Abstract
Viruses generally are defined as lacking the fundamental properties of living organisms in that they do not harbor an energy metabolism system or protein synthesis machinery. However, the discovery of giant viruses of amoeba has fundamentally challenged this view because of their exceptional genome properties, particle sizes and encoding of the enzyme machinery for some steps of protein synthesis. Although giant viruses are not able to replicate autonomously and still require a host for their multiplication, numerous metabolic genes involved in energy production have been recently detected in giant virus genomes from many environments. These findings have further blurred the boundaries that separate viruses and living organisms. Herein, we summarize information concerning genes and proteins involved in cellular metabolic pathways and their orthologues that have, surprisingly, been discovered in giant viruses. The remarkable diversity of metabolic genes described in giant viruses include genes encoding enzymes involved in glycolysis, gluconeogenesis, tricarboxylic acid cycle, photosynthesis, and β-oxidation. These viral genes are thought to have been acquired from diverse biological sources through lateral gene transfer early in the evolution of Nucleo-Cytoplasmic Large DNA Viruses, or in some cases more recently. It was assumed that viruses are capable of hijacking host metabolic networks. But the giant virus auxiliary metabolic genes also may represent another form of host metabolism manipulation, by expanding the catalytic capabilities of the host cells especially in harsh environments, providing the infected host cells with a selective evolutionary advantage compared to non-infected cells and hence favoring the viral replication. However, the mechanism of these genes' functionality remains unclear to date.
Introduction
Viruses are the most abundant biological entities in the biosphere (Baltimore, 1971; Suttle, 2005; Suttle, 2007) and have been the subject of persistent debates as to their place in the domains of life. Active energy metabolism pathways and the autonomous ability to reproduce independently are the most fundamental criteria associated with living cells and therefore distinguish the three domains of life – archaea, bacteria, and eukarya – from viral entities. Specifically, the fundamental hallmark of all living organisms is the self-maintenance of their own functional and structural integrity via the biochemical transformations of environmental resources. (Dupré and O’Malley, 2009).
Viruses originally were defined as filterable agents capable of passing through membrane filters with pore sizes of 0.22 µm. Most viruses possess small genomes that carry only a handful of genes supporting their replication and capsid production in a host cell. Viruses have thus been characterized as ‘molecular genetic parasites’ that take advantage of cellular functions to replicate (Luria, 1959). Generally, once viruses find their replication site they hijack the host cellular metabolic pathways to their own advantage with the ultimate goal to replicate (Villarreal, 2005). However, ambiguity regarding the definition of viruses appears in the concept of a ‘virocell’, defined by Forterre, 2013 as a ‘living form’ of the virus, consisting of a living cell transformed to produce more virions (Forterre, 2013). At the blurred boundaries between viruses and cellular organisms, intracellular bacteria such as Rickettsia and Chlamydia also are known to hijack host energy metabolism enabling their persistence and proliferation (Driscoll et al., 2017; Gehre, 2015), a characteristic shared with viruses. Ultimately, however, viruses lack many of the essential attributes of living organisms, such as the ability to capture and store free energy, as well as autonomous replication and self-repairing capacity (Dupré and O’Malley, 2009; Regenmortel, 2010).
In 2003, the discovery of the giant viruses of amoeba revolutionized the view of viruses (La Scola et al., 2003). Specifically, the size of the giant virus particles (reaching up to 2.5 μm) and of their genome sizes (up to 2.5 Mb), encoding almost a thousand proteins, fundamentally changed the landscape of virus research (Philippe et al., 2013). Notably, Acanthamoeba polyphaga Mimivirus, the first giant virus of amoeba discovered, has a 1.2-Mbp genome, and possesses several genes encoding proteins involved in transcription and translation (Raoult et al., 2004; Raoult and Forterre, 2008b). Phylogenetic analyses have demonstrated that some bacterial and eukaryotic genes found in the Mimivirus genome, and other giant virus genomes, were acquired by horizontal gene transfer from amoebal hosts, and from amoebal bacteria parasites (Moreira and Brochier-Armanet, 2008), although most giant virus genes still have enigmatic origins.
Giant viruses represent a group of ancient viruses that are termed the Nucleo–Cytoplasmic Large DNA Viruses (NCLDVs). While some are found in amoebae, other of these viruses infect hosts ranging from algae to animals and form an apparently monophyletic group in the phylum Nucleocytoviricota (Colson et al., 2013). The giant viruses are classified into several families: Mimiviridae, Pithoviridae, Pandoraviridae, Phycodnaviridae, Poxviridae, Iridoviridae, Asfarviridae, and others. Many, including newly discovered viruses in the Mollivirus genus are not yet classified (Abergel et al., 2015; Legendre et al., 2014; Rolland et al., 2019).
The genomes of giant viruses encode proteins that had never before been identified in viruses. These proteins have sequence identity with closely related eukaryotic homologs (Chelkha et al., 2020; Sun et al., 2020). For example, Cafeteria roenbergensis virus, Klosneuviruses and Tupanviruses encode several gene products that are involved in protein translation (Abrahão et al., 2018; Schulz et al., 2017); the presence of such translation machinery had not been previously described in viruses. This finding suggested that those viruses may have relative independence from their hosts (and hence are ‘quasi-autonomous’ viruses) (Claverie and Abergel, 2010). This has led to a reconsidering of the historical parasitic hallmark of viruses (Abrahão et al., 2017; Raoult and Forterre, 2008a). In addition to the translation components, giant virus gene products have been implicated in RNA maturation, DNA maintenance, and proteostasis (Fischer et al., 2010; Yutin et al., 2013). Recently, defined metabolic genes involved in specialized cellular biochemical pathways have also been discovered in some giant viruses (Aherfi et al., 2022; Moniruzzaman et al., 2020; Schulz et al., 2020; Schvarcz and Steward, 2018). Herein, we analyze and describe giant virus genes encoding enzymes that are potentially involved in traditional biochemical pathways, findings that challenge the last distinguishing barriers separating the traditional domains of life and the giant viruses.
Carbohydrate metabolism
Carbohydrates are key enzymatic substrates for many essential metabolic pathways in cells (Maughan, 2009). Generally, autotrophic organisms biosynthesize carbohydrates from carbon dioxide and water, via photosynthesis (Butterworth, 2005; Lehninger et al., 2004; Nelson and Cox, 2017). All organisms then break down stored or consumed carbohydrates in order to produce energy for cell functioning (Nelson and Cox, 2017). Both autotrophic and heterotrophic organisms temporarily store their generated energy in the form of high-energy molecules, for example adenosine triphosphate (ATP) and reduced nicotinamide adenine dinucleotide (NADH), for use in the various metabolic processes (Sanders, 2016).
Although some viruses in the Nucleocytoviricota phylum infect known hosts such as amoeba, it is not clear if these are actually the specific hosts. In any case, auxiliary metabolic genes (AMG) encoded by giant viruses appear to exercise control and modulation of host cellular energetic metabolism, especially during viral genome replication, gene expression, and virion assembly (Sun et al., 2020). Such control mechanisms can be based on transcriptional regulation of nuclear and mitochondrion-encoded genes that are involved in energy metabolism, as observed with coccolithoviruses (Ku et al., 2020). Single cell transcriptomics of the microalga Emiliania huxleyi during infection by its specific giant virus showed that either host or viral transcriptome dominated the generated mRNA pool, and that only very few cells were observed in intermediate states. The rapid shutdown of host transcript generation indicates that the E. huxleyi coccolithovirus massively takes over controlling synthesis of almost the entire mRNA pool.
Many giant viral genes and gene products related to energy production and metabolic pathways have been identified recently (Aherfi et al., 2022; Ha et al., 2021; Moniruzzaman et al., 2020; Figure 1). In a detailed study examining a database of 2436 annotated Nucleocytoviricota, including metatranscriptomic data from California’s surface waters, hundreds of viruses, mainly from the Mimiviridae and Phycodnaviridae families, were identified. Many expressed viral transcripts in this dataset are genes involved in glycolysis (e.g. phosphofructokinase [EC 2.7.1.11], glyceraldehyde 3-phosphate dehydrogenase [EC 1.2. 1.12], phosphoglycerate mutase [EC 5.4.2.11]), gluconeogenesis (e.g. phosphoenolpyruvate carboxykinase [EC 4.1.1.32] and fructose-1,6-bisphosphatase [EC 3.1.3.11]), the tricarboxylic acid (TCA) cycle (e.g. succinate dehydrogenase [EC 1.3.5.1]), and the pentose phosphate pathway (e.g. 6-phosphogluconate dehydrogenase EC 1.1.1.1; Ha et al., 2021; Ha et al., 2021).
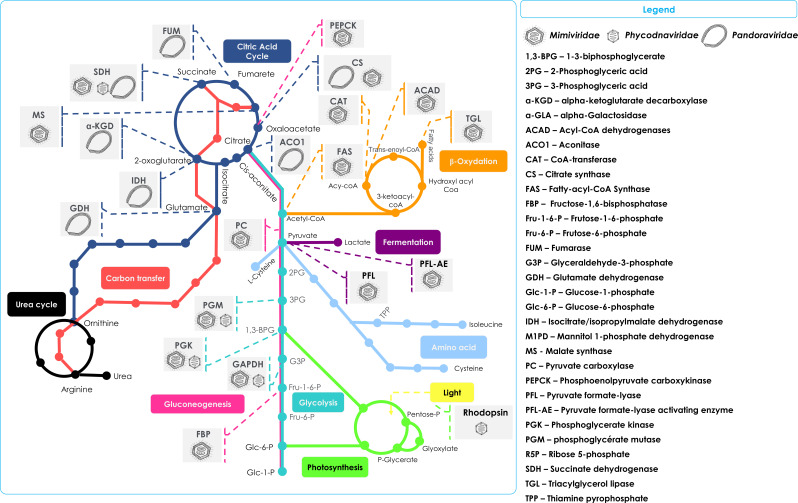
Figure 1. Schematic representation of the metabolic enzymes and pathways detected in NCLDVs.
