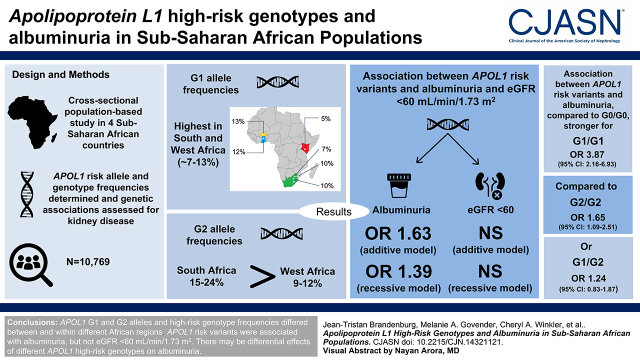
Visual Abstract
Keywords: albuminuria, glomerular filtration rate, molecular genetics, chronic kidney disease, apolipoprotein L1
Abstract
Background and objectives
Recessive inheritance of African-specific APOL1 kidney risk variants is associated with higher risk of nondiabetic kidney disease, progression to kidney failure, and early-onset albuminuria that precedes eGFR decline. The effect of APOL1 risk variants on kidney disease in continental Africans is understudied. Objectives of this study were to determine APOL1 risk allele prevalence and associations between APOL1 genotypes and kidney disease in West, East, and South Africa.
Design, setting, participants, & measurements
This cross-sectional population-based study in four African countries included 10,769 participants largely aged 40–60 years with sociodemographic and health information, anthropometry data, and blood and urine tests for biomarkers of kidney disease. APOL1 risk alleles were imputed from the H3Africa genotyping array, APOL1 risk allele and genotype frequencies were determined, and genetic associations were assessed for kidney disease. Kidney disease was defined as the presence of eGFR <60 ml/min per 1.73 m2, albuminuria, or a composite end point including eGFR <60 ml/min per 1.73 m2 and/or albuminuria.
Results
High G1 allele frequencies occurred in South and West Africa (approximately 7%–13%). G2 allele frequencies were highest in South Africa (15%–24%), followed by West Africa (9%–12%). Associations between APOL1 risk variants and albuminuria were significant for recessive (odds ratio, 1.63; 95% confidence interval, 1.25 to 2.12) and additive (odds ratio, 1.39; 95% confidence interval, 1.09 to 1.76) models. Associations were stronger for APOL1 G1/G1 genotypes versus G0/G0 (odds ratio, 3.87; 95% confidence interval, 2.16 to 6.93) compared with either G2/G2 (odds ratio, 1.65; 95% confidence interval, 1.09 to 2.51) or G1/G2 (odds ratio, 1.24; 95% confidence interval, 0.83 to 1.87). No association between APOL1 risk variants and eGFR <60 ml/min per 1.73 m2 was observed.
Conclusions
APOL1 G1 and G2 alleles and high-risk genotype frequencies differed between and within West and South Africa and were almost absent from East Africa. APOL1 risk variants were associated with albuminuria but not eGFR <60 ml/min per 1.73 m2. There may be differential effects of homozygous G1 and G2 genotypes on albuminuria that require further investigation.
Podcast
This article contains a podcast at https://www.asn-online.org/media/podcast/CJASN/2022_05_16_CJN14321121.mp3
Introduction
Prevalent CKD is estimated at 8%–16% worldwide, with high morbidity and mortality in resource-limited countries (1). The true prevalence of CKD and phenotypic associations with genetic risk factors are understudied in resident African populations (2). By contrast, CKD and associated genetic risk are well characterized in admixed African American populations, informed by a series of landmark studies. These studies showed that focal segmental glomerulosclerosis (FSGS), hypertension-attributed kidney disease, and human immunodeficiency virus-associated nephropathy (HIVAN) were strongly associated with three African-specific apolipoprotein L1 (APOL1) coding variants on chromosome 22 (3–8).
The chromosome 22 region was discovered by admixture mapping and found to harbor African-specific kidney risk variants that include two missense single nucleotide polymorphisms (SNPs) defining the G1 haplotype (rs73885319 and rs60910145) and a six-base–pair deletion removing two amino acids (rs71785313) defining the G2 haplotype (9). Most likely, G1 and G2 variants originated in West Africa, and their frequency increased due to recent positive selection by Trypanosoma brucei rhodesiense in East Africa and Trypanosoma brucei gambiense in West Africa (10).
Subsequent studies showed that APOL1 risk alleles were not associated with diabetic or IgA nephropathy but were associated with collapsing glomerulopathies due to systemic lupus erythematosus, exogenous interferon (IFN) administration, untreated HIV infection, and, more recently, severe acute respiratory syndrome coronavirus 2 (6,11–15). To date, the spectrum of APOL1-associated disease includes sickle cell nephropathy, preeclampsia, FSGS, and hypertension-attributed kidney disease (16). In the African American population, APOL1 allele frequencies are approximately 23% for G1 and 13% for G2 (6). Almost 50% of the African American population carries one risk allele, and approximately 13% of the African American population carries two risk alleles (G1/G1, G1/G2, and G2/G2), comprising the high-risk genotype for kidney disease (17). Although case-control studies contribute to understanding APOL1-associated kidney disease (often reporting high odds ratios [ORs]), these are limited by selection and survival biases and require corroboration in population-based studies (18).
Large community-based studies have confirmed higher risk for incident kidney disease (albuminuria; eGFR <60 ml/min per 1.73 m2), progression to kidney failure, and early-onset albuminuria that precedes decline in eGFR in African American populations with APOL1 high-risk genotypes compared with other US population groups (7,19–23). Most studies support a recessive model of inheritance with little or no differential effects between high-risk genotypes (5,6,11).
However, differential effects between G1 and G2 homozygotes have been reported. When compared with G2 homozygotes, G1 homozygotes in the Dallas Heart study had a higher incidence of kidney disease (eGFR <60 ml/min per 1.73 m2) overall (P=0.05) and for nondiabetic kidney disease (P=0.007), and G1 homozygotes in the Jackson Heart study had higher risks for CKD (P=0.04) and albuminuria (P=0.02) (7,24). Furthermore, G1/G0 heterozygosity has been associated with higher risk for hypertension-associated kidney failure, younger age at presentation of nondiabetic kidney failure, and higher risk for FSGS or HIVAN, whereas G2/G0 heterozygosity is present in all documented cases of IFN-associated nephropathy (5,6,15,25–27).
Although frequency distributions of APOL1 alleles have been reported, no population-based studies and only a handful of case-control studies have investigated the spectrum of APOL1-associated kidney disease in sub-Saharan Africa. Regarding APOL1 allele frequencies, some West African ethnic groups from Ghana and Nigeria have G1 frequencies >40% with lower G2 frequencies (6%–24%). Previously published studies reported lower combined G1 and G2 frequencies (approximately 18.4%) in South Africa, and absent or infrequent occurrence in East Africa (15,17,28).
