Summary
Background
Diabetes in childhood and adolescence includes autoimmune and non-autoimmune forms with heterogeneity in clinical and biochemical presentations. An unresolved question is whether there are subtypes, endotypes, or theratypes within these forms of diabetes.
Methods
The multivariable classification and regression tree (CART) analysis method was used to identify subgroups of diabetes with differing residual C-peptide levels in patients with newly diagnosed diabetes before 20 years of age (n=1192). The robustness of the model was assessed in a confirmation and prognosis cohort (n=2722).
Findings
The analysis selected age, haemoglobin A1c (HbA1c), and body mass index (BMI) as split parameters that classified patients into seven islet autoantibody-positive and three autoantibody-negative groups. There were substantial differences in genetics, inflammatory markers, diabetes family history, lipids, 25-OH-Vitamin D3, insulin treatment, insulin sensitivity and insulin autoimmunity among the groups, and the method stratified patients with potentially different pathogeneses and prognoses. Interferon-ɣ and/or tumour necrosis factor inflammatory signatures were enriched in the youngest islet autoantibody-positive groups and in patients with the lowest C-peptide values, while higher BMI and type 2 diabetes characteristics were found in older patients. The prognostic relevance was demonstrated by persistent differences in HbA1c at 7 years median follow-up.
Interpretation
This multivariable analysis revealed subgroups of young patients with diabetes that have potential pathogenetic and therapeutic relevance.
Funding
The work was supported by funds from the German Federal Ministry of Education and Research (01KX1818; FKZ 01GI0805; DZD e.V.), the Innovative Medicine Initiative 2 Joint Undertaking INNODIA (grant agreement No. 115797), the German Robert Koch Institute, and the German Diabetes Association.
Keywords: Diabetes in childhood, Diabetes endotypes, CART analysis, Islet autoantibody, Childhood autoimmune disease, C-peptide, Inflammation, Obesity, Type 1 diabetes genetic susceptibility
Abbreviations: aa, amino acid; BMI, body mass index; CART, classification and regression tree; GADA, glutamate decarboxylase (65-kDa isoform) autoantibodies; HbA1c, haemoglobin A1c; HLA, human leukocyte antigen; IA-2A, insulinoma-associated antigen-2 autoantibodies; IAA, insulin autoantibodies; IFNɣ, interferon-ɣ; IL, interleukin; IQR, interquartile range; MODY, maturity-onset diabetes of the young; RBA, radiobinding assay; SDS, standard deviation score; SNP, single nucleotide polymorphism; TGA, transglutaminase autoantibodies; TNF, tumour necrosis factor; TPOA, thyroid peroxidase autoantibodies; ZnT8A, zinc transporter-8 autoantibodies
Research in context.
Evidence before this study
We searched PubMed up to Dec 31, 2021, using terms “type 1 diabetes”, “endotype”, “subtype”, “phenotype”, “heterogeneity”, “childhood”, and “intervention”. In children and adolescents, at the time of clinical manifestation of type 1 diabetes, there are marked differences in the amount of insulin secretion still maintained, which can be determined by measuring C-peptide in the blood. There is an inverse relationship between disease severity and prognosis and the amount of remaining β-cell function. Age is an important factor influencing residual β-cell function with lower C-peptide concentrations in younger patients. Age-related heterogeneity has also been described in islet infiltrates of patients with type 1 diabetes with an inflammatory high/C-peptide low phenotype in young age and an inflammatory low/C-peptide intermediate phenotype in older age. Finally, there are variations documented for insulin sensitivity, BMI, and genetics in patients with and without islet autoantibodies and in relation to age of onset. Overall, the published data indicate that type 1 diabetes in children is not a uniform disease and therefore classifiers for subtyping patients and subtype-specific treatment options are needed to achieve improved therapeutic outcomes.
Added value of this study
A multivariable CART approach in a large representative cohort of children and adolescents with newly diagnosed diabetes identified subgroups of type 1 diabetes that differed in C-peptide and age and had substantially heterogeneous characteristics. Subgroups of young patients (age <8 years) with islet autoantibodies and low C-peptide had features of inflammation (TNF or IFNɣ signatures) and insulin autoimmunity. These were contrasted by subgroups of older patients with islet autoantibodies and higher C-peptide who had features usually associated with type 2 diabetes, including insulin insensitivity, or high BMI. Among the islet autoantibody-negative patients, the CART identified a further subgroup with distinct features of type 1 diabetes along with subgroups enriched for monogenic diabetes or type 2 diabetes. The robustness of the CART approach was confirmed in a second cohort of patients who were followed for a median of 7 years, and showed that the CART-defined subgroups had prognostic relevance.
Implications of all the available evidence
Childhood and adolescent type 1 diabetes can be classified into prognostically relevant subgroups with distinct features such as inflammation or insulin insensitivity. Subgroup-specific application of therapies that target such features should be considered.
Alt-text: Unlabelled box
Introduction
Diabetes is one of the most common chronic diseases in childhood. According to recent estimates, more than 1.1 million children and adolescents worldwide are living with diabetes.1 Diabetes in childhood and adolescence is classified into type 1,2 which is the most frequent form and is caused by autoimmune-mediated destruction of the insulin-producing pancreatic islet β-cells; type 2, which is caused by inadequate insulin secretion in a background of insulin resistance and obesity; and specific types of diabetes with other causes (e.g. monogenic diabetes syndromes).3 The classification is a basis for assigning therapy. However, there is considerable variability in the clinical presentation and severity that can cause uncertainty surrounding the type of diabetes and appropriate therapy at the time of diagnosis. This has led to the suggestion that additional parameters such as β-cell function and insulin sensitivity may help discriminate between autoimmune and non-autoimmune forms of diabetes,4 and the use of type 1 diabetes genetic risk scores and the combination of clinical characteristics could predict monogenic diabetes.5,6
An unresolved question is whether there are subtypes, endotypes, or theratypes within young-onset type 1 diabetes, as reported for type 2 diabetes in adults,7 and for allergy in childhood.8 Age-related endotypes of type 1 diabetes were recently proposed based on the T and B cell responses to autoantigens and the predominant types of cells that infiltrate the islets of patients.9, 10, 11, 12 The genetic load and functional β-cell reserve also vary with age at diabetes onset.13,14 Finally, there are age-related differences in the responses to immune therapy,15 further suggesting the possibility of endotypes of type 1 diabetes with distinct pathogenetic features and therapeutic responsiveness.
The objective of this study was to distinguish clinically relevant subgroups of patients with type 1 diabetes diagnosed before 20 years of age using a supervised multivariable approach. C-peptide, as an indicator of residual β-cell function, was chosen as the outcome measure because it is associated with mid- to long-term disease severity and prognosis,16, 17, 18 and is often used as an eligibility criterion in clinical trials of patients with new-onset type 1 diabetes.19,20 We used the classification and regression tree (CART) analysis method to search for algorithms that associate with C-peptide concentrations in 1192 patients with new-onset diabetes enrolled in the German DiMelli cohort study.21 The validity and prognostic relevance of the models were assessed in a large second cohort and additional biochemical measurements were used to search for potential pathogenetic and/or therapeutic heterogeneity among the identified groups. The validated algorithm defined diverse subgroups with prognostic and potential therapeutic relevance.
Methods
Patient cohort
DiMelli is a cohort and biobank study in Bavaria, Germany of incident cases of diabetes who are diagnosed before 20 years of age and enrolled within 6 months of diagnosis. DiMelli started recruitment in April 2009 and ended in December 2018. The study design has been described previously.21 At the time of enrolment, a fasting blood sample was collected and a structured questionnaire was completed by the local physician at the hospital or primary care centre. The blood sample was used to determine C-peptide, HbA1c, islet autoantibodies, thyroid peroxidase autoantibodies (TPOA), and transglutaminase autoantibodies (TGA), and samples of DNA, plasma, serum, and peripheral blood mononuclear cells were stored. Blood samples were sent to the Institute of Diabetes Research, Helmholtz Zentrum München, by overnight express courier where all parameters were measured centrally. The questionnaire included sex, the date of diagnosis of diabetes, first-degree family history, and current antidiabetic medications. Weight and height were measured by trained nurses or physicians. The main analysis was planned for participants enrolled by December 2016 (n=1192). Participants enrolled after this time (n=308) were included in a validation cohort.
Ethics
Each patient and/or parent provided written informed consent to participate in DiMelli. The DiMelli cohort study was approved by the medical ethics committee of Bavaria, Germany (Bayerische Landesaerztekammer, #08043).
Outcome and predictor variables
C-peptide concentrations, the outcome variable in the CART analysis, were measured using fasting, aprotinin-stabilized EDTA plasma samples on an automated immunoassay analyser (AIA 360; Tosoh, San Francisco, CA).
Sex, age at diagnosis, diabetes duration, BMI SDS, haemoglobin A1c (HbA1c), first-degree family history of type 1 (yes/no), type 2 (yes/no) or any other form of diabetes (yes/no) were considered as predictor variables. Additionally, the number of positive islet autoantibodies, TPOA or TGA (yes/no), islet autoantibody type, HLA-DR/DQ genotype or genetic risk score were used as predictor variables in a sensitivity analysis. Sex- and age-specific BMI percentiles were based on the national reference values,22 and were also categorized as overweight (>+1SD) and obese (>+2SD).23 HbA1c concentrations were measured in EDTA samples using a glycohaemoglobin analyser (TOSOH-723 G7; Tosoh). TGA and TPOA were measured using radiobinding assays (RBAs) as previously described.24,25 Autoantibodies to the islet antigens insulin (IAA), GAD65 (GADA), IA-2 (IA-2A), and zinc transporter 8 (ZnT8A) were measured using RBAs as previously described.26, 27, 28, 29 IAA were determined by a protein A/G-based RBA using recombinant human insulin labelled at amino acid (aa) 14 with [125I]tyrosine. GADA, IA-2A, and ZnT8A were determined by protein A-based RBAs using [35S]methionine-labelled in vitro transcribed/translated recombinant human GAD65 (aa 1–585), the intracellular portion of IA2 (aa 605–979), and COOH-terminal (aa 268–369) constructs of the ZnT8 R325 and W325 variants, respectively. Samples were considered positive for ZnT8A if autoantibodies to at least one of the ZnT8 variants (ZnT8RA and/or ZnT8WA) were found. The upper limit of normal for each assay was determined using Q–Q plots, as previously described,30 that corresponded to the 99th percentile of nondiabetic control children. The autoantibody assays were evaluated by the Diabetes Antibody Standardization Program,31, 32, 33 Their performances are shown as those of laboratory 121 in published reports and for GADA, IA-2A and ZnT8A are highly comparable to commercial tests such as the Kronus ELISA method. The number of positive islet autoantibodies was categorized as one, two, three, or four, and as yes/no for each of the four autoantibodies. IAA positivity was not included in the categorization of positive/negative autoantibody status if the sample was obtained >14 days after diagnosis. Samples negative for IAA, GADA, IA-2A, and ZnT8A in the RBAs were also tested by luciferase immunoprecipitation assays for autoantibodies to insulin, GAD65, IA-2, ZnT8, and tetraspanin 7,34 and confirmed as negative in these assays.
