Abstract
The selective effects of sulfur-containing hydrocarbons, with respect to changes in bacterial community structure and selection of desulfurizing organisms and genes, were studied in soil. Samples taken from a polluted field soil (A) along a concentration gradient of sulfurous oil and from soil microcosms treated with dibenzothiophene (DBT)-containing petroleum (FSL soil) were analyzed. Analyses included plate counts of total bacteria and of DBT utilizers, molecular community profiling via soil DNA-based PCR-denaturing gradient gel electrophoresis (PCR-DGGE), and detection of genes that encode enzymes involved in the desulfurization of hydrocarbons, i.e., dszA, dszB, and dszC.Data obtained from the A soil showed no discriminating effects of oil levels on the culturable bacterial numbers on either medium used. Generally, counts of DBT degraders were 10- to 100-fold lower than the total culturable counts. However, PCR-DGGE showed that the numbers of bands detected in the molecular community profiles decreased with increasing oil content of the soil. Analysis of the sequences of three prominent bands of the profiles generated with the highly polluted soil samples suggested that the underlying organisms were related to Actinomyces sp., Arthrobacter sp., and a bacterium of uncertain affiliation. dszA, dszB, and dszC genes were present in all A soil samples, whereas a range of unpolluted soils gave negative results in this analysis. Results from the study of FSL soil revealed minor effects of the petroleum-DBT treatment on culturable bacterial numbers and clear effects on the DBT-utilizing communities. The molecular community profiles were largely stable over time in the untreated soil, whereas they showed a progressive change over time following treatment with DBT-containing petroleum. Direct PCR assessment revealed the presence of dszB-related signals in the untreated FSL soil and the apparent selection of dszA- and dszC-related sequences by the petroleum-DBT treatment. PCR-DGGE applied to sequential enrichment cultures in DBT-containing sulfur-free basal salts medium prepared from the A and treated FSL soils revealed the selection of up to 10 distinct bands. Sequencing a subset of these bands provided evidence for the presence of organisms related to Pseudomonas putida, a Pseudomonas sp., Stenotrophomonas maltophilia, and Rhodococcus erythropolis. Several of 52 colonies obtained from the A and FSL soils on agar plates with DBT as the sole sulfur source produced bands that matched the migration of bands selected in the enrichment cultures. Evidence for the presence of dszB in 12 strains was obtained, whereas dszA and dszC genes were found in only 7 and 6 strains, respectively. Most of the strains carrying dszA or dszC were classified as R. erythropolis related, and all revealed the capacity to desulfurize DBT. A comparison of 37 dszA sequences, obtained via PCR from the A and FSL soils, from enrichments of these soils, and from isolates, revealed the great similarity of all sequences to the canonical (R. erythropolis strain IGTS8) dszA sequence and a large degree of internal conservation. The 37 sequences recovered were grouped in three clusters. One group, consisting of 30 sequences, was minimally 98% related to the IGTS8 sequence, a second group of 2 sequences was slightly different, and a third group of 5 sequences was 95% similar. The first two groups contained sequences obtained from both soil types and enrichment cultures (including isolates), but the last consisted of sequences obtained directly from the polluted A soil.
Terrestrial oil or petroleum deposits, which often contain high levels of sulfurous hydrocarbons, are being increasingly employed for the production of fuels. However, concerns regarding the emission of sulfur dioxide to the atmosphere, causing acid rain, have stimulated oil refiners to develop strategies for the reduction of the levels of sulfur in their products. Efforts have thus been made to develop an effective oxidative biocatalytic desulfurization approach in order to decrease the sulfur moiety in fuels through a specific nondestructive ring cleavage pathway (20, 28).
Soil sites contaminated with oil are a concern for both industry and regulators. Crude oil contains polycyclic aromatic hydrocarbons with up to 5% organically bound sulfur (42). The response of bacterial populations in soil to the stress imposed by the presence of such compounds and their intrinsic capacity to degrade them have been relatively unexplored, even though a recent study addressed the hydrocarbon degradation response of microbial communities in beach areas (25). Bacterial communities in soil are likely to respond to pollution with sulfurous oil by changing their structure to one that favors organisms that are able to thrive under the new selective conditions (18) at the expense of other organisms that are suppressed (25). Due to the selective pressure exerted by sulfurous hydrocarbons and given the possible need of microbial consortia in soil for sources of sulfur, bacterial populations with an enhanced presence of desulfurization genes may be selected.
A classical system for the study of microbial desulfurization of hydrocarbons has made use of Rhodococcus erythropolis strain IGTS8 because of its ability to specifically remove the sulfur atom from the model compound dibenzothiophene (DBT) (20). The plasmid-borne sox (7) or dsz (24, 33) gene, responsible for the oxidation of sulfur in DBT, has been cloned and sequenced (6, 7, 33). These desulfurization genes have been well studied, and considerable knowledge of the properties of the corresponding enzymes has been gained. It was shown that the dszC gene encodes a sulfide/sulfoxide mono-oxygenase (7, 23, 28), which catalyzes the stepwise S oxidation of DBT via DBT 5-oxide (DBTO) to DBT sulfone (DBTO2). DBTO2 is subsequently converted to 2-(2′-hydroxyphenyl)benzene (HBP) sulfinate (HBPSi-) by the product of the dszA gene, after which HBPSi- is desulfinated to give HBP and sulfite by the aromatic sulfinic acid hydrolase encoded by dszB (28).
The desulfurization genes have often been shown to be conserved among Rhodococcus species (5). Several strains of R. erythropolis, i.e., SY1 (29, 30), D-1 (16), Ni-36, Ni-43, and QIA-22 (53), have been reported to perform a nondegradative DBT-desulfurizing reaction, probably through the same metabolic pathway as that of strain IGTS8. Also, species from other bacterial genera, including Brevibacterium sp. strain DO (49), strains identified as Arthrobacter spp. (22, 40), thermophilic Paenibacillus strains (21), and, more recently, Gordona sp. strain CYKS1 (34), are able to convert DBT to HBP. However, the exact desulfurization mechanisms used by these organisms have remained relatively unexplored (40).
In spite of the recent knowledge about the desulfurization ability of pure and mixed cultures obtained from oil-polluted environments (Duarte et al., unpublished observations), no studies regarding the presence and diversity of oil desulfurization genes or microbial populations in polluted-soil habitats have been performed. This study aimed to determine the bacterial community structure as well as the prevalence and diversity of dsz genes in soil samples polluted with sulfurous oil in the open environment as well as in microcosms containing soil treated with petroleum supplemented with DBT. The bacterial community structure of the soil samples was analyzed by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rDNA fragments generated from soil DNA (14, 15, 25, 27) to obtain information about the effects of different levels of sulfurous oil on soil bacterial communities. Furthermore, the occurrence and diversity of the dszA, dszB, and dszC genes in oil-polluted soil samples were assessed via PCR with specific primers, in conjunction with DGGE analysis and sequencing. Finally, the two data sets were linked via the detection of dsz genes in isolates from enrichment cultures that matched the responders identified by molecular means in DGGE profiles.
MATERIALS AND METHODS
Bacterial strains and media.
Cells of R. erythropolis strain IGTS8 were maintained in basal salts medium (BSM) (7) containing (per liter) 2 g of glycerol, 4 g of NaH2PO4 · H2O, 4 g of K2HPO4 · 3H2O, 2 g of NH4Cl, 0.2 g of MgCl2.6H2O, 0.001 g of CaCl2 · 2H2O, and 0.001 g of FeCl3 · 6H2O) supplemented with 0.1 mM DBT as the sole sulfur source (added from a stock containing 10 mM DBT in ethanol). For plating, solidified (1.5% agar) BSM-DBT was used. Cells of Escherichia coli DH5 and Paenibacillus azotofixans ATCC 35681 were grown overnight in Luria-Bertani medium (38) at 30°C (200 rpm) and in TBN medium (39) at 32°C, respectively.
Field soil samples.
