Abstract
HP1 proteins traverse a complex and crowded chromatin landscape to bind with low affinity but high specificity to histone H3K9 methylation (H3K9me) and form transcriptionally inactive genomic compartments called heterochromatin. Here, we visualize single-molecule dynamics of an HP1 homolog, the fission yeast Swi6, in its native chromatin environment. By tracking single Swi6 molecules, we identify mobility states that map to discrete biochemical intermediates. Using Swi6 mutants that perturb H3K9me recognition, oligomerization, or nucleic acid binding, we determine how each biochemical property affects protein dynamics. We estimate that Swi6 recognizes H3K9me3 with ~94-fold specificity relative to unmodified nucleosomes in living cells. While nucleic acid binding competes with Swi6 oligomerization, as few as four tandem chromodomains can overcome these inhibitory effects to facilitate Swi6 localization at heterochromatin formation sites. Our studies indicate that HP1 oligomerization is essential to form dynamic, higher-order complexes that outcompete nucleic acid binding to enable specific H3K9me recognition.
Imaging HP1 reveals rules for H3K9 methylation recognition and binding in living cells.
INTRODUCTION
Despite having identical genomes, eukaryotic cells can establish distinct phenotypic states that remain stable and heritable throughout their lifetimes (1). In the context of a multicellular organism, the persistence of epigenetic states is vital to establish and maintain distinct cellular lineages (2). This process of phenotypic diversification depends, in part, on the posttranslational modifications of DNA packaging proteins called histones (3, 4). Proteins that can “read, write, and erase” histone modifications interact weakly and transiently with their histone substrates. Nevertheless, dynamic, low-affinity interactions between histone modifiers and their substrates can encode stable memories of gene expression that can be inherited following DNA replication and cell division (5).
H3K9 methylation (H3K9me) is a conserved epigenetic modification that is associated with transcriptional silencing and heterochromatin formation (6). Heterochromatin establishment is critical for chromosome segregation, sister chromatid cohesion, transposon silencing, and maintaining lineage-specific patterns of gene expression (7). These diverse cellular functions associated with heterochromatin are critically dependent on an evolutionarily conserved HP1 family of proteins that recognize and bind to H3K9me nucleosomes (8). HP1 proteins have a distinct architecture that consists of two conserved structural domains: (i) an N-terminal chromodomain (CD) that recognizes H3K9me nucleosomes and (ii) a C-terminal chromoshadow domain (CSD) that promotes dimerization (Fig. 1A) (9). The HP1 CD domain binds to H3K9me peptides with low micromolar affinity (1 to 10 μM) (10, 11). The HP1 CSD domain promotes protein-protein and protein-nucleosome interactions and oligomerizes to form higher-order, phase-separated HP1-containing chromatin complexes that exhibit liquid-like properties (12–14). In addition, HP1 proteins are posttranslationally modified, and these modifications affect both nucleosome binding and HP1 protein–protein interactions (15, 16). A flexible and unstructured hinge region connects the Swi6 CD and CSD domains. The hinge region binds to nucleic acids without any sequence specificity (17). At present, it is unclear how the competing demands of hinge-mediated nucleic acid binding, CD-dependent H3K9me recognition, and CSD-mediated oligomerization and nucleosome interactions influence HP1 enrichment at specific locations in the genome.
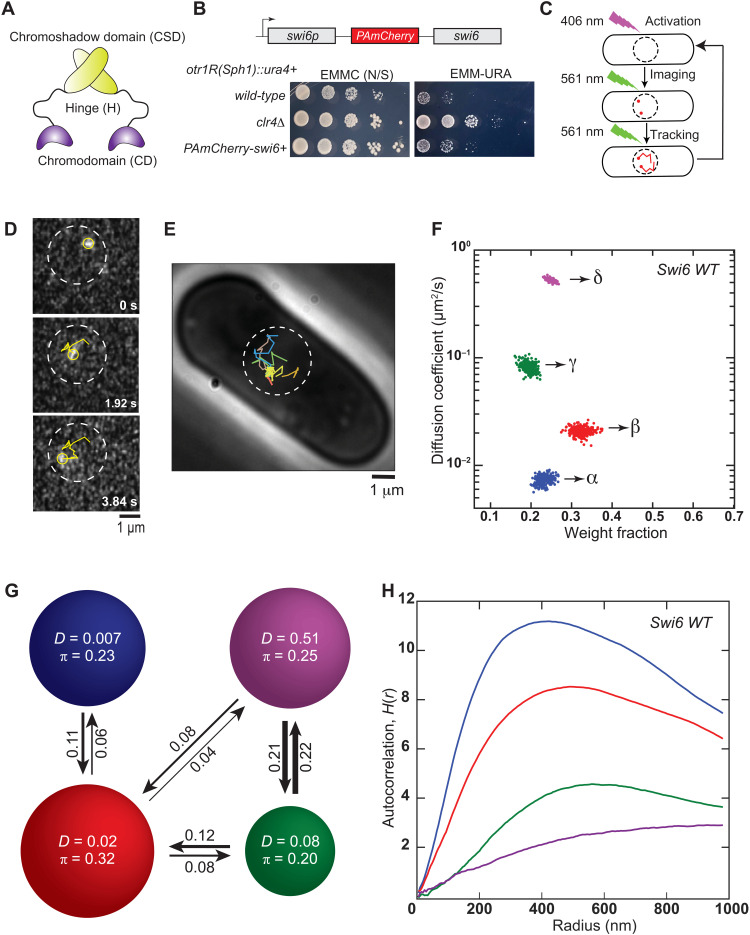
Fig. 1. Single-molecule tracking photoactivation localization microscopy of Swi6 in the S. pombe nucleus.
(A) Each Swi6 domain has a distinct biochemical role. CD: H3K9me recognition; H: nucleic acid binding; CSD: protein oligomerization. (B) Top: pamCherry-swi6 expressed from the endogenous swi6+ promoter. Bottom: Silencing assay using a ura4+ reporter inserted at the pericentromeric repeats (otr1R). (C) Single-molecule experiment workflow. PAmCherry-Swi6 molecules are photoactivated (406 nm, 1.50 to 4.50 W/cm2, 200 ms), then imaged, and tracked until photobleaching (561 nm, 71.00 W/cm2, 25 frames/s). The cycle is repeated 10 to 20 times per cell. (D) Image of a single photoactivated PAmCherry-Swi6 molecule at three time points. Yellow circle, molecule position; yellow line, Swi6-PAmCherry trajectory from photoactivation until the current frame; white circle, approximate nucleus position. (E) Representative single-molecule trajectories in a live S. pombe cell. Each trajectory is acquired after a new photoactivation cycle. (F) SMAUG identifies four distinct mobility states, (α, β, γ, and δ) for PAmCherry-Swi6 in WT cells. Each point is the average single-molecule diffusion coefficient, D, of Swi6 molecules in that state at a saved iteration of the Bayesian algorithm after convergence. Dataset: 10,095 steps from 1491 trajectories. (G) Average probabilities (arrows) of a PAmCherry-Swi6 molecule transitioning between the mobility states (circles) from (F). Each circle area is proportional to the weight fraction, π; D is in square micrometers per second. Low-frequency transition probabilities (below 0.04) are excluded. (H) Ripley’s analysis shows higher autocorrelation for PAmCherry-Swi6 in the slower (blue and red) states compared to H(r) ≤ 2 for the faster (green and purple) states.
In fission yeast (Schizosaccharomyces pombe), H3K9me is enriched at sites of constitutive heterochromatin, which includes the pericentromeric repeat sequences (dg and dh), the telomeres (tlh), and the mating-type locus (mat) (18). A conserved SET domain–containing methyltransferase, Clr4, is the sole enzyme that catalyzes H3K9me in S. pombe (19, 20). The major S. pombe HP1 homolog, Swi6, senses the resulting epigenetic landscape and binds to H3K9me chromatin with low affinity but high specificity (21). Swi6 is an archetypal member of the HP1 family of proteins (22). Its ability to simultaneously recognize H3K9me and oligomerize enables linear spreading across broad segments of the chromosome encompassing several hundred kilobases of DNA (23). Heterochromatin spreading subsequently leads to the silencing of genes that are distal from heterochromatin nucleation centers.
Only ~2% of nucleosomes in the S. pombe genome are marked with H3K9me (24). Given the limiting amount of substrate, most Swi6 molecules (~80 to 90%) are located elsewhere in the genome, potentially engaged in promiscuous DNA- and chromatin-dependent interactions (25, 26). Under conditions of acute heterochromatin misregulation, these promiscuous Swi6-chromatin interactions lead to epimutations that alter cellular fitness (27). Overexpressing Swi6 enhances epigenetic silencing of a reporter gene at sites of constitutive heterochromatin (28). Therefore, Swi6 functions as a dose-sensitive heterochromatin-associated protein. Altering the fractional occupancy of Swi6 at sites of H3K9me is likely to have a profound impact on transcriptional silencing, heterochromatin stability, and epigenetic inheritance.
On the basis of fluorescence recovery after photobleaching (FRAP) measurements, the turnover rates of Swi6 and other HP1 homologs from sites of heterochromatin range from a few hundred milliseconds to several seconds (29, 30). Point mutations that impair CD binding to H3K9me nucleosomes or CSD-mediated dimerization abolish Swi6 binding to nucleosomes. Hence, the rapid turnover of HP1 proteins from heterochromatin involves both (i) CD-dependent binding and unbinding of Swi6 from H3K9me nucleosomes and (ii) CSD-dependent association or dissociation of Swi6 oligomers. CSD-dependent oligomerization drives the formation of heterochromatin condensates, but we do not understand whether such interactions inhibit or enhance H3K9me recognition in vivo. In addition, reconstitution studies show that an increase in the on-rate for DNA binding enhances the interaction between HP1 proteins and nucleosomes (31). However, given a genome that is replete with nucleic acids (DNA and RNA), it is equally likely that the on-rate associated with nucleic acid binding prevents specific Swi6 binding to H3K9me and could titrate Swi6 away from sites of H3K9me. In essence, we lack a fundamental understanding of the extent of coupling between CSD-dependent oligomerization, CD-dependent H3K9me recognition, and hinge-dependent nucleic acid binding in the context of the fission yeast nucleus.
In this study, we use single-particle tracking photoactivated localization microscopy to measure the in vivo binding dynamics of Swi6 in real time as it samples the fission yeast nucleus (32, 33). In vitro binding measurements have thus far served as the gold standard to measure interactions between chromatin readers and modified histone peptides or recombinant nucleosome substrates (8, 13, 22, 34–37). However, these studies are typically carried out under dilute, noncompetitive solution conditions, which do not reflect the chromatin environment that Swi6 encounters in the nucleus. Rather, here, we analyze individual Swi6 molecule trajectories with high spatial and temporal resolution in living cells. Our studies determined the precise biochemical attributes of Swi6 that give rise to distinct mobility states. Our measurements enabled us to engineer precise degrees of multivalency within Swi6 that entirely circumvent the need for CSD-dependent oligomerization while suppressing the inhibitory effects of nucleic acid binding. We find that the simultaneous engagement of at least four H3K9me CD domains is both necessary and sufficient for the heterochromatin-specific targeting of Swi6, while nucleic acid binding competes with oligomerization. Overall, our results demonstrate that the evolutionarily conserved phenomenon of HP1 oligomerization may represent a tunable mechanism that compensates for weak, low-affinity H3K9me recognition that outcompetes promiscuous nucleic acid binding.
RESULTS
The single-molecule dynamics of Swi6 in live S. pombe indicate a heterogeneous environment
Because Swi6 molecules within the fission yeast nucleus undergo binding and unbinding events in a complex environment, we used single-molecule tracking to measure their heterogeneous dynamics. For instance, a Swi6 molecule bound to an H3K9me nucleosome is, on average, likely to exhibit slower, more confined motion compared to rapidly diffusing proteins. Hence, the biochemical properties of Swi6 will directly influence its mobility within the fission yeast nucleus. We transformed S. pombe cells with PAmCherry fused to the N terminus of Swi6 (38). This fusion protein replaces the wild-type (WT) endogenous Swi6 gene and serves as the sole source of Swi6 protein in fission yeast cells (Fig. 1B). To test the functionality of PAmCherry-Swi6 in heterochromatin assembly, we used strains where a ura4+ reporter is inserted within the outermost pericentromeric repeats (otr1R) (39). We concluded that PAmCherry-Swi6 is functional since cells expressing the fusion protein exhibit reduced growth on −URA medium consistent with ura4+ silencing (Fig. 1B). In contrast, cells lacking the H3K9 methyltransferase Clr4 (clr4Δ) exhibit no growth inhibition when plated on −URA-containing medium, consistent with ura4+ expression and the loss of H3K9me-dependent epigenetic silencing (Fig. 1B). In addition, chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP-qPCR) measurements show that H3K9me2 and Swi6 binding to sites of constitutive heterochromatin is preserved in cells expressing PAmCherry-Swi6 (fig. S1, A and B).
