection to develop a final logistic regression classifier model. First, we combined the PPMI and UPenn Cohorts (N = 508) and then we randomly split this combined dataset into a non‐overlapping Training dataset and Test dataset in a 2:1 ratio. To select the subset of variables to keep in our final model (providing the best fit to the data), we used the Training dataset only, first performing backward feature selection with fivefold cross‐validation repeated 100 times on the Training dataset (261 ICD− and 78 ICD+ PD participants). We fit the final model (which included two SNPs (rs1800497 and rs1799971) as well as cohort, age, sex, dopamine agonist use, levodopa use, disease duration, and ethnicity as predictors) to the Training dataset employing Bayesian logistic regression. We then evaluated the ability of the Bayesian logistic regression model to predict ICD in the held‐out Test dataset (130 ICD− and 39 ICD+ PD participants). The final classifier model achieved ROC‐AUC = 0.72 on the Test dataset. For each PD participant in the Test dataset, we calculated the risk score (log odds) and RR of developing an ICD behavior using the predictive model. ICD, impulse control disorder; PD, Parkinson's disease; GWAS, Genomewide Association Study; PPMI, Parkinson's Progression Markers Initiative; RR, risk ratio. [Colour figure can be viewed at wileyonlinelibrary.com]
Methods
Participants
23andMe cohort
The 23andMe, Inc. research cohort is derived from the research participant base of 23andMe, Inc., a direct‐to‐consumer genetic testing company. Consented research participants provided saliva samples for genotyping. Genotyping was performed on five genotyping platforms (Illumina, Inc., San Diego, CA, USA ) by Labcorp, Inc., and followed by a genotype imputation step using a reference panel combining the May 2015 release of the 1000 Genomes Phase 3 haplotypes 22 with the UK10K imputation reference panel. 23 Details of the analyses and variant quality controls have been published elsewhere. 24 A total of 5262 23andMe participants with a self‐reported diagnosis of PD and >97% European ancestry (determined through an analysis of local ancestry 25 ), completed the web‐administered modified QUIP‐short form for ascertainment of an ICD anytime during PD. These participants comprised the 23andMe cohort for ICD prediction.
The full Genomewide Association Study (GWAS) summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset‐access/ for more information and to apply to access the data.
University of Pennsylvania cohort
The parent study of the UPenn cohort is the Clinical Core of the Penn NIA U19 (Center On Alpha‐synuclein Strains In Alzheimer Disease & Related Dementias at the Perelman School of Medicine at the University of Pennsylvania (UPenn), formerly the Morris K. Udall Center at the Perelman School of Medicine at the University of Pennsylvania). Over the past 13 years, the Clinical Core of the UPenn NIA U19 has recruited PD subjects to participate in a longitudinal study that includes: (1) serial assessment with a battery of motor, non‐motor, and neuropsychological tests; (2) blood for plasma‐based biomarkers and DNA, cerebrospinal fluid, and structural and functional brain imaging; and (3) post‐mortem brain tissue, enrolling over 400 subjects in this time span, with approximately 180 active participants at any given time. Starting in 2015 the QUIP‐RS was administered to U19 Clinical Core participants annually/biennially. At the time of our ICD study, 188 PD subjects from the U19 Clinical Core parent study had complete demographic and clinical records, including QUIP‐RS scores, as well as DNA availability. For these individuals, genotypes were obtained, and they formed the UPenn cohort for ICD prediction. SNP genotyping was performed on a GSA chip (Illumina Infinium Global Screening Array) or by TaqMan SNP Genotyping Assays. We used proxies (R 2 = 1) for one SNP that was not directly genotyped (Table S4).
Parkinson's Progression Markers Initiative cohort
PPMI is a comprehensive longitudinal, observational, international, multi‐center study designed to identify PD progression biomarkers. The original PPMI cohort included 400+ recently diagnosed, untreated (at baseline) PD subjects. Biological samples include a longitudinal collection of blood for genotyping and biomarker measurement. The aims and methodology of the study have been published elsewhere 26 (www.ppmi‐info.org/studydesign). Participants complete the QUIP‐short form annually. Three hundred and twenty PPMI participants had complete genetic and QUIP‐short form data available, and they comprise the PPMI cohort for this ICD study. Genotypes for 17 SNPs included in our analysis were obtained from the PPMI database (http://www.ppmi‐info.org/data).
Ethics statement for human subjects research
For the UPenn Cohort, the Institutional Review Board of University of Pennsylvania approved the human subjects research in this study. Written informed consent was obtained from cohort participants. For the PPMI Cohort, all participants signed a written consent form to participate in the study. For the 23andMe cohort, participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP‐accredited IRB, Ethical & Independent Review Services (E&I Review). Participants were included in the analysis based on consent status as checked at the time data analyses were initiated.
Ascertainment of ICD symptoms
The 188 PD participants of the UPenn cohort, 5262 participants of 23andMe cohort, 22 , 23 , 24 , 25 and 320 participants of the PPMI cohort 26 were assessed for the four primary ICD behaviors: gambling, hypersexuality, buying, and eating. 16 Individuals were deemed ICD+ based on previously established QUIP and QUIP‐RS cut‐off scores. 16 , 17
For the 23andMe cohort, a modified QUIP‐short form for web‐based administration was used, ascertaining the presence/absence of ICD symptoms at any time during the course of PD. For the UPenn cohort, the QUIP‐RS is administered annually/biennially per study protocol and assesses current ICD symptoms. In the PPMI study, the QUIP‐short form is administered annually and assesses current ICD symptoms. In the UPenn and PPMI cohorts, for ICD+ participants with data from more than one research visit, we used clinical data from the visit coinciding with the first occurrence of the ICD behavior. For ICD participants with data from more than one research visit, we used clinical data from the most recent visit.
Genome‐wide association study for novel ICD variants
In the 23andMe cohort, we performed GWAS with 23andMe's GWAS pipeline, 24 in order to find variants associated with ICD. The logistic regression model included age, sex, PD duration, DA use, levodopa use, the first five genetic principal components, and variables representing the genotyping platform. The principal component analysis was performed using ~65,000 high‐quality genotyped and trans‐ethnic variants that are present on all five genotyping platforms. GWAS analyses were run independently for the genotyped and imputed variants. Approximately 15.2M variants passed the pre‐ and post‐GWAS quality control, and the genomic inflation factor for these variants was 1.03. The top four loci from this GWAS, with a suggestive p < 1.3e‐06 for association with ICD, were nominated for follow‐up investigation.