Schematic of the TCA cycle (dark blue) feeding into the Urea cycle (black); Carbon transfer (red); Gluconeogenesis (scarlet); Glycolysis (marine); Photosynthesis (green); Amino acid metabolism (blue); Fermentation (purple); and lipid β-oxidation (orange). Also shown in the Legend are the identified cellular enzymes and putative substrates which have been identified as being encoded in specific NCLDV genomes, here represented by Mimiviridae, Phycodnaviridae, and Pandoraviridae.
Glycolysis and gluconeogenesis
Glycolysis is an ancient metabolic pathway that occurs in the cytoplasm of the cell and converts glucose into pyruvate, which is accompanied by transfer of electrons to NAD+ in order to generate NADH2 (Potter and Fothergill-Gilmore, 1993; Kumari, 2018). Inversely, gluconeogenesis is a process occurring in animals, fungi, plants, and bacteria, which results in the generation of glucose from non-carbohydrate carbon substrates such as lactate, amino acids, and glycerol (Rodwell et al., 2015).
A genomic study using environmental metagenome-assembled genomes (MAGs), identified genes encoding the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate mutase, and phosphoglycerate kinase. These genes were considered to be particularly prevalent within the Mimiviridae (Figure 2A) and to a lesser extent in the Phycodnaviridae (Moniruzzaman et al., 2020). Studies of Mimivirus metagenomes identified a unique protein domain architecture, with a single fused gene encoding the enzymes glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase, which serve in two adjacent steps in glycolysis (Moniruzzaman et al., 2020). Such protein architecture is unique thus far and may reflect a more efficient process for enzymatic activity. Although this possibility requires experimental verification, it nevertheless highlights that giant viruses may drive evolutionary innovations of their metabolic genes.
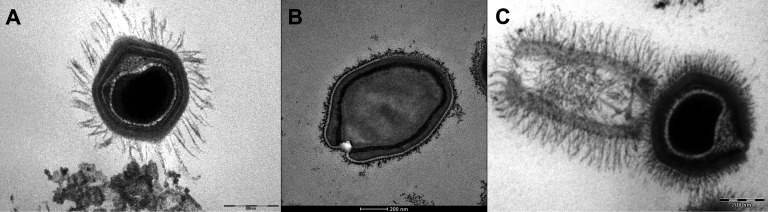
Figure 2. Transmission electron microscopy images of Mimivirus (A), Pandoravirus massiliensis (B) and Tupanvirus (C).
(A) Mimivirus particle is composed of an external layer of dense fibers surrounding an icosahedral capsid and an internal membrane sac enveloping the virus genomic material. (B) Pandoravirus massiliensis virion is ovoid-shaped with an ostiole-like apex, measuring 1.0 μm in length and 0.5 μm in diameter. (C) Tupanvirus exhibits an icosahedral capsid similar to those of Mimivirus measuring ~450 nm. However, Tupanvirus virion harbors a large cylindrical tail (550 nm extension;~450 nm diameter, including fibrils) attached to the base of the capsid. Electron micrographs were acquired on a Tecnai G2 transmission electron microscope (Scale bar, 200 nm).
This same study revealed that three genes associated with gluconeogenesis were present in Mimiviridae genomes (Moniruzzaman et al., 2020): fructose 1,6-bisphosphatase, which catalyzes the biotransformation of fructose-1,6-bisphosphate into fructose-6-phosphate; phosphoenolpyruvate carboxykinase that converts oxaloacetate to phosphoenolpyruvate; and pyruvate carboxylase that catalyzes the carboxylation of pyruvate to form oxaloacetate (Figure 1, Table 1). The presence of such genes in infecting viruses also suggests there could be reprogramming of the host glycolysis pathways.
Table 1. List of metabolic enzymes detected in NCLDVs.
The enzymes were grouped according to the metabolic pathway to which they belong and associated with the giant virus and/or the family in which they were identified.
| Pathway | Enzyme | Function | KEGG* | Detected in | Family | Reference(s) |
|---|---|---|---|---|---|---|
| Amino acid catabolism | Glutamate dehydrogenase | Reversible conversion of glutamate to α-ketoglutarate and ammonia | R00243 | Pandoravirus and others uncharacterized viruses† | Mimiviridae, Pandoraviridae and Phycodnaviridae |
Moniruzzaman et al., 2020; Aherfi et al., 2018 |
| Glutamine synthetase | Condensation of glutamate and ammonia to form glutamine: | R00253 | Uncharacterized viruses† | Mimiviridae | Ha et al., 2021 | |
| Glutaminase | Hydrolysis of glutamine into glutamate | R00256 | Uncharacterized viruses† | Mimiviridae |
Moniruzzaman et al., 2020; Ha et al., 2021 |
|
| Lipide catabolism and β-Oxydation | Triacylglycerol lipase | Degrades triacylglycerol into glycerol and fatty acids | R01369 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 |
| Fatty-acyl-CoA Synthase | Conversion of a acetyl-CoA and seven malonyl-CoA molecules to produce a Palmitoyl-CoA | R05190 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| CoA-transferase | Conversion acyl-CoA and acetate into fatty acid anion and acetyl-CoA. | R00393 | Prymnesium kappa virus RF01 | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| Acyl-CoA dehydrogenase | Desaturation of the acyl-CoA esters | R00392 | Prymnesium kappa virus RF01 and others uncharacterized viruses† | Mimiviridae | Blanc-Mathieu et al., 2021 | |
| Citric Acid Cycle | Succinate dehydrogenase | Conversion of succinate into fumarate | R02164 | Prymnesium kappa virus RF01, Pandoravirus massiliensis and others uncharacterized viruses† | Mimiviridae, Pandoraviridae and Phycodnaviridae | Moniruzzaman et al., 2020; Aherfi et al., 2022; Blanc-Mathieu et al., 2021; Ha et al., 2021; |
| Citrate synthase | Claisen condensation between acetyl CoA and oxaloacetate to yield, after hydrolysis of the thioester bond, citrate and CoA | R00351 | Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Aherfi et al., 2022; Moniruzzaman et al., 2020 | |
| Aconitase | Catalyzes the stereospecific isomerization of citrate to isocitrate via cis-aconitate in a non-redox reaction | R01324 | Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Moniruzzaman et al., 2020; Rodrigues et al., 2019; Aherfi et al., 2022 | |
| Isocitrate/isopropyl malate dehydrogenase | Oxidative decarboxylation of isocitrate, resulting in alpha-ketoglutarate and carbon dioxide. |
R00267 / R01652 |
Pandoravirus massiliensis and others uncharacterized viruses† | Pandoraviridae and Mimiviridae | Aherfi et al., 2022; Moniruzzaman et al., 2020 | |
| Malate synthase | Conversion of enzyme are acetyl-CoA, H2O, and glyoxylate into (S)-malate and CoA. | R00472 | Uncharacterized viruses† | Mimiviridae | Ha et al., 2021 | |
| Alpha-ketoglutarate decarboxylase | Conversion of α-ketoglutarate to succinyl-CoA and produces NADH directly providing electrons for the respiratory chain | R00272 | Pandoravirus massiliensis | Pandoraviridae | Aherfi et al., 2022 | |
| Fumarase | Conversion of fumarate to L-malate | R01082 | Pandoravirus massiliensis | Pandoraviridae | Aherfi et al., 2022 | |
| Fermentation | Pyruvate formate-lyase | Catalyzes the reaction of pyruvate +CoA acetyl-CoA +formate | R00212 | Tetraselmis virus | Phycodnaviridae | Müller et al., 2012; Schvarcz and Steward, 2018; Sun et al., 2020 |
| Formate-lyase activating enzyme | Converts pyruvate and CoA into acetyl CoA and formate | R04710 | Tetraselmis virus | Phycodnaviridae | Müller et al., 2012; Schvarcz and Steward, 2018 | |
| Gluconeogenesis | Fructose bisphosphatase | Converts fructose-1,6-bisphosphate to fructose 6-phosphate | R00762 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020; Ha et al., 2021 |
| Phosphoenolpyruvate carboxykinase | Converts oxaloacetate into phosphoenolpyruvate and carbon dioxide. | R00341 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020; Ha et al., 2021 | |
| Pyruvate carboxylase | Catalyzes the conversion of pyruvate to oxaloacetate | R00344 | Uncharacterized viruses† | Mimiviridae | Moniruzzaman et al., 2020 | |
| Glycolysis | Glyceraldehyde-3-phosphate dehydrogenase | Conversion of pyruvate to oxaloacetate | R01061 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020 |
| Phosphoglycerate mutase | Transfers the phosphate from 3-phosphoglyceric acid (3 PG) to the second carbon to form 2-phosphoglyceric acid (2 PG) | R01518 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020; Ha et al., 2021 | |
| Phosphoglycerate kinase | Catalyzes the formation of ATP from ADP and 1,3-diphosphoglycerate | R01512 | Uncharacterized viruses† | Mimiviridae and Phycodnaviridae | Moniruzzaman et al., 2020 | |
| Photosynthesis | Rhodopsin | Generating a proton motive force across the cell membrane (light dependent) | R02903 | Organic Lake Phycodnavirus 2 and Phaeocystis globosa virus | Phycodnaviridae | Needham et al., 2019; Yutin and Koonin, 2012; Schulz et al., 2020 |
| Mannitol metabolism | Mannitol 1-phosphate dehydrogenase | Converts D-mannitol 1-phosphate and NAD +into fructose 6-phosphate, NADH and H+. | R00758 | Tetraselmis virus | Phycodnaviridae | Schvarcz and Steward, 2018 |
| Saccharide degradation | Alpha-galactosidase | Catalyzes the removal of terminal α-galactose groups from substrates such as glycoproteins and glycolipids | R01101 | Tetraselmis virus | Phycodnaviridae | Schvarcz and Steward, 2018 |
KEGG codes for the biochemical reactions described (https://www.genome.jp/kegg/reaction/).
