Abstract
Prokaryotes evolved clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins as a kind of adaptive immune defense against mobile genetic elements including harmful phages. To counteract this defense, many mobile genetic elements in turn encode anti-CRISPR proteins (Acrs) to inactivate the CRISPR-Cas system. While multiple mechanisms of Acrs have been uncovered, it remains unknown whether other mechanisms are utilized by uncharacterized Acrs. Here, we report a novel mechanism adopted by recently identified AcrIF23. We show that AcrIF23 interacts with the Cas2/3 helicase-nuclease in the type I-F CRISPR-Cas system, similar to AcrIF3. The structure of AcrIF23 demonstrated a novel fold and structure-based mutagenesis identified a surface region of AcrIF23 involved in both Cas2/3-binding and its inhibition capacity. Unlike AcrIF3, however, we found AcrIF23 only potently inhibits the DNA cleavage activity of Cas2/3 but does not hinder the recruitment of Cas2/3 to the CRISPR RNA-guided surveillance complex (the Csy complex). Also, in contrast to AcrIF3 which hinders substrate DNA recognition by Cas2/3, we show AcrIF23 promotes DNA binding to Cas2/3. Taken together, our study identifies a novel anti-CRISPR mechanism used by AcrIF23 and highlights the diverse mechanisms adopted by Acrs.
Keywords: CRISPR/Cas, anti-CRISPR, crystal structure, protein–protein interaction, protein complex
Abbreviations: Acr, anti-CRISPR; Cas, CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR RNA; dsDNA, double-stranded DNA; EMSA, electrophoretic mobility shift assay; MGE, mobile genetic element; MST, microscale thermophoresis; ssDNA, single-stranded DNA
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins constitute thus far the only discovered prokaryotic adaptive immune system against the invasion of potentially harmful mobile genetic elements (MGEs) such as bacteriophages (1). The CRISPR-Cas systems are widespread among prokaryotes, distributed in about 40% bacterial and 85% archaeal phyla (2). Based on working by multisubunit surveillance complex or a single multidomain Cas effector, CRISPR-Cas systems are primarily classified into class 1 and class 2, which are further divided into six types (I-VI) and 33 subtypes based upon their specific Cas protein components (2).
The immunity strategies of CRISPR-Cas systems of different types are approximately convergent. First, the nucleic acids of the invasive MGEs are recognized, processed, and integrated into the CRISPR arrays as spacers in the prokaryotic genome. Next, the precursor CRISPR RNA (crRNA) containing CRISPR arrays was transcribed and further processed within the repeats region to generate mature crRNAs composed of both repeat and spacer segments. Last, the nucleic acids of the invading MGEs were recognized through protospacer adjacent motifs in their nucleic acids and cleaved by crRNA-guided Cas effector. Overall, all these steps could be summarized into three working stages: adaptation, crRNA maturation, and interference (3).
The subtype I-F (also named as I-F1 in the latest classification) CRISPR-Cas system, belonging to the most widely distributed type I system, is composed of Cas1, Cas2-3 fusion protein, Cas8f (Csy1), Cas5f (Csy2), Cas7f (Csy3), and Cas6f (Csy4) as protein components (2, 4, 5). In the type I-F system, the crRNA-guided surveillance complex (or the Csy complex) is generated through the assembly of one Cas5f, one Cas6f, six Cas7f subunits, and one Cas8f around the central crRNA backbone. The Csy complex recognizes, unwinds, and hybridizes with the target double-stranded DNA (dsDNA), resulting in conformational changes of both the Csy complex and dsDNA, which is further in favor of the recruitment of the Cas2/3 protein to degrade the target DNA to accomplish the immunity.
However, MGEs have developed counter-adaptations, such as anti-CRISPR (Acr) proteins, to fight against the CRISPR-Cas system and survive from this immune system. In 2013, five distinct ‘anti-CRISPR’ genes, i.e., acrIF1-5, inhibiting type I-F CRISPR system were first found in the genomes of bacteriophages infecting Pseudomonas aeruginosa (6). Until now, about 90 families of Acr proteins have already been experimentally verified to counteract 12 CRISPR-Cas subtypes (7). Presently, working strategies of all characterized Acr proteins could be summarized as four major types: interference of target DNA/RNA recognition, inhibition of target DNA/RNA cleavage, prevention of assembly of the surveillance complex (only for Class 2), and degradation of cyclic oligoadenylates for signal transduction (only for type III) (8). Notably, most Acr proteins have been characterized to achieve its inhibition through a stable interaction with corresponding Cas components (9).
For Acr proteins against subtype I-F CRISPR-Cas system, 24 families have been described until now (6, 10, 11, 12), and the structures of 11 of them have been determined for elucidating their inhibition mechanisms. Specifically, AcrIF1/2/4/6/7/8/9/10/14 played their roles though direct interaction with the Csy complex (13, 14, 15, 16, 17, 18), while AcrIF3 stably binds the Cas2/3 protein (19, 20, 21). Out of these, interestingly, other than sterically inhibiting the binding between the Csy complex and target DNA, AcrIF9, and AcrIF14 have also been found to mislead the Csy complex to absorb nonsequence–specific DNA, which has been demonstrated to contribute to the inhibition activity of the Acr proteins (18, 22, 23). Distinct from the above ten Acr proteins, AcrIF11 does not stably bind the Csy complex, instead, it has been reported to possess ADP-ribosylation activity on the N250 residue of the Cas8f subunit of the Csy complex, a key residue required for protospacer adjacent motif recognition (24).
