Abstract
Genetically modified microorganisms (GMMs) can enable a wide range of important applications including environmental sensing and responsive engineered living materials. However, containment of GMMs to prevent environmental escape and satisfy regulatory requirements is a bottleneck for real-world use. While current biochemical strategies restrict unwanted growth of GMMs in the environment, there is a need for deployable physical containment technologies to achieve redundant, multi-layered, and robust containment. We developed a hydrogel-based encapsulation system that incorporates a biocompatible multilayer tough shell and an alginate-based core. This DEployable Physical COntainment Strategy (DEPCOS) allows no detectable GMM escape, bacteria to be protected against environmental insults including antibiotics and low pH, controllable lifespan, and easy retrieval of genomically recoded bacteria. To highlight the versatility of DEPCOS, we demonstrated that robustly encapsulated cells can execute useful functions, including performing cell-cell communication with other encapsulated bacteria and sensing heavy metals in water samples from the Charles River.
Main
Genetically modified microorganisms (GMMs) are being developed and used for bioremediation1, agriculture2, and the production of engineered living materials and devices3. However, the potential for GMMs to escape into the environment has created a need for strategies to contain these organisms and prevent their uncontrolled release4,5.
Chemical biocontainment utilizes chemical barriers to impede the escape and survival of microorganisms in the environment4,6. Several strategies have been developed for chemical containment of GMMs7,8. For example, GMMs can carry genetic circuits that require specific chemical combinations to prevent cell death by inhibiting toxin production9, rescue cells from being killed by a constitutively expressed toxin by producing the corresponding antitoxin10, or multi-layered safeguards that modulate the expression of essential genes and toxins11. In addition, microbes can be engineered with auxotrophies so that they require synthetic amino acids for survival12. However, chemical strategies alone are imperfect for containment because mutation rates of GMMs, while low, are never zero, thus resulting in escape mutants. This implies that the number of chemically-contained GMMs that can be deployed is intrinsically limited by its mutation rate13. Thus, it would be beneficial to combine biological containment strategies and physical encapsulation such that functional redundancy further reduces any chance of inadvertent escape.
To address this challenge, we created a DEployable Physical COntainment Strategy (DEPCOS) that prevents GMM escape while providing a tunable protective environment in which GMMs can execute engineered functions. DEPCOS erects a physical barrier to prevent GMMs from escaping into their surroundings, limit horizontal gene transfer between GMMs and natural species in the environment, and allow for easy retrieval of bacterial communities.
Hydrogels are desirable materials for encapsulating living cells as they provide an aqueous environment that can be infused with nutrients, allowing for cell growth14 and sensing15, while also protecting against environmental hazards16. Alginate forms hydrogels in the presence of di-cationic solutions (e.g., Ca2+, Ba2+) and has been used in various biomedical applications17,18 because of its low cost, negligible cytotoxicity, and mild gelation conditions. However, weak mechanical properties and susceptibility to multiple chemical conditions (such as low pH, citrate, and phosphate) make alginate, as well as other traditional hydrogels, poor solutions for robust physical containment when used on their own19–21.
Core-shell designs that include an alginate core and a polymer-based protective shell have emerged as potential design for alginate-based microbial biosensors20,22. Nonetheless, there is a major need for a mechanically-tough shell that is also highly permeable to analytes for sensing. Our DEPCOS design for bacterial encapsulation consists of two parts: 1) an alginate-based hydrogel core and 2) a tough hydrogel shell (Fig. 1) that combines both a stretchy polymer network (polyacrylamide) and an energy dissipation network (alginate, through the unzipping of ionic crosslinking between polymer chains)23. This shell material is extremely tough and resistant to fracture, yet retains permeability for small molecules24. Herein, we test the biocompatibility of this hydrogel and further expand upon its physical characterization for core-shell particle form factors. By combining physical and chemical containment strategies, the DEPCOS platform enables near-perfect biocontainment and protects the encapsulated cells from environmental insults. Various engineered bacterial strains hosting genetic circuits for sensing, recording, and communication operate robustly in a plug-and-play fashion with DEPCOS. This study demonstrates the potential of using dual-mode biocontainment for the real-world deployment of GMMs powered by synthetic biology.
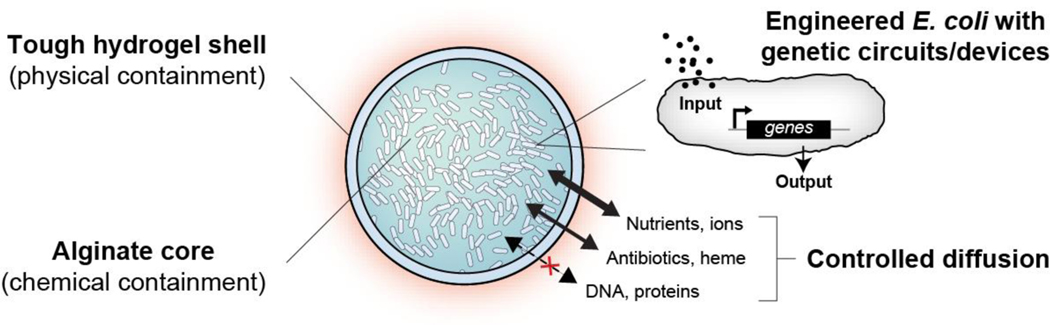
Fig. 1 |. Schematic of the DEPCOS platform.
The DEployable Physical COntainment Strategy (DEPCOS) platform provides secure biocontainment by using a core-shell hydrogel design. The tough and semipermeable hydrogel shell provides a confining barrier for physical containment and protection against insults. The alginate-based hydrogel core contains nutrients for microbial growth and survival, enabling chemical containment. Engineered cells equipped with genetic circuits receive environmental inputs and generate desirable outputs. Nutrients, biomolecules, and other analytes diffuse into and out of the bead selectively depending on molecular weight and charge.
Results
Manufacturing the DEPCOS hydrogel beads.
To incorporate living cells into the particle core, liquid cultures of E. coli were mixed with alginate in 50 μl or 100 μl droplets that were crosslinked with calcium ions to form spheres. The cell-containing alginate hydrogel was easily shaped by a mold or cut into different geometries (Supplementary Fig. 1). Cores were then coated with the tough polyacrylamide-alginate hydrogel layer23 (Fig. 2). In our core-shell system, the alginate core is pre-loaded with nutrients to support growth while the hydrogel shell provides mechanical protection for the entire bead. For downstream analyses after deployment, cells can easily be retrieved from the beads by removing the shell with a razor blade and homogenizing the core (Supplementary Fig. 2). Due to the observed toxicity of the chemical crosslinkers (Supplementary Fig. 3), a 12-hour outgrowth step was performed to replenish the CFU count before the following experiments (Supplementary Fig. 4).
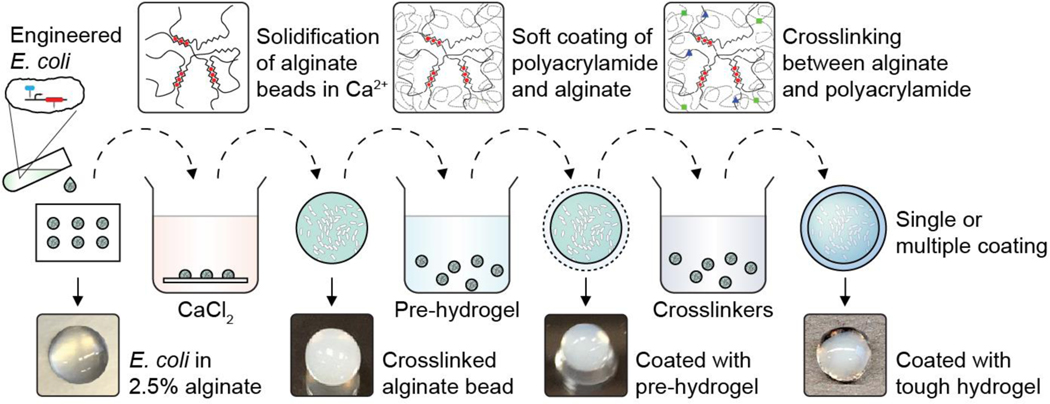
Fig. 2 |. Cell encapsulation in tough hydrogel capsules.