Investigation of novel ICD variants in UPenn and PPMI cohorts
To test for association between genotypes at the four nominated SNPs from the 23andMe ICD GWAS, and the presence or absence of ICD in the PPMI and UPenn cohorts, we used logistic regression models assuming a log‐additive (multiplicative) genetic model. The effect of each SNP on ICD was adjusted for age, sex, DA use (yes/no), levodopa use (yes/no), and PD duration.
Evaluation of SNPs nominated from the literature in UPenn, PPMI, and 23andMe cohorts
In addition to SNPs found by hypothesis‐free GWAS, we investigated 13 candidate SNPs nominated from the literature on ICDs 11 for their association with ICD in the PPMI, UPenn, and 23andMe cohorts. For these candidate SNPs, we again employed logistic regression with a log‐additive model of genetic inheritance, and the effect of each SNP on ICD was adjusted for age, sex, DA use, levodopa use, and disease duration.
Logistic regression model predicting ICD
We combined the PPMI (n = 320) and UPenn (n = 188) cohorts for a total of 508 individuals with PD. We then randomly assigned these individuals into Training (n = 339) and Test (n = 169) sets, only ensuring that the proportion of ICD+ individuals within the Training and Test sets was not significantly different.
In the Training set, we developed a classifier to predict which individuals were ICD+. Specifically, we used a logistic regression model with 25 potential predictor variables; 17 SNPs (Table S1, cohort (UPenn, PPMI), sex (male, female), age (years), ethnicity (White or non‐white, by self‐report), education (years), PD duration (years), current DA therapy (yes/no), and current levodopa therapy (yes/no). In order to select the best subset of predictor variables, we employed the R caret package. 27 We used the glmStepAIC function to perform backward variable selection, based on Akaike information criterion, with fivefold cross‐validation repeated 100 times, in order to select the best final variables to include in the model. To perform unbiased variable selection, we randomly sampled seeds for each of the 500 iterations. Our final classifier then used the selected variables under a Bayesian logistic regression model to avoid inflation of estimate effect sizes for rare variants.
Model performance at prediction of ICD was assessed in both the Training and Test sets using receiver operating characteristic (ROC) curves, generating an area under the curve (AUC) for both the Training and Test sets.
Development of the ICD‐RS
Having constructed a model (Bayesian logistic regression classifier) using the Training set, we evaluated the ability of this classifier to predict ICD behavior in an independent Test set of 130 ICD‐negative (ICD−) and 39 ICD‐positive (ICD+) participants. We used the final model, which included two SNPs (rs1800497, rs1799971) and adjusts for cohort, age, sex, ethnicity, disease duration, DA use, and levodopa use, to develop an ICD risk score (ICD‐RS) as a continuous measure of ICD risk by relating the output of the Bayesian logistic regression model to probabilities:
We additionally developed a more generalizable ICD risk score (genICD‐RS) by employing the same methodology, but removing cohort as an input variable, in order to test performance without adjustment for cohort. This genICD‐RS used data from the combined datasets of 320 PPMI and 188 UPenn PD individuals, as we sought a predictor that may be broadly tested for replication in many clinical cohorts.
Additional details regarding statistical analysis
We conducted all analyses in R (http://www.r‐project.org); R‐scripts are available as a supplemental file.
To test for significant differences between the ICD+ and ICD− groups across demographic and clinical variables we employed t‐tests or Fisher's exact tests, as indicated by variable distribution. Here we compared the means or proportions of the two groups (ICD+, ICD−) for each demographic and clinical variable individually. All statistical tests were two‐sided.
For cross‐validation and model generation we used the “caret” package. 27 We created and analyzed receiver operating characteristic (ROC) curves employing the “pROC” package. 28 The specificity versus sensitivity analysis was performed by employing the function coords from the R package pROC. 28 To estimate the best cutoff point we used closest.topleft method which chooses the point closest to the top‐left part of the curve as the optimal threshold.
We also conducted individual SNP association analysis for all 17 SNPs, separately in the PPMI and UPenn cohorts. For this analysis, we employed the R SNPassoc package. 29
RESULTS
Prevalence of ICD in each cohort
A detailed description of the 5770 PD individuals enrolled in the study is reported in Table 1. In all three PD cohorts studied, the prevalence of ICD observed greatly exceeded ICD prevalence reported in the general population. 30 However, we observed a wide range (21.2–37.6%) of ICD prevalence across the three cohorts (Table 1). Specifically, the ICD prevalence in the 23andMe cohort (37.6%) was much higher than the prevalence found in the UPenn (26.1%) and PPMI (21.2%) cohorts. We note, however, that ICD ascertainment in the 23andMe Cohort differed from the Upenn and PPMI cohorts in two key respects. First, in the 23andMe Cohort the QUIP was administered by web‐based survey, rather than in‐person with a research coordinator. Second, because the 23andMe Cohort was only accessed one time, the QUIP short form used in this cohort screened for impulsive‐compulsive behaviors at any time during the course of PD, whereas the QUIP‐RS used in the longitudinally followed Upenn and PPMI cohorts screened for current ICD behaviors.
Table 1.
Demographic and clinical characteristics of studied PD cohorts.