Elucidation of the inhibition mechanisms of uncharacterized Acr proteins can not only facilitate the development of new “brake” tools for CRISPR-Cas–based genome editing but also provide the weakness points of the CRISPR-Cas systems. Here, we determined the crystal structure of AcrIF23 and further elucidated its working mechanism through biochemical and mutagenesis assays. AcrIF23 interacts with the Cas2/3 protein but not the Csy complex, as AcrIF3 does. However, unlike AcrIF3, which binds Cas2/3 and prevents its recruitment by the Csy–target DNA complex, AcrIF23 binds Cas2/3 to inhibit its nuclease activity but does not hinder its recruitment. Further biochemical and mutagenesis analyses uncovered the key residues involved in AcrIF23-Cas2/3 binding, as well as the inhibition capacity of AcrIF23. In conclusion, our study reveals another Cas2/3-interacting AcrIF protein and a novel inhibition mechanism of solely inactivating the Cas2/3 nuclease. In all, this work provides new insights into the highly diverse mechanisms adopted by Acr proteins.
Results
AcrIF23 functions after the hybridization between the Csy complex and target DNA
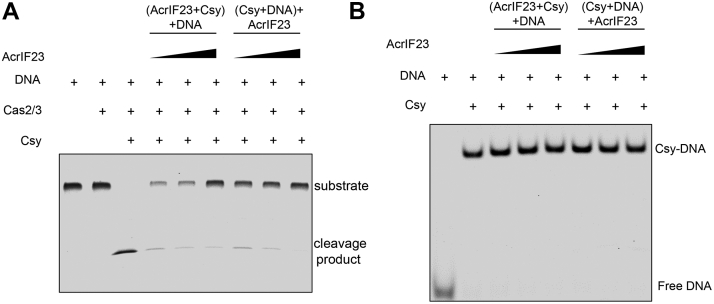
First, we tested the inhibition ability of AcrIF23 against the type I-F CRISPR-Cas system through an in vitro dsDNA cleavage assay. As indicated in Figure 1A, while target dsDNA was only slightly cleaved by the Cas2/3 alone, this cleavage was remarkably enhanced through a previous incubation of the Csy complex and target dsDNA (lanes 1–3, Fig. 1A). Consistent with the results of the in vivo assay (10), AcrIF23 could inhibit the cleavage of target DNA by the Csy complex and Cas2/3 (lanes 4–6, Fig. 1A). Interestingly, the inhibition was not affected whether AcrIF23 was added before or after the incubation of Csy and target dsDNA (lanes 4–9, Fig. 1A), suggesting that AcrIF23 may function after the hybridization between the Csy complex and target dsDNA. To further verify the interference stage of AcrIF23, we conducted an electrophoretic mobility shift assay (EMSA). As expected, an obvious super-shift of the band of the target dsDNA was observed with the addition of the Csy complex (lanes 1 and 2, Fig. 1B), suggesting their interaction. As we hypothesized based on the results of the DNA cleavage assay (Fig. 1A), addition of AcrIF23 did not influence the hybridization between the Csy complex and target DNA regardless of the incubation order (lanes 3–8, Fig. 1B). Taken together, AcrIF23 achieves its inhibition after the binding between the Csy complex and target DNA.
Figure 1.
AcrIF23 functions after the hybridization between the Csy complex and target DNA.A, AcrIF23 inhibits the DNA cleavage regardless of the incubation order. The reaction system contains 320 nM Csy complex, 50 nM dsDNA, and 0.5, 1, or 2 μM AcrIF23. The Csy complex is incubated first with AcrIF23 or DNA, respectively. After incubation of the Csy/DNA/AcrIF23, Cas2/3 was added with a final concentration of 200 nM and incubated for 20 min at 37 °C. B, EMSA to test the inhibition of Csy-DNA binding by AcrIF23. The reaction system contains 1.6 μM Csy complex, 0.1 μM dsDNA, and 0.5, 1, or 2 μM AcrIF23. Similarly, the Csy complex is incubated first with AcrIF23 or DNA, respectively. dsDNA, double-stranded DNA; EMSA, electrophoretic mobility shift assay.
AcrIF23 interacts with Cas2/3 but not the Csy complex
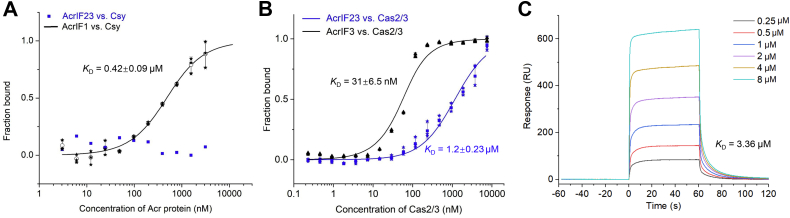
Next, we moved on to investigate the detailed inhibitory mechanism of AcrIF23. First, we tested whether AcrIF23 interacts with the Csy complex or the Cas2/3 protein like most characterized type I-F Acr proteins do. Microscale thermophoresis (MST) assay showed that while AcrIF1 exhibited a stable interaction with the Csy complex (binding KD of 0.42 μM), no obvious interaction was observed between AcrIF23 and the Csy complex (Fig. 2A). However, interaction between AcrIF23 and Cas2/3 was clearly observed with a binding KD calculated to be 1.2 μM by MST and 3.36 μM by surface plasmon resonance assay (Fig. 2, B and C). AcrIF3 was used as a positive control with a binding KD of 31 nM toward Cas2-3 by MST assay (Fig. 2B). In conclusion, AcrIF23 interacts with the Cas2/3 protein but not the Csy complex to fulfil its inhibition.