We used single-molecule microscopy to investigate the dynamics of Swi6 molecules with high spatial (20 to 40 nm) and temporal (40 ms) resolution. We photoactivated zero to two PAmCherry-Swi6 fusion proteins per activation with 406-nm light. Next, we imaged the photoactivated Swi6 molecules with 561-nm laser excitation until photobleaching, obtained a 5- to 15-step trajectory based on localizing molecules at 40-ms intervals, and then repeated the photoactivation/imaging cycle with another PAmCherry-Swi6 molecule ~10 times per cell across several individual cells (Fig. 1, C and D, and movie S1). Our experimentally measured localization accuracy is about ~36 nm (fig. S1C). We observed both stationary and fast-moving molecules (Fig. 1E). We hypothesized that each type of motion, which we term a “mobility state,” corresponds to a distinct biochemical property of Swi6 in the cell (e.g., bound versus unbound) and that molecules can transition between the different mobility states during a single trajectory. Thus, rather than assign a single diffusion coefficient to each single-molecule trajectory, we analyzed our data using Single-Molecule Analysis by Unsupervised Gibbs (SMAUG) sampling algorithm (Materials and Methods) (40). SMAUG estimates the biophysical descriptors of a system by embedding a Gibbs sampler in a Markov Chain Monte Carlo framework. This nonparametric Bayesian analysis approach determines the most likely number of mobility states and the average diffusion coefficient of single molecules in each state, the occupancy of each state, and the probability of transitioning between different mobility states between subsequent 40-ms frames. For each strain, we sampled over a thousand trajectories of PAmCherry-Swi6 within the nucleus of S. pombe cells (fig. S1D). SMAUG analyzed the ~10,000 steps from these trajectories in aggregate. For cells expressing fusions of PAmCherry-Swi6, the algorithm converged to four mobility states and estimated the diffusion coefficient, D, and the fraction of molecules in each state, π, for each iteration (each dot in Fig. 1F is the assignment from one iteration of the SMAUG analysis). We also compared our results from SMAUG to other single-particle tracking algorithms such as Spot-On and vbSPT (Supplementary Methods). Our comparisons with different single-molecule tracking methods reveal that for our single-particle tracking datasets, SMAUG and other analysis methods are consistent with respect to the number of states and the estimated diffusion coefficients (Davg) for each state.
We refer to the four mobility states as α, β, γ, and δ in order of increasing Davg (Fig. 1F). The clusters of points correspond to estimates of D and π for each of the four mobility states, and the spread in the clusters indicates the inferential uncertainty. The slowest mobility state comprises ~23% of the Swi6 molecules; its average diffusion coefficient (Davg,α = 0.007 ± 0.001 μm2/s) is close to the localization precision of the microscope, indicating no measurable motion for these molecules (error bars indicate the 95% credible interval). Given that Swi6 forms discrete foci at sites of constitutive heterochromatin (centromeres, telomeres, and the mating-type locus), the α mobility state likely corresponds to Swi6 molecules that are stably bound at these constitutive sites, which are enriched with H3K9me. The fastest diffusion coefficient that we estimated is Davg,δ = 0.51 ± 0.03 μm2/s. We tested the extent to which our experimental temporal resolution affects the estimation of Davg and found that using a shorter (20-ms) exposure time results in larger localization errors due to the limited brightness of PAmCherry, although it does not affect the weight fraction of molecules in the fast-diffusing δ state. Because of this reduced signal-to-noise ratio, 40 ms was chosen as an optimal temporal resolution. Although the 40-ms time resolution of our measurements is likely to result in uncertainty and potential underestimation of the Davg associated with the fast state (δ state), our measurements are appropriate to quantify the slower-diffusing α, β, and γ populations.
We also measured how often Swi6 molecules in one mobility state transition to another mobility state. We found that Swi6 molecules are most likely to transition between adjacent rather than nonadjacent mobility states. The transition probabilities reveal that a distinct hierarchy of biochemical interactions dictates how Swi6 interacts with H3K9me sites in the fission yeast genome (Fig. 1G). Notably, only molecules in the β intermediate-mobility state transition with high probability to the slow-mobility α state, which (as noted above) likely corresponds to Swi6 molecules stably bound at sites of H3K9me.
We used a spatial autocorrelation analysis to measure inhomogeneities in Swi6 diffusion (Fig. 1H). The Ripley H function, H(r), measures deviations from spatial homogeneity for a set of points and quantifies the correlation as a function of the search radius, r (41). We calculated H(r) values for the positions of molecules in each mobility state from the single-molecule tracking dataset. To eliminate the bias that would come from the spatial correlation between steps along the same trajectory, we compared the experimental observations to a null model by randomly simulating confined diffusion trajectories for each of the four mobility states (Materials and Methods). We represented the spatial autocorrelation values associated with each state to an H(r) function normalized using simulated trajectories (fig. S1, E and F). The real trajectories show substantially higher-magnitude and longer distance correlations than a realistic simulated dataset. We also observed low autocorrelation values for the δ and γ states (purple and green) and high autocorrelation values for the β and α states (blue and red) (Fig. 1F). The cross-correlation between the β, γ, and δ mobility states and the slow-moving α state further supports our model of spatial confinement: Molecules in the β state are most likely to be proximal to H3K9me-bound molecules in the α state (fig. S1G). In summary, the combination of transition plots and spatial homogeneity maps suggests that the β and γ intermediate-mobility states represent biochemical intermediates that sequester Swi6 molecules from transitioning between the H3K9me-bound α state and the fast-diffusing, chromatin-unbound δ state.
The slowest moving Swi6 molecules correspond to foci that are typically observed at sites of constitutive heterochromatin
Following the baseline characterization of the different mobility states associated with Swi6 and their relative patterns of spatial confinement, we used fission yeast mutants to assign individual biochemical properties to each mobility state. As a first approximation, we hypothesized that the major features likely to affect Swi6 binding within the nucleus are (i) CD-dependent H3K9me recognition, (ii) hinge-mediated nucleic acid binding, and (iii) CSD-mediated oligomerization (8).
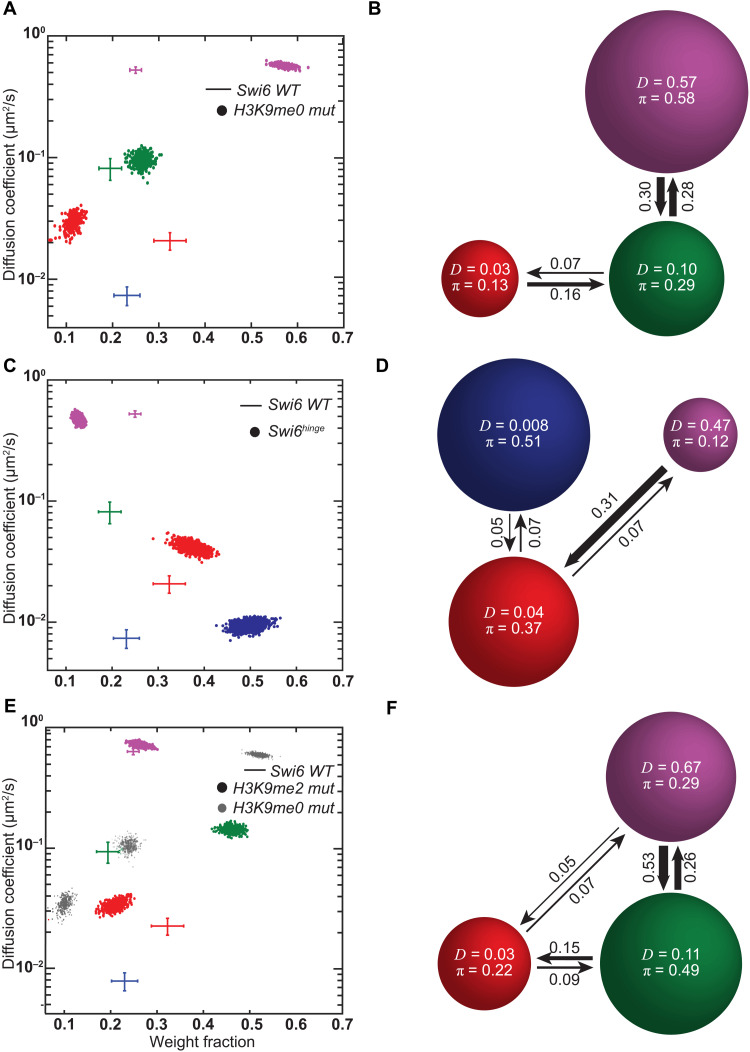
We deleted the sole S. pombe H3K9 methyltransferase, Clr4, to determine how the four mobility states associated with Swi6 diffusion respond to the genome-wide loss of H3K9 di- and trimethylation. The mobility of PAmCherry-Swi6 is substantially different in (H3K9me0) clr4Δ cells compared to clr4+ cells. Most prominently, most Swi6 molecules in clr4Δ cells move rapidly and show no subnuclear patterns of spatial confinement compared to WT cells. The slow-mobility α state is absent, and the fraction of molecules in the intermediate-mobility β state decreases twofold (Fig. 2A). In contrast, the weight fractions of the γ and δ states substantially increase in H3K9me0 clr4Δ cells, suggesting that these mobility states are exclusively H3K9me independent (Fig. 2A). The estimated transition probabilities between states show that in the absence of the H3K9me-dependent low-mobility α state, PAmCherry-Swi6 molecules predominantly reside in and transition between the γ and δ fast-mobility states with only rare transitions to the β state (Fig. 2B). Hence, both the α and β states depend on Swi6 binding to H3K9me nucleosomes, with α occurring exclusively in clr4+ cells and β representing a mixture of H3K9me-dependent and H3K9me-independent substates (present in clr4+ and clr4Δ cells).
Fig. 2. Individual Swi6 mobility states reflect distinct biochemical intermediates.
(A) SMAUG identifies three distinct mobility states for PAmCherry-Swi6 in H3K9me0 mut (clr4Δ) cells; the α mobility state is absent. Dataset: 10,432 steps from 2463 trajectories. (B) Average probabilities (arrows) of a PAmCherry-Swi6 molecule in H3K9me0 mut cells transitioning between the mobility states (circles) from (A). (C) SMAUG identifies three distinct mobility states for PAmCherry-Swi6hinge (swi6 KR25A) molecules; the γ mobility state is absent. Dataset: 12,788 steps from 1210 trajectories. (D) Average probabilities (arrows) of a PAmCherry-Swi6hinge molecule transitioning between the mobility states (circles) from (C). (E) SMAUG identifies three distinct mobility states for PAmCherry-Swi6 in H3K9me2 mut (clr4 F449Y mutant) cells; the α mobility state is absent. Dataset: 14,837 steps from 2308 trajectories. H3K9me0 mut clusters from (A) are provided as a reference (gray circles). (F) Average probabilities (arrows) of a PAmCherry-Swi6 molecule in H3K9me2 mut cells transitioning between the mobility states (circles) from (E). Each point in (A), (C), and (E) is the average single-molecule diffusion coefficient, D, of Swi6 molecules in that state at a saved iteration of the Bayesian algorithm after convergence; the average and SD of the WT Swi6 clusters (blue α, red β, green γ, and purple δ, respectively, from Fig. 1F) are provided as a reference (crosshairs). Each circle area in (B), (D), and (E) is proportional to the weight fraction, π; D is in square micrometers per second; low-frequency transition probabilities below 0.04 are excluded.
A tryptophan to alanine substitution (W104A) within the Swi6 CD attenuates H3K9me binding approximately 100-fold (Swi6 CDmut) (11, 42). We expressed PAmCherry-Swi6 CDmut in S. pombe cells. Our SMAUG analysis identified three mobility states for PAmCherry-Swi6 CDmut (fig. S2A), and the distribution of mobility states also showed depletion of the α state similar to the depletion of the α state of Swi6 in H3K9me0 clr4Δ cells (c.f. Fig. 2A). A leucine to glutamate substitution (L315E) within the Swi6 CSD disrupts dimerization and higher-order oligomerization (Swi6 CSDmut). We expressed PAmCherry-Swi6 CSDmut in S. pombe cells. Our SMAUG analysis identified three mobility states for PAmCherry-Swi6 CSDmut (fig. S2B). Notably, the low-mobility α state is absent in PAmCherry-Swi6 CSDmut cells. We also mutated two residues within the Swi6 CD domain (R93A K94A), which disrupts CD-CD–dependent Swi6 oligomerization (42), and measured the mobility states associated with PAmCherry-Swi6 Loop-X. We were unable to detect substantive changes in the fraction of molecules in the α state upon introducing alanine substitutions with the ARK loop (fig. S2C). This could be because CD-CD interactions have a more subtle effect on Swi6 oligomerization in vivo and play a minor role relative to CSD-dependent interactions. It is likely that additional destabilizing mutations within the CD-CD interface might be needed to further disrupt CD-CD–dependent Swi6 oligomerization in vivo. On the basis of our measurements, we conclude that the low-mobility α state depends on the H3K9me substrate, on CD-mediated H3K9me recognition, and on CSD-mediated dimerization.