Of the available patient-control studies, APOL1 high-risk genotypes have been associated with hypertension-attributed kidney disease in the Democratic Republic of Congo, specifically G1/G1 and G1/G2 (OR, 7.7; 95% confidence interval [95% CI], 1.5 to 39.7) (29); nondiabetic kidney disease in Nigeria (OR, 4.8; 95% CI, 1.6 to 14.9) (30); and HIVAN in South Africa (OR, 89; 95% CI, 18 to 912) (15). The APOL1 association with HIVAN differs remarkably between South Africans (OR, 89; 95% CI, 18 to 912) and African American participants (OR, 29; 95% CI, 13.1 to 68.5), although approximately the same number (78% and 72%, respectively) of cases carried high-risk genotypes (6,15). In the South African study (but not the US study), the G1/G0 genotype was strongly associated with HIVAN risk. In a Nigerian study of HIV-infected participants on antiretroviral therapy, APOL1 high-risk genotypes were associated with a higher likelihood of microalbuminuria (OR, 1.97; 95% CI, 1.37 to 2.82) and macroalbuminuria (OR, 3.96; 95% CI, 1.95 to 8.02); this was the first study from the region to investigate the effect of APOL1 high-risk genotypes on early kidney disease (31).
In our large population-based study from West, East, and South Africa, we performed an exploratory analysis to characterize APOL1 genetic variation, compare allele and genotype frequencies within and between study sites, and investigate associations between APOL1 genotypes and biomarkers of kidney disease: low eGFR, albuminuria, and a composite end point comprising low eGFR and/or albuminuria.
Materials and Methods
Study Participants
This study was part of the Africa Wits–International Network for the Demographic Evaluation of Populations and Their Health Partnership for Genomic Studies (referred to as AWI-Gen), a cross-sectional population-based study of over 12,000 participants largely aged 40–60 years at recruitment from six participating urban and rural centers in four sub-Saharan African countries. Pregnant women, first-degree relatives of existing participants, recent immigrants (with <10 years of residence in the region), and individuals with physical impairments preventing measurement of BP and other anthropometric indices were excluded. West Africa included two countries: Ghana (Navrongo) and Burkina Faso (Nanoro), East Africa included Kenya (Nairobi), and South Africa had three study sites (Soweto, Agincourt, and Dikgale). The overall aim of the AWI-Gen collaboration is to investigate genomic and environmental risk factors that affect cardiometabolic disease (32,33).
For this study, 10,769 adults with data for APOL1 genotypes were included. The Human Research Ethics Committee of the University of the Witwatersrand, South Africa, approved the study, with additional institutional review board approval from each participating site. All participants provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki. Study methods are detailed elsewhere (33), but briefly, cross-sectional demographic, health, and family history information was obtained through a researcher-administered questionnaire. Self-reported ethnicity was collected to assess diversity of participants. Anthropometric measurements, clinical data, fasting venous blood samples, and spot midstream urine samples were collected.
This study received the approval of the Human Research Ethics Committee (Medical), University of the Witwatersrand/South Africa (M121029 and M170880); the Centre Muraz Institutional Ethics Committee/Burkina Faso (015-2014/CE- CM); the National Ethics Committee for Health Research/Burkina Faso (2014-08-096); the Ghana Health Service Ethics Review Committee (identification no. GHS-ERC:05/05/2015); the Navrongo Institutional Review Board (identification no. NHRCIRB178); and the AMREF Health Ethics and Scientific Review Committee in Kenya (approval no. P114/2014). All of the participants provided informed consent before entering the study.
Biomarkers of Kidney Disease
Biomarkers of kidney disease were defined as low eGFR (eGFR <60 ml/min per 1.73 m2), albuminuria (random spot urine: urine albumin-creatinine ratio [UACR] >30 mg/g), and a composite end point including low eGFR and/or albuminuria. The Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI; creatinine) 2009 equation without adjustment for African American ethnicity (34) was used to estimate the GFR.
At each partner site, blood and urine specimens were processed and stored at −80°C. After completion of the study, each partner site transported samples on dry ice to a central laboratory in Johannesburg, South Africa. At the central laboratory, all analyses were batched and performed according to good laboratory practice with external monitoring for quality control. Serum creatinine and urine creatinine (milligrams per deciliter) were measured using the Jaffe kinetic method calibrated to an isotope dilution mass spectrometry–traceable standard. Urinary albumin concentration was measured with immunoturbidimetry. The UACR (milligrams per gram) was calculated from these measurements. DNA extracted from buffy coat samples was used for genotyping.
Comorbidities and Socioeconomic Status
Diabetes mellitus was defined as fasting blood glucose ≥126 mg/dl, random blood glucose >198 mg/dl, self-reported history of diabetes mellitus, or currently on treatment regardless of the glucose concentration. Hypertension was defined as mean systolic BP ≥140 mm Hg, mean diastolic BP ≥90 mm Hg, self-reported history of hypertension, or currently on treatment regardless of BP measurement. HIV status was self-reported or diagnosed with a rapid test. Socioeconomic status was assessed using a validated tool appropriate for assessing wealth in low- and middle-income settings. Household assets are scored, and on the basis of the score, data are represented as quintiles, with the poorest in the first quintile (33).
Genotyping and Imputation of APOL1 Genotypes
APOL1 G1 and G2 are formally haplotypes (Supplemental Table 1), but for ease of understanding, we refer to them as alleles in this publication. The APOL1 haplotypes carrying neither G1 nor G2 variants are termed G0. APOL1 high-risk genotypes comprise two risk alleles (G1/G1, G1/G2, and G2/G2), and low-risk genotypes comprise zero or one risk allele (G0/G0, G0/G1, and G0/G2).
Genomic DNA was genotyped using the H3Africa genotyping array (https://www.h3abionet.org/h3africa-chip) designed as an African common variant–enriched GWAS array (Illumina) with approximately 2.3 million SNPs. Preimputation quality control was applied to the entire AWI-Gen genotype dataset. Individuals with a missing SNP calling rate >0.05 were removed. SNPs with genotype missingness >0.05, minor allele frequency <0.01, and Hardy–Weinberg equilibrium P value <0.001 were removed. Nonautosomal and mitochondrial SNPs and ambiguous SNPs that did not match the GRCh37 reference alleles or strands were removed.
Imputation was performed on the cleaned dataset (with 1,729,661 SNPs and 10,903 individuals) with the UK10K and 1000 Genomes Project panel, using the Sanger imputation service (35). We extracted 187 positions between 35649056 and 37663576 on chromosome 22 around the APOL1 gene from AWI-Gen GWAS data and used three whole-genome sequencing datasets to assess imputation accuracy: AWI-Gen whole-genome sequencing (36), 1000 Genomes Project (37), and the African Genome Variation Project (38). Three genomics positions were used to define the APOL1 risk alleles G1 and G2: rs73885319 (p.S342G), rs60910145 (p.I384M), and insertion/deletion rs71785313 (p.N388Y389/−) (Supplemental Table 1). The imputation of these three variants was compared with the original sequences from the whole-genome sequencing datasets.
Statistical Analyses: Genetic Association with Biomarkers of Kidney Disease
We used mixed effects models using the ImerTest package (39) to compute associations between APOL1 alleles and biomarkers of kidney disease. Logistic mixed models were used for binary biomarkers (low eGFR, albuminuria, and the composite end point for kidney disease). For analyses stratified by site, we used logistic models for binary biomarkers. ORs with 95% CIs were calculated for the association between biomarkers of kidney disease and (1) zero or one versus two APOL1 risk alleles (recessive model), (2) zero versus one or two APOL1 risk alleles (dominant model), or (3) three genotype groups (zero versus one versus two risk alleles; additive model). The models were adjusted for age, sex, body mass index (BMI), diabetes mellitus, hypertension, and HIV status as fixed variables and study site as a random variable. ORs and statistical significance were assessed using mixed effects models for carriage of one or two APOL1 risk alleles in the recessive, dominant, and additive models. APOL1 analysis was stratified by diabetes mellitus, hypertension, and HIV status, and mixed effects models were used to assess associations with albuminuria in recessive models. Furthermore, GxE interaction terms were computed independently using interaction terms between APOL1 and diabetes mellitus, hypertension, and HIV in models adjusted for covariates.
