Abstract
The ability to regulate gene activity spatially and temporally is essential to investigate cell-type-specific gene function during development and in postembryonic processes and disease models. The Cre/lox system has been widely used for performing cell and tissue-specific conditional analysis of gene function in zebrafish. However, simple and efficient methods for isolation of stable, Cre/lox regulated zebrafish alleles are lacking. Here, we applied our GeneWeld CRISPR-Cas9 targeted integration strategy to generate floxed alleles that provide robust conditional inactivation and rescue. A universal targeting vector, UFlip, with sites for cloning short homology arms flanking a floxed 2A-mRFP gene trap, was integrated into an intron in rbbp4 and rb1. rbbp4off and rb1off integration alleles resulted in strong mRFP expression,>99% reduction of endogenous gene expression, and recapitulated known indel loss-of-function phenotypes. Introduction of Cre led to stable inversion of the floxed cassette, loss of mRFP expression, and phenotypic rescue. rbbp4on and rb1on integration alleles did not cause phenotypes in combination with a loss-of-function mutation. Addition of Cre led to conditional inactivation by stable inversion of the cassette, gene trapping and mRFP expression, and the expected mutant phenotype. Neural progenitor Cre drivers were used for conditional inactivation and phenotypic rescue to showcase how this approach can be used in specific cell populations. Together these results validate a simplified approach for efficient isolation of Cre/lox-responsive conditional alleles in zebrafish. Our strategy provides a new toolkit for generating genetic mosaics and represents a significant advance in zebrafish genetics.
Research organism: Zebrafish
Introduction
The ability to regulate gene activity spatially and temporally is essential to investigate cell-type-specific gene function during development and in postembryonic processes and disease models. Conditional gene inactivation can be achieved with the Cre/lox system in which the bacteriophage Cre recombinase promotes site-specific recombination at compatible loxP sites engineered in a gene of interest (Sauer and Henderson, 1988). Effective application of Cre/lox in vertebrates requires the isolation of transgenic lines expressing Cre, and gene modification to introduce loxP sequences at desired locations. Homologous recombination driven site-directed genetic modification in embryonic stem cells enabled creation of the mouse Cre/lox resource that contains more than 2500 Cre drivers and lox conditional gene alleles (Bult et al., 2019). In zebrafish, the lack of embryonic stem cell technology and inefficient homologous recombination in the early embryo necessitated alternative approaches to generate Cre/lox tools (Carney and Mosimann, 2018). Tol2 transposon transgenesis (Balciunas et al., 2006; Kawakami et al., 2000) combined with defined promoters (Mosimann et al., 2011) or BAC recombineering (Förster et al., 2017) has been used to develop Cre lines, and a rich source of zebrafish cell-type-specific Cre drivers has been generated by random Tol2 transposon enhancer trap screens (Jungke et al., 2015; Marquart et al., 2015; Tabor et al., 2019; Zhong et al., 2019). Cre/lox regulated conditional control of gene function in zebrafish was previously limited to generation of knock out alleles by random Tol2 transposon insertional mutagenesis with a floxed/FRT gene trap, which can be reverted by Cre or Flip-mediated recombination (Clark et al., 2011; Ni et al., 2012; Trinh et al., 2011). Only recently have gene editing methods been developed in zebrafish to simplify Cre driver isolation (Almeida et al., 2021; Kesavan et al., 2018) and generate genuine lox-regulated conditional alleles by targeted integration (Burg et al., 2018; Han et al., 2021; Hoshijima et al., 2016; Li et al., 2019; Sugimoto et al., 2017). These advances in zebrafish Cre/lox genetics are being driven by rapidly evolving methods for homology directed gene editing using CRISPR-Cas9 and TALEN endonucleases.
We and others recently demonstrated CRISPR-Cas9 targeted integration is an efficient method to isolate zebrafish proneural specific Cre and CreERT2 drivers that are expressed under the control of endogenous gene regulatory elements (Almeida et al., 2021; Kesavan et al., 2018). Efforts to introduce loxP sequences directly into the zebrafish genome by somatic gene targeting was first described using TALENs to target a double-strand break in the cdrh2 gene, followed by repair from a DNA oligonucleotide template containing homologous sequences flanking a loxP site (Bedell et al., 2012). This approach was expanded upon using CRISPR-Cas9 to create a floxed allele in two zebrafish genes by sequential loxP oligonucleotide targeting at sites flanking an exon, and demonstrated robust gene knockdown and loss of function after Cre-mediated exon excision (Burg et al., 2018). Although this approach is effective for creating a Cre/lox regulated allele, a single integration event of a floxed cassette with a linked reporter has distinct advantages, including only one generation of targeting and fluorescent genotyping of the integration allele. The first example of a germline floxed conditional allele with linked reporter used a single TALEN site downstream of an exon (Hoshijima et al., 2016). The repair template consisted of a linear cassette containing 1 kb long homology arms flanking a floxed exon plus reporter, to replace the endogenous exon by homologous recombination. A similar strategy using long homology arms was used to isolate a zebrafish conditional shha allele by integration of a modified Flex invertible gene trap (Ni et al., 2012), called Zwitch, by homologous recombination from an intact plasmid (Sugimoto et al., 2017). Stable inversion of the Zwitch cassette by Cre led to effective shha gene knockdown and defective shha signaling. More recently homology-independent targeting strategies have been described to generate conditional knockouts with a linked reporter, or bidirectional knockin of a dual reporter gene trap cassette, providing a method for labeling conditional gene knockout cells (Han et al., 2021; Li et al., 2019). As an alternative to isolation of stable germline conditional alleles, rapid tissue-specific gene knockdown and cell labeling can be achieved by somatic targeting with a floxed Cas9-2A-GFP; U6:gRNA transposon in a Cre-specific transgenic background, allowing for simultaneous bi-allelic inactivation with GFP cell labeling (Hans et al., 2021). Together, these approaches provide effective conditional gene knockdown strategies with varying degrees of complexity in design and execution. However, there is still a need for a simple method to efficiently recover robust conditional alleles through a single gene engineering event.
To address this need for a simple method to recover zebrafish germline conditional alleles in a single generation, we applied our GeneWeld CRISPR-Cas9 knock in strategy (Welker et al., 2021; Wierson et al., 2020) to target integration of a highly mutagenic floxed gene trap into an intron in two genes. We previously showed our GeneWeld strategy simplifies targeting vector assembly and enhances the efficiency of on target integration (Almeida et al., 2021; Wierson et al., 2020). In the present study, we built a universal vector, UFlip, which is derived from the GeneWeld pGTag and pPRISM targeted integration vectors (Almeida et al., 2021; Wierson et al., 2020), and the Tol2 Gene Breaking Transposon vectors (Clark et al., 2011; Ichino et al., 2020). The GeneWeld vectors contain sites for cloning short homology arms flanking a cargo of interest, and universal gRNA sites for efficient CRISPR-Cas9 cutting in vivo. In the UFlip vector, homology arm cloning sites flank a floxed gene trap cassette that contains a 2A-mRFP primary reporter based on the Tol2 RP2 gene trap (Clark et al., 2011; Ichino et al., 2020). The cassette also contains a heart or lens BFP secondary reporter for transgenic identification. Using 48 bp flanking short homology arms to drive integration at unique intron target gRNA sites, we recovered precise UFlip integration alleles in rbbp4 and rb1 with frequencies of 4–14%. We demonstrate that each molecular component in the floxed UFlip cassette functions as expected in vivo. Integration alleles with UFlip in the active, gene ‘off’ orientation show mRFP expression from the gene trap in the expected gene-specific pattern. UFlip ‘off’ alleles lead to >99% knock down of gene expression compared to wild-type levels and recapitulate indel mutant phenotypes, demonstrating highly effective loss of function by primary transcript splicing into the gene trap. Recombination of an ‘off’ allele by Cre injection or a transgenic Cre source leads loss of RFP expression, stable inversion of the UFlip cassette to the passive orientation, and robust conditional rescue of mutant phenotypes. UFlip ‘on’ alleles are phenotypically normal in combination with loss-of-function mutations. Recombination of an ‘on’ allele by Cre injection or a transgenic Cre source leads to induction of RFP expression, stable cassette inversion, and robust conditional inactivation. These results demonstrate GeneWeld UFlip CRISPR targeted integration is an effective approach to generate zebrafish Cre/lox conditional alleles that can be used for cell type-specific genetic mosaic analysis. The ability to perform conditional inactivation as well as conditional rescue provides a significant advance in zebrafish genetics.
Results
UFlip, a universal vector to generate stable Cre/lox regulated conditional alleles by CRISPR-Cas9 targeted integration
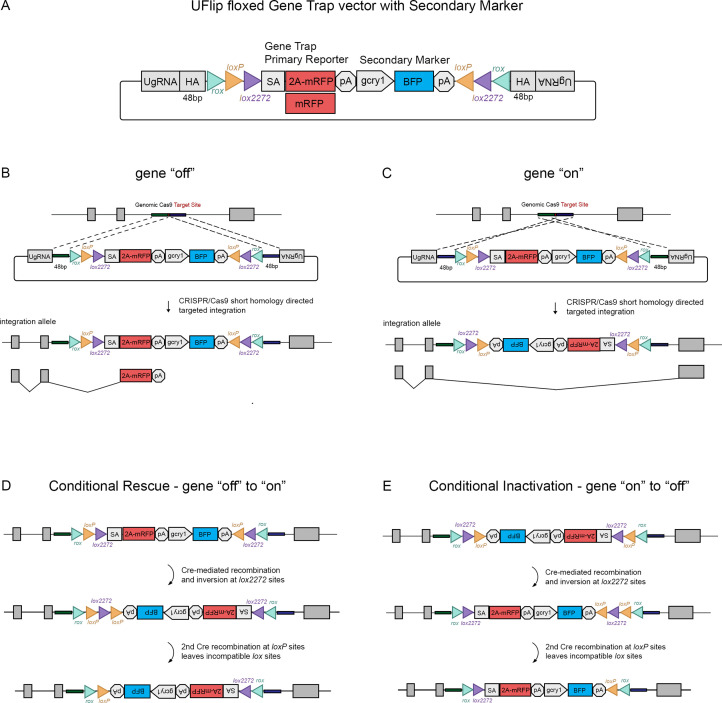
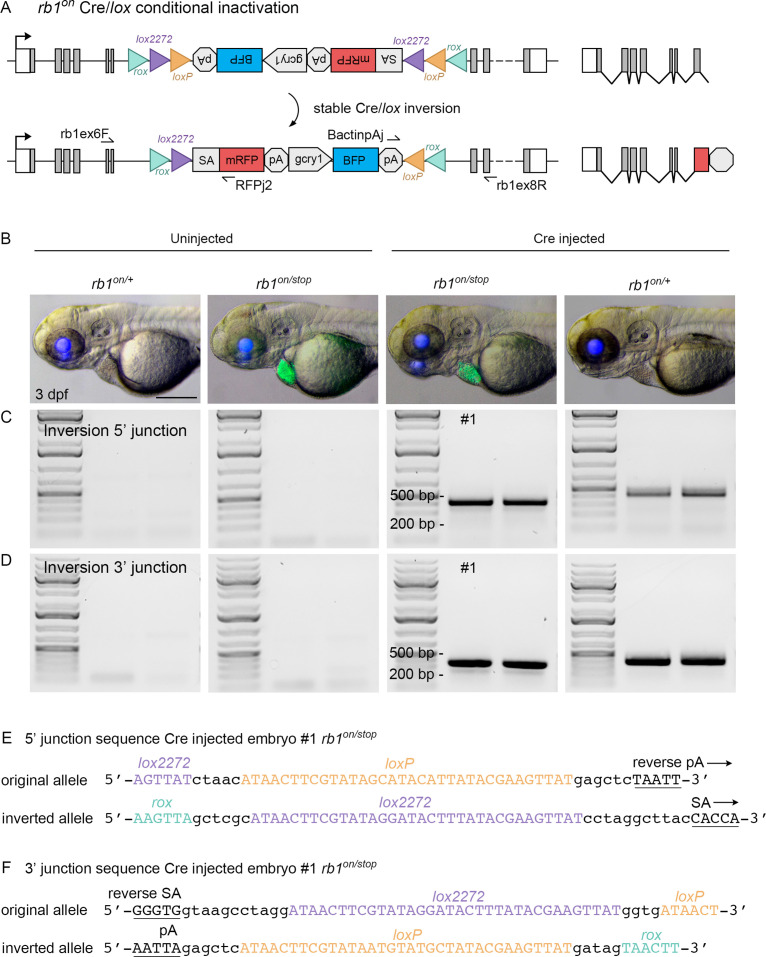
The Universal Flip (UFlip) vector was designed to be used with the GeneWeld strategy for CRISPR-Cas9 targeted integration driven by short homology (Wierson et al., 2020). The vector contains a cassette with a floxed gene trap plus secondary marker flanked by cloning sites for homology arms (HA) complementary to the genomic CRISPR target site, and universal gRNA sites (UgRNA) for in vivo liberation of the cassette after injection into zebrafish embryos (Figure 1A). The gene trap was derived from the gene-breaking Tol2 transposon RP2 that has previously been shown to lead to >99% knockdown of gene expression (Clark et al., 2011; Ichino et al., 2020). RP2 was modified by insertion of the porcine teschvirus-1 2A peptide sequence in between the splice acceptor and mRFP, which enables expression of a primary fluorescent reporter from the gene trap without creating a fusion protein. The ocean pout antifreeze gene transcriptional terminator follows the mRFP cDNA. Like our recently published endogenous Cre driver lines (Almeida et al., 2021), the primary gene trap reporter is followed by a secondary marker that drives tissue-specific expression of blue fluorescent protein (BFP) from the Xenopus gamma crystallin 1 (gcry1) lens promoter or the zebrafish myosin light chain 7 (myl7) cardiac muscle promoter, simplifying identification of an integration allele. Alternating pairs of loxP and lox2272 sites oriented head to head flanking the cassette are designed to drive two Cre recombination events that invert and lock the cassette in place, a design strategy described previously to generate floxed conditional alleles in mice (Robles-Oteiza et al., 2015; Schnütgen et al., 2003) and zebrafish (Sugimoto et al., 2017). rox sites flanking the entire cassette provide an alternative method for inversion using Dre recombinase (Figure 1A), which we demonstrate below provides highly efficient recovery of inverted alleles through the germline. Integration of UFlip into an intron in the active, gene ‘off’ orientation is expected to lead to premature transcriptional termination of the primary transcript in the gene trap and splicing of upstream exons into 2A-mRFP (Figure 1B), resulting in a loss-of-function allele. In contrast, integration of UFlip into an intron in the passive, gene ‘on’ orientation is not predicted to interrupt endogenous gene expression, since RNA polymerase will read through the cassette in the intron, which is then spliced out of the mature transcript (Figure 1C). For clarity, the UFlip alleles described here are referred to as ‘off’ when the cassette was integrated in the active, gene trap orientation. Alleles are designated ‘on’ when UFlip was integrated in the passive orientation, that leads to removal of the UFlip cassette during splicing and allows proper functioning of the gene.
Figure 1. The UFlip floxed gene trap vector for isolation of conditional gene alleles generated by GeneWeld CRISPR-Cas9 targeted integration.
(A) Diagram of the UFlip. The vector contains a floxed rox loxP lox2272 gene trap plus secondary marker loxP lox2272 rox cassette. The cassette is flanked by cloning sites for homology arms (HA) complementary to a genomic CRISPR target site, and universal gRNA sites (UgRNA) for in vivo liberation of the targeting cassette. (B) Gene ‘off’ alleles are generated by integration of the UFlip cassette into an intron in the active orientation, leading to transcription termination and splicing of the primary transcript in the mRFP gene trap. (C) Gene ‘on” alleles are generated by integration of the UFlip cassette into an intron in the passive orientation. This is driven by cloning the genomic 5’ homology arm downstream of the UFlip cassette, and cloning the genomic 3’ homology arm upstream of the UFlip cassette. Integration at the genomic CRISPR-Cas9 target site occurs in the opposite orientation. During transcription RNA polymerase reads through the integrated UFlip cassette, which is spliced out with the intron during processing of the primary transcript. (D) Cre-mediated recombination at an ‘off’ allele locks the cassette in the ‘on’ orientation. The first recombination occurs stochastically at either lox2272 or loxP sites. The diagram shows the intermediate that forms if the first recombination occurs at the lox2272 sites. (E) Cre-mediated recombination at an ‘on’ allele locks the cassette in the ‘off’ orientation. The first recombination occurs stochastically at either lox2272 or loxP sites. The diagram shows the intermediate that forms if the first recombination occurs at the lox2272 sites. BFP, blue fluorescent protein; gcry1, gamma crystallin 1 promoter; myl7, cardiac myosin light chain 7 promoter; 2 A, porcine teschvirus-1 2A peptide; mRFP, monomeric red fluorescent protein; pA, transcription termination and polyadenylation signal; SA, splice acceptor.