Oil-contaminated soil samples (A soil; silt loam, pH 5.0) were collected from a grass-covered petroleum storage area in the harbor of Amsterdam, The Netherlands, with an oil pollution history of 50 years. The crude oil that had entered the soil contained sulfur levels of up to 5% (R. Veenstra, personal communication). Sampling was done randomly in blocks (52). Four random samples were taken from the top layer (10 cm) of each of six plots in the area, which had different degrees of oil contamination (four characterized as low and two characterized as high). The individual samples from each plot were bulked and mixed, generating a total of four composite samples for low-pollution sites and two for high-pollution sites. The levels of oil in the low-pollution sites, denoted LP1 to LP4, averaged 710 mg kg−1 of soil, and those in the polluted sites, P1 and P2, averaged 5,500 mg kg−1 of soil (R. Veenstra, personal communication). The average total S and sulfate S contents determined by standard soil analysis techniques (Analytico, Lunteren, The Netherlands; NEN norms 7322 and 6654) were 840 and 240 mg kg−1, respectively, in the low-pollution soil and 920 and 350 mg kg−1, respectively, in the high-pollution samples, whereas 426 mg of organic S kg−1 was found in an adjacent, non-oil-polluted area of the soil. Hence, 600 and 670 mg of organic S kg−1 were estimated to be present in the low-pollution and high-pollution sites, respectively, of which, respectively, 174 and 246 mg kg−1 were hydrocarbon bound.
Samples from unpolluted soil, i.e., Flevo silt loam (FSL) and Ede loamy sand (ELS), both obtained from microplots at IPO-DLO, Wageningen, The Netherlands (50), Finnish organic soil (FOS) (51), and soils from three different Brazilian regions (36), i.e., Cerrado (C), Várzea (V), and São Paulo (SP) were used as controls. FSL soil was also used in the microcosm experiment described below.
Soil microcosm experiment.
The effect of DBT-containing petroleum in FSL soil on the indigenous bacterial communities was assessed in microcosms. Thus, to mimic a spill of sulfurous oil, duplicate 100-g FSL soil portions with soil moisture contents of around 20% were treated with 5 ml of petroleum (containing <0.4% S) supplemented with 200 mg of DBT and the soil-petroleum mixture was carefully homogenized using a sterile spatula. Control soil portions were left untreated. All soil portions were placed in plastic cups to yield individual soil microcosms. The microcosms were incubated in the dark at 20°C and sampled periodically, i.e., after zero (3 h), 7, 14, 30, 60, and 90 days. Samples were subjected to cultivation-based and direct molecular analyses of bacterial diversity and the prevalence of dsz genes, as described below.
Enrichment cultures from the polluted field and treated microcosm soils.
Soil samples from the highly polluted field sites (A soil; P1 and P2 plots) as well as samples of the petroleum-DBT-treated FSL soil microcosms after 15 days of incubation were selected for enrichment of DBT degraders. Both types of sample showed a prevalence of dszB genes by direct molecular assessment of soil DNA. Five-gram portions of soil were added to flasks containing 95 ml of BSM supplemented with 1 mM DBT as the sole source of sulfur. Flasks were incubated on a rotary shaker (150 rpm) at 20°C for 3 days or until turbid. Then, fresh flasks containing BSM-DBT were inoculated with 10% (first transfer) or 1% inocula (subsequent transfers), and the procedure was repeated (total of three transfers). Samples from each enrichment culture were subjected to analyses of culturable bacteria, of DBT utilizers, of dsz genes, and of the molecular community profile based on directly extracted DNA.
Colonies obtained on plates were screened for the presence of the dsz genes as well as for their 16S rDNA by colony PCR followed by regular or denaturing gel electrophoresis.
Enumeration of culturable bacterial communities in soil and screening for the presence of dsz genes.
To assess the size of the culturable bacterial populations in soil, 10-g portions of each soil sample were mixed with 95 ml of 0.1% sodium pyrophosphate and 10 g of gravel and shaken for 20 min at 200 rpm. Serial 10-fold dilutions of these suspensions were plated on 10% strength tryptone soy agar (0.1×TSA; BioMérieux, Marcy l'Etoile, France) supplemented with cycloheximide (50 μg/ml) to enumerate total indigenous bacterial CFUs and onto BSM agar (7) supplemented with 0.1% DBT to count bacteria able to grow with DBT as the sole sulfur source. Plates were incubated at 27°C, and colonies were enumerated after 48 (0.1× TSA) or 72 h (BSM-DBT).
Colonies from BSM-DBT agar plates were lifted and used as targets for the dszA, dszB, and dszC probes generated by PCR (see below) in a colony hybridization assay as described by Sambrook et al. (38). In addition, colonies were subjected to colony PCR using the dszA, dszB, and dszC primer sets (see below).
Isolation of strains and analysis of isolates.
Colonies from BSM-DBT plates that revealed the presence of dsz genes were purified by streaking on the same medium. Stocks for storage at −80°C were then prepared from single colonies. Isolates thus obtained were subjected to analyses of their 16S rDNA sequence, to fatty acid methyl ester (FAME) analysis using the MIDI microbial identification system, and to assays of their DBT-desulfurizing capacity. For the last analysis, a presumptive assay performed in microtiter plates assessed the release of fluorescent compounds from DBT (o,o′-biphenol or 2-hydroxybiphenyl) (53). After 24 h of incubation in BSM-DBT at 30°C, cultures that were positive for fluorescence under UV were analyzed by gas chromatography and high-pressure liquid chromatography (HPLC), as described by Duarte et al. (submitted for publication). Briefly, 25-ml cultures in BSM-DBT grown for 3 to 5 days at 30°C were spun down as described previously (16). The culture broth was then acidified to pH 2.0 and extracted with 0.8 volume of ethyl acetate. An aliquot of the ethyl acetate layer was removed and centrifuged, and 5 μl of supernatant was analyzed by gas chromatography (GC-14A; Shimadzu, Kyoto, Japan). HPLC analysis was performed using a 4-μm-pore-size phenyl Novapak column (Waters, Milford, Mass.); DBT was detected at 233 nm.
DNA extraction from strains.
An aliquot (1.5 ml) of an overnight culture was centrifuged for 2 min (12,000 × g), washed with Tris-EDTA (TE) (38) buffer, and centrifuged again, and the pellet was resuspended in 500 μl of TE buffer plus 0.5 g of glass beads (0.1 mm in diameter). After bead beating for 30 s, 30 μl of 10% sodium dodecyl sulfate (SDS) and 500 μl of Tris-buffered phenol (pH 8) (38) were added. The mixture was then centrifuged for 10 min in an Eppendorf centrifuge at 14,000 rpm, and the aqueous phase was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). Finally, the DNA was precipitated with 0.6 volume of isopropanol (5 min, room temperature), centrifuged (12,000 × g; 10 min), and washed with 70% ethanol. The pellet was air dried and resuspended in 50 μl of TE buffer. DNA concentrations in the purified extract were determined with a GeneQuant apparatus (Pharmacia, Uppsala, Sweden).
Genomic DNA of R. erythropolis IGTS8 was used as a positive control in PCR experiments involving dszA, dszB, and/or dszC primers, whereas genomic DNAs of E. coli and P. azotofixans ATCC 35681 were used as negative controls.
DNA extraction from soil and enrichment cultures.
Direct extractions of total microbial DNA from soil samples and from enrichment cultures were performed using a rapid bead beating protocol (51). Briefly, 2 g of soil, or the bacterial pellet from a 95-ml enrichment culture, plus 5 ml of 120 mM phosphate buffer and 3 g of glass beads (0.11 mm in diameter) were subjected to three 90-s bead beating steps followed by the addition of 180 μl of SDS (20%). The resulting suspensions were subjected to extractions with equal volumes of Tris-buffered phenol and chloroform-isoamyl alcohol (24:1), precipitation with ethanol, and resuspension of the pellet in TE buffer (38). Subsequent steps consisted of purification with CsCl (51) to precipitate impurities, followed by precipitation of the DNA with isopropanol, precipitation with potassium acetate (20 μl of an 8 M solution per 100 μl), precipitation with 0.6 volume of isopropanol, and, finally, purification over resin spin columns (Wizard DNA cleanup system; Promega, Madison, Wis.). Soil DNA was visualized on 0.8% (wt/vol) agarose gels (38) to check its purity and molecular size. The final DNA extracts obtained from all soils were amplifiable by PCR using 1 μl of extract (10 to 20 ng of DNA) per 50-μl reaction volume.
PCR.