Transient nucleic acid binding leads to an intermediate apparent diffusion coefficient for Swi6
HP1 proteins have a variable-length hinge region that connects the H3K9me recognition CD domain and the CSD oligomerization domain (31). In the case of Swi6, the hinge region has 25 lysine and arginine residues that modulate the interaction between Swi6 and nucleic acids (DNA and RNA). We replaced all 25 lysine and arginine residues with alanine (Swi6hinge) and imaged the mobility patterns of PAmCherry-Swi6hinge (25). Neutralizing the net positive charge within the hinge region results in fewer fast-diffusing molecules and a substantial increase in the proportion of low-mobility Swi6hinge molecules relative to the WT protein. These qualitative observations were consistent with our quantitative analysis by SMAUG. We detected three mobility states (as opposed to four in the case of the WT Swi6 protein), a substantial increase in the populations of the H3K9me-dependent α slow-mobility and β intermediate-mobility states, and a concomitant decrease in the population of the δ fast-diffusing state (Fig. 2C). Notably, the γ intermediate-mobility state is absent in the case of PAmCherry-Swi6hinge (Fig. 2C). Hence, we infer that the γ intermediate-mobility state corresponds to Swi6 bound to nucleic acids (DNA or RNA) via its hinge region. Furthermore, the twofold increase in the weight fraction of the slow α state suggests that, in the absence of DNA or RNA binding, PAmCherry-Swi6hinge molecules preferentially interact with H3K9me chromatin. Last, we noticed that the diffusion coefficient of the β mobility state exhibits a twofold increase, suggesting that the loss of nucleic acid binding additionally destabilizes this intermediate.
On the basis of the assignment of transition probabilities between the mobility states, we identified two critical features of Swi6 dynamics in the hinge mutant: (i) PAmCherry-Swi6hinge molecules in the slow-mobility α state transition less often to the remaining β and δ mobility states, and (ii) the probability of cross-transitions between the fast-diffusing δ state and the intermediate-mobility β state increases (Fig. 2D). Although negligible in the case of the WT PAmCherry-Swi6 protein, the cross-transitions (δ state to β state) become prominent in the case of PAmCherry-Swi6hinge mutant. Hence, by eliminating nucleic acid binding, we observed a substantial increase in H3K9me-dependent and H3K9me-independent chromatin association. Overall, these measurements strongly suggest that nucleic acid binding interactions compete with H3K9me localization and could potentially promote Swi6 unbinding from chromatin.
Weak chromatin interactions also result in an intermediate apparent diffusion coefficient for Swi6
Deleting Clr4 reduces but does not fully eliminate the population of the β intermediate-mobility state (Fig. 2A). Therefore, the β state must have both H3K9me-dependent and H3K9me-independent components. We hypothesized that the β intermediate-mobility state likely represents the transient sampling of chromatin by Swi6 (H3K9me or H3K9me0) before stable binding at sites of H3K9me (α state). Swi6 binds to H3K9me3 chromatin with higher affinity compared to H3K9me1/2. To eliminate the high-affinity Swi6 binding state and exclusively interrogate transient chromatin interactions, we replaced the H3K9 methyltransferase Clr4 with a mutant methyltransferase (Clr4 F449Y, referred to here as the Clr4 H3K9me2 mutant) that catalyzes H3K9 mono- and dimethylation (H3K9me1/2) but is unable to catalyze trimethylation (H3K9me3) due to a mutation within the catalytic SET domain (24). We verified the expression of the Clr4 mutant protein in fission yeast cells using a myc epitope tag.
Following single-particle tracking measurements of Swi6, we found that cells expressing the Clr4 H3K9me2 mutant exhibit only three mobility states (Fig. 2E), having lost the α state. These results are consistent with our expectations and previously published data where selectively eliminating H3K9me3 interrupts stable Swi6 association at sites of heterochromatin formation (24). Notably, we observed a twofold increase in the weight fraction of molecules residing in the β intermediate-mobility state relative to H3K9me0 cells, suggesting that H3K9me2 is sufficient to drive an increase in the occupancy of the Swi6 chromatin-bound population (Fig. 2E). Mapping the transition probabilities of Swi6 molecules between the remaining three mobility states further supports our conclusions. We observed an increase in the transition probability between the fast-mobility δ and γ mobility states, which is negligible or absent in H3K9me0 cells (Fig. 2F). Therefore, H3K9me2 enhances Swi6 chromatin binding but is incapable of driving Swi6 occupancy to the exclusively H3K9me3-dependent α state. We also tested how Swi6 mobility states change upon expression of a Clr4 CD–deficient protein, which leads to a substantial reduction in H3K9me3 levels and an increase in H3K9me2 levels (24). The mobility states of Swi6 measured in cells expressing the Clr4 ΔCD mutant notably resemble those in cells expressing Clr4 F449Y, further supporting our observations that H3K9me2 is sufficient to promote an increase in Swi6 chromatin occupancy (β state) (fig. S2D). Hence, we conclude that the β intermediate corresponds to a chromatin sampling state consisting of weak and unstable interactions between Swi6 and H3K9me or H3K9me0 nucleosomes and is an on-pathway intermediate to the lowest-mobility α state.
Simultaneously, we also tested how heterochromatin misregulation affects Swi6 dynamics. We deleted proteins that are involved in maintaining proper heterochromatin boundaries: (i) Epe1, a putative H3K9 demethylase that erases H3K9me, and (ii) Mst2, an H3K14 acetyltransferase that acetylates histones and promotes active transcription (43–45). We deleted either epe1 or mst2 individually in cells expressing PAmCherry-Swi6 (fig. S2, E and F). We observed relatively few changes in the weight fractions of the different Swi6 mobility states in these individual mutants. However, simultaneously deleting both epe1 and mst2 leads to a more marked rearrangement of the Swi6 mobility states (fig. S2G). We refer to this double mutant as an H3K9mespreading mutant. Unlike the H3K9me2 mutant, the fraction of molecules in the β intermediate-mobility state increases nearly twofold in the H3K9mespreading mutant, which also coincides with a near-complete depletion of Swi6 molecules from the unbound δ state (fig. S2G). As expected, deleting clr4 in this H3K9mespreading mutant (H3K9mespreading clr4Δ) collapses the β intermediate-mobility state from 48 to ~14%, similar to what we observed in epe1+ mst2+ clr4Δ cells (Fig. 2A and fig. S2H). Hence, transient chromatin interactions increase in the case of H3K9mespreading mutants, suggesting that heterochromatin misregulation leads to an increase in the chromatin-bound fraction of Swi6. Through measurements of PAmCherry-Swi6 expression levels across different mutant backgrounds, we determined that there is no correlation between protein expression and mobility state occupancy (fig. S2, I to M).
Fine-grained kinetic modeling transitions reveal H3K9me binding specificity in vivo
Our fission yeast mutants enabled us to assign biochemical properties to each of our experimentally measured mobility states (Fig. 3A). The state-to-state transitions inferred by SMAUG (Fig. 1F) estimate the probability with which a molecule of Swi6 assigned to one state during one 40-ms imaging frame will be assigned to some other state in the next frame (40 ms later). To infer the direct biochemical processes underlying the observed Swi6 dynamics, we implemented a high–temporal resolution model using a Bayesian synthetic likelihood (BSL) approach (see Materials and Methods for details) to determine the most consistent set of fine-scale chemical rate constants (46). Note that the model we have used is a statistical inference algorithm, not a generative model: As opposed to being trained using our experimental data to simulate other data, the model infers rate constants that can best describe the experimental results (as well as our uncertainties regarding those quantities).
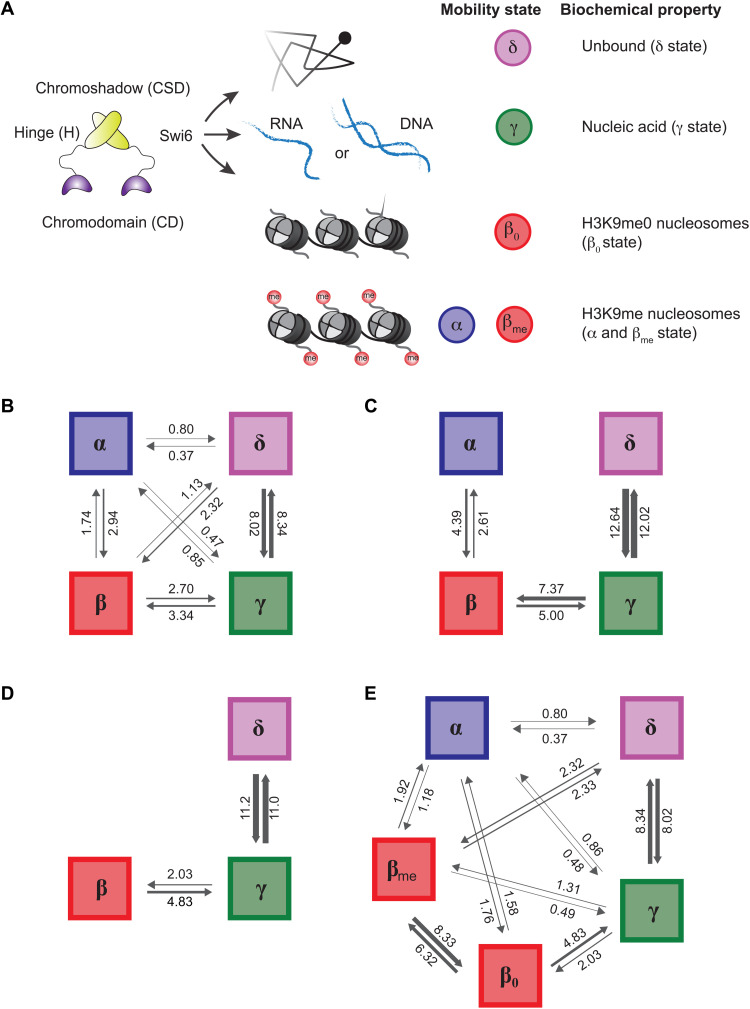
Fig. 3. Rate constants inferred from fine-grained chemical kinetic simulation and approximate Bayesian computation.
(A) Schematic showing the inferred biochemical nature of each state considered in our fine-grained chemical simulations. (B) Inferred rate constants for cells expressing WT Swi6, in units of 1/s, for transitions of Swi6 assuming that there are four biochemical states of Swi6 and that any state can chemically transition to any other. (C) Inferred rate constants (in units of 1/s) for the same situation as in (B) but assuming that states can only transition to the adjacent mobility states. (D) Inferred rate constants for clr4Δ cells, in units of 1/s, for the Swi6 transitions, assuming a three-state model as inferred by SMAUG. (E) Inferred rate constants, in units of 1/s, assuming five biochemical states of Swi6 in the WT cells. This model assumes that the β state corresponds to two chemical states: one with Swi6 bound to fully methylated H3K9me and one with Swi6 bound to unmethylated H3K9me; the latter is assigned parameters based on the clr4Δ simulation shown in (B).
We applied the BSL inference algorithm to the WT Swi6 data (shown in Fig. 1F) to simulate transition rates between the different mobility states (α, β, γ, and δ). We compared the complete simulated posterior distributions (violin plots in fig. S3A) to the naïve transition rates obtained by assuming that the SMAUG transitions were exclusively single chemical transitions (dashed lines in fig. S3A). When we simulate the experiment using the inferred chemical rate constants from BSL, the simulations agree qualitatively with the direct analysis of the experimental results, demonstrating that our SMAUG analysis indeed captures the relevant time scales underlying Swi6 interstate transitions. Formal model comparison (Materials and Methods) indicates that the rates inferred via BSL show a favorable value of the Bayesian information criterion (ΔBIC = −5.4 × 103) relative to the naïve rates. Hence, a detailed consideration of the chemical kinetics underlying the observed Swi6 transitions yields more quantitatively accurate information. Next, we compared our rate constant inferences between the models where (i) Swi6 transitions directly between nonadjacent mobility states (a dense model; Fig. 3B) and (ii) Swi6 transitions between adjacent mobility states (a sparse model; Fig. 3C). Since we have full posterior distributions for our inferences, we switch to the widely applicable information criterion (WAIC) for these comparisons. We found that the dense model is favored (ΔWAIC = −3.7 × 104), indicating that transitions between both adjacent and nonadjacent states are possible within a single (40-ms) experimental observation. We also confirmed that our algorithm inferred the absolute quantities of our rate constants, not only relative quantities (Supplementary Methods).
We next examined the effects of eliminating H3K9me (replicating the scenario observed in clr4Δ cells) on interstate transitions. Consistent with the experimental data from Fig. 2B, our simulations revealed that the transition rates between the γ and δ states do not appreciably change in clr4Δ cells relative to WT cells (Fig. 3D). However, the transitions involving the β state are significantly altered in clr4Δ cells, with far more rapid transitions from the β to the γ state, indicating that the β state is less stable (Fig. 3D): When we did a separate BSL inference on the clr4Δ experiments, the new inference agreed better with the experiments than the WT inference rates without the α state (ΔWAIC = −8.9 × 104). As previously noted, our single-molecule tracking experiments led us to infer that the β state consists of H3K9me-dependent and H3K9me-independent components, with only the H3K9me-independent component present in clr4Δ cells.