Figure 2.
AcrIF23 interacts with Cas2/3, but not the Csy complex.A, MST assay of the binding affinity between Csy and AcrIF23 or AcrIF1. Fitted binding curve and calculated binding KD value are marked for AcrIF1. Error bars indicate the s.d. of three independent measurements. B, MST assays of the binding affinity between Cas2/3 and AcrIF23 or AcrIF3. Fitted binding curve and calculated binding KD values are marked. Error bars indicate the s.d. of three independent measurements. C, SPR assay of the binding affinity between Cas2/3 and AcrIF23. Data were analyzed and fitted with BIAevaluation 4.1 software (GE Healthcare) using the affinity model. MST, microscale thermophoresis; SPR, surface plasmon resonance.
Crystal structure of AcrIF23 reveals a novel fold
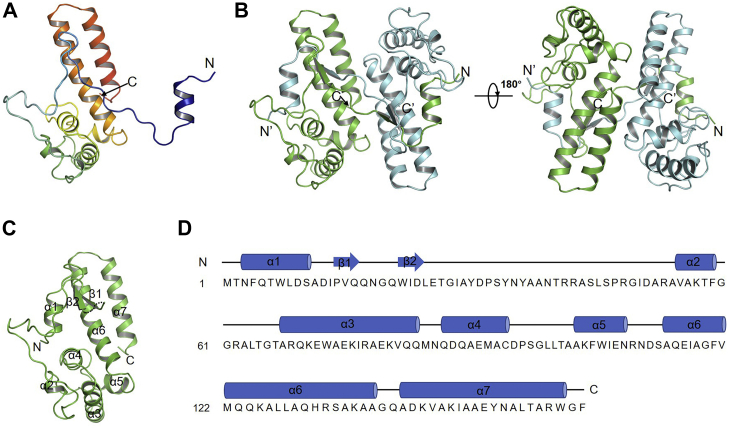
To further elucidate the working mechanism of AcrIF23, we solved its crystal structure at a resolution of 2.13 Å (Fig. 3A and Table 1) using single-wavelength anomalous diffraction. Data collection and refinement statistics are summarized in Table 1. The Rfree factor (0.2338) is a little high for the structure, which might be caused by some less well defined residues near the N terminus. The asymmetric unit contains only one AcrIF23 molecule; however, it forms a stable dimer with its symmetry-related molecule by swapping their N terminus (residues 1–18) with a buried surface area of 5869.6 Å2 (Fig. 3B). Nevertheless, our size-exclusion chromatography with multi-angle light scattering analysis showed that AcrIF23 is a monomer in solution (Fig. S1, 17.96 kDa monomer mass theoretically and 17.97 ± 1.40 kDa experimentally). This suggests that the dimeric assembly of AcrIF23 deduced from the crystal structure is a crystallographic artifact. Therefore, we hereafter describe the structure of the monomeric AcrIF23 form composed of the swapped N-terminal region (residues 1–17) of the symmetry-related AcrIF23 molecule and the C-terminal region (residues 20–159) of the AcrIF23 molecule in the asymmetric unit (Fig. 3C). AcrIF23 is almost exclusively composed of α helices, hereafter naming α1-α7. Typically, α1 is connected to α2 through a rather long loop (residues 11–54) with a short antiparallel two-stranded β sheet within it. Moreover, α6 (residues 114–137) and α7 (residues 140–157) are two long antiparallel α helices (Fig. 3C). Dali search revealed no entries with a sequence coverage over 50% of AcrIF23, suggesting that AcrIF23 adopts an overall novel fold (Table S1 and Fig. S2).
Figure 3.
Crystal structure of AcrIF23 reveals a novel overall fold.A, overall structure of one AcrIF23 protomer in the asymmetric unit. The AcrIF23 molecule is colored in a rainbow format. B, two views of the two AcrIF23 protomers in the asymmetric unit. The two protomers are colored in cyan and green, respectively. C, biologically relevant monomeric unit of AcrIF23 is shown. D, secondary structural elements are marked above the sequence of AcrIF23.
Table 1.
Data collection and refinement statistics
| Statistics | AcrIF23 (native, PDB: 7FIA) | AcrIF23 (selenourea-soaked) |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 65.569, 65.569, 95.053 | 65.986, 65.986, 96.424 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50–2.13 (2.21–2.13)a,b | 50–2.38 (2.47–2.38) |
| Rsym or Rmerge (%) | 10.4 (70.2) | 13.0 (150.9) |
| I/σ(I) | 34.8 (5.8) | 40.8 (2.6) |
| Completeness (%) | 100.0 (100.0) | 99.9 (99.2) |
| Redundancy | 24.6 (25.4) | 86.6 (35.4) |
| Refinement | ||
| Resolution (Å) | 30.99–2.13 | 46.66–2.38 |
| No. reflections | 12,148 | 8915 |
| Rwork/Rfreec | 0.2042/0.2338 | 0.2193/0.2321 |
| No. atoms | 1375 | 1337 |
| Protein | 1241 | 1235 |
| Ligand/ion | 0 | 20 |
| Water | 134 | 82 |
| B factors | 28.74 | 39.31 |
| Protein | 28.30 | 39.01 |
| Ligand/ion | 57.93 | |
| Water | 32.78 | 39.31 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.007 |
| Bond angles (°) | 0.60 | 1.03 |
| Ramachandran plot (%) | ||
| Favored | 98.10 | 98.09 |
| Allowed | 1.90 | 1.91 |
| Outliers | 0 | 0 |
| MolProbity score | 1.38 | 1.43 |
For each structure one crystal was used.