All PCR amplifications were performed in a thermal cycler (model 480; Perkin-Elmer/Cetus, Nieuwerkerk, The Netherlands). The PCR mixtures were prepared with 1 μl of target DNA (ca. 10 to 20 ng), 5 μl of 10× Stoffel buffer (10 mM Tris-HCl [pH 8.3], 10 mM KCl), 200 μmol of each deoxyribonucleoside triphosphate, 3.75 mM MgCl2, 20 pmol of each of the appropriate primers, 1% (vol/vol) formamide, 0.25 μg of T4 gene 32 protein (Boehringer, Ingelheim, Germany), 5 U of Taq DNA polymerase, and the Stoffel fragment (Perkin-Elmer/Cetus) in a 50-μl final volume. T4 gene 32 protein was added since it improved the efficiency of amplification of targets in soil DNA (45). The reaction mixtures were overlaid with mineral oil (Sigma, Zwijndrecht, The Netherlands). A hot-start procedure (5 min, 94°C) was used before the enzyme was added to prevent aspecific annealing of the primers (4). Negative controls (PCR mixture without added target DNA) were included in all PCRs.
To analyze the diversity of the soil bacterial communities via DGGE, PCR based on primers 968F and 1401R (E. coli numbering [3]) was performed (15). A GC clamp (27) was attached to the 968F primer. The PCR product obtained was about 450 bp long. The PCR used a touchdown protocol in which the annealing temperature was initially set at 65°C and decreased by 2°C every second cycle until 55°C, at which temperature 20 additional cycles were carried out. Melting was carried out at 94°C for 1 min, primer annealing was performed according to the above scheme for 1 min, and primer extension was at 72°C for 3 min. A final chase was carried out at 72°C (10 min). The amplification products were stored at −20°C until DGGE analysis. A portion was routinely analyzed by electrophoresis in 1.4% (wt/vol) agarose gels in 0.5× Tris-borate-EDTA buffer (38).
PCR with the dszB and dszC detection systems entailed 35 cycles (94°C for 1 min, 55°C for 1.5 min, 72°C for 2 min). For the dszA detection system, a touchdown PCR (9, 36) was performed, with the annealing temperature decreasing by 1°C every second cycle, from 65 to 55°C, with 15 additional cycles done at 55°C. A final extension was carried out at 72°C (10 min). Primers used for the dszA, dszB, and dszC systems (Table 1) were based on the sequences of the R. erythropolis strain IGTS8 dsz gene cluster (7, 33). Primers were designed using the PRIMER program of the CAMMSA suite at the CAOS/CAMM center (University of Nijmegen, Nijmegen, The Netherlands). The criterion used for primer selection was the absence of adventitious amplification of any nontarget sequences. A search of the EMBL database using FastA (32) did not reveal significant homologies to sequences other than those of the dsz gene regions, which validated the use of the dszA, dszB, and dszC PCR systems for direct screening of soil DNA. Also, all primer pairs were specific for the respective dsz region of strain IGTS8, as they only produced amplicons of the expected sizes with genomic DNA of this strain, not with genomic DNA of a suite of 25 other bacterial species (not shown). To allow an analysis by DGGE of the potential diversity in the dsz gene regions in soil organisms, a GC clamp (27) was added to all forward primers.
TABLE 1.
Specific PCR primers for the detection of dszA, dszB, and dszC genes based on the sequence of the R. erythropolis IGTS8 dsz operona
| Gene | Primer orientation | Position | Product size (bp) | Sequence |
|---|---|---|---|---|
| dszA | Forward | 371–388 | 5′-TCGATCAGTTGTCAGGGG-3′ | |
| Reverse | 898–917 | 547 | 3′-GGATGGACCAGACTGTTGAG-5′ | |
| dszB | Forward | 139–158 | 5′-ATCGAACTCGACGTCCTCAG-3′ | |
| Reverse | 541–560 | 422 | 3′-GGAACATCGACACCAGGACT-5′ | |
| dszC | Forward | 277–296 | 5′-CTGTTCGGATACCACCTCAC-3′ | |
| Reverse | 651–668 | 392 | 3′-ACGTTGTGGAAGTCCGTG-5′ |
The forward primers used for DGGE analysis contained a GC clamp (27).
DGGE analysis.
Denaturing gels (6% polyacrylamide) were prepared and run with 0.5× TAE buffer (20 mM Tris-acetate [pH 7.4], 10 mM sodium acetate, 0.5 mM disodium EDTA) using a phorU2 apparatus (Ingeny, Leiden, The Netherlands). These gels contained 25 to 75% or 35 to 65% gradients of urea and formamide solution (100% is 7 M urea plus 40% [vol/vol] formamide deionized with AG501-X8 mixed-bed resin [Bio-Rad, Veenendaal, The Netherlands]) for products generated with the bacterial 16S rDNA-based primers or with the dszA, dszB, and dszC primer pairs, respectively. DGGE was performed for 16 h at 60°C and 100 V. After electrophoresis, gels were stained for 30 min with 1 μl of SYBR Green I (Molecular Probes, Leiden, The Netherlands) stock solution diluted in 10 ml of 1× TAE (pH 8.3) and photographed under UV ilumination.
Gel pictures were digitized and used for analysis by the Molecular Analyst software (Bio-Rad), or profiles were manually converted to 1/0 matrices, which were subsequently used for clustering by the unweighted pair group method with mathematical averages (UPGMA; Dice coefficient of similarity), followed by tree inference (Treecon program; Y. van de Peer).
Blotting of DGGE gels onto nylon membranes (Boehringer) using 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) (38) was carried out overnight as described previously (40), except that 15-min intervals were used between the denaturation and neutralization steps. To prepare specific probes, PCR products generated with the dsz primers on DNA from R. erythropolis IGTS8 were excised from gel and purified using the GeneClean II kit (Bio 101, La Jolla, Calif.). Probes were labeled with a digoxigenin labeling kit (Boehringer). After hybridization under stringent conditions (hybridization solutions containing 50% formamide and 5% blocking reagent, hybridization temperature of 42°C [12]), blots were washed at increasing stringencies (twice for 5 min in 2× SSC–0.1% SDS at room temperature, twice for 5 min in 0.1× SSC–0.1% SDS at 50°C [36]), treated with a chemiluminescence detection kit (Boehringer Mannheim), and exposed to X-ray films for 15 to 30 min.
Cloning and sequencing of 16S rDNA and dsz PCR fragments.
PCR products obtained from soil, enrichment culture, or bacterial DNA were resolved via DGGE, and selected bands were either cloned directly or reamplified following scratching or a crush-soak protocol (38) (only bands from denaturing gels), purified, and cloned into the pCRII cloning vector (TA cloning kit, Invitrogen, Leek, The Netherlands). The nature of the bands obtained from DGGE gels was confirmed using PCR-DGGE. Competent E. coli Inv-alfa cells were transformed according to the protocol of the cloning kit. Plasmid DNA was then extracted from randomly picked recombinant clones as described previously (38) and purified using Wizard resin spin columns (Promega). This DNA was used as the template in sequencing reactions by applying the Thermo-Sequenase fluorescently labeled primer cycle sequencing kit, with 7-deaza-dGTP (Amersham, ‘s Hertogenbosch, The Netherlands) and an automatic sequence analyzer (ALF DNA sequencer; Pharmacia).
Analysis of sequence data.
DNA sequence data were analyzed by using FastA of the GCG (University of Wisconsin Genetics Computer Group) (8) software via the CAOS/CAMM center (University of Nijmegen). New sequences obtained were further aligned to database sequences by using BLAST-N (1). The sequences were also checked for chimeric molecules by using the CHECK_CHIMERA tool of the Ribosomal Database Project (26).
To infer phylogenies of the sequences obtained, alignments were prepared using CLUSTAL-W (47) and trees were constructed using neighbor joining (37) by the Treecon program (Y. van de Peer) or TreeView, version 1.5 (31) following the use of the PAUP* software package (44).
Nucleotide sequence accession numbers.
The sequences obtained in this study are available from GenBank under accession no. AF165524 through AF165526 (three selected bands from DGGE of A soil); AF322027 through AF322032 (six bands sequenced from soil enrichments); and AF322033 through AF322042 (10 of 37 sequences representative of groups I, II, and III).
RESULTS
Enumeration of bacterial populations in oil-polluted field soil.