To deconvolute the two β state components, we performed simulations in WT cells under the assumption that the β state observed in clr4Δ cells solely represents the H3K9me0 β state. Denoting the proposed substates of β as β0 (H3K9me0) and βme (H3K9me1/2/3), we repeated our BSL inference on the WT Swi6 data using a five-state model of the system. We constrained parameters involving transitions between the β0 and γ states and the β0 and δ states to match those for the clr4Δ cells while using the WT rates for the transitions between the γ and δ states. The resulting rate constants agree substantially better with the experimental data than did the original four-state model (ΔWAIC = −4.7 × 104 relative to the four-state model) (Fig. 3E). On the basis of these results, we conclude that the β0 state is an unstable intermediate, with high rates of β0 → γ transitions (H3K9me0 to nucleic acid binding) and β0 → βme transitions (H3K9me0 to H3K9me binding). Hence, despite the relative abundance of H3K9me0 chromatin in S. pombe cells, the fast dissociation of Swi6 from the β0 state ensures that Swi6 spends little time bound nonspecifically to H3K9me0 chromatin.
Last, on the basis of estimates that 1 to 2.5% of the S. pombe genome consists of H3K9me3 nucleosomes, we found that the preference of Swi6 to bind H3K9me versus H3K9me0 chromatin (ratio of rate constants between β0 and βme) is ~94-fold (24). While our inference regarding the equilibrium constant is sensitive to the proportion of H3K9, which is trimethylated, the most favorable plausible equilibrium for Swi6 still favors βme more than 35-fold (fig. S3B). Since the low-mobility α state depends on H3K9me recognition and oligomerization (fig. S2, A and B), we propose that CSD oligomerization amplifies the ability of Swi6 to discriminate between H3K9me0 and H3K9me chromatin in the nucleus. In summary, our analysis using the BSL-based model achieves three important goals: (i) validates the time resolution we use in our single-molecule imaging study, (ii) identifies rate constants for transitions between mobility states with virtually identical diffusion coefficients (β0 and βme3), and (iii) provides a direct measure of the H3K9me3 recognition specificity in cells.
Increased CD valency compensates the disruption of Swi6 oligomerization
To uncouple H3K9me recognition from CSD-dependent oligomerization, we replaced the Swi6 CSD oligomerization domain with a glutathione S-transferase (GST) tag (fig. S4A) (47). Overall, the GST fusion construct is expected to maintain Swi6 dimerization while eliminating higher-order CSD-mediated oligomerization. We refer to this hybrid protein construct as Swi61XCD-GST since the newly engineered protein has only one intact CD. GST homodimerization of the Swi61XCD-GST results in a complex that precisely consists of two CDs. We expressed PAmCherry-Swi61XCD-GST in fission yeast cells, which lack an endogenous copy of Swi6 (swi6Δ). Following a strong 406-nm activation pulse, we imaged the ensemble of PAmCherry-Swi61XCD-GST molecules in the S. pombe nucleus. We observed a diffuse distribution of PAmCherry-Swi61XCD-GST proteins within the nucleus in contrast to WT Swi6, which exhibits prominent clusters (fig. S4A). These observations differ from previous studies of the mammalian HP1 isoform, HP1β, in which case replacing the CSD domain with GST did not affect HP1β localization (34). However, unlike Swi6, HP1β exhibits a reduced oligomerization capacity and fails to form condensates in vitro (34, 48).
Next, we added a second CD domain to the existing PAmCherry-Swi61XCD-GST construct to generate PAmCherry-Swi62XCD-GST. GST homodimerization of the Swi62XCD-GST results in an engineered Swi6 protein that has a precise twofold increase in CD valency relative to WT Swi6 or PAmCherry-Swi61XCD-GST (Fig. 4A). We used high-intensity 406-nm illumination to activate the ensemble of PAmCherry-Swi62XCD-GST molecules and acquired z sections by imaging at 561 nm (Fig. 4C). We observed prominent foci of PAmCherry-Swi62XCD-GST molecules that qualitatively resemble WT Swi6 foci in cells (Fig. 4B). We measured expression levels of Swi61XCD and Swi62XCD-GST proteins and found no correlation between their expression levels and mobility state occupancy (fig. S4, B and C). We measured H3K9me2 levels and the binding of our PAmCherry fusion proteins at sites of constitutive heterochromatin (pericentromeric repeats, mating-type locus, and telomeres) using ChIP-qPCR (fig. S4, D and E). H3K9me2 levels are intact at dg and tel1 in cells expressing PAmCherry-Swi62XCD-GST. H3K9me2 is absent at the mat locus, consistent with previous studies that have shown that deleting Swi6 sensitizes the mating-type locus to the complete loss of H3K9me (28). Furthermore, using ChIP-qPCR, we confirmed that PAmCherry-Swi62XCD-GST binds to chromatin at dg and tel1 in a manner that is similar to that of WT PAmCherry-Swi6 (fig. S4E).
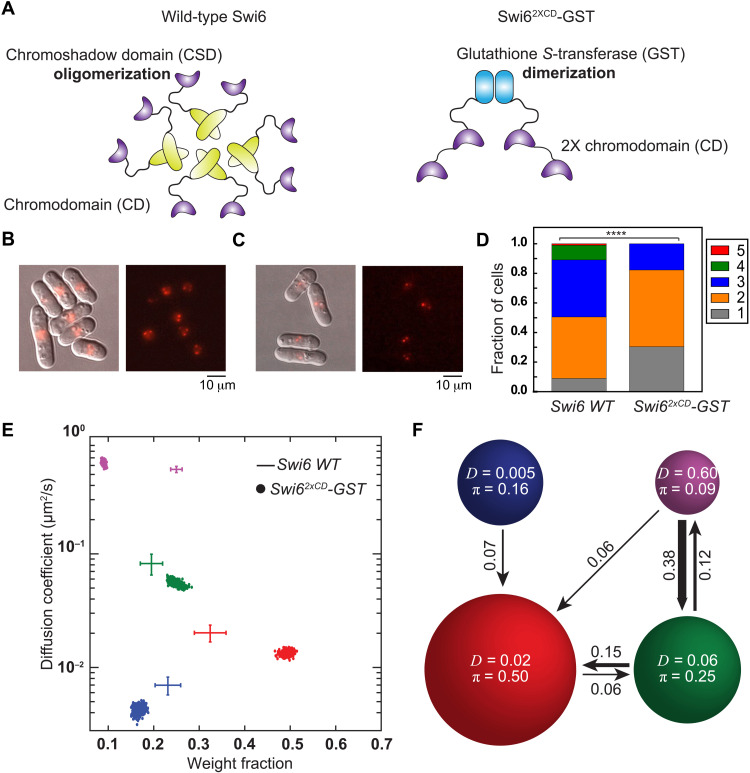
Fig. 4. Tandem CDs are both necessary and sufficient for H3K9me recognition and Swi6 localization.
(A) CSD-mediated oligomerization of Swi6 (left) and GST-mediated dimerization of an engineered Swi62XCD-GST mutant with two tandem CDs. (B) Overlaid differential interference contrast and epifluorescence images (left) and epifluorescence images alone (right) of PAmCherry-Swi6 simultaneously activated using high-power 405-nm excitation and imaged using 561-nm excitation. The images are a maximum intensity projection of a Z-stack consisting of 13 images acquired at 250-nm z-axis intervals. (C) Overlaid differential interference contrast and epifluorescence images (left) and epifluorescence images alone (right) of PAmCherry-Swi62XCD-GST simultaneously activated using high-power 405-nm excitation and imaged using 561-nm excitation. The images are a maximum intensity projection of a Z-stack consisting of 13 images acquired at 250-nm z-axis intervals. (D) Distribution of the number of photoactivated PAmCherry. ****P = 0.0005, Pearson’s chi-squared test. (E) SMAUG identifies four distinct mobility states for PAmCherry-Swi62XCD-GST molecules (dots); the average and SD of the WT Swi6 clusters from Fig. 1F in the same four states are provided as a reference (crosshairs). Each point is the average single-molecule diffusion coefficient, D, of molecules in that state at a saved iteration of the Bayesian algorithm after convergence. Dataset: 42,382 steps from 5182 trajectories. (F) Average probabilities (arrows) of a PAmCherry-Swi62XCD-GST molecule transitioning between the mobility states (circles) from (E).
Comparing the distribution of foci numbers per cell reveals a skew in the distribution with a more significant proportion of cells that exhibit three to five foci in the case of WT Swi6 compared to the 2XCD-GST fusion construct (Fig. 4D). To assess the localization and silencing capability of Swi62XCD-GST in the absence of any protein-mediated dimerization, we introduced seven mutations within the GST dimer interface to attenuate the high-affinity GST dimerization (49) to create PAmCherry-Swi62XCD-GSTmutant. The majority of cells exhibit zero or one cluster (71%) compared to the dimerization-competent GST allele (fig. S4, F and I). Hence, GST-mediated dimerization of two tandem CDs is necessary for H3K9me recognition, and mutations that affect dimerization reduce CD-mediated localization. In addition, we introduced CD binding mutations (W104A) to either one (PAmCherry-Swi62XCD-mut1-GST) or both CDs (PAmCherry-Swi62XCD-mut1 and 2) in the context of the 2XCD-GST fusion construct. Imaging cells where one CD was mutated resulted in ~66% of cells having no clusters, whereas mutating both CDs resulted in a complete loss of PAmCherry foci (fig. S4, G to I). Hence, the second tandem CD partially contributes to PAmCherry-Swi62XCD-GST localization even when one CD has been mutated.
Using glutaraldehyde-based cross-linking, we set up reactions where we added increasing amounts of recombinant 3XFLAG epitope–tagged WT Swi6, Swi6 L315E, and Swi62XCD-GST. We confirmed that the 2XCD-GST fusion construct fails to form higher-order oligomers unlike WT Swi6 (fig. S4J). Last, we tested whether the expression of the 2XCD-GST fusion construct in cells that lack Swi6 (swi6Δ) restores epigenetic silencing using an ade6+ reporter inserted at the pericentromeric repeats (otr1R) where we observe PAmCherry-Swi62XCD-GST localization. Unlike WT Swi6–expressing cells, which appear red due to silencing of the ade6+ reporter, PAmCherry-Swi62XCD-GST–expressing cells exhibit a loss of red pigmentation consistent with the absence of silencing (fig. S4K). Hence, restoring CD-dependent protein localization is insufficient to rescue epigenetic silencing, confirming that additional Swi6 CSD–mediated protein-protein interactions are required for this process.
We mapped the mobility states associated with PAmCherry-Swi62XCD-GST in S. pombe. Despite the differences in the overall number of foci, the twofold increase in CD valency completely circumvents the need for higher-order oligomerization: Swi62XCD-GST fully restores the localization of Swi6 to sites of heterochromatin at levels that rival those of WT Swi6 (Fig. 4E). We measured a slow-mobility state (α state) population of ~20%, similar to that of the WT Swi6 protein (Fig. 1F). Besides, there is a substantial increase in the β intermediate sampling state, indicating that PAmCherry-Swi62XCD-GST exhibits increased chromatin association. We confirmed that the localization of PAmCherry-Swi62XCD-GST within the genome depends exclusively on H3K9me by deleting Clr4 (PAmCherry-Swi62XCD-GST clr4Δ). PAmCherry-Swi62XCD-GST clr4Δ cells exhibit a complete loss of the slow-mobility α state and a concomitant decrease in the β intermediate-mobility state from 50 to 12% (fig. S4L). We also introduced previously characterized Swi6 CD Loop-X mutations to suppress Swi6 CD domain–dependent oligomerization, which could confound our interpretations (42). We determined that there are no quantitative differences in the mobility states associated with PAmCherry-Swi62XCD-GST Loop-X versus PAmCherry-Swi62XCD-GST constructs without the CD Loop-X mutation (fig. S4M). Hence, the recovery of the slow-mobility state in the case of PAmCherry-Swi62XCD-GST depends solely on CD-dependent H3K9me recognition. Since binding occurs in the context of a chimeric protein that acts as a dimer and fails to form higher-order oligomers, we concluded that four CD domains are necessary and sufficient for Swi6 to localize at sites of H3K9me.
Last, we inferred transition probabilities between the different mobility states in the case of PAmCherry-Swi62XCD-GST (Fig. 4F). Notably, we observed that molecules rarely exchange between the H3K9me-dependent α and β states, unlike what we detect in the case of the oligomerization-competent, WT Swi6 protein. The forward and reverse transition probabilities between the α and β states decrease approximately fourfold, suggesting that the PAmCherry-Swi62XCD-GST protein is less dynamic with regard to its chromatin-associated states. Thus, although engineered multivalent CD domains are sufficient to achieve target search in vivo, PAmCherry-Swi62XCD-GST molecules are less dynamic and exhibit fewer binding and unbinding transitions from chromatin. To understand the molecular basis for the enhanced binding of the Swi62XCD-GST fusion construct, we performed nucleosome binding assays. Using H3K9me0 and H3K9me3 nucleosomes, we measured the apparent binding affinity and binding specificity for recombinant WT Swi6, Swi6 CSDmut, Swi61XCD-GST, and Swi62XCD-GST. Although Swi62XCD-GST binds with similar affinity to H3K9me3 nucleosomes as WT Swi6, the fusion protein exhibits a twofold increase in H3K9me3 nucleosome binding specificity (fig. S5).