| Variable | 23andMe cohort | UPenn cohort | PPMI cohort | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ICD− | ICD+ | p | ICD− | ICD+ | p | ICD− | ICD+ | p | |
| n = 3286 | n = 1976 | n = 139 | n = 49 | n = 252 | n = 68 | ||||
| 62.45% | 37.55% | 73.90% | 26.10% | 78.80% | 21.20% | ||||
| Age (years), M (SD) | 70.2 (8.8) | 67.6 (9.5) | <0.001 1 | 71.5 (8.1) | 68.9 (6.3) | 0.02 1 | 66.9 (9.7) | 62.3 (9.9) | <0.001 1 |
| Education (years), M (SD) | 16.7 (2.7) | 16.4 (2.7) | <0.001 1 | 16.1 (2.4) | 16.1 (2.5) | 0.49 1 | 15.6 (3.0) | 15.6 (3.0) | 0.91 1 |
| Disease duration (years), M (SD) | 5.2 (4.7) | 7.5 (5.9) | <0.001 1 | 11.3 (5.9) | 9.9 (5.0) | 0.12 1 | 5.06 (1.17) | 3.24 (1.45) | <0.001 1 |
| Sex, n (%) | |||||||||
| Female | 1535 (46.7) | 775 (39.2) | <0.001 2 | 64 (46.0) | 12 (24.5) | 0.008 2 | 86 (34.1) | 20 (29.4) | 0.56 2 |
| Male | 1751 (53.3) | 1201 (60.8) | 75 (54.0) | 37 (75.5) | 166 (65.9) | 48 (70.6) | |||
| Ancestry, n (%) | |||||||||
| Other | 0 (0) | 0 (0) | na | 16 (11.5) | 0 (0) | 0.01 2 | 14 (5.6) | 3 (4.4) | 1 2 |
| European | 3286 (100) | 1976 (100) | 123 (88.5) | 49 (100) | 238 (94.4) | 65 (95.6) | |||
| Dopamine agonist use, n (%) | |||||||||
| No | 2008 (61.1) | 712 (36.0) | <0.001 2 | 86 (61.9) | 24 (49.0) | 0.12 2 | 173 (68.7) | 32 (47.1) | <0.001 2 |
| Yes | 1278 (38.9) | 1264 (64.0) | 53 (38.1) | 25 (51.0) | 79 (31.3) | 36 (52.9) | |||
| Levodopa use, n (%) | |||||||||
| No | 632 (19.2) | 334 (16.9) | 0.036 2 | 18 (12.9) | 6 (12.2) | 0.90 2 | 52 (20.6) | 36 (52.9) | <0.001 2 |
| Yes | 2654 (80.8) | 1642 (83.1) | 121 (87.1) | 43 (88.5) | 200 (79.4) | 32 (47.1) | |||
ICD, impulse control disorder; PD, Parkinson's disease; M, mean; SD, standard deviation.
t‐test
Fisher exact test.
Bold values indicate statistically significant differences.
Differences in PD individuals with versus without ICD
Because the three cohorts differ significantly in terms of stage of PD, medication exposure, disease duration, and age, we analyzed these additional factors within each cohort individually, employing t‐tests and Fisher's exact tests. Younger age was associated with ICD across all three cohorts. In the Upenn and 23andMe cohort, males were more likely to have ICD (p = 0.008, p = 0.001). Exposure to DA was associated with ICD in the 23andMe (p < 0.001) and PPMI (p < 0.001) cohorts. Levodopa use was also associated with ICD in the 23andMe (p = 0.036) and PPMI (p < 0.001) cohorts, with no association in the Upenn cohort (p = 0.90). Additional results are summarized for each cohort and each variable in Table 1.
Association of individual ICD risk SNPs with ICD
The previous report of Kraemmer et al 11 evaluated 13 SNPs to predict risk for ICD in PD, using data from 276 participants in the PPMI cohort, studied for an average follow‐up duration of 2.2 years. Of the 13 SNPs in the original report of Kraemmer et al, 11 only two (rs702764 and rs7305115) showed borderline associations with impulsivity when tested independently in models adjusted for age, medication use, and duration of follow‐up. However, all 13 SNPs were used for predictive model development. We investigated these 13 SNPs for association with ICD in the PPMI cohort with two additional years of follow‐up, as well as in the 23andMe and Upenn cohorts.
In the PPMI cohort, only one of these 13 SNPs (rs1800497, in the DRD2 locus), was associated with ICD behavior (nominal p = 0.002, Table S2). None of the 13 candidate SNPS individually associated with ICD behavior in either the 23andMe (Table S3) or the Upenn Cohort (Table S4).
GWAS to nominate novel ICD risk variants
Prior studies of genetic risk factors for ICD in PD have employed a candidate gene approach, which may limit the discovery of novel variants. Moreover, the lack of clear associations between ICD and each of the 13 candidate SNPs nominated from the prior literature in the 23andMe, PPMI, and Upenn cohorts underlines the need for a discovery approach. Thus, we next employed a GWAS approach in the 23andMe PD cohort. After quality‐control filtering, 15.2M SNPs were analyzed for association with ICD, as ascertained by QUIP. Four loci associated with ICD at suggestive p‐values of 4.9e‐07 to 1.3e‐06 (Fig. 2). Minor allele frequencies for each of the four SNPs ranged from 0.019 to 0.267. Three of these four suggestive association signals are localized within the following genes—PRKAG2, MEFV, and PRKCE (Fig. 3). Moreover, multiple linked SNPs associated with ICD are localized within PRKAG2 and PRKCE, adding confidence to these signals (Fig. 3).
Figure 2.
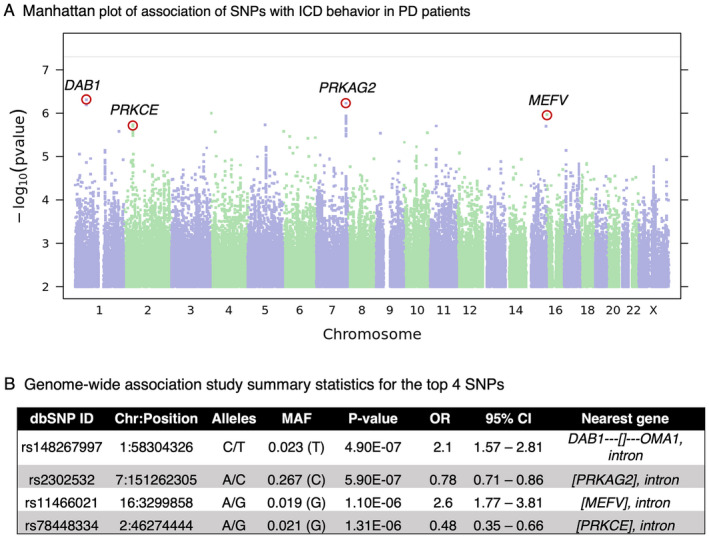
The top four SNPs revealed by 23andMe GWAS. (A) Manhattan plot of GWAS on 23andMe Cohort comparing PD subjects with and without ICD behavior. For each SNP, −log10 scaled p‐value is plotted against chromosomal position. The top 4 SNPs (p < 1.30e‐06) are labeled by the nearest gene. The horizontal solid line indicates the genome‐wide significant cutoff p‐value (p = 5.0e‐08). (B) GWAS summary statistics for the most highly associated variants. For each SNP we show: dbSNP build 146 rsid, chromosomal position (GRCh37 build), the two SNP alleles (A1/A2) in alphabetical order, OR for allele A2, the association test p‐value adjusted for genomic inflation, the confidence interval based on the standard error of the effect size, and the nearest gene. The nearest gene legend: [Gene1, Gene2,…] = The SNP is contained within the transcripts of the specified gene(s). Gene‐‐‐[] = The SNP is flanked by gene on the left and there is no gene within 1 Mb on the right. []‐‐‐Gene = The SNP is flanked by gene on the right and there is no gene within 1 Mb on the left. ICD, impulse control disorder; PD, Parkinson's disease; GWAS, Genomewide Association Study; OR, odds ratio. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 3.
