Abstract
Complement component C1q is a protein complex of the innate immune system with well-characterized binding partners that constitutes part of the classical complement pathway. In addition, C1q was recently described in the central nervous system as having a role in synapse elimination both in the healthy brain and in neurodegenerative diseases. However, the molecular mechanism of C1q-associated synapse phagocytosis is still unclear. Here, we designed monomer and multimer protein constructs, which comprised the globular interaction recognition parts of mouse C1q (globular part of C1q [gC1q]) as single-chain molecules (sc-gC1q proteins) lacking the collagen-like effector region. These molecules, which can competitively inhibit the function of C1q, were expressed in an Escherichia coli expression system, and their structure and capabilities to bind known complement pathway activators were validated by mass spectrometry, analytical size-exclusion chromatography, analytical ultracentrifugation, CD spectroscopy, and ELISA. We further characterized the interactions between these molecules and immunoglobulins and neuronal pentraxins using surface plasmon resonance spectroscopy. We demonstrated that sc-gC1qs potently inhibited the function of C1q. Furthermore, these sc-gC1qs competed with C1q in binding to the embryonal neuronal cell membrane. We conclude that the application of sc-gC1qs can reveal neuronal localization and functions of C1q in assays in vivo and might serve as a basis for engineering inhibitors for therapeutic purposes.
Keywords: complement inhibition, complement activation, hemolysis, CD spectroscopy, surface plasmon resonance, molecular cloning, multimers, neuronal pentraxins
Abbreviations: BeStSel, beta structure selection; BSA, bovine serum albumin; CLR, collagen-like tail region; CNS, central nervous system; CP, complement pathway; gC1q, globular part of C1q; gC1qR, globular C1q receptor; hIgG, human IgG; HRP, horseradish peroxidase; Ig, immunoglobulin; IgG, immunoglobulin G; IgM, immunoglobulin M; IMAC, immobilized metal ion affinity chromatography; mAb, monoclonal antibody; MD, molecular dynamics; MS, mass spectrometry; NPTX, neuronal pentraxin; PBS-T, Tween-20 containing PBS; PDB, Protein Data Bank; PTX3, pentraxin-3; sc-gC1q, single-chain mouse globular part of C1q; sc-gC1q2, dimer single-chain mouse globular part of C1q; sc-gC1q3, trimer single-chain mouse globular part of C1q; sc-gC1q2l, dimer single-chain mouse globular part of C1q with longer linker; sc-gC1q3l, trimer single-chain mouse globular part of C1q with longer linker; SpD, surfactant protein D; SPR, surface plasmon resonance; SRBC, sheep red blood cell; SRCD, synchrotron radiation CD; VBS, veronal-buffered saline
Complement component C1q, one of the three subunits of the C1 complex, is known as the recognition molecule of the classical complement pathway (CP). C1q (approximately 460 kDa) consists of 18 polypeptide chains, each containing a C-terminal globular head domain and an N-terminal collagen-like tail region (CLR). C1q assembles as a hexamer bouquet of heterotrimers. Trimeric subcomponents are composed of chains A, B, and C, forming the globular heads (gC1q), and collagen-like triple helical tails. The formation of six and three interchain disulfide bonds between the A–B and C–C chains, respectively, is responsible for the hexamerization of ABC trimers. CLRs of the A–B dimer form a triple helical structure with the similar region of one of the chains in a C–C dimer (1). C1r2–C1s2 tetrameric proenzyme binds to the CLR of C1q. Upon C1q interactions formed via gC1q, the conformation of the CLRs changes. These events activate the C1r, followed by the C1s serine proteases. This C1 activation is the first step of classical complement cascade amplification. C1q interacts with a broad range of ligands, and some of the most prominent partners are immune complexes formed by immunoglobulin G (IgG) and immunoglobulin M (IgM). Short pentraxins (PTXs; serum amyloid P-component and C-reactive protein) (2, 3) and pentraxin 3 (PTX3) (4) are also well-known binding partners of gC1q.
Versatile recognition properties of C1q are due to its globular head domains. In contrast to structurally similar homotrimers, each gC1q domain differs in surface patterns in terms of hydrophobic and charged patches. C1q has numerous interactions in which more gC1q subunits participate. According to gC1q crystal structure (Protein Data Bank [PDB] ID: 1PK6), each head domain consists of two 5-stranded antiparallel β-sheets making up a jelly-roll topology, which is reminiscent of the structure of tumor necrosis factor superfamily members (5).
Whereas gC1q serves as a recognition part of C1q, CLR is responsible for effector functions. Besides having a role in C1r2–C1s2 activation, CLR also binds to C1q receptors. A few cell surface receptors were identified as potential C1q receptors (6). Presumably, C1q exerts its diverse functions via more than one putative receptor. Calreticulin was identified on the cell surface of phagocytes, and it may contribute to C1q-mediated elimination of apoptotic cells and immune complexes (7). Another identified C1q receptor, gC1qR, binds the globular head region of C1q, and upon activation, it regulates immune processes and inflammation (8, 9). C1q and gC1qR also play a vital role in cancer cell migration and proliferation (10, 11, 12). C1q has been shown to exhibit a noncanonical function in the central nervous system (CNS) having a role in synaptic pruning both in the developing and adult brain (13, 14).