For tables of descriptive data, the proportion of missing data for each variable was indicated using the appropriate denominator (different from the stipulated sample size) or annotated as a footnote. Missing data were not imputed. For UACR, data were missing at random for the Agincourt and Soweto sites, South Africa. Of note, there were no UACR data for women participants in Soweto. To assess potential effects of these missing data on the association between high-risk APOL1 genotypes and albuminuria, fixed effects meta-analyses using results from linear mixed models by site and by sex confirmed that no significant bias was introduced from the missing data. To assess whether there was a site effect for the association between high-risk APOL1 genotypes and albuminuria, we tested for heterogeneity using the Cochrane test.
Results
Genotype Imputation Quality
The rs73885319 SNP defining the G1 allele was present in the H3Africa array, and there was full concordance between the genotyped allele calls and imputed calls. Imputation accuracy values using African samples for rs60910145 (in linkage disequilibrium with the functional G1 rs73885319 SNP) and rs71785313 (G2) were 99.9% and 99.0%, respectively, between whole-genome sequence and imputed data. Similarly, other populations showed concordance between G0, G1, and G2 alleles and imputed data assessed using sequence data (Supplemental Table 2).
Population Characteristics and APOL1 Allele and Genotype Frequencies in South, East, and West African AWI-Gen Studies Sites
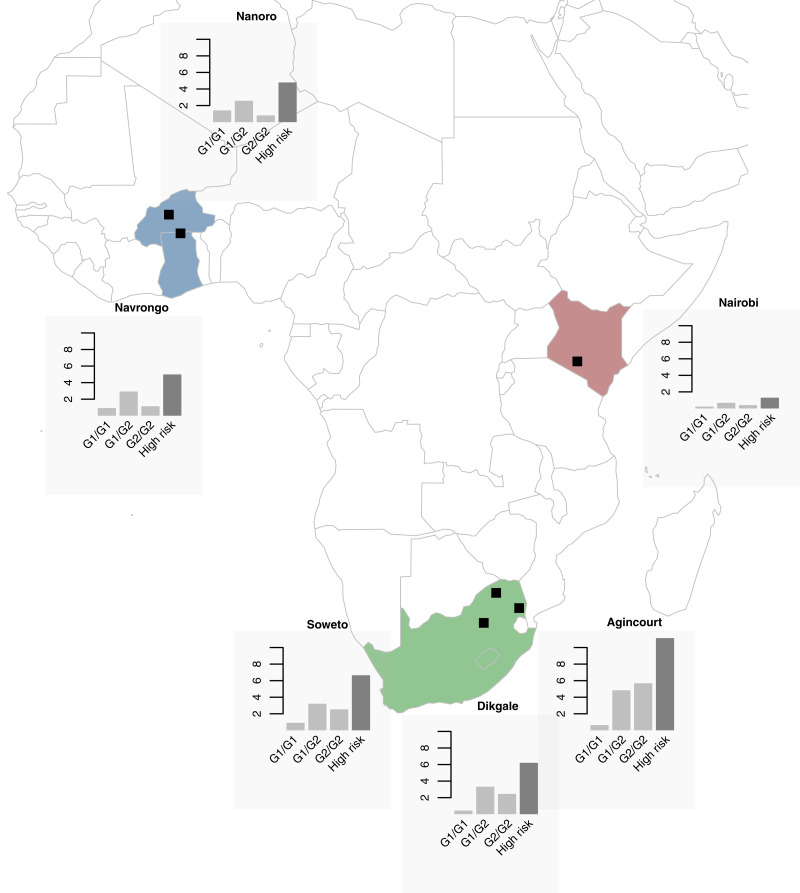
Demographic and clinical characteristics and APOL1 allele and genotype frequency distributions for each study site and overall are summarized in Table 1. The mean age of participants was similar across all sites, with some differences in ratios of men to women. Compared with East and West Africa, the three South African sites had the highest prevalence of diabetes mellitus, hypertension, HIV, obesity, and kidney disease. Overall, APOL1 allele frequencies were 10% for G1 and 14% for G2. G1 allele frequencies ranged from 5% to 13% across the sites, with the highest frequencies in West Africa. G2 allele frequencies varied widely from 7% to 24%, with the highest frequencies in South Africa. The lowest allele frequencies for G1 (5%) and G2 (7%) occurred in East Africa (Table 1). High-risk APOL1 genotype frequencies were 6% overall, with the lowest in East Africa (1%) and the highest in South Africa (11%). APOL1 allele and genotype frequencies were highly variable among self-reported ethnic groups (Figure 1, Supplemental Table 3).
Table 1.
Population characteristics and APOL1 allele and genotype frequencies in the Africa Wits–International Network for the Demographic Evaluation of Populations and Their Health Partnership for Genomic Studies
| Variablea | Africa Wits–International Network for the Demographic Evaluation of Populations and Their Health Partnership for Genomic Studies Site | ||||||
|---|---|---|---|---|---|---|---|
| South Africa | East Africa | West Africa | Overall | ||||
| Agincourt, Rural | Soweto, Urban | Dikgale, Rural | Nairobi, Urban | Nanoro, Rural | Navrongo, Rural | All Sites | |
| Sample size, N | 2253 | 1775 | 1143 | 1771 | 1983 | 1844 | 10,769 |
| Clinical phenotype | |||||||
| Age, yr | 58 (11) | 49 (6) | 52 (8) | 49 (6) | 50 (6) | 51 (6) | 52 (8) |
| Men, N (%) | 938 (42) | 922 (52) | 361 (32) | 804 (45) | 1009 (51) | 842 (46) | 4876 (45) |
| Women, N (%) | 1315 (58) | 853 (48) | 782 (68) | 967 (55) | 974 (49) | 1002 (64) | 5893 (55) |
| Height, m | 1.65 (0.85) | 1.65 (0.88) | 1.62 (0.84) | 1.64 (0.87) | 1.68 (0.87) | 1.62 (0.86) | 1.65 (0.89) |
| Weight, kg | 74 (17) | 78 (19) | 72 (20) | 68 (14) | 59 (12) | 57 (10) | 68 (17) |
| BMI, kg/m2b | 27.2 (6.7) | 28.9 (7.7) | 27.8 (8.2) | 25.4 (5.7) | 21.0 (3.5) | 21.6 (3.6) | 25.1 (6.7) |
| Diabetes mellitus, N (%)c | 194/2214 (9) | 167/1770 (9) | 113/1136 (10) | 124/1755 (7) | 72/1973 (4) | 25/1794 (1) | 695/10,642 (7) |
| Hypertension, N (%) | 1265/2253 (56) | 949/1775 (54) | 515/1143 (45) | 486/1771 (27) | 322/1983 (16) | 455/1844 (25) | 3992/10,769 (37) |
| HIV infection, N (%)d | 602/2251 (27) | 253/1207 (21) | 224/1124 (20) | 216/1561 (14) | 8/1983 (0.4) | 15/1844 (0.8) | 1318/9970 (13) |
| Biomarkers of kidney function, median (IQR) | |||||||
| Serum creatinine, mg/dle | 0.77 (0.22) | 0.79 (0.22) | 0.78 (0.24) | 0.75 (0.21) | 0.72 (0.21) | 0.75 (0.21) | 0.82 (0.2) |
| eGFR, ml/min per 1.73 m2e,f | 92 (21) | 100 (23) | 95 (25) | 102 (17) | 104 (13) | 100 (16) | 100 (20) |
| Urine albumin, mg/gg | 4 (10) | 1 (6) | 3 (8) | 4 (8) | 2 (3) | 1 (2) | 2 (6) |
| Biomarkers of kidney disease,h N (%) | |||||||
| Low eGFRi | 114/2197 (5.2) | 53/1724 (3.1) | 36/1140 (3.2) | 55/1768 (3.1) | 45/1970 (2.3) | 44/1843 (2.4) | 347/10,642 (3.3) |