Oligomerization directly competes with nucleic acid binding to promote Swi6 localization at sites of heterochromatin formation
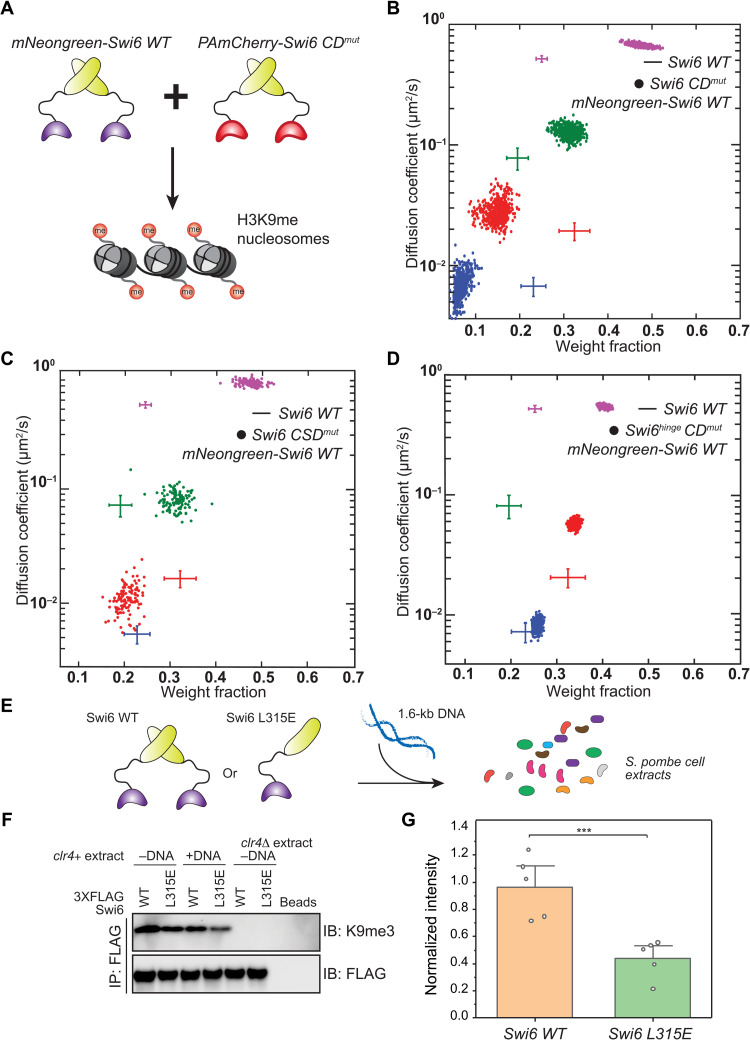
Since an engineered multivalent CD–containing construct (Swi62XCD-GST) is less dynamic, we hypothesized that the association and dissociation of Swi6 oligomers regulate H3K9me-dependent binding and turnover from heterochromatin. We tested our hypothesis by achieving oligomerization-dependent Swi6 localization that is independent of H3K9me recognition. We made strains that express combinations of WT Swi6 and a Swi6 CDmut protein that has an inactive CD domain but retains an intact CSD oligomerization domain (Fig. 5A). We coexpressed mNeonGreen-Swi6 protein in cells that also express PAmCherry-Swi6 CDmut. Since mNeonGreen and PAmCherry emissions are spectrally distinct, we used our photoactivation approach to image single PAmCherry-Swi6 CDmut molecules (red channel) after verifying the presence of discrete mNeonGreen-Swi6 foci at sites of constitutive heterochromatin (green channel). We previously showed that the lack of CD-mediated H3K9me recognition eliminates the low-mobility α state (fig. S2A). In contrast, our SMAUG analysis of strains coexpressing WT Swi6 and Swi6 CDmut revealed that 5% of PAmCherry-Swi6 CDmut proteins now reside in the slow-mobility α state (Fig. 5B). Therefore, CSD-mediated oligomerization alone can drive Swi6 localization independently of H3K9me recognition. We cannot eliminate the possibility of heterodimers consisting of WT and Swi6 CDmut proteins from our measurements. However, it is unlikely that heterodimers consisting of a single, functional CD domain will contribute significantly to the observed localization patterns since at least four CD domains are required for H3K9me binding (Fig. 4E). Consistent with the fraction being small, PAmCherry foci are not visible in our epifluorescence data where we imaged an ensemble of PAmCherry-Swi6 CDmut molecules, although mNeonGreen-Swi6 foci are intact (fig. S6A).
Fig. 5. Oligomerization competes with nucleic acid binding to promote Swi6 localization at heterochromatin formation sites.
(A) PAmCherry-Swi6 CDmut and mNeonGreen-Swi6 (WT) proteins are coexpressed; mobility states are measured using single-molecule tracking of PAmCherry-Swi6 CDmut. (B) SMAUG identifies four distinct mobility states for PAmCherry-Swi6 CDmut (swi6 W104A) in cells coexpressing mNeonGreen-Swi6. Dataset: 13,194 steps from 1900 trajectories. (C) SMAUG identifies three distinct mobility states for PAmCherry-Swi6 CSDmut (swi6 L315E) in cells coexpressing mNeonGreen-Swi6; the α mobility state is absent. Dataset: 3200 steps from 1270 trajectories. (D) SMAUG identifies three distinct mobility states for PAmCherry- Swi6hinge CDmut (swi6 KR25A W104A) in cells coexpressing mNeonGreen-Swi6; the γ mobility state is absent. Dataset: 15,462 steps from 3250 trajectories. Each point in (B) to (D) is the average single-molecule diffusion coefficient, D, of Swi6 molecules in that state at a saved iteration of the Bayesian algorithm after convergence; the average and SD of the WT Swi6 clusters (blue α, red β, green γ, and purple δ, respectively, from Fig. 1F) are provided as a reference (crosshairs). (E) Schematic of the competition between Swi6 oligomerization and nucleic acid binding with 3XFLAG-Swi6 or 3XFLAG-Swi6 L315. (F) FLAG IP assay to detect histone H3 and histone H3K9me3 bound to 3XFLAG-Swi6 or 3XFLAG-Swi6 L315E using an H3K9me3 antibody in extracts from WT (clr4+) or H3K9me0 (clr4Δ) cells. (G) Mean intensity of H3K9me3 histones detected upon addition of DNA relative to no DNA. Error bars: SD (N = 5, ***P = 0.009, Wilcoxon rank sum test).
To confirm that the recovery of the α slow-mobility state is due to CSD-dependent Swi6 interactions, we coexpressed a Swi6 CSD mutant (PAmCherry-Swi6 CSDmut), which is unable to oligomerize but has an intact CD domain, together with the WT mNeonGreen-Swi6 protein. The coexpression of WT Swi6 protein fails to restore any measurable occupancy of PAmCherry-Swi6 CSDmut protein in the low-mobility α state (Fig. 5C). Suppressing nucleic acid binding results in increased occupancy of Swi6 molecules in the α and β states (Fig. 2C). We hypothesized that eliminating nucleic acid binding might enhance oligomerization-mediated recruitment of PAmCherry-Swi6 CDmut. Therefore, we coexpressed PAmCherry-Swi6hinge CDmut proteins in cells that also express mNeonGreen-Swi6. Notably, we observed a substantial increase in the occupancy of PAmCherry-Swi6hinge CDmut molecules in the α mobility state (approximately 20%) (Fig. 5D). PAmCherry foci could be easily visualized in our epifluorescence data where we simultaneously imaged an ensemble of PAmCherry-Swi6hinge CDmut molecules and mNeonGreen-Swi6 (fig. S6B). Therefore, our results reveal that nucleic acid binding and Swi6 oligomerization are in direct competition. Disrupting nucleic acid binding promotes a CD-independent mode of Swi6 binding at sites of H3K9me with the CSD domain having a causal role in driving interactions between the differently labeled Swi6 molecules.
We performed a biochemical assay to directly measure the competition between nucleic acid binding, oligomerization, and H3K9me recognition using recombinant Swi6 proteins and multivalent H3K9me chromatin derived from fission yeast cell extracts as substrates. We incubated recombinant WT 3XFLAG Swi6 or 3XFLAG Swi6 L315E protein with fission yeast cell extracts. We then pulled down Swi6-bound chromatin in the WT and mutant protein context (Fig. 5E). In the absence of DNA, both WT Swi6 and Swi6 L315E pull down a similar amount of H3K9me chromatin as detected using an H3K9me3-specific antibody (Fig. 5F). Our pull-downs of H3K9me chromatin using recombinant Swi6 proteins are specific since extracts prepared using clr4Δ cells failed to recover any chromatin. We added an exogenous 1.6-kb DNA fragment and observed that the amount of H3K9me chromatin that we recovered substantially decreased in the case of Swi6 L315E (CSDmut) but remains unchanged in the case of WT Swi6 (Fig. 5G). Hence, our observations suggest that CSD-dependent interactions that promote Swi6 oligomerization can indeed resist the effects of promiscuous nucleic acid binding.
DISCUSSION
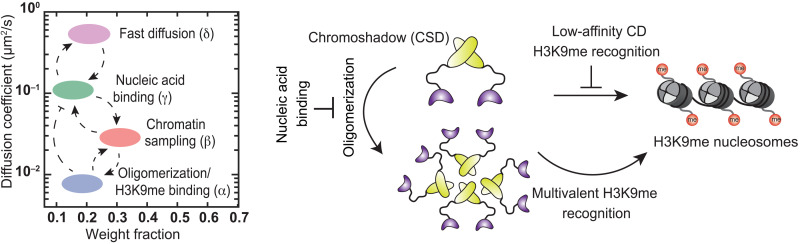
Our results reveal the molecular basis for how Swi6 identifies sites of H3K9me within the complex and crowded chromatin landscape of the S. pombe nucleus. Despite only ~2% of chromatin being marked with H3K9me, Swi6 readily discriminates between modified H3K9me chromatin and unmodified H3K9me0 chromatin; we found that Swi6 binds in vivo to H3K9me nucleosomes with 94-fold specificity. Our numbers most closely resemble in vitro measurements of Swi6 binding to H3K9me3 peptides as opposed to nucleosomes (22). Hence, modified H3K9me histone tails are the primary specificity determinants of Swi6 binding in the nucleus. The reduced specificity observed in in vitro studies is likely due to nucleic acid binding, which leads to Swi6-nucleosome interactions that are independent of the histone tails being modified (37). In contrast, our in vivo studies reveal that nucleic acids, given their large excess in a native chromatin context, promote Swi6 unbinding by directly competing with oligomerization. Swi6 dimers are incapable of staying bound at sites of H3K9me, which, in turn, places a unique emphasis on the coordination between oligomerization and H3K9me recognition (Fig. 6). We propose that Swi6 oligomerization stabilizes higher-order molecular configurations consisting of at least four CDs to promote cooperative and multivalent H3K9me recognition and binding.
Fig. 6. Model for how Swi6 locates sites of H3K9me within a complex and crowded genome.
Although CDs have the requisite specificity to localize at sites of H3K9me, nucleic acid binding titrates proteins away from sites of heterochromatin formation. Oligomerization stabilizes higher-order configurations of the Swi6 CD domain to ensure rapid and efficient localization of Swi6 at sites of heterochromatin formation and outcompetes nucleic acid binding.
Unlike earlier FRAP measurements, our model-independent superresolution assessment of Swi6 diffusion identifies four distinct mobility states (26, 29, 30). Using fission yeast mutants, we have validated the biochemical attributes associated with each mobility state. Note that in our mutants, the chromatin environment and sequence composition of Swi6 significantly change. Hence, our assignments of mobility states are based on the order of magnitude of the D value only, and the specific value of D for the same assigned state might differ between different strains; for example, Davg,δ, WT (0.51 μm2/s) is lower than Davg,δ H3K9me2 (0.67 μm2/s). Therefore, in our comparisons between SMAUG results of different strains, we focus on the depletion or emergence of a mobility state and on the change of weight fraction for each mobility state, not on subtle differences in D for the state in different mutants.
The primary drivers of Swi6 mobility are nucleic acid binding and weak and strong H3K9me-dependent interactions. The transition probabilities reveal how each mobility state functions to sequester or titrate Swi6 molecules, suggesting that altering their relative occupancy ultimately affects the H3K9me-bound population of the protein. Most prominently, we find that nucleic acid binding titrates Swi6 away from sites of H3K9me, while neutralizing nucleic acid binding promotes stable interactions at sites of H3K9me (α state), and it also increases the overall chromatin-bound population of the protein (β state). We propose that a significant function of Swi6 oligomerization is to counterbalance inhibitory and titratable molecular interactions that would otherwise wholly suppress Swi6 localization in cells.
