Abstract
Protein splicing in trans has been demonstrated both in vivo and in vitro by biochemical and immunological analyses, but in vivo production of a functional protein by trans-splicing has not been reported previously. In this study, we used the DnaE intein from Synechocystis sp. strain PCC6803, which presumably reconstitutes functional DnaE protein by trans-splicing in vivo, to produce functional herbicide-resistant acetolactate synthase II (ALSII) from two unlinked gene fragments in Escherichia coli. The gene for herbicide-resistant ALSII was fused in frame to DnaE intein segments capable of promoting protein splicing in trans and was expressed from two compatible plasmids as two unlinked fragments. Cotransformation of E. coli with the two plasmids led to production of a functional enzyme that conferred herbicide resistance to the host E. coli cells. These results demonstrate the feasibility of expressing functional genes from two unlinked DNA loci and provide a model for the design of nontransferable transgenes in plants.
Although the dnaE gene of Synechocystis sp. strain PCC6803 is encoded in two segments separated by 745 kb on the chromosome, it presumably yields a functional product, the catalytic α subunit of the replicative DNA polymerase, through protein splicing in trans mediated by intein segments that are fused to ends of the two DnaE fragments (16). The split DnaE intein has been shown to be capable of trans-splicing and cyclization of various proteins (4), but an ability to promote functional reconstitution of an enzyme in vivo has not been demonstrated. If this were possible, the split DnaE intein would provide a tool for generating organisms with unlinked bipartite transgenes which would yield functional products yet would have a low probability of joint horizontal transfer to other organisms. Such an attribute would be of special value for agricultural use of genetically modified plants, which has provoked a heated debate over potential threats to the environment due to the possible transfer of transgenes to other organisms (5, 6, 9, 13). In the case of herbicide resistance, it has been postulated that this could contribute to the emergence of “superweeds” (J. Bergelson, C. B. Purrington, and G. Wichmann, Letter, Nature 395:25, 1998).
To demonstrate production of a functional protein by trans-splicing in vivo, we chose the protein acetolactate synthase (ALS). The enzyme ALS (EC 4.1.3.18) in both bacteria and plants is the target for sulfonylurea herbicides, such as sulfometuron methyl (SM) (11, 12, 14, 17). Single point mutations in ALS genes that confer herbicide resistance have been identified, and sulfonylurea herbicide-tolerant corn carrying a mutant copy of the ALS gene, such as ICI 8532 IT and Pioneer 3180 IR, has been commercialized (2, 12, 17). ALS catalyzes the first common step in the biosynthesis of branched-chain amino acids in both plants and bacteria (3, 17). Escherichia coli has three ALS isoforms, two of which (ALSI and ALSIII) are sensitive to feedback inhibition by valine; consequently, the third isoform, ALSII, is essential for growth of E. coli in the presence of valine. A plasmid carrying the gene for ALSII has been shown to rescue E. coli ER2744, which lacks an active ALSII, from growth inhibition by valine (3, 11). Accordingly, we chose E. coli ER2744 and ALSII as the subjects of our initial model experiments (Fig. 1).
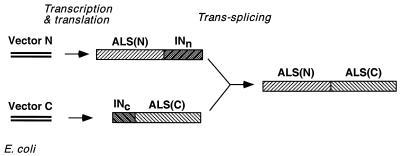
FIG. 1.
trans-Splicing of ALS protein in E. coli ER2744. The ALS gene carrying the herbicide resistance mutation Ala26 to Val26 is split by the Synechocystis sp. DnaE intein fragments (INn and INc) and is coexpressed as two inactive fusion proteins from two compatible vectors (4). Protein trans-splicing produces an active ALS protein that confers herbicide resistance to ER2744 host cells.
In this study, we demonstrated efficient in vivo production of E. coli and corn ALS (cALS) by protein trans-splicing. Reconstituted E. coli ALSII was shown to confer herbicide resistance to E. coli host cells.
MATERIALS AND METHODS
Bacterial strains and materials.