The process of core-shell encapsulation of cells. Droplets of 2.5% alginate with engineered E. coli were crosslinked in a calcium solution to form the soft core of the beads, which were then coated with a layer of alginate/polyacrylamide to form a tough hydrogel shell. The process can be repeated to achieve multiple coatings.
Tough hydrogel shell enables robust physical containment.
We hypothesized that the tough hydrogel layer would serve as a containment mechanism because its pore size (5–50 nm) is too small for E. coli to penetrate25. To test this hypothesis, we measured the containment efficiency of hydrogel beads by incubating E. coli-encapsulated beads at the optimal temperature for bacterial growth (37°C) with shaking. Specifically, we encapsulated a concentration of ~109 bacteria/mL in each bead. Beads lacking a tough shell allowed bacteria to escape into the surrounding media and to grow to high densities after overnight incubation, whereas there was no physical escape of bacteria from coated beads even after 72 hours of incubation (Fig. 3a and Supplementary Fig. 5). In this assay, we plated all of the media (5 mL) surrounding the beads, with a lower limit of detection (LLOD) of 1 CFU in 5 mL. Furthermore, the tough hydrogel shell maintained zero escape under physical insult such as prolonged shaking at 200 rpm, outperforming alginate and agarose, two common coating materials for core-shell cell encapsulation (Supplementary Fig. 6).
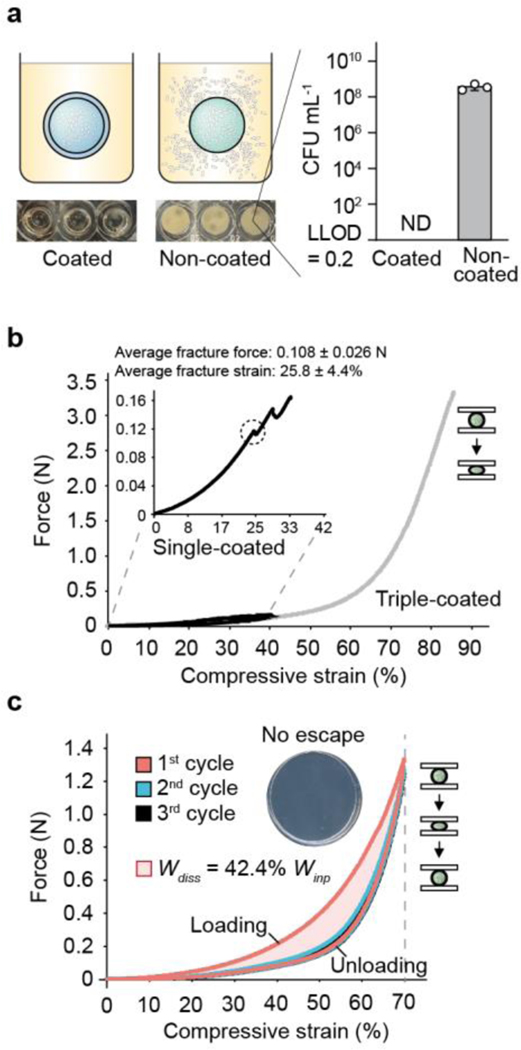
Fig. 3 |. Tough hydrogel shell provides robust biocontainment.
(a) Encapsulated bacteria escaped from non-coated beads at high rates but did not escape from tough-hydrogel-coated beads at detectable levels after 72h. Inset shows media in which non-coated and coated bead were grown for 24h (lower limit of detection (LLOD) = 1 CFU / 5 mL). ND = not detectable. Samples prepared in triplicate, data represent the mean ±1 SD. (b) Typical force-displacement curves of single-layer tough-hydrogel-coated beads subjected to 40% (black) compressive strain and triple-coated beads subjected to compression up to 85% (gray) compression. Inset zooms in on the single-coated bead stress-strain curve. The average maximum strain and force before fracture for the single-layer coating were 25.8 % and 0.108 N, respectively. Triple-coated beads showed no fracture under compression. Samples prepared and measured independently from n = 14 beads. (c) Cyclic compression of triple-layer coated beads showed hysteresis in the stress-strain curve between the first and second cycles due to plastic deformation. Work dissipated Wdiss in the first cycle was calculated as 42.4% of the total work Win. Stress-strain curves are representative of at least 6 independent experiments. Inset shows there was no escape: plating the surrounding media of a bead after cyclic compression yielded no colonies. Samples prepared in triplicate.
Next, we used compression testing to characterize the mechanical robustness of the hydrogel-bacteria beads (radius = 3–4 mm) with varying shell layers. We found that beads with a single-layer shell coating (Supplementary Fig. 7) could sustain 25% compressive strains and forces up to ~0.1 N before fracture occurred (Fig. 3b and Supplementary Fig. 8a). We further improved the mechanical properties of the beads by creating multilayer shells via repetitive coating. With beads that were coated with three layers of tough polyacrylamide-alginate hydrogel, we did not observe any fracture when the beads were subjected to up to 85% compressive strains and forces up to ~3.3 N (Fig. 3b and Supplementary Fig. 8a). The bead capsules were also subjected to cyclic compression at 70% strain, revealing a pronounced hysteresis due to plastic deformation and energy dissipation (Fig. 3c and Supplementary Fig. 8b). Based on the dimensions of the beads, the cyclic effective compressive stress was calculated as ~70 kPa (Supplementary Fig. 8b), which is equivalent to pressure at ~7 m depth under water and ~4 m depth under dry soil26, and is comparable to our previously published ingestible hydrogel device27. This result suggests that these beads could sustain much stronger stresses higher than the maximum gastric pressure (~10 kPa)28, highlighting their potential for in vivo biosensing. Thus, multilayer coating with elastic tough hydrogel around an alginate core provides mechanical robustness to the entire capsule, a phenomenon which is observed with other stiff polymer coatings29. Importantly, zero CFU counts were detected when plating the surrounding media that was incubated with compressed single- and triple-coated beads, suggesting that the capsules withstood successive compressions without fracturing and maintained perfect containment needed for safe environmental deployment (Fig. 3c, inset). In the liquid environment, the beads showed ~25% swelling after day 1 and remained stable over the course of 14 days (Supplementary Fig. 9a). This swelling has a limited impact on the beads’ mechanical properties measured by compression (Supplementary Fig. 9b). However, swelling might be a potential challenge for long-term deployment in the low-salt environment and could be minimized by replacing the salt-sensitive alginate with swelling-resistant PEG-containing hydrogels30.
Synergistic dual-mode containment ensures zero escape.
Since extreme forces could potentially compromise our hydrogels and permit bacterial escape, we hypothesized that chemical containment could be employed to enforce an additional layer of control over encapsulated cells. Genomically recoded organisms (GROs, microbes with synthetic autotrophies)13 can be contained because the growth of these microbes is dependent on the supply of synthetic amino acids (enabling a permissive environment). Here, we sought to combine physical and chemical strategies for biocontainment by encapsulating two GROs auxotrophic for the synthetic amino acid p-iodo-L-phenylalanine (pIF, β) in tough hydrogel beads. The E. coli strains rEc.β.dC.12’.ΔtY (mutation rate <4.9 × 10−12) and LspA.Y54β (mutation rate = 1.86 × 10−5) have amber codons (TAG) inserted in three (Lsp, DnaX, SecY) essential genes13, respectively, to restrict growth to permissive media (containing pIF). We showed that: 1) chemical containment in the beads enables programmable loss of cellular viability after 48 hours, which prevents undesirable growth once a given time frame has expired; and 2) physical containment adds another layer of protection over chemically contained microbes, which is necessary for applications that require extremely high standards of biocontainment.