The process by which Cre recombination leads to stable inversion of the UFlip cassette is illustrated in Figure 1D, E (Schnütgen et al., 2003). Introduction of Cre leads to stochastic recombination at either the loxP or the lox2272 pairs of recombinase sites. Figure 1D, E show the intermediate results after 1 recombination event at the lox2272 sites. In the intermediate the cassette has inverted from ‘off’ to ‘on’ (Figure 1D) or from ‘on’ to ‘off’ (Figure 1E), resulting in a pair of loxP sites flanking one of the lox2272 sites. Recombination at the head to tail oriented loxP sites removes the intervening lox2272, resulting in a cassette flanked by a single pair of incompatible loxP and lox2272 sites. This prevents further recombination and produces stable inversion of the cassette.
Isolation of rbbp4 and rb1 UFlip conditional alleles
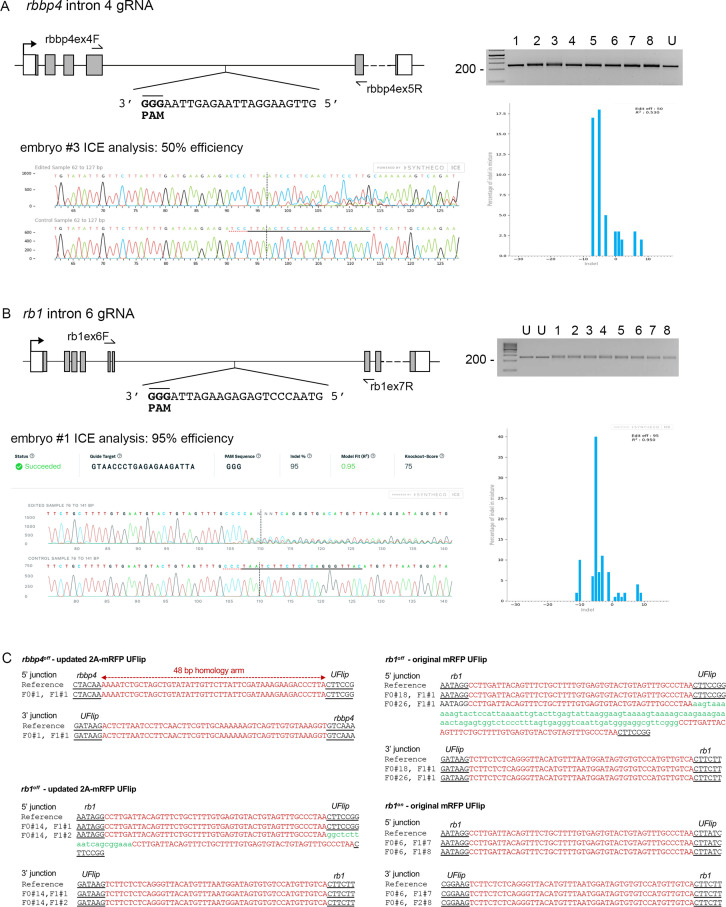
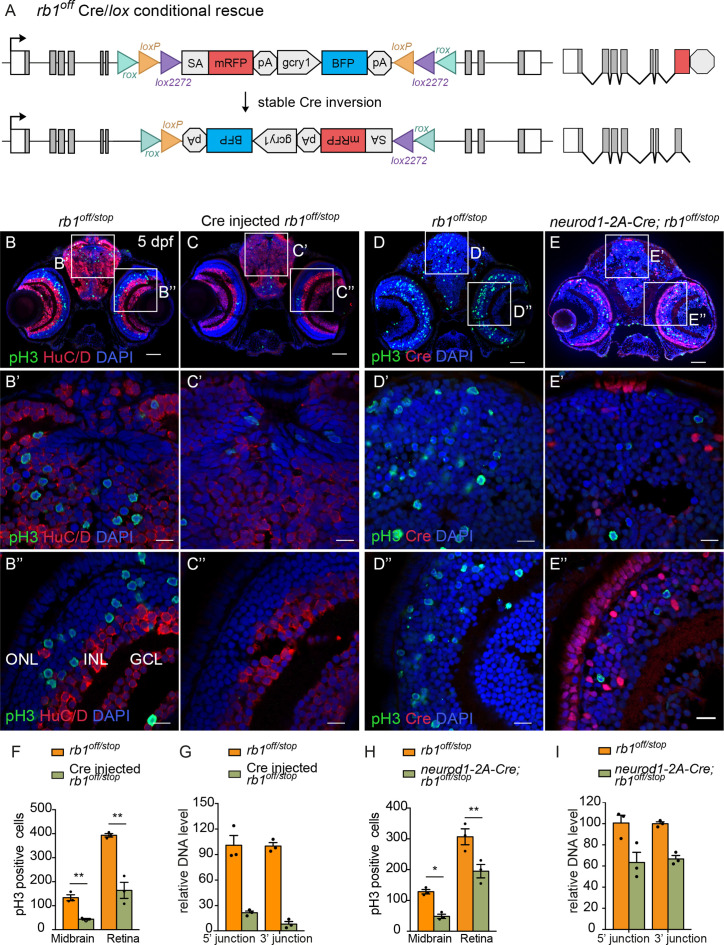
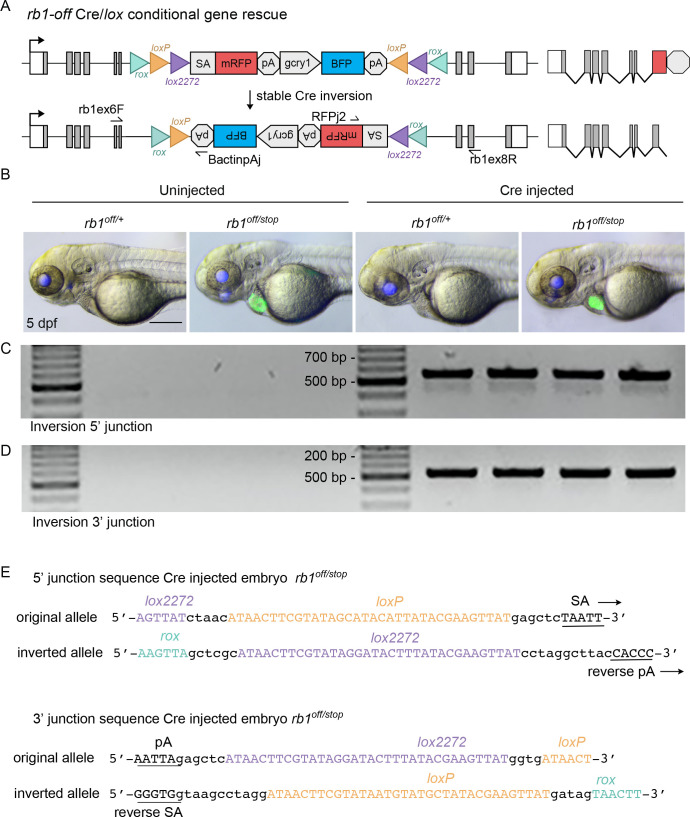
To test the ability of the UFlip cassette to generate zebrafish Cre-responsive conditional alleles, we chose to target integration of UFlip two genes, retinoblastoma binding protein 4 (rbbp4) and retinoblastoma 1 (rb1). We had previously isolated indel mutations in both genes that show strong loss-of-function phenotypes at the morphological and cellular level (Schultz et al., 2018; Schultz-Rogers et al., 2022; Solin et al., 2015). Intronic genomic DNA was amplified from wild-type WIK fish, cloned, sequenced, and analyzed to locate unique Cas9 gRNA sites that were shared among fish and that did not map to repetitive elements or retrotransposons. Cas9 target sites were identified in rbbp4 intron 4 and rb1 intron 6 (Table 1) and synthetic guides ordered from Synthego (https://www.synthego.com). Efficient indel formation at the intronic gRNA sites was confirmed by co-injection of gRNA and Cas9 mRNA into single-cell embryos, followed by extraction of genomic DNA at 2 dpf. A PCR amplicon surrounding the target site was directly sequenced followed by ICE analysis (https://ice.synthego.com/#/). The rbbp4 intron 4 and rb1 intron 6 guides showed 50% and 95% indel formation at the target site, respectively (Figure 2A, B). UFlip targeting vectors were assembled with 48 bp 5’ and 3’ homology arms complementary to the DNA flanking the Cas9 genomic DNA double strand break site (Table 1). One cell stage WIK embryos were co-injected with targeting vector (10 pg), Cas9 mRNA (150 pg), universal gRNA (25 pg), and gene-specific gRNA (25 pg). Four to six embryos with positive expression of the primary mRFP reporter and secondary BFP reporter were selected to test for evidence of on target integration by PCR amplification of the 5’ and 3’ junctions (data not shown). Primary and secondary reporter positive siblings were raised to adulthood. rbbp4 intron 4 was targeted with the UFlip-2A-mRFP cassette in the active orientation to recover an ‘off’ allele. Embryos expressing the primary reporter mRFP in the nervous system, where rbbp4 is normally expressed, and the lens:BFP secondary marker, were raised to adulthood. Two out of seven adult F0 fish transmitted embryos with widespread mRFP and lens BFP expression (Table 2; Table 2—source data 1). One adult transmitted an allele with precise 5’ and 3’ junctions at the intron target site. F1 embryos were raised to adulthood and the presence of the precise rbbp4-2A-mRFP-off, gcry1:BFP (rbbp4off) allele confirmed by fin clip and sequencing (Figure 2C). An F1 adult with the precise integration allele was outcrossed to WIK to establish an F2 family of the rbbp4off is61 allele.
Table 1. Genome intronic CRISPR gRNA sites and UFlip targeting vector homology arm sequences.
| Gene | Genomic sgRNA with PAM | 5' Homology arm | 3' Homology arm |
|---|---|---|---|
| rbbp4 | GTTGAAGGATTAAGAGTTAAGGG |
AAAATCTGCTAGCTGTATATTGTTCTTATTTGAT
GAAGAAGACCCTTA |
ACTCTTAATCCTTCAACTTCGTTGCAAAAAAGTCA
GTTGTGTAAAGGT |
| rb1 | ATTAGAAGAGAGTCCCAATGGGG |
CCTTGATTACAGTTTCTGCTTTTGTGAG
TGTACTGTAGTTTGCCCTAA |
TCTTCTCTCAGGGTTACATGTTTAATGGATAGTGTGT
CCATGTTGTCA |
Figure 2. rbbp4 and rb1 intronic gRNA efficiency and F1 UFlip allele junction analysis.
(A) rbbp4 gene model with sequence of the intron 4 reverse strand gRNA. Gel image of PCR amplicons surrounding the target site from 8 Cas9 plus gRNA injected and 1 uninjected (U) embryo. Amplicons from embryo #3 and the uninjected embryo were sequenced and analyzed with Synthego’s ICE software, and indicate 50% indel efficiency at the target site. Plot shows the range and percentage of indels present in the sequences. PAM sequence shown in bold and underlined. (B) rb1 gene model with sequence of the intron 6 reverse strand gRNA. Gel image of PCR amplicons surrounding the target site from eight embryos injected with Cas9 and the gRNA (1-8), and two uninjected embryos (U). Amplicons from embryo #1 and an uninjected embryo were sequenced and analyzed with Synthego’s ICE software, and indicate 95% indel efficiency at the target site. Plots show the range and percentage of indels present in the sequences. PAM sequences shown in bold and underlined. (C) 5’ and 3’ genomic-UFlip integration junctions were PCR amplified from F1 transgenic zebrafish fin clip genomic DNA. The PCR products were sequenced and aligned to the reference sequence expected for a precise integration at the genomic target site. Capitalized red nucleotides represent 48 bp homology arms. Lowercase green nucleotides represent random inserted sequences.
Table 2. Recovery of rbbp4 and rb1 UFlip floxed conditional alleles by GeneWeld CRISPR-Cas9 targeted integration.
| UFlip allele | Targeted Intron | Homology arm length bp/bp | Injected F0 embryo secondary marker expression | Germline transmission of secondary marker | Germline transmission of precise integration allele |
|---|---|---|---|---|---|
| rbbp4off is61 | 4 | 48/48 | 19% (26/134) | 29% (2/7) | 14% (1/7) |
| rb1off is58 | 6 | 48/48 | 47% (34/73) | 9% (3/33) | 6% (2/33) |
| rb1off is63 | 6 | 48/48 | 48% (60/125) | 25% (2/8) | 12.5% (1/8) |
| rb1on is57 | 6 | 48/48 | 43% (61/101) | 20% (5/25) | 4% (1/25) |
We initially targeted rb1 intron 6 with the original UFlip-mRFP primary reporter construct designed to create an in-frame fusion of the polypeptide with mRFP. We recovered rb1-mRFP-off, gcry1:BFP (rb1off) and rb1-mRFP-on, gcry1:BFP (rb1on) alleles with frequencies of 6% (2/33) for rb1off and 4% (1/25) for rb1on (Table 2; Table 2—source data 1). 5’ and 3’ junction PCR and sequence analysis of F1 embryos showed the integrations were precise, except for the 5’ junction in the allele transmitted by the second rb1off founder #26 (Figure 2C). The 5’ junction included a duplication of the 5’ Homology Arm sequence that flanked a 115 bp segment of the vector backbone. The rb1off founder #18 transmitting the precise allele was lost, therefore we moved forward with the allele from founder #26. As shown below, the 153 bp insertion on the 5’ side of the rb1off integration did not appear to affect the functionality of the allele. F1 adults were outcrossed to wildtype WIK to establish rb1off is58 and rb1on is57 families. We subsequently targeted the same rb1 intron six site with the updated UFlip-2A-mRFP vector, which allows expression of mRFP without creating a fusion protein with the targeted gene polypeptide. We recovered 2/8 (12.5%) founders transmitting lens:BFP +alleles with ubiquitous expression of the mRFP primary reporter (Table 2; Table 2—source data 1). Founder F0#14 transmitted two alleles, one with precise junctions, the other with a precise 3’ junction and a 5’ junction with a homology arm duplication and insertion of 19 bp (Figure 2C). F1s from this founder will be used to establish the line rb1off is63. For the experiments outlined below demonstrating Cre-meditated rb1 conditional rescue and inactivation, we used the established F3 generation rb1off is58 and rb1on is57 lines generated with the original UFlip construct, which did not show the mRFP gene trap reporter expression.
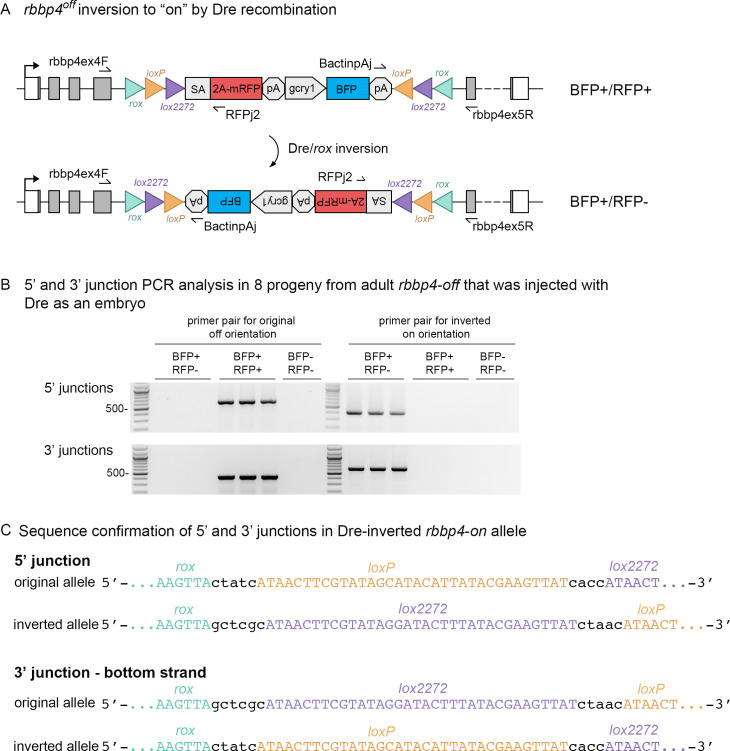
The UFlip construct contains rox sites flanking the entire cassette to allow the use of Dre recombination to recover an allele of the opposite orientation when starting with the rbbp4off allele (Figure 3A). To test this, we injected Dre synthetic mRNA into F1 and F2 rbbp4off/+ embryos. Injected embryos were raised to adulthood and five F1 and six F2 adults were outcrossed to wildtype WIK. Embryos were screened for expression of the primary mRFP reporter and the linked BFP secondary marker. Three of 5 F1 adults, and 4/6 F2 adults, transmitted both the mRFP/BFP positive original ‘off’ allele to their progeny, as well as the mRFP negative/BFP positive Dre-inverted ‘on’ allele (Table 3). The frequency of embryos inheriting the inverted allele vs. the original allele ranged from 7.7% to 43.2% (Table 3). We confirmed the rbbp4on allele recovered by Dre recombination contained the expected 5’ and 3’ junctions by PCR amplification and sequencing (Figure 3B, C). A single F2 adult was used to establish an F3 family of the rbbp4on is62 allele. These results demonstrate efficient Dre mediated recombination at rox sites in the germline, and a simple method for isolation of an alternative conditional allele starting with one UFlip allele.