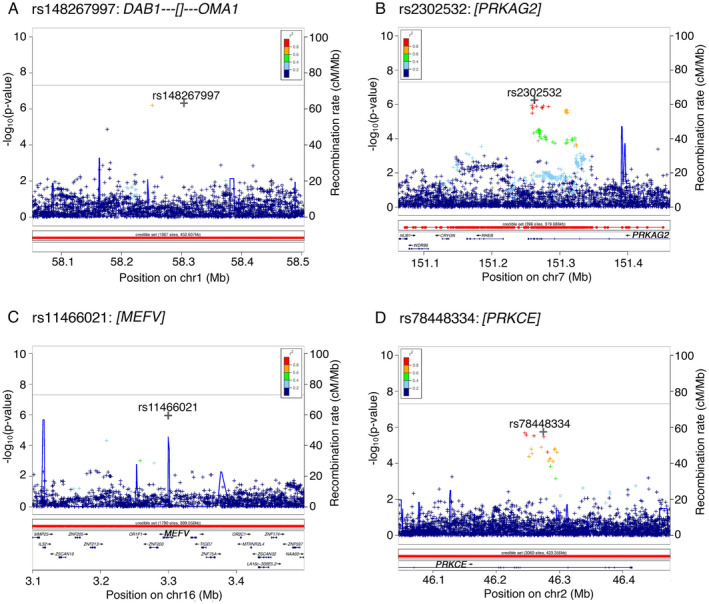
Regional association plots of both genotyped and imputed SNPs across four genomic regions linked to ICD behavior in PD subjects. (A) Region chr1p32.2 shows association of rs148267997 annotated to DAB1. (B) chr7q36.1 region with rs2302532 as a top associated SNP annotated to PRKAG2. (C) chr16p13.3 region with rs11466021 as a top associated SNP annotated to MEFV. (D) Region chr2p21 shows association of rs78448334 annotated to PRKCE. A –log10 p‐value for association between individual SNPs and ICD is plotted against the SNP's chromosomal position. X‐axis shows physical position based on NCBI genome Build 37. The right y axis shows the recombination rate (solid blue line on the plots) estimated from 1000 Genomes Project. A symbol “o” indicates a genotyped variant, a “◊” indicates a protein altering genotyped SNP, “+” is an imputed variant, and an “x” indicates a protein‐altering imputed SNP. Color represents the pairwise LD with the SNP with the most significant p‐value at each locus computed from a set of 10,000 23andMe samples. ICD, impulse control disorder; PD, Parkinson's disease. [Colour figure can be viewed at wileyonlinelibrary.com]
Clinico‐genetic predictor of ICD in PD
While single‐SNP associations with ICD in PD are worthy of mechanistic investigation, our primary goal in this study was to develop a clinico‐genetic predictor that might guide drug choice for PD patients at the individual level. Thus, we investigated the 13 SNPs previously nominated from the Kraemmer et al study, 11 the four SNPs newly associated with ICD in the 23andMe cohort, and eight clinical variables associated with ICD (age, sex, cohort, disease duration, education, ethnicity, DA use, and levodopa use) to develop such a predictor. Moreover, we sought to create a broadly applicable predictor of ICD risk informative in various types of clinical cohorts.
To do this, we turned to the PPMI and Upenn cohorts, which differ in stage of PD, with the PPMI study enrolling participants with PD at the time of diagnosis, while the Upenn study captures a typical subspecialty clinic population, with an average PD duration of 10.8 years at ICD assessment. Specifically, we combined these two cohorts and then split individuals randomly (2:1 ratio) into a Training dataset comprised of 339 subjects (261 ICD− and 78 ICD+) and a non‐overlapping Test dataset comprised of 169 subjects (130 ICD− and 39 ICD+).
In the Training dataset, we used backward stepwise logistic regression starting with our full model (25 variables: 17 SNPs +8 clinical). The optimal model included the following nine variables: cohort, age, sex, ethnicity, disease duration, DA therapy, levodopa therapy, and genotype at two SNPs; rs1800497 (in the dopamine receptor D2 gene, DRD2) and rs1799971 (in the Opioid Receptor Mu 1 gene, OPRM1). Fitting this nine‐variable model to the Training dataset by Bayesian logistic regression, we obtained estimates of coefficients for each variable. In our Training dataset, for the final model, all clinical covariates except ethnicity significantly associated with ICD, while each of the two SNPs showed borderline associations (p‐value 0.10–0.17) with ICD (Fig. 4A). The odds ratio for ICD for DA use was 1.8, while levodopa use was negatively associated with ICD.
Figure 4.
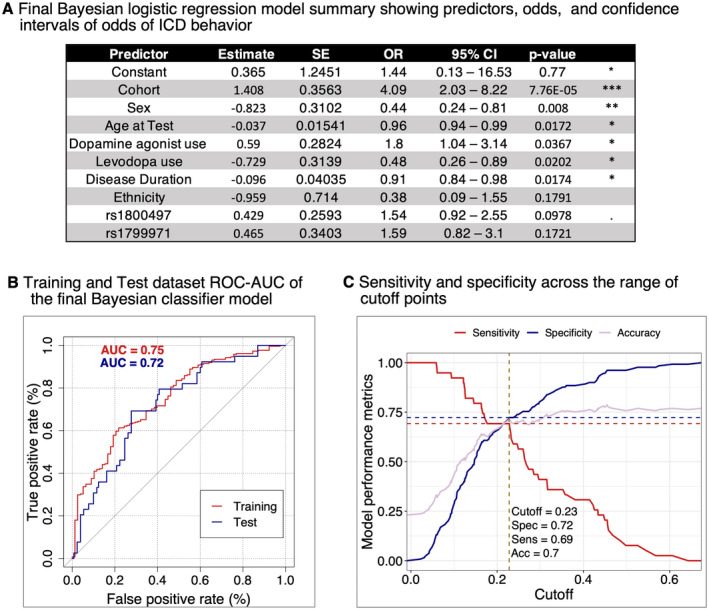
Development of ICD behavior classifier model. (A) The Bayesian logistic regression model estimates of the effects of two SNPs, adjusted for cohort, age at test, sex, dopamine agonist use, levodopa use, disease duration and ethnicity. We calculated the upper (UCL) and lower (LCL) confidence limits of odds of ICD behavior as: CL = odds ±1.96 SE (odds), where , and is a linear predictor of ICD. Cohort = UPenn versus PPMI, with UPenn associated with higher risk of ICD (positive estimate), Sex = female versus male, with females associated with lower risk of ICD (negative estimate). (B) The performance of the Bayesian classifier model measured in the Training dataset (261 ICD− and 78 ICD+ participants) by ROC‐AUC was 75%. The same model achieved ROC‐AUC = 72% when we performed prediction in the non‐overlapping Test dataset (130 ICD− and 39 ICD+ participants). (C) Estimating the best ROC‐AUC cutoff point in the Test dataset. Specificity and sensitivity of final Bayesian logistic regression model when predicting ICD behavior in the Test dataset across a range of cutoff points. We performed this analysis using the method closest.topleft (pROC package function coords), which revealed 0.23 as the best cutoff point, yielding an accuracy of 70%, sensitivity of 69% and specificity of 72% (dotted lines). ICD, impulse control disorder; ROC‐AUC, receiver operator characteristic curves‐area under the curve. *p < 0.05, **p < 0.01, ***p < 0.001. [Colour figure can be viewed at wileyonlinelibrary.com]
We assessed the performance of this model in our Training dataset by receiver operator characteristic curves (ROC), AUC, and accuracy metrics. Our predictor model showed a moderate ability to differentiate ICD+ from ICD− PD subjects with an AUC of 0.75 (Fig. 4B).