The numbers of culturable bacteria detected in the field soil samples with different degrees of oil pollution are shown in Table 2. Total viable counts averaged 2.5 × 106 CFU g−1 of soil, and there were no significant differences between counts obtained from the plots with different oil pollution levels. In contrast, the CFU counts on BSM agar supplemented with DBT were 10- to 100-fold lower than the total CFU counts, suggesting that organisms able to utilize DBT as a sulfur source made up only 1 to 10% of the total culturable bacterial populations. There was no significant difference in the counts of DBT utilizers between the plots with high and low oil pollution levels.
TABLE 2.
Enumeration of bacterial populations in oil-polluted field soil
| Growth medium | Log CFU g−1 of dry soil (SD)a in plot
|
|||||
|---|---|---|---|---|---|---|
| LP1 | LP2 | LP3 | LP4 | P1 | P2 | |
| 0.1 × TSA | 6.5 (0.25) | 6.45 (0.10) | 6.35 (0.30) | 6.40 (0.10) | 6.45 (0.10) | 6.34 (0.10) |
| BSM + DBT | 5.30 (0.30) | 5.25 (0.90) | 4.40 (0.10) | 4.65 (1.60) | 4.50 (0.50) | 4.60 (0.40) |
From duplicate samples.
Direct hybridization of >200 colonies grown on the BSM-DBT plates with the appropriate dszA, dszB, and dszC probes failed to unequivocally identify organisms that carried the dszA, dszB, or dszC gene.
PCR-DGGE profiling of bacterial diversity in oil-polluted field soil.
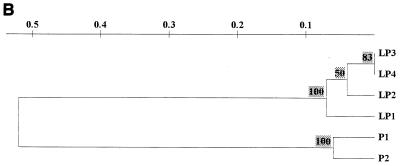
About 10 to 15 μg of DNA per g of soil was obtained for all samples from the A soil. Following purification, this DNA was amplifiable with the 16S rDNA-based bacterial primers. The 16S rDNA amplicons generated from the field soil samples were then subjected to DGGE profiling (Fig. 1A). A decrease in the number of dominant bacterial types detectable by DGGE was observed with increasing oil content, with a consistent concentration of amplicons in a few bands. The soil samples with oil contents averaging 710 mg of oil kg−1 of soil (lanes 1 to 4) had very similar profiles, with about 20 dominant bacterial types per lane. Samples P1 (lane 5) and P2 (lane 6), with oil contents averaging 5,500 mg of oil kg−1 of soil, revealed lower numbers of dominating bacterial types (respectively, about 14 and 10 bands). The clustering of the patterns obtained clearly indicated the existence of two clusters, one containing all plots with a low pollution level and the other one encompassing the two plots with a high pollution level (Fig. 1B).
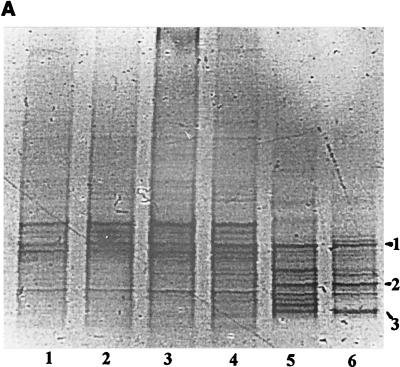
FIG. 1.
PCR-DGGE analysis of the bacterial communities in the oil-polluted A soil samples. (A) Molecular community profiles on denaturing gel (25 to 65% denaturants; SybrGold stained). Lanes: 1 to 4, profiles obtained from plots LP1 through LP4, respectively (710 mg of oil per kg of soil); 5 and 6, profiles from plots P1 and P2, respectively (5,500 mg of oil per kg of soil). Numbers 1 to 3 to the side indicate bands 1 to 3, which were selected for sequence analysis. (B) Dendrogram of gel obtained via 1/0 clustering using UPGMA (Dice coefficient of similarity) followed by treeing using the Treecon program.
Three strong bands present in the DGGE profiles of all samples, which strongly dominated the profiles of the highly polluted plots (P1 and P2) were reamplified, purified, and sequenced. DGGE band 1 had 91% similarity to its closest relative, unidentified bacterium rJ4, 90% similarity to the uncultured eubacterium SRang2.5 isolated from an aquatic sulfide-based microbial community (2), and lower but significant similarities to Beggiatoa spp., including strain 1401-13, a sulfide-oxidizing bacterium (46) and Beggiatoa alba ATCC 33555, a coal-desulfurizing bacterium. Band 2 was closely related to a range of different Arthrobacter species, at maximally 97% similarity. Band 3 was most closely related (90% similarity) to a cluster formed by an Actinomyces sp. (isolates TM146, TM220, and TM36), (35) and, at a lower level, to several Rhodococcus spp. (11).
Enumeration of bacterial populations in soil microcosms treated with DBT-containing petroleum.
The numbers of culturable bacteria detected in the FSL microcosms over time are shown in Table 3. The total CFU counts obtained on 0.1× TSA were generally on the order 106 to 107 CFU g−1 of dry soil. These counts were roughly stable over time, and they did not change significantly as a result of the treatment with petroleum-DBT. Counts on BSM agar supplemented with 0.1% DBT were also relatively stable over time in the untreated microcosms and were generally 5- to 20-fold lower than those obtained on 0.1× TSA. In the treated microcosms, these counts increased to roughly 50% of the total CFU counts (30 days), after which they dropped again to levels equalling those in the untreated microcosms. Unfortunately, direct-colony PCR and hybridization of colonies grown on the plates with 0.1× TSA and BSM plus DBT and the appropriate dsz gene-specific primer-probe combinations failed to unequivocally identify organisms that carried the dszA, dszB, or dszC gene.
TABLE 3.
Enumeration of bacterial populations in treated FSL soil in microcosmsa
| Condition | Growth medium | Avg log CFU/g of dry soil (SD) after:
|
|||||
|---|---|---|---|---|---|---|---|
| 3 h | 7 days | 15 days | 30 days | 60 days | 90 days | ||
| Untreated | 0.1 × TSA | 6.33 (0.13) | 6.30 (0.15) | 6.73 (0.12) | 6.85 (0.06) | 6.75 (0.10) | 6.62 (0.02) |
| BSM + DBT | 5.88 (0.22) | 5.31 (0.01) | 5.50 (0.03) | 6.20 (0.28) | 5.40 (0.06) | 5.34 (0.02) | |
| Treated | 0.1 × TSA | 6.56 (0.04) | 6.37 (0.15) | 6.87 (0.04) | 6.96 (0.05) | 6.78 (0.17) | 6.48 (0.21) |
| BSM + DBT | 6.19 (0.08) | 6.03 (0.04) | 6.20 (0.39) | 6.62 (0.20) | 5.40 (0.25) | 5.35 (0.10) | |
Counts in untreated microcosms were 2 × 107 for 0.1× TSA and 5.0 × 103 for BSM + DBT.
PCR-DGGE profiling of bacterial diversity in FSL soil microcosms treated with DBT-containing petroleum.
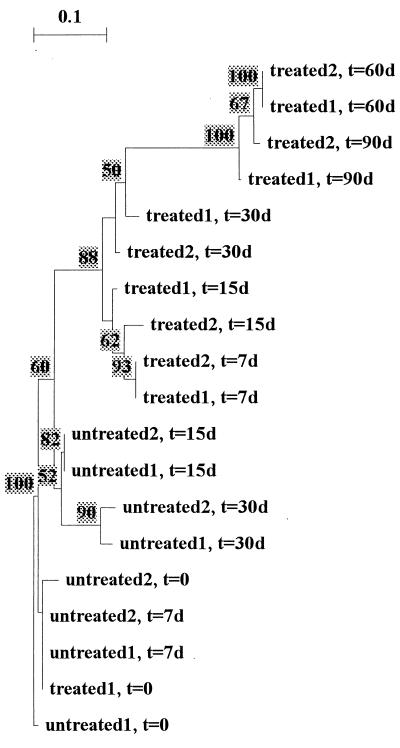
DGGE analysis of the PCR-amplified 16S rDNA fragments from the microcosm study provided a picture of relative stability of the bacterial community profiles both over time and between the treatments. Thirty to 45 bands were visible in all community profiles. Figure 2 shows the dendrogram obtained after the clustering of the profiles, revealing their relatedness. The profiles generated with the untreated soils were all very similar to each other, at >90% similarity, whereas the profiles generated with the treated soils fell in a separate cluster at roughly 10% difference. This cluster showed a divergent branch encompassing all day-60 and day-90 samples from the treated soil. Furthermore, there was no clear indication of a decreased “diversity” (richness and evenness of dominant bacterial types) over time due to the treatment, as a reduction in the numbers of bands was not found.