Figure 3. Dre mRNA injection into rbbp4off/+ embryos leads to inversion of the UFlip cassette and efficient germline recovery of an inverted rbbp4on allele.
(A) Diagram illustrating Dre-mediated inversion of the rbbp4off allele to the on orientation. Repeated inversion of the cassette will continue as long as Dre is present. The final allele is predicted to be in the inverted ‘on’ orientation at a frequency of 50%. (B) PCR junction analysis of 8 embryos from an F1 adult that had been injected with Dre mRNA at the one-cell stage. Three embryos positive for expression of the lens BFP secondary marker show the expected 5’ and 3’ junction PCR amplicons for the inverted rbbp4on allele. (C) Sequence analysis confirms Dre-mediated inversion of the cassette from the ‘off’ to ‘on’ orientation in BRP+/RFP - embryos.
Table 3. Germline recovery of rbbp4on allele by Dre-mediated inversion of rbbp4off.
| Adult rbbp4off/+ | BFP-/RFP- embryos | BFP+/RFP + embryos (Off) original allele | BFP+/RFP- embryos (On) inverted allele | % transmission of On inverted allele/total BFP+ |
|---|---|---|---|---|
| F1 Female #1 | 48 | 37 | 0 | 0 |
| F1 Female #2 | 76 | 62 | 11 | 15% (11/73) |
| F1 Female #3 | 25 | 16 | 5 | 24% (5/21) |
| F1 Female #4 | 20 | 13 | 2 | 13% (2/15) |
| F1 Female #5 | 19 | 12 | 0 | 0 |
| F2 Female #1 | 18 | 24 | 2 | 7.7% (2/26) |
| F2 Female #2 | 50 | 25 | 19 | 43.2% (19/44) |
| F2 Female #3 | 36 | 40 | 4 | 9.1% (4/44) |
| F2 Female #4 | 22 | 14 | 0 | 0 |
| F2 Male #1 | 139 | 87 | 14 | 13.9% (14/101) |
| F2 Male #2 | 37 | 28 | 0 | 0 |
Experimental group 1: F1 embryos from the F0 founder rbbp4off/+ adult crossed to WIK were injected with 15 pg Dre mRNA. Inversion was confirmed by PCR on genomic DNA isolated from -injected embryos . 5 sibling injected embryos were raised to adulthood and were outcrossed to WIK. F2 embryos were screened for Primary (RFP+) and Secondary (BFP+) marker expression. Embryos inheriting the original rbbp4off allele show expression of both markers. Embryos inheriting an allele that was inverted from “off” to “on” by Dre recombination show expression of the secondary marker (BFP+) but lose expression of the primary marker due to the inverted orientation of the cassette. Experimental group 2: The same experiment was performed with an adult F1 rbbp4off/+ female crossed to WIK. F2 Dre mRNA injected embryos were raised to adulthood, and 7 adults screened for transmission of the inverted rbbp4on allele to the F3 generation.
rbbp4off allele provides robust gene knockdown and loss of function
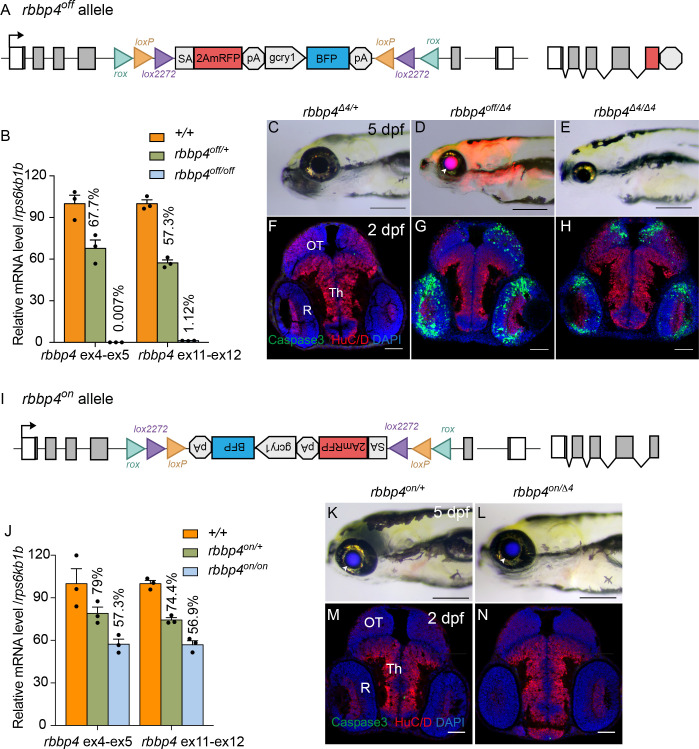
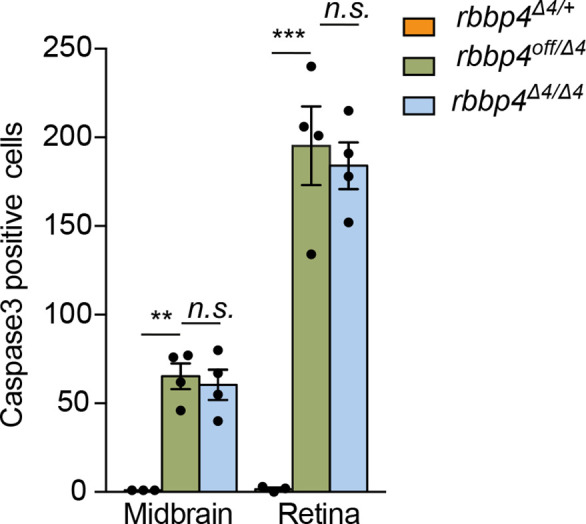
To demonstrate that integration of the UFlip cassette in the active, ‘off’ orientation leads to effective gene knockdown we examined gene expression by quantitative PCR in rbbp4off/off homozygotes. Homozygous rbbp4off/off 3 dpf larvae show a>99% reduction in rbbp4 mRNA levels in comparison to wild type sibling larvae (Figure 4A, B; Figure 4—source data 1). In comparison to wild-type larvae, the rbbp4off allele is easily identifiable by primary reporter gene trap RFP expression in the retina, brain, and mesodermal tissues, and secondary marker lens specific BFP expression (Figure 4C, D arrowhead). In trans-heterozygous combination with the established loss-of-function indel allele rbbp4Δ4 (Figure 4D, E), the rbbp4off allele recapitulates the microcephaly and microphthalmia phenotypes found in rbbp4Δ4/Δ4 homozygotes (Schultz et al., 2018; Schultz-Rogers et al., 2022). In comparison to control, anti-activated caspase 3 labeling and quantification in sectioned rbbp4off/Δ4 2 dpf head tissue reveals extensive apoptosis throughout the midbrain (** p<0.01) and retina (** p<0.01) (Figure 4F, G). This cellular phenotype is not significantly different from our previous description of apoptosis in rbbp4Δ4/Δ4 homozygotes (Figure 4F–H). Together these results demonstrate the rbbp4off allele causes robust gene knockdown by the primary reporter RFP gene trap, induces the expected pattern of RFP expression, and recapitulates the rbbp4 strong loss of phenotype at the molecular, cellular, and morphological levels.
Figure 4. Molecular and phenotypic characterization of rbbp4off and rbbp4on alleles.
(A) Diagram of the rbbp4off allele. (B) Plot of RT-qPCR results from wild type +/+ (n=3), heterozygous rbbp4off/+(n=3), and homozygous rbbp4off/off (n=3) larvae showing the relative level of rbbp4 mRNA transcript using reference gene rps6kb1b. Primer pairs were located in exons 4 and 5, or downstream exons 11 and 12. (C – E) Gross phenotype of rbbp4Δ4/+ (C), rbbp4off/Δ4 (D), and rbbp4Δ4/Δ4 (E) 5 dpf larvae. Arrowhead in (D) points to overlap of rbbp4off 2A-mRFP primary reporter and gcry1:BFP secondary reporter expression in the lens, which appears purple. (F – H) Caspase-3 and HuC/D labeling of sectioned head tissue from 2 dpf rbbp4Δ4/+ (F) rbbp4off/Δ4 (G) and rbbp4Δ4/Δ4 (H) embryos. (I) Diagram of the rbbp4on allele. (J) Plot of RT-qPCR results from wild type +/+ (n=3), heterozygous rbbp4on/+ (n=3), and homozygous rbbp4on/on (n=3) larvae showing the relative level of rbbp4 mRNA transcript using reference gene rps6kb1b. Primer pairs were located in exons 4 and 5, or downstream exons 11 and 12. (K, L) Gross phenotype of rbbp4on/+ (K) and rbbp4on/Δ4 (L) 5 dpf larvae. The rbbp4on allele secondary marker gcry1:BFP expression is visible in the lens. Caspase-3 and HuC/D labeling of sectioned head tissue from 2 dpf rbbp4Δ4/+ (M) and rbbp4off/Δ4 (N) embryos. OT, optic tectum; R, retina; Th, thalamic region. Error bars represent mean ± s.e.m. Scale bars: 200 μm (C–E, K, L), 50 μm (F–H, M,N).
Figure 4—figure supplement 1. Quantification of activated caspase-3a labeled cells in rbbp4Δ4/+, rbbp4off/Δ4, and rbbp4Δ4/Δ 2 dpf embryo midbrain and retina.
rbbp4on allele maintains rbbp4 function
In the rbbp4on allele, the UFlip cassette is predicted to be read through by RNA polymerase and then removed from the primary transcript when intron 4 is spliced out (Figure 4I). To test whether the rbbp4on allele alters normal rbbp4 mRNA expression levels RT-qPCR was performed on 3 dpf rbbp4on/+ heterozygous and rbbp4on/on homozygous larvae. rbbp4on/+ heterozygotes showed ~25% reduction in wild-type rbbp4 mRNA levels, while rbbp4on/on homozygotes showed ~40% reduction (Figure 4J, Figure 4—source data 1). This suggests the integrated UFlip cassette may impact splicing rate, stability, or other processing of the primary transcript. However, neither rbbp4on/+ heterozygotes nor rbbp4on/Δ4 trans-heterozygous show a gross phenotype in 5 dpf larvae (Figure 4K and L) or evidence of apoptosis in the developing midbrain and retina at 2 dpf (Figure 4M and N). Therefore, the level of processed, mature rbbp4 transcript is enough to maintain wild type levels of rbbp4 activity.
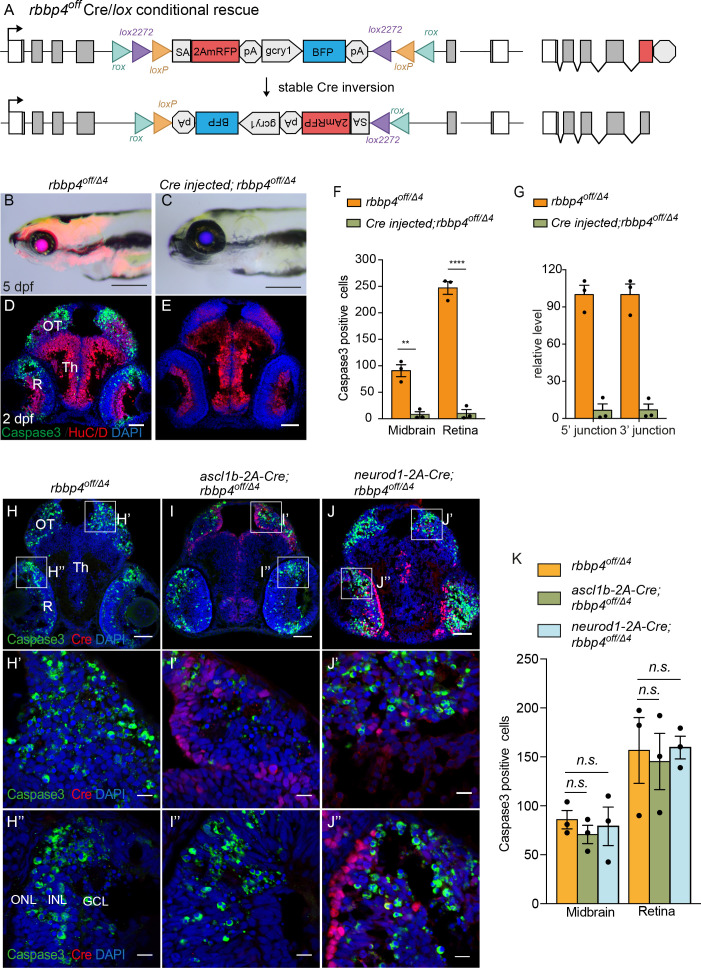
Cre/lox recombination of rbbp4off leads to stable inversion and conditional gene rescue
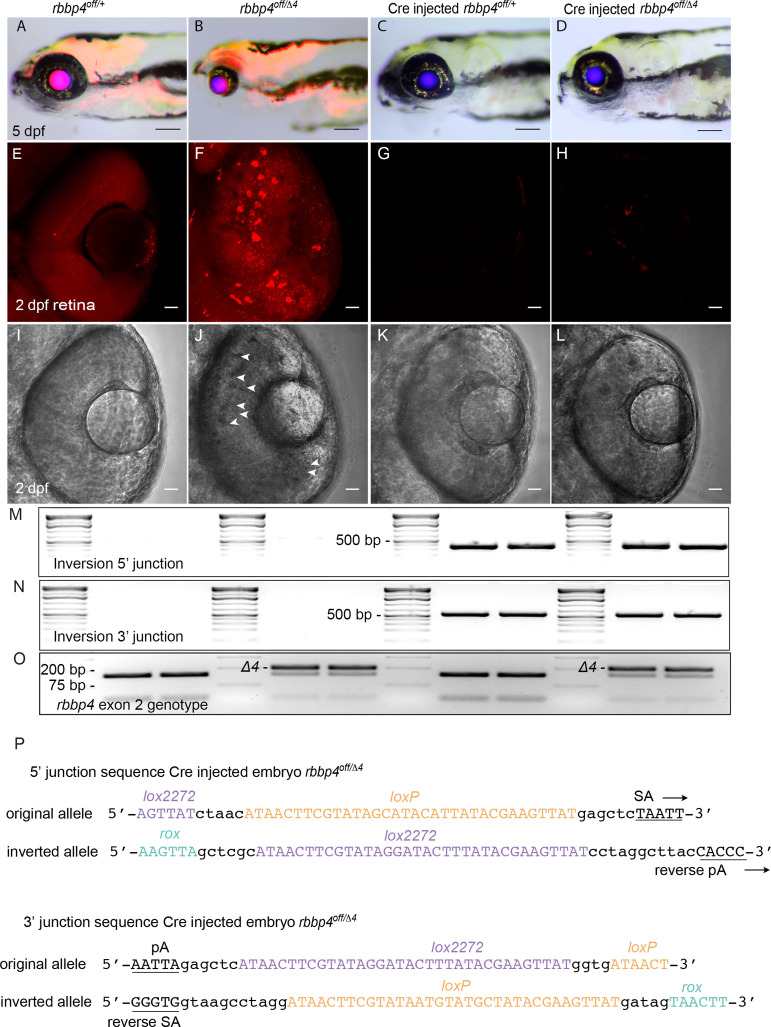
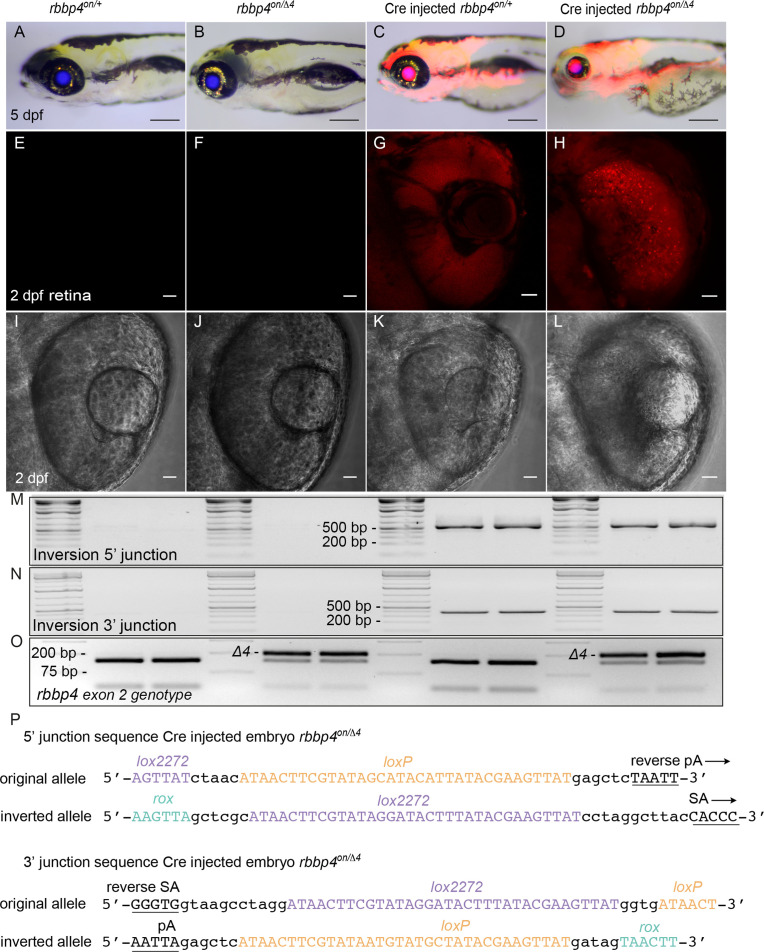
To test the ability of the UFlip cassette to undergo stable inversion and provide conditional rescue, we first used Cre mRNA injection. A total of 12.5 pg Cre synthetic mRNA was injected into embryos from a cross between rbbp4off/+ and rbbp4Δ4/+ heterozygous adults (Figure 5A–G). In comparison to uninjected rbbp4off/Δ4 larvae (Figure 5B), 5 dpf rbbp4off/Δ4 Cre injected larvae show normal morphology and an absence of RFP expression (Figure 5C). Confocal live imaging of 2 dpf in injected rbbp4off/Δ4 confirmed loss of RFP and rescue of the cell death phenotype in the retina (Figure 5—figure supplement 1A–L). Stable inversion of the rbbp4off cassette to the ‘on’ orientation was confirmed by PCR and sequence analysis, which showed the expected sequences at the 5’ and 3’ junctions after inversion at the lox sites (Figure 5—figure supplement 1M–P). Activated caspase 3 labeling in 2 dpf sectioned head tissue showed a nearly complete absence of apoptosis in the midbrain (** p<0.01) and retina (**** p<0.0001) compared to controls (Figure 5D–F, Figure 5—source data 1). Genomic DNA quantitative PCR of the original 5’ and 3’ rbbp4on allele junction fragments in the Cre injected embryos indicated a 93% reduction (Figure 5G, Figure 5—source data 1). Together these data demonstrate Cre mediated recombination at the incompatible pairs of loxP and lox2272 sites in the rbbp4off allele leads to efficient, stable inversion of the cassette to the ‘on’ orientation, which results in robust conditional rescue of the rbbp4 loss-of-function phenotype.