Additional variables
The following variables were assessed to characterize the subgroups of diabetes obtained by the CART analysis, or to perform cluster analyses for identification of the immune response and genetic phenotypes: ketonuria (yes/no), insulin treatment (yes/no), serum triglyceride concentrations, serum 25-OH-Vitamin D3 (vitamin D3) concentrations, insulin sensitivity, serum cytokine concentrations, inflammatory markers, and type 1 diabetes-associated genes (HLA-DR/DQ and genetic risk score). Triglyceride and vitamin D3 concentrations were measured using an enzymatic colorimetric test on a cobas 8000® analyser with a c502 module (Roche Diagnostics, Basel, Switzerland). Cytokines were measured by an electrochemiluminescent proinflammatory cytokine kit (Mesoscale Discovery, Rockville, MD). Inflammatory markers were measured using the OLINK inflammation panel (OLINK, Uppsala, Sweden) in islet autoantibody-positive patients. Insulin sensitivity was calculated using the formula: exp(4.64725 − 0.02032 × waistcircumference [cm] − 0.09779 × HbA1c [%] − 0.00235 ×triglycerides [mg/dL]) as previously described.35 HLA genotyping was performed by high-resolution sequencing-based typing of exons 2 and 3 of HLA-DRB1
and HLA-DQB1, including heterozygous ambiguity resolution (Conexio Genomics, Fremantle, Western Australia, Australia). The HLA genotype was categorized into six groups based on their association with type 1 diabetes (Table S1), as previously defined.36 SNP analysis was performed using the Illumina Immunochip and the genetic risk score was calculated as the sum of HLA DR/DQ genotype-weighted values and allele-weighted values from an additional 41 SNPs as previously defined.37,38 Exome sequencing was performed to identify mutations associated with monogenetic diabetes using SureSelect XT2 chemistry V7 Human All Exon (Agilent Technologies, Santa Clara, CA) as 2 × 101 bp paired-end on a NovaSeq S1 flowcell resulting in approximately 35 mio reads per sample. The reads were mapped with Burrows-Wheeler Alignment tool to human genome hg19 using default parameters.39 SNV analysis was done according to GenomeAnalysisToolkit best practices.40,41 Variants were annotated to the ClinVar database,42 and filtered by clinical condition or gene name and clinical significance.
Independent second cohort
The German Diabetes Prospective Follow-up Registry (DPV) collects data from children and adolescents who are diagnosed with diabetes at diabetes centres (hospitals and medical practices) throughout Germany via a centralized data management unit at the Institute of Epidemiology and Medical Biometry at Ulm University, Germany. The registry includes >90% of paediatric patients with diabetes nationwide.43,44 Between May 2008 and February 2013, patients diagnosed with diabetes <6 months earlier and before 20 years of age were asked to enrol in a central islet autoantibody testing program at the Institute of Diabetes Research, Helmholtz Zentrum München, Germany. Each patient and/or parent provided written informed consent to participate in the DPV central antibody testing (Bayerische Landesaerztekammer, #08043). HbA1c and random C-peptide concentrations were measured locally, and the results, together with the patient's BMI, were entered in the DPV database. DPV participants were followed longitudinally and data from the registry and the central islet autoantibody measurements were merged for analysis on 2414 participants. For the present analyses, data were retrieved from the DPV database in February 2021. In addition, 308 participants who were enrolled in the DiMelli study between January 2017 and December 2018 were used for data confirmation and were combined with the DPV patient cohort (Figure S1).
Statistical analysis
The CART analyses were performed in participants pre-stratified as islet autoantibody-positive or autoantibody-negative.45 The fasting C-peptide concentration, as a measure of endogenous insulin secretion, was used as the outcome variable. The maximum tree depth was set to 3. The robustness of the CART model was ensured by 25-fold cross-validation, with splits of 75/25% for training/validation sets. OLINK inflammatory marker data were normalized by calculating the difference from the median, categorized according to the 50th, 75th, 90th, 92.5th, 95th, and 97.5th percentiles, and subsequently scaled per analyte. Analytes with low variation were identified if their 97.5th centile was less than 1.5 CT values above the median value and were not included in the analysis. Clustering of the categorized OLINK data was then performed using the Euclidean distance and the ward.D2 method. Between-group comparisons were performed using the Kruskal–Wallis test for continuous variables and the χ2 test for categorical variables. To account for multiple comparisons of variables in the main cohort analyses, two-tailed P-values of <0.005 were considered significant. All statistical analyses were performed using R version 4.0.3 (http://cran.r-project.org) with the childsds, train, NbClust, and party packages.
Role of funders
The funders had no role in study design, data collection, data analysis, interpretation, or writing of report.
Results
The main analysis was performed on 1192 participants (665 males, 55.9%), who were enrolled in the DiMelli study between April 2009 and December 2016 (Figure S1). All of the subjects were diagnosed with diabetes before 20 years of age (median age at diagnosis: 10.4 years, interquartile range [IQR] 7.1–13.5). The participants were enrolled at a median of 9 days after diagnosis (IQR 6–13). Participants were classified as islet autoantibody-positive (n=1088, 91.3%) if they had one (n=103), two (n=265), three (n=360), or four (n=360) islet autoantibodies and as islet autoantibody-negative (n=104, 8.7%) (Table S2, Figure S1).
For confirmation and prognosis, a second cohort was included, which comprised 308 participants who were enrolled in the DiMelli study from January 2017, and 2414 participants who were enrolled - using similar eligibility criteria - in DPV, between May 2008 and February 2013 (Table S3, Figure S1).
CART analysis of patients with islet autoantibodies
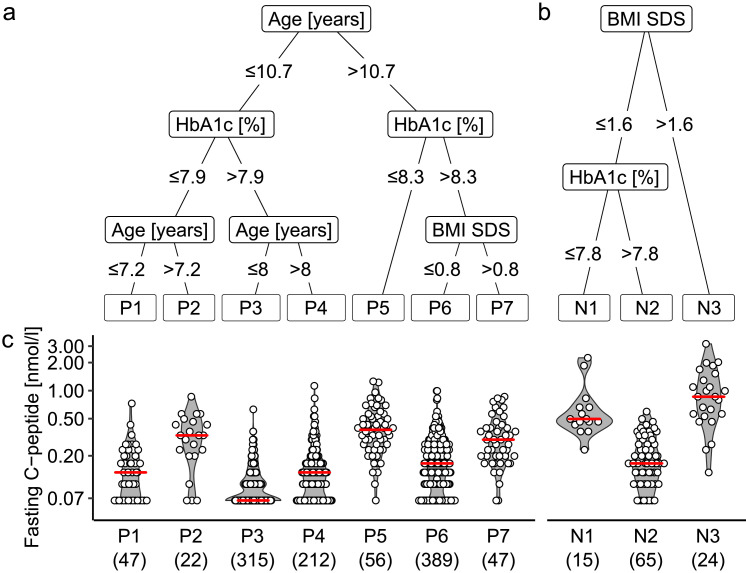
The fasting C-peptide concentrations in islet autoantibody-positive patients ranged from undetectable (<0.07 nmol/L) to 1.26 nmol/L. We searched for heterogeneity within these patients using the multivariable CART analysis method to identify groups based on fasting C-peptide concentrations. The CART analysis selected age as the first level predictor of the fasting C-peptide concentration, and discrimination was further improved by HbA1c, and BMI SDS (Figure 1a). The analysis identified seven groups of 47 (P1, 4.3%), 22 (P2, 2.0%), 315 (P3, 29.0%), 212 (P4, 19.5%) patients ≤10.7 years of age, and 56 (P5, 5.1%), 389 (P6, 35.7%), and 47 (P7, 4.3%) patients >10.7 years of age (Figure S2a). The median (IQR) C-peptide concentration was lowest in the P3 group patients (0.07 [<0.07–0.13] nmol/L), characterized by young age (≤8 years) and HbA1c >7.9% (Table 1, Figure 1a, c). The median (IQR) C-peptide concentration was highest in the older (>10.7 years) P5 group patients (0.38 [0.26–0.54] nmol/L) with HbA1c ≤8.3 and P7 group patients (0.30 [0.20–0.40] nmol/L) with HbA1c >8.3% and BMI SDS >0.8%, as well as the P2 group patients (0.33 [0.21–0.46] nmol/L) aged 7.2–10.7 years with HbA1c ≤7.9%. For sensitivity analyses, we included HLA genotype, genetic risk score, or the islet autoantibody phenotype, and observed only minor changes in the selected predictor variables with HLA genotype and genetic risk scores, and no selection of the added variables (Table S4).
Figure 1.
Multivariable CART model for classifying DiMelli participants with new-onset diabetes into subgroups. The model was applied to 1088 islet autoantibody-positive patients (a) and 104 islet autoantibody-negative patients (b) separately, using fasting C-peptide concentrations (c) as the outcome marker. Sex, age at diagnosis, days since diagnosis, HbA1c, BMI SDS, first-degree family history of type 1 diabetes, and first-degree family history of any other form of diabetes were included as possible predictor variables in the model. Of these variables, the CART model selected age, HbA1c, and BMI for the autoantibody-positive patients, and BMI and HbA1c for the autoantibody-negative patients. P values were ≤0.001 [CART] for each of the splits selected by the models. The red line displays the median. The numbers of patients are shown in parentheses. BMI, body mass index; HbA1c, haemoglobin A1c; SDS, standard deviation score.
Table 1.
Characteristics of the CART-defined groups of islet autoantibody-positive patients in the DiMelli cohort included in the main analysis.