E. coli MI162 was obtained from the E. coli Genetic Stock Center, Yale University, New Haven, Conn. E. coli ER2744 [fhuA2 glnV44 el4-rfbD1? relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10 lacZ::T7 gene1] was obtained from New England Biolabs (Beverly, Mass.). The herbicide SM was purchased from Supelco (Bellefonte, Pa.). Valine was purchased from Sigma (St. Louis, Mo.). All restriction enzymes were obtained from New England Biolabs.
Construction of expression vectors.
E. coli ALSII DNA was cloned by PCR amplification of DNA extracted from E. coli MI162; 5′ GGAGGGGGCATATGAATGGCGCACAGTGGG 3′ and 5′ GGGGGGTCATGATAATTTCTCCAAC 3′ were the primers used in these reactions. The DNA fragment encoding the N-terminal 327 amino acid residues of ALSII was amplified by using forward primer 5′ GGGGGTCATGAATGGCGCACAGTGGG 3′ and reverse primer 5′ GCGCGCTC GAGTTGATTTAACGGCTGCTGTAATG 3′ and was inserted into the NcoI and XhoI sites of pMEB16, a derivative of pMEB21 (4), which generated the ALSII(N)-INn fusion gene. The DNA fragment encoding the C-terminal 221 amino acid residues of ALSII was amplified by using forward primer 5′ GCGCGACCGGTTGTGACTGGCAGCAACACTGC 3′ and reverse primer 5′ GGGGGGCTGCAGTCATGATAATTTCTCCAAC 3′ and was inserted into the AgeI and PstI sites of pKEB1 (4), which created the INc-ALSII(C) fusion gene. Ala26 to Val26, the herbicide resistance mutation in ALSII, was introduced by site-directed mutagenesis performed with primers 5′ CCGGGTGGCGTAATTATGCCGGTTTACG 3′ and 5′ CGTAAACCGGCATAATTACGCCACCCGG 3′ and a Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The INc-ALSII(C) fusion gene was further modified by deleting the coding region for the six intein residues (VKVIGR) following the translation initiation codon which yielded a construct expressing INcΔ6-ALSII(C). Note that plasmids pMEB and pKEB are compatible in E. coli (4).
cALS cDNA was cloned by reverse transcription-PCR from mRNA prepared from corn leaves with an RNAqueous kit (Ambion, Austin, Tex.) by using reverse primer 5′ ATCAGTACACAGTCCTGCCATC 3′ and forward primer 5′ GAGACAGCCGCCGCAACCAT 3′. DNA encoding the N-terminal 397 amino acid residues of the cALS gene was amplified by PCR performed with forward primer 5′ GGGCCCATATGGCCACCGCCGCCGCCGCG 3′ and reverse primer 5′ GGGCCCTCGAGGCTTCCTTCAAGAAGAGC 3′ and was cloned into the NdeI and XhoI sites of pMEB10 (4) which yielded cALS(N)-INn. The DNA fragment encoding the C-terminal 241 amino acid residues of the cALS gene was amplified by PCR performed with forward primer 5′ GGGCCACCGGTACATCAAAGAAGAGCTTG 3′ and reverse primer 5′ GGGGCTGCATTCAGTACACAGTCCTGCCATC 3′ and was inserted into the AgeI and PstI sites of pKEB1, which resulted in INc-cALS(C). To enhance the ability to undergo protein splicing in trans, the nonnative residues LEKFAEY were included at the junction of the ALS and the intein N-terminal segments, and residues CFNKSTG were included at the junction of the intein and the ALS C-terminal segments.
Western blot analysis.
A single bacterial colony was inoculated into Luria-Bertani (LB) medium supplemented with 100 μg of ampicillin per ml and 50 μg of kanamycin per ml. After incubation for 4 h at 37°C, the cells were induced by adding 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for 16 h at 15°C or as indicated below. Culture samples (40 μl) were removed, mixed with 20 μl of 3× sodium dodecyl sulfate (SDS) loading buffer (New England Biolabs), and boiled for 5 min, and 2-μl samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE on 12% Tris-glycine gels (Invitrogen, Carlsbad, Calif.). Protein bands were transferred to a nitrocellulose membrane. After blocking with 5% skim milk powder for 1 h at room temperature, the blots were incubated overnight at 4°C with antiserum (1:20,000 dilution) in the presence of 1% skim milk powder and then washed three times (15 min each) and incubated with 1:10,000-diluted horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at room temperature. The reaction was visualized with a chemiluminescent Western detection kit (New England Biolabs). The antisera used were raised in rabbits against peptides corresponding to residues Ala4 to Tyr23 (CAQWVVHALRAQGVNTVFGYG) or Val530 to Ser548 (CVWPLVPPGASNSEMLEKLS) of E. coli ALSII or to residues Lys66 to Ala85 (CKGADILVESLERCGVRDVFA) or Ile619 to Tyr638 (CIPSGGAFKDMILDGDGRTVY) of cALS.