Figure 5. Ubiquitous and cell-type specific Cre-mediated conditional rescue with rbbp4-off.
(A) Diagram of expected Cre mediated inversion of rbbp4off to on orientation. (B) Gross morphological phenotype of microcephaly and microphthalmia in 5 dpf transheterozygous rbbp4off/Δlarva. (C) Cre injected 5 dpf transheterozygous rbbp4off/Δ4larva shows rescue of gross phenotype and loss of mRFP expression. (D) Activated caspase-3a labeling throughout midbrain and retina section from 2 dpf transheterozygous rbbp4off/Δ4embryo. (E) Absence of activated caspase-3a labeling in midbrain and retina of 2 dpf transheterozygous rbbp4off/Δ4embryo after Cre injection. (F) Quantification of caspase-3a labeling in control rbbp4off/Δ4(n=3) and Cre injected rbbp4off/Δ4 (n=3) midbrain (** p<0.01) and retina (**** p<0.0001). (G) Genomic DNA qPCR quantification of rbbp4off original orientation 5’ and 3 junctions in control rbbp4off/Δ4 (n=3) and Cre injected rbbp4off/Δ4 (n=3). Cre injection reduced the level of rbbp4off original orientation 5’ (>93%) and 3’ junctions (>93%). (H – J”) Activated caspase-3A and Cre labeling in sectioned head tissue from 2 dpf rbbp4off/Δ4 (H-H"), ascl1b-2A-Cre; rbbp4off/Δ4 (I-I"), and neurod1-2A-Cre; rbbp4off/Δ4 (J-J") embryos. (K) Quantification of caspase-3a labeling in rbbp4off/Δ4(n=3), ascl1b-2A-Cre; rbbp4off/Δ4 (n=3) and neurod1-2A-Cre; rbbp4off/Δ4 (n=3).rbbp4off/Δ4vs. ascl1b-2A-Cre; rbbp4off/Δ4 midbrain (n.s. p=0.3248) and retina (n.s. p=0.8153), and neurod1-2A-Cre; rbbp4off/Δ4 midbrain (n.s. p=0.7794) and retina (n.s. p=0.9365). OT, optic tectum; R, retina; Th, thalamic region. Error bars represent mean ± s.e.m. with two-tailed t-test. Scale bars: 200 μm (B, C), 50 μm (D, E, H – J). 10 μm (H’ – J”).
Figure 5—figure supplement 1. Live imaging and molecular analysis of rbbp4off conditional rescue by Cre injection.
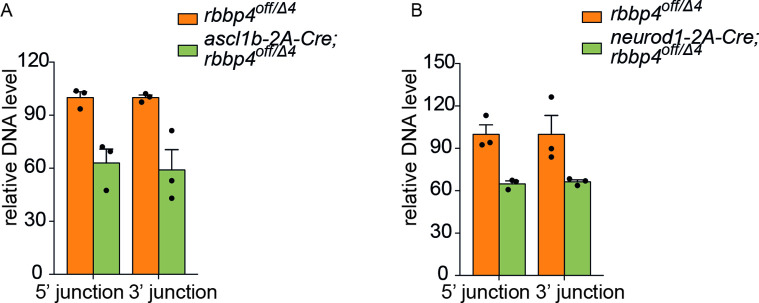
Figure 5—figure supplement 2. qPCR quantification of rbbp4off allele inversion by ascl1b-2A-Cre and neurod1-2A-Cre.
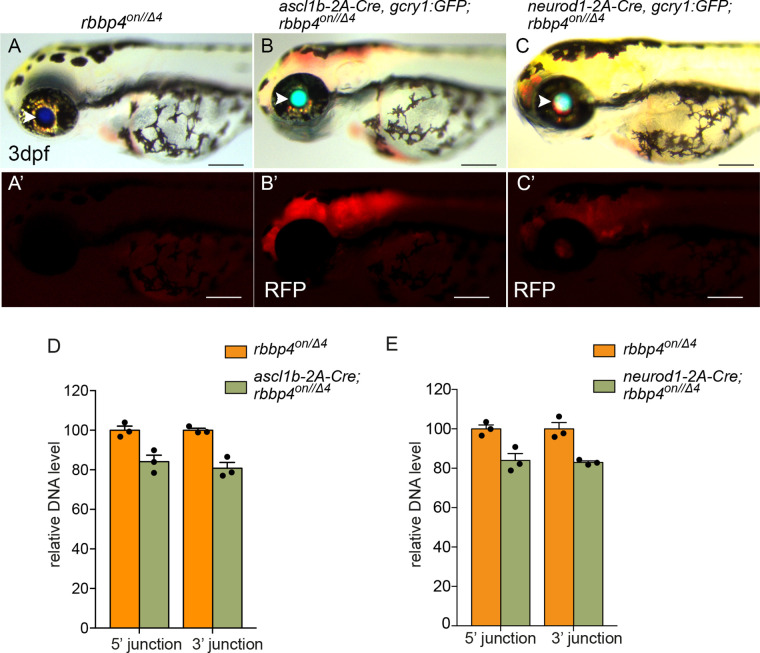
To test whether cell type-specific Cre sources could be used to rescue rbbp4 activity in distinct cell populations (Figure 5H–K), we used our previously published endogenous proneural Cre driver lines ascl1b-2A-Cre, gcry1:GFP is75 and neurod1-2A-Cre, gcry1:GFP is77 (Almeida et al., 2021). 2 dpf larvae from rbbp4off/+ crossed with ascl1b-2A-Cre/+; rbbp4Δ4/+ were sectioned and head tissue labeled to detect activated caspase 3. In comparison to control rbbp4off/Δ4 embryos, ascl1b-2A-Cre; rbbp4off/Δ4 embryos appeared to have reduced caspase labeling in the midbrain, where Cre is expressed (Figure 5H–I’), but the change was not significant (n.s. p=0.3248) (Figure 5K; Figure 5—source data 1). ascl1b isn’t expressed in the retina and consistent with this, Cre labeling was not detected in this tissue (Figure 5I”). As expected, there was no significant change in caspase labeling or levels in retinal sections from ascl1b-2A-Cre; rbbp4off/Δ4 embryos (n.s. p=0.8152) (Figure 5I" and K). To confirm expression of functional Cre recombinase and inversion of the cassette by ascl1b-2A-Cre, 5’ and 3’ junction genomic DNA qPCR showed >30% inversion of the cassette (Figure 5—figure supplement 2; Figure 5—figure supplement 2—source data 1). Testing for conditional rescue with the neurod1-2A-Cre driver showed there was no significant rescue of apoptosis in the brain (n.s. p=0.7794) and retina (n.s. p=0.9365) where neurod1-2A-Cre is expressed (Figure 5J, K; Figure 5—source data 1). 5’ and 3’ junction genomic DNA qPCR confirmed 30% inversion of the rbbp4off allele to the ‘on’ orientation (Figure 5—figure supplement 2; Figure 5—figure supplement 2—source data 1). Together, these results demonstrate a ubiquitous source of Cre can robustly rescue the loss-of-function phenotype of the rbbp4off allele. ascl1b-2A-Cre led to a modest rescue in the developing midbrain, possibly due to expression of ascl1b in a subset of proneural progenitors in this tissue. The complete absence of rescue by neurod1-2A-Cre indicates rbbp4 isn’t required for survival in this progenitor population. These interpretations are supported by the results of conditional gene inactivation experiments with the rbbp4on allele described below. rbbp4on cell-type-specific inactivation with ascl1b-2A-Cre leads to the expected apoptotic mutant phenotype in the midbrain, whereas inactivation with neurod1-2A-Cre did not result in apoptosis in either the midbrain or retina (Figure 6).
Figure 6. Ubiquitous and cell-type specific Cre-mediated conditional inactivation with rbbp4-on.
(A) Diagram of expected Cre-mediated inversion of rbbp4on to “off” orientation. (B) Normal morphological phenotype in 5 dpf transheterozygous rbbp4on/Δ4larva. (C) Induction of microcephaly and microphthalmia and mRFP expression in Cre injected 5 dpf transheterozygous rbbp4on/Δ4larva. (D) Absence of activated caspase-3a labeling in sectioned tissue from 2 dpf uninjected transheterozygous rbbp4on/Δ4embryo. (E) Activated caspase-3a labeling in the midbrain and retina of 2 dpf transheterozygous rbbp4on/Δ4embryo after Cre injection. (F) Quantification of caspase-3a labeling in control rbbp4on/Δ4(n=3) and Cre-injected rbbp4on/Δ4(n=3) midbrain (* p<0.05) and retina (* p<0.05). (G) Genomic DNA qPCR quantification of rbbp4on original orientation 5’ and 3 junctions in control rbbp4on (n=3) and Cre injected rbbp4on/Δ4 (n=3). (H – K”) Activated caspase-3a and Cre labeling in sectioned head tissue from 2 dpf ascl1b-2A-Cre; rbbp4D4/+ (H-H"), ascl1b-2A-Cre; rbbp4on/Δ4 (I-I"), neurod1-2A-Cre; rbbp4Δ4/+ (J-J"), and neurod1-2A-Cre; rbbp4on/Δ4 (K-K"), embryos. Green arrowheads, activated caspase-3a-positive cells. White arrowheads, hypercondensed and fragmented nuclei. (L) Quantification of caspase-3a labeling in ascl1b-2A-Cre; rbbp4on/+ (n=4) and ascl1b-2A-Cre; rbbp4on/Δ4 (n=6) midbrain (** p<0.01) and retina (n.s. p=0.8543). (M) Quantification of caspase-3a labeling in neurod1-2A-Cre; rbbp4on/+ (n=3) and neurod1-2A-Cre; rbbp4on/Δ4 (n=3) midbrain (n.s. p=0.3739) and retina (n.s. p=0.6433). OT, optic tectum; R, retina; Th, thalamic region. Error bars represent mean ± s.e.m. with two-tailed t-test. Scale bars: 200 μm (B, C), 50 μm (D, E, H – K). 10 μm (H’ – K”).
Figure 6—figure supplement 1. Live imaging and molecular analysis of rbbp4on conditional inactivation by Cre injection.
Figure 6—figure supplement 2. Induction of primary reporter RFP expression and qPCR quantification of rbbp4on allele inversion by ascl1b-2A-Cre and neurod1-2A-Cre.
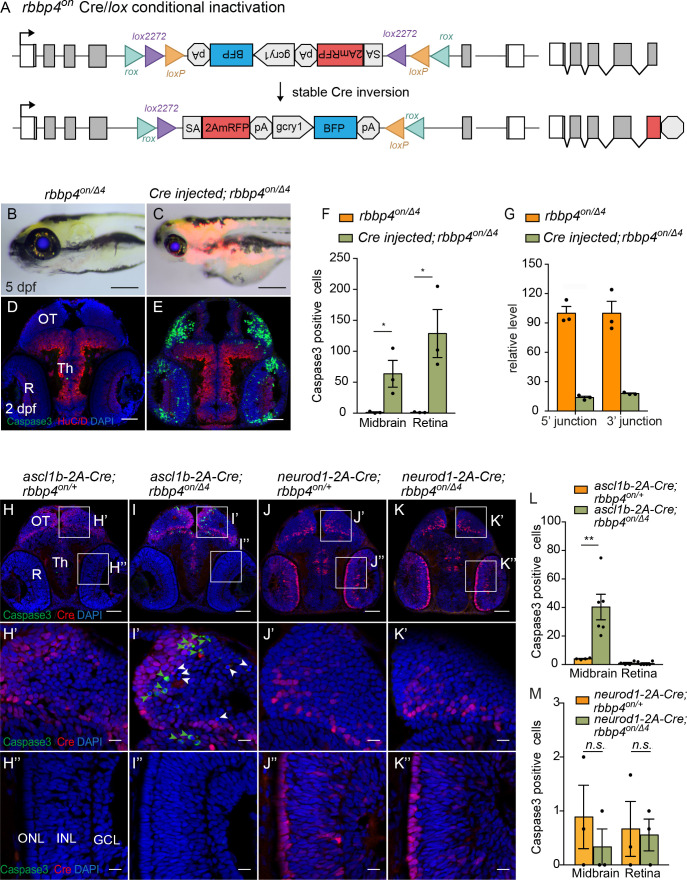
Cre/lox recombination of rbbp4on leads to efficient conditional gene inactivation and recapitulates morphological and cellular loss-of-function phenotypes
To assess whether Cre recombination of the rbbp4on allele to the ‘off’ orientation would lead to conditional inactivation and induction of expression of the gene trap mRFP, we first used 12.5 pg Cre mRNA injection into embryos from a cross between rbbp4on/+ and rbbp4Δ4/+ heterozygous adults (Figure 6). In comparison to uninjected rbbp4on/Δ4 larvae which appear morphologically normal (Figure 6B), 5 dpf rbbp4on/Δ4 Cre mRNA-injected larvae display the rbbp4 microcephaly and microphthalmia loss-of-function phenotype, and induction of RFP expression (Figure 6C). Sectioned tissue from 2 dpf embryos showed a significant increase in activated caspase 3 labeling throughout the midbrain (* p<0.05) and retina (* p<0.05) (Figure 6D–F; Figure 6—source data 1), consistent with the rbbp4 mutant apoptotic cellular phenotype. Confocal live imaging in 2 dpf control and injected rbbp4on/Δ4 embryos confirmed induction of the gene trap RFP expression and cell death in the retina (Figure 6—figure supplement 1A–L). PCR junction analysis and sequencing (Figure 6—figure supplement 1M–P), and genomic DNA qPCR analysis of 5’ and 3’ junctions (Figure 6G; Figure 6—source data 1) confirmed robust stable inversion of the rbbp4on allele to the ‘off’ orientation (82%–84% efficiency) correlated with induction of the rbbp4 loss-of-function phenotype.