Model performance in the Test dataset
We next applied the ICD‐predictor model developed in our Training dataset to the Test dataset of PD individuals whose data were not used to develop the predictor. Our model performed equally well in the Test dataset, with a ROC‐AUC of 0.72 (Fig. 4B). To refine ICD prediction in the Test dataset, we also compared sensitivity versus specificity across a range of thresholds (Fig. 4C).
Prediction of ICD‐RS in Test dataset
To develop a clinically useful tool, we used our Bayesian logistic regression model to generate a continuous risk score for ICDs in PD (ICD‐RS, Fig. 5A).
Figure 5.
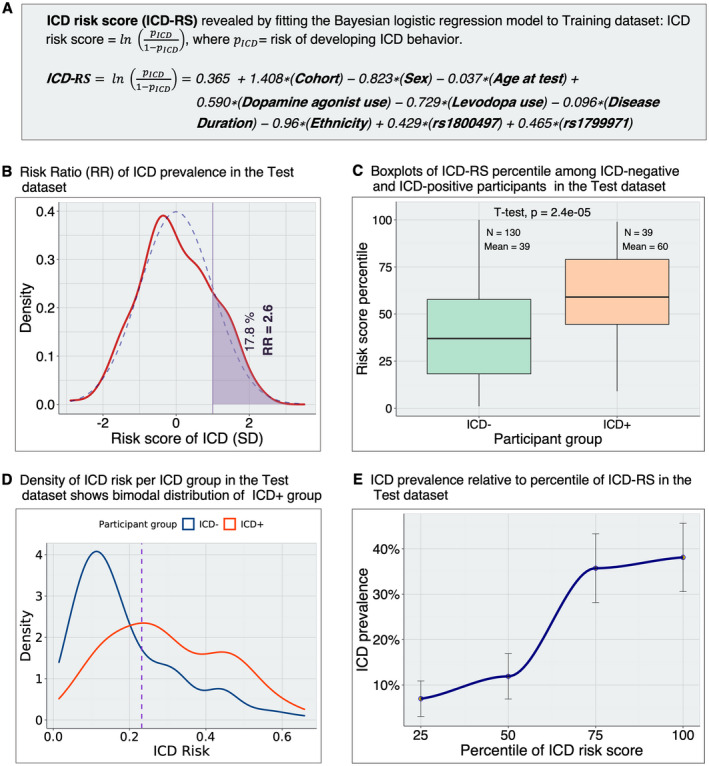
Risk scores for development of ICD behavior in PD subjects. (A) Calculation of ICD‐RS in the Test dataset. We calculate the ICD‐RSs for each participant in the Test dataset using the log odds (coefficient estimates) obtained by fitting the final Bayesian logistic regression model to the Training dataset. (B) Distribution of the RR in the Test dataset. The RR is the ratio of the empirical ICD prevalence within subgroups of the Test dataset. First, we calculated the ICD prevalence in the group of PD participants with ICD‐RS >1 SD above the mean of ICD‐RS. Then, we calculated ICD prevalence in the remainder of the PD participants (ICD‐RS below the cutoff of +1 SD): . The participants above 1 standard deviation of ICD‐RS have 2.6‐fold higher rates of ICD behavior than the rest of the participants in the Test dataset. Dashed blue and solid red lines represent normal and empirical distribution of ICD‐RS, respectively. (C) ICD‐RS (log odds) percentile among ICD+ versus ICD− PD participants in the Test dataset. While the horizontal line within the box indicates the median, we also show the percentile mean for each group. Both median and mean percentile are higher in the ICD+ group. (D) Distributions of risk (pICD) per ICD group. Both ICD+ PD and ICD− PD are skewed to the left because only ~23% of participants are ICD+. The purple dotted line indicates the best threshold (0.23) as estimated by the closest.topleft method. (E) The relationship between prevalence of ICD and ICD‐RS percentiles in the Test dataset. Error bars indicate standard errors (SE) generated by 1000 bootstrap replicates. ICD prevalence, binned according to percentiles of ICD‐RS, is highly correlated with ICD‐RS percentiles. Empirical ICD prevalence increases from 7% in individuals within the lowest quartile of ICD‐RS to 38% in the highest quartile. ICD, impulse control disorder; ICD‐RS, ICD risk scores; RR, risk ratio. [Colour figure can be viewed at wileyonlinelibrary.com]
We then evaluated the distribution of ICD‐RS in the Test dataset only. For individuals with ICD‐RS greater than +1 standard deviation above the mean, we found the empirical prevalence of ICD was 2.6‐fold higher than for individuals below that cutoff (Fig. 5B). Among the 39 ICD+ individuals in the Test dataset, the average ICD‐RS was significantly higher (p = 2.4e‐05) than for the 130 ICD− individuals (Fig. 5C). Moreover, we found that the ICD+ individuals demonstrated a bimodal distribution of ICD‐RS, with one subgroup of individuals slightly “right‐shifted” in ICD‐RS from the ICD‐group and a second subgroup of individuals with much higher ICD‐RS than the ICD‐group (Fig. 5D). Importantly, we found that individuals in the lowest quartile of ICD‐RS had minimal rates of ICD – ~7%, with bootstrap estimates SE in this quartile ranging from 3% to 11%—while ICD rates in the highest quartile of ICD‐RS approached 40% (Fig. 5E).