First, beads encapsulating the pIF-auxotroph GROs, rEc.β.dC.12’.ΔtY and LspA.Y54β were pre-soaked in lysogeny broth (LB) in the absence (non-permissive media) or presence (permissive media) of 1 mM pIF and 0.2% L-arabinose (L-ara), which is required for aminoacyl-tRNA synthetases (aaRS) expression in these strains13. We hypothesized that in non-permissive media, the GROs would be unable to synthesize functional essential proteins and thus, lose viability. Indeed, beads pre-soaked in non-permissive media failed to sustain cell growth and showed less than 10% survival after 12 hours in LB only (Fig. 4a, cells were plated on permissive solid media), with no survival detected at 24 h. On the other hand, pre-soaking encapsulated beads in permissive media greatly prolonged cell survival. Greater than 50% of the rEc.β.dC.12’.ΔtY population and >25% of the LspA.Y54β population remained viable after 24h of incubation. Nearly all cells (>99%) lost viability after 2 days of incubation, which we believe is due to pIF and L-ara depletion by cells, as well as passive diffusion of these molecules out of the encapsulated hydrogel. Because many chemical induction and sensing responses in E. coli require less than 24 hours to complete, this defined survival window could be used to prevent the undesirable growth of cells upon completion of tasks.
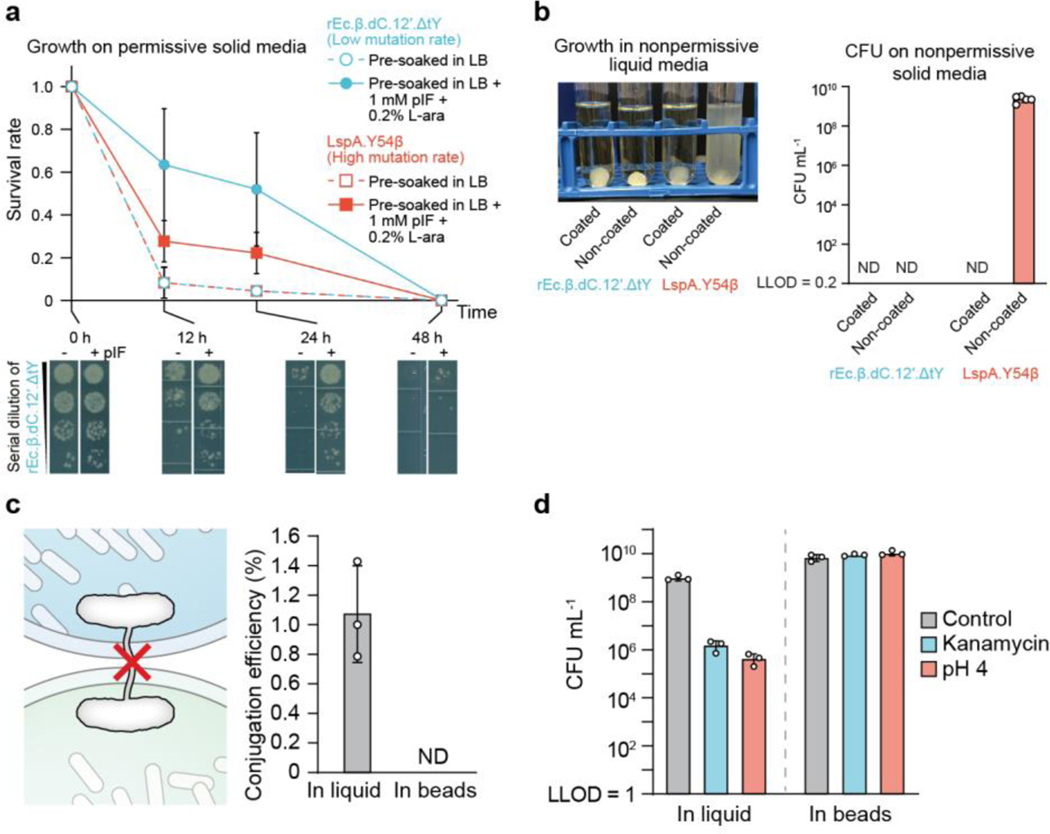
Fig. 4 |. Combining chemical and physical containment strategies for optimal biocontainment and protection.
(a) Comparison of cell survival in beads between two genomically recoded organism (GRO) strains with different containment efficiencies. Survival rate calculated by the number of encapsulated GRO in tough-hydrogel beads that were pre-soaked in permissive media (lysogeny broth (LB) + 1 mM pIF + 0.2% L-ara) (closed circles and solid lines) versus encapsulated GRO in tough-hydrogel beads that were pre-soaked in nonpermissive media (LB only) (open circles and dashed lines) before incubation of the beads in LB. Survival rates were calculated by normalizing colony forming units (CFU) from samples inside the beads, plated on permissive solid media at each time point to CFU at 0 h. Dilution series of the rEc.β.dC.12’.ΔtY at different incubation time points are shown at the bottom. Samples prepared and measured independently from triplicate, data represent the mean ±1 SD. (b) Left: Escape of GROs into 5 mL of nonpermissive media (LB only) surrounding the coated versus non-coated beads containing the GROs after shaking the tough-hydrogel beads at 200 rpm for 3 days. Right: The surrounding media was plated on nonpermissive solid media in order to obtain CFU counts (ND: not detectable with LLOD = 1 CFU/5 mL). Samples prepared and measured independently from n = 5 beads, data represent the mean ±1 SD. (c) The tough hydrogel shell prevents horizontal gene transfer by direct cell-to-cell conjugation. Conjugation efficiency is calculated as the ratio of recipient strain that acquired the F’ plasmid over the total number of recipient cells in media. ND = not detectable. Samples prepared in triplicate, data represent the mean ±1 SD. (d) Survival of bacteria after subjecting liquid bacterial cultures or bacteria in tough-hydrogel-coated beads to environmental challenges such as antibiotics (30 μg/ml kanamycin for 2 hours), low pH (pH 4 for 4 hours), and untreated controls (LLOD = 200 CFU/mL). Samples prepared and measured independently from triplicate, data represent the mean ±1 SD.
We demonstrated the benefit of combining physical and chemical containment (Fig. 3f) by placing beads in non-permissive liquid media and then plating samples from the liquid media on non-permissive solid media. This experiment allowed us to screen for escape mutants. For beads with tough hydrogel coating encapsulating either GRO strain (1.2 × 107 cells), no viable cells were observed in the surrounding non-permissive media at the end of a 3-day shaking incubation period, indicating complete containment (Fig. 4b). When rEc.β.dC.12’.ΔtY cells (low mutational escape rate, <4.9 × 10−12)13 were encapsulated in beads without the tough hydrogel coating, no viable cells were observed in the non-permissive media. On the other hand, when LspA.Y54β cells (higher mutational escape rate, ~1.86 × 10−5)13 were encapsulated in beads without the tough hydrogel coating, mutational escape was observed and cells grew in the non-permissive media. These results demonstrate that physical containment can complement chemical containment strategies to achieve near-zero escape rates (chemical plus physical). Furthermore, we can program a “biological timer” system that ceases to grow in the absence of artificial chemicals and eliminates potential bacterial growth outside the bead even when the hydrogel shell is compromised.
DEPCOS prevents DNA transfer and protects GMMs.
Horizontal gene transfer (HGT) of engineered genes into the environment and disruption of native ecosystems is a major regulatory concern regarding deployment of GMMs. Since DNA is much smaller than bacteria, we sought to explore whether DEPCOs could prevent HGT. We used a bacterial conjugation assay (Fig. 4c) where the conjugation efficiency of an F-plasmid carrying a chloramphenicol (Cm) resistance from an F+ donor strain was measured for transfer into a recipient bacteria strain (F-) that lacks Cm resistance. In liquid media, we measured conjugation efficiency to be ~1%. When the F+ donor strain was encapsulated within tough hydrogel beads and incubated with recipient bacteria in the surrounding media (2 mL), no transconjugants were detected after 24 hours of co-incubation (LLOD: 1 CFU/2 mL). In additional to conjugation, GMM-derived DNA might reach the environment through diffusion from decayed GMM after cell death. We encapsulated DNA molecules at high concentration (3 × 109 copy/μL) in the beads and measured DNA copy number in the surrounding media using quantitative PCR. There was no DNA molecules leakage as they were perfectly contained (Supplementary Fig. 10) by the alginate-containing DEPCOS, which effectively blocked the diffusion of large biomacromolecules such as DNA31.
