Abstract
Four bacterial phenolic acid decarboxylases (PAD) from Lactobacillus plantarum, Pediococcus pentosaceus, Bacillus subtilis, and Bacillus pumilus were expressed in Escherichia coli, and their activities on p-coumaric, ferulic, and caffeic acids were compared. Although these four enzymes displayed 61% amino acid sequence identity, they exhibit different activities for ferulic and caffeic acid metabolism. To elucidate the domain(s) that determines these differences, chimeric PAD proteins were constructed and expressed in E. coli by exchanging their individual carboxy-terminal portions. Analysis of the chimeric enzyme activities suggests that the C-terminal region may be involved in determining PAD substrate specificity and catalytic capacity. In order to test phenolic acid toxicity, the levels of growth of recombinant E. coli displaying and not displaying PAD activity were compared on medium supplemented with different concentrations of phenolic acids and with differing pHs. Though these acids already have a slight inhibitory effect on E. coli, vinyl phenol derivatives, created during decarboxylation of phenolic acids, were much more inhibitory to the E. coli control strain. To take advantage of this property, a solid medium with the appropriate pH and phenolic acid concentration was developed; in this medium the recombinant E. coli strains expressing PAD activity form colonies approximately five times smaller than those formed by strains devoid of PAD activity.
Phenolic acids, principally represented by p-coumaric and ferulic acids, are naturally abundant plant compounds which ensure cell wall rigidity by linking the polysaccharide xylan to lignin (17). Phenolic acids can be released from these complex structures by cinnamoyl esterase activities, which are expressed by various microorganisms (12, 16, 26). In their free form, these acids become substrates of phenolic acid decarboxylase (PAD) enzymes, which convert these compounds into their vinyl phenol derivatives. To date, the DNA constituting the gene pad has been cloned from five microorganisms: Bacillus pumilus (35), Lactobacillus plantarum (8), Bacillus subtilis (10), Pediococcus pentosaceus (5), and the yeast Saccharomyces cerevisiae (13). Although the four bacterial PADs have 61% amino acid sequence identity, they differ individually in structure and in biochemical characteristics. They are also different from the phenylacrylic acid decarboxylase PAD1 of S. cerevisiae. This enzyme displays no amino acid sequence identity with the bacterial PADs we have observed and has approximately 1,000-fold-lower activity as well.
There are two main reasons for improving our understanding of PADs. First, these enzymes are involved in the formation of useful volatile phenol derivatives (24) which contribute naturally to aroma in wine (20) and other fermented foods and beverages. However, some of these volatile phenols, such as vinyl and ethyl phenol (from p-coumaric acid) are most often considered off-flavors and are responsible for alterations in organoleptic properties of foods (19). Understanding structure-function relationships of the PADs may be useful for the future construction of recombinant bacterial starter cultures with appropriate substrate specificities for desirable aroma production in vegetable fermentations and wine.
The second reason is to understand the physiological function of the PAD in the growth of microorganisms in phenolic acid-supplemented media. We have previously demonstrated that PAD activity allows lactic acid bacterium L. plantarum to resist inhibitory effects of p-coumaric acid (4) and proposed that PAD synthesis could be considered a stress response induced by phenolic acids in the environment. Concerning the gram-negative bacteria, only Klebsiella oxytoca is known to display PAD activity (23). Escherichia coli, which is inhibited by phenolic acids (36), has three open reading frames in its genome which encode potential PAD enzymes (6, 30), yet no detectable PAD activity is displayed (8), suggesting that either the three genes are not expressed or their products are not functional.
In this work, we have constructed four chimeric bacterial PAD enzymes, which were functional and which displayed enzymatic activities different from those of the native PADs. Our results suggested that the C-terminal region in the bacterial PADs is involved in enzymatic activity, especially substrate specificity. In the course of our experiments, we have demonstrated that vinyl phenol derivatives produced by PAD activity have much higher inhibitory effects on the growth of E. coli than the phenolic acid forms. A medium which takes advantage of this fact was developed to screen recombinant E. coli strains which displayed various PAD activities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Bacterial strains and plasmids used in this study are listed in Table. 1. E. coli TG1 and B. pumilus ATCC 15884 strains were grown in Luria-Bertani (LB) medium containing 100 μg of erythromycin/ml as necessary.
DNA manipulation and transformation procedures.
Standard procedures described by Sambrook et al. (31) were used for DNA manipulation. PCR products were purified with the Jet Sorb kit (Genomed, Bioprobe Systems, Montreuil, France) and sequenced by the dideoxy chain termination method (32) with the Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science, Inc., Cleveland, Ohio) in accordance with the recommendations of the manufacturer. E. coli TG1 strain was transformed by electroporation as described by Dower et al. (18). PCR amplifications were performed using 0.1 μg of DNA a template with 0.5 U of Tfu DNA polymerase (Appligene) under standard conditions in an automated DNA thermocycler (Eppendorf, Hamburg, Germany).