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | N | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | p-value |
| Fasting C-peptide [nmol/l] | 47 | 0.13 [0.00;0.20] | 22 | 0.33 [0.21;0.46] | 315 | 0.07 [0.00;0.13] | 212 | 0.13 [0.07;0.17] | 56 | 0.38 [0.26;0.54] | 389 | 0.17 [0.10;0.23] | 47 | 0.30 [0.20;0.40] | <0.001 |
| Sex: Male | 47 | 26 (55.3%) | 22 | 13 (59.1%) | 315 | 158 (50.2%) | 212 | 104 (49.06%) | 56 | 36 (64.3%) | 389 | 235 (60.4%) | 47 | 35 (74.5%) | 0.003 |
| Age [years] | 47 | 5.02 [3.05;6.08] | 22 | 9.13 [8.14;10.18] | 315 | 5.42 [3.26;7.02] | 212 | 9.44 [8.89;10.16] | 56 | 14.09 [12.62;15.95] | 389 | 13.54 [12.07;14.87] | 47 | 13.18 [11.87;15.74] | <0.001 |
| BMI SDSa | 47 | -0.66 [-1.35;0.13] | 22 | -0.49 [-0.79;0.53] | 315 | -0.65 [-1.54;0.13] | 212 | -0.61 [-1.34;0.36] | 56 | 0.01 [-0.54;1.23] | 389 | -0.74 [-1.48;-0.11] | 47 | 1.42 [1.18;1.88] | <0.001 |
| HbA1c [%] | 47 | 7.20 [6.40;7.65] | 22 | 7.10 [6.45;7.60] | 315 | 10.50 [9.30;11.70] | 212 | 11.30 [10.20;12.80] | 56 | 7.30 [6.30;7.93] | 389 | 11.90 [10.50;13.50] | 47 | 10.80 [10.00;12.10] | <0.001 |
| Family History T1D | 47 | 6 (12.8%) | 22 | 3 (13.6%) | 315 | 22 (7.0%) | 212 | 10 (4.7%) | 56 | 7 (12.5%) | 389 | 26 (6.7%) | 47 | 4 (8.5%) | 0.16 |
| Family History T2D | 47 | 0 (0.0%) | 22 | 0 (0.0%) | 315 | 2 (0.6%) | 212 | 3 (1.4%) | 56 | 7 (12.5%) | 389 | 12 (3.7%) | 47 | 1 (2.1%) | 0.001 |
| Other autoimmunityb | 47 | 8 (17.0%) | 22 | 2 (9.1%) | 315 | 50 (15.9%) | 212 | 35 (16.5%) | 56 | 8 (14.3%) | 389 | 75 (19.3%) | 47 | 7 (14.9%) | 0.85 |
| Treatment: Insulin | 47 | 43 (91.5%) | 22 | 19 (86.4%) | 315 | 313 (99.4%) | 212 | 210 (99.1%) | 56 | 50 (89.3%) | 389 | 385 (99.0%) | 47 | 45 (95.7%) | <0.001 |
| Insulin dose [U/day/kg]c | 34 | 0.52 [0.15;0.83] | 14 | 0.46 [0.33;0.63] | 275 | 0.87 [0.63;1.18] | 189 | 0.99 [0.69;1.23] | 45 | 0.46 [0.31;0.64] | 338 | 0.98 [0.75;1.26] | 37 | 0.87 [0.60;1.09] | <0.001 |
| Ketonuria | 40 | 22 (55.0%) | 18 | 12 (66.7%) | 301 | 269 (89.4%) | 202 | 191 (94.5%) | 45 | 31 (68.9%) | 365 | 337 (92.3%) | 45 | 41 (91.1%) | <0.001 |
| Insulin sensitivityd | 27 | 15.82 [14.59;17.07] | 11 | 12.47 [11.51;13.70] | 163 | 10.46 [8.74;12.34] | 128 | 7.99 [6.61;9.45] | 27 | 9.97 [7.14;11.05] | 199 | 6.28 [5.06;7.69] | 24 | 4.71 [3.76;5.38] | <0.001 |
| Triglycerides [mg/dl] | 41 | 64.0 [53.0;81.0] | 19 | 62.0 [55.5;78.5] | 243 | 72.0 [57.5;92.5] | 171 | 75.0 [57.0;99.5] | 45 | 74.0 [57.0;106.0] | 291 | 85.0 [67.0;108.5] | 37 | 120.0 [83.0;157.0] | <0.001 |
| Vitamin D3 [ng/ml] | 42 | 27.6 [18.2;35.0] | 21 | 28.8 [20.6;37.3] | 292 | 27.1 [19.5;36.0] | 198 | 24.8 [15.6;33.2] | 51 | 20.3 [13.0;32.1] | 357 | 24.1 [15.0;32.6] | 45 | 22.2 [14.4;30.0] | 0.005 |
| Vitamin D3 deficiencye | 42 | 24 (57.1%) | 21 | 12 (57.1%) | 292 | 166 (56.8%) | 198 | 126 (63.6%) | 51 | 37 (72.5%) | 357 | 244 (68.3%) | 45 | 34 (75.6%) | 0.02 |
| IFNɣ [pg/ml] | 23 | 10.3 [6.5;21.2] | 15 | 6.3 [4.7;10.2] | 170 | 7.7 [5.0;13.7] | 133 | 5.8 [4.0;10.1] | 24 | 4.6 [3.2;8.7] | 217 | 4.7 [3.5;7.3] | 21 | 4.9 [3.3;6.2] | <0.001 |
| IL-10 [pg/ml] | 23 | 1.0 [0.6;1.4] | 15 | 0.8 [0.5;1.3] | 170 | 1.2 [0.7;2.5] | 133 | 0.7 [0.5;1.2] | 24 | 0.8 [0.5;1.3] | 217 | 0.6 [0.5;1.2] | 21 | 0.6 [0.4;1.0] | <0.001 |
| IL-12p70 [pg/ml] | 23 | 0.3 [0.2;0.6] | 15 | 0.3 [0.2;0.7] | 170 | 0.3 [0.2;0.8] | 133 | 0.3 [0.1;0.6] | 24 | 0.3 [0.1;0.4] | 217 | 0.3 [0.1;0.6] | 21 | 0.3 [0.1;0.5] | 0.10 |
| IL-1beta [pg/ml] | 23 | 1.0 [0.3;2.3] | 15 | 0.3 [0.2;5.1] | 170 | 1.2 [0.5;4.7] | 133 | 0.5 [0.2;2.5] | 24 | 0.6 [0.3;2.6] | 217 | 0.8 [0.3;2.9] | 21 | 0.8 [0.3;2.0] | 0.05 |
| IL-2 [pg/ml] | 23 | 0.3 [0.1;0.9] | 15 | 0.4 [0.1;0.8] | 170 | 0.4 [0.1;1.1] | 133 | 0.2 [0.1;1.0] | 24 | 0.1 [0.0;0.8] | 217 | 0.2 [0.1;0.7] | 21 | 0.2 [0.0;0.5] | 0.14 |
| IL-6 [pg/ml] | 23 | 1.9 [0.7;3.5] | 15 | 1.7 [0.5;9.3] | 170 | 2.3 [0.9;8.8] | 133 | 1.1 [0.6;4.6] | 24 | 1.3 [0.6;8.5] | 217 | 1.3 [0.6;4.8] | 21 | 1.4 [0.7;3.7] | 0.09 |
| IL-8 [pg/ml] | 23 | 70.3 [15.9;1393.3] | 15 | 98.1 [16.8;1973.6] | 170 | 524.5 [95.8;2618.1] | 133 | 147.1 [31.6;1303.9] | 24 | 146.5 [63.7;925.2] | 217 | 242.9 [53.9;1628.9] | 21 | 470.3 [112.5;1235.3] | 0.06 |
| TNF [pg/ml] | 23 | 4.6 [2.9;8.0] | 15 | 3.9 [2.0;6.9] | 170 | 4.3 [2.9;7.9] | 133 | 3.0 [1.9;4.8] | 24 | 3.1 [1.9;5.8] | 217 | 3.0 [1.7;5.4] | 21 | 3.4 [1.8;4.6] | <0.001 |
| HLAf - high risk | 36 | 12 (33.3%) | 18 | 7 (38.9%) | 270 | 89 (33.0%) | 181 | 50 (27.6%) | 45 | 12 (26.7%) | 347 | 99 (28.5%) | 39 | 7 (17.9%) | 0.78 |
| - moderate risk | 36 | 12 (33.3%) | 18 | 4 (22.2%) | 270 | 67 (24.8%) | 181 | 53 (29.3%) | 45 | 9 (20.0%) | 347 | 89 (25.6%) | 39 | 11 (28.2%) | |
| - neutral genotypes | 36 | 9 (25.0%) | 18 | 6 (33.3%) | 270 | 100 (37.0%) | 181 | 65 (35.9%) | 45 | 21 (46.7%) | 347 | 136 (39.2%) | 39 | 20 (51.3%) | |
| - protective genotypes | 36 | 3 (8.3%) | 18 | 1 (5.6%) | 270 | 14 (5.2%) | 181 | 13 (7.2%) | 45 | 3 (6.7%) | 347 | 23 (6.6%) | 39 | 1 (2.6%) | |
| Genetic Risk Scoreg | 34 | 10.93 [10.20;11.57] | 18 | 11.22 [9.97;11.61] | 258 | 10.73 [10.22;11.29] | 178 | 10.80 [10.13;11.20] | 42 | 10.52 [9.96;10.91] | 337 | 10.71 [10.15;11.19] | 35 | 10.76 [10.20;11.42] | 0.331 |
| N autoantibodies - 1 | 47 | 6 (12.8%) | 22 | 4 (18.2%) | 315 | 26 (8.2%) | 212 | 28 (13.2%) | 56 | 7 (12.5%) | 389 | 28 (7.2%) | 47 | 4 (8.5%) | 0.17 |
| - 2 | 47 | 9 (19.1%) | 22 | 4 (18.2%) | 315 | 80 (25.4%) | 212 | 48 (22.6%) | 56 | 12 (21.4%) | 389 | 104 (26.7%) | 47 | 8 (17.0%) | |
| - 3 | 47 | 12 (25.5%) | 22 | 5 (22.7%) | 315 | 106 (33.6%) | 212 | 62 (29.2%) | 56 | 26 (46.4%) | 389 | 133 (34.2%) | 47 | 16 (34.0%) | |
| - 4 | 47 | 20 (42.5%) | 22 | 9 (40.9%) | 315 | 103 (32.7%) | 212 | 74 (34.9%) | 56 | 11 (19.6%) | 389 | 124 (31.9%) | 47 | 19 (40.4%) | |
| IAA positiveh | 44 | 37 (84.1%) | 18 | 12 (66.7%) | 313 | 254 (81.1%) | 209 | 136 (65.1%) | 49 | 36 (73.5%) | 385 | 231 (60.0%) | 45 | 32 (71.1%) | <0.001 |
| GADA positive | 47 | 36 (76.6%) | 22 | 14 (63.6%) | 315 | 201 (63.8%) | 212 | 141 (66.5%) | 56 | 37 (66.1%) | 389 | 290 (74.5%) | 47 | 36 (76.6%) | 0.04 |
| IA-2A positive | 47 | 36 (76.6%) | 22 | 17 (77.3%) | 315 | 237 (75.2%) | 212 | 165 (77.8%) | 56 | 40 (71.4%) | 389 | 311 (79.9%) | 47 | 37 (78.7%) | 0.74 |
| ZnT8A positive | 47 | 31 (66.0%) | 22 | 20 (90.9%) | 315 | 224 (71.1%) | 212 | 164 (77.4%) | 56 | 40 (71.4%) | 389 | 299 (76.9%) | 47 | 39 (83.0%) | 0.09 |
SDS: age- and sex-adjusted standard deviation score.