Our studies highlight how the high-resolution tracking of the in vivo dynamics of single molecules in cells can fully recapitulate all the biochemical features of proteins despite their heterogeneous dynamics in a native chromatin context. Our studies represent a vital step toward the ultimate goal of in vivo biochemistry, where the on and off rates of proteins and their substrates can be reliably and directly measured in their cellular environment. Last, we used our inferred transition rates, combined with known biochemical parameters, to infer the precise chemical rate constants governing the behavior of Swi6. The main caveats of these inferred rate constants are that they cannot capture spatial effects, and they model binding reactions as pseudo first order. While modeling spatial effects will require further study, we can infer true binding rate constants from our inferred pseudo first-order rates by assuming that Swi6 is the limiting reagent of its binding reactions. Furthermore, the unbinding reactions are truly first order, and thus, our inferred rates for these reactions are true rate constants.
Preserving the same degree of nucleic acid binding but preventing oligomerization (Swi61XCD-GST) disrupts Swi6 localization at sites of H3K9me (fig. S4A). Simply adding a second CD restores H3K9me-specific localization. Although multivalency represents a longstanding principle for how HP1 proteins bind to chromatin, our results suggest well-defined stoichiometric configurations that enable stable and selective H3K9me binding. Our engineered constructs reveal that four tandem CDs are both necessary and sufficient for effective H3K9me-dependent localization in cells. Although recombinant Swi6 purified from Escherichia coli is predominantly a dimer (~83%) in vitro, about 10% of Swi6 molecules form tetramers (22). On the basis of our results, we hypothesize that oligomerization and subsequent phase separation might increase the local concentration of Swi6 molecules to shift their equilibrium distribution from dimers to tetramers. In this manner, the ability of HP1 proteins to form condensates could be vital to coordinate oligomerization and H3K9me recognition, as shown in our studies. The underlying mechanisms that promote Swi6 oligomerization in our experiments remain unclear since the Swi6 Loop-X mutant exhibits virtually identical mobility states relative to WT Swi6. It is possible that the Loop-X mutation has a subtle effect in vivo, and additional residues are needed to fully disrupt the CD-CD oligomerization interface. An alternative model that is also consistent with our data is that chromatin offers a multivalent surface not only for CD-H3K9me interactions but also for CSD-nucleosome interactions, leading to additional modes of Swi6 oligomerization (13). We cannot differentiate between oligomers being stabilized before H3K9me binding and oligomers being stabilized after binding to sites of H3K9me. A model where Swi6 dimers “probe” chromatin before oligomerizing would be entirely consistent with our data. We would expect that such probing would lead to likely encounters between additional Swi6 molecules at sites enriched with H3K9me.
In the case of Swi6 and its mammalian homolog, HP1α, oligomerization is essential to promote the formation of chromatin condensates that exhibit liquid-like properties (12, 13, 50). Our results suggest that the presence of low-affinity CD domains could be one reason why some classes of HP1 proteins are proficient in oligomerization. The ability of specific HP1 isoforms to oligomerize promotes the multivalent recognition of H3K9me nucleosomes since the dimeric state of a protein such as Swi6 is insufficient for stable heterochromatin binding. Our findings raise the question of why high-affinity CD domains are not more prevalent among HP1 proteins since this could represent the most straightforward solution to the localization question. It is noteworthy that an engineered version of Swi6 with two CDs and no oligomerization exhibits reduced binding and unbinding transitions to other intermediate states from the H3K9me-dependent low-mobility α state (Fig. 4F). Therefore, our engineered protein constructs, once engaged at sites of H3K9me, exhibit little to no protein turnover.
On the basis of our observations, we speculate that protein recruitment that is exclusively dependent on high-affinity CD binding lacks tunability. The lack of exchange and protein turnover from chromatin would also impede subsequent downstream, CD-dependent binding events. Most notably, in S. pombe, the H3K9 methyltransferase (Clr4) and the second HP1 protein (Chp2) both have CD domains that recognize and bind to H3K9me and are essential for epigenetic silencing (51, 52). Deleting Chp2 leads to a decrease in the fraction of the fast-moving Swi6 molecules and a concomitant increase in the fraction of molecules in chromatin sampling β state (fig. S6C). Instead, we propose that weak oligomerization and protein turnover ensure a time-sharing approach that provides opportunities for regulatory inputs either via protein-protein interactions or posttranslational modifications. The formation of heterochromatin condensates, in addition to serving as mechanisms that promote epigenetic silencing through physical changes to the genome, could be fundamentally involved in shifting the equilibrium states of Swi6 oligomerization to promote efficient and highly selective H3K9me target recognition in living cells (12, 14).
MATERIALS AND METHODS
Plasmids
Plasmids containing Swi6 WT and point mutants were constructed by modified existing pFA6a N-terminal tagging plasmids. We used Gibson-based gene assembly to make in-frame fusion constructs of Swi6 proteins and PAmCherry. Point mutations were introduced by designing primers using guidelines described in Quick Change mutagenesis protocols. The Swi6hinge mutant was amplified and subcloned into pFA6a vectors containing PAmCherry. In the case of Swi61XCD-GST, Swi62XCD-GST, and Swi62XCD-GST-mutant, we obtained synthetic DNA fragments from Twist Biosciences. The mutant Schistosoma japonicum GST allele has seven mutations (L50Q, R73L, P86A, A90K, M94E, G96K, and Y104A), which were introduced on the basis of structural similarity to human GST (49).
Strains
All strains were constructed using a PCR-based gene targeting approach (53). All strains expressing PAmCherry-Swi6 or Swi6 mutants were constructed by reintroducing PCR products in swi6Δ strains. All strains were genotyped using colony PCR assays. Strains expressing Swi62XCD-GST or mNeonGreen-Swi6 were made by digesting the pDual vectors with Not1 restriction enzyme to facilitate their insertion at the leu1+ locus in S. pombe cells. Cells were selected on the basis of their ability to restore leucine auxotrophy. Other deletions of heterochromatin-associated factors were achieved either by PCR-based gene targeting approaches or by a cross followed by random spore analysis and PCR-based screening to select for colonies that express PAmCherry-Swi6 or PAmCherry-Swi6 mutants. All strains used in this study are listed in table S1.
S. pombe live-cell imaging
Yeast strains containing a copy of PAmCherry-Swi6 or PAmCherry-Swi6 mutants under the control of the native Swi6 promoter were grown in standard complete YES media (US Biological, catalog no. Y2060) containing the full complement of yeast amino acids and incubated overnight at 32°C. The seed culture was diluted and incubated at 25°C with shaking to reach an optical density at 600 nm (OD600) of ∼0.5. To maintain cells in an exponential phase and eliminate extranuclear vacuole formation, the culture was maintained at OD600 ∼0.5 for 2 days, with dilutions performed at ~12-hour time intervals. To prepare agarose pads for imaging, cells were pipetted onto a pad of 2% agarose prepared in YES media, with 0.1 mM N-propyl gallate (Sigma-Aldrich, catalog no. P-3130) and 1% gelatin (Millipore, catalog no. 04055) as additives to reduce phototoxicity during imaging. S. pombe cells were imaged at room temperature (RT) with a 100× 1.40 numerical aperture (NA) oil-immersion objective in an Olympus IX-71 inverted microscope. First, the fluorescent background was decreased by exposure to 488-nm light (Coherent Sapphire, 200 W/cm2 for 20 to 40 s). A 406-nm laser (Coherent Cube, 405-100; 102 W/cm2) was used for photoactivation (200-ms activation time), and a 561-nm laser (Coherent Sapphire, 561-50; 163 W/cm2) was used for imaging. Images were acquired at 40-ms exposure time per frame. The fluorescence emission was filtered with Semrock LL02-561-12.5 filter and Chroma ZT488/561rpc 488/561 dichroic to eliminate the 561-nm excitation source and imaged using a 512 × 512 pixel Photometrics Evolve EMCCD camera.
For the epifluorescence images in Fig. 4 and fig. S4, a 405-nm light-emitting diode (LED) light source (Lumencor SpectraX) at 25 mW/nm (100% power) was used to photoactivate cells, and a 561-nm LED was used to image them subsequently. Images were collected with 100-ms exposure time per frame with a 100× 1.45 NA oil-immersion objective using a Photometrics Prime95B sCMOS camera.
Single-molecule trajectory analysis with SMAUG algorithm
Recorded Swi6-PAmCherry single-molecule positions were detected and localized with two-dimensional Gaussian fitting with home-built MATLAB software as previously described and connected into trajectories using the Hungarian algorithm (54, 55). These single-molecule trajectory datasets were analyzed by a nonparametric Bayesian framework to reveal heterogeneous dynamics (40). This SMAUG algorithm uses nonparametric Bayesian statistics and Gibbs sampling to identify the number of distinct mobility states, n, in the single-molecule tracking dataset in an iterative manner. It also infers the parameter such as weight fraction, πi, and effective diffusion coefficient, Di, for each mobility state (i = …,n), assuming a Brownian motion model. To ensure that even rare events would be captured, we collected more than 10,000 steps in our single-molecule tracking dataset for each measured strain, and we ran the algorithm over >10,000 iterations to achieve a thoroughly mixed state space. The state number and associated parameters were updated in each iteration of the SMAUG algorithm and saved after convergence. The final estimation (e.g., Fig. 1F) shows the data after convergence for iterations with the most frequent state number. Each mobility state, i, is assigned a distinct color, and for each saved iteration, the value of Di is plotted against the value of πi. The distributions of estimates over the iterations give the uncertainty in the determination of Di. Furthermore, the transition probabilities (e.g., Fig. 1G) give the average probability of transitioning between states from one step to the next in any given trajectory. For static molecules from imaging fixed S. pombe cells, SMAUG converges to a single state with Davg = 0.0041 ± 0.0003 μm2/s. The average localization error for single-molecule localizations in this fixed-cell imaging is 32.6 nm.
Clustering analysis for the Swi6 distributions
The spatial pattern (i.e., dispersed, clustered, or homogeneously distributed and at what scale) of each mobility state was investigated using Ripley’s K function (41)
where r is the search radius, n is the number of points in the set, λ is the point density, and rij is the distance between the ith and jth point. I is an indicator function (1 when true and 0 when false). For convenience, we further normalized K(r) to attain Ripley’s H function
where H(r) = 0 for a random distribution, H(r) > 0 for a clustered distribution pattern, and H(r) < 0 for a dispersed pattern. The maximum of H(r) approximately indicates the cluster size (56). The cross-correlation between different states was studied with the same method. In all analysis, the nucleus was approximated as a circle to determine the area and perform edge correction (57). We calculated H(r) for each cell, and then we consolidated data from different cells into an overall H(r) from the average across all cells weighted by the point density.
To eliminate effects from the intrinsic spatial correlation between steps that come from the same trajectories, we simulated diffusion trajectories with similar confined area size, average track length, and overall density as experimental trajectories by drawing step lengths from the step size distribution of the corresponding experiment steps. These trajectories are random in the initial position and step direction. We calculated a four-state H(r) distribution for trajectories simulated corresponding to the WT Swi6 dataset (fig. S1C). To eliminate the contribution of the in-track autocorrelation of steps in H(r), we subtracted H(r) of the randomly simulated trajectories from H(r) of the experimental data for each mobility state. The same H(r) simulation and subtraction were carried out for all Ripley autocorrelation analyses.
Fine-grained chemical rate constant inference
To calculate the chemical rate constants underlying the observed in vivo Swi6 transitions, we applied Bayesian inference by making use of an approximated likelihood function to account for the stochasticity of the interstate transitions occurring with the small numbers of Swi6 molecules present in living cells. This inference framework follows the principles of BSL (46, 58) using a distribution to approximate the likelihood that is appropriate for the problem at hand (multinomial instead of a Gaussian mixture). Similar approaches have previously been applied to other inference problems in chemical kinetics (59, 60). The overall framework for our BSL procedure is shown in fig. S6: At each iteration, rate constants are evaluated by simulating 105 possible experimental transition matrices. Multinomial distributions are then fit to the probabilities of transitions (details below). Calculation of the probability that the experimentally observed state-state transition data could have occurred given these distributions yields the approximated likelihood, which is then used in standard Bayesian inference with the posterior distribution on the parameters obtained via Metropolis Markov Chain Monte Carlo. We chose an improper Jeffries prior: P(rate) ∝ 1/rate because the real chemical rate constants in this system are unknown.