Levels of C1q correlate with various diseases. C1q deficiency is a rare immunodeficiency disorder that causes severe glomerulonephritis, systemic lupus erythematosus or systemic lupus erythematosus–like diseases (15, 16, 17). C1q deficiency–related problems are well-characterized conditions with a clear genetic background or are caused by anti-C1q autoantibodies. Occasionally, excessive activation of a complement causes problems (e.g., xenograft rejection) (18). In addition, C1q is associated with disorders in which the molecular mechanisms are not fully understood. Recently, C1q and CP members are related to a number of neurodegenerative and mental diseases by their deposition onto synapses that should be eliminated (19, 20, 21, 22).
C1q serves as an “eat-me” signal for microglia cells, although the binding partners of C1q on these synapses are still unknown. Scott-Hewitt et al. (23) have shown that C1q binds to externalized phosphatidylserine during developmental synaptic pruning. According to our recent results, synaptic neuronal pentraxins (NPTXs) are potential C1q-binding partners (24). NPTXs, which show extensive structural homology with PTX3, are expressed in the CNS and play a pivotal role in synaptic plasticity (25). Alzheimer’s disease–associated β-amyloid protein is also recognized by C1q (26); however, it is not well established whether CLR or gC1q interacts with it (27, 28). Upon this interaction, C1q exerts neuroprotective effects via activating survival signal expression (29, 30).
As described, dysregulated complement activation generates undesired damage. Specific inhibition of complement elements either in the CNS or in the periphery is an urgent need. Furthermore, complement inhibitors may help us understand the molecular mechanisms of unexplored disorders. Despite a growing number of candidates, there are only two complement inhibitor drugs approved: C1 inhibitor used in hereditary angioedema and anti-C5 monoclonal antibody (mAb; eculizumab, sold as Soliris). Eculizumab is used in diseases where impaired complement regulation is pathological, such as atypical hemolytic uremic syndrome and in autoimmune disorders where the overactivated complement drives the disease, such as neuromyelitis optica spectrum disorder and generalized myasthenia gravis (31). However, C5-targeting molecules cannot prevent either C1q deposition or upstream activation.
Here, we created a single-chain protein construct of the head domains of mouse C1qA, C1qC, and C1qB (sc-gC1q) to reproduce the globular head of the C1q molecule similarly to the work of Moreau et al. (32) on human C1q. In order to mimic the avidity effect of the globular heads of the original C1q, we designed multimeric sc-gC1q proteins (dimers and trimers), as well. Our aim for the recombinant expression of these constructs was to use them as competitive inhibitors of C1q function. We performed structural and functional analysis of sc-gC1q constructs in vitro and in cell culture and showed that they can compete with the full-length C1q in binding to partners and inhibit its function. We believe that mouse sc-gC1q proteins can help us understand the diverse function of C1q, especially those related to the CNS that cannot be carried out in humans and might be a basis for therapeutic purposes.
Results
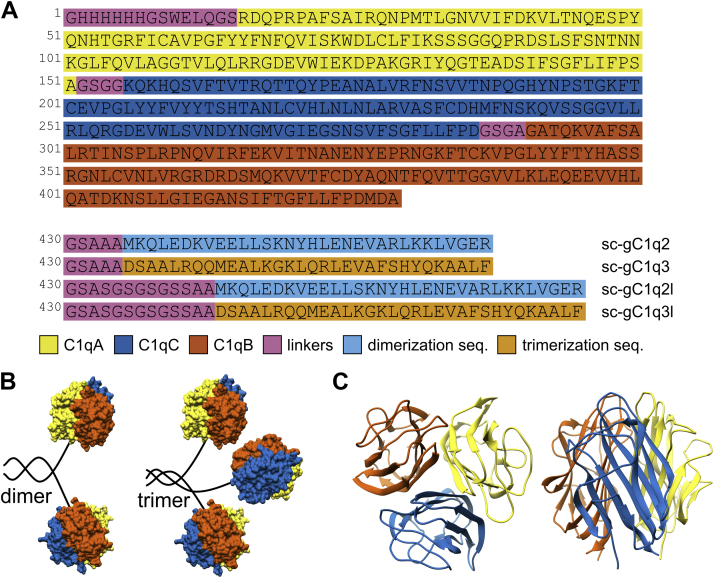
Single-chain globular head, the recognition part of C1q, was created by linking sequentially the head domain regions of C1qA, C1qC, and C1qB chains using four residue linkers (see Experimental procedures section). We created multimers using either a 5-residue-long (sc-gC1q2 and sc-gC1q3) or a more flexible 13-residue-long linker (sc-gC1q2l and sc-gC1q3l) between the dimerizing GCN4 leucine zipper (sc-gC1q2 and sc-gC1q2l) or trimerizing α-helical neck region of surfactant protein D (SpD) (sc-gC1q3 and sc-gC1q3l), and sc-gC1q. Detailed construction of the sc-gC1q molecule and its multimeric forms is presented in Figure 1A.