Positive for either thyroid peroxidase or tissue transglutaminase autoantibodies.
The insulin dose was calculated for insulin-treated patients.
Insulin sensitivity score.
Defined as <30 ng/ml.
High-risk (DR3/4-DQ8 or DR4-DQ8/DR4-DQ8), moderate, neutral, and protective HLA genotypes, as defined by Walter et al.36 (Table S1).
Remaining genetic risk score calculated without the HLA-DR/DQ genotype.
IAA positive status was only assigned to samples collected <14 days after starting insulin.
IQR, interquartile range; BMI, body mass index; HbA1c, haemoglobin A1c; T1D, type 1 diabetes; T2D, type 2 diabetes; IFNɣ, interferon-ɣ; IL, interleukin; TNF, tumour necrosis factor; HLA, human leukocyte antigen; IAA, insulin autoantibodies; IA-2A, insulinoma-associated antigen-2 autoantibodies; GADA, glutamate decarboxylase (65-kDa isoform) autoantibodies; ZnT8A, zinc transporter-8 autoantibodies.
The seven groups differed in terms of vitamin D3 (P=0.005 [Kruskal–Wallis]), interferon-ɣ (IFNɣ; P<0.001 [Kruskal–Wallis]), IL-10 (P<0.001 [Kruskal–Wallis]), and tumour necrosis factor (TNF; P<0.001 [Kruskal–Wallis]) concentrations, the family history of type 2 diabetes (P=0.001 [χ2]), insulin sensitivity (P<0.001 [Kruskal–Wallis]), insulin demand (P<0.001 [Kruskal–Wallis]), ketonuria (P<0.001), triglyceride concentrations (P<0.001 [Kruskal–Wallis]), and frequency of IAA (P<0.001 [χ2]) (Table 1, Figure 2a).
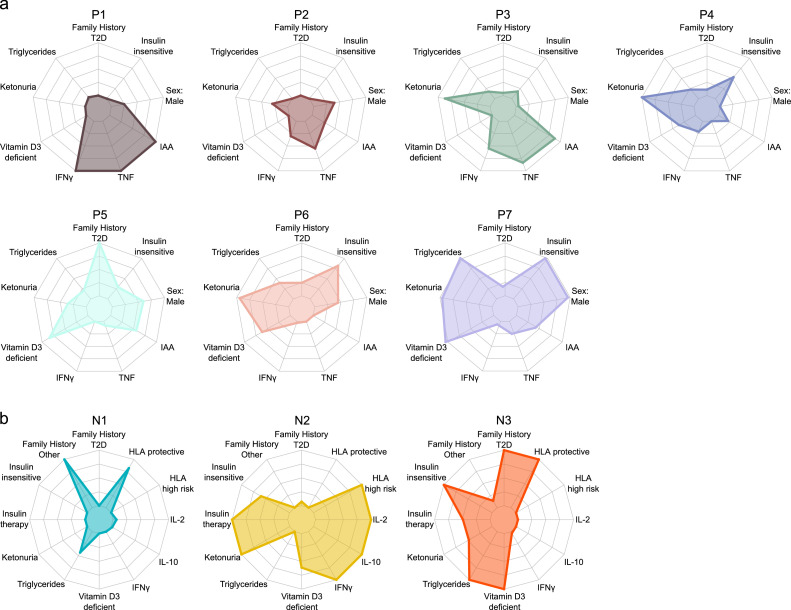
Figure 2.
Features of the CART-defined subgroups derived from DiMelli participants included in the main analysis. The radar plots show the CART groups for 1088 islet autoantibody-positive patients (a) and 104 islet autoantibody-negative patients (b) at diagnosis. The indicated variables were highly significant between the groups, as shown in Tables 1 and 2. The radar plots convert the differences observed between the groups to the full scale of the plot for each variable. HLA, human leukocyte antigen; IFNɣ, interferon-ɣ; IL, interleukin; T2D, type 2 diabetes; TNF, tumour necrosis factor.
The robustness of the CART categories to distinguish groups with different C-peptide concentrations was tested in 2536 islet autoantibody-positive patients, including 968 with an available C-peptide concentration in the second cohort. The proportion of patients in each subgroup was comparable to the distribution in the main analysis (Figure S2a). Similar to the main analysis, the median (IQR) C-peptide concentration was lowest in group p3 (0.10 [0.07-0.17] nmol/L), and highest in group p5 (0.50 [0.23–0.84] nmol/L), followed by group p7 (0.20 [0.11–0.33] nmol/L), and group p2 (0.19 [0.12–0.34] nmol/L; Figure S3; Table S5).
CART analysis of islet autoantibody-negative patients
The CART analysis of the autoantibody-negative patients selected BMI SDS as the first level predictor of the fasting C-peptide concentration, and discrimination was further improved by HbA1c in patients in the low BMI SDS category (Figure 1b). The analysis identified three groups of 15 (N1, 14.4%), 65 (N2, 62.5%), and 24 (N3, 23.1%) patients. The median (IQR) C-peptide concentration was lowest in the N2 patients (0.17 [0.10–0.26] nmol/L), characterized by a BMI SDS of ≤1.6 and HbA1c >7.8, and highest in the N3 patients (0.86 [0.55–1.35] nmol/L) with a BMI SDS >1.6 (Figure 1c). There were substantial differences among the three groups in terms of IL-2 concentrations (P=0.001 [Kruskal–Wallis]), insulin treatment (P<0.001 [χ2]), HLA genotype (P<0.001 [χ2]), family history of type 2 diabetes (P=0.002 [χ2]), triglyceride concentrations (P=0.001 [Kruskal–Wallis]), and vitamin D3 concentrations (P<0.001 [Kruskal–Wallis]) (Table 2, Figure 2b). In sensitivity analyses, the inclusion of HLA genotype, genetic risk score, or other autoimmunity in the model did not alter the CART stratification (Table S6).
Table 2.
Characteristics of CART-defined groups of islet autoantibody-negative patients in the DiMelli cohort included in the main analysis.
| N1 | N2 | N3 | |||||
|---|---|---|---|---|---|---|---|
| Variable | n | median [IQR] or n(%) | n | median [IQR] or n(%) | n | median [IQR] or n(%) | p-value |
| Fasting C-peptide [nmol/l] | 15 | 0.50 [0.45;0.66] | 65 | 0.17 [0.10;0.26] | 24 | 0.86 [0.55;1.35] | <0.001 |
| Sex: Male | 15 | 5 (33.3%) | 65 | 41 (63.1%) | 24 | 12 (50.0%) | 0.09 |
| Age [years] | 15 | 11.27 [10.23;15.82] | 65 | 12.55 [8.11;15.32] | 24 | 14.29 [13.43;15.63] | 0.09 |
| BMI SDSa | 15 | 0.34 [-0.47;0.81] | 65 | -0.29 [-1.28;0.50] | 24 | 2.29 [1.79;2.84] | <0.001 |
| HbA1c [%] | 15 | 6.60 [6.10;6.95] | 65 | 10.80 [9.30;13.00] | 24 | 10.05 [7.90;10.90] | <0.001 |
| Family History T1D | 15 | 3 (20.0%) | 65 | 8 (12.3%) | 24 | 1 (4.2%) | 0.33 |
| Family History T2D | 15 | 1 (6.7%) | 65 | 6 (9.2%) | 24 | 10 (41.7%) | 0.002 |
| Family History other | 15 | 3 (20.0%) | 65 | 1 (1.5%) | 24 | 1 (4.2%) | 0.02 |
| Other autoimmunityb | 15 | 0 (0.0%) | 65 | 10 (15.4%) | 24 | 1 (4.2%) | 0.18 |
| Treatment: Insulinc | 15 | 5 (33.3%) | 65 | 64 (98.5%) | 24 | 17 (70.8%) | <0.001 |
| Ketonuria | 12 | 3 (25.0%) | 57 | 47 (82.5%) | 19 | 10 (52.6%) | <0.001 |
| Insulin sensitivityd | 9 | 9.68 [9.19;15.50] | 44 | 6.73 [5.30;8.97] | 19 | 3.33 [2.32;3.99] | <0.001 |
| Systolic blood pressure SDSa | 14 | 0.34 [-0.31;1.22] | 62 | 0.76 [-0.48;1.76] | 24 | 1.44 [0.45;2.22] | 0.14 |
| Diastolic blood pressure SDSa | 14 | -0.14 [-0.62;0.72] | 62 | 0.15 [-0.57;1.26] | 24 | 0.88 [-0.77;2.08] | 0.40 |
| Triglycerides [mg/dl] | 13 | 120.0 [65.0;150.0] | 58 | 92.5 [70.2;129.0] | 23 | 156.0 [106.0;277.5] | 0.001 |
| Vitamin D3 [ng/ml] | 15 | 37.8 [27.0;49.0] | 63 | 22.5 [15.9;35.3] | 23 | 10.9 [7.7;25.8] | <0.001 |
| Vitamin D3 deficiencye | 15 | 5 (33.3%) | 63 | 40 (63.5%) | 23 | 19 (82.6%) | 0.009 |
| IFNɣ [pg/ml] | 13 | 4.1 [3.4;8.0] | 37 | 6.1 [3.8;10.2] | 13 | 4.1 [3.0;5.0] | 0.04 |
| IL-10 [pg/ml] | 13 | 0.5 [0.4;0.6] | 37 | 0.9 [0.5;1.6] | 13 | 0.4 [0.3;0.6] | 0.02 |
| IL-12p70 [pg/ml] | 13 | 0.2 [0.1;0.8] | 37 | 0.4 [0.2;1.0] | 13 | 0.3 [0.2;0.4] | 0.37 |
| IL-1beta [pg/ml] | 13 | 0.4 [0.3;3.6] | 37 | 1.1 [0.4;6.3] | 13 | 0.6 [0.2;1.0] | 0.23 |
| IL-2 [pg/ml] | 13 | 0.0 [0.0;0.4] | 37 | 0.3 [0.1;1.1] | 13 | 0.0 [0.0;0.1] | 0.001 |
| IL-6 [pg/ml] | 13 | 0.9 [0.5;1.5] | 37 | 3.0 [0.7;10.3] | 13 | 1.0 [0.9;1.7] | 0.42 |
| IL-8 [pg/ml] | 13 | 94.9 [12.4;258.8] | 37 | 374.2 [19.6;3871.3] | 13 | 17.3 [8.2;438.3] | 0.09 |
| TNF [pg/ml] | 13 | 3.2 [2.4;4.5] | 37 | 4.0 [3.0;7.8] | 13 | 2.9 [2.2;4.1] | 0.08 |
| HLAf - high risk | 12 | 0 (0.0%) | 58 | 11 (19.0%) | 21 | 0 (0.0%) | <0.001 |
| - moderate risk | 12 | 1 (8.3%) | 58 | 18 (31.0%) | 21 | 1 (4.7%) | |
| - neutral genotypes | 12 | 4 (33.3%) | 58 | 17 (29.3%) | 21 | 6 (28.6%) | |
| - protective genotypes | 12 | 7 (58.3%) | 58 | 12 (20.7%) | 21 | 14 (66.7%) | |
| Genetic Risk Scoreg | 13 | 10.10 [9.78;10.69] | 55 | 10.30 [9.80;11.09] | 19 | 10.28 [9.72;10.76] | 0.95 |
SDS: age- and sex-adjusted standard deviation score.
