Abstract
Purpose:
Persistent sustained attention deficit (SAD) after continuous positive airway pressure (CPAP) treatment is a source of quality of life and occupational impairment in obstructive sleep apnea (OSA). However, persistent SAD is difficult to predict in patients initiated on CPAP treatment. We performed secondary analyses of brain magnetic resonance (MR) images in treated OSA participants, using deep learning, to predict SAD.
Methods:
26 middle-aged men with CPAP use of more than 6 hours daily and MR imaging were included. SAD was defined by psychomotor vigilance task lapses of more than 2. 17 participants had SAD and 9 were without SAD. A Convolutional Neural Network (CNN) model was used for classifying the MR images into +SAD and −SAD categories.
Results:
The CNN model achieved an accuracy of 97.02±0.80% in classifying MR images into +SAD and −SAD categories. Assuming a threshold of 90% probability for the MR image being correctly classified, the model provided a participant-level accuracy of 99.11±0.55% and a stable image level accuracy of 97.45±0.63%.
Conclusion:
Deep learning methods, such as the proposed CNN model, can accurately predict persistent SAD based on MR images. Further replication of these findings will allow early initiation of adjunctive pharmacologic treatment in high-risk patients, along with CPAP, to improve quality of life and occupational fitness. Future augmentation of this approach with explainable artificial intelligence methods may elucidate the neuroanatomical areas underlying persistent SAD to provide mechanistic insights and novel therapeutic targets.
Introduction
Daytime sleepiness is a disabling neurocognitive consequence of obstructive sleep apnea (OSA). Despite continuous positive airway pressure (CPAP) treatment adherence of more than 4 hours daily, 12–65% of patients with treated OSA experience persistent sleepiness1, which adversely affects the quality of life and occupational fitness. Self-reported sleepiness is limited in occupational evaluation. It does not correlate well with objective measurements of sleepiness2 that are either time-intensive such as the multiple sleep latency test (MSLT), or lack large-scale normative data, for e.g., the Psychomotor Vigilance Task (PVT), which measures sustained attention deficit (SAD)3. Importantly, the objective measures of sleepiness fail to elucidate the neurological basis of neurocognitive dysfunction. Brain magnetic resonance imaging (MRI) provides anatomical details of grey and white matter and has been used to identify differences in brain structures between CPAP-treated OSA patients with and without sleepiness4. Our group has previously4,10 examined the differences in whole brain Diffusion-Weighted Imaging (DWI) between the groups. In an earlier study10, fractional anisotropy, mean diffusivity, axial diffusivity and radial diffusivity were used to reveal the differences in white matter fiber tracts between the sleepy and non-sleepy groups following CPAP-treatment. In a more recent study4, common white matter fiber tracts were determined by tract-based spatial statistics (TBSS)5, followed by assessment of whole brain and regional white matter differences between groups using a continuous-time random-walk (CTRW) diffusion model6. The sleepy group showed significantly higher temporal diffusion heterogeneity (α) and anomalous diffusion coefficient (Dm) globally, and regional differences in α and spatial diffusion heterogeneity (β) within twelve fiber tracts compared to the non-sleepy group. The parameters, α and Dm, in the right superior corona radiata were positively correlated, and β was negatively correlated with SAD (defined by PVT-lapses). Another study reported reversible white matter changes associated with improvements in memory, attention, and executive-function after CPAP treatment7. While these studies provide potential mechanistic insights into OSA associated SAD, the standard biostatistical approaches have limited power to predict SAD.
Deep learning, a subtopic of the field of machine learning, has been applied to brain MRI analysis for improving the classification of neurocognitive outcomes in various neurological and psychiatric disorders8,9. Based on our previous publications4,10, we hypothesized that machine learning, a powerful model-free computational approach increasingly used in biomedical research, will accurately predict SAD based on brain MRI in treated OSA patients. Further, we explored if explainable artificial intelligence techniques could highlight the neuroanatomical structures that drive this classification power.