Cloning of the pad gene from B. pumilus ATCC 15884.
This strain of B. pumilus displayed inducible decarboxylase activity on p-coumaric, ferulic, and caffeic acids. Two oligonucleotides which introduced PstI (BPPAD1) and HindIII (BPPAD2) restriction sites into the PCR product (Table 2) were deduced from the sequence of the fdc gene of B. pumilus strain PS213 (35). One, located approximately 350 bp upstream of the start codon of the fdc gene (BPPAD1) and the other located approximately 150 bp from the stop codon (BPPAD2), were used for the amplification of a 925-bp DNA fragment, which was then cloned into the PstI/HindIII sites of pJDC9. The resulting clone, pJPADBP, expressed in E. coli TG1, displayed a constitutive PAD activity of approximately 10 μmol min−1 mg−1 in ferulic acid, confirming that the original amplified DNA fragment contained the B. pumilus pad gene. The DNA sequence of this fragment allowed the identification of one open reading frame, which encoded a polypeptide which displayed 98.5% identity with BPFDC from B. pumilus PS213 (35).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′)a | Created restriction site |
|---|---|---|
| LPΔ1 | AGCCTGCAGACCGACACTGATCCACTC | PstI |
| LPΔ4 | GGCAAGCTTGCAGAGCAAGGTAAG | HindIII |
| PPPAD8 | CAGGTACCGATCTACTGGAGGGGCAACC | KpnI |
| PPPAD9 | CAGAGCTCTTTACCAGACATCATTCCG | SacI |
| BSFP3 | TCGAAAACAAGCGGCA | |
| BPPAD1 | AGCTGCAGTGAGATAAACCTTCTTC | PstI |
| BPPAD2 | CGAAGCTTGTTATCTCAAAAGACGTTAGG | HindIII |
| Chim2 | GCACTAGTTTYGGATACGTTTCATATTTTTCRCGGG | SpeI |
Specific restriction sites are underlined.
Construction of chimeric LP113PP and PP113LP genes.
The two pdc and padA genes from L. plantarum and P. pentosaceus, respectively, contained a ClaI restriction site at the same position, located in the last third of the nucleotide sequence. The pJDC9 vector, used for cloning the pdc gene (leading to pJPDC1) and the padA gene (leading to pJPADP1), also possesses a unique ClaI site. The two recombinant plasmids, containing their respective pad genes in the same orientation, were digested with ClaI, generating two DNA fragments each, which were subsequently ligated. In order to select for the expected chimeric construction, PCR amplifications were performed using the ligation mixture as the template and primers LPΔ1, located upstream of the pdc promoter, and PPPAD9, located downstream of the padA gene stop codon (Table 2). The 1,024-bp amplified DNA fragment was purified and later cloned into pJDC9 (digested with PstI/SacI) to generate pJLP113PP. In order to obtain the second chimeric gene, a similar method was employed. PCR amplification was performed using the same ligation mixture previously used as the template with primers PPPAD8, located upstream the padA promoter, and LPΔ4, located downstream of the pdc gene stop codon (Table 2). The 1,165-bp amplified fragment was purified and cloned into pJDC9 (digested with KpnI/HindIII) to generate pJPP113LP. The in-frame fusions of these two constructions were verified by DNA sequencing.
Construction of chimeric BP121LP and BS121LP genes.
Primers LPΔ1 and LPΔ4 (Table 2) were used to amplify a segment of pJPDC1 DNA while incorporating PstI and HindIII sites at either end. The 1,230-bp fragment contained the pdc gene preceded by its putative promoter and followed by its putative transcriptional terminator. This fragment was cloned into pJDC9 predigested with PstI/HindIII to generate pJPAD14. Using pJPADBP as a template, a 700-bp fragment was amplified with primers BPPAD1 and Chim2 (Table 2) to make a construct which replaced the pdc promoter and the first 384 nucleotides of the pdc gene with the pad promoter from B. pumilus, followed by the gene segment encoding the first 121 amino acids of BPPAD. This DNA fragment was purified and cloned in pJPDC14 prerestricted with PstI and SpeI to generate pJBP121LP. Using plasmid pHPADBS as a template, a 610-bp fragment was amplified with primers BSFP3 and Chim2 (Table 2) in order to replace the pdc promoter and the first 384 nucleotides of the pdc gene with the B. subtilis pad promoter followed by the gene segment encoding the first 121 amino acids of PAD. This fragment was cloned into pJPAD14, prerestricted with SmaI and SpeI, to generate pJBS121LP. In-frame fusions generated in the two chimeric genes were verified by DNA sequencing as described above.
Preparation of whole-cell suspensions and cell extracts and assay of PAD activity.