Values in parentheses are for highest-resolution shell.
Rfree was calculated with 5% of the reflections selected.
Interaction between AcrIF23 and Cas2/3 is essential for its inhibition activity
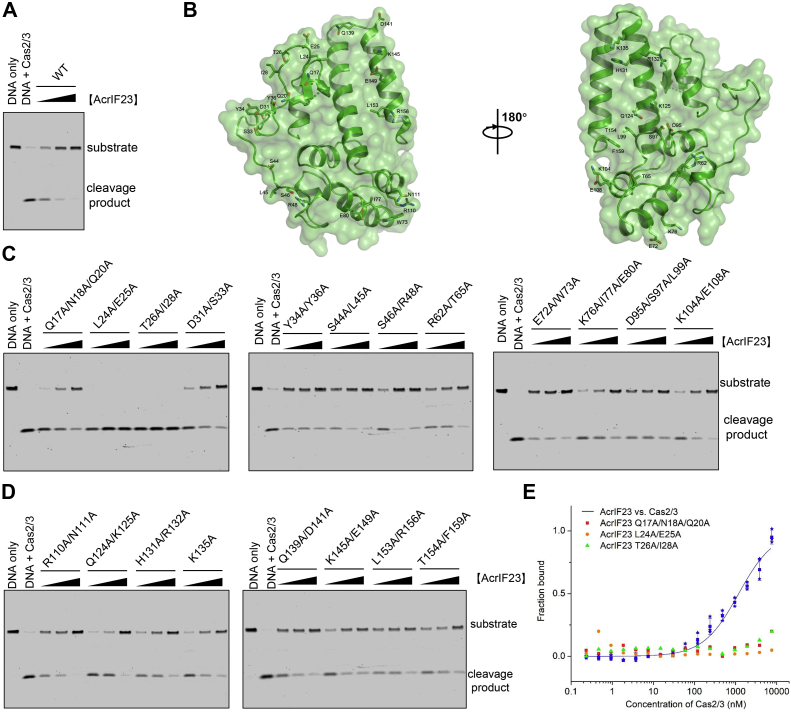
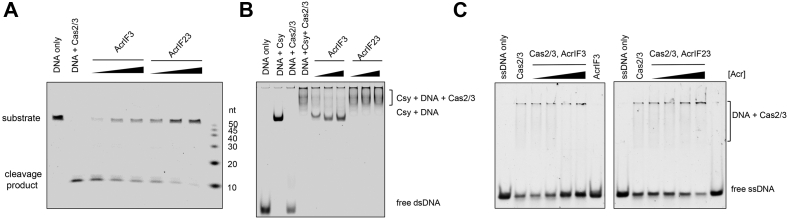
Since AcrIF23 interacts with Cas2/3 but not the Csy complex, we speculated that the interaction between AcrIF23 and Cas2/3 might be essential for its inhibitory activity. First, we tested whether AcrIF23 could inhibit the activity of Cas2/3 alone. It has been reported that Cas2/3 alone could efficiently degrade single-stranded DNA (ssDNA) and dsDNA with an inner bubble in the absence of the Csy complex (25). Hence, we performed an in vitro ssDNA cleavage assay with Cas2/3 and AcrIF23. The results showed that the DNA cleavage was markedly reduced with the addition of AcrIF23 (Fig. 4A), suggesting that AcrIF23 can inhibit the nuclease activity of Cas2/3. To determine the key AcrIF23 residues involved in its inhibition ability, we performed a structure-guided mutagenesis study through mutating the surface residues of AcrIF23 (Fig. 4B) and testing their inhibitory activities toward Cas2/3. Results of the in vitro ssDNA cleavage assays revealed that out of the 20 mutants we designed, three AcrIF23 mutants, i.e., Q17A/N18A/Q20A, L24A/E25A, and T26A/I28A exhibit markedly reduced inhibition of the ssDNA cleavage compared with wildtype AcrIF23 (Fig. 4, C and D). Moreover, further MST assay indicated that none of the three AcrIF23 mutants interact with Cas2/3 (Fig. 4E). Notably, the 20 mutants were expressed and purified well as WT AcrIF23 (Fig. S3). The mutations might decrease the binding between AcrIF23 and Cas2/3 to an affinity unable to be detected by the MST assay, at least for Q17A/N18A/Q20A, which mutant still retained weak inhibitory effect (Fig. 4C). Interestingly, the mutagenesis study indicated that the interacting residues of AcrIF23 are all located at the surface loop region (residues 17–28) between α1 and α2, suggesting the important role of this surface in Cas2/3 binding (Fig. 4B). Notably, all these residues, except Q17 and Q20, are also highly conserved among AcrIF23 homologs (Fig. S4). That is, AcrIF23 interacts with Cas2/3 predominantly by a surface loop region, which is essential for the inhibitory ability of AcrIF23.
Figure 4.