Materials and Methods
We performed secondary analyses of a previously reported study of 26 middle-aged men with severe OSA treated with CPAP; use ≥6 hours daily4. Similar to our previous work, the presence of SAD was defined by PVT lapses ≥2 (17 participants with SAD) and PVT lapses <2 was used to identify the absence of SAD (9 without SAD)2. The baseline characteristics of the two groups are listed in Table 1. Regarding comorbidities, none of the participants had chronic obstructive lung disease, 6 participants had asthma (3 in each group), 1 participant had history of myocardial infarction, 2 participants had heart failure (1 in each group), none had stroke or seizures. The more common comorbidities are listed in Table 1, where no significant differences were noted between the groups. There was no relationship between hours of CPAP use per day and the number of PVT lapses (Pearson’s correlation, r = −0.03, p=0.83), probably due to the overall high CPAP adherence given the inclusion criteria of CPAP use ≥6 hours daily.
Table 1. Baseline Characteristics.
Baseline characteristics of the groups with and without Sustained Attention Deficit. The study sample inclusion criteria were men with AHI ≥30/hour and CPAP use ≥ 6 hours per day.
| Parameters | +SAD (N=17) | −SAD (N=9) | *p-value |
|---|---|---|---|
| Age | 45.73±8.29 | 43.23±8.49 | 0.41 |
| Body Mass Index (Kg/m 2 )_ | 34.4±4.7 | 32.9±5.6 | 0.47 |
| AHI per hour | 45.9±28.8 | 40.1±23.6 | 0.60 |
| CPAP nightly use (Hours) | 7.01±1.02 | 6.64±0.65 | 0.25 |
| Smoking (%) | 6 (36) | 3 (33) | 0.84 |
| Diabetes | 6 (36) | 4 (44) | 0.89 |
| Hypertension | 10 (58) | 6 (66) | 0.83 |
| Depression | 7 (42) | 3 (33) | 0.62 |
Sustained Attention Deficit = SAD, Apnea Hypopnea Index = AHI, Continuous Positive Airway Pressure = CPAP.
The groups were compared using t-tests or two sample Z test of proportions, using STATA v15.
Dataset generation for signal processing
The MRI data consisted of 13025 diffusion tensor images (DTI) covering the entire brain with 27 diffusion directions and a b-value of 1000 s/mm2. To remove the skull from the images, we assumed a bimodal histogram of the pixels in the Magnetic Resonance (MR) images, generated a binary mask using Otsu’s method11, and overlaid the binary mask on the original image (see examples in Figure 1). To optimize model training with a small sample size, we used an 80–20 DTI data split (80% training, 10% validation, and 10% testing) and evaluated the model performance using a 5-fold cross-validation technique.
Figure 1:
Three examples of segmented brain images from the original MR images.
Proposed model
Convolutional Neural Network (CNN, a class of deep neural networks) is a widely used architecture for imaging-based classification tasks due to its high performance8,12. Thus we used a CNN model for classifying the MR images into +SAD and −SAD. The model had three key components: (1) four convolutional blocks, (2) two fully-connected layers with ReLU activations, and (3) an output layer with two categorical nodes. Table 2 outlines the number of channels and neurons per layer in the CNN framework.
Table 2.
The convolutional neural network architectures for the Brain Magnetic Resonance Image classification task. There were 12 channels in each convolutional layer. Note that each “conv” layer shown in the table corresponds to the sequence Conv-BN-ReLU.
| Input | Layer Configuration | Output size |
|---|---|---|
| Convolutional Block (1) | [3×3 conv] × 12 2×2 max pool |
12 × 112 × 112 |
| Convolutional Block (2) | [3×3 conv] × 12 2×2 max pool |
12 × 56 × 56 |
| Convolutional Block (3) | [3×3 conv] × 12 2 × 2 max pool |
12 × 28 × 28 |
| Convolutional Block (4) | [3×3 conv] × 12 2 × 2 max pool |
12 × 14 × 14 |
| Flatten Layer | [1×2352] | 1 × 2352 |
| Fully Connected Layer (1) | 120-unit fully connected layer ReLU | 1 × 120 |
| Fully Connected Layer (2) | 64-unit fully connected layer ReLU | 1 × 64 |
| Classification Layer | [1×2] softmax | 1 × 2 |
Training and Inference
An iteration is defined as a single pass of the entire dataset, both forward and backward through the CNN. The proposed model was trained using Adam optimizer for 10 iterations13 and a binary cross-entropy loss.