Cells of recombinant E. coli grown in LB medium were harvested by centrifugation, washed with 25 mM potassium phosphate buffer (pH 6.0), and resuspended in the same buffer at final concentrations of 0.5 g (dry weight) per liter to measure high PAD activities (1 to 50 μmol min−1 mg−1) and 5 g per liter to measure the weaker PAD activities (10 to 500 nmol min−1 mg−1). Reactions were started by addition of substrate. During incubation, samples were centrifuged and supernatants were diluted 50-fold in Stop buffer (20 mM Tris-HCl, 0.3% sodium dodecyl sulfate [SDS], pH 6) to stop activity prior to analysis. For cell extract preparation, cells were harvested as described above and disrupted with a French press at 1.2 × 108 Pa. Kinetic reactions were started by addition of the substrate, and samples were diluted 50-fold in Stop buffer. Phenolic acid degradation and derivative production were monitored by UV spectrophotometry as previously described (4). The total protein concentration in cell extract was determined using a protein assay kit (Bio-Rad, Richmond, Calif.) with bovine serum albumin as the standard. The specific activity was expressed as micromoles or nanomoles of substrate degraded per minute per milligram of protein. For whole cells, protein concentration was deduced from the dry biomass in the cell suspension (1 g of dry biomass per liter corresponded to 0.4 g of total protein per liter).
4-Vinyl phenol synthesis.
One liter of overnight culture of E. coli TGI (pJPADP6) (5) overexpressing PAD from P. pentosaceus was harvested by centrifugation, washed twice with 25 mM phosphate buffer (pH 6.0), and resuspended in the same buffer at a final concentration of 25 g (dry weight) of cells per liter. Cells were incubated for 1 h at 37°C with 12 mM (2 g/liter) p-coumaric acid. The bioconversion of available p-coumaric acid in a solution of 4-vinyl phenol was checked by UV spectrophotometry. Cells were then discarded by centrifugation. The buffer supernatant, containing 4-vinyl phenol, was sterilized by filtration (0.22-μm pore size) prior to use.
PAGE analysis.
The protein extracts were resolved by denaturing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (12% resolving gel) as previously described (9) with molecular mass markers (low-range SDS-PAGE standards; Bio-Rad) as standards.
RESULTS
Amino acid sequences and substrate specificities of the four native PADs.
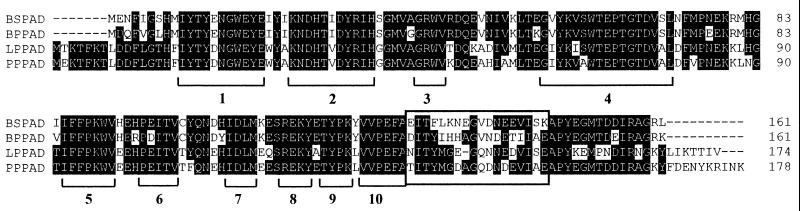
The amino acid sequences of L. plantarum PAD (LPPAD), P. pentosaceus PAD (PPPAD), B. subtilis PAD, (BSPAD), and B. pumilus PAD (BPPAD) have an average identity of 61%. The identity is highest (71%) in the central portion of the enzymes, which contains several highly conserved regions (1 to 10) (Fig. 1). When amino acid residues with similar side chains are taken into consideration, the four enzymes are 66% similar for the total amino acid sequence and 80% similar for the central, more-conserved portion. The amino acid sequences flanking the central region are the most divergent (Fig. 1). Upon greater examination it was observed that the similarity of the carboxy termini of the four PADs decreased significantly. One particular cluster of 18 amino acid residues displays only 22% identity and 44% similarity (Fig. 1). We observed that the LPPAD and PPPAD enzymes exhibited high sequence similarity (87%), as did the BPPAD and BSPAD enzymes (89%).
FIG. 1.
Comparison of the amino acid sequences of LPPAD (accession no. U63827), PPPAD (accession no. AJ276891), BPPAD (accession no. AJ278683), and BSPAD (accession no. AF017117). The sequences were aligned using the Clustal program. Identical residues are shaded. The cluster of 18 amino acids which corresponds to the variable region is boxed. The conserved regions are indicated (1 to 10). The numbers on the right correspond to the amino acid position in the protein sequence.