Figure 1.
Single-chain gC1q (sc-gC1q) and its multimeric constructs.A, sequence of monomer and multimer sc-gC1q proteins. Color coding is marked below the sequences. Dimer proteins consist of a sc-gC1q and GCN4 leucine zipper as dimerizing module, whereas trimeric constructs contain the trimerizing neck region of surfactant protein D (SpD). Globular head domains of C1q are linked by 4 to 4 amino acid long linkers (GTGG between C1qA and C1qC chains; GSGA: between C1qC and C1qB chains). Both dimer and trimer constructs were designed with shorter (five residues: GSAAA) and longer (13 residues: GSASGSGSGSSAA) linkers fused to sc-gC1q sequence. B, schematic structure of the multimeric constructs. Two or three globular gC1q heads are connected by dimerizing or trimerizing coiled-coil sequences, respectively. C, representative structure from two viewpoints of sc-gC1q: β-strands of the three domains of jelly-roll fold are shown in blue, red, and yellow. gC1q, globular part of C1q.
In silico modeling of sc-gC1q structure
First, we carried out an in silico modeling study on the structure of the sc-gC1q molecule to verify that the designed single polypeptide chain will be compatible with the expected structure of the C1q head and will form a stable fold. Moreover, because of the lack of X-ray structures of this construct with a 72% sequence identity to human C1q head, a reliable structural model is needed to understand its function and molecular interactions, and for it to be used as a basis for designing specific mutations in the molecule. An initial homology model of sc-gC1q was generated by the Phyre2 server (33) using the highest scoring template of the human sc-gC1q head structure (32). The model was successfully created on 94% of the sequence with 100% confidence. The N-terminal sequence was built into the model manually using the SwissPDBViewer program (https://spdbv.unil.ch/) (34). This model revealed that the polypeptide chain of sc-gC1q probably folds without steric clashes into a structure similar to the available C1q globular head structures deposited in the PDB. This model was subjected to molecular dynamics (MD) simulations of 1 μs using the GROMACS software (https://www.gromacs.org) (35). The MD trajectory showed that beside a fast conformational adjustment of the starting model (Fig. S1), the structure was well defined and stable (Fig. S2A). The final structural model after the MD simulation for sc-gC1q shows the typical fold of the three jelly-roll folds consisting of sandwiches of antiparallel β-sheets (Fig. 1C). During the MD simulations, the β-sheets were rather rigid, whereas loop regions proved to be more flexible (Fig. S2B).
Protein expression
The sc-gC1q constructs were expressed in Escherichia coli BL21 (DE3) cells and purified by Ni2+-affinity chromatography followed by cation exchange yielding 1 to 2 mg purified protein per liter of LB medium. The purity and the presence of the disulfide bridges were checked by SDS-PAGE (Fig. S3) and mass spectrometry (MS; Fig. S4) on the intact protein variants. To investigate if the two disulfides (C1qB: 179C–198C; C1qC: 180C–194C; and C1qA: no SS bond) were formed properly, we found MS more suitable than SDS-PAGE. Using LC–MS, we found a difference of ∼4 Da in molecular mass between the reduced and nonreduced samples indicating that the two intrachain disulfide bridges were formed (Fig. S4). The overall molecular masses proved to be the expected masses calculated from the sequence of the constructs, revealing that our purified proteins are indeed the C1q constructs. A low amount of oxidized form was also observable in each sample. Using MS-peptide mapping, we gained further proof for the identity of the purified proteins. In the peptide mapping experiments, sc-gC1q, scgC1q2, and sc-gC1q2l protein variants were digested with bovine trypsin, the proteolytic fragments were then separated on reversed-phase column, and subjected to MSE experiments using fragmentation in the trap cell with collision voltage ramping. Data analysis showed 100% sequence coverage for sc-gC1q as well as nearly 100% for sc-gC1q2 and sc-C1q2l indicating that the expressed proteins are identical with the designed protein constructs (Fig. S5).
Analysis of protein multimerization by gel filtration
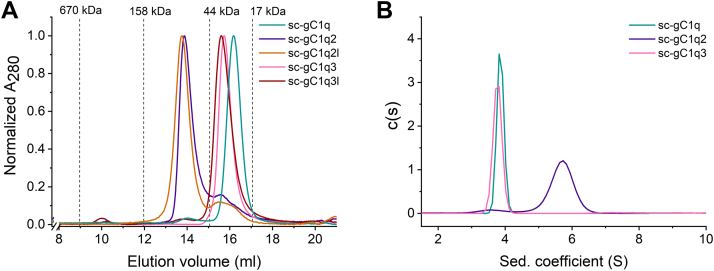
Analytical gel filtration was performed in order to investigate the multimerization of the expressed, purified, and soluble sc-gC1q proteins. We also applied gel filtration protein standard series in order to have reference points. The presented chromatograms clearly indicate the dimerization of the sc-gC1q2 and sc-gC1q2l constructs by the introduction of the GCN4 leucine zipper (Fig. 2A). For the dimers, a smaller peak could be observed with an elution volume similar to sc-gC1q, which might be a small fraction of the monomeric form in the sample. The trimeric constructs had only slightly lower elution volume than the monomer suggesting that the trimer assembly has failed.