Results
We addressed the following three questions regarding the clinical relevance of the proposed CNN model: (1) Is the model accurate in classifying participants with SAD? (2) Does the model learn MR image features unique to each category (+SAD and −SAD)? and (3) Can we identify the MR image features underlying the classification decision made by the model?
Performance:
The CNN model achieved an accuracy of 97.02±0.80% in classifying MR images into +SAD and −SAD categories. We extended the image-level analysis by employing a voting technique to obtain participant-level accuracy. For each participant, we considered the CNN model’s classification of the MR images as correct only if the prediction probability was higher than 0.9. The majority class predicted across all MR images from a participant was then calculated. Following this technique, the CNN model correctly classified participants into +SAD and −SAD categories with an accuracy of 99.11±0.55% and a stable image level accuracy of 97.45±0.63%.
Feature representation:
Our goal was to project the high-dimension MR image features learned by the CNN to a lower-dimensional space. Given the high-classification accuracy, we hypothesized that majority of the MR images belonging to either +SAD or −SAD categories would be correctly placed in a two-dimensional feature space. We used t-SNE dimensionality reduction14 to visualize the features in a two-dimensional feature space. As shown in Figure 2, the (+SAD and −SAD categories were clearly distinguished, thus validating the potential predictive performance of the model at the participant level.
Figure 2:
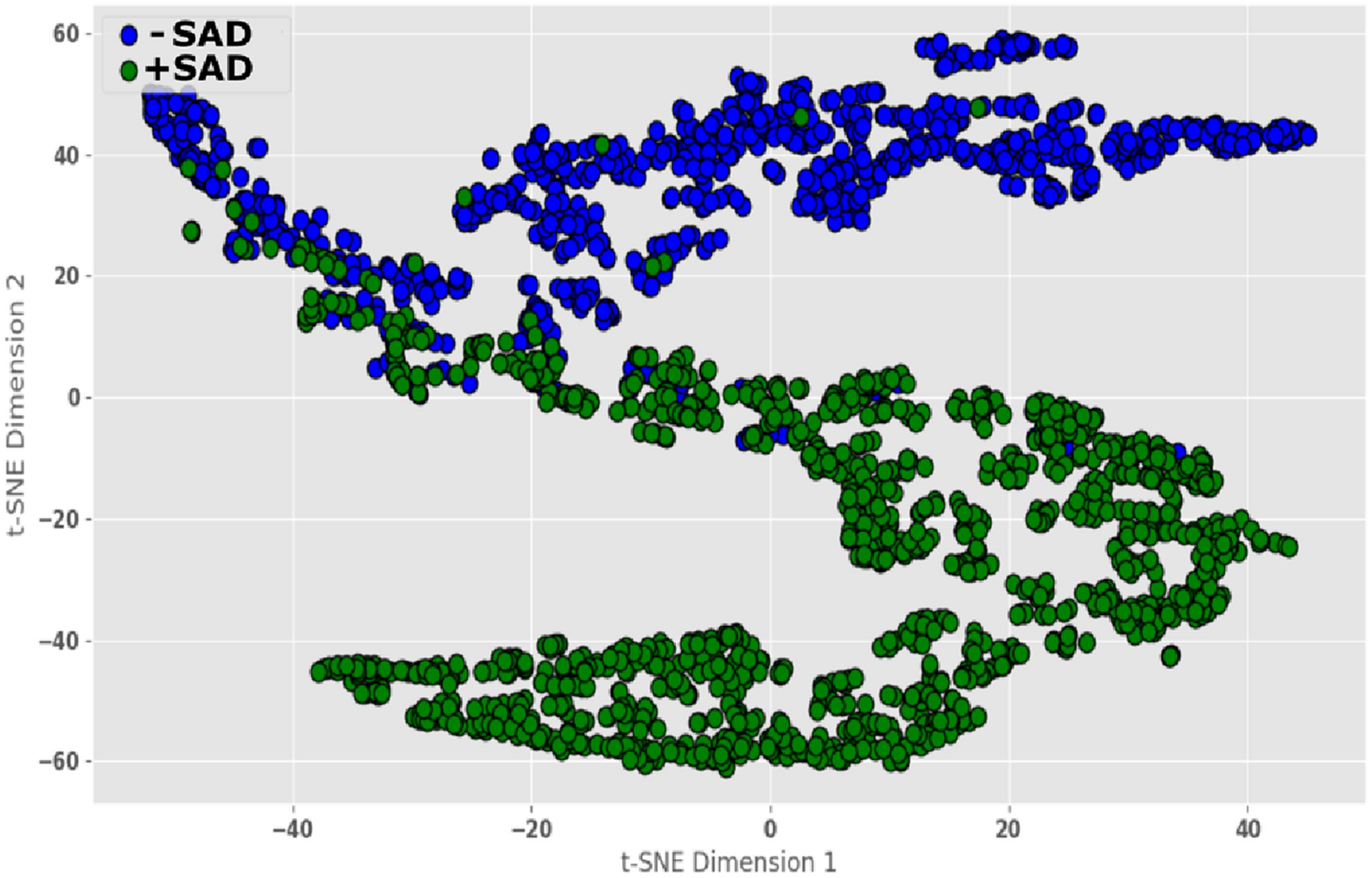
Visualization of the features learned by the CNN model showing two distinct clusters for the +SAD and −SAD categories. *SAD= sustained attention deficit.
Model explanation:
We used perturbation attribution methods to generate heatmaps that highlighted the MR image regions providing evidence for or against the CNN’s classification decision. Following Fong et al., we learned the smallest “mask” (by blurring an input image), which caused a significant decrease in the probability of correct classification15. The heatmaps of two participants with highest and lowest number of PVT lapses (84.5 in +SAD and 0 in −SAD categories) are shown in Figure 3, highlighting the brain regions which the model considered salient for accurate prediction (yellow represents salient regions).
Figure 3:
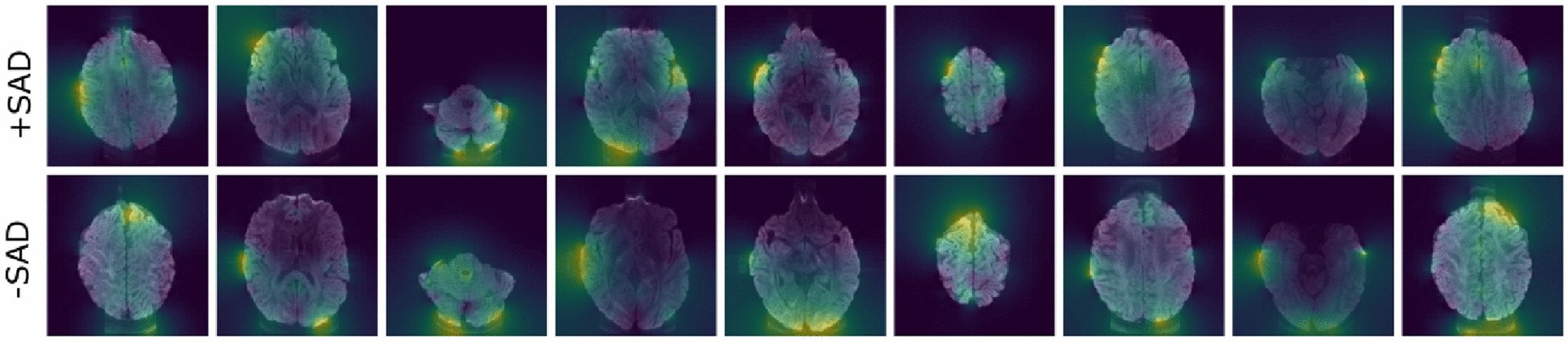
Attribution maps for MRI. The top panel depicts MRI from a +SAD participant (number of PVT lapses = 84.5). The bottom panel depicts MRI from a −SAD participant (number of PVT lapse = 0). Yellow represents more salient and purple represents less salient regions. *SAD= sustained attention deficit.