We then investigated the protective effects of the beads on bacterial cells by comparing the resistance of encapsulated cells versus planktonic cells (without bead encapsulation) to a series of chemical and biological stresses (Fig. 4d). Encapsulated bacteria survived to a much greater extent than planktonic cells in the presence of the aminoglycoside antibiotic kanamycin. Surprisingly, encapsulation also helped cells survive acidic environments (pH 4). Such protection became more prominent as the size of the alginate core increase because the killing is localized near the surface of the alginate core (Supplementary Fig. 11). Thus, our robust hydrogels can prevent bacterial conjugation-based HGT and enhance GMM survivability in certain stressful conditions.
Sensing, recording, and communication by genetic devices.
During the outgrowth step (Supplementary Fig. 4), the number of cells in the beads increased by ~105 fold (~16–17 doublings) and reached stationary phase after 12 hours of incubation, corresponding to a doubling time of ~40 minutes. These data indicate that bacterial cells within the beads are metabolically active and able to divide in the alginate core, which is important for GMMs that must carry out active biological functions32.
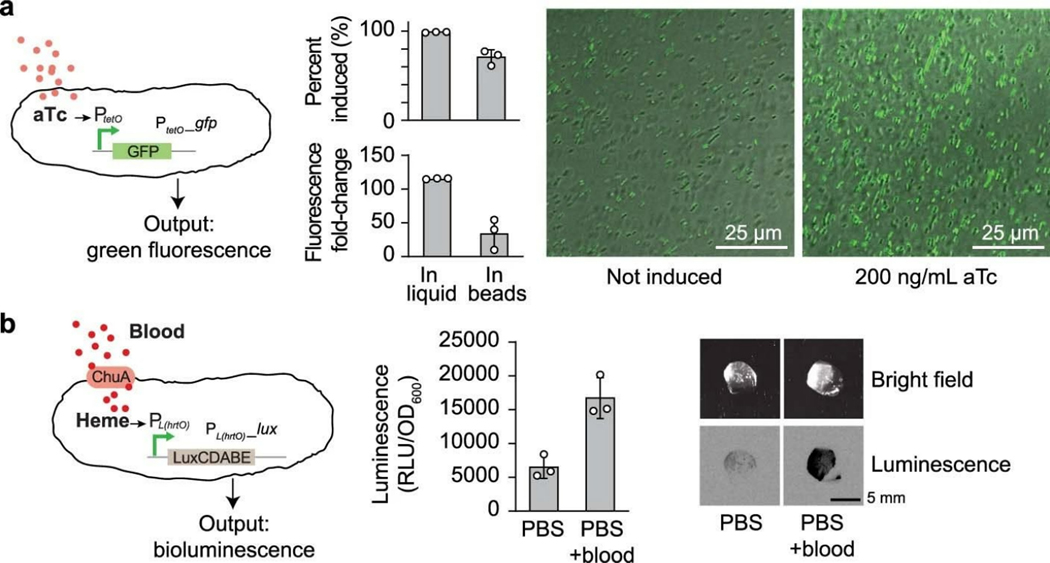
The nanoporous structures of the hydrogel shell and alginate core should allow rapid diffusion of small molecules and ions17,33 while blocking out large biopolymers such as DNA (Supplementary Fig. 10) and proteins17. The anionic nature of alginate in both components further restrict the diffusion of highly charged molecules such as tobramycin34 and kanamycin (Fig. 4d). Combining the tough hydrogel shell and the alginate core as a whole system, we observed that mildly charged small molecules could quickly diffuse into the beads (Supplementary Fig. 12), which also suggests that the H+ ions could diffuse at an even faster rate. To determine whether encapsulated bacteria can respond to these stimuli, we encapsulated bacteria containing a genetic construct that expresses GFP in response to aTc induction. We then incubated the beads at 37°C in the absence of aTc or in the presence of 200 ng/mL aTc. We found that encapsulated cells exposed to aTc exhibited a 35-fold increase in green fluorescence compared with encapsulated cells not exposed to aTc, which was lower than the fold-induction seen in liquid cultures (Extended Data Fig. 1a), potentially due to the limited diffusion within the core. Thus, gene expression in cells encapsulated in tough hydrogels can be exogenously controlled by chemical inducers. In addition, activation of gene expression could still be observed in ready-to-use beads stored at 4°C for 14 days (Supplementary Fig. 13), which is comparable to current state-of-the-art whole-cell biosensors for field applications35,36, such as hydrogel-based21,37 and liquid-in-a-cartridge devices38. Additionally, to demonstrate the sensing versatility of our system, we showed that a larger and more charged molecule (heme, physiological charge: 3+) with physiological importance could be easily detected using an engineered probiotic E. coli39 (Extended Data Fig. 1b).
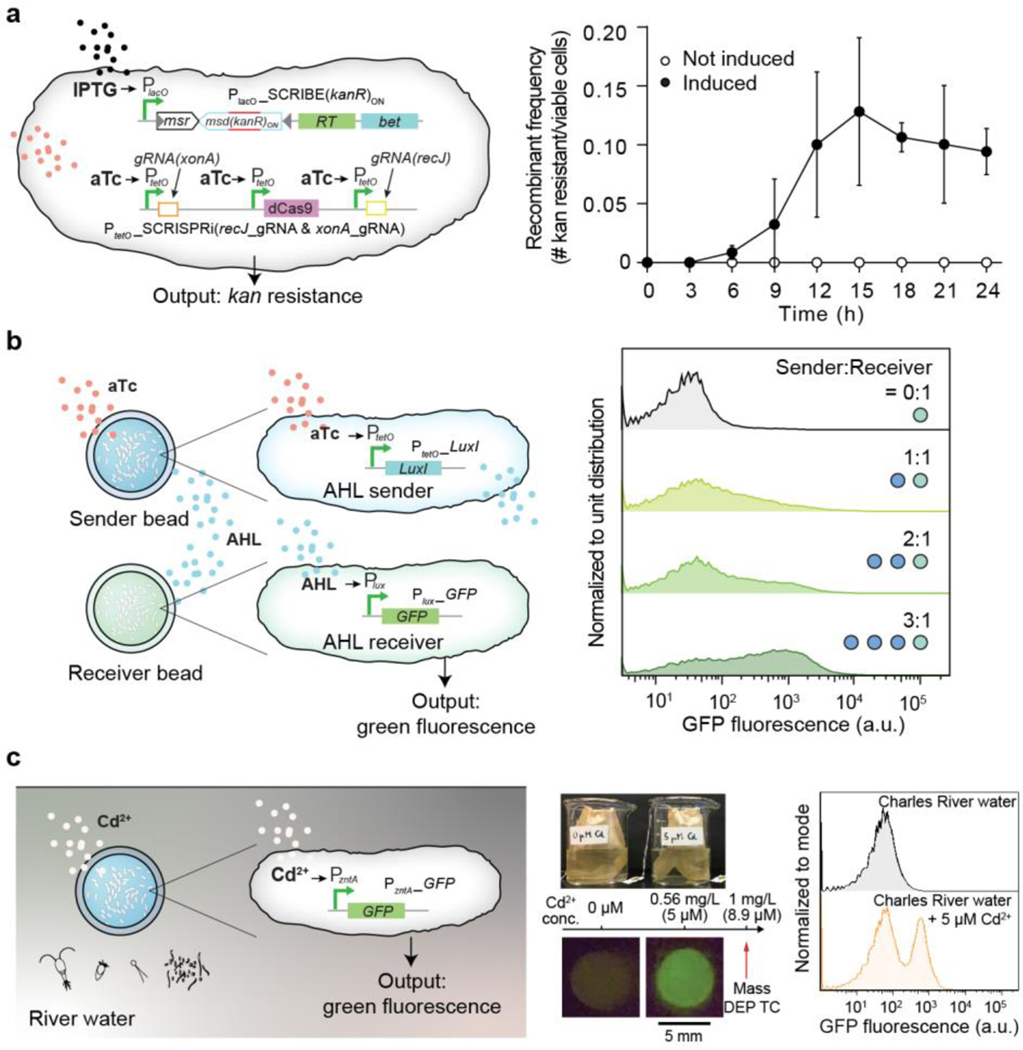
We then tested whether bacteria containing a genomically encoded memory system that requires cell division to function would be able to record information within the beads. Recording information on genomic DNA is advantageous in that DNA is a highly stable information storage medium (turnover time up to weeks in aquatic environments and years in soil41), information can be retrieved after cell death, and is amenable to multiplexing42. We used our SCRIBE platform32,40 for targeted in vivo genome editing to record information in encapsulated GMMs. The SCRIBE circuit was designed so that IPTG and aTc controlled the expression of Beta recombinase and the CRISPRi system, respectively; in this design, gene editing of the kanR gene records chemical exposure (Fig. 5a, left). We exposed beads containing SCRIBE bacteria to IPTG and aTc over 48 hours and found increasing numbers of bacteria acquired kanamycin resistance over the first 12 hours (Fig. 4c, right). The high recombinant frequency (~10%) by 12 hours is comparable to results obtained using liquid cultures of non-encapsulated bacteria32,40, and the plateau in recombination frequency after 12 hours corresponds to growth saturation (Supplementary Fig. 4). This DNA-encoded memory is stable and can be retrieved at the end of the testing period and even after cell death without constant monitoring by electronics.