Figure 2.
Investigation of molecular size of single-chain gC1q (sc-gC1q) variants.A, analytical size-exclusion chromatography of sc-gC1q, sc-gC1q2, sc-gC1q2l, sc-gC1q3, and sc-gC1q3l marked with teal, purple, orange, pink, and dark red colors, respectively. Dashed lines indicate the elution volumes of the gel filtration protein weight standard components. All absorbances (measured at 280 nm) were normalized to a maximal value of 1.0. B, distribution of sedimentation coefficients in analytical ultracentrifuge experiments of sc-gC1q (teal), sc-gC1q2 (purple), and sc-gC1q3 (pink). gC1q, globular part of C1q.
Analytical ultracentrifugation
To avoid interfering effects, such as interactions with the matrix of the gel filtration column, we verified the correct fold of the globular head and investigated the apparent molecular masses of sc-gC1q, sc-gC1q2, and sc-gC1q3 by analytical ultracentrifugation sedimentation velocity experiments. In the case of sc-gC1q, distribution of the sedimentation coefficient showed a sharp peak indicating a homogeneous protein solution with a calculated molecular mass of 49.9 ± 0.4 kDa, which is in a good agreement with the theoretical 47,811 Da molecular weight calculated from the amino acid sequence (Figs. 2B and S6). This result also indicates that sc-gC1q is well folded into its expected globular structure (Figs. 2B and S6). In the case of the dimer sc-gC1q2, a main peak in the sedimentation coefficient distribution is observed with a related molecular mass of 94.9 ± 0.6 kDa, which clearly indicates the presence of dimers in the solution. A small component with a corresponding 47.7 ± 1.0 kDa molecular mass is also observed, which might be a small portion of monomeric form in the solution (Figs. 2B and S6). In the case of sc-gC1q3, a sedimentation profile similar to the monomer sc-gC1q was observed with an apparent molecular mass of 51.1 ± 0.5 kDa, suggesting that sc-gC1q3 is monomeric in the solution and the trimerizing SpD neck region was not able to trimerize the C1q heads (Figs. 2B and S6).
Structure validation by CD spectroscopy
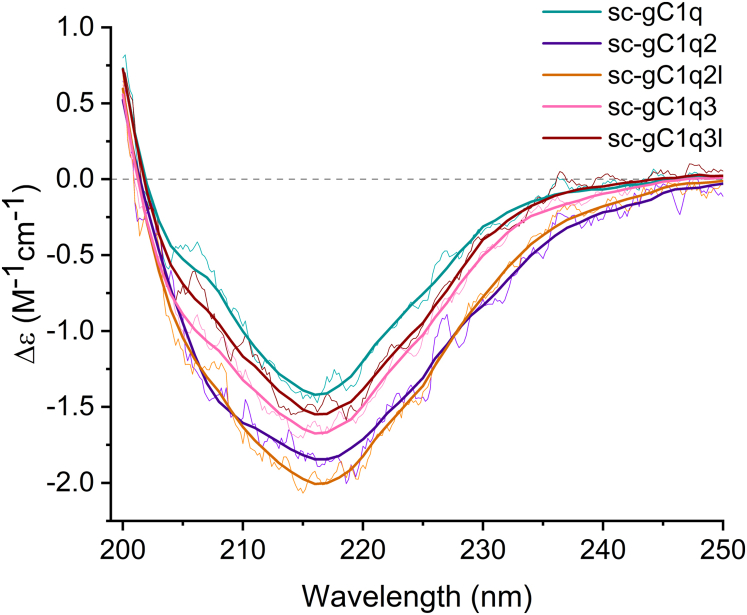
CD spectroscopy is a valuable method for investigating secondary structural content and proper protein folding. The beta structure selection (BeStSel) method provides a reliable analysis of CD spectra for secondary structure estimation and fold prediction (36). We collected the spectra of all the sc-gC1q variants using a conventional CD instrument in the 200 to 250 nm wavelength range (Fig. 3). All these spectra showed a similar shape with a positive band at 200 nm and a negative peak at 216 nm, which are characteristic features of the antiparallel β-structure (Fig. 3). However, some differences were observed, which, according to the BeStSel analysis, correspond to higher α-helical content in the case of the multimeric proteins, especially for the dimer (Table 1). This confirms the proper folding of the dimerization sequence into a helical coiled-coil structure. In the case of the trimer construct, α-helix was observed to a lower extent, suggesting the failure of trimer assembly.
Figure 3.