In order to determine the substrate specificity of each recombinant enzyme, resting cells and cell extracts from the two controls, E. coli TG1(pJDC9) and E. coli TG1(pHT315), as well as the four recombinant E. coli clones carrying pad genes, were prepared and tested for PAD activity. No PAD activity was detected in the two controls. Each recombinant enzyme displayed PAD activity on p-coumaric acid at approximately the same levels in vivo and in vitro with or without ammonium sulfate addition (Table 3). The high activity on p-coumaric acid displayed by each PAD expressed in E. coli indicates that the four bacterial pad promoters were accurately recognized by the RNA polymerase from E. coli. Ferulic and caffeic acids were decarboxylated by BSPAD and BPPAD at nearly the same level as p-coumaric acid in the three samples, while LPPDC and PPPAD displayed activities approximately 100 to 1,000 times lower on ferulic and caffeic acids than on p-coumaric acid. As previously observed, a weak activity on ferulic acid was detected in whole cells of E. coli (pJPDC1) and only in corresponding cell extract supplemented with ammonium sulfate (4). The p-coumarate decarboxylase (PDC) from L. plantarum (8) was renamed LPPAD due to its activity on p-coumaric, ferulic, and caffeic acids (4).
TABLE 3.
PAD activities of whole cells and corresponding cell extracts from the four bacterial PADs
| Enzyme | Activity (μmol min−1 mg−1) of indicated samplea in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
p-Coumaric acid
|
Ferulic acid
|
Caffeic acid
|
|||||||
| Cells (−) | Ext (−) | Ext (+) | Cells (−) | Ext (−) | Ext (+) | Cells (−) | Ext (−) | Ext (+) | |
| LPPAD | 5 | 8.5 | 12 | 0.04 | <10−4 | 0.03 | 0.04 | 0.03 | 0.06 |
| PPPAD | 60 | 42 | 48 | 0.03 | 0.08 | 0.14 | 0.05 | 0.02 | 0.09 |
| BPPAD | 3.5 | 8.8 | 9.2 | 7.5 | 10.2 | 9.7 | 6 | 8.5 | 9.1 |
| BSPAD | 3 | 2.5 | 2.7 | 3.2 | 2.5 | 2.6 | 1.5 | 1.1 | 1.1 |
Cells, whole cells; ext., cell extract; +, ammonium sulfate (0.75 M) addition; −, no ammonium sulfate addition.
Taken together, these observations indicate that LPPAD is more like PPPAD than BSPAD or BPPAD. Thus, the major differences within the four PAD sequences located in the carboxy-terminal region are likely to be involved in substrate specificity.
Exchange of C-terminal portions between PADs induces modifications in substrate specificity and enzymatic activity of the chimeric proteins.
Four chimeric proteins were constructed from the four bacterial PADs. Each pad promoter tested yielded good expression of each of the four pad genes in E. coli. Each chimeric gene was expressed under the control of the promoter carried with each 5′ portion of the fusions (Fig. 2). Three of these four chimeric proteins, designated PP113LP, BS121LP, and BP121LP, contain the first 113, 121 and 121 amino acids from PPPAD, BSPAD, and BPPAD, respectively, followed by the last 61, 46, and 46 amino acids, respectively, from LPPAD. The fourth chimeric protein, which contains the first 113 amino acids from LPPAD followed by the last 65 amino acids from PPPAD, was designated LP113PP (Fig. 2).
FIG. 2.
(a) Structure of the PAD proteins expressed in E. coli under the control of their own promoters. The positions of the ClaI sites of pdc and padA and the SpeI site of pdc are indicated. (b) Structure of the four chimeric proteins. The promoter which controls chimeric gene expression in E. coli is indicated. Restriction sites are indicated as follows: H, HindIII; P, PstI; Sa, SacI; K, KpnI; S, SmaI. Names of genes are on the right.
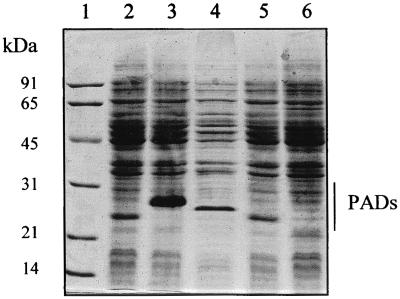
Cell extracts of the resulting clones E. coli TG1(pJLP113PP) and TG1(pJPP113LP) were prepared and analyzed by SDS-PAGE. An intense protein band of about 26 kDa was detected in the recombinant E. coli carrying the LP113PP gene, and a smaller protein band was detected in the recombinant E. coli carrying the PP113LP gene (Fig. 3). PAD activities of these two new proteins were then tested on p-coumaric, ferulic, and caffeic acids (Table 4). LPPAD and PPPAD metabolized p-coumaric acid with activities about 400-fold higher than those for ferulic and caffeic acids. Interestingly, the corresponding chimeric proteins, LP113PP and PP113LP, respectively, displayed activities only 65- to 90-fold higher on p-coumaric acid than on ferulic acid. Moreover, LP113PP displayed significant activity on ferulic acid in vitro, without requiring ammonium sulfate addition, contrary to what was found for the wild LPPAD enzyme. However, E. coli TG1(pJPP113LP) cell extract displayed no detectable activity on ferulic acid in phosphate buffer. A weak ferulic acid decarboxylase activity was detected in this cell extract supplemented with ammonium sulfate (Table 4).