Proneural Cre/lox rbbp4on conditional inactivation reveals cell-type-specific requirement for rbbp4 in ascl1b progenitor survival
We conducted the same experimental approach as described above using the endogenous proneural Cre driver lines ascl1b-2A-Cre, gcry1:GFP is75 and neurod1-2A-Cre, gcry1:GFP is77, but in combination with the rbbp4on allele to test for cell-type-specific conditional inactivation (Figure 6H–M). Embryos from rbbp4on/+ + crossed with ascl1b-2A-Cre/+; rbbp4Δ4/+ adults were examined for induction of gene trap RFP expression and assessed for Cre-mediated allele inversion by 5’ and 3’ junction genomic qPCR (Figure 6—figure supplement 2; Figure 6—figure supplement 2—source data 1). Three dpf ascl1b-2A-Cre/+; rbbp4on/Δ4 larvae show RFP expression throughout the brain and neural tube and a 20% inversion rate (Figure 6—figure supplement 2 B, B’, D). Although the larvae appeared morphologically normal, compared to controls the sectioned tissue from 2 dpf embryos showed a significant increase in activated caspase 3 labeling in the midbrain tectum and dorsal thalamic region (** p<0.01) (Figure 6H–I and L; Figure 6—source data 1). Activated caspase 3 labeling was not detected in the retina (n.s. p=0.08543), consistent with the lack of retinal expression of ascl1b and Cre (Figure 6H”–I” and L). Conditional inactivation of rbbp4on with neurod1-2A-Cre also lead to induction of RFP expression in both the brain and the retina, albeit at lower levels, and 20% cassette inversion efficiency (Figure 6—figure supplement 2 C, C’, E; Figure 6—figure supplement 2—source data 1), However, in contrast to conditional inactivation with ascl1b-2A-Cre, inactivation with neurod1-2A-Cre did not lead to significantly different levels of caspase 3 labeling in the 2 dpf brain (n.s. p=0.3739) or retina (n.s. p=0.6434) compared to controls (Figure 6J–K, M; Figure 6—source data 1). These results suggest Cre-mediated conditional inactivation of rbbp4 in the ascl1b neural progenitor population leads to apoptosis. Although rbbp4 is expressed in the neurod1 population as shown by induction of RFP, the absence of apoptosis indicates rbbp4 may not be required for survival in committed progenitors or newborn neurons. These results support two critical conclusions: First, conditional inactivation of rbbp4on with ubiquitous Cre leads to efficient stable inversion of the cassette from ‘on’ to ‘off’, and replicates the rbbp4 loss-of-function phenotype. Second, rbbp4on can be inactivated in a cell-type-specific manner that reveals a potential cell-specific function for rbbp4 in ascl1b-neural progenitor cell survival.
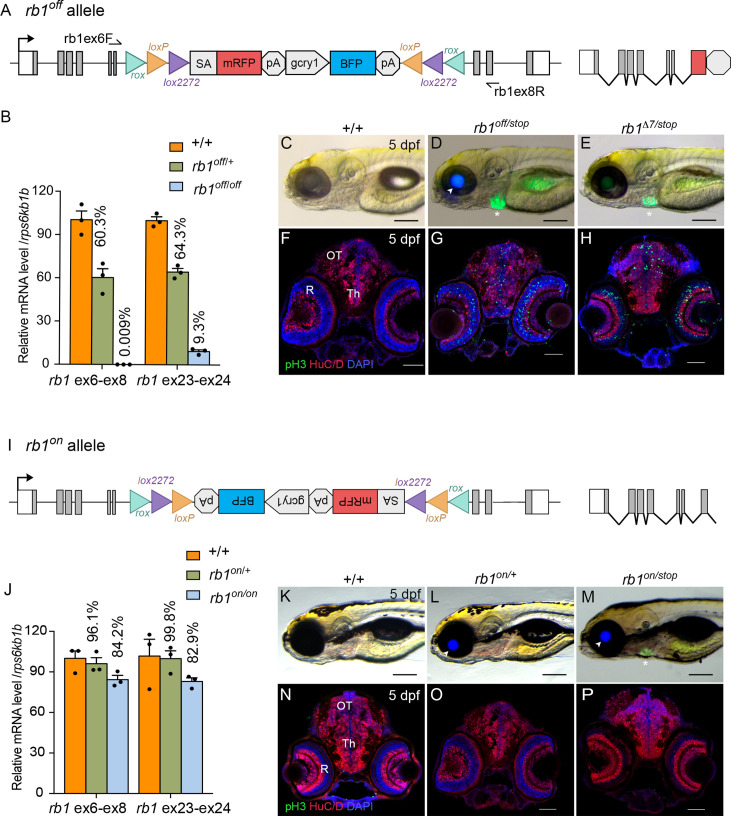
rb1off allele provides robust gene knockdown and loss of function
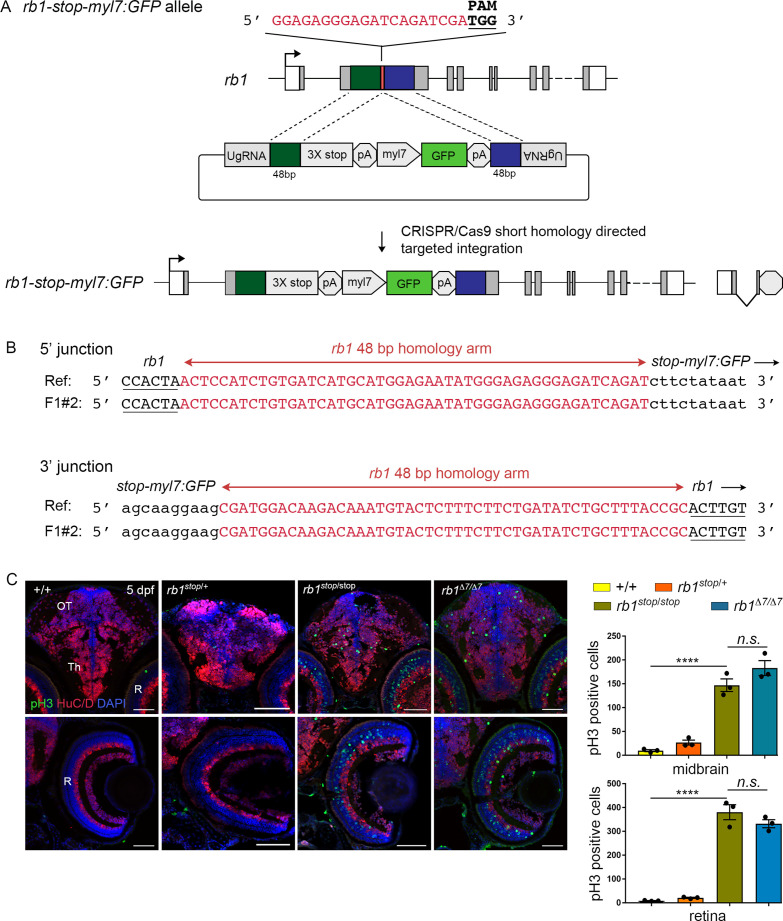
To validate the UFlip approach for generating conditional alleles at a second locus, we characterized the activity of the rb1off is58 and rb1on is57 alleles (Figure 7). RT-qPCR of homozygous rb1off 3 dpf larvae with primers spanning rb1 exons 6–8 showed >99% knockdown of rb1 mRNA levels in comparison to wild-type sibling larvae (Figure 7A, B; Figure 7—source data 1). To simplify identification of trans heterozygous rb1 mutant larvae, we isolated an rb1 exon 2 CRISPR integration allele, rb1-stop-myl7:GFPis59 (rb1-stop) using a targeting vector design similar to our recently published method to isolate endogenous Cre lines in proneural genes (Almeida et al., 2021). The stop-myl:GFP cassette contains three translation termination codons, in each of the three reading frames, followed by a transcription terminator plus a myl7:GFP secondary marker (Figure 7—figure supplement 1A, B). The rb1stop allele is larval lethal in combination with the rb1Δ7 indel mutation and homozygous rb1stop/stop 5 dpf larvae show the same level of increased numbers of pH3 labeled cells throughout the midbrain (n.s. p=0.2951) and retina (n.s. P=0.1534) (Figure 7—figure supplement 1C; Figure 7—figure supplement 1—source data 1) as rb1Δ7/Δ7 homozygotes (Schultz et al., 2018). Trans-heterozygous rb1off/stop larvae from a cross between rb1off/+ and rb1stop/+ heterozygotes are easily distinguished from siblings by the expression of lens BFP by the rb1off allele (Figure 7D arrowhead) and heart GFP by the rb1stop allele (Figure 7D asterisk). rb1off/stop 5 dpf larvae appear morphologically normal compared to wild type, with the exception of the lack of a swim bladder (Figure 7C, D), similar to rb1Δ7/stop larvae (Figure 7E). In contrast to wild type (Figure 7F), 5 dpf sectioned head tissue shows pH3-labeled cells throughout the rb1off/stop midbrain (*** p<0.001) and retina (**** p<0.0001), like rb1Δ7/stop trans-heterozygotes (Figure 7F–H; Figure 7—figure supplement 2A; Figure 7—source data 1) and rb1stop/stop and rb1Δ7/Δ7 homozygotes (Figure 7—figure supplement 1C). Together these results demonstrate the rb1off allele provides robust gene knockdown and recapitulates the zebrafish rb1 loss-of-function morphological and cellular phenotypes.
Figure 7. Molecular and phenotypic characterization of rb1off and rb1on alleles.
(A) Diagram of the rb1off allele. (B) Plot of RT-qPCR results from wild type +/+ (n=3), heterozygous rb1off/+ (n=3), and homozygous rb1off/off (n=3) larvae showing the relative level of rb1 mRNA transcript using reference gene rps6kb1b. Primer pairs were located in exons 6 and 8, or downstream exons 23 and 24. (C – E) Gross phenotype of wildtype +/+ (C), rb1off/stop (D), and rb1Δ7/stop (E) 5 dpf larvae. Arrowhead in D points to rb1off allele gcry1:BFP secondary reporter expression in lens. Asterisk marks the rb1stop allele mly7:GFP secondary reporter expression in heart. (F – H) pH3 and HuC/D labeling of sectioned head tissue from 5 dpf +/+ (F), rb1off/stop (G), and rb1Δ7/stop (H). (I) Diagram of the rb1on allele. (J) Plot of RT-qPCR results from wild type +/+ (n=3), heterozygous rb1on/+ (n=3), and homozygous rb1on/on (n=3) larvae showing the relative level of rb1 mRNA transcript using reference gene rps6kb1b. Primer pairs were located in exons 6 and 8, or downstream exons 23 and 24. (K, L) Gross phenotype of wildtype +/+ (K), heterozygous rb1on/+ (L) and transheterozygous rb1on/stop (M) 5 dpf larvae. Arrowhead in L, M points to rb1off allele gcry1:BFP secondary reporter expression in lens. Asterisk in M marks the rb1stop allele mly7:GFP secondary reporter expression in heart. pH3 and HuC/D labeling of sectioned head tissue from 5 dpf +/+ (N), heterozygous rb1on/+ (O) and transheterozygous rb1on/stop (P).OT, optic tectum; R, retina; Th, thalamic region. Error bars represent mean ± s.e.m. Scale bars: 200 μm (C–E, K–M), 50 μm (F–H, N–P).
Figure 7—figure supplement 1. The rb1-stop integration allele recapitulates the rb1Δ7 indel loss-of-function phenotype.
Figure 7—figure supplement 2. Quantification of pH3 labeled cells in rb1off/stop, rb1Δ7/stop, and rb1off/stop mutant larvae.
rb1on allele maintains rb1 function
The rb1on allele (Figure 7I) was examined for its effect on rb1 mRNA expression and gene activity. RT-qPCR of rb1on/+ heterozygotes and rb1on/on homozygotes showed 84% and 83% levels, respectively, of rb1 mRNA compared to wild-type controls (Figure 7J; Figure 5—source data 1). Like wildtype, heterozygous rb1on/+ and trans-heterozygous rb1on/stop 5 dpf larvae appear morphologically normal (Figure 7K–M). pH3 labeling in 5 dpf sectioned head tissue showed no significant difference between wild type (+/+) and rb1on/+ (Figure 7N, O; Figure 7—figure supplement 2B; Figure 7—figure supplement 2—source data 1) in the midbrain (n.s. p=0.1118) and a slightly significant increase in the retina (* p>0.05). In rb1on/stop transheterozygous larvae an increase in pH3 labeled cells was detected in the midbrain (** p<0.01) and retina (** p0.01) (Figure 7P; Figure 7—figure supplement 2B; Figure 7—source data 1). However, the actual increase in number was minor, from 10 to ~30, in contrast to hundreds of pH3-positive cells in homozygous mutant rb1 tissues. Together, these results demonstrate the rb1off allele knocks down rb1 mRNA expression and gene activity as expected, while the rb1on allele does not significantly disrupt either.
Cre/lox recombination of rb1off leads to stable inversion and conditional gene rescue
To test the rb1off and rb1on alleles for Cre-mediated conditional rescue and inactivation, we used the same approach as outlined above for the conditional rbbp4off and rbbp4on alleles. First, 12.5 pg Cre mRNA injection was used to test for recombinase mediated inversion and conditional gene rescue in embryos from a cross between rb1off/+and rb1stop/+adults (Figure 8A–G). Stable inversion of the rb1off cassette to the ‘on’ orientation was confirmed by PCR and sequence analysis, which showed the expected sequences at the 5’ and 3’ junctions after inversion at the lox sites (Figure 8—figure supplement 1). In comparison to uninjected rb1off/stop larvae (Figure 8B), 5 dpf rb1off/stop Cre injected larvae showed a significant reduction in the number of pH3-labeled cells in the midbrain (** p<0.01) and retina (** p<0.01) (Figure 8C, F; Figure 8—source data 1). Genomic DNA quantitative PCR of the original 5’ and 3’ rb1off allele junction fragments in the Cre injected embryos indicated a>78% inversion rate (Figure 8G; Figure 8—source data 1). Together these data demonstrate Cre mediated recombination of the rb1off allele leads to efficient, stable inversion of the cassette to the ‘on’ orientation and conditional rescue of the rb1 loss-of-function phenotype.
Figure 8. Ubiquitous and proneural neurod1-specific Cre-mediated conditional rescue with rb1off.
(A) Diagram of expected Cre-mediated inversion of rb1off to on orientation. (B, C) pH3 and HuC/D labeling of larval sectioned head tissue from 5 dpf transheterozygous rb1off/stop (B – B”) and Cre injected rb1off/stop (C – C”). (D, E) pH3 and Cre labeling of larval sectioned head tissue from 5 dpf transheterozygous rb1off/stop (D – D”) and neurod1-2A-Cre; rb1off/stop (E – E”). (F) Quantification of pH3-positive cells in control rb1off/stop (n=3) and Cre injected rb1off/stop (n=3) midbrain (** p<0.01) and retina (** p<0.01). (G) Genomic DNA qPCR quantification of rb1off original orientation DNA 5’ and 3’ junctions in control rb1off/stop (n=3) and Cre injected rb1off/stop (n=3). (H) Quantification of pH3-positive cells in rb1off/stop (n=3) and neurod1-2A-Cre; rb1off/stop (n=3) midbrain (** p<0.01) and retina (* p<0.05). (I) Genomic DNA qPCR quantification of rb1off original orientation DNA 5’ and 3’ junctions in control rb1off (n=3) and neurod1-2A-Cre; rb1off/stop (n=3). Error bars represent mean ± s.e.m. with two-tailed t-test. Scale bars: 50 μm (B - E), 10 μm (B’ – E”).
Figure 8—figure supplement 1. Molecular analysis of rb1off inversion and conditional rescue by Cre injection.
neurod1-specific rb1off conditional rescue demonstrates rb1 is required to suppress neurod1 neural progenitor cell cycle entry
The proneural neurod1-2A-Cre, gcry1:GFP is77 driver was used to test for conditional gene rescue of rb1 in the neurod1 progenitor population. Five dpf larvae from adult rb1off/+ crossed with neurod1-2A-Cre/+; rb1stop/+ was labeled with antibodies to Cre and pH3 (Figure 8D–I). In comparison to control rb1off/stop larvae, neurod1-2A-Cre/+; rb1off/stop larvae showed a significant reduction in pH3 positive cells in the midbrain (* p<0.05) and retina (** p<0.01) where Cre is expressed (Figure 8D–H; Figure 8—source data 1). Rescue of the phenotype correlated with >34% Cre mediated inversion of rb1off to the ‘on’ orientation (Figure 8I; Figure 8—source data 1). The results demonstrate cell-type-specific conditional rescue and a requirement for rb1 to suppress cell cycle entry in neurod1 progenitors.