Secondary structure analysis by CD spectroscopy. Far-UV CD spectra of recombinant single-chain gC1q (sc-gC1q) proteins were recorded in triplicates and averaged (thin lines). Experimental data were fitted by BeStSel software (36) (thick lines). Sc-gC1q, sc-gC1q2, sc-gC1q2l, sc-gC1q3, and sc-gC1q3l were marked with teal, purple, orange, pink, and dark red colors, respectively. Spectra showed a positive band at 200 nm and a negative band at 216 nm, which indicates the abundance of antiparallel β-structure. Secondary structure contents are shown in Table 1. gC1q, globular part of C1q.
Table 1.
Secondary structure composition of sc-gC1q variants
| Secondary structural elements | Human C1q head X-raya | Mouse sc-gC1q MD | sc-gC1q SRCD |
sc-gC1q |
sc-gC1q2 |
sc-gC1q2l |
sc-gC1q3 |
sc-gC1q3l |
|---|---|---|---|---|---|---|---|---|
| 190–250 nmb | 200–250 nmc | |||||||
| α-Helix | 0 | 0 | 1.9 | 3.1 | 10.9 | 8.3 | 6.5 | 2.8 |
| Antiparallel-β | 49.3 | 49.2 | 45.6 | 38.5 | 36.7 | 33.7 | 34.5 | 36.9 |
| Parallel-β | 0 | 0 | 0 | 0 | 0 | 0.7 | 0 | 0 |
| Turn | 10.1 | 9.8 | 12.3 | 10.3 | 10.6 | 11.1 | 11.3 | 11.7 |
| Others | 38.3 | 38.6 | 40.2 | 48.1 | 41.7 | 46.3 | 47.7 | 48.6 |
Calculated from the X-ray structure of human C1q head (PDB ID: 5HKJ).
SRCD measured at SOLEIL synchrotron.
Conventional CD measurements in the 200 to 250 nm wavelength range. We have to note that SRCD provided a secondary structure composition similar to that of the X-ray structure and structural model.
Sc-gC1q monomer was also measured by synchrotron radiation CD (SRCD) spectroscopy. The advantage of SRCD is that we could record the CD spectrum over a wider wavelength range compared with the conventional instrument, which has a more limited spectral range because of the high background absorption of the 250 mM NaCl buffer. Comparison of the human X-ray structure (PDB ID: 5HZF) with our mouse MD model and the BeStSel analysis of the SRCD spectrum of mouse sc-gC1q reveals that the solution structure of the expressed sc-gC1q protein is practically identical (Table 1). According to these measurements, all sc-gC1qs are properly folded proteins. These data from the CD spectroscopy strengthen the results of the analytical ultracentrifugation analysis and gel filtration experiments; therefore, the further measurements will be performed using the correctly folded monomer and dimers.
Binding of sc-gC1qs to Igs and PTXs
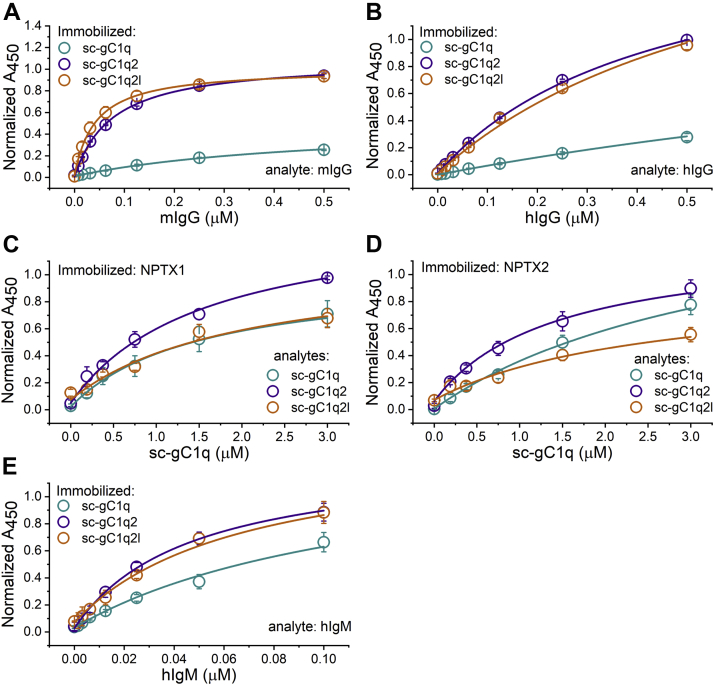
The functionality of recombinant sc-gC1qs was tested by ELISA binding assays. In addition, our aim was to prove that our mouse constructs equally bind to mouse and human IgG (hIgG). In the case of interaction with Igs, sc-gC1q variants with short and long linkers were coated, followed by the detection of the human IgM, IgG, and mouse IgG using either antihuman or antimouse secondary antibodies. Dimers showed similar half-saturation concentrations, whereas the monomer bound an order of magnitude weaker to Igs (Fig. 4). NPTXs are homologs of PTX3, the well-known interaction partner of C1q. According to our previous results, C1q binds to NPTXs (NPTX1 and NPTX2), but it was unexplored whether the globular head domain or the collagen-like tail part of C1q is the interacting partner (24). Using our new recombinant sc-gC1qs, we revealed that the head domain contains the binding sites able to interact with NPTXs similar to that reported for PTX3 (37). Both dimers and monomer showed comparable half-saturation concentrations when binding to NPTXs (Fig. 4). All sc-gC1q proteins showed binding to human proteins, and for this reason, further experiments were performed using hIgG, IgM, and NPTXs.