Enzymes detected in NCLDVs from metagenome-assembled genome analysis.
Fermentation
Fermentation is a heterotrophic anaerobic redox process that uses an organic compound as the terminal electron acceptor (Müller et al., 2012; Zhou et al., 2017). It is considered as an ancient metabolic pathway. Genes encoding for two key fermentation genes – pyruvate formate-lyase and pyruvate formate-lyase activating enzyme – have been found in Tetraselmis virus (TetV) (Chlorodendrephycae) (Figure 1, Table 1), a mimivirus infecting the green algae Tetraselmis and land plants of the lineage Viridiplantae Müller et al., 2012. This viral host utilizes anaerobic energy metabolism in low-oxygen conditions, suggesting horizontal gene transfer (Müller et al., 2012; Schvarcz and Steward, 2018; T.-W. Sun et al., 2020, p. 1). Pyruvate formate-lyase catalyzes the reversible conversion of pyruvate and coenzyme-A into formate and acetyl-CoA (Knappe et al., 1974). It is thought that in low oxygen conditions the host may use these viral genes to generate the required energy via manipulation of anaerobic energy metabolism. In the absence of light, algae and bacteria deplete dissolved oxygen in the superficial water. In anoxic conditions, the fermentation process thus may favor viral spreading, with the metabolism of infected cells being potentially maintained by virally encoded fermentation genes, an advantage for the viral host (Schvarcz and Steward, 2018).
Other putative viral fermentation genes that have been identified include the mannitol metabolism protein, mannitol 1-phosphate dehydrogenase and the saccharide degradation enzyme alpha-galactosidase in the Tetraselmis virus (Table 1). However, the role of these genes in the virus is less clear (Schvarcz and Steward, 2018).
Tricarboxylic acid cycle
The tricarboxylic acid (TCA) cycle – also known as the Krebs cycle – is a linked series of biochemical reactions used by all aerobic organisms to release stored energy via the oxidation of acetyl-CoA, derived from carbohydrates, fats, and proteins (Smith and Morowitz, 2004). It is the central metabolic hub for many biochemical pathways. It begins from acetate (in the form of acetyl-CoA) and water, to reduce NAD +into NADH, with the concomitant release of carbon dioxide (Krebs and Johnson, 1937; Meléndez-Hevia et al., 1996; Noor et al., 2010). The NADH generated by this cycle enters the oxidative phosphorylation (electron transport) pathway. As a result of these two closely linked metabolic pathways, several molecules of ATP are produced.
In the previously cited study based on MAGs, predicted TCA-related gene products were the most represented, especially aconitase and succinate dehydrogenase, encoded in Mimiviridae and Phycodnaviridae genomes (Moniruzzaman et al., 2020). Aconitase was previously reported in Tupanvirus as well (Rodrigues et al., 2019; Figures 1 and 2.C Table 1). In cells, aconitase catalyzes the stereospecific isomerization of citrate to isocitrate via cis-aconitate in a non-redox reaction. Succinate dehydrogenase is a complex enzyme that catalyzes the conversion of succinate into fumarate, which generates electrons used to reduce oxygen into water (Oyedotun and Lemire, 2004). The viral TCA enzymes may be utilized to boost host TCA cycle steps and hence energy production, possibly providing competitive advantages to the host.
The alga-infecting giant virus Prymnesium kappa virus RF01, which belongs to the Mimiviridae family, harbors six energy metabolism gene homologs (Blanc-Mathieu et al., 2021). Among them, there are the four succinate dehydrogenase subunits (A-D), an enzyme involved in oxidative phosphorylation pathway and the TCA cycle (Figure 1, Table 1). The gene encoding subunit A of this complex protein is transcribed during infection, again intimating that host energy metabolism could be modulated by the virus (Blanc-Mathieu et al., 2021). Putative succinate dehydrogenase genes detected by metatranscriptomic analysis were associated with Mimiviridae, suggesting the gene product to be highly expressed in the virosphere of surface oceanic waters of California (USA) (Ha et al., 2021), and may be widespread among marine Mimiviridae (Blanc-Mathieu et al., 2021). In the same study, malate synthase and isocitrate lyase, other key enzymes of the TCA cycle, were found expressed in different time points in viruses in the Mimiviridae (Ha et al., 2021; Figure 1, Table 1).
Several predicted proteins recently found in Pandoravirus massiliensis (Figure 2B) are homologues of enzymes involved in the TCA cycle, albeit with low % identities: citrate synthase, aconitase, isocitrate/isopropyl malate dehydrogenase, alpha-ketoglutarate decarboxylase, succinate dehydrogenase, and fumarase (Figure 1, Table 1). The predicted isocitrate dehydrogenase of P. massiliensis was functional both in crude viral particles and following in vitro reconstitution of the purified recombinant protein. In addition, it was also demonstrated experimentally that P. massiliensis can generate an electrochemical gradient, an essential component of all living cells (Aherfi et al., 2022). The membrane voltage in P. massiliensis may be involved in the amoeba infection process, notably in the early stages of infection (Aherfi et al., 2022), helping the virus to release its DNA into host cells. Besides, it has been shown previously that the inhibition of potassium ion channels of Chlorella viruses, proteins related to membrane voltage, causes the depolarization of host cells and consequently the inhibition of viral DNA release into the host cell (Frohns et al., 2006; Neupärtl et al., 2008). Whether similar mechanisms are involved in P. massiliensis is unknown.
These findings potentially suggest that these viruses use such genes either to produce energy autonomously, or to stimulate the host energy metabolism in the aim to confer a replicative advantage to infected cells. Further investigations such as expression of these genes, followed by enzymatic activity assays are needed for a detailed comprehension of these enzymes.
Lipids
Lipid metabolism consists of the catabolism of fatty acids and steroids/sterols in order to generate energy, and anabolic processes used to synthesize new lipids from smaller constituent molecules. Generally, lipids classified as fatty acids, triacylglycerols, phospholipids, sterols and sphingolipids are utilized in energy generation. Lipidomic β-oxidation is the main metabolic pathway for the degradation of fatty acid molecules to generate acetyl-CoA, which then enters in the TCA cycle.
A recent metatranscriptomic time-series study from the California Current surface waters has identified many novel transcripts putatively for lipid metabolism enzymes in viruses in the Nucleocytoviricota. Identified viral genes included those predicted to be involved in lipid β-oxidation (Ha et al., 2021). Genes encoding acyl-CoA dehydrogenases (EC 1.3.8.7), which catalyze the first committed enzymatic step of lipid β-oxidation, were consistently expressed across sampled timepoints, especially in the Mimiviridae. This finding again suggests that virus-mediated reprogramming of host central carbon metabolism is functional and occurs in oceanic surface waters (Ha et al., 2021).
In Prymnesium kappa virus RF01 (PCV RF01), numerous genes were found that are predicted to encode enzymes known to be involved in cellular lipid metabolism, such as a triacylglycerol lipase, which degrades triacylglycerol into glycerol and fatty acids (Figure 1, Table 1). These metabolites are used as precursors for ATP production via glycolysis and β-oxidation, respectively. In addition, it has been found that PCV RF01 encodes the key β-oxidation enzymes fatty acyl-CoA synthetase (EC 6.2.1.1), CoA-transferase (EC 2.8.3.8), and acyl-CoA dehydrogenase (EC 1.3.8.7) (Blanc-Mathieu et al., 2021). However, enzymes involved in the two intermediate steps immediately following each oxidation, enoyl-CoA hydratase (EC 4.2.1.17) or a β-ketothiolase (EC2.3.1.16), have not been detected in any giant virus to-date.
Amino-acids / proteins
In cells, protein and/or amino acid utilization as a source of energy release mainly occurs when the intake of carbohydrates or lipids is insufficient to supply the required energy demand (Hothersall and Ahmed, 2013). In prokaryotes, and in mitochondria, glutamate dehydrogenase (EC 1.4.1.2) catalyzes the reversible conversion of glutamate to α-ketoglutarate and ammonia while reducing NAD(P)+to NAD(P)H. The α-ketoglutarate is consumed by the TCA cycle to produce ATP (Glevarec et al., 2004; Miller and Magasanik, 1990; Plaitakis et al., 2017). MAGs studies of members of multiple NCLDV families report the presence of putative glutamate dehydrogenase, glutamine synthase (EC 6.3.1.2), and glutaminase (EC 3.5.1.2) enzymes (Moniruzzaman et al., 2020). Other analyses confirmed the presence of glutamate dehydrogenase in members of the Pandoraviridae family (Figure 1, Table 1; Aherfi et al., 2018; Hosokawa et al., 2021; Legendre et al., 2018; Philippe et al., 2013).
Glutaminase and glutamine synthase, two cellular enzymes that regulate primary energy metabolism, found in Mimiviridae were expressed upon infection (Ha et al., 2021). The glutaminase catalyzes the hydrolysis of glutamine into glutamate, regulating cellular energy metabolism via the increase of glutamate and α-ketoglutarate. This results in enhanced mitochondrial respiration and ATP generation (Campos-Sandoval et al., 2007; Hu et al., 2010). Glutamine synthetase plays a key role in the cellular utilization of carbon and nitrogen sources, helps modulate the energy budget in bacteria and mediates the release of energy stored in glutamine, that increases especially in stressed cells (Aldarini et al., 2017). Thus, the presence of such glutaminolysis enzymes in giant viruses may help reprogram metabolism by maximizing glutamine catabolism and thus increase available energy to promote virus replication and virion production (Ha et al., 2021).