FIG. 2.
Dendrogram of PCR-DGGE analysis of the bacterial communities in FSL soil treated with petroleum-DBT in microcosms. The dendrogram was constructed by UPGMA (Dice coefficient of similarity), applied to a 1/0 matrix based on the primary gel. The tree was inferred using the Treecon program. t, time; d, days.
After 7 and 15 days of incubation, a distinct novel band became visible, at about 52.5% denaturant, in the profiles generated with total community DNA from the treated-soil microcosms, and this band was not visible in the profiles obtained from the untreated soils. All attempts to isolate and sequence this band failed. As this extra band might be linked to a dominant organism involved in the community response to the petroleum-DBT treatment, comparative PCR-DGGE was applied to products obtained from about 120 colonies from 0.1× TSA plates. Unfortunately, no colony whose 16S rDNA product showed the migration behavior of the putative responder was found (not shown).
Screening for dsz genes in control and oil-polluted soils.
The prevalence of dszA, dszB, and dszC genes was first assessed in a suite of soils with no known history of pollution with sulfurous oil. Total microbial community DNA from the ELS, FOS, C, V, and SP soils did not yield any product with the dszA, dszB, or dszC primer pair, whereas genomic DNA of R. erythropolis strain IGTS8 (102 to 103 genome equivalents per reaction) was readily amplified with the respective primers in these soil DNA backgrounds. However, products of the same size as those obtained with DNA from R. erythropolis IGTS8, i.e., about 422 bp, were consistently obtained with the dszB detection system when applied to the unpolluted FSL soil. This dszB product hybridized under high-stringency conditions (indicating >90% similarity) with the dszB probe.
dszA-, dszB-, and dszC-specific PCR with total community DNA obtained from the A soil samples with different oil pollution levels yielded products of sizes similar to those obtained with genomic DNA from R. erythropolis IGTS8, i.e., about 547, 422, and 392 bp for dszA, dszB, and dszC, respectively. The blotting of the gels containing the dszA-, dszB-, and dszC-related PCR products followed by hybridization under high-stringency conditions with the appropriate probes generated on strain IGTS8 revealed substantial (≥90%) homology between the PCR products generated from the oil-polluted soil samples and the respective probes. DGGE analysis of the products generated with dszA, dszB, and dszC primer pairs (not shown) also showed that the fragments amplified from the oil-polluted field soil samples resembled those generated with R. erythropolis IGTS8, since the respective bands migrated to the same positions in the denaturing gels. The blotting of the gels and hybridization with the appropriate probes under high-stringency conditions provided evidence that the melting behavior of the soil-derived products was similar to that of those generated with R. erythropolis IGTS8. This result suggested a limited sequence divergence from the canonical dsz operon in the dsz genes from the investigated soil in the regions spanned by the primers.
Enrichment cultures: selection of bacterial types and presence of dsz genes.
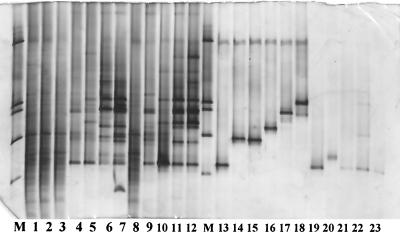
Enrichment cultures were set up using the A (sample P2) and treated FSL soils (15 days) in BSM-DBT to more strongly select for DBT-desulfurizing organisms. Analysis of the bacterial populations in these enrichment cultures via PCR-DGGE clearly showed the selection of a limited number (5 to 10) of dominant bacterial types from the highly complex soil bacterial communities (Fig. 3; results for FSL soil enrichments are shown). Whereas the communities selected still shifted their compositions along the first two enrichments, there was considerable stability in the community profiles between enrichment cultures 3 and 4. As judged from the community profiles, the selection of dominant bands was further quite consistent among the replicate enrichment cultures (Fig. 3) and partly consistent among soil sample types (not shown). Six major bands from the profiles of the fourth cultures were successfully isolated for sequence determinations (Fig. 3). Analysis of the sequences of these bands provided evidence for the putative selection of the following bacterial types (from top to bottom; the closest neighbors from the database are indicated in the figure legend [similarity levels, >95%]): Buttiauxella brennerae DSM9396, Stenotrophomonas maltophilia, Pseudomonas rhodesiae, Pseudomonas putida, Pseudomonas aureofaciens, and R. erythropolis.
FIG. 3.
PCR-DGGE analysis of the bacterial diversity in enrichment cultures of FSL soils treated with petroleum supplemented with DBT (duplicate day-15 microcosms used). Lanes: M, marker (products of, from top to bottom, Enterobacter cloacae BE1, Listeria innocua ALM105, Rhizobium leguminosarum bv. trifolii R62, Arthrobacter sp., Burkholderia cepacia P2); 1, FSL soil, untreated; 2, FSL soil, treated, time zero; 3, FSL soil, treated, 15 days (replicate 1); 4 to 7, enrichment culture from FSL, treated, 15 days (replicate 1), from first to fourth culture, respectively; 8, FSL, treated, 15 days (replicate 2); 9 to 12, enrichment culture from FSL, treated, 15 days (replicate 2), from first to fourth culture, respectively; 13, band 3 (sequence similarity to R. erythropolis); 14, band 7 (similar to P. aureofaciens); 15, band 2 (similar to P. putida); 16, band 6 (similar to P. rhodesiae); 17, band 5 (similar to S. maltophilia); 18, band 4 (similar to B. brennerae DSM9396); 19, product of strain M39; 20, product of strain M41; 21, product of strain A36; 22, product of strain A69; 23, product of strain A96.
In addition, molecular evidence for the selection of genes of the dsz operon was obtained via PCR with the dszA, dszB, and dszC primer pairs on community DNA extracted from the enrichment cultures (Table 4). In both the A and treated FSL soils (day-7 and day-15 samples), all three dsz genes could be detected following extended selection in BSM with DBT. While these genes were already detectable in the second enrichments from the A soil, the FSL soil showed the consistent presence of all three dsz genes only from the third enrichment culture (Table 4).
TABLE 4.
Appearance of dszA, dszB, and dszC genes in soils from different geographical locations, as well as in enrichment cultures, as evidenced by dsz gene-specific PCR followed by hybridizationa
| Soil(s) or enrichment culture | Origin | Pollution level | Site or time | Signalb in indicated culture for:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
dszA
|
dszB
|
dszC
|
||||||||||||||||
| D | E1 | E2 | E3 | E4 | D | E1 | E2 | E3 | E4 | D | E1 | E2 | E3 | E4 | ||||
| ELS | Ede, The Netherlands | Pristine | − | − | − | |||||||||||||
| FOS | Finland | Pristine | − | − | − | |||||||||||||
| C, V, SP | Brazil | Pristine | − | − | − | |||||||||||||
| FSL | Flevopolder, The Netherlands | Pristine | − | +/− | − | |||||||||||||
| FSL + DBT | Flevopolder, The Netherlands | Petroleum with DBT added | 7 days | − | − | + | + | + | +/− | − | − | + | + | − | + | + | + | + |
| 7 days | − | − | − | + | + | +/− | − | − | + | + | − | − | − | + | + | |||
| 15 days | − | + | + | + | + | +/− | + | + | + | + | − | + | + | + | + | |||
| 15 days | − | − | + | + | + | +/− | + | + | + | + | − | + | + | + | + | |||
| A | Amsterdam, The Netherlands (soil below oil storage tanks) | Long term; low-level-pollution | LP1 | +/− | +/− | + | + | + | + | + | + | + | + | +/− | − | + | + | + |
| LP2 | +/− | + | − | |||||||||||||||
| Long term; high-level pollution | P1 | +/− | + | − | ||||||||||||||
| P2 | + | + | + | |||||||||||||||
All products were verified by direct gel electrophoresis followed by blotting and hybridization with the appropriate probes generated by PCR with R. erythropolis strain IGTS8.