Figure 4.
Investigation of the binding properties of single-cell gC1q (sc-gC1q) variants by ELISA. Human and mouse IgG proteins (hIgG and mIgG, respectively) and human IgM were immobilized and titrated with each form of sc-gC1q variants (A, B, and E). In order to use the same primary antibody in assays examining neuronal pentraxins, we coated the wells with NPTX1 or NPTX2 and titrated with the different forms of sc-gC1q (C and D; marked with different colors). Hill equations (lines) were fitted to the data (circles) using Origin8 software. Values represent mean (from three replicates) ± SEM. gC1q, globular part of C1q; IgG, immunoglobulin G; IgM, immunoglobulin M; NPTX, neuronal pentraxin.
Investigation of binding affinities by surface plasmon resonance
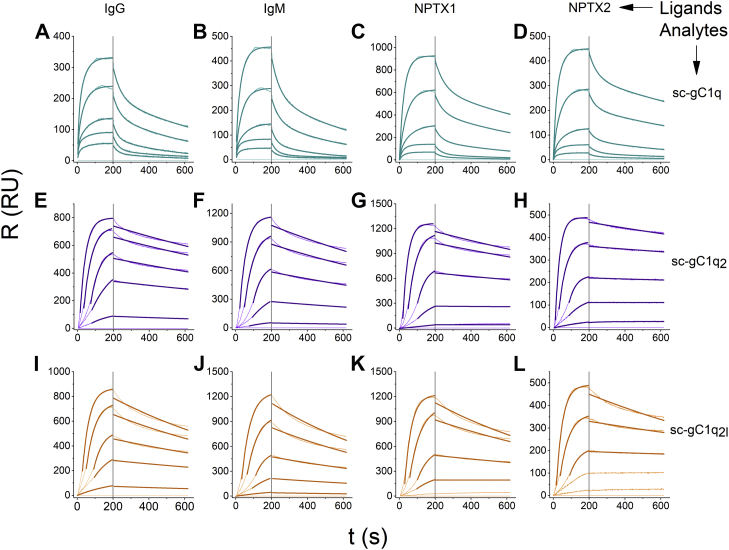
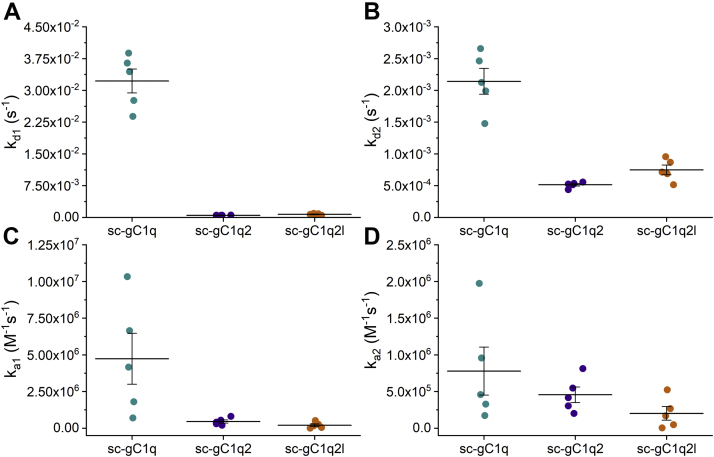
To quantitatively assess the binding capability of sc-gC1q proteins to the various C1q partners, we carried out surface plasmon resonance (SPR) experiments, where IgG, IgM, NPTX1, and NPTX2 were covalently immobilized in high density to the sensor chip surface. The sensorgrams of monomer injections differ markedly from that of the dimers. When scC1q was applied as analyte, a pronounced biphasic binding was found, so these data were fitted with the assumption that two binding events took place. Fitting the sc-gC1g2 and sc-gC1q2l sensorgrams with the aforementioned model resulted in an unrealistically small dissociation constant for the slow binding event, which hindered the fitting of the association phase. Consequently, for the dimers, a 1:1 Langmuir model with single exponential fitting was used for data analysis. Because of potential ligand heterogeneity, which may cause biphasic binding curves, and differences in data processing, we refer to the calculated constants as “apparent” values. The apparent dissociation rates of dimer sc-gC1qs were shown to be two orders of magnitude lower than that of the sc-gC1q, whereas the apparent association rate constant of the monomeric construct is one order of magnitude higher than that of the dimers (Figs. 5, 6 and Table 2). These tendencies indicate that avidity took place in the case of the dimer constructs, which increases their apparent binding affinity compared with sc-gC1q.