Positive for either thyroid peroxidase or tissue transglutaminase autoantibodies.
Two of the insulin-treated patients were also treated with metformin.
Insulin sensitivity score.
Defined as <30 ng/ml.
High-risk (DR3/4-DQ8 or DR4-DQ8/DR4-DQ8), moderate, neutral, and protective HLA genotypes, as defined by Walter et al.36 (Table S1).
Remaining genetic risk score calculated without the HLA-DR/DQ genotype.
IQR, interquartile range; BMI, body mass index; HbA1c, haemoglobin A1c; T1D, type 1 diabetes; T2D, type 2 diabetes; IFNɣ, interferon-ɣ; IL, interleukin; TNF, tumour necrosis factor; HLA, human leukocyte antigen.
The CART categories were examined in 186 islet autoantibody-negative patients, including 80 with available C-peptide from the second cohort. As in the main analysis, the median (IQR) C-peptide concentration was lowest in the n2 patients (0.13 [0.07–0.24] nmol/L) and was highest in the n3 patients (0.60 [0.39–0.93] nmol/L; P<0.001 [Kruskal–Wallis]) (Figure S3, Table S7). The proportion of patients in each subgroup (n1, 12.9%; n2, 60.2%; n3, 26.9%) was comparable to the distribution in the main analysis (Figure S2b).
Features of the CART subgroups
The CART groups were examined for evidence of subtypes. Characteristic features were observed in most islet autoantibody-positive groups (Figure 2a, Table 1). Inflammatory signatures and insulin autoimmunity were dominant features in subgroups of children aged <8 years at type 1 diabetes diagnosis. Group P1 was distinguished from group P3 in terms of lower frequencies of ketonuria and higher IFNɣ concentrations. In contrast, higher BMI and characteristics associated with type 2 diabetes were frequent in children diagnosed at older ages (groups P5, P6, and P7). P5 had the highest frequency of patients with a family history of type 2 diabetes and vitamin D3 deficiency; P6 had a high proportion of insulin insensitivity, and group P7, had the highest triglyceride concentration, highest frequency of patients with insulin insensitivity and vitamin D3 deficiency, and a higher frequency of male patients.
The three islet autoantibody-negative groups also had characteristic features (Figure 2b). Group N2 displayed characteristics of type 1 diabetes, which included a predominance of patients with susceptible HLA genotypes, high proportions of patients with ketonuria and on insulin therapy, higher concentrations of IFNɣ, IL-10, and IL-2, and low vitamin D3 concentrations. Typical of patients with monogenic forms of diabetes, group N1 was characterized by a relatively high prevalence of a first-degree family history of diabetes other than type 1 or type 2, low HLA-associated genetic risk with a high proportion of patients who had protective HLA genotypes, a low proportion of patients on insulin therapy, and relatively few patients with vitamin D3 deficiency. Group N3 had features of type 2 diabetes, including the highest proportion of patients with a first-degree family history of type 2 diabetes, low insulin sensitivity, high triglyceride concentrations, and the highest proportion of patients with vitamin D3 deficiency. Whole-exome sequencing or maturity-onset diabetes of the young (MODY) typing was performed in 93 islet autoantibody-negative patients and identified 11 (11.8%) patients who were carrying variants of recognized forms of monogenic diabetes, including 5 of 13 (38.5%) patients in group N1, 3 of 57 (5.3%) in group N2, and 3 of 21 (14.3%) in group N3 (Table S8).
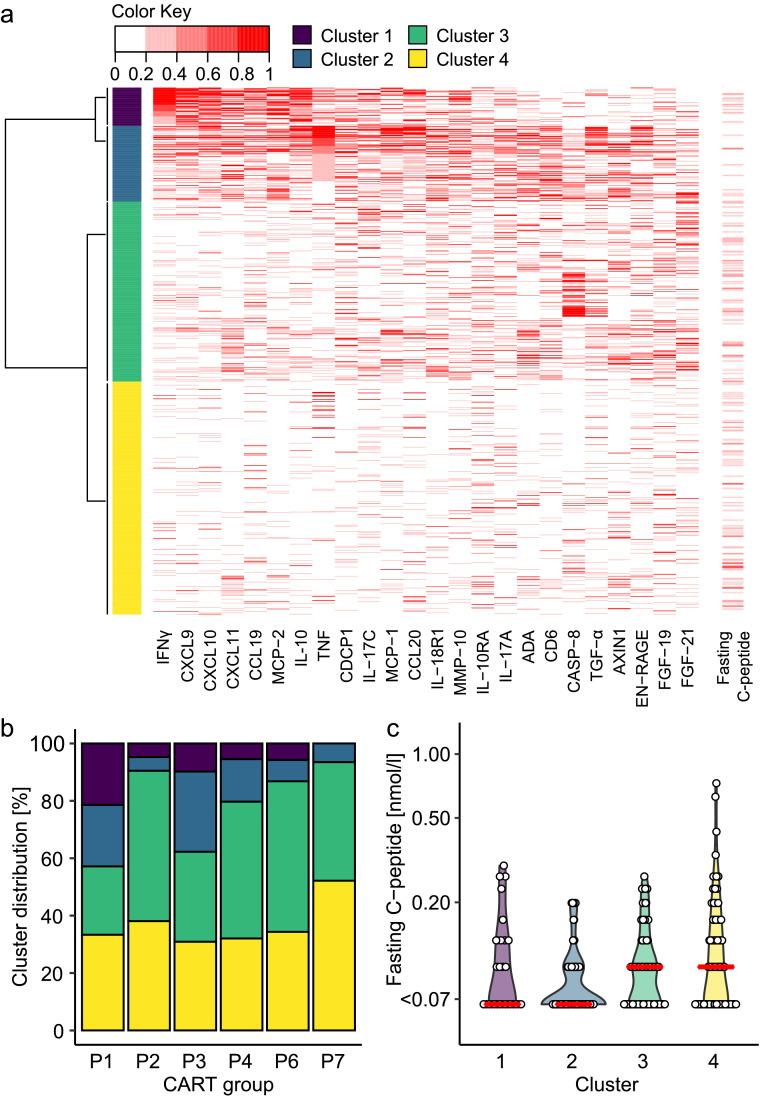
Inflammatory markers
The cytokine concentrations differed among the islet autoantibody-positive groups. Therefore, we measured an expanded panel of inflammatory markers in 805 islet autoantibody-positive patients from groups P1, P2, P3, P4, P6, and P7 where a suitable sample was available. Unsupervised clustering identified four clusters of patients (Figure 3a). Cluster 1 (58 patients) was characterized by an IFNɣ signature with increased concentrations of IFNɣ, IL-10, and the IFNɣ-inducible chemokines CXCL9, CXCL10, CXCL11, and CCL19. Cluster 2 (116 patients) had increased concentrations of TNF, proteins that may be inducible by TNF such as CCL20, proteins that may be associated with T cell activation such as CD6, and moderate concentrations of IFNɣ and IFNɣ-inducible chemokines. Cluster 3 (275 patients) was an intermediate, heterogeneous group with elevated concentrations of some inflammatory proteins such as fibroblast growth factor 21, but without a consistent pattern. Cluster 4 (356 patients) generally had low concentrations of the inflammatory proteins. The patients in clusters 1 and 2 were younger (P<0.001 [Kruskal–Wallis]) and had lower median C-peptide concentrations (P<0.001 [Kruskal–Wallis]) than the patients in clusters 3 and 4 (Table S9).
Figure 3.
Inflammatory markers (OLINK inflammation panel) at diagnosis of diabetes in 805 islet autoantibody-positive patients in DiMelli. (a) The data were used to generate heatmaps and identify four clusters of patients. The most informative proteins in these clusters are indicated in the heatmap. Fasting C-peptide is shown on the right. (b) Frequency distribution of the four clusters within the CART-defined patient groups. (c) Fasting C-peptide concentrations in the group P1 and P3 patients stratified by inflammatory cluster. ADA, adenosine deaminase; CASP-8, caspase-8; FGF, fibroblast growth factor; IFNɣ, interferon-ɣ; IL, interleukin; MCP, monocyte chemoattractant protein; MMP, matrix metalloproteinase; TGF, transforming growth factor; TNF, tumour necrosis factor.