FIG. 3.
SDS-PAGE of crude cell extracts from recombinant clones of E. coli TG1 expressing native and chimeric PADs. Lanes: 1, molecular mass standards (SDS-PAGE standards; Bio-Rad); 2, TG1(pJPAD14); 3, TG1(pJPLP113PP); 4, TG1(pJPADP6); 5, TG1(pJPP113LP); 6, TG1(pJDC9). Molecular size markers are indicated on the left.
TABLE 4.
PAD activities of cell extracts from the four chimeric proteins
| Enzyme | Activity (μmol min−1 mg−1)a in:
|
|||||
|---|---|---|---|---|---|---|
|
p-Coumaric acid
|
Ferulic acid
|
Caffeic acid
|
||||
| − | + | − | + | − | + | |
| LP113PP | 17.5 | 25 | 0.14 | 0.28 | 0.05 | 0.07 |
| PP113LP | 0.2 | 1.3 | <10−4 | 0.02 | 0.06 | 0.08 |
| BP121LP | 0.04 | 0.05 | 0.07 | 0.52 | 0.03 | 0.04 |
| BS121LP | 0.05 | 0.07 | 0.04 | 0.06 | 0.05 | 0.04 |
+, ammonium sulfate (0.75 M) addition; −, no ammonium sulfate addition.
Cell extracts of resulting clones E. coli TG1(pJBS121LP) and TG1(pJBP121LP) were also tested on the three phenolic acids (Table 4). BS121LP metabolized the three substrates with a drastic reduction of activity compared to the wild BSPAD activity, but substrate specificities of the two proteins for the three acids were comparable. A comparably significant reduction of activity between BPPAD and chimeric BP121LP was also observed. Nevertheless, BPPAD metabolized p-coumaric and ferulic acids at almost the same levels while BP121LP displayed activity on ferulic acid about 10-fold higher than that on p-coumaric acid.
Taken together, these results indicate that the exchange of the C-terminal region between the four PADs results in the synthesis of functional enzymes which exhibit some differences from the native enzyme PADs, especially with respect to substrate specificity and the enzymatic activity.
PAD activity inhibits E. coli growth due to the high toxicity of vinyl phenol derivatives.
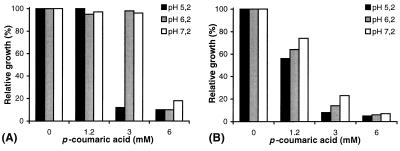
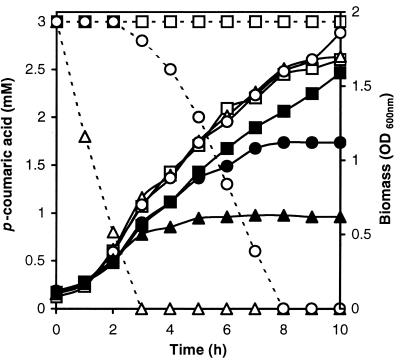
In order to develop a phenotypic test on petri dishes to distinguish colonies of E. coli TG1 with and without a functional PAD, the effect of phenolic acid on the control strain of E. coli TG1 and recombinant PAD strains was examined. Increasing concentrations of p-coumaric acid (0, 1.2, 3, and 6 mM) were tested against three initial pHs (5.2, 6.2, and 7.2). Optical density at 600 nm (OD600) was measured after 20 h of incubation to compare relative levels of growth of the recombinant E. coli strains (Fig. 4). The toxicity of p-coumaric acid was highest at pH 5.2 and decreased with an increase in the initial pH of growth medium. Surprisingly, p-coumaric acid was more toxic for the growth of E. coli TG1(pJPAD14), expressing LPPAD enzyme, than for the growth of control strain TG1(pJDC9). This suggests that the p-coumaric acid degradation product is more toxic for E. coli growth than p-coumaric acid itself. Similar results were observed with E. coli TG1(pHPAD), expressing BSPAD, with p-coumaric and ferulic acids, compared to E. coli TG1(pHT315) grown under the same conditions (data not shown). The levels of growth of E. coli strains TG1(pJDC9), TGI(pJPAD14), and TG1(pJBS121LP) in LB medium supplemented or not with p-coumaric acid (3 mM) at pH 6.2 were then examined. TG1(pJBS121LP) displayed PAD activity 100-fold lower than TG1(pJPAD14) on this substrate. The residual p-coumaric acid concentration was measured during the growth cycle by UV spectrophotometry (Fig. 5). Addition of 3 mM p-coumaric acid reduced the growth of E. coli TG1(pJDC9) slightly, while the growth of E. coli TG1(pJPAD14) and that of TG1(pJBS121LP) were completely inhibited after 3 and 8 h of incubation, respectively. At this stage of the growth, each clone had degraded all available p-coumaric acid into 4-vinyl phenol. These results suggest that 4-vinyl phenol has a high inhibitory effect on E. coli growth. To further confirm 4-vinyl phenol toxicity, clone E. coli TG1(pJPAD14) was incubated with various concentrations of 4-vinyl phenol; 0.3 mM (0.4 g/liter) 4-vinyl phenol addition was shown to totally inhibit its growth (data not shown). 4-Vinyl phenol displayed the same inhibitory effect on the growth of E. coli TG1 which was grown without erythromycin (data not shown). These results allowed us to develop a phenotypic test, based on high vinyl phenol derivative sensitivity, to distinguish E. coli TG1 colonies expressing a functional, cloned PAD. Solid LB medium supplemented with 3 mM p-coumaric acid at pH 6.2 reduced colony size of functional chimeric PAD clones to approximately one-fifth that of colonies typically formed by the control strain, displaying no PAD activity (data not shown).