Figure 5.
SPR-binding studies. After covalent immobilization of IgG, IgM, NPTX1, and NPTX2) via amine coupling, the ligand channels were covalently blocked with bovine serum albumin (BSA) along with an empty channel that served as negative control. The sensorgrams (pale lines) are generated with the subtraction of the responses detected on the BSA channel from those detected on specific ligand channels. Dark lines represent the data fitting with a single exponential (sc-gC1q2 and sc-gC1q2l) or a double exponential (sc-gC1q). A–D, sc-gC1q analyte interactions with IgG, IgM, NPTX1, and NPTX2 ligands, respectively. E–H, sc-gC1q2 analyte interactions with IgG, IgM, NPTX1, and NPTX2 ligands, respectively. I–L, sc-gC1q2l analyte interactions with IgG, IgM, NPTX1, and NPTX2 ligands, respectively. gC1q, globular part of C1q; IgG, immunoglobulin G; IgM, immunoglobulin M; NPTX, neuronal pentraxin; sc-gC1q, single-chain gC1q; SPR, surface plasmon resonance.
Figure 6.
Kinetic constants of IgG–sc-gC1q interaction. Dissociation rate (A and B) and association rate (C and D) constants of SPR measurements are presented (mean ± SEM, five dilutions of IgG-sc-gC1q SPR measurement). Note that the experimental data of dimers were fitted with the one-phase model resulting in a single ka and kd value, which are shown in both A–B and C–D for comparison. gC1q, globular part of C1q; IgG, immunoglobulin G; sc-gC1q, single-chain gC1q; SPR, surface plasmon resonance.
Table 2.
Kinetic binding parameters of sc-gC1q variants measured by SPRa
| Protein variants | Binding parameters | IgG | IgM | NPTX1 | NPTX2 |
|---|---|---|---|---|---|
| sc-gC1q | kd1 (s−1) | 3.2 × 10−2 | 2.9 × 10−2 | 2.6 × 10−2 | 2.6 × 10−2 |
| ka1 (M−1 s−1) | 4.7 × 106 | 3.4 × 106 | 2.7 × 106 | 4.5 × 106 | |
| Kd1 (M) | 6.8 × 10−9 | 8.5 × 10−9 | 9.8 × 10−9 | 5.7 × 10−9 | |
| kd2 (s−1) | 2.1 × 10−3 | 2.3 × 10−3 | 1.8 × 10−3 | 1.4 × 10−3 | |
| ka2 (M−1 s−1) | 7.8 × 105 | 9.4 × 105 | 1.2 × 106 | 4.5 × 106 | |
| Kd2 (M) | 2.8 × 10−9 | 2.4 × 10−9 | 1.5 × 10−9 | 3.1 × 10−9 | |
| sc-gC1q2 | kd (s−1) | 5.2 × 10−4 | 6.6 × 10−4 | 4.1 × 10−4 | 1.9 × 10−4 |
| ka (M−1 s−1) | 4.6 × 105 | 4.4 × 105 | 4.0 × 105 | 4.5 × 105 | |
| Kd (M) | 1.1 × 10−9 | 1.5 × 10−9 | 1.0 × 10−9 | 4.1 × 10−10 | |
| sc-gC1q2l | kd (s−1) | 7.5 × 10−4 | 9.5 × 10−4 | 7.6 × 10−4 | 4.2 × 10−4 |
| ka (M−1 s−1) | 3.2 × 105 | 3.0 × 105 | 2.9 × 105 | 2.9 × 105 | |
| Kd (M) | 2.3 × 10−9 | 3.1 × 10−9 | 2.7 × 10−9 | 1.5 × 10−9 |
Apparent dissociation rate (kd), apparent association rate (ka), and apparent dissociation constants (Kd) are shown for interactions with IgG, IgM, NPTX1, and NPTX2.
Inhibition of complement-induced hemolysis
Sheep red blood cells (SRBCs) coated with rabbit anti-SRBC antibody (hemolysin) are traditionally applied to examine CP activation. When hemolysin-coated SRBCs are incubated with serum containing all CP elements, C1q facilitates the lysis of erythrocytes. This system is sensitive to the amounts of all CP factors, inhibition of any component resulting in decreased hemolysis (38).
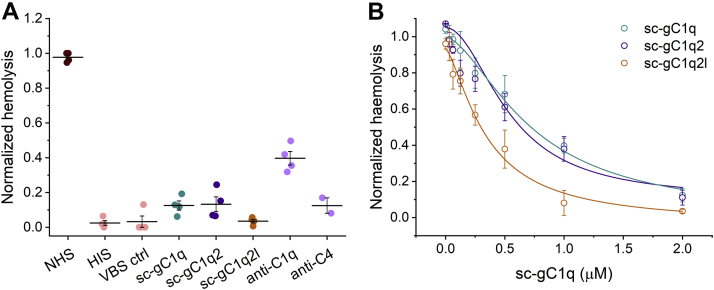
The effects of sc-gC1qs were compared to two commercially available antibodies specific to complement factors C1q and C4. All recombinant sc-gC1qs exerted similar or better inhibition of hemolysis compared with the antibodies in comparable molar concentrations. All types of sc-gC1qs showed >90% inhibition at micromolar concentration (Fig. 7).