The IFNɣ inflammatory cluster 1 was most prevalent in group P1 (21.4% of patients versus 5.5% in the remaining groups; range 0% in P7 to 9.8% in P3; P<0.001 [χ2]), and the TNF inflammatory cluster 2 was most prevalent in groups P3 and P1 (27.9% and 21.4% of patients, respectively, versus 7.2% in the remaining groups; range 4.8% in P2 to 14.8% in P4; P<0.001 [χ2]; Figure 3b). The C-peptide concentration at diagnosis was associated with age (P<0.001) and was inversely correlated with the serum TNF concentration (P=0.003) in a multivariable linear regression model. Moreover, within groups P1 and P3, the C-peptide concentrations were lower in patients with the TNF cluster 2 profile than in patients in the intermediate (cluster 3) and low (cluster 4) inflammatory profiles (median, <0.07 nmol/L vs 0.10 nmol/L; P=0.001 [Kruskal–Wallis]; Figure 3c).
Genetic and autoimmune features fail to stratify autoantibody-positive patients
We observed no additional contributions of genetics or autoantibodies to the CART analysis, as well as very similar genetic and autoimmune features among the islet autoantibody-positive CART groups (Table 1). To exclude the possibility that important genetic- or autoantibody-based subgroups were missed by the CART analysis, we re-examined the patients and classified them using genetics (Table S10) or autoantibodies. An inverse relationship between the hierarchy of genetic risk and C-peptide was observed in the islet autoantibody-negative patients (Table S11, Figure S4b), However, the genetic risk was not associated with the C-peptide concentrations within CART subgroups of islet autoantibody-negative patients, and also not in the islet autoantibody-positive patients (Table S10 & S12, Figure S4a).
Endotypes based on differences in the first-appearing islet autoantibodies (IAA versus GADA) or rates of disease progression have been proposed.27,46,47 Therefore, we categorized the participants into four groups with and without IAA and/or GADA (Table S13). As expected, the patients in the IAA without GADA group were youngest and had a higher frequency of the DR4-DQ8 haplotype than the other groups. Patients in the GADA without IAA group were older and had a higher frequency of the DR3-DQ2 haplotype than the other groups. The median HbA1c was lower in patients with IAA than in patients without IAA (10.60% [9.10%–12.20%] vs 11.50% [9.83%–13.10%]; P=0.001 [Kruskal–Wallis]). There were no differences in C-peptide concentrations among the four groups. The associations of the autoantibody pattern with age and HbA1c, and the lack of an association with the C-peptide concentration was confirmed in the second cohort (Table S14).
Prognostic relevance of CART subgroups
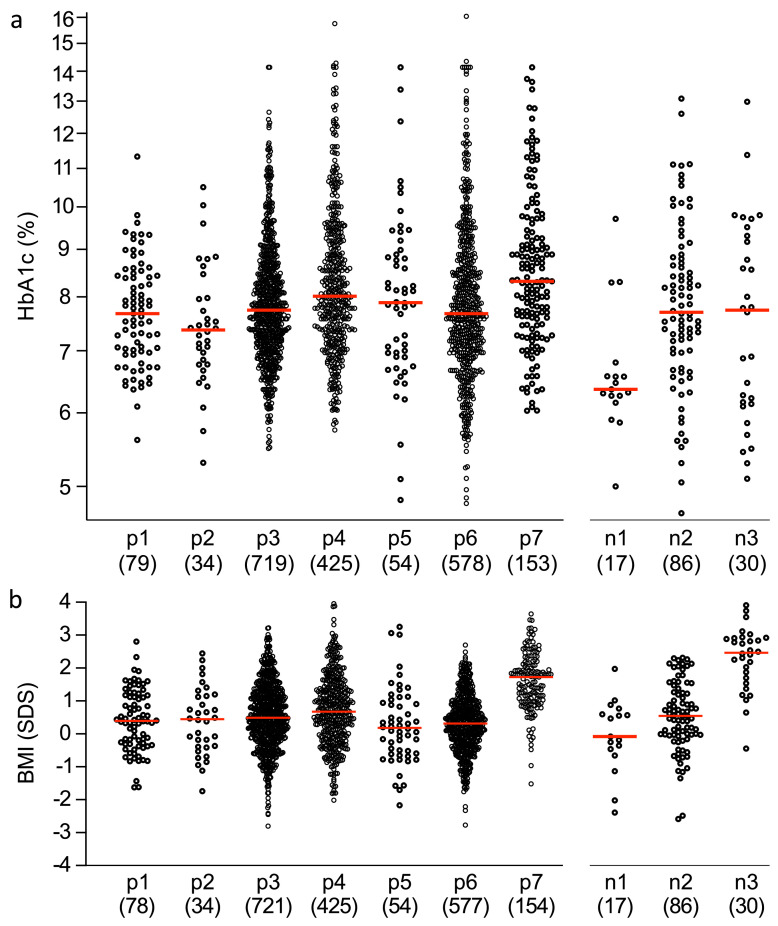
The mid- to longer-term outcomes of the subgroups were analysed in 2196 participants from the DPV cohort. These patients were followed longitudinally (total follow-up 16,460 years) for documentation of metabolic parameters and thereby provided an opportunity to assess the prognostic relevance of each subgroup. There was substantial variability in HbA1c at a median follow-up of 7.0 years (IQR 4–8) after the diagnosis of diabetes (Figure 4). Within the islet autoantibody-positive subgroups, the median HbA1c values were 7.37% in group p2, 8.32% in group p7 (P<0.001 vs other subgroups [Kruskal–Wallis]), and ranged from 7.68% to 8.01% in the other groups (Figure 4, Table S15). Similarly, the proportion of patients with an HbA1c <8% was highest in group p2 (26 of 34; 76.5%) and lowest in group p7 (61 of 153; 39.9%). The majority (116 of 154; 75.3%) of patients in group p7 were overweight or obese at follow-up visits (Table S15). Only minor differences in the frequency of insulin treated patient or insulin dose at follow-up were observed between the islet autoantibody-positive groups. Within the autoantibody-negative subgroups, 84 of 86 (98%) patients in group n2 required insulin supporting the diagnosis of ‘autoantibody-negative type 1 diabetes’ (Figure 4, Table S16). The n1 group showed the best long-term metabolic control (HbA1c 6.36% [6.26–6.57]), had a normal weight (BMI SDS −0.08 [−0.46 to 0.60]), and only seven of 17 (41%) patients required insulin therapy. Patients in group n3 remained overweight (BMI SDS 2.47 [1.74; 2.90]) and seven of 30 (23%) received treatments other than insulin, including five (17%) treated with oral antidiabetic therapies.
Figure 4.
HbA1c (a) and BMI (b) at a median follow-up of 7.0 years after the diagnosis of diabetes in the second cohort. The patients were classified according to their islet autoantibody status and the CART group from the parameters recorded at disease onset (islet autoantibody-positive groups: p1–p7; islet autoantibody-negative groups: n1, n2, and n3). HbA1c (a) and BMI expressed as the age- and sex-adjusted SDS (b) at follow-up are shown in each of the subgroups. The red line displays the median. The numbers of patients are shown in parentheses.
Discussion
A clinically relevant classifier of subtypes that was based on islet autoantibodies, age, HbA1c, and BMI defined 10 subgroups of diabetes among children and adolescents with new-onset diabetes. These included seven groups in islet autoantibody-positive patients and one group within the islet autoantibody-negative patients that had features consistent with type 1 diabetes. The subgroups differed in terms of their residual C-peptide concentrations, inflammatory markers, insulin sensitivity and other type 2 diabetes associated characteristics, and genetics, as well as their long-term metabolic control. The approach provides a first step to address the heterogeneity of young-onset diabetes with high resolution and prognostic relevance.
For subgroup classification, we used the CART analysis approach, which has been successfully applied in cancer research,48 and other autoimmune diseases.49 Previous approaches to the classification of diabetes in children and adolescents, such as those used in the SEARCH for Diabetes in Youth (SEARCH) study,4,50,51 mainly aimed at distinguishing type 1 and type 2 diabetes and were based on predetermined variables and thresholds. The CART analysis is a supervised multivariable approach that is used to discriminate a target outcome measure. We chose the fasting C-peptide concentration because of its prognostic value.2,16, 17, 18 The classification based on CART was robust, as demonstrated by its consistent selection of the group defining parameters in 25-fold cross validation or the inclusion of additional patient characteristics and confirmation in a second cohort of patients. It uses parameters readily available to clinicians soon after onset and, therefore, can be easily applied in the clinic. An important consideration is that the CART analysis was successful when patients were first classified as islet autoantibody-positive or autoantibody-negative. It is also notable that, although the islet autoantibody status, age, BMI and HbA1c are known to be associated with C-peptide,4,52 the CART multivariable analysis yielded a decision tree from these variables that was remarkably associated with C-peptide concentration and defined clinically relevant subgroups.
The CART analysis subgrouping of cases will be valuable in the future if it helps to define different disease forms, disease endotypes, or theratypes,8 or to identify patients with particularly poor prognosis. In islet autoantibody-positive patients, the CART analysis could identify subgroups of young patients (<8 years) with insulin autoimmunity and increased prevalences of TNF and IFNɣ inflammatory profiles (P1 and P3), and subgroups of older children (>10.7 years) with features of insulin insensitivity and type 2 diabetes characteristics (P5, P6, P7). The CART analysis also distinguished a group of islet autoantibody-negative patients who were more likely to have type 1 diabetes (N2), and who may be subjected to further tests, such as evaluation of the type 1 diabetes genetic risk score or autoantibody testing on follow-up. Although the scope of our study was not to diagnose monogenic diabetes, the CART analysis could identify a subgroup of islet autoantibody-negative patients (N1, 14.4%), where screening for monogenic diabetes was most effective. None of the patients in group N1 had high-risk HLA genotypes or elevated genetic risk scores, and around one-third of patients in this group had monogenic forms of diabetes. The CART grouping may, therefore, supplement existing MODY probability calculators.53
The inflammatory markers were particularly informative. Group P1 included the highest proportion of patients with an IFNɣ inflammatory cluster profile, and over 20% of patients in groups P1 and P3 had a TNF inflammatory profile, which was associated with the lowest C-peptide concentrations in these groups. This may suggest an aggressive phenotype of early-onset type 1 diabetes characterized by inflammatory autoimmunity that defines a portion of patients with young onset. The groups with inflammatory features have similarities to a young-onset endotype suggested by histological examination of the pancreas of patients and characterized by young age (<7 years), a predominance and higher number of B lymphocytes in and around pancreatic islets and low β-cell function.12 The same study described a second endotype characterized by an older age (>13 years) and a paucity of B lymphocytes in the infiltrate, which may correspond to groups P5, P6 and P7. Although the more extensive inflammatory panel was not applied to the islet autoantibody-negative patients, it is notable that IFNɣ concentration was slightly elevated in group N2 patients as compared to concentration in the N1 and N3 groups. A potential limitation of these data is that the values for some of the measured markers may have been affected by overnight shipping of samples at room temperature.