Fig. 5 |. Sensing, recording, and communication capabilities of encapsulated bacterial cells.
(a) Left: An improved SCRIBE strain using CRISPRi to knock down cellular exonucleases (xonA and recJ) for enhanced genome editing efficiency via SCRIBE in DH5αPRO40. Right: Recombinant frequencies of beads containing the high-efficiency SCRIBE strain induced for a total of 24 hours with or without aTc and IPTG. Samples prepared in triplicate, data represent the mean ±1 SD. (b) Left: An AHL sender strain responds to aTc and produces AHL as an output, which later reaches an AHL receiver strain through diffusion and induces GFP expression. Right: Cells retrieved from receiver beads showed various levels of induction corresponding to different AHL sender bead to AHL receiver bead ratios. The data is representative of three independent experiments and normalized to unit distribution (area under the curve). The sender:receiver numbers represent actual number of beads, each contains ~109 cells. (c) Left: Schematic of GFP expression under the control of a cadmium-inducible promoter. Center: Photograph of the heavy metal sensing experiment setup (top). Tea bags containing five beads each were incubated in beakers containing Charles River water with and without 5 μM CdCl2. The Massachusetts Department of Environmental Protection toxic limit for CdCl2 is 1 mg/L, corresponding to 8.9 μM. Beads retrieved after 6 hours showed green fluorescence (bottom). Right: Flow cytometry analysis of encapsulated cells responding to cadmium ions in Charles River water. The flow cytometry panels are representative of samples prepared in triplicate. Data are normalized to mode (peak value).
Communication between GMMs in beads can be used to implement computation with higher complexity, division of labor, and signal integration/amplification43,44. To demonstrate this capability, we showed that different E. coli strains contained within beads could communicate with each other via quorum-sensing molecules. An acyl homoserine lactone (AHL) sender strain and an AHL receiver strain were encapsulated in separate beads and incubated together in 1 mL of Luria-Bertani (LB) media plus carbenicillin (Fig. 5b, left). Upon receiving externally added aTc, the sender bead produced AHL, which induced GFP expression in the neighboring receiver bead. The receiver beads exhibited intensified fluorescence (4-, 12-, and 21-fold-increase for 1, 2, and 3 sender beads to receiver beads ratio, respectively) as more sender beads were used (Fig. 5b right, and Supplementary Fig. 14). These results demonstrate that DEPCOS can enable a modular and distributed strategy for the collective execution of complex tasks based on cell-to-cell communication using multiple beads with different GMMs.
DEPCOS bead can sense contaminants in river water.
Finally, to demonstrate that encapsulated bacteria can function in a real-world setting, we used an E. coli strain to detect the presence of metal ions in water samples from the Charles River, such as cadmium. Cd2+ is a well-known and widespread environmental contaminant that can adversely affect human health45. Specifically, we used ZntR, a transcriptional regulator activated by metal ions (Zn2+, Pb2+, Cd2+) and activates the promoter PzntA, to express GFP46. We characterized the induction of PzntA by Zn2+, Pb2+, and Cd2+ in liquid cultures of E. coli harboring the plasmid pEZ074 (PzntA-GFP construct) (Fig. 5c, Supplementary Fig. 15a, and Supplementary Fig. 16). While encapsulated in hydrogel beads and incubated in LB media for a total of three hours, cells produced green fluorescence intensities proportional to Cd2+ concentrations (Supplementary Fig. 15b and 16).
Next, hydrogel-bacteria beads (pre-soaked in 4x LB) were incubated in water samples extracted from the Charles River having exogenously added Cd2+ (Figure 5c, center). The hydrogel-bacteria beads were placed in tea bags to facilitate easy deployment and retrieval. Exposure to 5 μM CdCl2 resulted in the emergence of a cell population expressing high levels of GFP (Figure 5c, right), indicating successful detection of cadmium ions. These results were confirmed visually under blue light: beads exposed to 5 μM CdCl2 exhibited strong green fluorescence (Fig. 5c, center and right, and Supplementary Fig. 15c). Importantly, the high sensitivity of this system to detect 5 μM CdCl2 is relevant to real-world use, as it is below the 8.9 μM (1 mg/L) standard defined by the Massachusetts Department of Environmental Protection as the maximum concentration of cadmium allowed in waste water. Thus, these results highlight the potential of physically biocontained bacteria to detect toxic levels of heavy metals in environmental settings.
Discussion
Traditionally, GMMs used as environmental sensors are confined in sealed vials into which water samples are manually injected36,38. To enable the environmental deployment of GMMs as biosensors and bioremediation devices, new strategies are needed that allow for interactions with the surrounding environment while maintaining containment of GMMs. Tough hydrogel scaffolds provide a highly hydrated environment that can sustain cell growth, protect cells from external stresses, and allow small molecules to diffuse between the interior and exterior of the device. Although previous work showed the long-term physical containment of bacteria by core-shell hydrogel microparticles, it did not demonstrate biological activity, robust sensing, or high mechanical toughness20,34,47. To the best of our knowledge, no reports have demonstrated robust physical containment while still permitting sensing and cell growth, thus overcoming the major limitations for the deployment of GMMs into the real world.
By combining two types of hydrogels into a core-shell structure, we have developed a reliable strategy for the physical containment and protection of microbes that are genetically engineered with heterologous functions. We showed that encapsulated cells could sense environmental and biomedical stimuli, record exogenous signals into genomically encoded memory, and communicate with each other via quorum-sensing molecules. Finally, we showed that heavy-metal-sensing bacteria can be incorporated into our hydrogel beads and successfully detect cadmium ions in Charles River water samples.
We anticipate that the DEPCOS containment platform can enable the deployment of microbes engineered with synthetic genetic circuits into real-world scenarios. For example, encapsulated GMMs could be used to detect explosives48 or monitor exposure time to toxic chemicals49 without potential escape into the wild. In addition, the geometry of DEPCOS could be adapted to meet the design specifications of desired applications, including wearables24 as well as other living materials and devices3. Future work will be focused on automating the manufacturing process to provide precise control over the device size and geometries in order to accommodate various physical environments and improve the miniaturization and scalability of the platform. We will also explore the incorporation of selective diffusion barriers and extreme pH resistance capabilities into the hydrogels to enable encapsulated microbial populations to survive in harsh environments, such as during transit through the human GI tract for detecting disease-relevant biomarkers. Another future challenge lies in devising large-scale standardized tests to determine whether encapsulated organisms can be contained, yet function robustly in harsh real-world scenarios, and not just in simulated laboratory settings.
Methods
Bacterial Strains and Plasmids
A complete listing of bacterial strains and plasmids including their sources can be found in Supplementary Table 1 and Supplementary Table 2. Specifically, pEZ055, pEZ058, and pEZ074 (Supplementary Fig. 17) were constructed on a high copy number plasmid (pZE12) backbone carrying a green fluorescence protein (GFP) reporter gene and transformed into DH5αPRO cells. For the aTc-inducible plasmid (pEZ055), the original pZE12 PLlacO-1 promoter was substituted by PLtetO-1. For the AHL-sensing plasmid (pEZ058), the Plux promoter was PCR amplified and cloned into pZE12 by substituting PLlacO-1 promoter via Gibson Assembly. For the heavy-metal-sensing plasmid (pEZ074), the PzntA promoter was PCR amplified from DH5αPRO E. coli genomic DNA and cloned into pZE12 by substituting PLlacO-1 promoter via Gibson Assembly.