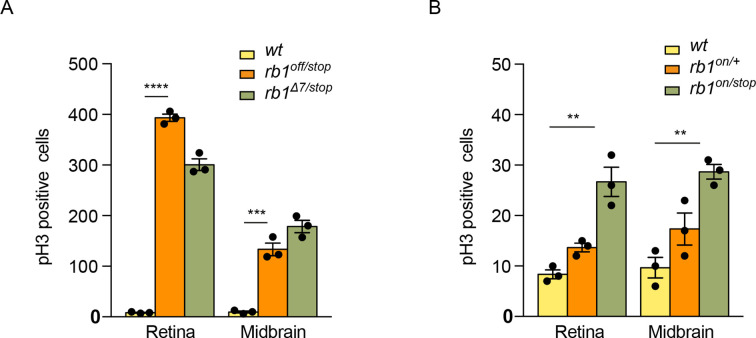
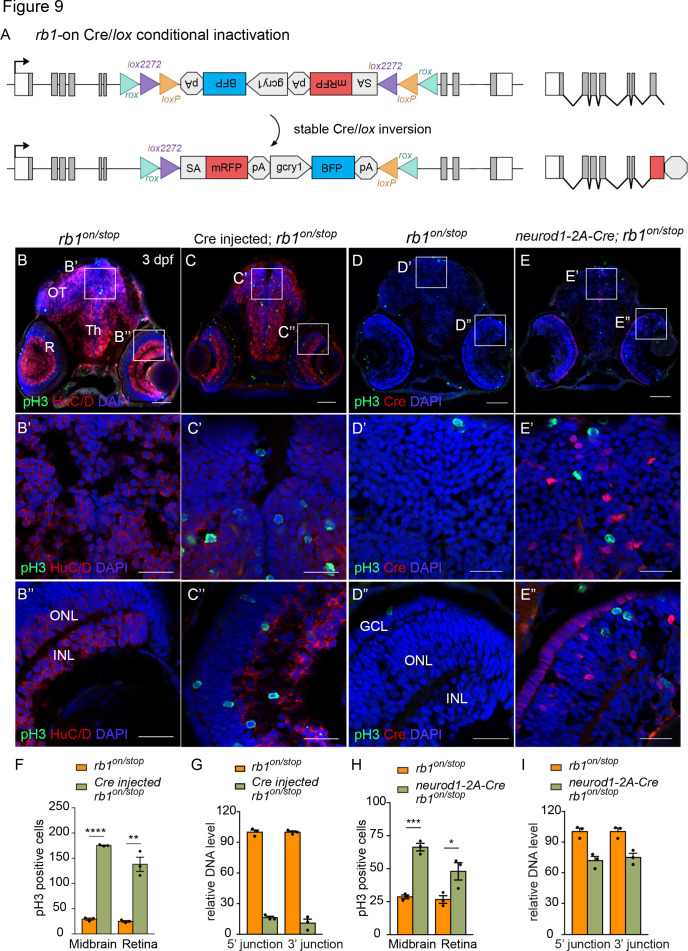
Cre/lox recombination of rb1on leads to efficient conditional gene inactivation and recapitulates cellular rb1 loss-of-function phenotype
Conditional rescue using the rb1on allele was first tested by injection of 12.5 pg Cre mRNA into embryos from a cross between heterozygous rb1on/+ and rb1stop/+ adults to induce inversion from the passive to the active orientation (Figure 9A). At 3 dpf rb1on/stop uninjected and injected larvae appear morphologically normal like control rb1on/+ larvae (Figure 9—figure supplement 1 A, B). However, in comparison to uninjected control sectioned head tissue (Figure 9B), Cre injected rb1on/stop showed increased levels of pH3 labeling throughout the brain and retina (Figure 9C), similar to the loss-of-function phenotype of rb1Δ7/stop and rb1off/stop (Figure 7D and E). Quantification showed a significant increase in the number of pH3-positive cells in injected larvae in both the midbrain (**** p<0.0001) and retina (** p<0.01) (Figure 9F; Figure 9—source data 1). Induction of the mutant phenotype correlated with >80% inversion rate by Cre recombination (Figure 9G; Figure 9—source data 1). Stable inversion of the UFlip cassette in Cre injected embryos was confirmed by PCR junction analysis and sequencing of the junction amplicons (Figure 9—figure supplement 1 C–F). These results demonstrate Cre recombination leads to stable inversion of the UFlip cassette and conditional gene inactivation which replicates the rb1 loss-of-function phenotype at the cellular level.
Figure 9. Ubiquitous and proneural neurod1-specific Cre-mediated conditional inactivation with rb1-on.
(A) Diagram of expected Cre mediated inversion of rb1on to ‘off’ orientation. (B, C) pH3 and HuC/D labeling of larval sectioned head tissue from 5 dpf transheterozygous rb1on/stop (B – B”) and Cre injected rb1on/stop (C – C”). (D, E) pH3 and Cre labeling of larval sectioned head tissue from 5 dpf transheterozygous rb1on/stop (D – D”) and neurod1-2A-Cre; rb1on/stop (E – E”). (F) Quantification of pH3-positive cells in control rb1on/stop (n=3) and Cre rb1on/stop (n=3) injected midbrain (**** p<0.0001) and retina (** p<0.01). (G) Genomic DNA qPCR quantification of rb1onoriginal orientation DNA 5’ and 3’ junctions in control rb1on/stop (n=3) and Cre injected rb1on/stop (n=3). (H) Quantification of pH3-positive cells in control rb1on/stop (n=3) and neurod1-2A-Cre; rb1on/stop (n=3) midbrain (*** p<0.001) and retina (* p<0.05). (I) Genomic DNA qPCR quantification of rb1on original orientation DNA 5’ and 3’ junctions in control rb1on/stop (n=3) control and neurod1-2A-Cre; rb1on/stop (n=3). Error bars represent mean ± s.e.m. with two-tailed t-test. Scale bars: 50 μm (B - E), 20 μm (B’ – E”).
Figure 9—figure supplement 1. Molecular analysis of rb1-on inversion and conditional inactivation by Cre injection.
neurod1-specific rb1on conditional inactivation leads to neural progenitor cell cycle entry
The proneural neurod1-2A-Cre, gcry1:GFP is77 driver was used to test for conditional inactivation of rb1 in the neurod1 progenitor population. 3 dpf larvae from adult rb1on/+ crossed with neurod1-2A-Cre/+; rb1stop/+ was labeled with antibodies to Cre and pH3 (Figure 9D–I). In comparison to control rb1on/stop larvae (Figure 9D), neurod1-2A-Cre/+; rb1on/stop larvae showed a significant increase in pH3 positive cells in the midbrain (*** p<0.001) and retina (* p<0.05) where Cre is expressed (Figure 9E and H; Figure 9—source data 1). Induction of the mutant phenotype correlated with >25% inversion of rb1on to the ‘off’ orientation (Figure 9I; Figure 9—source data 1). The results are consistent with the rb1off conditional rescue experiment, and demonstrate a requirement for rb1 in suppressing cell cycle entry in the neurod1 progenitor cell population in the developing midbrain and retina.
Overall, these analyses show conditional alleles generated by targeted integration of the floxed UFlip cassette provide robust control of gene activity. Combined with cell-type-specific Cre drivers, the alleles demonstrate cell-type-specific requirements for gene activity through both conditional inactivation and rescue.
Discussion
In this study, we used GeneWeld CRISPR-Cas9 targeting to isolate Cre/lox regulated conditional alleles with linked reporters by integration of a floxed gene trap cassette, UFlip, at a single gRNA site in an intron. Gene ‘off’ and gene ‘on’ germline UFlip alleles were recovered for two genes, rbbp4 and rb1. Our results showed the UFlip cassette led to >99% gene knockdown at the molecular level, did not disrupt gene activity in the passive orientation, allowed stable Cre/lox mediated inversion for conditional inactivation or rescue, and recapitulated expected loss-of-function phenotypes at the morphological and cellular levels. We further demonstrated using proneural Cre drivers that conditional gene inactivation or rescue can be restricted to distinct neural progenitor cell populations. These studies revealed cell-type-specific requirements for gene activity that had previously only been inferred by global loss-of-function homozygous mutant analysis. Our approach is simple in design, efficient, generates effective Cre/lox regulated alleles for genetic mosaic analysis and can be applied to any gene of interest.
We assembled UFlip vectors with 48 bp homology arms to generate integrations in rbbp4 and rb1 in both the ‘off’ and ‘on’ orientations. Of the four potential targeted integration alleles, we recovered three precise alleles at a frequency of 4–14% of founders screened. This frequency is lower than our previous reports using GeneWeld to knock in fluorescent reporters (22–100%) (Wierson et al., 2020) or 2A-Cre (10–100%) (Almeida et al., 2021; Wierson et al., 2020). In those studies, injected embryos showing expression of the fluorescent tag, or Cre-mediated switching of a floxed reporter, were selected and raised to adulthood to screen for germline transmission. In this report, some lines were generated with vectors that only enabled embryo selection based on expression of the secondary reporter, which does not reflect on target integration. In support of this, only 7 and 8 adults needed to be screened to recover the rbbp4off and rb1off 2A-mRFP UFlip alleles, respectively, which showed mRFP expression in the injected embryos. This is in comparison to 25 adults that were screened to recover the original rb1on mRFP UFlip alleles. Moreover, starting with an allele in one orientation, it was straightforward to use Dre recombination at rox sites flanking the UFlip cassette to recover an allele in the opposite orientation. Dre/rox recombination has been shown to be highly efficient in somatic tissue (Park and Leach, 2013). By injection of Dre mRNA into rbbp4off embryos we readily recovered the rbbp4on allele in the next generation from 7/11 adults, demonstrating efficient Dre recombinase activity in the zebrafish germline. Previously reported methods for recovery of intron targeted floxed conditional alleles varied widely in frequency, from 12% for homology directed integration from a linear template with long 1–1.5 kb homology arms (Hoshijima et al., 2016), 53% by homologous recombination from a circular plasmid with 1 kb long homology arms (Sugimoto et al., 2017), to 56–60% for integration by Non-Homologous End Joining after preselection of a primary reporter (Han et al., 2021; Li et al., 2019). The simplicity of UFlip vector construction and ability to recover precise integrations, in either orientation, underscore the power of our approach for isolation of stable Cre/lox regulated conditional alleles.
Similar to our previous findings when isolating endogenous Cre drivers (Almeida et al., 2021), the UFlip founders transmitted both precise and imprecise integration events. In some cases, the imprecise event was a simple homology arm duplication or short insertion at either the 5’ or 3’ junction, which would be predicted not to affect gene function since the integrated cassette is located in an intron. As predicted, the rb1off allele, from which we were able to establish a line, had a 5’ homology arm duplication separated by a 115 bp insertion. This did not affect the ability of the cassette to efficiently invert in response to Cre recombinase. When inverted into the passive orientation the cassette allowed effective conditional rescue, indicating the additional sequences in the intron flanking the cassette didn’t disrupt splicing or rb1 gene activity.
For effective conditional gene inactivation, it is essential that the method leads to robust knockdown of wildtype mRNA expression and recapitulates loss-of-function phenotypes for genetic analysis. Our UFlip conditional construct is based on the Tol2 transposon RP2 gene trap, which contains a transcriptional terminator that effectively blocks transcription readthrough and reduces transcript levels by >99% (Clark et al., 2011; Ichino et al., 2020). The efficacy of gene knockdown by the UFlip gene trap was demonstrated by quantitative RT-qPCR and revealed >99% reduction in mRNA levels. Cre/lox-mediated inversion of the UFlip cassette from the passive to active orientation was highly efficient after Cre mRNA injection or exposure to a transgenic source of Cre, generating expected phenotypes in combination with loss-of-function mutations. The recently published Cre-regulated CRISPR indel targeting study provides a rapid method for conditional mutagenesis and phenotypic knockout in embryos (Hans et al., 2021). In contrast, a significant advantage of our stable conditional UFlip alleles rests on the strength of transcriptional termination by the UFlip cassette in the active orientation. In addition, Cre induction of conditional inactivation leads to mRFP fluorescent cell labeling, as described in other recent studies (Han et al., 2021; Hans et al., 2021; Li et al., 2019), which is critical for lineage tracing and capturing mutant cells for downstream genomics applications.
We further validated effective UFlip conditional gene inactivation at the cellular level. We previously demonstrated that rb1 is required to prevent neural progenitor cell cycle re-entry in the developing zebrafish brain and rbbp4 is necessary for neural progenitor survival and differentiation (Schultz et al., 2018). Using our ascl1b-2A-Cre and neurod1-2A-Cre drivers in combination with rbbp4on and rb1on conditional alleles, we demonstrated the requirement for each gene could be examined specifically in the ascl1b or neurod1 neural progenitor populations. Rb1 is required to prevent cell cycle re-entry in ascl1b and neurod1 neural progenitors. Rbbp4 appears to be required for neural progenitor survival only in ascl1b progenitors. Further analysis with tamoxifen regulated CreERT2 drivers will refine our understanding of the requirement for these genes in neural progenitor survival and proliferation. These conditional genetic tools provide a foundation for future mechanistic studies investigating the interaction of rb1 and rbbp4 specifically in neural progenitor cell cycle regulation and differentiation but can also be used with other cell-type-specific Cre drivers. Our results illustrate the robustness of the UFlip conditional alleles for cell type-specific gene knockout analysis and their potential application in different developmental or disease model contexts.
In summary, building on our previous streamlined GeneWeld approach for CRISPR-Cas short homology directed targeted integration, we describe an effective method for generating zebrafish floxed conditional alleles. We present a thorough molecular and phenotypic validation of the robustness of the UFlip floxed gene trap cassette for effective knockdown of gene expression and Cre/lox regulated conditional inactivation and rescue. UFlip alleles can be combined with any available cell or tissue-specific Cre driver for spatial and temporal gene knock out, enabling the investigation of gene function in specific developmental and post embryonic stages or disease models. The UFlip vector can be adapted to introduce a variety of gene modification cassettes, such as alternative exons and complete cDNAs. In contrast to Tol2 transposon binary UAS overexpression systems, targeted integration leads to control by endogenous gene regulatory elements and should increase phenotype consistency. The UFlip integration approach provides a powerful platform for generating new Cre/lox genetic tools to modify gene function at endogenous loci in zebrafish.