| Albuminuriag,j | 258/1789 (14) | 107/857 (13) | 115/1024 (11) | 184/1599 (12) | 114/1913 (6) | 114/1701 (7) | 892/8883 (10) |
| Composite end pointk | 295/1740 (17) | 120/856 (14) | 142/1021 (14) | 222/1596 (14) | 144/1902 (8) | 151/1700 (9) | 1074/8815 (12) |
| APOL1 allele distributions, N (%) | |||||||
| G0 allele | 2999 (67) | 2666 (75) | 1730 (76) | 3108 (88) | 3098 (78) | 2793 (76) | 16,394 (76) |
| G1 allele | 437 (10) | 355 (10) | 167 (7) | 177 (5) | 500 (13) | 437 (12) | 2073 (10) |
| G2 allele | 1070 (24) | 529 (15) | 389 (17) | 257 (7) | 368 (9) | 458 (12) | 3071 (14) |
| APOL1 genotype distributions, N (%) | |||||||
| 0 risk alleles (G0/G0) | 997 (44)l | 1009 (57)l | 658 (58)l | 1360 (77)l | 1210 (61)l | 1041 (57)l | 6275 (58)l |
| 1 risk allele | 1005 (45)l | 648 (37)l | 414 (36)l | 388 (22)l | 678 (34)l | 711 (39)l | 3844 (36)l |
| G0/G1 | 300 (13) | 266 (15) | 119 (10) | 157 (9) | 393 (20) | 349 (19) | 1584 (15) |
| G0/G2 | 705 (31) | 382 (22) | 295 (26) | 231 (13) | 285 (14) | 362 (20) | 2260 (21) |
| 2 risk alleles | 251 (11)l | 118 (7)l | 71 (6)l | 23 (1)l | 95 (5)l | 92 (5)l | 650 (6)l |
| G1/G1 | 14 (0.6) | 16 (0.9) | 5 (0.4) | 4 (0.2) | 28 (1.4) | 17 (0.9) | 84 (0.8) |
| G1/G2 | 109 (5) | 57 (3) | 38 (3) | 12 (0.7) | 51 (3) | 54 (3) | 321 (3) |
| G2/G2 | 128 (6) | 45 (3) | 28 (3) | 7 (0.4) | 16 (0.8) | 21 (1) | 245 (2) |
BMI, body mass index; IQR, interquartile range.
All data are reported as mean (SD) unless stated otherwise. Categories are reported as number (N) and percentage; percentages may sum to ±100 from rounding up.
BMI missing data overall n=456 (4%): 453 of 456 Agincourt, two of 456 Soweto, and one of 456 Nanoro.
Diabetes mellitus missing data overall n=127 (1%): 50 of 127 Navrongo, 39 of 127 Agincourt, 16 of 127 Nairobi, ten of 127 Nanoro, seven of 127 Dikgale, and five of 127 Soweto.
HIV infection missing data overall n=799 (7%): 568 of 799 Soweto, 210 of 799 Nairobi, 19 of 799 Dikgale, and two of 799 Agincourt.
Serum creatinine eGFR missing data overall n=239 (2%): 91 of 239 Agincourt, 63 of 239 Soweto, 31 of 239 Nanoro, 22 of 239 Navrongo, 21 of 239 Nairobi, and 11 of 239 Dikgale.
eGFR is calculated using the Chronic Kidney Disease Epidemiology Collaboration (creatinine) equation 2009 without adjusting for the African American coefficient.
Urine albumin and albuminuria missing data overall n=1886 (18%): 918 of 1886 Soweto, 464 of 1886 Agincourt, 172 of 1886 Nairobi, 143 of 1886 Navrongo, 119 of 1886 Dikgale, and 70 of 1886 Nanoro.
For biomarkers of kidney disease, repeat measures for those with albuminuria or low eGFR were not performed, preventing confirmation of CKD. The category “Biomarkers of kidney disease” was used to define these measures on a single screening.
Low eGFR is eGFR <60 ml/min per 1.73 m2 calculated using the Chronic Kidney Disease Epidemiology Collaboration (creatinine) equation 2009 without adjusting for the African American coefficient.
Albuminuria is random spot urine albumin-creatinine ratio >30 mg/g.
The composite end point is low eGFR and/or albuminuria.
Corresponds at main section and in genotype at the total of individual with zero risk allele, one risk allele, and two risk alleles.
Figure 1.
Combined “high-risk” genotype frequencies are highest in West Africa or South Africa and lowest in East Africa, with distinct differences with regard to the G1/G1, G1/G2 and G2/G2 genotypes across regions. APOL1 genotype (G1/G1, G1/G2, and G2/G2) frequencies (percentages) and combined “high-risk” genotypes (G1/G1, G1/G2, and G2/G2) for each sub-Saharan African region: Nanoro and Navrongo in West Africa; Nairobi in East Africa; and Soweto, Dikgale, and Agincourt study sites in South Africa.
APOL1 Genotypes and Associated Risk for Kidney Disease across All AWI-Gen Studies Sites
Associations between APOL1 status (stratified by zero, one, or two risk alleles) and kidney disease risk factors are summarized in Supplemental Table 4. For the study outcomes—low eGFR, albuminuria, and the composite end point—associations with APOL1 stratified by zero, one, or two risk alleles and using additive, recessive, and dominant models of inheritance are summarized in Table 2. Overall, for low eGFR, there was no association with APOL1 for any of the models tested. For albuminuria, the association with APOL1 was significant for the recessive (OR, 1.63; 95% CI, 1.25 to 2.12) and additive models (OR, 1.39; 95% CI, 1.09 to 1.76); for the composite end point, the association was significant for the recessive model (OR, 1.37; 95% CI, 1.06 to 1.78), likely driven by the association with albuminuria. There was no significant heterogeneity (P=0.78) when comparing the association with APOL1 and albuminuria among West, East, and South Africa, and the sample size was likely underpowered to reach statistical significance in East Africa (Supplemental Figure 1, Supplemental Table 5).
Table 2.