Age remained a dominant determinant of the heterogeneity of islet autoantibody-positive patients, including the inflammatory markers. This finding could be relevant to the recent success of anti-TNF therapy in patients with type 1 diabetes,54 and this may indicate that patients in group P3 or patients with a TNF inflammatory profile may benefit most from such therapy if applied early enough. Similarly, patients from the relatively small group P1 (<5% of patients) may benefit from therapies that interfere with IFNɣ signalling such as JAK/STAT inhibitors and/or anti-TNF therapies. Despite relatively high C-peptide concentration at diabetes onset, patients in the older group P7 experienced the worst metabolic outcomes in the follow-up. This group represented around 5% of patients who were defined by islet autoantibody positivity and high BMI. These patients had high triglyceride concentrations and were relatively insulin insensitive, but had clear genetic and antibody features of type 1 diabetes and, apart from their increased BMI and low insulin sensitivity, had no distinguishing features of type 2 diabetes. Nevertheless, their poor prognosis suggests they may be particularly suited for additional therapies for reducing lipid concentrations or obesity and/or increasing insulin sensitivity. Feature of type 2 diabetes such as BMI, glycaemic index, and obesity have been previously associated with progression to type 1 diabetes in people with islet autoantibodies.55,56 Finally, patients in the N3 subgroup may also benefit from therapies for type 2 diabetes such as metformin, glucagon-like peptide-1 receptor analogues, and sodium-glucose cotransporter-2 inhibitors rather than insulin.
Our findings are also relevant to the selection of patients for clinical trials. Recent trials of patients with type 1 diabetes have often excluded islet autoantibody-negative patients and patients with very low C-peptide concentrations. Our CART analysis suggests that many of the islet autoantibody-negative patients in the N2 subgroup should be considered for such trials and could be selected by genetic typing for high-risk type 1 diabetes genotypes. Genetic risk scores have been shown to be valuable for classification of islet autoantibody-negative patients in the SEARCH study.57 The CART analysis also showed how patient selection will be biased if based on the C-peptide concentrations. The inclusion of patients with higher residual C-peptide concentrations will show bias towards overweight or obese patients, older patients, and patients with the lowest HbA1c. This bias may exclude potentially important effects in, for example, the large P3 group where almost one-third of patients have a TNF inflammatory profile and who may benefit most from combination therapies that target immune and β-cell recovery.
In addition to CART, we also classified patients by their genetic susceptibility for type 1 diabetes, and by the islet autoantibody type. However, none of these provided additional value. Type 1 diabetes genetic susceptibility was inversely correlated with the fasting C-peptide concentration in islet autoantibody-negative patients. The lack of association of autoantibody and genetic risk profiles with fasting C-peptide in the islet autoantibody-positive patients is consistent with previous work from SEARCH.4,50 It is likely, however, that type 1 diabetes or type 2 diabetes genetic risk profiles may provide discrimination well after diabetes onset when autoantibodies may no longer be detected or in single islet autoantibody cases with features of type 2 diabetes as previously reported.58
A strength of the study is the relatively large number of participants representing one geographical region in the main analysis and the large number of participants in the confirmation and prognosis cohort. A limitation of our study is that C-peptide concentration was only measured soon after onset. Residual β-cell function may be impaired in the presence of DKA and that onset C-peptide values or factors associated with onset values may not be predictive of C-peptide trajectories after onset.4 Another important limitation is that results are representative of a Western European population. Therefore, the findings may not be representative of childhood and adolescent diabetes in other populations, especially where the prevalence of other forms of diabetes or obesity differ. The US-based SEARCH study, for example, where 35% of cases are from other ethnic groups, described a larger proportion (26.1%) of patients without islet autoantibodies and 35%–50% (depending on ethnicity) who are overweight or obese.50,51 C-peptide trajectories after onset were unavailable in the patients and it is possible that the fasting values soon after diabetes onset may not accurately reflect true residual β-cell functional capacity. Moreover, the CART predictors age, HbA1c, and BMI change in the disease course and CART subgroup classification may, therefore, vary depending upon when it is applied. Further limitations include an insufficient follow-up duration of the second cohort to allow the assessment of diabetes complications within the different subgroups, and the lack of a comparative control population for inflammatory markers. The study has not addressed heterogeneity of type 1 diabetes in adults.
In conclusion, this supervised multivariable analysis of patients with youth-onset diabetes has demonstrated clear disease type-related heterogeneity within islet autoantibody-negative patients and less, but nevertheless potential, pathogenetic and therapeutic heterogeneity within islet autoantibody-positive patients.
Contributors
P.A. and M.H. contributed to data collection and were responsible for phenotyping and laboratory analyses.
A.G.Z. is the principal investigator of the DiMelli study and was responsible for study design and the acquisition of samples.
P.A. and E.B. oversaw the islet autoantibody measurements for the DiMelli and DPV studies.
A.P. was responsible for OLINK measurements.
M.H., J.Z.G. and E.B. performed the statistical analyses.
B.K. and R.W.H. are principal investigators of the DPV registry and provided access to longitudinal DPV data from patients providing serum samples at diagnosis.
M.H., P.A., E.B. and A.G.Z. contributed to data interpretation and drafted the manuscript.
A.G.Z. and E.B. are guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
All authors read and approved the final version of the manuscript.
Data sharing statement
The de-identified individual participant data that underlie the results (text, tables, figures, and appendix, excluding genetic data) reported in this article can be shared. Requests will be honoured from researchers who provide a methodologically sound proposal and who complete a Data Use Agreement with the Helmholtz Zentrum München. Requests should be directed by email to the corresponding author.
Declaration of interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank all of the patients who participated in this research study. We thank Andreas Beyerlein (Technische Universität München) and Fabian Theis (Helmholtz Zentrum München) for statistical consultancy.
Funding
Phenotyping and this analysis were supported by funds from the German Federal Ministry of Education and Research (01KX1818), and the Innovative Medicine Initiative 2 Joint Undertaking (IMI2-JU) INNODIA under grant agreement No. 115797. This Joint Undertaking receives support from the Union's Horizon 2020 research and innovation program and ‘EFPIA’, ‘JDRF’ and ‘The Leona M. and Harry B. Helmsley Charitable Trust’. The DiMelli cohort study and determination of islet autoantibodies in the DPV were supported by funds from the German Federal Ministry of Education and Research to the Competence Network Diabetes Mellitus (FKZ 01GI0805), and to the German Center for Diabetes Research (DZD e.V.). The DPV registry is funded by the DZD e.V., the German Robert Koch Institute as part of the national diabetes surveillance, and the German Diabetes Association (DDG).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104118.