Manufacturing the Alginate Cores
5 wt % alginate solution was made by dissolving medium viscosity alginate (Sigma-Aldrich A2033) in MilliQ water followed by autoclaving at 120 °C for 20 minutes to ensure sterility. A fresh bacterial culture (~109 cells/ml in LB plus antibiotics) was then mixed with the alginate solution in a one to one volume ratio to reach a final alginate concentration of 2.5 wt %. This bacteria-alginate premix was loaded into a syringe and disposed onto parafilm to form bead-like droplets. The droplets were solidified by immersing them in 5 wt % CaCl2 (an ionic crosslinker, Sigma-Aldrich 223506) solution for 15 minutes.
Coating with Tough Hydrogel
A precursor solution composed of 2 wt % alginate, 30 wt % acrylamide (AAm; Sigma-Aldrich A8887), 0.046 wt % ammonium persulphate (APS; Sigma-Aldrich A3678), and 0.015 wt % N,N-methylenebisacrylamide (MBAA; Sigma-Aldrich 146072) was thoroughly de-gassed. Before the coating process, the viscous precursor solution was mixed with an accelerator, N,N,N’,N’-tetramethylethylenediamine (TEMED; Sigma-Aldrich T9281; 0.1% times the volume of the precursor solution) to form a fast-curable pre-gel solution. Alginate cores from the previous section were dipped into the pre-gel solution to form a tunable thin shell layer of 100~1000 microns surrounding the core under a nitrogen atmosphere. To stabilize the shell layer, the hydrogel then was immersed in a MES buffer (0.1 M MES and 0.5 M NaCl, pH 6.0) together with cross-linkers and catalysts including 0.00125 wt % 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), 0.000375 wt % N-hydroxysuccinimide (NHS), and 0.00075 wt % adipic acid dihydrazide (AAD) to form the covalent bonding between the alginate and polyacrylamide network for 3 hours.
Retrieval of Bacterial Cells
After experiments described in following sections, beads were retrieved from liquid and the tough shell around the alginate core was carefully removed with razor blade and tweezers. The cores were then placed in tubes containing 1 mL phosphate-buffered solution (PBS; Research Products International) plus 55 mM sodium citrate (Sigma-Aldrich S4641) and homogenized with 5 mm stainless steel beads on a TissueLyzer II (Qiagen 85300) at 30 Hz for 30 minutes. To quantify cell density, homogenized samples were serially diluted (10x) and plated on LB plus antibiotics agar plates. Colony forming units (CFU) were counted after overnight incubation at 37°C.
Growth of Bacteria in Beads
All the beads containing E. coli cells underwent an outgrowth step after they were taken out of the crosslinking solution to replenish their CFU counts. Each bead was placed in a well on a 24-well plate and incubated in LB plus antibiotics and 20 mM of CaCl2 at 37°C. Any beads that showed bacterial growth in the surrounding media were discarded. To quantify the bacterial growth kinetics inside the alginate core, at each time point (every 3 hours for a total duration of 24 hours, see Supplementary Fig. 4), cells were retrieved from beads and plated on LB plus antibiotics agar plates for CFU counting.
Comparison with Alginate and Agarose Shell
Alginate cores containing EZ074 cells were prepared as described previously. For coating with alginate shell34, alginate cores were dipped in 2.5 wt% alginate solution and crosslinked in 0.1 M CaCl2 for 30 minutes. For coating with agarose shell30, alginate cores were covered by 2 wt% agarose solution (~40°C) and let solidified for 30 minutes. Coated beads were then incubated in 1 mL LB plus antibiotics and shaken at 200 rpm for 12 hours at 37°C.
Swelling Test
Hydrogel beads containing EZ074 cells were incubated in PBS plus 20 mM of CaCl2 at 37°C for 14 days. At given time points, the beads were retrieved and weighted (normalized to D0) using an electronic scale. Day 0 was defined as 12 hours post manufacturing of the beads.
Compression Test
The compression of hydrogel beads was carried out using a mechanical testing machine (Z2.5 with testXpert Ill Vl.11; Zwick/Roell) with a 20-N load cell. The samples were compressed in air or in PBS plus 20 mM of CaCl2 by two rigid flat substrates at a loading speed of 2 mm min−1. As the beads will be immersed in liquid in all practical applications, the mechanical properties (force and displacement) in air were determined in the swollen state. This was carried out by keeping the bead immerged in PBS plus 20 mM of CaCl2 up until the measurement. The approximate engineering stress is defined as:
where r0, and P are the initial radius of the bead, and the magnitude of the compressive load, respectively27,50. After testing, beads were incubated in LB overnight and the surrounding media were plated to detect potential cell leakage from compression.
Controlling GRO Life Span
GROs (rEc.β.dC.12’.ΔtY and LspA.Y54β) were grown in LB plus 1 mM p-iodo-L-phenylalanine (pIF; Sigma-Aldrich I8757), 0.02% L-arabinose (L-ara), and carbenicillin at 30°C overnight13, washed twice with PBS to remove pIF and L-ara, and encapsulated in hydrogel beads. GRO beads were then incubated in LB plus carbenicillin with or without 1 mM pIF at 4°C overnight to allow pIF infusion. At t = 0, beads were placed in 50 mL LB medium and incubated at 30°C for 12 h, 24 h, and 48 h. Cells were retrieved at given time points and plated on LB plus carbenicillin with 1 mM pIF and 0.02% L-ara agar plates for CFU counting. Survival rates were calculated by normalizing CFU counts to t = 0.
Detecting GRO Escape
GROs (rEc.β.dC.12’.ΔtY and LspA.Y54β) were grown in LB plus 1 mM pIF, 0.02% L-ara, and carbenicillin at 30°C overnight, washed twice with PBS to remove pIF and L-ara, and encapsulated in alginate beads with or without the tough hydrogel coating. The beads were then incubated in 5 mL LB plus carbenicillin at 30°C with 200 rpm shaking for 3 days. Media from each tube was plated on LB plus carbenicillin plates for CFU counts.
Environmental Insult Experiments
For antibiotics and acidic condition treatments, beads containing EZ074 cells were incubated in 1 mL of LB plus carbenicillin at 37°C for 12 to bring cell densities in the different beads to a similar level (~109 per bead). At t = 0, culture media was switched to LB plus 30 μg/ml kanamycin and LB at pH 4, respectively. At the end of the experiments, beads were retrieved from liquid media and cells were harvested for CFU counting.
Microscopy of Hydrogel Beads
Hydrogel beads containing EZ074 cells post-outgrowth were incubated in LB at pH 4 and shaken at 120 rpm at 37°C for 4 hours. At the end of treatment, the alginate cores of the beads were retrieved and washed in PBS twice. The cores were cut in half with a razorblade and stained using Live/dead™ BacLight™ Bacterial Viability Kit, for microscopy & quantitative assays (Thermo Fisher Scientific L7012). Microscopy was performed using a confocal microscope (Leica SP 8 with LAS X 3.1.5.16308) with excitation wavelength at 495 nm and emission wavelength at 515 nm for living cells; and excitation wavelength at 495 nm and emission wavelength at 635 nm for dead cells. Z-stack was performed at a fixed step size of 17 μm.
Bacterial Conjugation
The F’ plasmid (containing chloramphenicol resistance) donor strain CJ236 was encapsulated in beads and underwent overnight outgrowth in LB without antibiotics. The donor beads were placed in 2 mL of LB and co-cultured with recipient strain rcF453 (with streptomycin resistance). After 24 hours of incubation at 37°C (shaking at 100 rpm), the surrounding media was plated on LB plus streptomycin (Sm, 25 μg/mL) and LB plus streptomycin (25 μg/mL) and chloramphenicol (Cm, 12.5 μg/mL). The conjugation efficiency was calculated as:
DNA Escape Test
A linear DNA fragment encoding a GFP transcriptional unit (~1k bp) was amplified using PCR and encapsulated in the hydrogel beads at 3e9 copy/μL. Soluble DNA in the surrounding media after 72-hr incubation was quantified using qPCR (Roche LightCycler 96 with LightCycler 96 Instrument Software Vl.1) with an optimized amplicon (314 bp). A standard curve was constructed using a serial dilution (10x) of the same fragment.