FIG. 4.
Effect of p-coumaric acid addition in LB medium at different pHs on growth of E. coli TG1(pJDC9) (A) and TG1(pJPAD14) (B). Cultures were inoculated to an initial density of 0.1 OD600 unit and incubated for 20 h at 37°C with shaking. A growth of 100% refers to the control cultures lacking p-coumaric acid in which final biomass levels at pH 5.2, 6.2, and 7.2 resulted in 1.8, 2, and 2.1 OD600 units, respectively.
FIG. 5.
Growth of E. coli TG1(pJDC9) (■), E. coli TG1(pJBS121LP) (●), and E. coli TG1(pJPAD14) (▴) supplemented (filled symbols) or not (open symbols) with p-coumaric acid (3 mM) at pH 6.2. Residual p-coumaric acid concentrations in cultures of E. coli TG1(pJDC9) (–□–), E. coli TG1(pJBS121LP) (–○–), and E. coli TG1(pJPAD14) (–▵–) were measured by UV spectrophotometry.
DISCUSSION
Expression of the four native bacterial PADs in E. coli reveals that p-coumaric acid was the main substrate for each PAD. Ferulic acid is metabolized by LPPAD and PPPAD with an activity about 500-fold lower than that for p-coumaric acid. BSPAD and BPPAD display similar activities on either substrate. Since p-coumaric acid decarboxylation results in production of phenol derivatives that yield “smoky” and aromatic odors and flavors (10, 19) and since ferulic acid derivatives are considered useful aromatic products for foods (25), it is of interest to investigate the PAD enzyme structure to characterize the regions involved in catalytic activity and substrate specificity. It may also be useful to produce PADs displaying new substrate specificities. Chimeric enzyme construction was shown to be useful for combining properties not typically found in any naturally occurring enzyme (29). Construction of chimeric PADs was initiated based on the comparison of PAD amino acid sequences, and this suggested that the PAD C-terminal region could be involved in enzyme substrate specificity. Our results demonstrate that different combinations of homologous C-terminal regions of PADs from four bacteria result in the formation of enzymatically active chimeric species that display catalytic activities different from those of the native PADs. Although the chimeric PADs displayed enzymatic characteristics different from those of the native enzymes, LP113PP, PP113LP, and BS121LP still displayed a greater activity on p-coumaric acid than on ferulic and caffeic acids. The fourth chimeric PAD, BP121LP is very interesting, as this enzyme possesses a characteristic that has never been observed in any native microbial PAD. Ferulic acid is decarboxylated by this enzyme with a relative activity approximately 10-fold higher than that for p-coumaric acid. Nevertheless, BP121LP differs from BS121LP by only seven amino acids. Of these amino acids, five are identical among LPPAD, PPPAD, and BSPAD sequences (A at position 39, E at position 55, N at position 77, H at position 94, and H at position 105; Fig. 1), indicating that these five amino acids should be implicated in the substrate site specificity, especially for ferulic acid metabolism. These results are the first step in the analysis of structure-function relationships for PADs.
Further-modified PADs with improved enzymatic properties might be produced by selective mutagenesis of the C-terminal and also of the N-terminal parts. A random-mutagenesis method with selection for specific catalytic properties should also be used to identify substrate specificity sites. In order to facilitate the screening of such modified enzymes, we have developed a medium to distinguish E. coli recombinant strains expressing PAD activity from those that do not display PAD activity. This screening medium is based on our results, which revealed a strong inhibition effect on E. coli growth induced by 4-vinyl phenol. We have demonstrated several issues through this work. The first is that phenolic acids inhibit E. coli growth in a manner similar to that which was previously demonstrated for some rumen bacteria (7, 34) and strains of S. cerevisiae (3). We observed that this inhibitory effect increased with a decrease of initial pH on LB medium. We also observed that phenolic acid addition has a much greater inhibitory effect on recombinant E. coli strains expressing xenobiotic PAD activity than on the control strain (E. coli TG1 containing pJDC9). This result suggests that vinyl phenol derivatives resulting from PAD activity are much more toxic for E. coli than their corresponding phenolic acid substrates. Addition of a low concentration (<0.3 mM) of vinyl phenol derivatives to the growth medium results in total inhibition of the growth of E. coli strains.