Overview of all results. Products of about 547 (dszA), 422 (dszB), and 392 bp (dszC) were scored. D, direct; E1 through E4, enrichment cultures 1 to 4 respectively. +, strong signal; +/−, weak or varying signal; −, no signal.
As the selected populations could thus encompass dominant organisms that carried active desulfurization genes including those of the dsz gene cluster, we tested a suite of isolates (26 per soil type) obtained on BSM-DBT agar plates for (i) the presence of dsz genes, (ii) the migration behavior of their 16S rDNA product, and (iii) their identity by FAME analysis. Organisms whose 16S rDNA products matched those of bands of the directly obtained molecular community profiles (irrespective of the presence of dsz genes) and organisms showing evidence for the presence of dsz genes were tested for their capacities to desulfurize DBT and for their identities. Twelve isolates, i.e., 7 from the A soil enrichments and 5 from the FSL soil, that matched dominant bands of the molecular community fingerprints and that contained dszB genes were thus obtained (Table 5). These isolates were identified as P. putida, a Pseudomonas sp., S. maltophilia, and R. erythropolis. For a subset of seven isolates, i.e., for strains A36, A61, A69, A74, and A96 (isolated from A soil enrichments) and M39 and M41 (isolated from FSL soil enrichments), evidence obtained by PCR indicated the presence of, in addition to dszB, the dszA and dszC genes (except for strain A74). All of these isolates were shown to desulfurize DBT, some at high rates (Table 5). With one exception, their 16S rDNA products comigrated with R. erythropolis products in DGGE (Fig. 4), and FAME analysis showed that they were related to R. erythropolis, albeit sometimes at low similarities (Table 5).
TABLE 5.
Strains isolated from enrichment cultures and their capacity to desulfurize DBT
| Strain | Enriched and isolated from: | Species bya:
|
SIb | dsz genes presentc | Desulfurization capacity (%)d | |
|---|---|---|---|---|---|---|
| Sequence | FAME | |||||
| A36 | A soil | R. erythropolis | R. erythropolis | 0.503 | dszABC | 63 |
| A61 | A soil | R. erythropolis | dszABC | 25 | ||
| A69 | A soil | R. erythropolis | R. erythropolis | 0.489 | dszABC | 78 |
| A74 | A soil | R. erythropolis | 0.584 | dszAB | 64 | |
| A96 | A soil | R. erythropolis | R. erythropolis | 0.489 | dszABC | 20 |
| M39 | FSL + DBT | R. erythropolis | R. erythropolis | 0.136 | dszABC | 53 |
| M41 | FSL + DBT | R. erythropolis | R. erythropolis | 0.136 | dszABC | 14 |
| A66 | A soil | NI∗ | dszAB | 33 | ||
| A73 | A soil | NI∗ | dszAB | ND | ||
| M16 | FSL + DBT | S. maltophilia | dszB | 4 | ||
| M26 | FSL + DBT | S. maltophilia | dszB | 3 | ||
Sequence, match on DGGE with products of R. erythropolis and S. maltophilia; FAME, closest relative from FAME database; NI, not identified.
SI, similarity index, the level of similarity to closest hit of FAME database.
As evidenced by PCR with dszA-, dszB-, and dszC-specific PCR systems.
The percentage of DBT transformed, under standard conditions, into products by the IGTS8 pathway. R. erythropolis IGTS8 used as the positive control, transformed >25% of DBT under these conditions. ND, not determined.
FIG. 4.
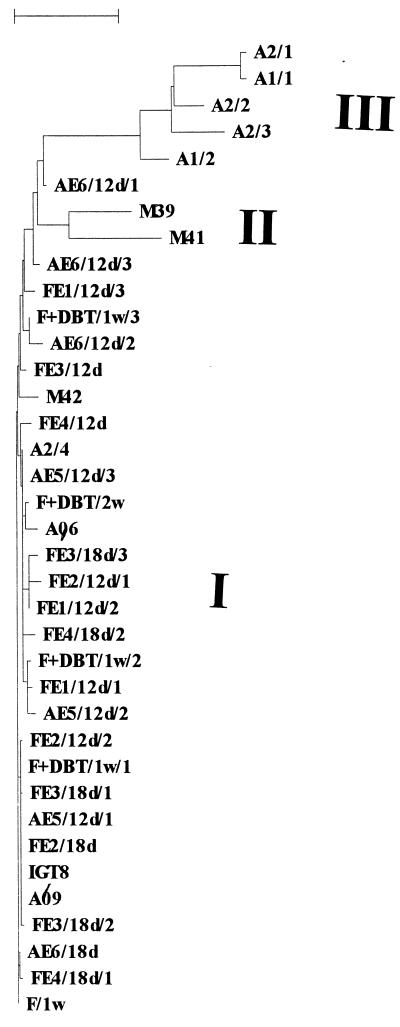
Cluster analysis of 37 dszA sequences obtained from the A and treated FSL soils, as well as from enrichment cultures and isolates obtained from these soils. I, II, and III, clusters I, II, and III, as described in the text. Bar, 0.05 distance. IGT8, canonical sequence of R. erythropolis IGTS8; A96 and A69, strains obtained from A soil; M39, M41, and M42, strains obtained from treated FSL soil; A1 and A2, sequences directly amplified from A soil (replicate sample number/sequence number); AE/d, sequences directly amplified from enrichment cultures of A soil (number, enrichment culture; d, days of incubation); F+DBT/w, sequences obtained from petroleum-DBT-treated FSL soil incubated 1 or 2 weeks (w); FE/d, sequences obtained from enrichment cultures from treated FSL soil.
The remaining isolates showed no to low DBT-desulfurizing activity and belonged to species other than R. erythropolis, e.g., S. maltophilia (Table 5). None of these isolates produced signals with all three dsz-specific PCR systems, although some appeared to contain dszB.
Screening for and analysis of dsz genes obtained from soils, enrichments, and isolates.
Initial analysis of the sequences of the PCR products generated with the dszA, dszB, and dszC primers using total microbial community DNA from the A soil sample (plot P2) revealed that all three products (one clone per gene analyzed) from this soil showed ≥98% similarity to their counterpart sequences from R. erythropolis IGTS8 (not shown).
In a more extensive analysis, the potential diversity in the 547-bp dszA fragment was analyzed for the A and FSL soils, as well as in enrichment cultures and isolates obtained from these soils. Thirty-six sequences, representing 3 or more sequences per habitat and/or treatment, were analyzed (Fig. 4). All sequences clustered at over 90% similarity with each other and with the canonical dszA sequence of R. erythropolis IGTS8 (Fig. 4). Three clusters, I, II, and III, could be distinguished. Cluster I consisted of 30 sequences (including the IGTS8 sequence) that were >98% similar to the canonical sequence; all of the sequences in this cluster had been derived either directly from the A and FSL soils or from enrichments or strains obtained from these soils. Cluster II consisted of two sequences derived from two rhodococcal FSL soil isolates that were slightly divergent from the canonical sequence. Cluster III, at roughly 95% similarity to clusters I and II, contained five internally relatively conserved sequences that had all been directly obtained from the two highly polluted plots of the A soil, P1 and P2, independent of cultivation. The cluster was most related to a sequence obtained by enrichment from the same soil. Sequences of this cluster all contained about 20 defined mutations, which clearly differentiated them from the canonical sequence. An analysis of changes in the putative amino acid sequence between the canonical sequence of strain IGTS8 and one representative sequence (A2/1) suggested that, over the 182-amino-acid region, only 4 amino acids had changed. More precisely, Asn/Asp, Glu/Asp, Ala/Thr, and His/Arg shifts had taken place. These changes did not result in a substantial modification of the ratio of strongly basic, strongly acidic, hydrophobic, and strongly polar amino acids in the 182-amino-acid region examined, although they might affect the protein function if present at critical locations of its structure.