Interaction between AcrIF23 and Cas2/3 is essential for its inhibition activity.A, AcrIF23 inhibits in vitro ssDNA cleavage by Cas2/3. The reaction system contains 200 nM Cas2/3, 25 nM ssDNA, and 0.5, 1, or 2 μM AcrIF23. B, AcrIF23 is shown in cartoon and surface model with views of its two sides. Mutated residues are shown in sticks. Residues whose mutations markedly reduce the inhibition of the ssDNA cleavage are marked with red texts. C and D, inhibition of in vitro ssDNA cleavage of the 20 AcrIF23 mutants. The experiment was performed as in A. E, MST assay of the binding between Cas2/3 and AcrIF23 and its mutants. Error bars indicate the s.d. of three independent measurements. Fitted binding curve is shown for AcrIF23-Cas2/3 binding.
The inhibition mechanism of AcrIF23
Interestingly, AcrIF3 has also been identified to interact with Cas2/3 to inhibit the CRISPR-Cas system. Therefore, we moved on to investigate whether the inhibition mechanisms of these two Acrs are similar. First, we compared the inhibition activities of AcrIF23 and AcrIF3 on the cleavage of ssDNA by Cas2/3. The results showed that while addition of AcrIF3 moderately reduced the cleavage activity of Cas2/3 (lanes 3–5, Fig. 5A), AcrIF23 displays a much stronger inhibition of the cleavage activity (lanes 6–8, Fig. 5A). Next, we tested whether AcrIF23 can inhibit the Cas2/3 recruitment by the Csy complex as AcrIF3 does. As indicated in the EMSA in Figure 5B, the band of Csy-DNA complex shifted again with the addition of Cas2/3, indicating the formation of a ternary complex Csy-DNA-Cas2/3 (lanes 1–3), which could be markedly inhibited by the addition of AcrIF3 (lanes 4–6), as indicated in previous studies (19). Notably, AcrIF23 showed no obvious inhibition on this recruitment (lanes 7–9), suggesting that AcrIF23 differs from AcrIF3 in mechanism. Previous study of the structure of Cas2/3-AcrIF3 complex proposed that AcrIF3 can prevent the substrate recognition of Cas2/3 due to the blocking of DNA binding tunnel by AcrIF3 (21). We confirmed this hypothesis through an EMSA experiment (Fig. 5C). Interestingly, under the same experimental conditions, AcrIF23 did not prevent substrate DNA binding but instead promote substrate DNA binding to Cas2/3, although AcrIF23 itself did not bind DNA under the AcrIF23 concentration used in the assay (Fig. 5C). Taken together, our results showed that AcrIF23 does not prevent substrate DNA binding or Cas2/3 recruitment by the Csy complex but promotes substrate DNA binding to Cas2/3.
Figure 5.
AcrIF23 does not prevent Cas2/3 recruitment by the Csy-DNA complex.A, comparison of the inhibition capacity of ssDNA cleavage by Cas2/3 between AcrIF23 and AcrIF3. The reaction system contains 200 nM Cas2/3, 25 nM ssDNA, and 0.5, 1, or 2 μM AcrIF23 or AcrIF3. B, AcrIF3 but not AcrIF23 prevents Cas2/3 recruitment by the Csy-DNA complex. Reactions were performed with 1.6 μM Csy complex, 0.1 μM dsDNA, 1 μM Cas2/3 and 0.5, 1, or 2 μM AcrIF23 or AcrIF3. C, EMSA experiments to test the effect on ssDNA binding of Cas2/3 by AcrIF23 and AcrIF3. The reaction system contains 1.5 μM Cas2/3, 100 nM ssDNA, and 0.03125, 0.125, 0.5 or 2 μM AcrIF23 or AcrIF3. In the last lanes of the two panels, AcrIF3 or AcrIF23 was added at 2 μM. MST, microscale thermophoresis; ssDNA, single-stranded DNA.
Discussion
Acr proteins exhibit highly variable protein sequences and structures. Up to now, the type I-F CRISPR-Cas system still harbors the largest Acr family, totally 24 Acr proteins, in which 11 Acr proteins have been characterized structurally and mechanically. Among these 11 families, targeting the Csy complex is a much more prevalent inhibition strategy which is adopted by AcrIF1/2/4/6/7/8/9/10/11/14 (13, 14, 15, 16, 17, 18). On the contrary, AcrIF23 is the second reported type I-F Acr protein (the other is AcrIF3) (19) and the third type I Acr protein (including AcrIE1) (26), which accomplishes its suppression through interacting with Cas2/3. However, unlike AcrIF3 and AcrIE1, which have been proposed to inhibit the recruitment of Cas2/3 by the Cascade complex (26, 27), AcrIF23 does not hinder Cas2/3 recruitment but solely inhibits the nuclease activity of Cas2/3. The result that AcrIF23 does not inhibit substrate DNA binding but promotes DNA binding to Cas2/3 (Fig. 5C) suggests that AcrIF23 does not function as a competitive inhibitor but more like an uncompetitive inhibitor. However, the fact that AcrIF23 can bind the apo Cas2/3 enzyme also suggests that it is not a canonical uncompetitive inhibitor. Taken together, the present data suggest that AcrIF23 might not directly bind to and block the nuclease active site of Cas2/3, but to a different region and allosterically inhibit the enzymatic activity of Cas2/3. We propose that AcrIF23 might function as a mixed-type inhibitor and can bind both the enzyme and enzyme–substrate complex. Therefore, this also suggests a possibility of formation of a quaternary complex, i.e., Csy-DNA-Cas2/3-AcrIF23, which means that AcrIF23 might not only accomplish the inhibition through interference with the nuclease activity of Cas2/3 but also occupy the Csy complex through the formation of the invalid quaternary complex. Future structural studies into the AcrIF23-Cas2/3 and the potential AcrIF23-Cas2/3-DNA complexes will further provide insights into the inhibition mechanism of AcrIF23.