Energy from inorganic compounds including photosynthesis
Energy from inorganic compounds
Chemolithotrophy is a metabolic process whereby energy is derived from the oxidation of inorganic compounds such as hydrogen (Friedrich and Schwartz, 1993), reduced sulfur compounds (Friedrich, 1997), hydrogen sulfide, thiosulfate, ferrous iron, and ammonia (Jetten et al., 1998). Microbial oxidation of inorganic compounds is governed by chemical and enzymatic reactions to generate energy (ATP) and reducing power (NADH).
The most common chemotrophic organisms that oxidize inorganic compounds are prokaryotic. Nonetheless, diverse deep-sea viruses including members of the Podoviridae, Siphoviridae, and Myoviridae, have been reported to contain genes putatively encoding the α and γ subunits of the reverse-acting dissimilatory sulfite reductase (Rdsr), an enzyme that oxidizes the element sulfur. It is assumed that this gene is used to maintain or augment host cellular processes during infection and to redirect energy and resources towards viral production (Anantharaman et al., 2014).
A recent metagenomic study found that predicted ferric reductase enzymes were encoded in several NCLDV genomes (Schulz et al., 2020). In cellular organisms, ferric reductases function as a terminal reductase in an electron transport chain, by reducing ferric ion Fe3+ into ferrous ion Fe2+. Ferric reductase enzymes are also critical for the assimilatory iron pathway in organisms (Lovley, 2002; Lovley and Coates, 2000; Lovley et al., 1998). The reduction of ferric iron combined with a proton gradient through the cell membrane, is used by membrane-bound ATP synthase to generate the ATP (Schröder et al., 2003). This proton gradient can also be used to reduce NAD(P)+ in chemolithotrophs for several biosynthetic reactions (Lovley, 2002; Lovley and Coates, 2000). The presence of these enzymes in giant viruses and their functioning biochemistry may support infected host cell(s) metabolism and confer on them a competitive advantage in suboxic environments (Márquez et al., 2007). Moreover, these enzymes may also play a significant role in modifying the composition of the surrounding chemical environment, which can impact other microorganisms, most notably those that use iron respiration (Schulz et al., 2020).
Photosynthesis
Photosynthesis is arguably the most important biological process functioning in nature, and is responsible for the existence of most life on Earth. It is essential for producing and maintaining the oxygen content of the atmosphere and supplies most of the energy necessary for life on the planet (Bryant and Frigaard, 2006). Photosynthesis is a pivotal process used by many autotrophic organisms, enabling them to convert light energy into chemical energy which is stored in the form of carbohydrates synthesized from carbon dioxide and water (Blankenship, 2010; Olson, 2006). Subsequently, the carbohydrates can be released to fuel the organism’s cellular metabolic activities. However, viral infections can potentially affect and/or redirect metabolic pathways in photosynthetic host organisms. For example, infection with a mimivirus has been shown to suppress transcripts related to photosynthesis as well as cytoskeleton formation in brown-tide forming pelagophyte Aureococcus anophagefferens (Moniruzzaman et al., 2018; see below).
In marine environments, viruses are the most abundant biological entities and drive organism population control by consistently impacting nutrient and biogeochemical cycling in these ecosystems (Hurwitz et al., 2013; Juneau et al., 2003; Suttle, 2016; Villarreal and Witzany, 2010). Marine viruses also affect algal blooms and their diversity as well as species distribution (Juneau et al., 2003). Marine viral genes that encode for numerous proteins involved in photosynthetic biology have already been found and characterized, especially in cyanophages (Alperovitch-Lavy et al., 2011; Coutinho et al., 2017; Lindell et al., 2005; Sharon et al., 2009; Sullivan et al., 2006). These proteins include photosystems I and II that drive the complete photosynthetic process during phage infection, with the overall effect of promoting maximal phage replication (Fridman et al., 2017).
Most NCLDV groups appear to be present ubiquitously in marine environments, based on metagenomics (Endo et al., 2020). The majority of these are closely related to the families Mimiviridae and Phycodnaviridae: of the more than 6,700 polymerase B genes (PolB; a conserved marker of NCLDVs) assembled from the Tara Oceans dataset, 5091 were related to Mimiviridae and 981 to Phycodnaviridae (Endo et al., 2020). However, other studies suggest that certain NCLDVs may be endemic to the regions in which they were identified (Needham et al., 2019), and there is considerable undiscovered diversity in marine giant viruses.
Many genes detected in these reassembled NCLDV genomes were predicted to encode for proteins with putative roles in photosynthesis, suggesting that reprogramming host metabolism may be a common phenomenon employed by NCLDVs (Schulz et al., 2020). Giant viruses thus may fundamentally impact photosynthetic processes in marine and freshwater protists (Needham et al., 2019; Endo et al., 2020; Moniruzzaman et al., 2017; Short, 2012). However, that NCLDV apparently endogenize into various green algae, so the metagenome assemblies may not be accurate in this case (Moniruzzaman et al., 2020).
Another example of the biochemical consequences of viral infections in marine ecosystems is represented by a recent transcriptome study, providing new insights regarding transcriptional remodeling in Aureococcus cells when infected by the giant virus Aureococcus anophagefferens. Immediately after A. anophagefferens infection, Aureococcus cellular genes related to light harvesting, photosystem structure, and isoprenoid biosynthesis are down-regulated while porphyrin biosynthesis genes are up-regulated (Moniruzzaman et al., 2018). This over-expression, by increasing intracellular porphyrin concentrations in infected cells, may increase cellular oxidative stress and may represent a host defense mechanism against the viral attack.
The choanoviruses in the phylum Nucleocytoviricota parasitize choanoflagellates, protistan predators. Choanovirus genomes assembled from metagenomic sequences encode the complete rhodopsin-based photosystem (Needham et al., 2019). This virally-induced light-driven energy transfer is closely connected with host ATP synthases and elegantly illustrates how giant viruses can modulate and alter nutrition and use of organic compounds by their unicellular eukaryotic hosts in marine environments (Needham et al., 2019). The photosystem genes in the choanoviruses may be the result of unique (or multiple) horizontal gene transfers events (Needham et al., 2019).
In another example of viral manipulation of host primary metabolism, genes encoding for proteorhodopsin were identified in the genomes of NCLDV Organic Lake Phycodnavirus (OLPV) 2 and Phaeocystis globosa virus (PGV) (Yutin and Koonin, 2012). Proteorhodopsins are photoreceptors found in marine planktonic bacteria, archaea, and eukaryotes (Béjà et al., 2000; Buhr et al., 2015; Frigaard et al., 2006; Lin et al., 2010; Slamovits et al., 2011). Metagenomic studies of extended sampling of NCLDV genomes revealed further that NCLDVs encode a large panel of diverse rhodopsins, representing one quarter of the current total diversity of rhodopsin proteins known to date (Schulz et al., 2020). Their role as sensory rhodopsins may complement the host’s rhodopsin function, or conversely confer a new functionality to the host. It has been suggested that their expression may induce host phototaxis, and stimulate host relocation to nutrient-rich areas that are necessary for viral replication (Yutin and Koonin, 2012). The origins of viral proteorhodopsin genes remain unknown, but they may have been captured from their unicellular eukaryotic hosts. Phylogenetic analyses based on sequences retrieved from metagenomic data argue for a monophyletic group formed by NCLDV rhodopsins, suggesting that these genes represent an ancestral trait shared by these viruses and which were then subsequently lost (Schulz et al., 2020). Furthermore, genes encoding putative carotenoid oxygenases have also been detected in these metagenomes. Viral carotenoid oxygenases may have the ability to modulate the host’s capacity to capture light and/or synthesize bioactive compounds. Thus, they may act in association with rhodopsins in order to provide metabolic advantages to infected populations (Schulz et al., 2020).
Others
In addition to primary metabolism energetic enzymes described above, a remarkable diversity of auxiliary metabolic genes involved in (i) carbon metabolism, nitrogen and nutrient cycling and (ii) soil organic matter degradation, have been found in marine viral communities (Table 2). It is hypothesized that these metabolic genes can complement deficient host metabolic pathways in order to sustain their host under environmental stressful conditions, with the overall goal to increase or maintain viral replication (Coutinho et al., 2017; Howard-Varona et al., 2020; Hurwitz and U’Ren, 2016). Such host / virus interactions appear to play an important role in the global ecosystem (Brum and Sullivan, 2015; Hurwitz and U’Ren, 2016; Suttle, 2007; Zimmerman et al., 2020).
Table 2. List of enzymes with other biological roles detected in NCLDVs.