Compared to microorganisms such as B. pumilus (15, 35), S. cerevisiae (13, 22), and L. plantarum (4), in which PAD activity confers resistance to the toxic effects of phenolic acids, E. coli apparently uses a different system to counteract phenolic acid toxicity. E. coli, especially the soil-dwelling strains, may have developed another system to resist these naturally occurring compounds. Most of the time, detoxification involves active efflux of the toxic compound from the cell by highly specific systems (21, 28). Interestingly, E. coli possesses a detoxification system encoded by the mar regulon, which can be induced by antibiotics and other chemicals containing aromatic rings, such as salicylate, benzoate, and cinnamate, a molecule very closely related to phenolic acids used in this study (1, 14). These compounds lead to the inactivation of the MarR repressor, resulting in mar operon transcription and MarA protein synthesis. This protein is a transcriptional activator which induces expression of mechanisms involved in the elimination of toxic compounds from the cell (27). The MarR repressor belongs to a family of bacterial regulatory proteins modulated by plant-derived phenolics (33). To our knowledge, phenolic acids have not been tested for their ability to inactivate the MarR repressor, but these acids could be involved in the induction of the mar operon.
TABLE 1.
Strains, vectors, and their characteristics
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TG1 | supE hsdΔ5thi Δ(lac-proAB)F′ [traD36 proAB+lacIqlacZΔM15] | 21a |
| B. pumilus ATCC 15884 | Wild type, gram positive | American Type Culture Collection |
| Plasmids | ||
| PJDC9 | Emr, ΔlacZ | 11 |
| pHT315 | Emr, ΔlacZ | 2 |
| PJPDC1 | pJDC9 containing the 2.3-kp Sau3AI fragment of L. plantarum CHL2 DNA containing the pdc gene | 8 |
| pJPAD14 | pJDC9 containing the 1,230-bp PstI-HindIII fragment of pJPDC1 with the pdc gene | This study |
| PHPADBS | pHT315 containing the 5-kb Sau3AI fragment of B. subtilis DNA with the pad gene | 10 |
| PJPADBP | pJDC9 containing the 925-bp fragment of B. pumilus with the pad gene | This study |
| PJPADP1 | PJDC9 containing the 6-kb HindIII fragment of P. pentosaceus with the padA gene | 5 |
| pJPADP6 | pJDC9 containing the 970-bp fragment of P. pentosaceus with the pad gene | 5 |
| pJLP113PP | pJDC9 containing pdc promoter and the DNA fragment from L. plantarum coding for the first 113 amino acids of LPPAD followed by the DNA fragment from P. pentosaceus coding for the last 65 amino acids of PPPAD | This study |
| pJPP113LP | pJDC9 containing padA promoter and the DNA fragment from P. pentosaceus coding for the first 113 amino acids of PPPAD followed by the DNA fragment from L. plantarum coding for the last 61 amino acids of LPPAD | This study |
| pJBP121LP | pJDC9 containing pad promoter and the DNA fragment from B. pumilus coding for the first 121 amino acids of BPPAD followed by the DNA fragment from L. plantarum coding for the last 46 amino acids of LPPAD | This study |
| pJBS121LP | pJDC9 containing pad promoter and the DNA fragment from B. subtilis coding for the first 121 amino acids of BSPAD followed by the DNA fragment from L. plantarum coding for the last 46 amino acids of LPPAD | This study |
ACKNOWLEDGMENTS
We are very grateful to Torey Arvik (Cornell University, Geneva, N.Y.) for critical review of the manuscript and to Christine Bernard-Rojas for technical assistance.
This study was supported by the Ministère de l'Education Nationale, de la Recherche et de la Technologie and the Conseil Régional de Bourgogne.