As for the CRISPR-Cas stages hindered by Acrs, only AcrIF3 has been verified to function after target DNA binding by the Csy complex before our study. A recent study showed that the binding of AcrIF4 to the Csy complex does not affect target DNA hybridization (16), suggesting that it may function after Csy-DNA hybridization. However, a previous in vivo assay suggested that AcrIF4 prevents the Csy complex to interact with its target DNA. Therefore, it still remains unknown how AcrIF4 exerts its inhibition capacity in vivo. In all, our studies uncover a novel inhibition mechanism of type I-F Acrs and provide new insights into the ongoing molecular arms race between viral parasites and the immune systems of their hosts.
Experimental procedures
Protein expression and purification
The full-length AcrIF3 and AcrIF23 genes from P. aeruginosa were synthesized by GenScript and cloned into pGEX-6p-1 to produce GST-tagged fusion proteins with a PreScission site. The proteins were expressed in E. coli strain Rosetta at 37 °C until the cell density reached an A600nm of 0.8. The culture was cooled and grew at 18 °C for 12 h and induced by 0.2 mM isopropyl-β-D-thiogalactopyranoside. The cells were then harvested, resuspended in lysis buffer (1× PBS, 2 mM DTT, and 1 mM PMSF) and lysed through a cell disruptor. The cell lysate was centrifuged at 20,000g for 1 h at 4 °C to remove cell debris. The supernatant was applied onto a self-packaged GST-affinity column (2 ml glutathione Sepharose 4B; GE Healthcare), and contaminant proteins were removed with wash buffer (lysis buffer plus 200 mM NaCl). The fusion protein was then digested with PreScission protease at 18 °C for 2 h. The eluant was further purified using a Superdex-200 (GE Healthcare) column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. The purified protein was analyzed by SDS-PAGE. The AcrIF23 mutants were generated by two-step PCR and were purified in the same way as wildtype protein.
The Csy complex from P. aeruginosa was purified as follows. The Cas8f/Cas5f and Cas7f/Cas6f genes were cloned into pETDuet-1 and pACYCDuet-1, respectively. The crRNA fragment was cloned into pRSFDuet-1 vector. After co-expression of the three plasmids in E. coli strain BL21, the Csy complex was purified through Ni-column, anion exchange chromatography, and gel filtration. The Cas2/3 gene was cloned into a modified pETDuet-1 vector with a GST tag at the N terminus of the protein and transformed into E. coli strain BL21. The protein was purified through GST column as stated above and then by heparin chromatography and gel filtration.
Crystallization, data collection, and structure determination
The protein of AcrIF23 was concentrated to 15 mg/ml in 10 mM Tris-HCl pH 7.5, 500 mM NaCl, and 5 mM DTT. We screened crystals by using the method of hanging-drop vapor diffusion. Crystals of AcrIF23 were grown at 18 °C by mixing an equal volume of the protein (15 mg/ml) with solution consist of 0.027 M sodium fluoride, 0.027 M sodium bromide, and 0.027 M sodium iodide; 0.1 M Tris (base), 0.1 M BICINE, pH 8.5; 33% v/v PEG 500× MME; and 17% w/v PEG 20000. The crystals grew to full size in about 6 to 7 days. The crystals were cryoprotected in the reservoir solution containing 10% glycerol. We used selenourea (SeU, Sigma, Product Number: 230499) to help solve the phase problem (28). The SeU crystalline powder was added into the solution above mentioned, into which crystals were transferred and soaked for 5 min, and then flash frozen in liquid nitrogen. After crystal diffraction tests at home and beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility, SeU-treated and native crystals suitable for structure determination were finally obtained, respectively.
All the data were collected at Shanghai Synchrotron Radiation Facility beamlines BL17U1 (29) and BL19U1 (30), integrated and scaled using the HKL2000 package (31). The initial model was solved by Autosol in PHENIX (32) and refined manually with COOT (33) using the data from a SeU-treated crystal. The overall phasing power of selenourea derivative compared to the native data is 0.94, as estimated by mlphare in the CCP4 package (34). The heavy atom sites found by SHELXD suggested that there are five strong selenium atoms in the asymmetric unit. The structure was further refined with PHENIX (32) against the data from a native crystal using stereochemistry information as restraints. The final structure was obtained through several rounds of refinement. MolProbity score for the structure from the native crystal was 1.43. Data collection and structure refinement statistics are summarized in Table 1. All of the structural illustrations were generated using the software PyMOL (L.L.C. Schrodinger).
Multi-angle light scattering analysis
Multi-angle light scattering experiment was performed in 20 mM Hepes pH 7.5 and 200 mM NaCl using a Superdex-200 10/300 GL size-exclusion column from GE Healthcare. 1.5 mg/ml AcrIF23 was used. The chromatography system was connected to a Wyatt DAWN HELEOS Laser photometer and a Wyatt Optilab T-rEX differential refractometer. Wyatt ASTRA 7.3.2 software was used for data analysis.