The enzymes were grouped according to the biological process to which they belong and associated with the giant virus and/or the family in which they were identified.
| Biological process | Enzyme | Function | Detected in | Family |
|---|---|---|---|---|
| Oxidative stress regulation | Superoxide dismutase | Catalyzes the dismutation of the superoxide radical into ordinary molecular oxygen and hydrogen | Emiliania huxleyi virus, Megavirus chiliensis, and others uncharacterized viruses* | Mimiviridae and Phycodnaviridae |
| Glutathione peroxidase | Reduces free hydrogen peroxide to water. | Emiliania huxleyi virus and others uncharacterized viruses* | Mimiviridae and Phycodnaviridae | |
| Ion’s transport and assimilation | Ammonium transporter | Mediates the transport of ammonium ions | Ostreococcus virus 6 | Phycodnaviridae |
| Phosphate transporter | Mediates the transport of phosphate ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Sulfur transporter | Mediates the transport of sulfur ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Magnesium transporter | Mediates the transport of magnesium ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Iron transporter | Mediates the transport of iron ions | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Ferritin | Iron storage protein | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Ferric reductases | Oxidation of NADPH and transference the electron to reduce metals like iron and copper | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Multicopper oxidases | Oxidation of different substrates by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre. | Uncharacterized viruses | Mimiviridae and Phycodnaviridae | |
| Biosynthesis of glycosphingolipids | Serine palmitoyltransferase | Catalyzes the decarboxylative condensation of L-serine and palmitoyl coenzyme A to 3-ketodihydrosphingosine. | Coccolitho virus | Phycodnaviridae |
| Polysaccharide biosynthesis | Hyaluronan synthase | Produces the glycosaminoglycan hyaluronan from UDP-α-N-acetyl-D-glucosamine and UDP-α-D-glucuronate | Chlorovirus CVK2 | Phycodnaviridae |
| Chitin synthase | Produces Uridine diphosphate (UDP) and [[[1,4-(N-acetyl-beta-D-glucosaminyl)]n+1]] from UDP-GlcNAc and [[[1,4-(N-acetyl-beta-D-glucosaminyl)]n]] | Chlorovirus CVK2 | Phycodnaviridae | |
| Sugar metabolism | GDP-D-mannose 4,6 dehydratase | Conversion of GDP-(d)-mannose to GDP-4-keto, 6-deoxy-(d)-mannose | Paramecium bursaria Chlorella virus 1 | Phycodnaviridae |
| GDP-4-keto-6-deoxy-D-mannose epimerase/reductase | Converts GDP-4-keto-6-deoxy-d-mannose into GDP-l-fucose | Paramecium bursaria Chlorella virus 1 | Phycodnaviridae | |
| Polysaccharides degradation | Chitinase | Chitin degradation by cleaves the disaccharide to its monomer subunits | Chlorella virus PBCV-1 | Phycodnaviridae |
| 1–3-beta glucanase | Successive hydrolysis at the nonreducing end of the glucan, resulting in the formation of oligosaccharides and glucose | Chlorella virus PBCV-1 | Phycodnaviridae | |
| Pectate lyase | Randomly cleaves α–1,4-polygalacturonic acid via a β-elimination reaction | Aureococcus anophagefferens virus | Phycodnaviridae |
Enzyme’s genes were detected in NCLDVs from metagenome-assembled genome analysis.
Other gene products have been implicated in the reprograming of cellular physiology during giant virus infection, especially in nutrient processing and oxidative stress (Monier et al., 2017; Moniruzzaman et al., 2018; Sheyn et al., 2016). For example, it has been reported that Mimiviridae and Phycodnaviridae families exhibit superoxide dismutase and glutathione peroxidase activities encoded by crucial enzymes involved in oxidative stress regulation (Table 2). A superoxide dismutase encoding gene, involved in the breakdown of reactive oxygen species (ROS), was found encoded in the Megavirus chiliensis genome (Lartigue et al., 2015). These enzymes likely help to protect the viral replication machinery from damage by ROS generated during viral infection (Moniruzzaman et al., 2018; Moniruzzaman et al., 2020).
Sheyn et al., 2016. have shown that during lytic infection by Emiliana huxleyi virus, which infects the cosmopolitan unicellular eukaryotic algal host Emiliania huxleyi, glutathione was overproduced and that hydrogen peroxide (H2O2) was the major ROS during the onset of the lytic phase of infection. Moreover, the concomitant production of GSH and H2O2 occurred in the same cellular subpopulations that exhibited a higher rate of infection compared with cells that had little or no GSH and H2O2. Interestingly, the inhibition of ROS production significantly reduced virion production and inhibited host cell death (Sheyn et al., 2016).
Ostreococcus virus 6, a virus of the green alga Ostreococcus tauri, harbors a gene encoding an ammonium transporter that is expressed during viral infection. The activity of this protein may increase host NH4 +uptake rates to fulfill the increased nitrogen requirements of infected cells undergoing viral replication (Monier et al., 2017). Moreover, numerous genes that encode for phosphate, sulfur, magnesium and iron transporters, ferric reductases and multicopper oxidases were identified recently in several NCLDV clades (Moniruzzaman et al., 2020; Schulz et al., 2017). These genes may boost the acquisition of these essential nutrients for host cell metabolism during virion production, notably in marine environments where nutrient availability may be limiting for cellular growth (Behrenfeld and Kolber, 1999; Herbik et al., 2002; Hogle et al., 2018; Saikia et al., 2014).
In addition to enzymes implicated in carbohydrate metabolism for energy generation as described above, some giant virus genes also encode proteins involved in the biosynthesis and manipulation of carbohydrate, lipid, and nucleotide metabolism. Coccolithoviruses, belonging to the family Phycodnaviridae, possess a cluster of biosynthetic genes, including a serine palmitoyltransferase encoding gene, involved in the biosynthesis of glycosphingolipids, a major component of the virion membrane envelopes. High glycosphingolipid producing strains of E. huxleyi virus (EhV), which are extremely virulent and harbor a greater infectivity at high host densities, provide a selective advantage under laboratory conditions. However, field data obtained from natural environments suggest a better survival rate of slow glycosphingolipid producing EhVs, where lower host densities are encountered. Viral glycosphingolipid biosynthesis impacts on ecological balance in natural oceanic environments, where E. huxleyi plays an essential role in the global carbon cycle (Nissimov et al., 2019). Moreover, it has been shown that viral glycosphingolipids are able to suppress host cell growth by inducing programmed cell death (Monier et al., 2009; Rosenwasser et al., 2016; Vardi et al., 2009; Wilson et al., 2005).
Chloroviruses encode enzymes involved in (i) polysaccharide biosynthesis for example hyaluronan synthase, chitin synthase (Mohammed Ali et al., 2005). (b); (ii) sugar metabolism, for example GDP-D-mannose 4,6 dehydratase and GDP-4-keto-6-deoxy-D-mannose epimerase/reductase (Tonetti et al., 2003); and (iii) polysaccharides degradation, for example chitinase and 1–3-beta glucanase (Sun et al., 1999; Sun et al., 2000). These enzymes may be necessary for the infection cycle, for the ability to enter and exit the host cell (Van Etten et al., 2017). Polysaccharide lyases for example, pectate lyase, were also found to be encoded in the A. anophagefferens virus (AaV) genome (Moniruzzaman et al., 2014). It is probable that the genes encoding these putative enzymes were acquired by lateral gene transfer from either bacteria or from their host A. anophagefferens (Moniruzzaman et al., 2014). AaV genes encoding polysaccharide lyases have been mainly detected in ocean sampling throughout the world suggesting that these viruses play important role(s) in shaping the biogeochemical potential, in global marine system communities (Gann et al., 2020).
Analysis of the A. polyphaga Mimivirus genome revealed the presence of genes potentially involved in the biosynthesis of viosamine, which may have a role in the formation of the long fibers surrounding the virions (Piacente et al., 2012). Moreover, several giant viruses harbor the metabolic machinery that is involved in the production of glycoconjugate substrates (Fischer et al., 2010; Philippe et al., 2013; Piacente et al., 2015; Santini et al., 2013). Such genes include those encoding for nucleotide sugars enzymes and glycosyltransferases (Piacente et al., 2014). These viral enzymes may play structural role(s), which seems obvious when considering the highly glycosylated surface of the virions that protect them from the environment. They also may play a role in the phagocytic vacuole (Lairson et al., 2008; Legendre et al., 2014; Legendre et al., 2015; Piacente et al., 2014).
Finally, even steroid metabolism may be manipulated by NCLDV infection. Genomes of many mimiviruses and pandoraviruses contain cytochrome P450 genes, which encode P450 monooxygenase enzymes (Lamb et al., 2019). P450s are key enzymes in the metabolism of numerous endogenous regulatory molecules and xenobiotics in Bacteria, Archaea, and Eukarya. To support this suggestion, multiple genes involved in other aspects of steroid metabolism, notably steroid reductases, are also present and expressed (typically early) during the course of NCLDV infection (De Souza et al., 2021).
Conclusion
Previously, viruses were traditionally defined as molecular genetic parasites, accessories to cellular life, and lacking many of the essential criteria that define living organisms, such as the ability to capture and store free energy. However, this strict paradigm has now been fundamentally challenged by the identification of a large number of energy-linked metabolic genes encoded in some NCLDV genomes, with sequence identity to cellular orthologs, including those possibly involved in energy generation from organic and inorganic compounds. These genes are thought to have been acquired by NCLDV from diverse sources, and especially from their hosts through lateral gene transfer. Notably, a number of the genes considered here were identified in metagenomic studies, which raises a possibility that some of these genes inferred as being in viral genomes might instead represent genes from bacteria, and emphasize the need for viromics benchmarking (Pratama et al., 2021). Indeed, the filtration process to discard non-viral sequences does not completely exclude the possibility of a contamination. Recognizing that caveat, we expect that sequences analysis in such metagenomic studies cited here employed benchmarking and curation approaches sufficient to discriminate between viral and microbial sequences. However, it should be noted that NCLDV apparently endogenize into various green algae, complicating the accurate assignment of partial metagenome assemblies, but also emphasizing the role of HGT in host and virus evolution (Moniruzzaman et al., 2020). Isolation of these viruses by culture and direct sequencing of viral strains combined with further analysis in molecular biology and biochemistry should be a complementary approach to elucidate their presence in the viral genomes.