Association between APOL1 and biomarkers of kidney disease by risk allele status (zero, one, or two) and recessive, dominant, and additive models
| Biomarkers of Kidney Diseasea | Low eGFRb | Albuminuriac | Composite End Pointd |
|---|---|---|---|
| Frequency of APOL1 risk alleles, N (%) | |||
| 0 | 202/6210 (3.3) | 507/5259 (10) | 627/5225 (12) |
| 1 | 122/3797 (3.2) | 306/3105 (10) | 363/3083 (12) |
| 2 | 23/635 (3.6) | 79/519 (15) | 84/507 (17) |
| All | 347/10,642 (3.0)e | 892/8883 (10)f | 1074/8815 (12) |
| APOL1 models odds ratio (95% confidence interval) | |||
| Sample size, N | 9340g | 8526g | 8486g |
| Additive | 0.78 (0.52 to 1.19) | 1.39 (1.09 to 1.76) | 1.16 (0.93 to 1.45) |
| Dominant, 0 versus 1 or 2 risk alleles | 0.86 (0.67 to 1.11) | 1.12 (0.97 to 1.31) | 1.03 (0.90 to 1.19) |
| Recessive, 0 or 1 versus 2 risk alleles | 0.87 (0.52 to 1.47) | 1.63 (1.25 to 2.12) | 1.37 (1.06 to 1.78) |
For biomarkers of kidney disease, repeat measures for those with albuminuria or low eGFR were not performed, preventing confirmation of CKD; therefore, we used “biomarkers of kidney disease” to define these measures on a single screening.
Low eGFR is eGFR <60 ml/min per 1.73 m2 calculated using the Chronic Kidney Disease Epidemiology Collaboration (creatinine) equation (2009) without adjusting for the African American coefficient.
Albuminuria is a random spot urine albumin-creatinine ratio >30 mg/g.
The composite end point is low eGFR and/or albuminuria.
Missing data n=127 of 10,967 (1%).
Missing data n=1886 of 10,967 (17%).
Corresponds at main section and in genotype at the total of individual with zero risk allele, one risk allele, and two risk alleles.
The prevalence of obesity was higher in the group with two APOL1 risk alleles (Supplemental Table 4). As obesity can be causal for albuminuria and we had adjusted our models for BMI, we tested whether the associations with APOL1 changed without adjusting for BMI and found no difference (Supplemental Table 6). There was no effect of socioeconomic status on the association between APOL1 and low eGFR, albuminuria, and the composite end point (Supplemental Table 7). We tested for associations with APOL1 and low eGFR, albuminuria, and the composite end point stratified by individuals with comorbidity (diabetes mellitus, hypertension, and HIV). APOL1 was associated with albuminuria in individuals who did not have diabetes mellitus, HT, or HIV, and the interaction term was only significant in the minimally adjusted model (Table 3, Supplemental Tables 9 and 10).
Table 3.
APOL1 associations with albuminuria stratified by diabetes, hypertension, and HIV status
| Comorbidity Status | Albuminuria,a n/N (%) | Albuminuriaa Odds Ratio (95% Confidence Interval), Age+Sex–Adjusted Modelb | Albuminuriaa Odds Ratio (95% Confidence Interval), Fully Adjusted Modelc | Pinteraction term Value, Age+Sex–Adjusted Modelb |
|---|---|---|---|---|
| Diabetes | 0.31 | |||
| Absent | 745/8259 (9.0) | 1.68 (1.28 to 2.20) | 1.71 (1.30 to 2.26) | |
| Present | 131/523 (25.0) | 1.20 (0.53 to 2.74) | 1.06 (0.45 to 2.49) | |
| Hypertension | 0.04 | |||
| Absent | 383/5781 (6.6) | 1.94 (1.34 to 2.82) | 2.09 (1.43 to 3.05) | |
| Present | 509/3102 (16.4) | 1.30 (0.91 to 1.85) | 1.33 (0.92 to 1.92) | |
| HIV | 0.40 | |||
| Negative | 655/7508 (8.7) | 1.64 (1.22 to 2.19) | 1.72 (1.28 to 2.33) | |
| Positive | 202/1129 (17.9) | 1.28 (0.74 to 2.23) | 1.36 (0.78 to 2.40) |
Albuminuria is random spot urine albumin-creatinine ratio >30 mg/g.
Logistic mixed models were adjusted for age and sex as fixed variables.
Logistic mixed models were adjusted for site as a random variable and age, sex, body mass index, diabetes mellitus status, hypertension status, and HIV status as fixed variables. Some participants were excluded from the fully adjusted model due to missing data.
We performed exploratory analyses with different genotype combinations for their association with kidney disease biomarkers. Although there was no association with low eGFR, the association with albuminuria was strongest for G1 homozygotes overall (OR, 3.87; 95% CI, 2.16 to 6.93) and in West Africa (OR, 4.93; 95% CI, 2.15 to 10.26) and South Africa (OR, 3.67; 95% CI, 1.38 to 8.82), and a significant, but smaller effect was seen with G2 homozygotes overall (OR, 1.65; 95% CI, 1.09 to 2.51) and in South Africa (OR, 1.62; 95% CI, 1.01 to 2.51). For the composite end point, only the association with G1 homozygotes remained significant overall (OR, 2.73; 95% CI, 1.51 to 4.96) and in West Africa (OR, 3.45; 95% CI, 1.52 to 7.13). There were no significant associations with the G1/G2 genotype and low eGFR, albuminuria, and the composite end point (Table 4). The differential effect sizes between the two homozygous high-risk genotypes (G1/G1 compared with G2/G2) on the three biomarkers of kidney disease are shown in Supplemental Table 8.
Table 4.
Association between APOL1 genotype and biomarkers of kidney disease stratified by region
| Genotypea | All | East Africa | South Africa | West Africa |
|---|---|---|---|---|
| Low eGFR b | ||||
| G0/G1 | 1.03 (0.73 to 1.47) | 1.00 (0.29 to 2.63) | 1.06 (0.61 to 1.76) | 1.01 (0.58 to 1.69) |
| G0/G2 | 0.76 (0.55 to 1.05) | 0.77 (0.26 to 1.88) | 0.82 (0.53 to 1.25) | 0.74 (0.37 to 1.34) |
| G1/G1 | 0.52 (0.07 to 3.83) | NAc | 0 (0 to 35,130.28) | 0.93 (0.05 to 4.47) |
| G1/G2 | 0.81 (0.39 to 1.69) | 4.0 (0.2 to 26.4) | 1.13 (0.46 to 2.38) | NAc |
| G2/G2 | 0.89 (0.41 to 1.94) | 15.09 (0.69 to 125.23) | 0.99 (0.38 to 2.18) | NAc |
| Albuminuria d | ||||
| G0/G1 | 0.97 (0.77 to 1.22) | 0.79 (0.38 to 1.48) | 0.82 (0.58 to 1.14) | 1.3 (0.9 to 1.8) |
| G0/G2 | 1.09 (0.9 to 1.31) | 1.21 (0.73 to 1.95) | 1.06 (0.84 to 1.35) | 1.1 (0.7 to 1.6) |
| G1/G1 | 3.87 (2.16 to 6.93)e | NAc | 3.67 (1.38 to 8.82)e | 4.93 (2.15 to 10.26)e |
| G1/G2 | 1.24 (0.83 to 1.87) | 3.24 (0.16 to 23.79) | 1.20 (0.72 to 1.92) | 1.28 (0.52 to 2.66) |
| G2/G2 | 1.65 (1.09 to 2.51)e | 4.63 (0.62 to 23.40) | 1.62 (1.01 to 2.51)e | 1.09 (0.17 to 3.71) |
| Composite end point f | ||||
| G0/G1 | 0.97 (0.79 to 1.19) | 0.84 (0.44 to 1.50) | 0.83 (0.61 to 1.13) | 1.22 (0.89 to 1.65) |
| G0/G2 | 0.99 (0.83 to 1.17) | 1.12 (0.69 to 1.77) | 0.98 (0.78 to 1.22) | 0.95 (0.67 to 1.34) |
| G1/G1 | 2.73 (1.51 to 4.96)e | NAc | 2.47 (0.87 to 6.18) | 3.45 (1.52 to 7.13)e |
| G1/G2 | 1.06 (0.72 to 1.57) | 2.56 (0.12 to 19.26) | 1.11 (0.69 to 1.73) | 0.89 (0.37 to 1.83) |
| G2/G2 | 1.40 (0.93 to 2.11) | 4.32 (0.59 to 21.73) | 1.41 (0.89 to 2.17) | 0.74 (0.12 to 2.52) |
The associations between APOL1 genotype and biomarkers of kidney disease are presented as odds ratios and 95% confidence intervals. For biomarkers of kidney disease, repeat measures for those with low eGFR or albuminuria were not performed, thus preventing confirmation of CKD; therefore, we used “biomarkers of kidney disease” to define these measures on a single screening. Linear and logistic mixed models were adjusted for site as a random variable and age, sex, body mass index, diabetes mellitus status, hypertension status, and HIV status as fixed variables. NA, not applicable.