Small Molecule Diffusion Assay
Two fluorescent dyes, rhodamine B and fluorescein, were used as surrogates for small molecule diffusion assay. Coated beads were soaked in dye solutions for various time periods, weighted, and transferred into 2 mL of PBS and incubated in dark for 24 hr. The fluorescence of the PBS at equilibrium was measured (494/521 nm and 540/625 nm with a Synergy H1 Hybrid Multi-Mode Reader with Gens Vl.11.5, BioTek Instruments), calibrated by the total weight of PBS plus bead, and normalized to the saturated maximum incubation period in dyes (24 hours).
aTc Induction in Beads
Beads containing EZ055 were incubated in LB plus carbenicillin and 200 ng/mL aTc at 37°C for 8 hours. The bead was then retrieved and sliced with a sharp razor blade at thickness of ~0.5 mm. The sliced sample was then imaged with a Zeiss LSM 700 confocal microscope with excitation wavelength at 488 nm and emission wavelength at 515 nm. To test inducibility after long-term storage, beads were kept at 4°C over the course of 30 days/At each time point, beads were retrieved and induced in LB plus carbenicillin and 200 ng/mL aTc at 37°C for 8 hours. Fluorescence profiles were characterized using a Synergy H1 Hybrid Multi-Mode Reader (488 nm excitation, 530/30 detection).
Heme Sensing in Beads
Defibrinated horse blood (Hemostat Laboratories DHB030) was used as the source of blood and was lysed by first diluting 1:10 in simulated gastric fluid (SGF) (0.2% NaCl, 0.32% pepsin, 84 mM HCl, pH 1.2) to release heme. This stock solution was diluted to 300 ppm in PBS right before experiments. Beads were placed in PBS or PBS + blood for 8 hours at 37°C. Cells were retrieved and measured for luminescence using a Synergy H1 plate reader. The relative luminescence units were normalized by CFU (measured through plating). Luminescence images of intact beads were acquired using a ChemiDoc Touch Imaging System (Bio-Rad).
Memory of Chemical Exposure (SCRIBE) in Beads
An higher efficiency version of SCRIBE (Synthetic Cellular Recorders Integrating Biological Events) was used in this study32,40. The SCRIBE strain was encapsulated in tough hydrogel beads and incubated in LB media with carbenicillin (100 μg/mL), chloramphenicol (25 μg/mL), aTc (100 ng/ml), and IPTG (1 mM) at 37°C. A control group was incubated using the same conditions but without the inducers (aTc and IPTG). At given time points, cells were retrieved from the beads and plated on LB plus kanamycin (50 μg/mL) agar plates as well as LB plus carbenicillin and chloramphenicol agar plates. The recombinant frequency was calculated by dividing the colony count on the LB plus kanamycin plate (kan-resistant cells) by the colony count on the LB plus carbenicillin and chloramphenicol plate (total viable cells).
Quorum Sensing between Beads
Beads containing the AHL sender strain (AYC261) and AHL receiver strain (EZ058) were placed in 1 mL LB plus 250 ng/ml aTc in a 12-well plate at specific ratios (sender:receiver = 0:1, 1:1, 2:1, and 3:1). After 24 hours of incubation at 37°C, we retrieved and diluted AHL receiver cells 1:20 into phosphate-buffered solution (PBS, Research Products International) and ran them on a BD-FACS LSRFortessa-HTS cell analyzer (BD Biosciences) with BD FACSDiva 8.0.1. We measured at least 20,000 cells for each sample and consistently gated by forward scatter and side scatter for all cells in an experiment. GFP intensity was measured on the FITC channel (488-nm excitation laser, 530/30 detection filter). Data from flow cytometry is normalized to unit distribution (normalized to area under the curve). Cells were gated using log forward scatter area (FSC-A) by log side scatter area (SSC-A), followed by gating on log forward scatter height (FSC-H) by log side scatter height (SSC-H), and subsequent gating on log forward scatter width (FSC-W) by log side scatter width (SSC-W), as exemplified in Supplementary Fig. 18.
Heavy Metal Sensing
To test inducibility of the Zn/Pb/Cd sensing strain (EZ074) in liquid, cells were grown overnight and diluted 200x in fresh LB plus 300 μM ZnCl2, 100 μM Pb(NO3)2, and 10 μM CdCl2, respectively on a 96-well plate. After 3 hours of incubation at 37°C, we retrieved bacterial cells and analyzed their GFP profile with flow cytometry. To build the dose-response curves for Cd2+ induction, overnight culture of EZ074 was diluted 200x in fresh LB plus antibiotics and induced with different concentrations of CdCl2. The GFP expression profiles were measured using a Synergy H1 Hybrid Multi-Mode Reader and normalized to their OD600 values. For testing inducibility in beads, EZ074 was encapsulated in tough hydrogel bead and incubated overnight in LB media with carbenicillin at 4°C. Before the experiment, beads were incubated at 37°C for 12 hours for bacterial cell outgrowth. Hydrogel beads were then placed in fresh LB medium with carbenicillin and corresponding metal ions at given concentrations and incubated at 37°C for 3 hours. Bacterial cells were retrieved and analyzed with flow cytometry. Data from flow cytometry is normalized to mode (normalized to peak value), which allows the visualization of differences in relative percentages of cell populations of interest.
Metal Sensing in Charles River Water
Beads containing EZ074 were incubated in 4x LB media at 4°C overnight to reach equilibrium. At t = 0, beads were placed in teabags and transferred to beakers containing 100 mL of fresh Charles River water with or without 5 mM CdCl2. After 6 hours of incubation at room temperature, cells were retrieved and analyzed with flow cytometry. Data from flow cytometry is normalized to mode.
Data Analysis
All data were processed and plotted using GraphPad Prism for Windows 64-bit V8.4.3 (686) and Microsoft Excel for Mac V16.16.17.
Data availability
Data supporting this study are presented in the main text and Supplementary Information, and are available from the corresponding authors upon request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1. Responses of encapsulated bacterial cells to external stimuli.
(a) Left: Schematic of GFP expression under the control of an aTc-inducible promoter. Center: Flow cytometry analysis of GFP expression in liquid culture and in hydrogel beads. Samples prepared in triplicate, data represent the mean ±1 s.d. based on analyses of 30000 events. The percentage data were calculated by dividing the numbers of GFP ON cells with the total cell counts. The fold-change data were derived from the mean of fluorescence. Right: Confocal microscopy images of beads encapsulating the aTc-sensing E. coli strain with and without 200 ng/mL aTc. (b) Left: A heme sensing strain which sense heme and generate bioluminescence as an output. The heme released from blood transported into the cell by ChuA. Middle: Cells retrieved from beads showed a significant increase in luciferase activity. Right: The resulting bioluminescence can be detected with high sensitivity from intact beads. Samples prepared in triplicate, data represent the mean ±1 s.d.
Supplementary Material
Acknowledgments
The authors thank Dr. Fahim Farzadfard for proving the SCRIBE strains and Dr. Mark Mimee for providing the heme sensing strain. We thank Dr. Shaoting Lin, Dr. Nathaniel Roquet, Dr. Robert Citorik, and Dr. Sebastien Lemire for useful discussions. T.K.L. is grateful for funding received from the National Institutes of Health (NIH) New Innovator Award (1DP2OD008435), NIH National Centers for Systems Biology (1P50GM098792), the U.S. Office of Naval Research (N00014-13-1-0424), and the Defense Advanced Research Projects Agency (HR0011-15-C-0091). X.Z. is grateful for funding received from the NIH (1R01HL153857-01), the National Science Foundation (EFMA-1935291), and the U.S. Army Research Office through the Institute for Soldier Nanotechnologies at MIT (W911NF-13-D-0001). T.-C.T. gratefully acknowledge the support from The Abdul Latif Jameel Water and Food Systems Lab (J-WAFS) Graduate Student Fellowship.