Plate and liquid assays.
Plate assays were conducted to examine the ability of ALSII or its variants to rescue E. coli ER2744 from growth inhibition by valine (100 μg/ml) or valine plus the herbicide SM (50 μg/ml) on M9 minimum medium plates supplemented with 2 μg of thiamine per ml, 2 mM MgSO4, 0.1 mM CaCl2, 0.2% glucose, 50 μg of kanamycin per ml, 100 μg of ampicillin per ml, and 0.3 mM IPTG. Overnight cultures of the strains to be tested were streaked on M9 plates with or without valine and/or SM. The plates were incubated at various temperatures (see below) for 48 to 72 h before photographs were taken.
Growth in liquid media was examined as follows. A single colony was inoculated into LB medium supplemented with ampicillin and kanamycin. After incubation for 4 h at 37°C, protein expression was induced by 0.3 mM IPTG, and the cultures were shifted to 30°C and incubated for another 2 h. Culture samples (optical density at 600 nm, 0.8) were spun down, washed once with M9 medium, resuspended in the original volume of M9 medium, and inoculated into 50 volumes of LB medium containing 0.3 mM IPTG and supplemented with valine (100 μg/ml) and SM (50 μg/ml) as indicated below. The culture optical density at 600 nm was measured after 24 to 72 h.
RESULTS
Construction of E. coli ALSII-intein fusions.
The ALS genes of bacteria, yeasts, and higher plants have substantial sequence homology, but some highly variable regions can be identified. The region near residue Glu327 in the E. coli ALSII gene (Fig. 2) has a 10-amino-acid gap and, by analogy with the crystal structure of a homolog, pyruvate oxidase, appears to be part of a linker between two folding domains (10). We reasoned that the insertion of intein segments between Glu327 and Cys328 of ALSII might provide sufficient flexibility for trans-splicing.
FIG. 2.
Alignment of ALS protein sequences adjacent to the split sites. The E. coli ALSII (residues 288 to 367; accession no. S48893) and cALS (residues 357 to 445; accession no. S22490) sequences were aligned with corresponding sequences of E. coli ALSIII (residues 298 to 387; accession no. P27819) and tobacco ALSI (residues 386 to 474; accession no. P09342) and ALSII (residues 383 to 471; accession no. P09114). Residues that are identical in half or more of the proteins are highlighted. The arrowheads indicate the sites at which E. coli ALSII and cALS were split and fused to intein segments as described in the text.
The SM-resistant E. coli ALSII gene possessing an Ala26→Val26 mutation (ALSIIm) (8) was split and fused in frame to the DnaE intein coding sequences (Fig. 1). The ALSIIm N-terminal sequence (residues 1 to 327) was fused to the 123-residue N-terminal intein segment to obtain ALSIIm(N)-INn (49 kDa). Similarly, the ALSIIm C-terminal sequence (residues 328 to 548) was fused in frame to the 36-residue C-terminal intein segment to obtain INc-ALSII(C) (27 kDa). Seven additional amino acids were included at each junction of ALSII and intein sequences. These residues were shown to enhance the efficiency of trans-splicing in vivo and in vitro (4). Control experiments showed that a derivative of ALSIIm in which the same 14 amino acids were inserted between residues 327 and 328 (ALSIIm-14) was as effective as ALSIIm in rescuing E. coli ER2744 from growth inhibition by valine and SM (data not shown).
trans-Splicing of ALSIIm-intein fusion proteins.