DISCUSSION
An understanding of the ecology of microbial communities in soil requires knowledge on how these communities are structured under prevailing and changing environmental conditions. Since chemical pollutants can exert drastic effects on microbial diversity in soil, there is a need to understand the response of the indigenous communities to this trigger with respect to (i) putative shifts in the community structure caused by selective pressure and (ii) the selection of functional genes important for the catabolism of soil pollutants. The approach taken in this study represents an attempt to simultaneously address the possible selective effects of sulfurous hydrocarbon compounds in soil on bacterial community structure and on the prevalence of selected genes involved in hydrocarbon desulfurization pathways, i.e., those of the dsz operon. With respect to the sources of sulfur apparently available to bacteria in the soil systems studied (in particular the A soil), it is evident that organically (oil-) bound S made up a substantial part of the total S measured in the system. However, the data also showed that this fraction was not the only available source of S. Aspects of spatial location of bacteria in the soil, in particular in relation to the oil moiety, can have a drastic effect on the selective pressure sensed by the bacterial communities that are locally present. Hence, it is difficult to ascertain the relative contributions of the oil- or organically bound S to the in situ bacterial S provision. Nevertheless, it was thought to be likely that, within the bacterial communities with affinity to the ecological niche offered by oil, organisms that had the capacity to desulfurize DBT might prevail. It is obvious that by focusing on the dsz genes, this study excluded organisms with other desulfurizing capacities.
PCR-DGGE using primers based on conserved regions of the 16S rRNA sequence yields complex molecular profiles when used to analyze the soil microbiota (10, 14, 15, 25). The profiles thus obtained reflect the composition of the dominant soil microbiota, which includes the nonculturable fractions (10, 14). The DGGE patterns obtained in this study with total community DNA from soils with different levels of oil are to be interpreted as a first approximation to showing shifts in the composition of the dominant bacterial populations related to the oil pollution levels. Whereas the numbers of culturable bacteria present in these samples were not drastically affected, the molecular profiling data (Fig. 1) indicated that specific bacterial populations were selected with increasing oil levels, most likely by increasing their numbers at the expense of other (sensitive) bacterial groups. The analysis of the sequences of three selected DGGE bands revealed that these were <97% similar to any database sequence, suggesting that none was typical for any known bacterial species (43). However, the closest relatives often fell in bacterial groups, such as Arthrobacter, Rhodococcus, and Beggiatoa spp. and bacterium SRang2.5, which are potentially able to grow and metabolize in contaminated environments and to desulfurize DBT (16, 20, 22, 30, 53). These direct molecular data thus provide an indication as to the nature of the bacterial communities selected under long-term sulfurous hydrocarbon pressure in the A soil without conclusively showing the involvement of the putative organisms in any steps of the dissimilatory pathways.
The response of the bacterial community in soil to sulfurous hydrocarbons was evident in the soil microcosm experiment. In the first place, the counts of putative desulfurizing organisms were, albeit temporarily, affected as a result of the treatment, indicating a shift to growth in these communities. Although it is theoretically possible that the petroleum in soil affected plate counts by influencing desorption of cells from soil, from an analysis of the data it was felt that this effect was probably minor. In the second place, the molecular bacterial community profiles obtained from the FSL soil also showed an effect of the petroleum-DBT treatment, as evidence for the growth of an organism responding to the treatment was already found after 7 and 15 days. The effect of the treatment became more apparent over time, as the community profiles showed increasing divergence from those in untreated systems. However, a considerable level of overall similarity between all profiles persisted even after 90 days of incubation. This high similarity was illustrative of the difficulty often encountered in PCR-DGGE profiling with eubacterial primers in assessing short-term (short-lived) bacterial community shifts in soil (14). The discrepancy between the inferences made from the molecular community profiles observed in the A soil, with over 50 years of pressure by sulfurous oil, and the inferences made from those observed in the treated FSL soil, with a maximum of 90 days of such pressure, can also be related to this time factor.
This study further showed that the genes of the dsz operon are not easily found in (agricultural) soils with no known history of pollution with sulfurous hydrocarbons (Table 4). This suggests that agricultural soils do not commonly select organisms by virtue of their carrying the dsz operon. The finding of evidence for the occurrence of dszB in the pristine FSL soil was, therefore, surprising. A possible explanation is that natural, or even anthropogenic, sulfur-containing organic substrates, such as sulfonates and sulfate esters (19), might have selected for organisms carrying dsz genes, as suggested for organisms carrying tfd genes, which were also found to be present in soils of noncontaminated ecosystems (13, 17, 48).
On the other hand, relatively conserved dsz genes were shown to occur in the two soils containing sulfurous hydrocarbons obtained from geographically distinct regions in The Netherlands. dszA, dszB, and dszC gene sequences were amplified from DNA obtained from the field (A) soil and petroleum-DBT-treated microcosm (FSL soil) samples (albeit after enrichment). The conservation of these genes in A soil and the resemblance of their sequences to the canonical dszA, dszB, and dszC sequences were initially indicated by DGGE analysis of the PCR fragments generated from soil. PCR-DGGE applied to these genes was a first approach to analyzing their diversity in soil, as it allows the rapid, sequencing-independent detection of similarities and differences between sequences of the same functional genes (36, 54). The conservative nature of the genes was then confirmed by sequencing three clones, representative of the dszA, dszB, and dszC genes, obtained directly from the highly polluted A soil, as these showed >98% similarity with the respective genes from R. erythropolis IGTS8. Moreover, a more thorough study of the diversity of the dszA gene also showed it to be relatively conserved among 37 sequences recovered from soils, enrichments in BSM-DBT, and strains. Sequences obtained from the A and FSL soils, from soil enrichments, and from bacterial isolates fell into three groups with >90% internal similarity. Surprisingly, the highly polluted A soil also contained a group of sequences with limited but internally highly consistent divergence from the canonical (IGTS8) sequence. This potentially novel member group of the dszA gene family clearly warrants further study. An analysis of the putative amino acid sequence of the gene product of one representative sequence already indicated that only four changes seemed to have taken place, with virtually no change in protein hydrophobicity, compared to the canonical sequence. At this point, one can only speculate about the significance of the finding of these divergent putative gene products of dszA sequences directly from polluted soil, possibly originating from a difficult-to-culture, as yet uncultured, or even nonculturable organisms. Clearly, there is a need to obtain isolates carrying these genes for a study of their functionality.
The conserved nature of the dsz genotype has previously also been reported for Rhodococcus strains isolated from different geographic locations (5). In this study, we extend this observation, albeit in a nuanced form, to soils with oil pollution pressure. The results indicate that populations of bacteria related to Rhodococcus and harboring the dsz operon are present and selected in oil-contaminated soils. These bacteria can be isolated via enrichments in BSM-DBT, and some of them show vigorous desulfurization of DBT. In addition, evidence for the presence of uncultured organisms with divergent dszA gene sequences in the oil-polluted A soil was found.
The data obtained with isolates from the enrichment cultures clearly provided a link between the direct molecular observations on bacterial community structure in the polluted soils and enrichments from these and the data on the prevalence of genes of the dsz gene cluster. A number of key bacterial isolates whose specific 16S rDNA PCR products matched bands in the directly obtained molecular community profiles that represented putative responders were thus found to contain the dsz operon and to be capable of vigorously desulfurizing DBT. These isolates all fell into the R. erythropolis cluster. On the other hand, other isolates selected by the treatment, without clear desulfurizing capacity, fell into other taxons. It is thus likely that the treatment applied to soil and enrichment cultures selected for bacterial consortia in which the key desulfurization function was represented by the R. erythropolis-like organisms, whereas other functions were present in other coselected taxa. The presence of S. maltophilia as a consistent member of the selected consortia is remarkable, as this organism can be regarded as an avid colonizer of disturbed or stressed ecosystems and thus might play a role in hydrocarbon degradation processes in soils. S. maltophilia was also a member of two consortia with DBT-desulfurizing capacity that have previously been obtained from crude oil fields in Brazil (G. F. Duarte et al., submitted for publication). Even without a full understanding of all possible interactions between the members of the consortia, it can be said that those identified likely play an important role in hydrocarbon degradation and desulfurization processes in soil. Given the data provided in this paper, future work in this area should more specifically address the population dynamics and functional aspects of the response by these organisms to sulfurous oil in soil.
ACKNOWLEDGMENTS
We thank Ruud Veenstra for providing the oil-contaminated soil samples from Amsterdam and their analysis. We are greatful to Monica M. Linhares (CENPES, Rio de Janeiro, Brazil) for performing the tests of desulfurization and to Mark Bailey (IVEM/NERC, Oxford, United Kingdom) for help with the FAME determinations. Leo van Overbeek is acknowledged for critical reading of the manuscript.