APOL1 genotypes G0/G1, G0/G2, G1/G1, G1/G2, and G2/G2 were compared with G0/G0 as the reference.
Low eGFR is eGFR <60 ml/min per 1.73 m2 calculated using the Chronic Kidney Disease Epidemiology Collaboration (creatinine) equation 2009 without adjusting for the African American coefficient.
Sample size was too small to perform the statistical test.
Albuminuria is random spot urine albumin-creatinine ratio >3.0 mg/g.
Corresponds at main section and in genotype at the total of individual with zero risk allele, one risk allele, and two risk alleles.
The composite end point is low eGFR and/or albuminuria.
Discussion
We determined population frequencies of APOL1 alleles and genotypes in multiple ethnic groups from sub-Saharan Africa and investigated their association with biomarkers of kidney disease, leading to several important findings. First, the overall population frequencies of G1 (10%) and G2 (14%) alleles and high-risk genotypes (6%) were lower than reported in African American populations but similar to a recent study from Nigeria, West Africa (high-risk genotype: 6%) (17). We observed distinct differences in population frequencies of APOL1 alleles and high-risk genotypes when comparing West, East, and South African regions and among ethnic groups within each region, with the lowest frequencies among East African ethnic groups.
Second, APOL1 high-risk genotypes were strongly associated with albuminuria but not with low eGFR. The association with albuminuria is consistent with continental African (29) and African American studies (7,40,41). The absence of association with low eGFR might be explained by the use of GFR estimating equations that have limited validation in sub-Saharan Africa, including little difference between the CKD-EPI (creatinine) 2009 and 2021 equations (Supplemental Appendix 1), and laboratory use of the Jaffe rather than the enzymatic assay for creatinine—both of which tend toward overestimating eGFR. It is also plausible that a survival or frailty bias resulting from the near absence of access to KRT for those with kidney failure accounts for the lack of association with low eGFR because those with progressive kidney disease due to APOL1 might be under-represented in our sample. Finally, the population-based study design did not select for groups with severe or progressive kidney disease.
Third, regarding kidney disease risk, we showed an association with obesity and two APOL1 risk alleles but no demonstrable effect of obesity on the association of APOL1 with albuminuria. Our findings corroborate those of a similar-sized study of 11,930 African American participants, where each risk allele was associated with greater obesity odds (1.13-fold) (42), but this association has not been demonstrated in other large cohorts, such as the Coronary Artery Risk Development in Young Adults and the Atherosclerosis Risk in Communities studies (19,43). There was no effect of socioeconomic status on the association between APOL1 and albuminuria. Although this finding is similar to those observed in African American participants (19,43), the assets-based score differs from scoring systems used in high-income settings, and quintile differences may be difficult to detect when the overall status of African participants is one of widespread poverty. There was a significant interaction when testing the APOL1 association with albuminuria for the minimally adjusted model between participants with and without hypertension, but no significant interactions between established risk factors (diabetes, hypertension, and HIV) and APOL1 risk alleles in the fully adjusted model. This lack of interaction could be due to the limited sample size of subgroups of individuals with HIV and diabetes.
Lastly, differential effects of G1 and G2 homozygous genotypes on albuminuria are similar to those reported in the Dallas and Jackson Heart studies, but contradict other studies in which high-risk genotypes have near-equivalent genetic associations in the homozygous or compound heterozygous states (7,24). The absence of association with APOL1 G1/G2 and albuminuria has not been previously described and is difficult to explain, especially because G1/G2 represents the largest group of high-risk genotypes. Studies have shown functional differences between G1 and G2, with their proteins showing differential binding affinities to the trypanosome-encoded serum resistance–associated protein and associations with susceptibility to T.b. rhodiense and T.b. gambiense infection and disease (27). Proposed mechanisms of APOL1-induced cytotoxicity in kidney tissue using in vitro and mouse models include enhanced local APOL1 expression and reduced degradation, increased cation transport, downstream intracellular activation of the inflammasome, defects in autophagy, and mitochondrial dysfunction. However, none of these demonstrate differential effects among high-risk genotypes (16).
Our study has important strengths and limitations. Strengths lie in the well-characterized cohort; the large population-based sampling frame; inclusion of multiple ethnic groups from countries at different stages of the health and epidemiologic transition in West, East, and South Africa; and the availability of GFR estimates and albuminuria as biomarkers of kidney disease. The cross-sectional study design is a limitation as single measurements of eGFR and albuminuria preclude CKD diagnosis. Although albuminuria is a marker for CKD progression, it can also be reversible or regress with time, which is particularly relevant in sub-Saharan Africa where acute, chronic, and endemic infectious disease burdens are high, as are heavy metal and toxin exposures. Without longitudinal follow-up, the effect of high-risk APOL1 genotypes on incident albuminuria, incident CKD, and CKD progression cannot be evaluated. As our analyses were exploratory and sample sizes were relatively small for some subgroup comparisons, our findings should be considered hypothesis generating and will hopefully inform future work for validation.
In conclusion, population frequencies of APOL1 G1 and G2 alleles and high-risk genotypes differed between and within West and South Africa and were almost absent from East Africa. APOL1 kidney risk variants were associated with albuminuria but not eGFR. We noted differential effects of homozygous G1 and G2 genotypes on albuminuria, which might result from genetic or environmental modifiers. Compared with findings in African American populations, the relatively low frequencies and variable penetrance of APOL1 high-risk genotypes, regional differences in the association with albuminuria, and absence of association with low eGFR may reflect the vast genetic diversity of African populations, differences in gene-environment interactions or genetic modifiers in resident African populations, and the absence of European admixture. Further studies in sub-Saharan Africa with data from different regions and ethnic groups are required to confirm and refine the effect of APOL1 risk variants on kidney phenotypes, and longitudinal studies are needed to assess the role of APOL1 and CKD incidence and progression.
Disclosures
M. Ramsay reports consultancy agreements with Roche/Genentech (The Health Equity and Population Science). M. Ramsay is a South African Research Chair in Genomics and Bioinformatics of African Populations hosted by the University of the Witwatersrand, funded by the Department of Science and Technology, and administered by the National Research Foundation. All remaining authors have nothing to disclose.