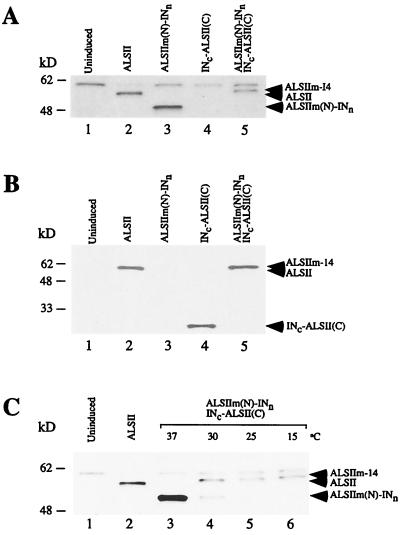
We predicted that when the two fusion proteins were coexpressed, trans-splicing of the two fragments of ALSIIm would produce a 59-kDa mature protein, ALSIIm-14, that differed from ALSIIm by the presence of 14 nonnative residues at the splicing junction. Indeed, coexpression of the two ALSIIm-intein fusion proteins at 25°C in E. coli ER2744 resulted in production of a 59-kDa protein that reacted with antibodies against either N- or C-terminal segments of ALSII, indicating that trans-splicing had occurred (Fig. 3A and B). As expected, this protein had a slightly higher molecular weight than the wild-type ALSII protein due to the presence of 14 extra amino acid residues. In cells expressing a single fusion protein [ALSIIm(N)-INn or INc-ALSII(C)], only the unspliced precursor protein was observed (Fig. 3A and B). trans-Splicing of the ALSIIm-intein fusion proteins appeared to be more efficient at lower temperatures (15 to 25°C) and was inhibited at 37°C (Fig. 3C), findings which are consistent with previous studies on the DnaE intein (4). At 15 and 25°C, ALSIIm(N)-INn was completely converted to the spliced product, ALSIIm-14, but at 30°C residual ALSIIm(N)-INn was observed, indicating a lower splicing efficiency. No spliced product was detected at 37°C.
FIG. 3.
Immunoblot analysis of production of ALSIIm-14 by protein trans-splicing. Cells transformed with plasmids were induced with 0.3 mM IPTG for 12 h at 25°C. Whole-cell lysates from uninduced cells (lane 1) or cells expressing ALSII (lane 2), ALSIIm(N)-INn (lane 3), INc-ALSII(C) (lane 4), or ALSIIm(N)-INn and INc-ALSII(C) (lane 5) were resolved by SDS-PAGE in 12% Tris-glycine gels and immunoblot analysis using antibodies raised against the N-terminal peptide (A) or C-terminal peptide (B) of ALSII. (C) Effect of temperature on trans-splicing. Whole-cell lysates were prepared from uninduced cells (lane 1) and cells expressing ALSII (lane 2) and ALSIIm(N)-INn and INc-ALSII(C) after induction with 0.3 mM IPTG at 37°C (lane 3), 30°C (lane 4), 25°C (lane 5), and 15°C (lane 6) and were subjected to immunoblotting with antiserum against the N-terminal peptide of ALSII. The 60-kDa band in panels A and C is an unknown protein that cross-reacts with the antiserum. kD, kilodaltons.
In vivo activity of reconstituted ALS.