Phylogenetic analysis reveals that these metabolic genes have diversified into virus-specific lineages. However, the origin of many of these genes still remains obscure, particularly as these NCLDV metabolism sequences tend to cluster together phylogenetically in deep-branching clades. This finding suggests that they diverged from their cellular orthologs in the distant past (Aherfi et al., 2022; Moniruzzaman et al., 2020; Needham et al., 2019; Schvarcz and Steward, 2018). These complex metabolic genes are thought to play a role(s) in manipulating host energy metabolism pathways during infection and help to ensure optimal intracellular host environments required for viral replication. Indeed, viruses are able to reshape the virocell metabolism not only by reprogramming host-encoded metabolic networks, but also by expanding the virocell metabolic abilities / needs by introducing new viral encoded auxiliary metabolic genes. This concept is particularly well illustrated by marine viruses, which play key role(s) in ecology, biogeochemistry and evolution of the marine environment, by impacting nutrient recycling and driving species composition. The massive amounts of viral macromolecules and viral particles that are synthesized in an infected cell impose heavy demands on the host. Biosynthesis of the elements that make up a viral particle, namely nucleotides, amino acids and sometimes fatty acids, requires energy in the form of ATP. The recruitment of cellular compartments involved in the morphogenesis and transport of new viral particles such as the Golgi apparatus and the membrane endoplasmic reticulum also requires energy that cannot totally be provided by host cell metabolism, perhaps particularly in harsher environments. A suggestion of a virus aiding thermal tolerance of a host has been made (Márquez et al., 2007). Therefore, the NCLDV auxiliary metabolic genes may be involved in energy generation by boosting, in either a dependent or independent manner, the energy metabolism enzymes of the host cell to create an appropriate environment for viral replication.
At this time, it is difficult to ascertain whether these viral primary metabolism enzymes function according to the exact mechanisms as their corresponding host orthologs. Further efforts are urgently required to elucidate whether these viral metabolism enzymes function independently. Such findings will help discern if the virus becomes autonomous and does not parasitize the metabolism of the host cell, or whether they are just enzymes that manipulate the host energetic metabolic system. Genetic and biochemical approaches must be considered going forward, including exploring individual viral enzymes functioning in a heterologous background, including complementation of knockout mutants (either in bacteria or yeast) to determine if the viral enzymes modulate host energy production.
The detection of the viral primary metabolic genes (with the potential to be involved in energy production) runs contrary to our traditional view of virus biology. Until recently, viruses have been regarded as parasites of the host cell’s energy machinery rather than encoding their own virus metabolic machinery (Moreira and López-García, 2009) and supporting the proposed virocell concept (Forterre, 2013). The notion of viral manipulation and expansion of the host metabolic network suggests that host cells with highest metabolic activity may be more permissive to viral infection. Metabolic modulation is thus the central hub of the host–virus dynamics. Further research in the coming years will help shed light on such unprecedented findings.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Bernard La Scola, Email: bernard.la-scola@univ-amu.fr.
Sarah Aherfi, Email: sarah.aherfi@ap-hm.fr.
Ron AM Fouchier, Erasmus Medical Center, Netherlands.
Jos W van der Meer, Radboud University Medical Centre, Netherlands.
Funding Information
This paper was supported by the following grants:
Royal Society to David C Lamb.
Woods Hole Center for Oceans and Human Health to John J Stegeman.
National Institutes of Health P01ES021923 to John J Stegeman.
National Science Foundation OCE-1314642 to John J Stegeman.
Agence Nationale de la Recherche "Investments for the Future" program Méditerranée-Infection 10-IAHU-03 to Djamal Brahim Belhaouari, Gabriel Augusto Pires De Souza, Philippe Colson, Sarah Aherfi.
Additional information
Competing interests
No competing interests declared.
Author contributions
Formal analysis, Writing – original draft.
Formal analysis, Writing – original draft.
Formal analysis, Supervision, Writing – original draft, Writing – review and editing.
Writing – original draft.
Writing – review and editing.
Writing – original draft, Writing – review and editing.
Validation.
Validation, Writing – review and editing.
Formal analysis, Supervision, Validation, Writing – original draft, Writing – review and editing.
References
- Abergel C, Legendre M, Claverie JM. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiology Reviews. 2015;39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- Abrahão JS, Araújo R, Colson P, La Scola B. The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLOS Genetics. 2017;13:e1006532. doi: 10.1371/journal.pgen.1006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahão J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, La Scola B. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nature Communications. 2018;9:1–12. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherfi S, Andreani J, Baptiste E, Oumessoum A, Dornas FP, Andrade A, Chabriere E, Abrahao J, Levasseur A, Raoult D, La Scola B, Colson P. A Large Open Pangenome and A Small Core Genome for Giant Pandoraviruses. Frontiers in Microbiology. 2018;9:1486. doi: 10.3389/fmicb.2018.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherfi S, Brahim Belhaouari D, Pinault L, Baudoin J-P, Decloquement P, Abrahao J, Colson P, Levasseur A, Lamb DC, Chabriere E, Raoult D, La Scola B. Incomplete tricarboxylic acid cycle and proton gradient in Pandoravirus massiliensis: is it still a virus? The ISME Journal. 2022;16:695–704. doi: 10.1038/s41396-021-01117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldarini N, Alhasawi AA, Thomas SC, Appanna VD. The role of glutamine synthetase in energy production and glutamine metabolism during oxidative stress. Antonie van Leeuwenhoek. 2017;110:629–639. doi: 10.1007/s10482-017-0829-3. [DOI] [PubMed] [Google Scholar]
- Alperovitch-Lavy A, Sharon I, Rohwer F, Aro E-M, Glaser F, Milo R, Nelson N, Béjà O. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I and photosystem-II gene suites inferred from oceanic metagenomic datasets. Environmental Microbiology. 2011;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]
- Anantharaman K, Duhaime MB, Breier JA, Wendt KA, Toner BM, Dick GJ. Sulfur oxidation genes in diverse deep-sea viruses. Science (New York, N.Y.) 2014;344:757–760. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Expression of animal virus genomes. Bacteriological Reviews. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrenfeld null, Kolber null. Widespread iron limitation of phytoplankton in the south pacific ocean. Science (New York, N.Y.) 1999;283:840–843. doi: 10.1126/science.283.5403.840. [DOI] [PubMed] [Google Scholar]
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science (New York, N.Y.) 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Blanc-Mathieu R, Dahle H, Hofgaard A, Brandt D, Ban H, Kalinowski J, Ogata H, Sandaa RA. A persistent giant algal virus, with A unique morphology, encodes an unprecedented number of genes involved in energy metabolism. Journal of Virology. 2021;20:JVI.02446-20. doi: 10.1128/JVI.02446-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship RE. Early evolution of photosynthesis. Plant Physiology. 2010;154:434–438. doi: 10.1104/pp.110.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nature Reviews. Microbiology. 2015;13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Frigaard NU. Prokaryotic photosynthesis and phototrophy illuminated. Trends in Microbiology. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Buhr ED, Yue WWS, Ren X, Jiang Z, Liao HWR, Mei X, Vemaraju S, Nguyen MT, Reed RR, Lang RA, Yau KW, Van Gelder RN. Neuropsin (OPN5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. PNAS. 2015;112:13093–13098. doi: 10.1073/pnas.1516259112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth PJ. Lehninger: Principles of Biochemistry Cell Biochemistry and Function. Lehninger 2005
- Campos-Sandoval JA, López de la Oliva AR, Lobo C, Segura JA, Matés JM, Alonso FJ, Márquez J. Expression of functional human glutaminase in baculovirus system: affinity purification, kinetic and molecular characterization. The International Journal of Biochemistry & Cell Biology. 2007;39:765–773. doi: 10.1016/j.biocel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chelkha N, Levasseur A, La Scola B, Colson P. Host-virus interactions and defense mechanisms for giant viruses. Annals of the New York Academy of Sciences. 2020;1486:39–57. doi: 10.1111/nyas.14469. [DOI] [PubMed] [Google Scholar]
- Claverie JM, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends in Genetics. 2010;26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng X-W, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Archives of Virology. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho FH, Silveira CB, Gregoracci GB, Thompson CC, Edwards RA, Brussaard CPD, Dutilh BE, Thompson FL. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nature Communications. 2017;8:15955. doi: 10.1038/ncomms15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza FG, Abrahão JS, Rodrigues RAL. Comparative Analysis of Transcriptional Regulation Patterns: Understanding the Gene Expression Profile in Nucleocytoviricota. Pathogens (Basel, Switzerland) 2021;10:935. doi: 10.3390/pathogens10080935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. Wholly Rickettsia! Reconstructed Metabolic Profile of the Quintessential Bacterial Parasite of Eukaryotic Cells. MBio. 2017;8:e00859-17. doi: 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré J, O’Malley MA. Varieties of Living Things: Life at the Intersection of Lineage and Metabolism. Philosophy and Theory in Biology. 2009;1:e003. doi: 10.3998/ptb.6959004.0001.003. [DOI] [Google Scholar]