Obtaining accurate likelihoods for the BSL procedure described above requires a simulation framework capable of running individual stochastic simulations of how the system might behave over a single experimental time step given a particular set of starting populations of transition rates. We first simulate our system by dividing the experimental time (0.04 s) into 100 time steps δt = 4 × 10–4 s. We confirmed that this time step was small enough to satisfy our assumption of one transition per time step by computing an approximate probability that two transitions would occur to the same molecule over this time step: A molecule starting at the most mobile state (the most likely state to undergo a transition per unit time) had a probability of 0.58 of being in that state after 40 ms, which implies that the probability density of leaving was exponentially distributed with parameter 13.6/s. To give an upper bound on the probability that a molecule would transition at least twice over a time step, we calculated this probability assuming that all states were equally mobile, which gave us the probability of observing at least two transitions in a time step asP(at least two transitions) = 1−exp(−13.6 × ẟt) × (1 + 13.6 × ẟt) = 1.47 × 10−5
The probability of two transitions occurring in one time step even once in an experimental simulation with 1500 molecules is therefore roughly 2% (and this is an upper bound that would only be achieved if all molecules were in the most mobile state, which is vanishingly unlikely).
At the beginning of each simulation, we assumed that the Swi6 molecules were partitioned across states according to their equilibrium populations from SMAUG. For each partition, we executed each time step based on Gillespie’s method for converting deterministic chemical rate constants to stochastic molecular behavior (61), assuming that our time step would be small enough that every particle could transition at most once. First, at each time step, for a given diffusion state A, we calculated the probability that a given particle would transition out of that state as p = exp(−δt × ∑X≠A(rA→X)), where rA→X is the chemical rate constant of transition from state A to state X in units of 1/s. We sampled from a binomial distribution with transition probability p as given above and n = the number of molecules in a state to determine the number of molecules m that transitioned. We then determined which state each of these molecules transitioned to by generating a sample from a multinomial distribution of a size equal to m, where the probability that a given transitioning molecule transitions to state B is rA→B/∑X≠A(rA→X). For each time step, we repeated this procedure for each Swi6 diffusion state, moving forward in time until reaching 0.04 s, the observation time frame of diffusion experiments (schematized in fig. S7A). For each calculation of the likelihood, we repeated this simulation procedure 105 times and then used the resulting matrices to fit one multinomial distribution for each initial state partition using the R package MGLM, version 0.2.0 (fig. S7B). To calculate the log-likelihood of the rates, we calculated the log probability that the transition matrices observed by the experiment would have occurred given our constructed multinomial distributions and then summed them (fig. S7C). Details of parameter inference for each case are given in text S1. To assess our models relative to each other, we calculate the Widely Applicable Information Criterion (62); lower values of the information criterion indicate a preferred model. For subsequent kinetic simulation of the time required for Swi6 to transition between fully unbound and fully bound states, we calculated the mean first passage time from the δ to the α state by running our simulation 100 times with one molecule in the δ state and stepping in time with δt = 4 × 10–4 s until our molecule ended in the α state and then calculating the mean of the result across the replicate simulations.
To ensure that the algorithm inferred absolute rates and not relative rates, we reran our inference algorithm with the initial condition of the posterior means of the dense model’s rates multiplied by five. These multiplied rates were far less consistent with the data than original rates (log likelihood of −1.3 × 105 instead of −1.2 × 104), although they had identical equilibrium values, and our algorithm rapidly converged to a set of rates that were nearly identical to our posterior means.
Nucleosome electrophoretic mobility shift assays
Swi6 proteins were cloned into N-terminal 6XHis-tag–containing pET vectors, and mutants were generated using ligase-independent cloning. All Swi6 proteins were purified from BL21(DE3)-RIPL E. coli cells. Cells were grown at 37°C to OD 0.5 to 0.8 in LB media with ampicillin (100 μg/ml), induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and were grown for 16 hours at 18°C. Cells were harvested and resuspended in lysis buffer [1× phosphate-buffered saline (PBS) buffer (pH 7.3), 300 mM NaCl, 10% glycerol, 0.1% Igepal CA-630, 1 mM phenylmethylsulfonyl fluoride (PMSF), aprotonin (1 μg/ml), pepstatin A, and leupeptin] and sonicated. Cell debris was removed by centrifugation at 25,000g for 35 min. Cell lysates were incubated with HisPur NiNTA resin (Thermo Fisher Scientific) at 4°C for at least 2 hours. The resin was washed with lysis buffer, and protein was eluted [20 mM Hepes (pH 7.5), 100 mM KCl, 10% glycerol, and 500 mM imidazole] and incubated with the corresponding protease (Ulp1 or TEV) overnight at 4°C. After cleavage of 6XHis-tag, the products were further isolated by anion exchange chromatography using a HiTRAP Q HP column (Cytiva). Proteins were dialyzed into storage buffer [20 mM Hepes, 100 mM KCl, 10% glycerol, and 1 mM dithiothreitol (DTT)]. Protein concentrations were determined using ultraviolet (UV) absorption measurements at 280 nm and molecular weights (MWs) and extinction coefficients computationally determined for WT Swi6 (MW = 37,292.6 g/mol, ε = 41,035 M−1 cm−1), Swi6 L315E (MW = 37,308.6 g/mol, ε = 41,035 M−1 cm−1), Swi61XCD-GST (MW = 47,675 g/mol, ε = 68,675 M−1 cm−1), and Swi62XCD-GST (MW = 54,704.7 g/mol, ε = 51,715 M−1 cm−1) using the Expasy ProtParam tool.
Reaction samples were prepared with varying concentrations of Swi6 and 10 nM mononucleosomes (H3K9me0 and H3K9me3, Epicypher catalog nos. 16-0006 and 16-0315-20) in binding buffer [20 mM Hepes (pH 7.5), 4 mM tris, 80 mM KCl, 0.1% Igepal CA-630, 0.2 mM EDTA, 2 mM DTT, and 10% glycerol]. Reactions were incubated at 30°C for 45 min. A 0.5× tris-borate EDTA 6% acrylamide:bis-acrylamide 37.5:1 gel was pre-run at RT for at least 1 hour at 75 V. Reactions were loaded on the gel and ran under the same conditions for 3 hours. Gels were poststained for 2 hours with polyacrylamide gel electrophoresis (PAGE) GelRed DNA stain (Biotium) and imaged using a Typhoon Imager. The unbound nucleosome band was quantified using ImageJ, and binding curves were fit using nonlinear regression (Prism 9).
Glutaraldehyde protein cross-linking oligomerization assay
A Swi62XCD-GST vector was synthesized (Twist Biosciences) to contain two copies of the Swi6 CD and one hinge domain fused to GST. Kinase, ligase, and DpnI cloning was used to delete one CD from this construct to make a Swi61XCD-GST construct. Full-length WT Swi6, Swi6 L315E, Swi61XCD-GST, and Swi62XCD-GST inserts were cloned into a pGEX vector, which introduces a 5′-GST-3XFLAG with a PreScission protease site upstream of the 3XFLAG sequence. All Swi6 proteins were purified from BL21(DE3)-RIPL E. coli cells. Cells were grown at 37°C to OD 0.5 to 0.8 in LB media with ampicillin (100 μg/ml), induced with 0.4 mM IPTG, and were grown for 16 hours at 18°C. Cells were harvested and resuspended in lysis buffer [1× PBS buffer (pH 7.8), 300 mM KCl, 10% glycerol, 1 mM DTT, 1 mM PMSF, aprotonin (1 μg/ml), pepstatin A, and leupeptin] and sonicated. Cell debris was removed by centrifugation at 25,000g for 35 min. Cell lysates were incubated with Pierce Glutathione Agarose resin (Thermo Fisher Scientific) for 4 hours at 4°C. The resin was washed with lysis buffer, and PreScission protease was incubated with resin overnight at 4°C. Cleaved protein elutions were collected and dialyzed into storage buffer [20 mM Hepes (pH 7.5), 150 mM KCl, 10% glycerol, and 1 mM DTT] and stored at −80°C. Protein concentrations were determined using UV absorption measurements at 280 nm and MWs and extinction coefficients computationally determined for 3XFLAG-Swi6 (MW = 40,900 g/mol, ε = 45,505 M−1 cm−1), 3XFLAG-Swi6L315E (MW = 40,916 g/mol, ε = 45,505 M−1 cm−1), 3XFLAG-Swi61XCD-GST (MW = 50,821.1 g/mol, ε = 73,145 M−1 cm−1), and 3XFLAG-Swi62XCD-GST (MW = 57,851 g/mol, ε = 97,220 M−1 cm−1) using the Expasy ProtParam tool.
Protein samples were prepared in reaction buffer [20 mM Hepes (pH 7.5), 100 mM KCl, and 10% glycerol] and were incubated at RT for 30 min with 0.01% glutaraldehyde; the total reaction volume was 20 μl. Glutaraldehyde reacts with primary amines and the ε- amino group of lysine residues. Reactions were quenched with 2 μl of 1 M tris (pH 8). SDS loading dye was added to the quenched reactions and incubated at 99°C for 5 min. Protein samples were separated on a 4 to 20% tris-glycine gradient PAGE gel (Bio-Rad) under SDS denaturing conditions, blotted onto polyvinylidene difluoride membranes (Bio-Rad), and probed with monoclonal anti-FLAG M2–horseradish peroxidase antibody (Sigma-Aldrich). Protein amounts were equalized using non–cross-linked input samples, and reaction samples were normalized by chemiluminescent signal (quantified using ImageJ) to account for any glutaraldehyde-induced epitope damage.
Biochemical measurements of the competition between Swi6 oligomerization, nucleic acid binding, and H3K9me recognition
WT S. pombe cells (250 ml) were grown to an OD600 of 3.5. Cells were harvested by pelleting, and lysis was carried out by bead beating using Omni Bead Ruptor at 3000 rpm × 30 s × 10 cycles, in 20 mM Hepes (pH 7.5), 100 mM NaCl, 0.25% Triton X-100, 5 mM MgCl2, 10% glycerol, 1 mM PMSF, and protease inhibitors using glass beads. The lysate was centrifuged, and the pellet was discarded. The supernatant consisting of the total S. pombe cell lysate was transferred to a 1.5-ml tube. Either recombinant 3XFLAG-Swi6 or 3XFLAG-Swi6 L315E [purified using methods previously described in (42)] was added at a final concentration of 85 nM to the cell extract. Swi6 is in large excess compared to the amount of H3K9me chromatin present in the extract. Hence, the IP reactions are performed under limiting substrate conditions. After incubating Swi6 proteins with the lysate for 2 hours, we added agarose beads cross-linked with a FLAG antibody (Sigma-Aldrich, catalog no. A8592) to pull down 3XFLAG-Swi6 or 3XFLAG-Swi6 L315E. In reactions where DNA was added, we used a 1.6-kb DNA fragment at 1 nM final concentration (DNA-to-Swi6 ratio = 1:85). The beads were subsequently washed using a wash buffer [20 mM Hepes (pH 7.5), 300 mM NaCl, 0.25% Triton X-100, 5 mM MgCl2, 10% glycerol, 1 mM PMSF, and protease inhibitors] three times followed by the addition of SDS loading buffer to elute Swi6 and all bound chromatin substrates. A total of 30% of the elution was loaded on a 4 to 20% gradient gel followed by detection of Swi6 protein (using an M2 FLAG antibody, Sigma-Aldrich, catalog no. A8592), histone H3 (catalog no. ab1791), and histone H3K9me3 (Abcam, catalog no. ab1898). We ensured that equal amounts of lysates were added to each reaction by measuring histone H3 in the input samples. We measured the amount of H3K9me3 in the IP samples using ImageJ. Last, H3K9me3 levels measured in the presence of DNA were normalized relative to H3K9me3 intensities measured in the absence of DNA.
Chromatin immunoprecipitation
Cells were grown to log phase (OD600 1.3 to 1.8) in yeast extract supplemented with adenine (YEA) or YEA containing tetracycline (2.5 μg/ml) medium and fixed with 1% formaldehyde for 15 min at RT. Glycine (130 mM) was then added to quench the reaction and incubated for 5 min at RT. The cells were harvested by centrifugation and washed twice with tris-buffered saline [50 mM tris (pH 7.6) and 500 mM NaCl]. Cell pellets were resuspended in 300 μl of lysis buffer [50 mM Hepes-KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, and protease inhibitors] to which 500 μl of 0.5-mm glass beads was added, and cell lysis was carried out by bead beating using Omni Bead Ruptor at 3000 rpm × 30 s × 10 cycles. Tubes were punctured, and the flow-through was collected in a new tube by centrifugation, which was subjected to sonication to obtain fragment sizes of roughly 100 to 500 base pairs long. After sonication, the extract was centrifuged for 15 min at 13,000 rpm at 4°C. The soluble chromatin was then transferred to a fresh tube and normalized for protein concentration by the Bradford assay. For each normalized sample, 25 μl of lysate was saved as input, to which 225 μl of 1× TE/1% SDS was added [TE: 50 mM tris (pH 8.0) and 1 mM EDTA]. Dynabeads Protein A were preincubated with anti-H3K9me2 antibody (Abcam, catalog no. ab1220; PRID:AB_449854). For each IP, 2 μg of antibody coupled to 30 μl of beads was added to 400 μl of soluble chromatin, and the final volume of 500 μl was achieved by adding lysis buffer. Samples were incubated for 2 hours at 4°C, and the beads were collected on magnetic stands and washed three times with 1 ml of lysis buffer and once with 1 ml of TE. For eluting bound chromatin, 100 μl of elution buffer I [50 mM tris (pH 8.0), 10 mM EDTA, and 1% SDS] was added, and the samples were incubated at 65°C for 5 min. The eluate was collected and incubated with 150 μl of 1× TE/0.67% SDS in the same way. Input and immunoprecipitated samples were lastly incubated overnight at 65°C to reverse cross-link for more than 6 hours. Glycogen (60 μg), 100 μg of proteinase K (Roche), 44 μl of 5 M LiCl, and 250 μl of 1× TE were added to each sample, and incubation was continued at 55°C for 1 hour. Phenol/chloroform extraction was carried out for all the samples followed by ethanol precipitation. Immunoprecipitated DNA was resuspended in 100 μl of 10 mM tris (pH 7.5) and 50 mM NaCl and was used for qPCR (SYBR Green) using a Bio-Rad 384-well real-time PCR instrument. For extra cross-linking to detect Swi6 using an mCherry antibody (catalog no. M11217), before fixing with 1% formaldehyde, the cultures were incubated at 18°C for 2 hours in a shaking incubator. The cells were pelleted and resuspended in 4.5 ml of 1× PBS. To this, 1.5 mM ethylene glycol bis(succinimidylsuccinate), Pierce (Thermo Fisher Scientific), was added, and the samples were incubated at RT for 20 min with mild shaking before adding 1% formaldehyde. The samples were then processed as mentioned above. All ChIP experiments were reproduced N = 2.