Discussion
This pilot study demonstrates that deep learning methods can be used to analyze brain MRI for accurate classification of OSA phenotypes based on persistent SAD after CPAP treatment with good adherence. In this study, we employed DTI as an example to demonstrate the efficiency of our CNN model. DTI is sensitive to diffusion anisotropy in brain tissues, and particularly useful for probing white-matter structural integrity10. Recent studies show that differences in white-matter fiber tracts may explain persistent sleepiness after CPAP treatment in OSA4,10. Previous studies have examined white and grey matter differences in brain MRI between OSA patients and controls, as well as changes in neuronal structures after CPAP treatment of OSA7,16. Some reports include specific regional differences that correlate with neurocognitive function, mood, and sleepiness17. These studies support ischemia-reperfusion neuronal and glial injury as a mechanism of neurocognitive dysfunction in OSA, and provide insight into regional reversibility of such injury after CPAP treatment. However, the insights gained from DTI depend on the analytical model. Because no existing analytical model can fully describe the underlying brain microstructures, any selected model is limited with respect to neuroanatomical details. Another limitation is that DTI typically focuses on white-matter characterization, although voxel-based morphometry can partially overcome this limitation16.
Deep learning is a sub-field of machine learning that allows computer-aided quantitative analysis of large MR image datasets, overcoming issues of model-dependency, inter-rater reliability, and efficiency. In this pilot study, the CNN model was trained in a supervised manner to generate empirical evidence. Our results highlight deep learning algorithm’s benefits over classical statistical methods by demonstrating an accurate classification of persistent SAD, an important source of morbidity in OSA. This benefit may be attributed to classical statistical algorithms’ limited generalizability due to variations in image acquisition and quality, and inter-individual variations in normal and pathologic brain structures. In future work, deep learning algorithms should be utilized with multiple layers of neural networks that can “self-learn” by training on larger datasets and offer the potential for discovery of novel findings from quantitative analysis of brain MR images in OSA. As noted in our previous publications, the persistence of sleepiness and neurocognitive dysfunction in some patients is not explained by OSA severity, CPAP adherence, sleep duration (by actigraphy), comorbidity, or use of sedating medications, and only partially explained by age4,10. Explainable artificial intelligence techniques can advance analytical results from black-box deep learning algorithms to interpretable segmentation of brain regions that provides critical input for the deep learning algorithm performance18. Due to the small sample size and lack of manual segmentation of the MR images in this pilot study, determination of neuroanatomical areas that were most helpful in predicting persistent sleepiness after OSA treatment was limited. However, visualization of the heatmaps in Figure 3 suggests that the right temporal area contributed significantly to the machine learning model performance. Previous research has shown that the right hippocampus, in the region deemed salient by the model, is vulnerable to hypoxic injury and reduced functional connectivity in patients with moderate to severe OSA, which may explain our findings19. Our previous study on white matters also suggested alterations of selected non-Gaussian diffusion parameters in the posterior limb of right internal capsule, right anterior corona radiata, and right superior longitudinal fasciculus4. However, changes in other diffusion parameters have also been reported in several white matter fiber tracts on the left hemisphere16. Overall, studies on brain anatomy associated with OSA are limited and inconclusive, particularly those concerning laterality. Further studies are needed to replicate and extend these preliminary findings to elucidate the neuroanatomical substrates of neurocognitive dysfunction in OSA.
Despite the promising results of this pilot study, the design and methodological limitations underscore the need to replicate the results with larger datasets. The classification accuracy of our current CNN model may reduce in larger datasets with greater participant heterogeneity, and will require improved methods. Additional steps for future studies with improved methods should include standardized imaging protocols, comprehensive preprocessing steps beyond skull stripping, atlas-based automated brain segmentation or combining manual segmentation of images by multiple experts using methods such as label fusion algorithms, and increasing the size of training datasets by applying random transformations (data augmentation that prevents overfitting). Finally, a generalizable deep learning algorithm requires advanced methods like transfer learning18, where the initial network is trained on a large dataset (population), then modified to work on similar, smaller datasets (subpopulations). For example, a network trained on larger datasets of treated OSA patients with residual SAD could be used for transfer learning applications to predict persistent SAD or sleepiness despite effective CPAP treatment. Persistent SAD or sleepiness is seen in more than a third of OSA patients and modifiable by pharmacologic therapy.