Footnotes
Competing interests
T.-C.T., E.T., X.L., H.Y., X.Z., and T.K.L. have filed a patent application based on the hydrogel encapsulation technologies with the US Patent and Trademark Office. T.K.L. is a co-founder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, Eligo Biosciences, Bota.Bio, Avendesora, and NE47Bio. TKL also holds financial interests in nest.bio, Armata, IndieBio, MedicusTek, Quark Biosciences, Personal Genomics, Thryve, Lexent Bio, MitoLab, Vulcan, Serotiny, Avendesora, and Pulmobiotics.
References
- 1.Singh JS, Abhilash PC, Singh HB, Singh RP & Singh DP Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene 480, 1–9 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Farrar K, Bryant D & Cope-Selby N Understanding and engineering beneficial plant-microbe interactions: Plant growth promotion in energy crops. Plant Biotechnol. J 12, 1193–1206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang T-C et al. Materials design by synthetic biology. Nat. Rev. Mater (2020) doi: 10.1038/s41578-020-00265-w. [DOI] [Google Scholar]
- 4.Dana GV, Kuiken T, Rejeski D & Snow AA Synthetic biology: Four steps to avoid a synthetic-biology disaster. Nature 483, 29 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Torres L, Krüger A, Csibra E, Gianni E & Pinheiro VB Synthetic biology approaches to biological containment: Pre-emptively tackling potential risks. Essays Biochem. 60, 393–410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein MM & Vermeire T Scientific Opinion on Risk Assessment of Synthetic Biology. Trends Biotechnol. 34, 601–603 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wright O, Stan GB & Ellis T Building-in biosafety for synthetic biology. Microbiol. (United Kingdom) 159, 1221–1235 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Chan CTY, Slomovic S & Collins JJ Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol 14, 530–537 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Chan CTY, Lee JW, Cameron DE, Bashor CJ & Collins JJ ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol 12, 82–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright O, Delmans M, Stan GB & Ellis T GeneGuard: A modular plasmid system designed for biosafety. ACS Synth. Biol 4, 307–316 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Gallagher RR, Patel JR, Interiano AL, Rovner AJ & Isaacs FJ Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rovner AJ et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rovner AJ et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman AS Hydrogels for biomedical applications. Adv. Drug Deliv. Rev 64, 18–23 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Choi M et al. Light-guiding hydrogels for cell-based sensing and optogenetic synthesis in vivo. Nat. Photonics 7, 987–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anselmo AC, McHugh KJ, Webster J, Langer R & Jaklenec A Layer-by-Layer Encapsulation of Probiotics for Delivery to the Microbiome. Adv. Mater 28, 9486–9490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KY & Mooney DJ Alginate: Properties and biomedical applications. Prog. Polym. Sci 37, 106–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearney CJ & Mooney DJ Macroscale delivery systems for molecular and cellular payloads. Nat. Mater 12, 1004–1017 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Billiet T, Vandenhaute M, Schelfhout J, Van Vlierberghe S & Dubruel P A review of trends and limitations in hydrogel-rapid prototyping for tissue engineering. Biomaterials 33, 6020–6041 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Kim BJ et al. Cytoprotective alginate/polydopamine core/shell microcapsules in microbial encapsulation. Angew. Chemie - Int. Ed 53, 14443–14446 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Li P, Müller M, Chang MW, Frettlöh M & Schönherr H Encapsulation of Autoinducer Sensing Reporter Bacteria in Reinforced Alginate-Based Microbeads. ACS Appl. Mater. Interfaces 9, 22321–22331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang BB, Wang L, Charles V, Rooke JC & Su BL Robust and Biocompatible Hybrid Matrix with Controllable Permeability for Microalgae Encapsulation. ACS Appl. Mater. Interfaces 8, 8939–8946 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Sun JY et al. Highly stretchable and tough hydrogels. Nature 489, 133–136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X et al. Stretchable living materials and devices with hydrogel-elastomer hybrids hosting programmed cells. Proc. Natl. Acad. Sci. U. S. A 114, 2200–2205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valade D, Wong LK, Jeon Y, Jia Z & Monteiro MJ Polyacrylamide hydrogel membranes with controlled pore sizes. J. Polym. Sci. Part A Polym. Chem 51, 129–138 (2013). [Google Scholar]
- 26.Atkinson J The Mechanics of Soils and Foundations, Second Edition. The Mechanics of Soils and Foundations, Second Edition (CRC Press, 2007). doi: 10.1201/9781315273549. [DOI] [Google Scholar]
- 27.Liu X et al. Ingestible hydrogel device. Nat. Commun 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houghton LA et al. Motor activity of the gastric antrum, pylorus, and duodenum under fasted conditions and after a liquid meal. Gastroenterology 94, 1276–1284 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Zarket BC & Raghavan SR Onion-like multilayered polymer capsules synthesized by a bioinspired inside-out technique. Nat. Commun 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eun YJ, Utada AS, Copeland MF, Takeuchi S & Weibel DB Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol 6, 260–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong HJ, Kim ES, Huang YC & Mooney DJ Design of biodegradable hydrogel for the local and sustained delivery of angiogenic plasmid DNA. Pharm. Res 25, 1230–1238 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Farzadfard F & Lu TK Genomically encoded analog memory with precise in vivo dna writing in living cell populations. Science (80-. ) 346, 1256272–1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golmohamadi M & Wilkinson KJ Diffusion of ions in a calcium alginate hydrogel-structure is the primary factor controlling diffusion. Carbohydr. Polym 94, 82–87 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Li Z et al. Biofilm-inspired encapsulation of probiotics for the treatment of complex infections. Adv. Mater 30, 1803925 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Bjerketorp J, Håkansson S, Belkin S & Jansson JK Advances in preservation methods: Keeping biosensor microorganisms alive and active. Curr. Opin. Biotechnol 17, 43–49 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Roggo C & van der Meer JR Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol 45, 24–33 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Kim BC & Gu MB A bioluminescent sensor for high throughput toxicity classification. Biosens. Bioelectron 18, 1015–1021 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Cevenini L, Calabretta MM, Tarantino G, Michelini E & Roda A Smartphone-interfaced 3D printed toxicity biosensor integrating bioluminescent ‘sentinel cells’. Sensors Actuators, B Chem. 225, 249–257 (2016). [Google Scholar]
- 39.Mimee M et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science (80-. ) 360, 915–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farzadfard F, Gharaei N, Citorik RJ & Lu TK Efficient retroelement-mediated DNA writing in bacteria. bioRxiv 2020.02.21.958983 (2020) doi: 10.1101/2020.02.21.958983. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen MW et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. B Biol. Sci 370, 20130383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheth RU & Wang HH DNA-based memory devices for recording cellular events. Nat. Rev. Genet 19, 718–732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss R, Knight TF Jr.., Weiss, R. & Knight, T. F. ,Jr. . Cellular Computation and Communication Using Engineered Genetic Regulatory Networks. Cell. Comput 120–121 (2004) doi: 10.1093/oso/9780195155396.003.0012. [DOI] [Google Scholar]
- 44.Chen MT & Weiss R Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol 23, 1551–1555 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Järup L & Åkesson A Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol 238, 201–208 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Brocklehurst KR et al. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol 31, 893–902 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Knierim C, Greenblatt CL, Agarwal S & Greiner A Blocked bacteria escape by ATRP grafting of a PMMA shell on PVA microparticles. Macromol. Biosci 14, 537– 545 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Belkin S et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol 35, 308–310 (2017). [DOI] [PubMed] [Google Scholar]
- 49.De Las Heras A, Carreño CA & De Lorenzo V Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol 10, 3305–3316 (2008). [DOI] [PubMed] [Google Scholar]
Method-only References
- 50.Tomović NS, Trifković KT, Rakin MP, Rakin MB & Bugarski BM Influence of compression speed and deformation percentage on mechanical properties of calcium alginate particles. Chem. Ind. Chem. Eng. Q 21, 411–417 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting this study are presented in the main text and Supplementary Information, and are available from the corresponding authors upon request. Source data are provided with this paper.