- Endo H, Blanc-Mathieu R, Li Y, Salazar G, Henry N, Labadie K, de Vargas C, Sullivan MB, Bowler C, Wincker P, Karp-Boss L, Sunagawa S, Ogata H. Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nature Ecology & Evolution. 2020;4:1639–1649. doi: 10.1038/s41559-020-01288-w. [DOI] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. PNAS. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The virocell concept and environmental microbiology. The ISME Journal. 2013;7:233–236. doi: 10.1038/ismej.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman S, Flores-Uribe J, Larom S, Alalouf O, Liran O, Yacoby I, Salama F, Bailleul B, Rappaport F, Ziv T, Sharon I, Cornejo-Castillo FM, Philosof A, Dupont CL, Sánchez P, Acinas SG, Rohwer FL, Lindell D, Béjà O. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nature Microbiology. 2017;2:1350–1357. doi: 10.1038/s41564-017-0002-9. [DOI] [PubMed] [Google Scholar]
- Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annual Review of Microbiology. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- Friedrich CG. In: Advances in Microbial Physiology. Poole RK, editor. Academic Press; 1997. Physiology and Genetics of Sulfur-oxidizing Bacteria; pp. 235–289. [DOI] [PubMed] [Google Scholar]
- Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Frohns F, Käsmann A, Kramer D, Schäfer B, Mehmel M, Kang M, Van Etten JL, Gazzarrini S, Moroni A, Thiel G. Potassium ion channels of Chlorella viruses cause rapid depolarization of host cells during infection. Journal of Virology. 2006;80:2437–2444. doi: 10.1128/JVI.80.5.2437-2444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gann ER, Xian Y, Abraham PE, Hettich RL, Reynolds TB, Xiao C, Wilhelm SW. Structural and Proteomic Studies of the Aureococcus anophagefferens Virus Demonstrate a Global Distribution of Virus-Encoded Carbohydrate Processing. Frontiers in Microbiology. 2020;11:2047. doi: 10.3389/fmicb.2020.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehre L. Université Pierre et Marie Curie; 2015. Chlamydia trachomatis hijacks energy stores from the host and accumulates glycogen in the inclusion lumen through a dual pathway.https://tel.archives-ouvertes.fr/tel-01333744/document [Google Scholar]
- Glevarec G, Bouton S, Jaspard E, Riou MT, Cliquet JB, Suzuki A, Limami AM. Respective roles of the glutamine synthetase/glutamate synthase cycle and glutamate dehydrogenase in ammonium and amino acid metabolism during germination and post-germinative growth in the model legume Medicago truncatula. Planta. 2004;219:286–297. doi: 10.1007/s00425-004-1214-9. [DOI] [PubMed] [Google Scholar]
- Ha AD, Moniruzzaman M, Aylward FO. High Transcriptional Activity and Diverse Functional Repertoires of Hundreds of Giant Viruses in a Coastal Marine System. MSystems. 2021;6:e0029321. doi: 10.1128/mSystems.00293-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A, Bölling C, Buckhout TJ. The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiology. 2002;130:2039–2048. doi: 10.1104/pp.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle SL, Dupont CL, Hopkinson BM, King AL, Buck KN, Roe KL, Stuart RK, Allen AE, Mann EL, Johnson ZI, Barbeau KA. Pervasive iron limitation at subsurface chlorophyll maxima of the California Current. PNAS. 2018;115:13300–13305. doi: 10.1073/pnas.1813192115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Takahashi H, Aoki K, Takemura M. Draft Genome Sequence of Pandoravirus japonicus Isolated from the Sabaishi River, Niigata, Japan. Microbiology Resource Announcements. 2021;10:19. doi: 10.1128/MRA.00365-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothersall JS, Ahmed A. Metabolic fate of the increased yeast amino Acid uptake subsequent to catabolite derepression. Journal of Amino Acids. 2013;2013:461901. doi: 10.1155/2013/461901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang H, Andreopoulos B, Brewer HM, Glavina Del Rio T, Adkins JN, Paul S, Sullivan MB, Duhaime MB. Phage-specific metabolic reprogramming of virocells. The ISME Journal. 2020;14:881–895. doi: 10.1038/s41396-019-0580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. PNAS. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biology. 2013;14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz BL, U’Ren JM. Viral metabolic reprogramming in marine ecosystems. Current Opinion in Microbiology. 2016;31:161–168. doi: 10.1016/j.mib.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Jetten MS, Strous M, van de Pas-Schoonen KT, Schalk J, van Dongen UG, van de Graaf AA, Logemann S, Muyzer G, van Loosdrecht MC, Kuenen JG. The anaerobic oxidation of ammonium. FEMS Microbiology Reviews. 1998;22:421–437. doi: 10.1111/j.1574-6976.1998.tb00379.x. [DOI] [PubMed] [Google Scholar]
- Juneau P, Lawrence JE, Suttle CA, Harrison PJ. Effects of viral infection on photosynthetic processes in the bloom-forming alga Heterosigma akashiwo. Aquatic Microbial Ecology. 2003;31:9–17. doi: 10.3354/ame031009. [DOI] [Google Scholar]
- Knappe J, Blaschkowski HP, Gröbner P, Schmitt T. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. European Journal of Biochemistry. 1974;50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- Krebs HA, Johnson WA. Acetopyruvic acid (alphagamma-diketovaleric acid) as an intermediate metabolite in animal tissues. The Biochemical Journal. 1937;31:772–779. doi: 10.1042/bj0310772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, Sheyn U, Sebé-Pedrós A, Ben-Dor S, Schatz D, Tanay A, Rosenwasser S, Vardi A. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Science Advances. 2020;6:eaba4137. doi: 10.1126/sciadv.aba4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A. In: Sweet Biochemistry. Kumari A, editor. Academic Press; 2018. Chapter 1 - Glycolysis; pp. 1–5. [DOI] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. A giant virus in amoebae. Science (New York, N.Y.) 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annual Review of Biochemistry. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Lamb DC, Follmer AH, Goldstone JV, Nelson DR, Warrilow AG, Price CL, True MY, Kelly SL, Poulos TL, Stegeman JJ. On the occurrence of cytochrome P450 in viruses. PNAS. 2019;116:12343–12352. doi: 10.1073/pnas.1901080116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue A, Burlat B, Coutard B, Chaspoul F, Claverie JM, Abergel C. The megavirus chilensis Cu,Zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme. Journal of Virology. 2015;89:824–832. doi: 10.1128/JVI.02588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Couté Y, Rivkina E, Abergel C, Claverie JM. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. PNAS. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic JM, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie JM. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. PNAS. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Fabre E, Poirot O, Jeudy S, Lartigue A, Alempic J-M, Beucher L, Philippe N, Bertaux L, Christo-Foroux E, Labadie K, Couté Y, Abergel C, Claverie J-M. Diversity and evolution of the emerging Pandoraviridae family. Nature Communications. 2018;9:1–12. doi: 10.1038/s41467-018-04698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Lehninger Principles of Biochemistry. Wort Publishers; 2004. [Google Scholar]
- Lin S, Zhang H, Zhuang Y, Tran B, Gill J. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. PNAS. 2010;107:20033–20038. doi: 10.1073/pnas.1007246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Fraga JL, Blunt-Harris EL, Hayes LA, Phillips EJP, Coates JD. Humic Substances as a Mediator for Microbially Catalyzed Metal Reduction. Acta Hydrochimica et Hydrobiologica. 1998;26:152–157. doi: 10.1002/(SICI)1521-401X(199805)26:3<152::AID-AHEH152>3.0.CO;2-D. [DOI] [Google Scholar]
- Lovley DR, Coates JD. Novel forms of anaerobic respiration of environmental relevance. Current Opinion in Microbiology. 2000;3:252–256. doi: 10.1016/s1369-5274(00)00085-0. [DOI] [PubMed] [Google Scholar]
- Lovley D. Dissimilatory Metal Reduction: From Early Life to Bioremediation. Bepress. 2002 https://works.bepress.com/derek_lovley/189/
- Luria SE. Virus Growth and Variation. Cambridge University Press; 1959. [Google Scholar]
- Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in A fungus in A plant: three-way symbiosis required for thermal tolerance. Science (New York, N.Y.) 2007;315:513–515. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- Maughan R. Carbohydrate metabolism. Surgery (Oxford) 2009;27:6–10. doi: 10.1016/j.mpsur.2008.12.002. [DOI] [Google Scholar]
- Meléndez-Hevia E, Waddell TG, Cascante M. The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. Journal of Molecular Evolution. 1996;43:293–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]
- Miller SM, Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. Journal of Bacteriology. 1990;172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed Ali AM, Kawasaki T, Yamada T. Genetic rearrangements on the Chlorovirus genome that switch between hyaluronan synthesis and chitin synthesis. Virology. 2005;342:102–110. doi: 10.1016/j.virol.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Monier A, Pagarete A, de Vargas C, Allen MJ, Read B, Claverie J-M, Ogata H. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Research. 2009;19:1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Chambouvet A, Milner DS, Attah V, Terrado R, Lovejoy C, Moreau H, Santoro AE, Derelle É, Richards TA. Host-derived viral transporter protein for nitrogen uptake in infected marine phytoplankton. PNAS. 2017;114:E7489–E7498. doi: 10.1073/pnas.1708097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, LeCleir GR, Brown CM, Gobler CJ, Bidle KD, Wilson WH, Wilhelm SW. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host-virus coevolution. Virology. 2014;466–467:60–70. doi: 10.1016/j.virol.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M, Wurch LL, Alexander H, Dyhrman ST, Gobler CJ, Wilhelm SW. Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nature Communications. 2017;8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, Gann ER, Wilhelm SW. Infection by A Giant Virus (AaV) Induces Widespread Physiological Reprogramming in Aureococcus anophagefferens CCMP1984 - A Harmful Bloom Algae. Frontiers in Microbiology. 2018;9:752. doi: 10.3389/fmicb.2018.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M, Martinez-Gutierrez CA, Weinheimer AR, Aylward FO. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nature Communications. 2020;11:1710. doi: 10.1038/s41467-020-15507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evolutionary Biology. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, López-García P. Ten reasons to exclude viruses from the tree of life. Nature Reviews. Microbiology. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu R-Y, van der Giezen M, Tielens AGM, Martin WF. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiology and Molecular Biology Reviews. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Yoshizawa S, Hosaka T, Poirier C, Choi CJ, Hehenberger E, Irwin NAT, Wilken S, Yung CM, Bachy C, Kurihara R, Nakajima Y, Kojima K, Kimura-Someya T, Leonard G, Malmstrom RR, Mende DR, Olson DK, Sudo Y, Sudek S, Richards TA, DeLong EF, Keeling PJ, Santoro AE, Shirouzu M, Iwasaki W, Worden AZ. A distinct lineage of giant viruses brings A rhodopsin photosystem to unicellular marine predators. PNAS. 2019;116:20574–20583. doi: 10.1073/pnas.1907517116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. W.H. Freeman; 2017. [Google Scholar]