Materials and methods
Key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | ascl1b | ensemble | ENSDARG00000009702 | |
| Gene (Danio rerio) | neurod1 | ensemble | ENSDARG00000019566 | |
| Gene (Danio rerio) | rb1 | ensemble | ENSDARG00000006782 | |
| Gene (Danio rerio) | rbbp4 | ensemble | ENSDARG00000029058 | |
| Strain, strain background (Danio rerio) | WIK | Zebrafish International Resource Center | ZIRC:ZL84 | Wildtype strain of zebrafish |
| Genetic reagent (Danio rerio) | Tg(ascl1b-2A-Cre) | McGrail lab | Tg(ascl1b-2A-Cre; gcry1:EGFP)is75 | Maintained in the lab of M. McGrail (Almeida et al., 2021) |
| Genetic reagent (Danio rerio) | Tg(neurod1-2A-Cre) | McGrail lab | Tg(neurod1-2A-Cre; gcry1:EGFP)is77 | Maintained in the lab of M. McGrail (Almeida et al., 2021) |
| Genetic reagent (Danio rerio) | rb1Δ7 | McGrail lab | rb1Δ7is54 | Maintained in the lab of M. McGrail (Solin et al., 2015) |
| Genetic reagent (Danio rerio) | rbbp4Δ4 | McGrail lab | rbbp4Δ4is60 | Maintained in the lab of M. McGrail (Schultz et al., 2018) |
| Genetic reagent (Danio rerio) | Tg(rb1-UFlip-on) | This paper | Tg(rb1-i6-UFlip-rox-lox2272-loxP-inverted<RFP;gcry1:BFP >lox2272-loxP-rox-on)is57 | Available from M. McGrail lab |
| Genetic reagent (Danio rerio) | Tg(rb1-UFlip-off) | This paper | Tg(rb1-i6-UFlip-rox-loxP-lox2272<RFP;gcry1:BFP >loxP-lox2272 -rox-off)is58 | Available from M. McGrail lab |
| Genetic reagent (Danio rerio) | Tg(rb1-UFlip-off) | This paper | Tg(rb1-i6-UFlip-rox-loxP-lox2272<2A-RFP;gcry1:BFP >loxP-lox2272 -rox-off)is63 | Available from M. McGrail lab |
| Genetic reagent (Danio rerio) | Tg(rbbp4-UFlip-off) | This paper | Tg(rbbp4-i4-UFlip-rox-lox2272-loxP-<2A-RFP;gcry1:BFP >lox2272-loxP-rox-off)is61 | Available from M. McGrail lab |
| Genetic reagent (Danio rerio) | Tg(rbbp4-UFlip-on) | This paper | Tg(rbbp4-i4-UFlip-rox-lox2272-loxP-<2A-RFP;gcry1:BFP >lox2272-loxP-rox-on)is62 | Available from M. McGrail lab |
| Genetic reagent (Danio rerio) | Tg(rb1-stop-PRISM) | This paper | Tg(rb1-3XSTOP; myl7:GFP)is59 | Available from M. McGrail lab |
| Recombinant DNA reagent | pT3TS-nCas9n | Wenbiao Chen | Addgene:46,757 | Plasmid for in vitro synthesis of Cas9 mRNA |
| Recombinant DNA reagent | pT3TS-Cre | Karl Clark | Plasmid for in vitro synthesis of Cre mRNA, available from K. Clark lab (Clark et al., 2011) | |
| Recombinant DNA reagent | pT3TS-Dre | Karl Clark | Dre cDNA (Anastassiadis et al., 2009) expression vector for in vitro mRNA synthesis, available from K. Clark lab | |
| Recombinant DNA reagent | p-CS2-KalTA4 | Martin Distel | Available from M. Distel lab (Distel et al., 2009) | |
| Recombinant DNA reagent | pUFlip-floxed-mRFP, gcry1:BFP | This paper | pUFlip(UgRNA-rox-loxP-lox2272<RFP; gcry1:BFP >loxP-lox2272-rox-UgRNA) | decommissioned |
| Recombinant DNA reagent | pUFlip-floxed-mRFP, myl7:BFP | This paper | pUFlip(UgRNA-rox-loxP-lox2272<RFP; myl7:BFP >loxP-lox2272-rox-UgRNA) | decommissioned |
| Recombinant DNA reagent | pUFlip-floxed-2A-mRFP, gcry1:BFP | This paper | pUFlip(UgRNA-rox-loxP-lox2272<2A-RFP; gcry1:BFP >loxP-lox2272-rox-UgRNA) | available from M. McGrail lab; Deposited at Addgene |
| Recombinant DNA reagent | pUFlip-floxed-2A-KalTA4, gcry1:BFP | This paper | pUFlip(UgRNA-rox-loxP-lox2272<2A-KalTA4; gcry1:BFP >loxP-lox2272-rox-UgRNA) | available from M. McGrail lab; Deposited at Addgene |
| Recombinant DNA reagent | pSTOP-PRISM-3Xstop-myl7:GFP | This paper | pPRISM(UgRNA-3XSTOP; myl7:GFP-UgRNA) | available from J. Essner lab; Deposited at Addgene |
| Sequence-based reagent | This paper | PCR primers and oligos | See Table 1 | |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Cloning kit | New England Biolabs | Catalog # E5520S | |
| Commercial assay or kit | PureYield Plasmid Miniprep System | Promega | Catalog # A1223 | |
| Commercial assay or kit | mMessage mMACHINE T3 Transcription Kit | Ambion | Catalog # AM1348 | |
| Commercial assay or kit | RNA Clean and Concentrator Kit (RCC) | Zymo | Catalog # R1013 | |
| Commercial assay or kit | pCR4 TOPO TA Cloning Kit | ThermoFisher/ Invitrogen | ThermoFisher:K457502 | |
| Commercial assay or kit | Superscript III | Invitrogen | Catalog # 18080093 | |
| Commercial reagent | SYBR Green | BioRad | Catalog # 1725271 | |
| Commercial reagent | Tissue-Tek O.C.T. Compound | Fisher | Catalog # 4,583 | |
| Software, algorithm | ICE | Synthego | Inference of CRISPR Edits (ICE) https://www.synthego.com/products/bioinformatics/crispr-analysis | Indel analysis of Sanger sequenced DNA |
| Software | Graphpad PRISM | Statistical analyses | ||
| Antibody | Rabbit polyclonal anti -Caspase-3a | BD Pharmingen | Catalog # 559,565 | (1:500) |
| Antibody | Mouse monoclonal anti-Cre Recombinase | Chemicon | Catalog # MAB3120 | (1:500) |
| Antibody | Mouse monoclonal anti-HuC/D | Invitrogen | Catalog # A21271 | (1:500) |
| Antibody | Mouse monoclonal anti-phospho-Histone H3 (Ser10) | Millipore | Catalog # 05–806 | (1:500) |
| Antibody | Rabbit polyclonal anti-phospho-Histone H3 | Millipore | Catalog # 06–570? | (1:1000) |
| Antibody | Rabbit polyclonal anti-Rbbp4 | Bethyl Laboratories | Catalog # A301-206A | (1:200) |
| Chemical compound, drug | DAPI | Invitrogen | Catalog # D1306 | (0.5 ug/ml) |
Ethics declarations and approval for animal experiments
Use of zebrafish for research in this study was performed according to the Guidelines for Ethical Conduct in the Care and Use of Animals (APA, 1986), and carried out in accordance with Iowa State University Animal Care and Use Committee IACUC-18–279 and IACUC-20–058 approved protocols. All methods involving zebrafish were in compliance with the American Veterinary Medical Association (2020), ARRIVE (Percie du Sert et al., 2020) and NIH guidelines for the humane use of animals in research.
Zebrafish strains and maintenance
Zebrafish (Danio rerio) were maintained on an Aquaneering aquaculture system at 26 °C on a 14 hr light/10 hr dark cycle. The WIK strain of wild type zebrafish was obtained from the Zebrafish International Resource Center (https://zebrafish.org/home/guide.php). Other zebrafish lines used in this study were previously described: Tg(ascl1b-2A-Cre; gcry1:EGFP)is75 and Tg(neurod1-2A-Cre; gcry1:EGFP)is77 (Almeida et al., 2021); rb1Δ7is54 (Solin et al., 2015); rbbp4Δ4is60 (Schultz et al., 2018).
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to Maura McGrail (mmcgrail@iastate.edu).
Floxed UFlip vector for CRISPR-targeted integration to generate conditional alleles
The UFlip version 1.0 floxed mRFP gene trap vector for targeted integration (Figure 1A) was designed to be compatible with our previously published GeneWeld CRISPR-Cas9 targeted integration strategy (Wierson et al., 2020). The UFlip cassette was assembled in the pPRISM parent vector (Almeida et al., 2021) which has Universal UgRNA sequences on and BfuAI and BspQI type II restriction enzyme sites for cloning 5’ and 3’ homology arms on either side of the cassette, as previously described (Wierson et al., 2020). In the UFlip vector, the homology arm cloning sites flank head to head oriented rox sites that sit outside alternating pairs of head to head loxP and lox2272 sites for stable inversion after Cre mediated recombination (Robles-Oteiza et al., 2015; Schnütgen et al., 2003). Internal to the rox and lox sequences is the RP2 gene trap (Clark et al., 2011), consisting of a gene trap Splice Acceptor-mRFP-ocean pout (Zoarces americanus) antifreeze gene transcriptional termination and polyadenylation sequence (Gibbs and Schmale, 2000), followed by a tissue-specific lens gcry1:BFP or heart myl7:BFP secondary reporter. To assemble UFlip an intermediate vector was created by adding EagI and ClaI restriction sites to the pPRISM backbone by PCR. Six pairs of oligos, each containing the sequence of one of the rox, loxP or lox2272 sites, with complementary overhangs were ligated into the EagI and ClaI digested backbone. All three reading frames of the RP2 gene trap plus secondary reporter was directionally cloned into AvrII and SacI restriction sites located inside the pairs of rox/loxP/lox2272 sites. The UFlip version 2.0 vector was generated by mRFP primary reporter in the first version of the UFlipv1.0 vector was subsequently replaced with the porcine teschovirus-1 polyprotein 2 A peptide fused to mRFP. A UFlip version with a gene trap primary reporter containing the 2A-KalTA4 cDNA (Distel et al., 2009) for signal amplification using with a UAS:reporter is also available.
Intronic sgRNA target site selection, UFlip homology arm design and targeted integration
To identify CRISPR gRNA sites in introns, intronic sequences from four adult female and four adult make WIK fish were amplified from fin clip DNA using the proofreading enzyme KOD and sequenced to identify non-repetitive sequences that were shared among adults in the population. Gene specific and vector Universal UgRNA synthetic gRNAs with 2’-O-Methyl at first and last bases, 3’ phosphorothioate bonds between first 3 and last 2 bases, were ordered from Synthego . gRNA efficiency was determined by co-injection of 25 pg gRNA plus 300 pg Cas9 mRNA into one-cell stage embryos, followed by PCR amplification of the targeted intron and analysis of heteroduplex formation by gel electrophoresis. Amplicons were Sanger sequenced and the sequences analyzed for indel efficiency using Synthego’s Inference of CRISPR Edits (ICE) analysis software (https://ice.synthego.com/#/). The UFlip targeting vectors were built following the GeneWeld protocol (Welker et al., 2021). 48 bp 5’ and 3’ homology arms were designed to sequences flanking the genome intronic CRISPR-Cas9 target site. The homology arms were assembled by annealing complementary oligonucleotides with appropriate overhangs for cloning into the BfuAI and BspQI type IIS restriction enzyme sites that flank the UFlip cassette. To generate an active, gene ‘off’ allele, the 5’ and 3’ homology arms were cloned into the UFlip BfuAI and BspQI sites, respectively. To integrate the UFlip cassette in the passive, gene ‘on’ orientation, the position of the cloned 5’ and 3’ homology arms was reversed, with the 3’ homology arm cloned into the BfuAI site upstream of the cassette, and the 5’ homology arm cloned in the BspQI site downstream of the cassette. gRNA and homology arm oligonucleotide sequences are listed in Table 1.
For synthesis of Cas9 mRNA the expression vector pT3TS-nCas9n (Addgene #46757) (Jao et al., 2013) was linearized with Xba I (New England Biolabs, R0145S). One μg linearized vector was purified with the PureYield Plasmid Miniprep System (Promega, A1223) and used as template for in vitro synthesis of capped mRNA with the Ambion mMessage Machine T3 Transcription Kit (Thermo Fisher, AM1348). in vitro synthesized mRNA was purified with the RNA Clean and Concentrator Kit RCC (Zymo, R1013).
To target CRISPR-Cas9 driven integration of the UFlip cassette into intronic sites, a 2 nl volume containing 25 pg of genomic gRNA, 25 pg of UgRNA, 10 pg of UFlip targeting vector, and 300 pg Cas9 mRNA was co-injected into one-cell stage embryos from crosses between sequence validated adult fish. Larvae were screened at 3 dpf for expression of the lens gcry1:BFP or heart myl7:BFP secondary marker. Three to four BFP-positive embryos were selected for genomic DNA isolation and confirmation of on target integration by PCR sequence analysis of 5’ and 3’ junctions. BFP-positive sibling embryos were raised to adulthood. Primers, gRNA and homology arm oligonucleotide sequences are listed in Table 1 and Table 4.
Table 4. Primer oligonucleotide sequences.
| Primer name | Sequence | Purpose |
|---|---|---|
| rbbp4e × 4 F | ACCAAACACCCCTCCAAACCAG | intron sequencing, sgRNA analysis and genome/vector 5' junction analysis |
| rbbp4e × 5 R | AGTGCACTCTCCAGAGGGGT | intron sequencing, sgRNA analysis and genome/vector 3' junction analysis |
| rb1e × 6 F | CATGTTCCTCCTGGCCAAG | genome/vector 5' junction analysis |
| rb1e × 7 R | CACAAGGTCATCTTCCATCTG | genome/vector 3' junction analysis |
| rb1e × 2 F | GAGGAGCTCCAGTCCACTAAC | genotyping and genome/vector 5' junction analysis |
| rb1e × 2 R | CCCAAAACACAAGTGCGGTAA | genotyping and genome/vector 3' junction analysis |
| R-5'-junc-Stop-pPRISM | CGGTGGCTGAGACTTAATTACT | stop-PRISM genome/vector 5' junction analysis |
| F-3'-junc-(all)-pPRISM | TTCAGATCAATTAACCCTCACC | stop-PRISM genome/vector 3' junction analysis |
| RFPj | ATGACGTCCTCGGAGGAGGC | UFlip genome/vector junction analysis |
| RFPj2 | CCTTGGTCACCTTCAGCTTG | UFlip genome/vector junction analysis |
| BactinpAj | GCAAACGGCCTTAACTTTCC | UFlip genome/vector junction analysis |
| 2AR | CATAGGACCGGGGTTTTCTT | UFlip and 2A-Cre genome/vector junction analysis |
| rb1e × 6 F | CAGCTGGACCATGTTCCTCC | QPCR |
| rb1e × 8 R | CCCTGATTACGGCGTGATGT | QPCR |
| rbbp4e × 4 F | TAGTGACGTGCTGGTCTTTG | QPCR |
| rppb4e × 5 R | CAGGACAGACCATAACCTTCTT | QPCR |
| rbbp4e × 11_1 F | CTCTGTGTCTGAGGACAACATC | QPCR |
| rbbp4e × 12_1 R | TATCCCTGAACCTCAGTGTCT | QPCR |
| rps6kb1b_1 F | TCCTGATGACTCCACACTGA | QPCR |
| rps6kb1b_1 R | GGCGAGGTGAACGGATTT | QPCR |
| BFP_F | CTGCCTCATCTACAACGTCAA | genotyping |
| BFP_R | CTTAGCGGGTTTCTTGGATCTAT | genotyping |
| neujF | TCCAACTGAACCCCAGAACT | genotyping |
| ascjF | GTCAACATGGGCTTCCAGAC | genotyping |
| rbbp4e × 2 F | GCGTGATGACAGATCTCATATTGTTTTCCC | genotyping |
| rbbp4e × 2 R | CTGGTGACATCTGGCAACCACT | genotyping |
Isolation of stable zebrafish rbbp4 and rb1 UFlip and stop-PRISM integration alleles
To identify founder fish transmitting a UFlip allele, adults were outcrossed to wild-type WIK and embryos were screened for expression of the UFlip secondary reporter expressing BFP in the lens (gcry1:BFP) or heart (myl7:BFP). To identify on target integration alleles, genomic DNA was extracted from individual BFP-positive embryos by digestion in 50 mM NaOH at 95 °C for 30 min and neutralization by addition of 1/10th volume 1 M Tris-HCl pH 8.0. Genomic DNA/UFlip cassette 5’ and 3’ junctions were amplified by PCR with gene specific and UFlip primers listed in Table 4, followed by direct Sanger sequencing. BFP-positive sibling embryos from a founder that was transmitting a precise UFlip integration allele were raised to adulthood. F1 adult animals were fin clipped and the genotype of individuals confirmed by PCR and sequencing. Confirmed F1 adults were outcrossed to wild-type WIK, and F2 adults again were confirmed for the presence of a precise UFlip integration allele. Individual confirmed F2 adults were outcrossed to WIK to establish independent transgenic lines.
The rb1-stop-PRISM-myl7:GFP allele was generated with a pPRISM (PRecise Integration with Secondary Marker) GeneWeld targeted integration vector containing a cassette with splice acceptor followed by three copies of TGA, TAA, TAG, the ocean pout (Zoarces americanus) antifreeze gene transcriptional termination and polyadenylation sequence (Gibbs and Schmale, 2000), and a myl7:eGFP-βactin polyadenylation secondary reporter. Previously described 5’ and 3’ homology arms complementary to the rb1 exon 2 CRISPR target site (Schultz et al., 2018; Wierson et al., 2020) were cloned into the BfuAI and BspQI type IIS restriction enzyme sites flanking the stop-PRISM-myl7:eGFP cassette. 25 pg of rb1 exon 2 genomic gRNA, 25 pg of UgRNA, 10 pg of rb1-stop-PRISM targeting vector, and 300 pg Cas9 mRNA were coinjected into one-cell stage embryos in a volume of 2 nl, and adult founders screened for transmission of the rb1-stop-PRISM-myl7:GFP allele. Precise 5’ and 3’ junctions were confirmed in heart myl7:eGFP expressing F2 fin clipped adults.