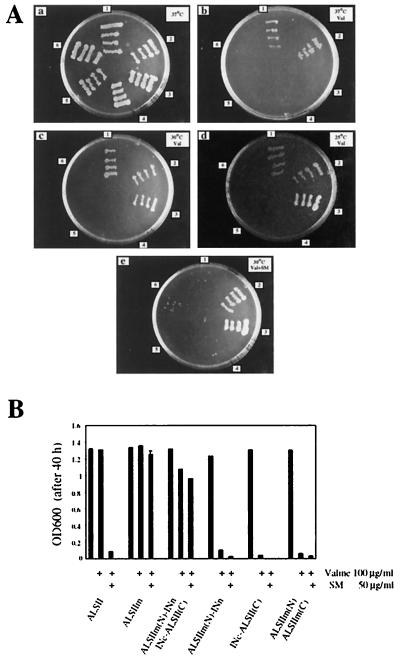
To determine whether the product of trans-splicing is indeed a functional, herbicide-resistant enzyme, we examined the effect of coexpression of the ALSIIm-intein fusion proteins on the growth response of E. coli ER2744 to valine and SM (Fig. 4A). Cells expressing the ALSIIm-intein fusion proteins or intact ALSIIm grew on agar media supplemented with valine and SM at 30°C. Coexpression of ALSIIm-intein fusions also rescued cell growth from valine inhibition at 25°C (Fig. 4A). However, coexpression of the ALSIIm-intein fusion proteins at 37°C did not rescue cells from valine inhibition, indicating that reconstitution of ALSIIm activity was temperature dependent. Expression of just one of the ALSIIm-intein fusion proteins or of a mixture of ALSIIm N- and C-terminal fragments not fused to the corresponding intein segments did not allow growth in the presence of valine. The results of the agar plate assay were confirmed by similar experiments performed in liquid culture (Fig. 4B). In the presence of valine, cells coexpressing the ALSIIm-intein fusion proteins grew at a rate similar to that of cells expressing ALSIIm protein (Fig. 4B). The slightly higher growth rate observed with cells expressing wild-type ALSII may have been due to the higher enzymatic activity of wild-type ALSII reported previously (8). The observation that functional reconstitution of ALSIIm occurred only when the ALSIIm fragments were fused to the appropriate intein segments shows that this process involved protein splicing rather than noncovalent complementation of the enzyme fragments. Furthermore, deletion of N-terminal residues 2 to 7 from the intein segment in the INc-ALSII(C) fusion resulted in no splicing activity, as shown by immunoblot analysis and the failure to rescue cells from valine (data not shown).
FIG. 4.
Effect of reconstitution of the ALSIIm-14 protein by trans-splicing on the growth of E. coli ER2744. (A) E. coli ER2744 transformed with plasmids expressing ALSII (sector 1), ALSIIm (sector 2), ALSIIm(N)-INn and INc-ALSII(C) (sector 3), ALSIIm(N)-INn (sector 4), INc-ALSII(C) (sector 5), or ALSIIm(N) and ALSII(C) (sector 6) were plated on M9 agar medium and incubated at 37°C (plate a), at 37°C with 100 μg of valine per ml (plate b), at 30°C with 100 μg of valine per ml (plate c), at 25°C with 100 μg of valine per ml (plate d), and at 30°C with 100 μg of valine per ml and 50 μg of SM per ml (plate e). All plates contained 0.3 mM IPTG. (B) E. coli ER2744 transformed with expression plasmids for fusion proteins as indicated on the figure were cultured in M9 medium containing 0.3 mM IPTG supplemented with valine and SM as indicated at the bottom. The cell optical density at 600 nm (OD600) was measured after incubation for 40 h at 30°C.
trans-Splicing of cALS-intein fusion proteins.
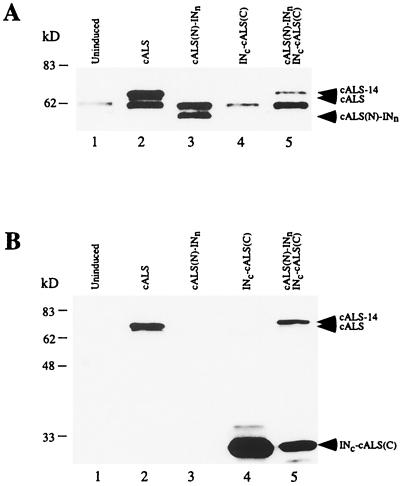
Successful reconstitution of functional ALS in E. coli suggested the possibility of utilizing this trans-splicing technology for containment of transgenes. As the next step, it was of interest to demonstrate intein-mediated trans-splicing of a plant gene in E. coli. To do this, we selected cALS, which is the basis of commercial herbicide-resistant corn hybrids (2). The bond between Ser397 and Thr398 of the 638-residue cALS was chosen as the split site, based on its sequence similarity with the split site of E. coli ALSII (Fig. 2). Two cALS-intein fusion proteins, cALS(N)-INn and INc-cALS(C), were constructed by using compatible E. coli expression vectors (4). Immunoblot analysis of extracts from cells coexpressing the cALS-intein fusion proteins at 25°C revealed protein having a molecular mass of approximately 69 kDa, the expected size of spliced cALS-14, which differed from cALS by having 14 additional amino acids (Fig. 5). The absence of residual cALS(N)-INn showed that trans-splicing occurred with high efficiency. The spliced product was not detected in cells expressing either of the cALS-intein fusion proteins alone. cALS is sensitive to valine that is present in growth medium to suppress bacterial ALSI and ALSIII activity (7). Therefore, as predicted, coexpression of the cALS-intein fusions did not rescue the growth of host cells in the presence of valine (data not shown). Thus, a plant model needs to be used to test the activity of trans-spliced cALS.