Development and performance of a genICD‐RS
Because a real‐world tool for use in identifying high‐ versus low‐ICD risk PD individuals would not depend on the cohort of origin, we finally sought to develop a genICD‐RS that omits Cohort as a predictive variable, testing its performance for risk stratification in the combined PPMI (n = 320) and UPenn (n = 188) PD individuals.
As shown in Figure S1, the remaining input variables for the genICD‐RS showed the same direction and similar magnitude as the ICD‐RS, and the performance of the genICD‐RS was similar as well. Specifically, for individuals with genICD‐RS greater than 1+ standard deviation above the mean, the empirical prevalence of ICD was 2.7‐fold higher than for individuals below that cutoff (Fig. S1B). Moreover, we found that individuals in the lowest quartile of genICD‐RS had much lower rates of ICD (12%), while ICD rate in the highest quartile of genICD‐RS was 43% (Fig. S1E).
Discussion
In the current study, we investigated multiple PD cohorts comprising 5770 PD individuals, employing a discovery‐replication design to better understand and predict ICD development in PD. We identified four novel genetic variants through GWAS for ICD in the 23andMe PD cohort. We then combined these newly discovered variants with genetic variants previously reported to associate with ICDs, developing a clinico‐genetic predictor for ICD development. Testing this predictor in PD individuals from both the Penn‐based cohort and the international PPMI cohort, we found that it achieved moderately high performance (AUC 0.75–0.72) in both the Training dataset in which it was developed and a held‐out Test dataset. From our predictor model, we developed a continuous metric, the ICD‐RS, demonstrating that this tool, incorporating just seven easily obtained clinical variables and genotypes at two SNPs, could risk‐stratify PD individuals with respect to ICD. Specifically, in the Test cohort individuals in the highest quartile of ICD‐RS had 38% prevalence of ICD, whereas PD individuals in the lowest quartile of ICD‐RS had only 7% ICD prevalence, a more than fivefold difference. Moreover, a generalized version of the ICD‐RS (the genICD‐RS), which omits cohort as an input variable and can thus in principle be tested in any clinical cohort, performed similarly.
Our findings have implications for pharmacogenetic decision‐making in PD. In particular, multiple previous reports, corroborated by our current study, suggest that a large proportion of PD individuals may suffer from ICD during their disease course. 2 , 4 , 5 , 14 , 31 These impulsive behaviors are strongly associated with the use of DAs. In clinical practice, the clear utility of DAs in managing PD symptoms may support their continued use despite the concern for triggering an ICD, and from a practical perspective the widespread use of DAs to control motor symptoms in PD patients largely precludes the adoption of strategies to eliminate DA exposure entirely. Thus, an easily enacted risk stratification strategy to determine, on an individual basis, which PD patients may be at highest versus lowest risk for ICD development could impact the field substantially. Simply put, high‐risk PD individuals might be counseled to use levodopa for motor symptom control, while DA use could still be considered in low‐risk PD individuals.
The ICD‐RS reported here can inform exactly such a risk‐stratification strategy. Clinical variables could be easily obtained on routine care visits, while a blood or saliva sample could serve as a source for DNA for genotyping. Because medication decision‐making in PD is rarely an emergent task, an ICD‐RS, obtainable within hours to days, could then inform drug choice, in a first example of “precision medicine” approaches in PD. While the exact cut‐offs for ICD‐RS that might be used for decision‐making regarding DA use, or any medication choice (since our model is agnostic with respect to use), remain to be determined, we note that our observed ICD HR of 2.6 for individuals with ICD‐RS >1 SD above the mean is considerably larger than the HR seen for one example of pharmacogenetic decision‐making already used in clinical medicine. Specifically, CYP2C19 genotypes, known to affect efficacy of clopidogrel for inhibition of platelet aggregation, are used for antiplatelet drug selection in cardiology, 32 despite the fact that the HR for major cardiovascular events observed with clopidogrel use is only 1.55 for carriers of one CYP2C19 loss‐of‐function allele and 1.76 for carriers of two alleles. 33
Strengths of this study, which support downstream efforts to translate findings into the clinical setting, include ascertainment of ICD by standardized, validated instruments, the discovery‐replication design, and reproducibility of findings across multiple large, multi‐site PD cohorts. Specifically, we employed versions of the QUIP and QUIP‐RS, developed in 2009 and 2012, and both recommended by the International Parkinson and Movement Disorder Society for evaluation of ICD behaviors, 18 for ascertainment of ICD in all three cohorts, ensuring that this important clinical measure was obtained in the most rigorous fashion possible for a large‐scale study. Moreover, our study had multiple phases, with the discovery of novel variants associating with ICD by GWAS in the 23andMe cohort, followed by testing of these variants in the PPMI and UPenn cohorts; and development of the ICD‐RS in a Training dataset, followed by validation of predictor performance in a non‐overlapping Test dataset of PD individuals. This iterative discovery‐replication design guards against overfitting (and indeed, the equal performance of our predictor in both the Training and Test datasets confirms that overfitting did not occur) and increases confidence in our findings. Finally, our use of the highly characterized UPenn and international PPMI cohorts, alongside the large, geographically dispersed 23andMe PD cohort, to develop and test our predictor greatly increases the likelihood that our findings will be widely applicable in PD. Indeed, our sample size of 5770 PD patients is the largest screening study of ICD behaviors in PD to date and the number of patients screening positive for an ICD at any point during the course of PD across the three studies included here is the largest sample size of affected patients known to date.