G. F. Duarte and A. S. Rosado were awarded fellowships by National Research Council of Brasil (CNPq).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angert E R, Northup D E, Reysenbach A-L, Peek A S, Goebel B M, Pace N R. Molecular phylogenetic analysis of a sulfide-based microbial community in Sulphur River, Parker Cave, Kentucky. Am Mineral. 1998;83:1583–1592. [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Chou Q, Russel M, Birch D, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denis-Larosse C, Labbé D, Bergeron H, Jones A M, Greer C W, Al-Hawari J, Grossman M J, Sankey B M, Lau P C K. Conservation of plasmid-encoded dibenzothiophene desulfurization genes in several rhodococci. Appl Environ Microbiol. 1997;63:2915–2919. doi: 10.1128/aem.63.7.2915-2919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denome S A, Olson E S, Young K D. Identification and cloning of genes involved in specific desulfurization of dibenzothiophene by Rhodococcus sp. strain IGTS8. Appl Environ Microbiol. 1993;59:2837–2853. doi: 10.1128/aem.59.9.2837-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denome S A, Oldfield C, Nash L J, Young K D. Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. J Bacteriol. 1994;176:6707–6716. doi: 10.1128/jb.176.21.6707-6716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte G F, Rosado A S, Seldin L, Keijzer-Wolters A C, van Elsas J D. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J Microbiol Methods. 1998;32:21–29. [Google Scholar]
- 11.Embley T M, Stackebrandt E. The molecular phylogeny and systematics of the Actinomycetes. Annu Rev Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 12.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulthorpe R R, Rhodes A N, Tiedje J M. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl Environ Microbiol. 1996;62:1159–1166. doi: 10.1128/aem.62.4.1159-1166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelsomino A, Keijzer-Wolters A C, Cacco G, van Elsas J D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J Microbiol Methods. 1999;38:1–15. doi: 10.1016/s0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 15.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In: van Elsas J D, Wellington E M H, Trevors J T, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 353–373. [Google Scholar]
- 16.Izumi Y, Ohshiro T, Ogino H, Hine Y, Shimao M. Selective desulfurization of dibenzothiophene by Rhodococcus erythropolis D-1. Appl Environ Microbiol. 1994;60:223–226. doi: 10.1128/aem.60.1.223-226.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanaly R, Bartha R, Fogel S, Findlay M. Biodegradation of [14C]benzo[a]pyrene added in crude oil to uncontaminated soil. Appl Environ Microbiol. 1997;63:4511–4515. doi: 10.1128/aem.63.11.4511-4515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kertesz M A. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol Rev. 2000;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 20.Kilbane J J. Desulfurization of coal: the microbial solution. Trends Biotechnol. 1989;7:97–101. [Google Scholar]
- 21.Konish J, Yoshitaka I, Onaka T, Okumura K, Suzuki M. Thermophilic carbon-sulfur-bond-targeted biodesulfurization. Appl Environ Microbiol. 1997;63:3164–3169. doi: 10.1128/aem.63.8.3164-3169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M K, Senius J D, Grossman M J. Sulfur-specific microbial desulfurization of sterically hindered analogs of dibenzothiophene. Appl Environ Microbiol. 1995;61:4361–4366. doi: 10.1128/aem.61.12.4362-4366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei B, Tu S-C. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J Bacteriol. 1996;178:5699–5705. doi: 10.1128/jb.178.19.5699-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M Z, Squires C H, Monticello D J, Childs J D. Genetic analysis of the dsz promoter and associated regulatory regions of Rhodococcus erythropolis IGTS8. J Bacteriol. 1996;178:6409–6418. doi: 10.1128/jb.178.22.6409-6418.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNaughton S J, Stephen J R, Venosa A D, Davis G A, Chang Y-J, White D C. Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol. 1999;65:3566–3574. doi: 10.1128/aem.65.8.3566-3574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. DGGE in microbial ecology, chapter 3.4.4. In: Akkermans A, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht. The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 28.Oldfield C, Pogrebinsky O, Simmonds J, Olson E S, Kulpa C F. Elucidation of the metabolic pathway for dibenzothiophene desulfurization by Rhodococcus sp. strain IGTS8 (ATCC53968) Microbiology. 1997;143:2961–2973. doi: 10.1099/00221287-143-9-2961. [DOI] [PubMed] [Google Scholar]
- 29.Omori T, Monna L, Saiki Y, Kodama T. Desulfurization of dibenzothiophene by Corynebacterium sp. strain SY1. Appl Environ Microbiol. 1992;58:911–915. doi: 10.1128/aem.58.3.911-915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omori T, Saiki Y, Kasuga K, Kodama T. Desulfurization of alkyl and aromatic sulfides and sulfonates by dibenzothiophene-desulfurizing Rhodococcus sp. strain SY1. Biosci Biotechnol Biochem. 1995;59:1195–1198. [Google Scholar]
- 31.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 32.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piddington C S, Kovacevich B R, Rambosek J. Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Appl Environ Microbiol. 1995;61:468–475. doi: 10.1128/aem.61.2.468-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee S-K, Chang J H, Chang Y K, Chang H N. Desulfurization of dibenzothiophene and diesel oils by a newly isolated Gordona strain, CYKS1. Appl Environ Microbiol. 1998;64:2327–2331. doi: 10.1128/aem.64.6.2327-2331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 36.Rosado A S, Duarte G F, Seldin L, van Elsas J D. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl Environ Microbiol. 1998;64:2770–2779. doi: 10.1128/aem.64.8.2770-2779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Seldin L, Penido E G C. Identification of Bacillus azotofixans using API tests. Antonie Leeuwenhoek. 1986;52:403–409. doi: 10.1007/BF00393468. [DOI] [PubMed] [Google Scholar]
- 40.Serbolisca L, de Ferra F, Margarit I. Manipulation of the DNA coding for the desulphurizing activity in isolate of Arthrobacter sp. Appl Microbiol Biotechnol. 1999;52:122–126. doi: 10.1007/s002530051498. [DOI] [PubMed] [Google Scholar]
- 41.Simonich S L, Hites R A. Global distribution of persistent organochlorine compounds. Science. 1995;269:1851–1854. doi: 10.1126/science.7569923. [DOI] [PubMed] [Google Scholar]
- 42.Speight J G. The chemistry and technology of petroleum. New York, N.Y: Marcel Dekker, Inc; 1980. [Google Scholar]
- 43.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 44.Swofford, D. L. PAUP∗ (phylogenetic analysis using parsimony) software version 4.0 d55. Smithsonian Institution, Washington, D. C.
- 45.Tebbe C, Vajhen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teske A P, Ramsing N B, Kuever J, Fossing H. Phylogeny of Thioplaca and related filamentous sulfide-oxidizing bacteria. Syst Appl Microbiol. 1995;18:517–526. [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallaeys T, Fulthorpe R R, Wright A M, Solas G. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families to tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol Ecol. 1996;20:163–172. [Google Scholar]
- 49.van Afferden M, Schacht S, Klein J, Trüper H G. Degradation of dibenzothiophene by Brevibacterium sp. DO Arch Microbiol. 1990;161:266–271. [Google Scholar]
- 50.van Elsas J D, Dijkstra A F, Govaert J M, van Veen J A. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soils of different texture in field microplots. FEMS Microbiol Ecol. 1986;38:151–160. [Google Scholar]
- 51.van Elsas J D, Mäntynen V, Wolters A C. Soil DNA extraction and assessment of the fate of Mycobacterium chlorophenolicum strain PCP-1 in different soils by 16S ribosomal RNA gene sequence based most-probable-number PCR and immunofluorescence. Biol Fertil Soils. 1997;24:188–195. [Google Scholar]
- 52.van Elsas J D, Smalla K. Methods for sampling soil microbes. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: American Society for Microbiology; 1997. pp. 383–390. [Google Scholar]
- 53.Wang P, Krawiec S. Desulfurization of dibenzothiophene to 2-hydroxybiphenyl by some newly isolated bacterial strains. Arch Microbiol. 1994;161:266–271. [Google Scholar]
- 54.Watanabe K, Teramoto M, Futamata H, Harayama S. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl Environ Microbiol. 1998;64:4396–4402. doi: 10.1128/aem.64.11.4396-4402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]