In vitro dsDNA cleavage assay
We mixed target with nontarget DNA strand at a molar ratio of 1.5: 1 to form a dsDNA. FAM was labeled on the 5′ end of the nontarget DNA strand, synthesized by Sangon, Shanghai. For testing the activity of AcrIF23 and its mutants, reactions were performed in a 20 μl buffer system containing 320 nM Csy complex, 50 nM dsDNA, and 0.5, 1, and 2 μM AcrIF23 or its mutants. We add AcrIF23 before or after incubating dsDNA with the Csy complex in ice in the reaction buffer (20 mM Hepes pH 7.5, 100 mM KCl, 5% glycerol, 1 mM TCEP) for 30 min, and AcrIF23 was added and incubated for 20 min at 37 °C. Then, Cas2/3 was added to a final concentration of 200 nM for 20 min, along which 5 mM MgCl2, 75 μM NiSO4, 5 mM CaCl2, and 1 mM ATP were added into the buffer. The reaction was further incubated for 30 min and quenched with 1% SDS and 50 mM EDTA. The products were separated by electrophoresis over 14% polyacrylamide gels containing 7 M urea and visualized by fluorescence imaging.
For testing the activity of AcrIF23 and other Acr proteins, Acr proteins were added with concentrations of 0.5 μM, 1 μM, and 2 μM, respectively. The experimental procedure was basically the same as above.
Nontarget DNA sequence (54 bp; 5′-FAM fluorescein labeled)
AGCAGCTGCACCTTCACGGCGGGCTTGATGTCCGCGTCTACCTGGATGGCTTCC
ssDNA cleavage assay
Acr proteins AcrIF3, AcrIF23, and its mutants were preincubated with 200 nM Cas2/3 for 30 min at 37 °C in a reaction buffer containing 20 mM Hepes, pH 7.5, 100 mM KCl, 5% glycerol, 5 mM MnCl2, 75 μM NiSO4, 5 mM CaCl2, 1 mM TCEP, and 1 mM ATP. The concentration of AcrIF3, AcrIF23, and its mutants is 0.5 μM, 1 μM, and 2 μM. Then, we added 25 nM ssDNA at 37 °C for another 30 min. The products were separated by electrophoresis over 14% polyacrylamide gels containing 7 M urea and visualized by fluorescence imaging.
ssDNA sequence (54 bp; 5′-FAM fluorescein labeled)
GGAAGCCATCCAGGTAGACGCGGACATCAAGCCCGCCGTGAAGGTGCAGCTGCT
Electrophoretic mobility shift assay
We mixed target with nontarget DNA strand at a molar ratio of 1:1.5 to form a dsDNA. FAM was labeled on 5′ end target DNA strand, synthesized by Sangon, Shanghai. Reactions were performed in a 20 μl buffer system containing 1.6 μM Csy complex, 0.1 μM dsDNA, 0.5 μM, 1 μM, and 2 μM AcrIF23 or AcrIF3. All binding reactions were conducted at 37 °C for 30 min in the buffer containing 20 mM Hepes pH 7.5, 100 mM KCl, and 5% glycerol. Products of the reaction were separated using 5% native polyacrylamide gels and visualized by fluorescence imaging.
Target DNA sequence (54 bp; 5′-FAM fluorescein labeled)
GGAAGCCATCCAGGTAGACGCGGACATCAAGCCCGCCGTGAAGGTGCAGCTGCT
Microscale thermophoresis assay
MST experiments were performed using a Monolith NT.115 instrument (NanoTemper). AcrIF23, its mutants, or Csy were fluorescently labeled with a Red-NHS labeling kit, respectively. For Csy binding, varying concentrations of native AcrIF23 or AcrIF1 were mixed with labeled Csy (50 nM) in buffer (20 mM Hepes pH 7.5, 150 mM KCl, 5% v/v glycerol) supplemented with 0.05% Tween-20. For Cas2/3 binding, varying concentrations of Cas2/3 were mixed with labeled AcrIF3, AcrIF23, or its mutants (50 nM) in the same buffer as above. Protein samples were loaded into capillaries, and MST measurements were performed using 20% MST power and 20% LED power. For each of binding experiments, we did three independent MST measurements carried out at 647 nm. Each protein KD value was obtained with a signal-to-noise ratio higher than 7. Datasets were processed with the MO.Affinity Analysis v2.3 software.
Surface plasmon resonance assay
The surface plasmon resonance analysis was performed using a Biacore T100 instrument at room temperature (25 °C). The Cas2/3 was immobilized on a CM5 chip and a concentration series of AcrIF23 in binding buffer (20 mM Hepes pH 7.5, 200 ml NaCl, and 0.05% (v/v) Tween-20) were injected over the chip. AcrIF23 was allowed to associate for 60 s and dissociate for 60 s. Data were analyzed and fitted with BIAevaluation 4.1 software (GE Healthcare) using the affinity model.
Data availability
The accession number for the coordinate and structure factor of AcrIF23 is PDB: 7FIA.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank the staff at beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility for their assistance with data collection. We would like to thank the Tsinghua University Branch of China National Center for Protein Sciences Beijing and Shilong Fan for providing facility support for X-ray diffraction of the crystal samples. This work was supported by the National Natural Science Foundation of China (32171274 and 32000901), the National Key Research and Development Program of China (2017YFA0506500, 2019YFC1200500 and 2019YFC1200502), Beijing Natural Science Foundation (5204038), Beijing Nova Program, and the Fundamental Research Funds for the Central Universities (XK1802-8).