The limitations of our study should be considered alongside the previously mentioned strengths. First, all clinical data from the 23andMe cohort is by self‐report, so there is potential for bias both in terms of who chose to participate in this study and through error in self‐report. We believe, however, that the benefit afforded by our ability to access such a large cohort of PD participants, which enables research approaches such as the GWAS for ICD in PD reported here, outweighs this liability. Additionally, the four novel variants conferring risk for ICD in PD found by GWAS here associate at p‐values that are suggestive of true association (p‐values 4.9e‐07 to 1.3e‐06) but fall slightly short of genomewide significance. However, our strategy of subsequent validation of preliminary results from the 23andMe cohort in the PPMI and UPenn cohorts ensures that our final conclusions are not overly influenced by any one cohort. Second, the AUC for ICD‐RS was ~0.72 in the Test cohort, suggesting moderately high, but far from perfect, performance. It is possible that multiple underlying biological substrates may contribute to the clinical phenomenon of ICDs in PD, putting a biologically based “ceiling” on performance for any global ICD predictor. Moreover, the three cohorts studied here differ significantly from each other, in rates of ICDs, disease duration, and other parameters, introducing heterogeneity that may also limit predictor performance. That said, our goal in this study was to create a clinically useful pharmacogenetic tool. As such, basing our analyses on PD individuals across a wide spectrum of disease, dispersed across countries and cohorts, is most likely to result in widely applicable findings. We additionally point to the ability of the ICD‐RS, as well as the more generalizable genICD‐RS, to separate quartiles of PD individuals whose actual ICD prevalence differed substantially and to a clinically meaningful extent (~fourfold increase in ICD comparing the highest vs. lowest quartiles of risk), supporting the utility of this score. We note, moreover, that while additional clinical information, such as a history of psychiatric disease or substance use disorder, may further refine estimates of ICD risk, the tool presented here requires minimal clinical data, with all variables easily obtained and objective in nature, allowing for its use in a wide variety of settings.
In summary, we present our findings from a study of 5770 PD individuals from the UPenn, PPMI, and 23andMe cohorts, demonstrating that an ICD risk predictor composed of seven easily obtained clinical variables and genotype at two SNPs can identify PD individuals at extremes of risk for ICD development. Our findings have clinical implications for pharmacogenetic decision‐making in PD: identification of high‐ICD‐risk individuals may allow for avoidance of DA use in this group, sparing them considerable ICD‐related morbidity. More generally, the development of molecular tools, such as the ICD‐RS reported here, may permit a new “precision medicine” approach to the care of patients with neurodegenerative disease.
Author Contributions
The author contributions to this work are as follows. D. W., T. F. T., P. C., and A. S. C.‐P. designed the study. D. W., P. F., E. M., E. S., J. Q. T., and V. M. V. D. acquired the data. M. P., P. F., and A. S. C.‐P. analyzed the data. D. W., M. P., and A. S. C.‐P. drafted the majority of the manuscript, and all authors edited and approved the final manuscript.
Conflict of Interest
Alice S. Chen‐Plotkin, Marijan Posavi, and Daniel Weintraub declare that they are the inventors of a University of Pennsylvania patent (pending) covering prediction of impulsivity in Parkinson's Disease.
Pierre Fontanillas and Paul Cannon are employed by and hold stock or stock options in 23andMe, Inc.
Supporting information
Figure S1. Risk scores for development of ICD behavior in PD subjects for clinical use.
Table S1. Characteristics of 17 nominated SNPs that were tested for association with ICD behavior
Table S2. Summary statistics of 17 SNPs association with ICD behavior in PPMI cohort (n = 320).
Table S3. Summary statistics of 13 SNPs association with ICD behavior in 23andMe cohort (n = 5262).
Table S4. Summary statistics of 17 SNPs association with ICD behavior in UPenn cohort (n = 188).
Acknowledgments
We thank Travis Unger for technical assistance. We additionally thank our patients and their families for their generosity in contributing to this research. We thank the 23andMe research participants who made this study possible. Members of the 23andMe Research Team are: Michelle Agee, Stella Aslibekyan, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah K. Clark, Sarah L. Elson, Kipper Fletez‐Brant, Pierre Fontanillas, Nicholas A. Furlotte, Pooja M. Gandhi, Karl Heilbron, Barry Hicks, David A. Hinds, Karen E. Huber, Ethan M. Jewett, Yunxuan Jiang, Aaron Kleinman, Keng‐Han Lin, Nadia K. Litterman, Marie K. Luff, Jennifer C. McCreight, Matthew H. McIntyre, Kimberly F. McManus, Joanna L. Mountain, Sahar V. Mozaffari, Priyanka Nandakumar, Elizabeth S. Noblin, Carrie A. M. Northover, Jared O'Connell, Aaron A. Petrakovitz, Steven J. Pitts, G. David Poznik, J. Fah Sathirapongsasuti, Madeleine Schoetter, Anjali J. Shastri, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Robert J. Tunney, Vladimir Vacic, Xin Wang, Amir S. Zare. This research was supported by the NIH (RO1 NS115139, U19 AG062418, P50 NS053488), a Biomarkers Across Neurodegenerative Diseases (BAND) grant from the Michael J. Fox Foundation/Alzheimer's Association/Weston Institute, the Penn Center for Precision Medicine, and a Copp Foundation grant. Alice Chen‐Plotkin is additionally supported by the Parker Family Chair, the AHA/Allen Institute Brain Health Initiative, and the Chan Zuckerberg Initiative Neurodegeneration Challenge.
Funding Information
This research was supported by the NIH (RO1 NS115139, U19 AG062418, P50 NS053488), a Biomarkers Across Neurodegenerative Diseases (BAND) grant from the Michael J. Fox Foundation/Alzheimer's Association/Weston Institute, the Penn Center for Precision Medicine, and a Copp Foundation grant. Alice Chen‐Plotkin is additionally supported by the Parker Family Chair, the AHA/Allen Institute Brain Health Initiative, and the Chan Zuckerberg Initiative Neurodegeneration Challenge.
Funding Statement
This work was funded by Allen Institute ; Alzheimer's Association ; Chan Zuckerberg Initiative ; Michael J. Fox Foundation for Parkinson's Research ; National Institute of Neurological Disorders and Stroke grants P50 NS053488 and RO1 NS115139; National Institute on Aging grant U19 AG062418; American Heart Association ; Parker Family Chair; Copp Foundation; Penn Center for Precision Medicine ; Weston Institute; National Institutes of Health .