FIG. 5.
Production of full-length cALS by protein trans-splicing. Whole-cell lysates were prepared from uninduced E. coli ER2744 cells (lane 1) or from cells transformed with plasmids expressing cALS (lane 2), cALS(N)-INn (lane 3), INc-cALS(C) (lane 4), or cALS(N)-INn and INc-cALS(C) (lane 5) and were induced with 0.3 mM IPTG for 12 h at 25°C. Immunoblots were prepared after SDS-PAGE and were probed with antibodies against the N-terminal peptide (A) or C-terminal peptide (B) of cALS. A nonspecific 60-kDa protein species was detected by the antibody against the N terminus (A). The product of trans-splicing (cALS-14) has a slightly higher molecular weight than cALS because 14 additional amino acids are inserted at the splicing junction. kD, kilodaltons.
DISCUSSION
The data presented here demonstrated that active ALSII was reconstituted in E. coli by trans-splicing of two inactive intein fusions from different plasmids. In general, this approach may also be used to reconstitute other proteins that are not normally generated by a trans-splicing mechanism. Although this work was carried out with a bacterial model system, it showed the feasibility of splitting a gene of interest and inserting its fragments as intein fusions into unlinked regions of a genome to obtain a functional protein by protein splicing in trans. This method has an advantage over cis-splicing since it may permit expression of a gene from two different loci of a genome or two cellular compartments. Furthermore, trans-splicing joins two polypeptide chains via a covalent bond, which may result in an enzyme more stable than that produced by other methods, such as dimerization with affinity domains (1).
It was shown previously that the two Ssp DnaE intein segments have a strong noncovalent interaction (4, 5). It is conceivable that dimerization of the intein segments may be separable from the splicing activity and may provide an alternative approach for reconstitution of proteins. In this study, dimerization could have contributed under certain conditions to reconstitution of active ALSII, since incomplete splicing activity was observed at 30°C (Fig. 3). On the other hand, since no residual ALSII(N)- INn was detected by immunoblot assays at 25 or 15°C, it is likely that the trans-splicing product ALSIIm-14 is responsible for rescuing cell growth in the presence of valine (Fig. 3 and 4). Furthermore, the failure of the expressed ALSII-intein fusions to rescue cells from valine inhibition at 37°C is in agreement with the temperature-dependent trans-splicing activity (Fig. 3 and 4). This feature of temperature sensitivity may provide a controllable switch for regulating protein function in vivo.
The approach described here may have broad applications to gene manipulation in both prokaryotic and eukaryotic organisms. For example, the trans-splicing technology can be utilized for containment of a transgene. In the case of genetically modified plants, if one of the transgene fragments is fused to an appropriate chloroplast transit peptide, the fragments can be inserted separately into the nuclear and chloroplast genomes (or into the DNA of any two cellular compartments) to produce fusion proteins that undergo trans-splicing in the chloroplast. Since neither the nucleus nor the chloroplast would carry an intact transgene and since chloroplast genes are generally not transferred through pollen, this technology would essentially eliminate the possibility of transferring functional foreign genes from transgenic plants to closely related species by cross-pollination. The technology described in this paper should therefore go a long way towards reducing the potential environmental hazards of genetically modified plants.