- Neupärtl M, Meyer C, Woll I, Frohns F, Kang M, Van Etten JL, Kramer D, Hertel B, Moroni A, Thiel G. Chlorella viruses evoke a rapid release of K+ from host cells during the early phase of infection. Virology. 2008;372:340–348. doi: 10.1016/j.virol.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Nissimov JI, Talmy D, Haramaty L, Fredricks HF, Zelzion E, Knowles B, Eren AM, Vandzura R, Laber CP, Schieler BM, Johns CT, More KD, Coolen MJL, Follows MJ, Bhattacharya D, Van Mooy BAS, Bidle KD. Biochemical diversity of glycosphingolipid biosynthesis as a driver of Coccolithovirus competitive ecology. Environmental Microbiology. 2019;21:2182–2197. doi: 10.1111/1462-2920.14633. [DOI] [PubMed] [Google Scholar]
- Noor E, Eden E, Milo R, Alon U. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Molecular Cell. 2010;39:809–820. doi: 10.1016/j.molcel.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Olson JM. Photosynthesis in the Archean era. Photosynthesis Research. 2006;88:109–117. doi: 10.1007/s11120-006-9040-5. [DOI] [PubMed] [Google Scholar]
- Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase Homology modeling, cofactor docking, and molecular dynamics simulation studies. The Journal of Biological Chemistry. 2004;279:9424–9431. doi: 10.1074/jbc.M311876200. [DOI] [PubMed] [Google Scholar]
- Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie J-M, Abergel C. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science (New York, N.Y.) 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- Piacente F, Marin M, Molinaro A, De Castro C, Seltzer V, Salis A, Damonte G, Bernardi C, Claverie J-M, Abergel C, Tonetti M. Giant DNA virus mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine. The Journal of Biological Chemistry. 2012;287:3009–3018. doi: 10.1074/jbc.M111.314559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, Bernardi C, Marin M, Blanc G, Abergel C, Tonetti MG. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology. 2014;24:51–61. doi: 10.1093/glycob/cwt089. [DOI] [PubMed] [Google Scholar]
- Piacente F, Gaglianone M, Laugieri ME, Tonetti MG. The Autonomous Glycosylation of Large DNA Viruses. International Journal of Molecular Sciences. 2015;16:29315–29328. doi: 10.3390/ijms161226169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaitakis A, Kalef-Ezra E, Kotzamani D, Zaganas I, Spanaki C. The Glutamate Dehydrogenase Pathway and Its Roles in Cell and Tissue Biology in Health and Disease. Biology. 2017;6:E11. doi: 10.3390/biology6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S, Fothergill-Gilmore LA. Molecular evolution: The origin of glycolysis. Biochemical Education. 1993;21:45–48. doi: 10.1016/0307-4412(93)90018-U. [DOI] [Google Scholar]
- Pratama AA, Bolduc B, Zayed AA, Zhong Z-P, Guo J, Vik DR, Gazitúa MC, Wainaina JM, Roux S, Sullivan MB. Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation. PeerJ. 2021;9:e11447. doi: 10.7717/peerj.11447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie J-M. The 1.2-megabase genome sequence of Mimivirus. Science (New York, N.Y.) 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nature Reviews. Microbiology. 2008a;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- Raoult D, Forterre P. What makes a virus a virus: reply from Raoult and Forterre. Nature Reviews Microbiology. 2008b;6:643. doi: 10.1038/nrmicro1858-c2. [DOI] [Google Scholar]
- Regenmortel MHVV. The Nature and Classification of Viruses, Topley & Wilson’s Microbiology and Microbial Infections. American Cancer Society; 2010. [DOI] [Google Scholar]
- Rodrigues RAL, Arantes TS, Oliveira GP, Dos Santos Silva LK, Abrahão JS. The Complex Nature of Tupanviruses. Advances in Virus Research. 2019;103:135–166. doi: 10.1016/bs.aivir.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Rodwell VW, Bender DA, Botham KM, Kennelly PJ, Weil PA. Harper’s Illustrated Biochemistry. McGraw Hill Medical; 2015. [Google Scholar]
- Rolland C, Andreani J, Louazani AC, Aherfi S, Francis R, Rodrigues R, Silva LS, Sahmi D, Mougari S, Chelkha N, Bekliz M, Silva L, Assis F, Dornas F, Khalil JYB, Pagnier I, Desnues C, Levasseur A, Colson P, Abrahão J, La Scola B. Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere. Viruses. 2019;11:E312. doi: 10.3390/v11040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Ziv C, Creveld SG, Vardi A. Virocell Metabolism: Metabolic Innovations During Host-Virus Interactions in the Ocean. Trends in Microbiology. 2016;24:821–832. doi: 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Saikia S, Oliveira D, Hu G, Kronstad J. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infection and Immunity. 2014;82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LM. In: Encyclopedia of Food and Health. Caballero B, Finglas PM, Toldrá F, editors. Oxford: Academic Press; 2016. Carbohydrate: Digestion, Absorption and Metabolism; pp. 643–650. [DOI] [Google Scholar]
- Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Claverie JM. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. PNAS. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder I, Johnson E, de Vries S. Microbial ferric iron reductases. FEMS Microbiology Reviews. 2003;27:427–447. doi: 10.1016/S0168-6445(03)00043-3. [DOI] [PubMed] [Google Scholar]
- Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, Kyrpides NC, Koonin EV, Woyke T. Giant viruses with an expanded complement of translation system components. Science (New York, N.Y.) 2017;356:82–85. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- Schulz F, Roux S, Paez-Espino D, Jungbluth S, Walsh DA, Denef VJ, McMahon KD, Konstantinidis KT, Eloe-Fadrosh EA, Kyrpides NC, Woyke T. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578:432–436. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarcz CR, Steward GF. A giant virus infecting green algae encodes key fermentation genes. Virology. 2018;518:423–433. doi: 10.1016/j.virol.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, Pinter RY, Partensky F, Koonin EV, Wolf YI, Nelson N, Béjà O. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. The ISME Journal. 2016;10:1742–1754. doi: 10.1038/ismej.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM. The ecology of viruses that infect eukaryotic algae. Environmental Microbiology. 2012;14:2253–2271. doi: 10.1111/j.1462-2920.2012.02706.x. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Okamoto N, Burri L, James ER, Keeling PJ. A bacterial proteorhodopsin proton pump in marine eukaryotes. Nature Communications. 2011;2:183. doi: 10.1038/ncomms1188. [DOI] [PubMed] [Google Scholar]
- Smith E, Morowitz HJ. Universality in intermediary metabolism. PNAS. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLOS Biology. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Adams B, Gurnon JR, Ye Y, Van Etten JL. Characterization of two chitinase genes and one chitosanase gene encoded by Chlorella virus PBCV-1. Virology. 1999;263:376–387. doi: 10.1006/viro.1999.9958. [DOI] [PubMed] [Google Scholar]
- Sun L, Gurnon JR, Adams BJ, Graves MV, Van Etten JL. Characterization of a beta-1,3-glucanase encoded by chlorella virus PBCV-1. Virology. 2000;276:27–36. doi: 10.1006/viro.2000.0500. [DOI] [PubMed] [Google Scholar]
- Sun TW, Yang CL, Kao TT, Wang TH, Lai MW, Ku C. Host Range and Coding Potential of Eukaryotic Giant Viruses. Viruses. 2020;12:E1337. doi: 10.3390/v12111337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses--major players in the global ecosystem. Nature Reviews. Microbiology. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Environmental microbiology: Viral diversity on the global stage. Nature Microbiology. 2016;1:16205. doi: 10.1038/nmicrobiol.2016.205. [DOI] [PubMed] [Google Scholar]
- Tonetti M, Zanardi D, Gurnon JR, Fruscione F, Armirotti A, Damonte G, Sturla L, De Flora A, Van Etten JL. Paramecium bursaria Chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. The Journal of Biological Chemistry. 2003;278:21559–21565. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Agarkova I, Dunigan DD, Tonetti M, De Castro C, Duncan GA. Chloroviruses Have a Sweet Tooth. Viruses. 2017;9:E88. doi: 10.3390/v9040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, Haramaty L, Bidle KD. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science (New York, N.Y.) 2009;326:861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- Villarreal LP. Viruses and the Evolution of Life. John Wiley & Sons, Inc; 2005. [DOI] [Google Scholar]
- Villarreal L, Witzany G. Viruses are essential agents within the roots and stem of the tree of life. Journal of Theoretical Biology. 2010;262:698–710. doi: 10.1016/j.jtbi.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJ, Holden MTG, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Proteorhodopsin genes in giant viruses. Biology Direct. 2012;7:34. doi: 10.1186/1745-6150-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Colson P, Raoult D, Koonin EV. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virology Journal. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Swamy KBS, Leu JY, McDonald MJ, Galafassi S, Compagno C, Piškur J. Coevolution with bacteria drives the evolution of aerobic fermentation in Lachancea kluyveri. PLOS ONE. 2017;12:e0173318. doi: 10.1371/journal.pone.0173318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, Waldbauer JR, Coleman ML. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nature Reviews. Microbiology. 2020;18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]