REFERENCES
- 1.Alekshun M, Levy S. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Baranowski J D, Davidson P M, Nagel C, Branen A L. Inhibition of Saccharomyces cerevisiae by naturally occurring hydroxycinnamates. J Food Sci. 1980;45:592–594. [Google Scholar]
- 4.Barthelmebs L, Diviès C, Cavin J-F. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existance of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl Environ Microbiol. 2000;66:3368–3375. doi: 10.1128/aem.66.8.3368-3375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthelmebs L, Lecomte B, Diviès C, Cavin J-F. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J Bacteriol. 2000;182:6724–6731. doi: 10.1128/jb.182.23.6724-6731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Borneman W S, Akin D E, van Eseltine W P. Effect of phenolic monomers on ruminal bacteria. Appl Environ Microbiol. 1986;52:1331–1339. doi: 10.1128/aem.52.6.1331-1339.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavin J-F, Barthelmebs L, Diviès C. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl Environ Microbiol. 1997;63:1939–1944. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavin J-F, Barthelmebs L, Guzzo J, van Beeumen J, Samyn B, Travers J-F, Diviès C. Purification and characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum. FEMS Microbiol Lett. 1997;147:291–295. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavin J-F, Dartois V, Diviès C. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:1466–1471. doi: 10.1128/aem.64.4.1466-1471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 12.Christov L P, Prior B A. Esterases of xylan-degrading microorganisms: production, properties and significance. Enzyme Microbiol Technol. 1993;15:460–475. doi: 10.1016/0141-0229(93)90078-g. [DOI] [PubMed] [Google Scholar]
- 13.Clausen M, Lamb C J, Megnet R, Doerner P W. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene. 1994;142:107–112. doi: 10.1016/0378-1119(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 14.Cohen S, Levy S, Foulds J, Rosner J. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degrassi G, Polverino de Laureto P, Bruschi C V. Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Appl Environ Microbiol. 1995;61:326–332. doi: 10.1128/aem.61.1.326-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries R P, Michelsen B, Poulsen C H, Kroon P A, van den Heuvel R H H, Faulds C B, Williamson G, van den Hombergh J P T W, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries R P, Poulsen C H, Madrid S, Visser J. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J Bacteriol. 1998;180:243–249. doi: 10.1128/jb.180.2.243-249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dower W J, Miller F, Ragsdale C W. Highly efficient transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edlin D A N, Narbad A, Gasson M J, Dickinson J R, Lloyd D. Purification and characterization of hydroxycinnamate decarboxylase from Brettanomyces anomalus. Enzyme Microbiol Technol. 1998;22:232–239. [Google Scholar]
- 20.Etiévant P X, Issanchou S N, Marie S, Ducruet V, Flanzy C. Sensory impact of volatile phenols on red wine aroma: influence of carbonic maceration and time of storage. Sci Aliments. 1989;9:19–33. [Google Scholar]
- 21.George A M. Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol Lett. 1996;139:1–10. doi: 10.1111/j.1574-6968.1996.tb08172.x. [DOI] [PubMed] [Google Scholar]
- 21a.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, United Kingdom: Cambridge University; 1984. [Google Scholar]
- 22.Goodey A R, Tubb R S. Genetic and biochemical analysis of the ability of Saccharomyces cerevisiae to decarboxylate cinnamic acids. J Gen Microbiol. 1982;128:2615–2620. [Google Scholar]
- 23.Hashidoko Y, Tahara S. Stereochemically specific proton transfer in decarboxylation of 4-hydroxycinnamic acids by 4-hydroxycinnamate decarboxylase from Klebsiella oxytoca. Arch Biochem Biophys. 1998;359:225–230. doi: 10.1006/abbi.1998.0911. [DOI] [PubMed] [Google Scholar]
- 24.Huang Z, Dostal L, Rosazza J P N. Microbial transformation of ferulic acid by Saccharomyces cerevisiae and Pseudomonas fluorescens. Appl Environ Microbiol. 1993;59:2244–2250. doi: 10.1128/aem.59.7.2244-2250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, Dostal L, Rosazza J P N. Purification and characterization of a ferulic acid decarboxylase from Pseudomonas fluorescens. J Bacteriol. 1994;176:5912–5918. doi: 10.1128/jb.176.19.5912-5918.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McSweeney C, Dulieu A, Webb R I, del Dot T, Blackall L L. Isolation and characterization of a Clostridium sp. with cinnamoyl esterase activity and unusual cell envelope ultrastructure. Arch Microbiol. 1999;172:139–149. doi: 10.1007/s002030050753. [DOI] [PubMed] [Google Scholar]
- 27.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 29.Nixon A E, Ostermeier M, Benkovic S J. Hybrid enzymes: manipulating enzyme design. Trends Biotechnol. 1998;16:258–264. doi: 10.1016/s0167-7799(98)01204-9. [DOI] [PubMed] [Google Scholar]
- 30.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. J Biol Chem. 1987;262:12209–12217. [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 34.Theodorou M K, Gascoyne D J, Akin D E, Hartley R D. Effect of phenolic acids and phenolics from plant cell walls on rumenlike fermentation in consecutive batch culture. Appl Environ Microbiol. 1987;53:1046–1050. doi: 10.1128/aem.53.5.1046-1050.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zago A, Degrassi G, Bruschi C V. Cloning, sequencing, and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid degradation. Appl Environ Microbiol. 1995;61:4484–4486. doi: 10.1128/aem.61.12.4484-4486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaldivar J, Ingram L O. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;66:203–210. doi: 10.1002/(sici)1097-0290(1999)66:4<203::aid-bit1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]