ACKNOWLEDGMENTS
We thank Donald G. Comb, Thomas C. Evans, Jr., Sriharsa Pradhan, Lixin Chen, Eric Cantor, and Fana Mersha for valuable discussions and helpful comments and Emma Naylor, David Nathan, and Robert Maunus for technical assistance. We also thank Elisabeth Raleigh and Marion Sibley for providing E. coli ER2744.
REFERENCES
- 1.Behncken S N, Billestrup N, Brown R, Amstrup J, Conway-Campbell B, Waters M J. Growth hormone (GH)-independent dimerization of GH receptor by a leucine zipper results in constitutive activation. J Biol Chem. 2000;275:17000–17007. doi: 10.1074/jbc.275.22.17000. [DOI] [PubMed] [Google Scholar]
- 2.Bernasconi P, Woodworth A R, Rosen B A, Subramanian M V, Siehl D L. A naturally occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J Biol Chem. 1995;270:17381. doi: 10.1074/jbc.270.29.17381. [DOI] [PubMed] [Google Scholar]
- 3.Dailey F E, Cronan J E., Jr Acetohydroxy acid synthase I, a required enzyme for isoleucine and valine biosynthesis in Escherichia coli K-12 during growth on acetate as the sole carbon source. J Bacteriol. 1986;165:453–460. doi: 10.1128/jb.165.2.453-460.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans T C, Jr, Martin D, Kolly R, Panne D, Sun L, Ghosh I, Chen L, Benner J, Liu X Q, Xu M-Q. Protein trans-splicing and cyclization by a naturally split intein from the dnaE gene of Synechocystis species PCC6803. J Biol Chem. 2000;275:9091–9094. doi: 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- 5.Ferber D. Ecology. New corn plant draws fire from GM food opponents. Science. 2000;287:1390. doi: 10.1126/science.287.5457.1390. [DOI] [PubMed] [Google Scholar]
- 6.Ferber D. GM crops in the cross hairs. Science. 1999;286:1662–1666. doi: 10.1126/science.286.5445.1662. [DOI] [PubMed] [Google Scholar]
- 7.Hervieu F, Vaucheret H. A single amino acid change in acetolactate synthase confers resistance to valine in tobacco. Mol Gen Genet. 1996;251:220–224. doi: 10.1007/BF02172921. [DOI] [PubMed] [Google Scholar]
- 8.Hill C M, Duggleby R G. Mutagenesis of Escherichia coli acetohydroxyacid synthase isoenzyme II and characterization of three herbicide-insensitive forms. Biochem J. 1998;335:653–661. doi: 10.1042/bj3350653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodgson J. Monarch Bt-corn paper questioned. Nat Biotechnol. 1999;17:627. doi: 10.1038/10834. [DOI] [PubMed] [Google Scholar]
- 10.Ibdah M, Bar-Ilan A, Livnah O, Schloss J V, Barak Z, Chipman D M. Homology modeling of the structure of bacterial acetohydroxy acid synthase and examination of the active site by site-directed mutagenesis. Biochemistry. 1996;35:16282–16291. doi: 10.1021/bi961588i. [DOI] [PubMed] [Google Scholar]
- 11.LaRossa R A, Schloss J V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J Biol Chem. 1984;259:8753–8757. [PubMed] [Google Scholar]
- 12.LaRossa R A, Smulski D R. ilvB-Encoded acetolactate synthase is resistant to the herbicide sulfometuron methyl. J Bacteriol. 1984;160:391–394. doi: 10.1128/jb.160.1.391-394.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poppy G. GM crops: environmental risks and non-target effects. Trends Plant Sci. 2000;5:4–6. doi: 10.1016/s1360-1385(99)01514-9. [DOI] [PubMed] [Google Scholar]
- 14.Ray T B. Plant Physiol. 1984;18:262–266. doi: 10.1104/pp.75.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott C P, Abel-Santos E, Wall M, Wahnon D C, Benkovic S J. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Hu Z, Liu X Q. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav N, McDevitt E R, Benard S, Falco C S. Single amino acid substitutions in the enzyme acetolactate synthase confer resistance to the herbicide sulfometuron methyl. Proc Natl Acad Sci USA. 1986;83:4418–4422. doi: 10.1073/pnas.83.12.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]