Funding
The AWI-Gen Collaborative Centre is funded by National Human Genome Research Institute award U54HG006938 and its supplements as part of the H3Africa Consortium. Additional funding came from Department of Science and Technology, South Africa award DST/CON 0056/2014. The project has been supported in part by NIH Clinical Center contract 75N91019D00024.
Supplementary Material
Acknowledgments
We thank all AWI-Gen participants and acknowledge our field workers, laboratory scientists, administrators, data manager, and other staff who contributed to data collection at each study site (Supplemental Appendix 2). In particular, we acknowledge Cassandra Soo, Freedom Mukomana, and Scott Hazelhurst from the Sydney Brenner Institute for Molecular Bioscience; Stephen Tollman; Kathleen Kahn (Agincourt Principal Investigator [PI]); Marianne Alberts (Dikgale PI, deceased in 2020); Catherine Kyobutungi (Nairobi PI); Halidou Tinto (Nanoro PI); Abraham R. Oduro (Navrongo PI); and Shane Norris (Soweto PI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The content of this publication does not necessarily reflect the view or policy of the funders, nor does mention of trade names, commercial products, or organizations imply endorsement by the US or South African governments.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
Author Contributions
J.-T. Brandenburg, J. Fabian, and M. Ramsay conceptualized the study; J.-T. Brandenburg, J. Fabian, M.A. Govender, M. Ramsay, and C.A. Winkler were responsible for formal analysis; J.-T. Brandenburg and M.A. Govender wrote the original draft; and G. Agongo, P.R. Boua, J.-T. Brandenburg, J. Fabian, M.A. Govender, M. Ramsay, and C.A. Winkler reviewed and edited the manuscript.
Data Sharing Statement
The phenotype and genotype data are available on request to the H3Africa Data and Biospecimen Access Committee (European Genome-Phenome Archive accession nos. EGAD00001006425 and EGAD00001001996, respectively).
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.14321121/-/DCSupplemental.
Supplemental Appendix 1. Comparison of models between eGFR using CKD-EPI 2009 and CKD-EPI 2021.
Supplemental Appendix 2. The H3Africa Consortium AWI-Gen Study.
Supplemental Table 1. APOL1 haplotypes (referred to as alleles G0, G1, and G2 for ease of comparison with other literature) on the basis of three polymorphic loci on chromosome 22.
Supplemental Table 2. Quality of imputation for the three variant positions and APOL1 G0, G1, and G2 risk alleles on chromosome 22.
Supplemental Table 3. APOL1 alleles and high-risk genotypes using recessive models by ethnicity (data used for Figure 1).
Supplemental Table 4. APOL1 risk alleles and associated risk factors for kidney disease across the combined dataset of the Africa Wits–International Network for the Demographic Evaluation of Populations and Their Health Partnership for Genomic Studies.
Supplemental Table 5. APOL1 associations with albuminuria across the Africa Wits–International Network for the Demographic Evaluation of Populations and Their Health Partnership for Genomic Studies study sites.
Supplemental Table 6. Association (OR; 95% CI) between APOL1 using additive, dominant, and recessive models and albuminuria alone or as a composite end point.
Supplemental Table 7. Association (OR; 95% CI) between APOL1 using additive, dominant, and recessive models and albuminuria, low eGFR, and the composite end point.
Supplemental Table 8. Effect of genotypes (G1/G1 compared with G2/G2) on biomarkers of kidney disease.
Supplemental Table 9. APOL1 associations with low eGFR stratified by diabetes, hypertension, and HIV status.
Supplemental Table 10. APOL1 associations with the composite end point stratified by diabetes, hypertension, and HIV status.
Supplemental Figure 1. Forest plot: association between high-risk APOL1 genotypes (OR; 95% CI) and albuminuria by region.
References
- 1.Ashuntantang G, Osafo C, Olowu WA, Arogundade F, Niang A, Porter J, Naicker S, Luyckx VA: Outcomes in adults and children with end-stage kidney disease requiring dialysis in sub-Saharan Africa: A systematic review. Lancet Glob Health 5: e408–e417, 2017 [DOI] [PubMed] [Google Scholar]
- 2.Kaze AD, Ilori T, Jaar BG, Echouffo-Tcheugui JB: Burden of chronic kidney disease on the African continent: A systematic review and meta-analysis. BMC Nephrol 19: 125, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, Berns JS, Briggs W, Cho ME, Dart RA, Kimmel PL, Korbet SM, Michel DM, Mokrzycki MH, Schelling JR, Simon E, Trachtman H, Vlahov D, Winkler CA: MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet 40: 1175–1184, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS; Family Investigation of Nephropathy and Diabetes Research Group : MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40: 1185–1192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR: Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol 22: 2098–2105, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR: APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol 22: 2119–2128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Pérez-Morga D: The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol 12: 575–584, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Papeta N, Kiryluk K, Patel A, Sterken R, Kacak N, Snyder HJ, Imus PH, Mhatre AN, Lawani AK, Julian BA, Wyatt RJ, Novak J, Wyatt CM, Ross MJ, Winston JA, Klotman ME, Cohen DJ, Appel GB, D’Agati VD, Klotman PE, Gharavi AG: APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol 22: 1991–1996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen CP, Beggs ML, Saeed M, Walker PD: Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol 24: 722–725, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollak MR, Friedman DJ: Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int 87: 332–342, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velez JCQ, Caza T, Larsen CP: COVAN is the new HIVAN: The re-emergence of collapsing glomerulopathy with COVID-19. Nat Rev Nephrol 16: 565–567, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T, Limou S, Sezgin E, Nelson GW, Fogo AB, Goetsch S, Kopp JB, Winkler CA, Naicker S: APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol 26: 2882–2890, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freedman BI, Kopp JB, Sampson MG, Susztak K: APOL1 at 10 years: Progress and next steps. Kidney Int 99: 1296–1302, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limou S, Nelson GW, Kopp JB, Winkler CA: APOL1 kidney risk alleles: Population genetics and disease associations. Adv Chronic Kidney Dis 21: 426–433, 2014. 10.1053/j.ackd.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruzel-Davila E, Wasser WG, Aviram S, Skorecki K: APOL1 nephropathy: From gene to mechanisms of kidney injury. Nephrol Dial Transplant 31: 349–358, 2016 [DOI] [PubMed] [Google Scholar]