Funding:
The MR images were acquired on a research facility supported in part by the National Institutes of Health (Grant Number 1S10RR028898). The original data collection was supported by a grant from TEVA Pharmaceuticals, Inc.
Footnotes
Conflict of Interest: All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee (University of Illinois at Chicago Institutional Review Board) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Informed consent was obtained from all individual participants included in the study.
COI statement: The authors have no conflict of interest relevant to this manuscript.
References:
- 1.Javaheri S, Javaheri S. Update on Persistent Excessive Daytime Sleepiness in OSA. Chest. Aug 2020;158(2):776–786. [DOI] [PubMed] [Google Scholar]
- 2.Prasad B, Steffen AD, Van Dongen HPA, et al. Determinants of sleepiness in obstructive sleep apnea. Sleep. Feb 1 2018;41(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basner M, Dinges DF. Maximizing sensitivity of the psychomotor vigilance test (PVT) to sleep loss. Sleep. May 1 2011;34(5):581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Weaver TE, Zhong Z, et al. White matter structural differences in OSA patients experiencing residual daytime sleepiness with high CPAP use: a non-Gaussian diffusion MRI study. Sleep medicine. Jan 2019;53:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SM, Jenkinson M, Johansen-Berg H, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. Jul 15 2006;31(4):1487–1505. [DOI] [PubMed] [Google Scholar]
- 6.Karaman MM, Sui Y, Wang H, Magin RL, Li Y, Zhou XJ. Differentiating low- and high-grade pediatric brain tumors using a continuous-time random-walk diffusion model at high b-values. Magn Reson Med. Oct 2016;76(4):1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castronovo V, Scifo P, Castellano A, et al. White matter integrity in obstructive sleep apnea before and after treatment. Sleep. Sep 1 2014;37(9):1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan R, Williams BG, Ferreira da Silva M, et al. Deep Convolutional Neural Networks With Ensemble Learning and Generative Adversarial Networks for Alzheimer’s Disease Image Data Classification. Frontiers in aging neuroscience. 2021;13:720226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Li G, Xu Y, Tang X. Application of Artificial Intelligence in the MRI Classification Task of Human Brain Neurological and Psychiatric Diseases: A Scoping Review. Diagnostics (Basel). Aug 3 2021;11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Y, Zhou XJ, Nisi RA, et al. Brain white matter changes in CPAP-treated obstructive sleep apnea patients with residual sleepiness. Journal of magnetic resonance imaging : JMRI. May 2017;45(5):1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsu N A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics 1979;9:62–66. [Google Scholar]
- 12.Hamwood J, Schmutz B, Collins MJ, Allenby MC, Alonso-Caneiro D. A deep learning method for automatic segmentation of the bony orbit in MRI and CT images. Scientific reports. Jul 1 2021;11(1):13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinton G, Srivastava N, Swersky K. Neural networks for machine learning. Coursera. 2012;Video Lectures(264). [Google Scholar]
- 14.van der Maaten LHG. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 15.Fong RVA. Interpretable explanations of black boxes by meaningful perturbation. Proceedings of the IEEE International Conference on Computer Vision. 2017:3429–3437. [Google Scholar]
- 16.Shi Y, Chen L, Chen T, et al. A Meta-analysis of Voxel-based Brain Morphometry Studies in Obstructive Sleep Apnea. Scientific reports. Aug 30 2017;7(1):10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canessa N, Castronovo V, Cappa SF, et al. Obstructive sleep apnea: brain structural changes and neurocognitive function before and after treatment. American journal of respiratory and critical care medicine. May 15 2011;183(10):1419–1426. [DOI] [PubMed] [Google Scholar]
- 18.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. Journal of digital imaging. Aug 2017;30(4):449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L, Liu G, Luo H, et al. Aberrant Hippocampal Network Connectivity Is Associated With Neurocognitive Dysfunction in Patients With Moderate and Severe Obstructive Sleep Apnea. Front Neurol. 2020;11:580408. [DOI] [PMC free article] [PubMed] [Google Scholar]