Acknowledgments
We are grateful to A. Khalil and R. Baldridge for comments on the manuscript. We thank D. Dasgupta for help in identifying GST monomerizing mutations, U. Cho for help with Swi6 cross-linking assays, A. T. Blanco for help with Swi6 purification, and A. Larkin for help in preparing figures. We also thank all members of the Biteen, Freddolino, and Ragunathan laboratories for invaluable support. We thank M. Bühler for sharing the Swi6 hinge mutant plasmid and G. Narlikar for sharing the Swi6 ARK mutant plasmid. We thank D. Moazed for sharing fission yeast strains that have been used in this study.
Funding: This work was funded by an NSF Understanding Rules of Life Award (1921677) to J.S.B., P.L.F., and K.R. and NIH award R35GM137832 to K.R. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) comet resource at the San Diego Supercomputing Center through allocation TG-MCB140220 to P.L.F. (63).
Author contributions: Conceptualization: P.L.F., J.S.B., and K.R. Methodology: Z.C., J.D.K., A.F., P.L.F., J.S.B., and K.R. Investigation: S.B., Z.C., J.D.K., A.F., A.A., G.R., P.L.F., J.S.B., and K.R. Formal analysis: S.B., Z.C., J.D.K., A.F., A.A., G.R., P.L.F., J.S.B., and K.R. Visualization: S.B., Z.C., A.F., A.A., P.L.F., J.S.B., and K.R. Validation: S.B., Z.C., and A.F. Writing—original draft: S.B., Z.C., A.F., and K.R. Writing—review and editing: S.B., Z.C., A.F., A.A., K.R., J.S.B., and P.L.F. Supervision: K.R., J.S.B., and P.L.F. Funding acquisition: K.R., J.S.B., and P.L.F. Project administration: K.R., J.S.B., and P.L.F.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
Supplementary Methods
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bonasio R., Tu S. J., Reinberg D., Molecular signals of epigenetic states. Science 330, 612–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T., Allis C. D., Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Strahl B. D., Allis C. D., The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Stewart-Morgan K. R., Petryk N., Groth A., Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 22, 361–371 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Grewal S. I. S., Moazed D., Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Nicetto D., Zaret K. S., Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 55, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canzio D., Larson A., Narlikar G. J., Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 24, 377–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowieson N. P., Partridge J. F., Allshire R. C., McLaughlin P. J., Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10, 517–525 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Hughes R. M., Wiggins K. R., Khorasanizadeh S., Waters M. L., Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc. Natl. Acad. Sci. U.S.A. 104, 11184–11188 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs S. A., Khorasanizadeh S., Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Ohnston J. B. J., Burlingame A. L., Agard D. A., Redding S., Narlikar G. J., Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanulli S., Trnka M. J., Dharmarajan V., Tibble R. W., Pascal B. D., Burlingame A. L., Griffin P. R., Gross J. D., Narlikar G. J., HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., Karpen G. H., Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iglesias N., Paulo J. A., Tatarakis A., Wang X., Edwards A. L., Bhanu N. V., Garcia B. A., Haas W., Gygi S. P., Moazed D., Native chromatin proteomics reveals a role for specific nucleoporins in heterochromatin organization and maintenance. Mol. Cell. 77, 51–66.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeRoy G., Weston J. T., Zee B. M., Young N. L., Plazas-Mayorca M. D., Garcia B. A., Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell. Proteomics 8, 2432–2442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smothers J. F., Henikoff S., The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 21, 2555–2569 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allshire R. C., Ekwall K., Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7, a018770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekwall K., Nimmo E. R., Javerzat J. P., Borgstrom B., Egel R., Cranston G., Allshire R., Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109 ( Pt. 11), 2637–2648 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Ivanova A. V., Bonaduce M. J., Ivanov S. V., Klar A. J. S., The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Ekwall K., Javerzat J. P., Lorentz A., Schmidt H., Cranston G., Allshire R., The chromodomain protein Swi6: A key component at fission yeast centromeres. Science 269, 1429–1431 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Canzio D., Chang E. Y., Shankar S., Kuchenbecker K. M., Simon M. D., Madhani H. D., Narlikar G. J., Al-Sady B., Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haldar S., Saini A., Nanda J. S., Saini S., Singh J., Role of Swi6/HP1 self-association-mediated recruitment of Clr4/Suv39 in establishment and maintenance of heterochromatin in fission yeast. J. Biol. Chem. 286, 9308–9320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jih G., Iglesias N., Currie M. A., Bhanu N. V., Paulo J. A., Gygi S. P., Garcia B. A., Moazed D., Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature 547, 463–467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller C., Adaixo R., Stunnenberg R., Woolcock K. J., Hiller S., Buhler M., HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol. Cell 47, 215–227 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Stunnenberg R., Kulasegaran-Shylini R., Keller C., Kirschmann M. A., Gelman L., Buhler M., H3K9 methylation extends across natural boundaries of heterochromatin in the absence of an HP1 protein. EMBO J. 34, 2789–2803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J. Y., Reddy B. D., Jia S. T., Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife 4, e06179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama J., Klar A. J. S., Grewal S. I. S., A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101, 307–317 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Cheutin T., Gorski S. A., May K. M., Singh P. B., Misteli T., In vivo dynamics of Swi6 in yeast: Evidence for a stochastic model of heterochromatin. Mol. Cell. Biol. 24, 3157–3167 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., Misteli T., Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Bryan L. C., Weilandt D. R., Bachmann A. L., Kilic S., Lechner C. C., Odermatt P. D., Fantner G. E., Georgeon S., Hantschel O., Hatzimanikatis V., Fierz B., Single-molecule kinetic analysis of HP1-chromatin binding reveals a dynamic network of histone modification and DNA interactions. Nucleic Acids Res. 45, 10504–10517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manley S., Gillette J. M., Patterson G. H., Shroff H., Hess H. F., Betzig E., Lippincott-Schwartz J., High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Tuson H. H., Biteen J. S., Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal. Chem. 87, 42–63 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Hiragami-Hamada K., Soeroes S., Nikolov M., Wilkins B., Kreuz S., Chen C., De La Rosa-Velazquez I. A., Zenn H. M., Kost N., Pohl W., Chernev A., Schwarzer D., Jenuwein T., Lorincz M., Zimmermann B., Walla P. J., Neumann H., Baubec T., Urlaub H., Fischle W., Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 7, 11310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaac R. S., Sanulli S., Tibble R., Hornsby M., Ravalin M., Craik C. S., Gross J. D., Narlikar G. J., Biochemical basis for distinct roles of the heterochromatin proteins Swi6 and Chp2. J. Mol. Biol. 429, 3666–3677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilic S., Bachmann A. L., Bryan L. C., Fierz B., Multivalency governs HP1α association dynamics with the silent chromatin state. Nat. Commun. 6, 7313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishibuchi G., Machida S., Osakabe A., Murakoshi H., Hiragami-Hamada K., Nakagawa R., Fischle W., Nishimura Y., Kurumizaka H., Tagami H., Nakayama J., N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity. Nucleic Acids Res. 42, 12498–12511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subach F. V., Patterson G. H., Manley S., Gillette J. M., Lippincott-Schwartz J., Verkhusha V. V., Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekwall K., Olsson T., Turner B. M., Cranston G., Allshire R. C., Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91, 1021–1032 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Karslake J. D., Donarski E. D., Shelby S. A., Demey L. M., DiRita V. J., Veatch S. L., Biteen J. S., SMAUG: Analyzing single-molecule tracks with nonparametric Bayesian statistics. Methods 193, 16–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripley B. D., The second-order analysis of stationary point processes. J. Appl. Probab. 13, 255–266 (1976). [Google Scholar]
- 42.Canzio D., Liao M. F., Naber N., Pate E., Larson A., Wu S. P., Marina D. B., Garcia J. F., Madhani H. D., Cooke R., Schuck P., Cheng Y. F., Narlikar G. J., A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 496, 377–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayoub N., Noma K., Isaac S., Kahan T., Grewal S. I. S., Cohen A., A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol. 23, 4356–4370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy B. D., Wang Y., Niu L. F., Higuchi E. C., Marguerat S. B., Bahler J., Smith G. R., Jia S. T., Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 25, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zofall M., Grewal S. I. S., Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Wood S. N., Statistical inference for noisy nonlinear ecological dynamic systems. Nature 466, 1102–1104 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Maru Y., Afar D. E., Witte O. N., Shibuya M., The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 271, 15353–15357 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Keenen M. M., Brown D., Brennan L. D., Renger R., Khoo H., Carlson C. R., Huang B., Grill S. W., Narlikar G. J., Redding S., HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 10, e64663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdalla A. M., Bruns C. M., Tainer J. A., Mannervik B., Stenberg G., Design of a monomeric human glutathione transferase GSTP1, a structurally stable but catalytically inactive protein. Protein Eng. 15, 827–834 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Larson A. G., Narlikar G. J., The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motamedi M. R., Hong E. J. E., Li X., Gerber S., Denison C., Gygi S., Moazed D., HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32, 778–790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadaie M., Kawaguchi R., Ohtani Y., Arisaka F., Tanaka K., Shirahige K., Nakayama J., Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 28, 6973–6988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., Steever A. B., Wach A., Philippsen P., Pringle J. R., Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Munkres J., Algorithms for the assignment and transportation problems. J. Soc. Indust.Appl. Math. 5, 32–38 (1957). [Google Scholar]
- 55.Rowland D. J., Biteen J. S., Measuring molecular motions inside single cells with improved analysis of single-particle trajectories. Chem. Phys. Lett. 674, 173–178 (2017). [Google Scholar]
- 56.Kiskowski M. A., Hancock J. F., Kenworthy A. K., On the use of Ripley’s K-Function and its derivatives to analyze domain size. Biophys. J. 97, 1095–1103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goreaud F., Pelissier R., On explicit formulas of edge effect correction for Ripley’s K-function. J. Veg. Sci. 10, 433–438 (1999). [Google Scholar]
- 58.Price L. F., Drovandi C. C., Lee A., Nott D. J., Bayesian synthetic likelihood. J. Comput. Graph. Stat. 27, 1–11 (2018). [Google Scholar]
- 59.Beaumont M. A., Zhang W. Y., Balding D. J., Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Csillery K., Blum M. G., Gaggiotti O. E., Francois O., Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 25, 410–418 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Gillespie D. T., Exact stochastic simulation of coupled chemical-reactions. J. Phys. Chem. 81, 2340–2361 (1977). [Google Scholar]
- 62.Watanabe S., A widely applicable bayesian information criterion. J. Mach. Learn. Rese 14, 867–897 (2013). [Google Scholar]
- 63.Towns J., Cockerill T., Dahan M., Foster I., Gaither K., Grimshaw A., Hazlewood V., Lathrop S., Lifka D., Peterson G. D., Roskies R., Scott J. R., Wilkins-Diehr N., XSEDE: Accelerating scientific discovery. Comput. Sci. Eng. 16, 62–74 (2014). [Google Scholar]
- 64.Persson F., Lindén M., Unoson C., Elf J., Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods 10, 265–269 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Hansen A. S., Woringer M., Grimm J. B., Lavis L. D., Tjian R., Darzacq X., Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife 7, e33125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karslake J. D., Donarski E. D., Shelby S. A., Demey L. M., DiRita V. J., Veatch S. L., Biteen J. S., SMAUG: Analyzing single-molecule tracks with nonparametric Bayesian statistics. Methods, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gelman A., Rubin D. B., Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472 (1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3
Supplementary Methods
References
Movie S1