- 19.Grams ME, Rebholz CM, Chen Y, Rawlings AM, Estrella MM, Selvin E, Appel LJ, Tin A, Coresh J: Race, APOL1 risk, and eGFR decline in the general population. J Am Soc Nephrol 27: 2842–2850, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer ND, Freedman BI: APOL1 and progression of nondiabetic nephropathy. J Am Soc Nephrol 24: 1344–1346, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH: APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 24: 1484–1491, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipkowitz MS, Freedman BI, Langefeld CD, Comeau ME, Bowden DW, Kao WH, Astor BC, Bottinger EP, Iyengar SK, Klotman PE, Freedman RG, Zhang W, Parekh RS, Choi MJ, Nelson GW, Winkler CA, Kopp JB; SK Investigators : Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int 83: 114–120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT Jr., Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ; AASK Study Investigators; CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito K, Bick AG, Flannick J, Friedman DJ, Genovese G, Parfenov MG, Depalma SR, Gupta N, Gabriel SB, Taylor HA Jr., Fox ER, Newton-Cheh C, Kathiresan S, Hirschhorn JN, Altshuler DM, Pollak MR, Wilson JG, Seidman JG, Seidman C: Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res 114: 845–850, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanji Z, Powe CE, Wenger JB, Huang C, Ankers E, Sullivan DA, Collerone G, Powe NR, Tonelli M, Bhan I, Bernhardy AJ, Dibartolo S, Friedman D, Genovese G, Pollak MR, Thadhani R: Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J Am Soc Nephrol 22: 2091–2097, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzur S, Rosset S, Skorecki K, Wasser WG: APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant 27: 1498–1505, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Friedman DJ, Pollak MR: APOL1 and kidney disease: From genetics to biology. Annu Rev Physiol 82: 323–342, 2020 [DOI] [PubMed] [Google Scholar]
- 28.Behar DM, Kedem E, Rosset S, Haileselassie Y, Tzur S, Kra-Oz Z, Wasser WG, Shenhar Y, Shahar E, Hassoun G, Maor C, Wolday D, Pollack S, Skorecki K: Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol 34: 452–459, 2011 [DOI] [PubMed] [Google Scholar]
- 29.Sumaili EK, Shemer R, Kruzel-Davila E, Cohen EP, Mutantu PN, Bukabau JB, Makulo JRR, Mokoli VM, Luse JL, Pakasa NM, Cavalier E, Wumba RD, Reiner-Benaim A, Boner G, Lifschitz M, Nseka NM, Skorecki K, Wasser WG: G1 is the major APOL1 risk allele for hypertension-attributed nephropathy in Central Africa. Clin Kidney J 12: 188–195, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulasi II, Tzur S, Wasser WG, Shemer R, Kruzel E, Feigin E, Ijoma CK, Onodugo OD, Okoye JU, Arodiwe EB, Ifebunandu NA, Chukwuka CJ, Onyedum CC, Ijoma UN, Nna E, Onuigbo M, Rosset S, Skorecki K: High population frequencies of APOL1 risk variants are associated with increased prevalence of non-diabetic chronic kidney disease in the Igbo people from south-eastern Nigeria. Nephron Clin Pract 123: 123–128, 2013 [DOI] [PubMed] [Google Scholar]
- 31.Wudil UJ, Aliyu MH, Prigmore HL, Ingles DJ, Ahonkhai AA, Musa BM, Muhammad H, Sani MU, Nalado AM, Abdu A, Abdussalam K, Shepherd BE, Dankishiya FS, Burgner AM, Ikizler TA, Wyatt CM, Kopp JB, Kimmel PL, Winkler CA, Wester CW: Apolipoprotein-1 risk variants and associated kidney phenotypes in an adult HIV cohort in Nigeria. Kidney Int 100: 146–154, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsay M, Crowther N, Tambo E, Agongo G, Baloyi V, Dikotope S, Gómez-Olivé X, Jaff N, Sorgho H, Wagner R, Khayeka-Wandabwa C, Choudhury A, Hazelhurst S, Kahn K, Lombard Z, Mukomana F, Soo C, Soodyall H, Wade A, Afolabi S, Agorinya I, Amenga-Etego L, Ali SA, Bognini JD, Boua RP, Debpuur C, Diallo S, Fato E, Kazienga A, Konkobo SZ, Kouraogo PM, Mashinya F, Micklesfield L, Nakanabo-Diallo S, Njamwea B, Nonterah E, Ouedraogo S, Pillay V, Somande AM, Tindana P, Twine R, Alberts M, Kyobutungi C, Norris SA, Oduro AR, Tinto H, Tollman S, Sankoh O: H3Africa AWI-Gen Collaborative Centre: A resource to study the interplay between genomic and environmental risk factors for cardiometabolic diseases in four sub-Saharan African countries. Glob Health Epidemiol Genom 1: e20, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali SA, Soo C, Agongo G, Alberts M, Amenga-Etego L, Boua RP, Choudhury A, Crowther NJ, Depuur C, Gómez-Olivé FX, Guiraud I, Haregu TN, Hazelhurst S, Kahn K, Khayeka-Wandabwa C, Kyobutungi C, Lombard Z, Mashinya F, Micklesfield L, Mohamed SF, Mukomana F, Nakanabo-Diallo S, Natama HM, Ngomi N, Nonterah EA, Norris SA, Oduro AR, Somé AM, Sorgho H, Tindana P, Tinto H, Tollman S, Twine R, Wade A, Sankoh O, Ramsay M: Genomic and environmental risk factors for cardiometabolic diseases in Africa: Methods used for phase 1 of the AWI-Gen population cross-sectional study. Glob Health Action 11[Suppl2]: 1507133, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R; Haplotype Reference Consortium : A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48: 1279–1283, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhury A, Aron S, Botigué LR, Sengupta D, Botha G, Bensellak T, Wells G, Kumuthini J, Shriner D, Fakim YJ, Ghoorah AW, Dareng E, Odia T, Falola O, Adebiyi E, Hazelhurst S, Mazandu G, Nyangiri OA, Mbiyavanga M, Benkahla A, Kassim SK, Mulder N, Adebamowo SN, Chimusa ER, Muzny D, Metcalf G, Gibbs RA, Rotimi C, Ramsay M, Adeyemo AA, Lombard Z, Hanchard NA; TrypanoGEN Research Group; H3Africa Consortium : High-depth African genomes inform human migration and health. Nature 586: 741–748, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR; 1000 Genomes Project Consortium : A global reference for human genetic variation. Nature 526: 68–74, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, Ritchie GR, Xue Y, Asimit J, Nsubuga RN, Young EH, Pomilla C, Kivinen K, Rockett K, Kamali A, Doumatey AP, Asiki G, Seeley J, Sisay-Joof F, Jallow M, Tollman S, Mekonnen E, Ekong R, Oljira T, Bradman N, Bojang K, Ramsay M, Adeyemo A, Bekele E, Motala A, Norris SA, Pirie F, Kaleebu P, Kwiatkowski D, Tyler-Smith C, Rotimi C, Zeggini E, Sandhu MS: The African Genome Variation Project shapes medical genetics in Africa. Nature 517: 327–332, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuznetsova A, Brockhoff PB, Christensen RH: lmerTest package: Tests in linear mixed effects models. J Stat Softw 82: 1–26, 2017 [Google Scholar]
- 40.Peralta CA, Bibbins-Domingo K, Vittinghoff E, Lin F, Fornage M, Kopp JB, Winkler CA: APOL1 genotype and race differences in incident albuminuria and renal function decline. J Am Soc Nephrol 27: 887–893, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen TK, Tin A, Peralta CA, Appel LJ, Choi MJ, Lipkowitz MS, Winkler CA, Estrella MM: APOL1 risk variants, incident proteinuria, and subsequent eGFR decline in blacks with hypertension-attributed CKD. Clin J Am Soc Nephrol 12: 1771–1777, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadkarni GN, Fei K, Galarneau G, Gao Y, Wilson JG, Cooper R, Madden EB, Denny JC, Richardson LD, Pollak M, Loos RJF, Horowitz CR: APOL1 renal risk variants are associated with obesity and body composition in African ancestry adults: An observational genotype-phenotype association study. Medicine (Baltimore) 100: e27785, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen TK, Estrella MM, Vittinghoff E, Lin F, Gutierrez OM, Kramer H, Lewis CE, Kopp JB, Allen NB, Winkler CA, Bibbins-Domingo KB, Peralta CA: APOL1 genetic variants are not associated with longitudinal blood pressure in young black adults. Kidney Int 92: 964–971, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.