Author contributions
J. R., H. W., and Y. F. methodology; J. R., F. L., Y. W., Z. L., and H. W. investigation; L. Y. software; L. Y. data analysis; Z. C. and Y. Z. supervision; Y. F. conceptualization; Y. F. writing-review & editing.
Edited by Wolfgang Peti
Contributor Information
Zeliang Chen, Email: zeliangchen@yahoo.com.
Yi Zhang, Email: zhangyishirly@hotmail.com.
Yue Feng, Email: fengyue@mail.buct.edu.cn.
Supporting information
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 4.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Aravind L., Wolf Y.I., Koonin E.V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L., Yang B., Yi H., Asif A., Wang J., Lithgow T., et al. AcrDB: a database of anti-CRISPR operons in prokaryotes and viruses. Nucleic Acids Res. 2021;49:D622–D629. doi: 10.1093/nar/gkaa857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia N., Patel D.J. Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat. Rev. Mol. Cell Biol. 2021;22:563–579. doi: 10.1038/s41580-021-00371-9. [DOI] [PubMed] [Google Scholar]
- 9.Wiegand T., Karambelkar S., Bondy-Denomy J., Wiedenheft B. Structures and strategies of anti-CRISPR-mediated immune suppression. Annu. Rev. Microbiol. 2020;74:21–37. doi: 10.1146/annurev-micro-020518-120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinilla-Redondo R., Shehreen S., Marino N.D., Fagerlund R.D., Brown C.M., Sorensen S.J., et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat. Commun. 2020;11:5652. doi: 10.1038/s41467-020-19415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marino N.D., Zhang J.Y., Borges A.L., Sousa A.A., Leon L.M., Rauch B.J., et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362:240–242. doi: 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pawluk A., Staals R.H., Taylor C., Watson B.N., Saha S., Fineran P.C., et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016;1:16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury S., Carter J., Rollins M.F., Golden S.M., Jackson R.N., Hoffmann C., et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169:47–57.e11. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo T.W., Bartesaghi A., Yang H., Falconieri V., Rao P., Merk A., et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell. 2017;171:414–426.e12. doi: 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng R., Xu Y., Zhu T., Li N., Qi J., Chai Y., et al. Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures. Cell Res. 2017;27:853–864. doi: 10.1038/cr.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabel C., Li Z., Zhang H., Chang L. Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14. Nucleic Acids Res. 2021;49:584–594. doi: 10.1093/nar/gkaa1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K., Wang S., Li S., Zhu Y., Pintilie G.D., Mou T.C., et al. Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7176–7182. doi: 10.1073/pnas.1922638117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschi M., Lu W.T., Santiago-Frangos A., Wilkinson R., Golden S.M., Davidson A.R., et al. AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nat. Commun. 2020;11:2730. doi: 10.1038/s41467-020-16512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondy-Denomy J., Garcia B., Strum S., Du M.J., Rollins M.F., Hidalgo-Reyes Y., et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Ma J., Cheng Z., Meng X., You L., Wang M., et al. A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses. Cell Res. 2016;26:1165–1168. doi: 10.1038/cr.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Yao D., Xu J.G., Li A., Xu J., Fu P., et al. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 2016;23:868–870. doi: 10.1038/nsmb.3269. [DOI] [PubMed] [Google Scholar]
- 22.Lu W.T., Trost C.N., Muller-Esparza H., Randau L., Davidson A.R. Anti-CRISPR AcrIF9 functions by inducing the CRISPR-Cas complex to bind DNA non-specifically. Nucleic Acids Res. 2021;49:3381–3393. doi: 10.1093/nar/gkab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Zhang L., Xiu Y., Gao T., Huang L., Xie Y., et al. Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex. Nucleic Acids Res. 2021;49:10178–10191. doi: 10.1093/nar/gkab738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu Y., Yang L., Gao T., Dong C., Zhang B., Yin P., et al. A type I-F anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-ribosylation. Mol. Cell. 2020;80:512–524.e15. doi: 10.1016/j.molcel.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Rollins M.F., Chowdhury S., Carter J., Golden S.M., Wilkinson R.A., Bondy-Denomy J., et al. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E5113–E5121. doi: 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawluk A., Shah M., Mejdani M., Calmettes C., Moraes T.F., Davidson A.R., et al. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio. 2017;8 doi: 10.1128/mBio.01751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollins M.F., Chowdhury S., Carter J., Golden S.M., Miettinen H.M., Santiago-Frangos A., et al. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol. Cell. 2019;74:132–142.e35. doi: 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Z. Selenourea: a convenient phasing vehicle for macromolecular X-ray crystal structures. Sci. Rep. 2016;6:37123. doi: 10.1038/srep37123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q.S., Zhang K.H., Cui Y., Wang Z.J., Pan Q.Y., Liu K., et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018;29:1–7. [Google Scholar]
- 30.Zhang W.Z., Tang J.C., Wang S.S., Wang Z.J., He J.H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 2019;30:170. [Google Scholar]
- 31.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Adams P.D., Grosse-Kunstleve R.W., Li W.H., Ioerger T.R., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Lohkamp B., Scott W., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey S.M. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the coordinate and structure factor of AcrIF23 is PDB: 7FIA.