REFERENCES
- 1. Voon V, Fox SH. Medication‐related impulse control and repetitive behaviors in Parkinson disease. Arch Neurol. 2007;64(8):1089‐1096. [DOI] [PubMed] [Google Scholar]
- 2. Weintraub D, Siderowf AD, Potenza MN, et al. Association of dopamine agonist use with impulse control disorders in Parkinson disease. Arch Neurol. 2006;63(7):969‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weintraub D, Papay K, Siderowf A; Parkinson's Progression Markers Initiative . Screening for impulse control symptoms in patients with de novo Parkinson disease: a case‐control study. Neurology. 2013;80(2):176‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weintraub D, Koester J, Potenza MN, et al. Impulse control disorders in Parkinson disease: a cross‐sectional study of 3090 patients. Arch Neurol. 2010;67(5):589‐595. doi: 10.1001/archneurol.2010.65 [DOI] [PubMed] [Google Scholar]
- 5. Corvol J‐C, Artaud F, Cormier‐Dequaire F, et al. Longitudinal analysis of impulse control disorders in Parkinson disease. Neurology. 2018;91(3):e189‐e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biundo R, Weis L, Abbruzzese G, et al. Impulse control disorders in advanced Parkinson's disease with dyskinesia: the ALTHEA study. Mov Disord. 2017;32:1557‐1565. [DOI] [PubMed] [Google Scholar]
- 7. Voon V, Sohr M, Lang AE, et al. Impulse control disorders in Parkinson disease: a multicenter case‐control study. Ann Neurol. 2011;69(6):986‐996. [DOI] [PubMed] [Google Scholar]
- 8. Fantini ML, Figorilli M, Arnulf I, et al. Sleep and REM sleep behaviour disorder in Parkinson's disease with impulse control disorder. J Neurol Neurosurg Psychiatry. 2018;89:305‐310. [DOI] [PubMed] [Google Scholar]
- 9. Marín‐Lahoz J, Sampedro F, Martinez‐Horta S, Pagonabarraga J, Kulisevsky J. Depression as a risk factor for impulse control disorders in Parkinson disease. Ann Neurol. 2019;86(5):762‐769. [DOI] [PubMed] [Google Scholar]
- 10. Evans AH, Pavese N, Lawrence AD, et al. Compulsive drug use linked to sensitized ventral striatal dopamine transmission. Ann Neurol. 2006;59:852‐858. [DOI] [PubMed] [Google Scholar]
- 11. Kraemmer J, Smith K, Weintraub D, et al. Clinical‐genetic model predicts incident impulse control disorders in Parkinson's disease. J Neurol Neurosurg Psychiatry. 2016;87(10):1106‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cormier‐Dequaire F, Bekadar S, Anheim M, et al. Suggestive association between OPRM1 and impulse control disorders in Parkinson's disease. Mov Disord. 2018;33(12):1878‐1886. [DOI] [PubMed] [Google Scholar]
- 13. Papay K, Mamikonyan E, Siderowf AD, et al. Patient versus informant reporting of ICD symptoms in Parkinson's disease using the QUIP: validity and variability. Parkinsonism Relat Disord. 2011;17(3):153‐155. doi: 10.1016/j.parkreldis.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bastiaens J, Dorfman BJ, Christos PJ, Nirenberg MJ. Prospective cohort study of impulse control disorders in Parkinson's disease. Mov Disord. 2013;28(3):327‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Antonini A, Chaudhuri KR, Boroojerdi B, et al. Impulse control disorder related behaviours during long‐term rotigotine treatment: a post hoc analysis. Eur J Neurol. 2016;23(10):1556‐1565. doi: 10.1111/ene.13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weintraub D, Stewart S, Shea JA, et al. Validation of the questionnaire for impulsive‐compulsive behaviors in Parkinson's disease (QUIP). Mov Disord. 2009;24:1461‐1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weintraub D, Mamikonyan E, Papay K, et al. Questionnaire for impulsive‐compulsive disorders in Parkinson's Disease‐Rating Scale. Mov Disord. 2012;27(2):242‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans AH, Okai D, Weintraub D, et al. Scales to assess impulsive and compulsive behaviors in Parkinson's disease: critique and recommendations. Mov Disord. 2019;34(6):791‐798. [DOI] [PubMed] [Google Scholar]
- 19. Seppi K, Ray Chaudhuri K, Coelho M, et al. Update on treatments for nonmotor symptoms of Parkinson's disease‐an evidence‐based medicine review. Mov Disord. 2019;34:180‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mamikonyan E, Siderowf AD, Duda JE, et al. Long‐term follow‐up of impulse control disorders in Parkinson's disease. Mov Disord. 2008;23:75‐80. PMCID: PMC17960796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabinak CA, Nirenberg MJ. Dopamine agonist withdrawal syndrome in Parkinson disease. Arch Neurol. 2010;67:58‐63. [DOI] [PubMed] [Google Scholar]
- 22. Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walter K, Min JL, Huang J, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian C, Hromatka BS, Kiefer AK, et al. Genome‐wide association and HLA region fine‐mapping studies identify susceptibility loci for multiple common infections. Nat Commun. 2017;8(1):599. doi: 10.1038/s41467-017-00257-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durand EY, Do CB, Mountain JL, Macpherson JM. Ancestry composition: a novel, efficient pipeline for ancestry deconvolution. bioRxiv. 2014;010512. Accessed May 21, 2021. http://biorxiv.org/content/early/2014/10/18/010512.abstract [Google Scholar]
- 26. Marek K, Jennings D, Lasch S, et al. The Parkinson progression marker initiative (PPMI). Prog Neurobiol. 2011;95:629‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28(5):1‐26.27774042 [Google Scholar]
- 28. Robin X, Turck N, Hainard A, et al. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(Swets 1973):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. González JR, Armengol L, Solé X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23(5):654‐655. doi: 10.1093/bioinformatics/btm025 [DOI] [PubMed] [Google Scholar]
- 30. Weintraub D, David AS, Evans AH, Grant JE, Stacy M. Clinical spectrum of impulse control disorders in Parkinson's disease. Mov Disord. 2015;30(2):121‐127. [DOI] [PubMed] [Google Scholar]
- 31. Voon V, Hassan K, Zurowski M, et al. Prospective prevalence of pathological gambling and medication association in Parkinson disease. Neurology. 2006;66:1750‐1752. [DOI] [PubMed] [Google Scholar]
- 32. Tuteja S, Glick H, Matthai W, et al. Prospective CYP2C19 genotyping to guide antiplatelet therapy following percutaneous coronary intervention. Circ Genom Precis Med. 2020;13(1):e002640. doi: 10.1161/CIRCGEN.119.002640 [DOI] [PubMed] [Google Scholar]
- 33. Mega JL, Simon T, Collet J‐P, et al. Reduced‐function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta‐analysis. JAMA. 2010;304(16):1821‐1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Risk scores for development of ICD behavior in PD subjects for clinical use.
Table S1. Characteristics of 17 nominated SNPs that were tested for association with ICD behavior
Table S2. Summary statistics of 17 SNPs association with ICD behavior in PPMI cohort (n = 320).
Table S3. Summary statistics of 13 SNPs association with ICD behavior in 23andMe cohort (n = 5262).
Table S4. Summary statistics of 17 SNPs association with ICD behavior in UPenn cohort (n = 188).