Appendix. Supplementary materials
References
- 1.Patterson CC, Karuranga S, Salpea P, et al. Worldwide estimates of incidence, prevalence and mortality of type 1 diabetes in children and adolescents: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157 doi: 10.1016/j.diabres.2019.107842. [DOI] [PubMed] [Google Scholar]
- 2.DiMeglio LA, Evans-Molina C, Oram RA. Type 1 diabetes. Lancet North Am Ed. 2018;391(10138):2449–2462. doi: 10.1016/S0140-6736(18)31320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Diabetes Association. 2 Classification and diagnosis of diabetes: standards of medical care in diabetes—2020. Diabetes Care. 2020;43(suppl 1):S14–S31. doi: 10.2337/dc20-S002. [DOI] [PubMed] [Google Scholar]
- 4.Dabelea D, Mayer-Davis EJ, Andrews JS, et al. Clinical evolution of beta cell function in youth with diabetes: the SEARCH for Diabetes in Youth study. Diabetologia. 2012;55(12):3359–3368. doi: 10.1007/s00125-012-2719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel KA, Oram RA, Flanagan SE, et al. Type 1 diabetes genetic risk score: a novel tool to discriminate monogenic and type 1 diabetes. Diabetes. 2016;65(7):2094–2099. doi: 10.2337/db15-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields BM, McDonald TJ, Ellard S, Campbell MJ, Hyde C, Hattersley AT. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia. 2012;55(5):1265–1272. doi: 10.1007/s00125-011-2418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlqvist E, Storm P, Karajamaki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6(5):361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 8.Agache I, Akdis CA. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J Clin Invest. 2019;129(4):1493–1503. doi: 10.1172/JCI124611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battaglia M, Ahmed S, Anderson MS, et al. Introducing the endotype concept to address the challenge of disease heterogeneity in type 1 diabetes. Diabetes Care. 2020;43(1):5–12. doi: 10.2337/dc19-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arif S, Leete P, Nguyen V, et al. Blood and islet phenotypes indicate immunological heterogeneity in type 1 diabetes. Diabetes. 2014;63(11):3835–3845. doi: 10.2337/db14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leete P, Willcox A, Krogvold L, et al. Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes. 2016;65(5):1362–1369. doi: 10.2337/db15-1615. [DOI] [PubMed] [Google Scholar]
- 12.Leete P, Oram RA, McDonald TJ, et al. Studies of insulin and proinsulin in pancreas and serum support the existence of aetiopathological endotypes of type 1 diabetes associated with age at diagnosis. Diabetologia. 2020;63(6):1258–1267. doi: 10.1007/s00125-020-05115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao W, Woodwyk A, Beam C, Bahnson HT, Palmer JP, Greenbaum CJ. Assessment of beta cell mass and function by airmax and intravenous glucose in high-risk subjects for type 1 diabetes. J Clin Endocrinol Metab. 2017;102(12):4428–4434. doi: 10.1210/jc.2017-01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karges B, Prinz N, Placzek K, et al. A comparison of familial and sporadic type 1 diabetes among young patients. Diabetes Care. 2021;44(5):1116. doi: 10.2337/dc20-1829. [DOI] [PubMed] [Google Scholar]
- 15.Herold KC, Gitelman SE, Willi SM, et al. Teplizumab treatment may improve C-peptide responses in participants with type 1 diabetes after the new-onset period: a randomised controlled trial. Diabetologia. 2013;56(2):391–400. doi: 10.1007/s00125-012-2753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubitosi-Klug RA, Braffett BH, Hitt S, et al. Residual β cell function in long-term type 1 diabetes associates with reduced incidence of hypoglycemia. J Clin Invest. 2021;131(3) doi: 10.1172/JCI143011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachin JM, McGee P, Palmer JP. Impact of c-peptide preservation on metabolic and clinical outcomes in the diabetes control and complications trial. Diabetes. 2014;63(2):739–748. doi: 10.2337/db13-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeyam A, Colhoun H, McGurnaghan S, et al. Clinical impact of residual C-peptide secretion in type 1 diabetes on glycemia and microvascular complications. Diabetes Care. 2021;44(2):390–398. doi: 10.2337/dc20-0567. [DOI] [PubMed] [Google Scholar]
- 19.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, et al. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361(22):2143–2152. doi: 10.1056/NEJMoa0904452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haller MJ, Gitelman SE, Gottlieb PA, et al. Anti-thymocyte globulin/G-CSF treatment preserves β cell function in patients with established type 1 diabetes. J Clin Invest. 2015;125(1):448–455. doi: 10.1172/JCI78492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thumer L, Adler K, Bonifacio E, et al. German new onset diabetes in the young incident cohort study: DiMelli study design and first-year results. Rev Diabetic Stud. 2010;7(3):202–208. doi: 10.1900/RDS.2010.7.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kromeyer-Hauschild K, Moss A, Wabitsch M. Referenzwerte für den Body-Mass-Index für Kinder, Jugendliche und Erwachsene in Deutschland. Adipositas - Ursachen, Folgeerkrankungen, Therapie. 2015;09(03):123–127. [Google Scholar]
- 23.WHO . 2020. Body Mass Index - BMI.https://www.euro.who.int/en/health-topics/disease-prevention/nutrition/a-healthy-lifestyle/body-mass-index-bmi2020 [Google Scholar]
- 24.Bonifacio E, Mayr A, Knopff A, Ziegler AG. Endocrine autoimmunity in families with type 1 diabetes: frequent appearance of thyroid autoimmunity during late childhood and adolescence. Diabetologia. 2009;52(2):185–192. doi: 10.1007/s00125-008-1206-6. [DOI] [PubMed] [Google Scholar]
- 25.Hummel M, Bonifacio E, Stern M, Dittler J, Schimmel A, Ziegler AG. Development of celiac disease-associated antibodies in offspring of parents with type I diabetes. Diabetologia. 2000;43(8):1005–1011. doi: 10.1007/s001250051483. [DOI] [PubMed] [Google Scholar]
- 26.Naserke HE, Bonifacio E, Ziegler AG. Immunoglobulin G insulin autoantibodies in BABYDIAB offspring appear postnatally: sensitive early detection using a protein A/G-based radiobinding assay. J Clin Endocrinol Metab. 1999;84(4):1239–1243. doi: 10.1210/jcem.84.4.5597. [DOI] [PubMed] [Google Scholar]
- 27.Achenbach P, Koczwara K, Knopff A, Naserke H, Ziegler AG, Bonifacio E. Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J Clin Invest. 2004;114(4):589–597. doi: 10.1172/JCI21307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Achenbach P, Lampasona V, Landherr U, et al. Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia. 2009;52(9):1881–1888. doi: 10.1007/s00125-009-1438-0. [DOI] [PubMed] [Google Scholar]
- 29.Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J Clin Endocrinol Metab. 2010;95(7):3360–3367. doi: 10.1210/jc.2010-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegler AG, Hummel M, Schenker M, Bonifacio E. Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: the 2-year analysis of the German BABYDIAB study. Diabetes. 1999;48(3):460–468. doi: 10.2337/diabetes.48.3.460. [DOI] [PubMed] [Google Scholar]
- 31.Torn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ. Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia. 2008;51(5):846–852. doi: 10.1007/s00125-008-0967-2. [DOI] [PubMed] [Google Scholar]
- 32.Schlosser M, Mueller PW, Torn C, Bonifacio E, Bingley PJ. Diabetes Antibody Standardization Program: evaluation of assays for insulin autoantibodies. Diabetologia. 2010;53(12):2611–2620. doi: 10.1007/s00125-010-1915-5. [DOI] [PubMed] [Google Scholar]
- 33.Lampasona V, Schlosser M, Mueller PW, et al. Diabetes antibody standardization program: first proficiency evaluation of assays for autoantibodies to zinc transporter 8. Clin Chem. 2011;57(12):1693–1702. doi: 10.1373/clinchem.2011.170662. [DOI] [PubMed] [Google Scholar]
- 34.Walther D, Eugster A, Jergens S, et al. Tetraspanin 7 autoantibodies in type 1 diabetes. Diabetologia. 2016;59(9):1973–1976. doi: 10.1007/s00125-016-3997-1. [DOI] [PubMed] [Google Scholar]
- 35.Dabelea D, D'Agostino RB, Mason CC, et al. Development, validation and use of an insulin sensitivity score in youths with diabetes: the SEARCH for Diabetes in Youth study. Diabetologia. 2011;54(1):78–86. doi: 10.1007/s00125-010-1911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter M, Albert E, Conrad M, et al. IDDM2/insulin VNTR modifies risk conferred by IDDM1/HLA for development of Type 1 diabetes and associated autoimmunity. Diabetologia. 2003;46(5):712–720. doi: 10.1007/s00125-003-1082-z. [DOI] [PubMed] [Google Scholar]
- 37.Bonifacio E, Beyerlein A, Hippich M, et al. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: a prospective study in children. PLoS Med. 2018;15(4) doi: 10.1371/journal.pmed.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57(12):2521–2529. doi: 10.1007/s00125-014-3362-1. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017:201178.
- 41.Klapproth E, Dickreuter E, Zakrzewski F, et al. Whole exome sequencing identifies mTOR and KEAP1 as potential targets for radiosensitization of HNSCC cells refractory to EGFR and β1 integrin inhibition. Oncotarget. 2018;9(26):18099–18114. doi: 10.18632/oncotarget.24266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamrath C, Mönkemöller K, Biester T, et al. Ketoacidosis in children and adolescents with newly diagnosed type 1 diabetes during the COVID-19 pandemic in Germany. JAMA. 2020;324(8):801–804. doi: 10.1001/jama.2020.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karges B, Schwandt A, Heidtmann B, et al. Association of insulin pump therapy vs insulin injection therapy with severe hypoglycemia, ketoacidosis, and glycemic control among children, adolescents, and young adults with type 1 diabetes. JAMA. 2017;318(14):1358–1366. doi: 10.1001/jama.2017.13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Statist. 2006;15(3):651–674. [Google Scholar]
- 46.Krischer JP, Liu X, Vehik K, et al. Predicting islet cell autoimmunity and type 1 diabetes: an 8-year TEDDY study progress report. Diabetes Care. 2019;42(6):1051–1060. doi: 10.2337/dc18-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziegler AG, Kick K, Bonifacio E, et al. Yield of a public health screening of children for islet autoantibodies in Bavaria, Germany. JAMA. 2020;323(4):339–351. doi: 10.1001/jama.2019.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang W, Dulaimi E, Devarajan K, et al. Intratumoral heterogeneity analysis reveals hidden associations between protein expression losses and patient survival in clear cell renal cell carcinoma. Oncotarget. 2017;8(23):37423–37434. doi: 10.18632/oncotarget.16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banchereau R, Hong S, Cantarel B, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(3):551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dabelea D, Pihoker C, Talton JW, et al. Etiological approach to characterization of diabetes type: the SEARCH for Diabetes in Youth Study. Diabetes Care. 2011;34(7):1628–1633. doi: 10.2337/dc10-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamman RF, Bell RA, Dabelea D, et al. The SEARCH for Diabetes in Youth study: rationale, findings, and future directions. Diabetes Care. 2014;37(12):3336–3344. doi: 10.2337/dc14-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redondo MJ, Rodriguez LM, Escalante M, Smith EO, Balasubramanyam A, Haymond MW. Types of pediatric diabetes mellitus defined by anti-islet autoimmunity and random C-peptide at diagnosis. Pediatr Diabetes. 2013;14(5):333–340. doi: 10.1111/pedi.12022. [DOI] [PubMed] [Google Scholar]
- 53.Shields BM, Shepherd M, Hudson M, et al. Population-based assessment of a biomarker-based screening pathway to aid diagnosis of monogenic diabetes in young-onset patients. Diabetes Care. 2017;40(8):1017–1025. doi: 10.2337/dc17-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quattrin T, Haller MJ, Steck AK, et al. Golimumab and beta-cell function in youth with new-onset type 1 diabetes. N Engl J Med. 2020;383(21):2007–2017. doi: 10.1056/NEJMoa2006136. [DOI] [PubMed] [Google Scholar]
- 55.So M, O'Rourke C, Ylescupidez A, et al. Characterising the age-dependent effects of risk factors on type 1 diabetes progression. Diabetologia. 2022;65(4):684–694. doi: 10.1007/s00125-021-05647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamb MM, Yin X, Barriga K, et al. Dietary glycemic index, development of islet autoimmunity, and subsequent progression to type 1 diabetes in young children. J Clin Endocrinol Metab. 2008;93(10):3936–3942. doi: 10.1210/jc.2008-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oram RA, Sharp SA, Pihoker C, et al. Utility of diabetes type-specific genetic risk scores for the classification of diabetes type among multiethnic youth. Diabetes Care. 2022;45(5):1124–1131. doi: 10.2337/dc20-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redondo MJ, Geyer S, Steck AK, et al. TCF7L2 genetic variants contribute to phenotypic heterogeneity of type 1 diabetes. Diabetes Care. 2018;41(2):311–317. doi: 10.2337/dc17-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.