Quantitative RT-PCR
Experiments to measure endogenous gene mRNA levels by Reverse Transcription-quantitative PCR were designed and performed according to MIQE and updated guidelines (Bustin et al., 2009; Taylor et al., 2019). Three biological replicates were performed, with each replicate representing embryos from a different mating pair of fish. At 3 dpf 20 randomly selected larvae from incrosses of heterozygous rbbp4off/+, rbbp4on/+, rb1off/+, and rb1on/+ were collected for RNA extraction and genotyping. Individual dissected head tissue was placed in RNAlater (Qiagen/Thermo Fisher AM7020) or DNA/RNA shield (Zymo Research R1100-50) and individual trunk tissue was placed in 50 mM NaOH for genotyping. Five heads of each genotype were pooled and total RNA extracted using the Direct-zol RNA Microprep kit (Zymo Research, R2060) and the quality determined using a Bioanalyzer 2100 (Agilent) at the Iowa State University DNA Facility. RNA samples with a RIN >5 were normalized to the same concentration, and first strand cDNA was synthesized using SuperScript III First-Strand Synthesis SuperMix (ThermoFisher, 11752050) containing random hexamer and oligo dT primers. Primers were designed to amplify ~200 bp amplicons with an annealing temperature of 60 °C. Primer optimization and validation was performed with 3 primer concentrations (100, 200, and 400 nM) and 3 cDNA amounts (5, 25, and 215 ng) with two replicates per condition. Primer efficiency was calculated as described (Bustin et al., 2009; Taylor et al., 2019) and primer pairs with 90–100% efficiency were used for qPCR of control and test samples. The sequence of rbbp4, rb1 and reference gene rps6kb1b qPCR primers are listed in Table 4. qPCR was performed on each sample in triplicate using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, 1725270) on a CFX Connect Real-Time System (Bio-Rad).
Cre and Dre-mediated inversion of UFlip alleles
For synthesis of Cre and Dre mRNAs, the expression vectors pT3TS-Cre (Clark et al., 2011) and pT3TS-Dre were linearized with SalI (New England Biolabs, R0138S) and BamHI (New England Biolabs, R0136S), respectively. 1 μg linearized vector was purified with the PureYield Plasmid Miniprep System (Promega, A1223) and used as template for in vitro synthesis of capped mRNA with the Ambion mMessage Machine T3 Transcription Kit (Thermo Fisher, AM1348). In vitro synthesized mRNA was purified with the RNA Clean and Concentrator Kit RCC (Zymo, R1013). A total of 12.5 pg Cre or 15 pg Dre mRNA was injected into one-cell stage embryos to promote recombination-mediated inversion of the UFlip cassette at lox or rox sites. UFlip cassette inversion in Cre or Dre injected 3 dpf larvae was confirmed by digestion of individual larvae in 50 mM NaOH at 95 °C for 30 min and neutralization by addition of 1/10th volume 1 M Tris-HCl pH 8.0. Genomic DNA/UFlip cassette 5’ and 3’ junctions were amplified by PCR with gene specific and UFlip primers listed in Table 4, followed by direct Sanger sequencing.
Zebrafish tissue embedding, sectioning, immunolocalization, and imaging
Zebrafish embryo and larvae fixation, embedding, sectioning, and immunolabeling was as described previously (Schultz et al., 2018). Zebrafish embryos and larvae were anesthetized in 160 µg/ml Ethyl 3-aminobenzoate methanesulfonate (Tricaine, MS-222) C9H11NO2·CH4SO3 (Sigma-Aldrich, 886-86-2) in E3 embryo media (Westerfield, 1995) and head and trunk dissected. Trunk tissue was placed in 20 µl 50 mM NaOH for genotyping. Heads were fixed in 4% paraformaldehyde overnight at 4 °C, incubated in 30% sucrose overnight at 4 °C, then processed and embedded in Tissue-Tek OCT (Fisher, 4583). Tissues were sectioned at 14–16 µm on a Microm HM 550 cryostat. Antibodies used for labeling: rabbit polyclonal anti-phospho-Histone H3 PH3 1:1000 (Cell Signaling Technology; 9701); mouse monoclonal anti-phospho-Histone H3 (Ser10), clone 3H10 1:500 (Millipore 05–806); mouse monoclonal anti-HuC/D 1:500 (Invitrogen A-21271); mouse monoclonal Anti-Cre recombinase 1:250 (Millipore-Sigma MAB3120); rabbit polyclonal anti-Caspase-3a 1:500 (BD Biosciences 559565); Alexa-594 (Invitrogen A-11005) and Alexa-488 (Invitrogen A-11008) conjugated secondary antibodies 1:500. Tissues were counterstained with 5 µg/ml DAPI, mounted in Fluoro-Gel II containing DAPI (Electron Microscopy Sciences 17985–50) and imaged on a Zeiss LSM800 laser scanning confocal microscope.
For live imaging, some embryos and larvae were treated with 0.003% 1-phenyl 2-thiourea (Sigma, P7629) to inhibit pigment synthesis, anesthetized in Tricaine in embryo media (Westerfield, 1995) and mounted on slides in 1.2% low-melt agarose/embryo media/Tricaine. Fluorescence and bright field imaging were performed on a Zeiss SteREO Discovery V12 microscope equipped with an X-Cite 120 W Metal Halide lamp (Excilitas Technologies, X-Cite 120Q). Images were captured with a Cannon Rebel T3 camera using EOS Utility software (Cannon). Bright field and fluorescence images were merged in Photoshop (Adobe). Live imaging was also performed on a Leica M165 FC stereomicroscope equipped with EL 6000 light source and K5 passive cooled sCMOS camera using Leica LAS X Multi Channel Acquisition software. Live imaging of larval retina was performed on a Zeiss LSM800 laser scanning confocal microscope.
Quantification and statistical analyses
Quantification of proliferation and apoptosis was performed on three sections of immunolabeled head tissue for each individual, from three biological replicates of zebrafish embryos or larvae. Prism (GraphPad) software was used for two-tailed unpaired Student’s t-test with mean ± s.e.m. statistical analyses and production of bar graphs.
Acknowledgements
The authors thank Dr Raquel Espin (Iowa State University) for the KalTA4 cDNA, and Jon Luiken and Tanya Schwab for technical assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Maura McGrail, Email: mmcgrail@iastate.edu.
Tanya T Whitfield, University of Sheffield, United Kingdom.
Richard M White, Memorial Sloan Kettering Cancer Center, United States.
Funding Information
This paper was supported by the following grants:
NIH Office of the Director R24 OD 020166 to Stephen C Ekker, Karl J Clark, Jeffrey J Essner, Maura McGrail.
Conselho Nacional de Desenvolvimento Científico e Tecnológico to Maira P Almeida.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
has competing interests with LifEngine and LifEngine Animal Health.
Reviewing editor, eLife and has competing interests with LifEngine and LifEngine Animal Health.
has competing interests with Recombinetics Inc, LifEngine and LifEngine Animal Health.
has competing interests with Recombinetics Inc, Immusoft Inc, LifEngine and LifEngine Animal Health.
Author contributions
Conceptualization, Investigation, Methodology, Validation, Visualization, Writing – review and editing.
Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – review and editing, Conceptualization.
Conceptualization, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization.
Conceptualization, Investigation, Methodology, Resources, Writing – review and editing.
Formal analysis, Investigation, Methodology, Validation, Writing – review and editing.
Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review and editing.
Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review and editing.
Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review and editing.
Ethics
Use of zebrafish for research in this study was performed according to the Guidelines for Ethical Conduct in the Care and Use of Animals (APA, 1986), and carried out in accordance with Iowa State University Animal Care and Use Committee IACUC-18-279 and IACUC-20-058 approved protocols. All methods involving zebrafish were in compliance with the American Veterinary Medical Association (2020), ARRIVE (Percie du Sert et al., 2020) and NIH guidelines for the humane use of animals in research.
Additional files
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files. Source data files have been provided for Table 2, Figure 4, Figure 4 - figure supplement 1, Figure 5, Figure 5 - figure supplement 2, Figure 6, Figure 6 - figure supplement 2, Figure 7, Figure 7 - figure supplement 1, Figure 7 - figure supplement 2, Figure 8, Figure 9. DNA constructs reported in this study have been deposited at Addgene in the Jeffrey Essner lab list (Plasmid IDs: 173886, 173887, 173888, 173889, 173890, 173891, 180007, 180008, 180009, 180010, 180011, 18012). DNA constructs and transgenic zebrafish lines are available on request to M. McGrail.
References
- Almeida MP, Welker JM, Siddiqui S, Luiken J, Ekker SC, Clark KJ, Essner JJ, McGrail M. Endogenous zebrafish proneural Cre drivers generated by CRISPR/Cas9 short homology directed targeted integration. Scientific Reports. 2021;11:1732. doi: 10.1038/s41598-021-81239-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Disease Models & Mechanisms. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- APA Guidelines for ethical conduct in the care and use of animals. Journal of the Experimental Analysis of Behavior. 1986;45:127–132. doi: 10.1901/jeab.1986.45-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLOS Genetics. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, Tan W, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Mouse Genome Database G. Mouse Genome Database (MGD) 2019. Nucleic Acids Research. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg L, Palmer N, Kikhi K, Miroshnik ES, Rueckert H, Gaddy E, MacPherson Cunningham C, Mattonet K, Lai SL, Marín-Juez R, Waring RB, Stainier DYR, Balciunas D. Conditional mutagenesis by oligonucleotide-mediated integration of loxP sites in zebrafish. PLOS Genetics. 2018;14:e1007754. doi: 10.1371/journal.pgen.1007754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Mosimann C. Switch and Trace: Recombinase Genetics in Zebrafish. Trends in Genetics. 2018;34:362–378. doi: 10.1016/j.tig.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Clark KJ, Balciunas D, Pogoda H-M, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, Ni J, Nielsen AL, Patowary A, Scaria V, Sivasubbu S, Xu X, Hammerschmidt M, Ekker SC. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nature Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Köster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. PNAS. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster D, Arnold-Ammer I, Laurell E, Barker AJ, Fernandes AM, Finger-Baier K, Filosa A, Helmbrecht TO, Kölsch Y, Kühn E, Robles E, Slanchev K, Thiele TR, Baier H, Kubo F. Genetic targeting and anatomical registration of neuronal populations in the zebrafish brain with a new set of BAC transgenic tools. Scientific Reports. 2017;7:5230. doi: 10.1038/s41598-017-04657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PD, Schmale MC. GFP as a Genetic Marker Scorable Throughout the Life Cycle of Transgenic Zebra Fish. Marine Biotechnology (New York, N.Y.) 2000;2:107–125. doi: 10.1007/s101269900014. [DOI] [PubMed] [Google Scholar]
- Han B, Zhang Y, Bi X, Zhou Y, Krueger CJ, Hu X, Zhu Z, Tong X, Zhang B. Bi-FoRe: an efficient bidirectional knockin strategy to generate pairwise conditional alleles with fluorescent indicators. Protein & Cell. 2021;12:39–56. doi: 10.1007/s13238-020-00747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Zöller D, Hammer J, Stucke J, Spieß S, Kesavan G, Kroehne V, Eguiguren JS, Ezhkova D, Petzold A, Dahl A, Brand M. Cre-Controlled CRISPR mutagenesis provides fast and easy conditional gene inactivation in zebrafish. Nature Communications. 2021;12:1125. doi: 10.1038/s41467-021-21427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima K, Jurynec MJ, Grunwald DJ. Precise Editing of the Zebrafish Genome Made Simple and Efficient. Developmental Cell. 2016;36:654–667. doi: 10.1016/j.devcel.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichino N, Serres MR, Urban RM, Urban MD, Treichel AJ, Schaefbauer KJ, Tallant LE, Varshney GK, Skuster KJ, McNulty MS, Daby CL, Wang Y, Liao H-K, El-Rass S, Ding Y, Liu W, Anderson JL, Wishman MD, Sabharwal A, Schimmenti LA, Sivasubbu S, Balciunas D, Hammerschmidt M, Farber SA, Wen X-Y, Xu X, McGrail M, Essner JJ, Burgess SM, Clark KJ, Ekker SC. Building the vertebrate codex using the gene breaking protein trap library. eLife. 2020;9:e54572. doi: 10.7554/eLife.54572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. PNAS. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungke P, Hammer J, Hans S, Brand M. Isolation of Novel CreERT2-Driver Lines in Zebrafish Using an Unbiased Gene Trap Approach. PLOS ONE. 2015;10:e0129072. doi: 10.1371/journal.pone.0129072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. PNAS. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan G, Hammer J, Hans S, Brand M. Targeted knock-in of CreER T2 in zebrafish using CRISPR/Cas9. Cell and Tissue Research. 2018;372:41–50. doi: 10.1007/s00441-018-2798-x. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang Y, Han B, Li L, Li M, Lu X, Chen C, Lu M, Zhang Y, Jia X, Zhu Z, Tong X, Zhang B. One-step efficient generation of dual-function conditional knockout and geno-tagging alleles in zebrafish. eLife. 2019;8:e48081. doi: 10.7554/eLife.48081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, Mueller T, Burgess SM, Higashijima S-I, Burgess HA. A 3D Searchable Database of Transgenic Zebrafish Gal4 and Cre Lines for Functional Neuroanatomy Studies. Frontiers in Neural Circuits. 2015;9:78. doi: 10.3389/fncir.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development (Cambridge, England) 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni TT, Lu J, Zhu M, Maddison LA, Boyd KL, Huskey L, Ju B, Hesselson D, Zhong TP, Page-McCaw PS, Stainier DY, Chen W. Conditional control of gene function by an invertible gene trap in zebrafish. PNAS. 2012;109:15389–15394. doi: 10.1073/pnas.1206131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JT, Leach SD. TAILOR: transgene activation and inactivation using lox and rox in zebrafish. PLOS ONE. 2013;8:e85218. doi: 10.1371/journal.pone.0085218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, Petersen OH, Rawle F, Reynolds P, Rooney K, Sena ES, Silberberg SD, Steckler T, Würbel H. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. BMC Veterinary Research. 2020;16:242. doi: 10.1186/s12917-020-02451-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Oteiza C, Taylor S, Yates T, Cicchini M, Lauderback B, Cashman CR, Burds AA, Winslow MM, Jacks T, Feldser DM. Recombinase-based conditional and reversible gene regulation via XTR alleles. Nature Communications. 2015;6:8783. doi: 10.1038/ncomms9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. PNAS. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nature Biotechnology. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Schultz LE, Haltom JA, Almeida MP, Wierson WA, Solin SL, Weiss TJ, Helmer JA, Sandquist EJ, Shive HR, McGrail M. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Disease Models & Mechanisms. 2018;11:dmm034124. doi: 10.1242/dmm.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz-Rogers LE, Thayer ML, Kambakam S, Wierson WA, Helmer JA, Wishman MD, Wall KA, Greig JL, Forsman JL, Puchhalapalli K, Nair S, Weiss TJ, Luiken JM, Blackburn PR, Ekker SC, Kool M, McGrail M. Rbbp4 loss disrupts neural progenitor cell cycle regulation independent of Rb and leads to Tp53 acetylation and apoptosis. Developmental Dynamics. 2022;9:467. doi: 10.1002/dvdy.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solin SL, Shive HR, Woolard KD, Essner JJ, McGrail M. Rapid tumor induction in zebrafish by TALEN-mediated somatic inactivation of the retinoblastoma1 tumor suppressor rb1. Scientific Reports. 2015;5:13745. doi: 10.1038/srep13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Hui SP, Sheng DZ, Kikuchi K. Dissection of zebrafish shha function using site-specific targeting with a Cre-dependent genetic switch. eLife. 2017;6:e24635. doi: 10.7554/eLife.24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor KM, Marquart GD, Hurt C, Smith TS, Geoca AK, Bhandiwad AA, Subedi A, Sinclair JL, Rose HM, Polys NF, Burgess HA. Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife. 2019;8:e42687. doi: 10.7554/eLife.42687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, Fenrich J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends in Biotechnology. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, Chow E, Gonzales C, Leung H-Y, Solomon I, Bronner-Fraser M, Megason SG, Fraser SE. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes & Development. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker JM, Wierson WA, Almeida MP, Mann CM, Torrie ME, Ming Z, Ekker SC, Clark KJ, Dobbs DL, Essner JJ, McGrail M. GeneWeld: Efficient Targeted Integration Directed by Short Homology in Zebrafish. Bio-Protocol. 2021;11:e4100. doi: 10.21769/BioProtoc.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) 3rd edn. University of Oregon Press; 1995. [Google Scholar]
- Wierson WA, Welker JM, Almeida MP, Mann CM, Webster DA, Torrie ME, Weiss TJ, Kambakam S, Vollbrecht MK, Lan M, McKeighan KC, Levey J, Ming Z, Wehmeier A, Mikelson CS, Haltom JA, Kwan KM, Chien C-B, Balciunas D, Ekker SC, Clark KJ, Webber BR, Moriarity BS, Solin SL, Carlson DF, Dobbs DL, McGrail M, Essner J. Efficient targeted integration directed by short homology in zebrafish and mammalian cells. eLife. 2020;9:e53968. doi: 10.7554/eLife.53968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Huang W, Du J, Wang Z, He J, Luo L. Improved Tol2-mediated enhancer trap identifies weakly expressed genes during liver and β cell development and regeneration in zebrafish. The Journal of Biological Chemistry. 2019;294:932–940. doi: 10.1074/jbc.RA118.005568. [DOI] [PMC free article] [PubMed] [Google Scholar]