Figure 7.
Inhibition of SRBC hemolysis by sc-gC1qs.A, antibody-coated SRBCs were treated with human serum with or without inhibitors (including control antibodies against CP components), and hemolysis was detected via the hemoglobin content of SRBC supernatant. NHS, HIS, and VBS are abbreviations for normal human serum, heat inactivated serum, and veronal-buffered saline, respectively. B, effect of sc-gC1q was investigated in a concentration-dependent manner. Curves of sc-gC1q, sc-gC1q2, and sc-gC1q2l are marked with teal, purple, and orange, respectively. Data were fitted with Hill equations. Values represent mean ± SEM (from four independent experiments and two independent experiments for the anti-C4 control). CP, complement pathway; gC1q, globular part of C1q; sc-gC1q, single-cell gC1q; SRBC, sheep red blood cell.
In order to calculate half inhibitory concentrations of sc-gC1qs, we performed hemolysis assay applying various protein concentrations. We concluded that both monomer and dimer proteins fully inhibit CP-caused hemolysis. The calculated IC50 values of sc-gC1q, sc-gC1q2, and sc-gC1q2l are 0.72, 0.54, and 0.34 μM, respectively. sc-gC1q2 showed similar IC50 value as sc-gC1q, whereas sc-gC1q2l was a more potent inhibitor of SRBC hemolysis (Fig. 7).
Detection of CP inhibition by C4 ELISA
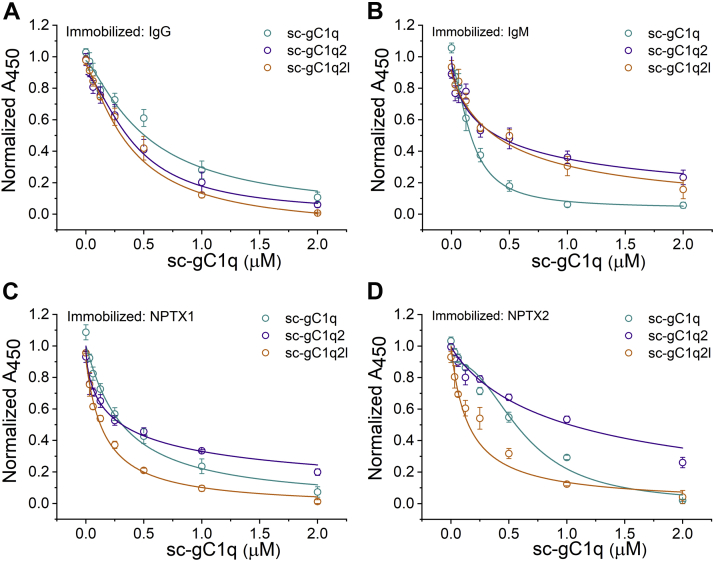
Inhibitory effect of sc-gC1q proteins on CP activation was investigated by C4 ELISA on microtiter plates coated with Igs (IgG and IgM) or NPTXs (NPTX1 or NPTX2) as CP activators (24, 39). Almost all inhibitors exhibited submicromolar IC50 values (Table 3). In the case of the monomeric activators (IgG, NPTX1, and NPTX2), sc-gC1q2l seemed to be the most potent inhibitor of CP. Interestingly, the IgM-induced CP activation was inhibited the most by the monomeric inhibitor sc-gC1q. Dimeric constructs had a similar effect on Ig-induced activation, whereas the inhibition of NPTX-induced activation was sensitive to the length of the dimerization linker (Fig. 8).
Table 3.
Half inhibition values obtained from C4 ELISA
| Binding partner | sc-gC1q | sc-gC1q2 | sc-gC1q2l |
|---|---|---|---|
| IgG | 0.53a | 0.42 | 0.39 |
| IgM | 0.17 | 0.39 | 0.47 |
| NPTX1 | 0.31 | 0.31 | 0.16 |
| NPTX2 | 0.62 | 1.03 | 0.15 |
Concentrations in micromolar.
Figure 8.
CP inhibition detected by C4 ELISA. Around 96-well microtiter plates were coated with IgG (A), IgM (B), neuronal pentraxin 1 and two (C and D, respectively). CP activation was detected by antibody against C4 deposited in the wells. Half inhibition constants were obtained from fitting Hill equations. Curves of sc-gC1q, sc-gC1q2, and sc-gC1q2l are marked with black, red, and purple, respectively. Values represent mean ± SEM. Two independent experiments were performed, each with at least 2-2 replicates. Estimated IC50 values are presented in Table 3. CP, complement pathway; gC1q, globular part of C1q; IgG, immunoglobulin G; IgM, immunoglobulin M; sc-gC1q, single-cell gC1q.
Inhibition of C1q binding to cell surface
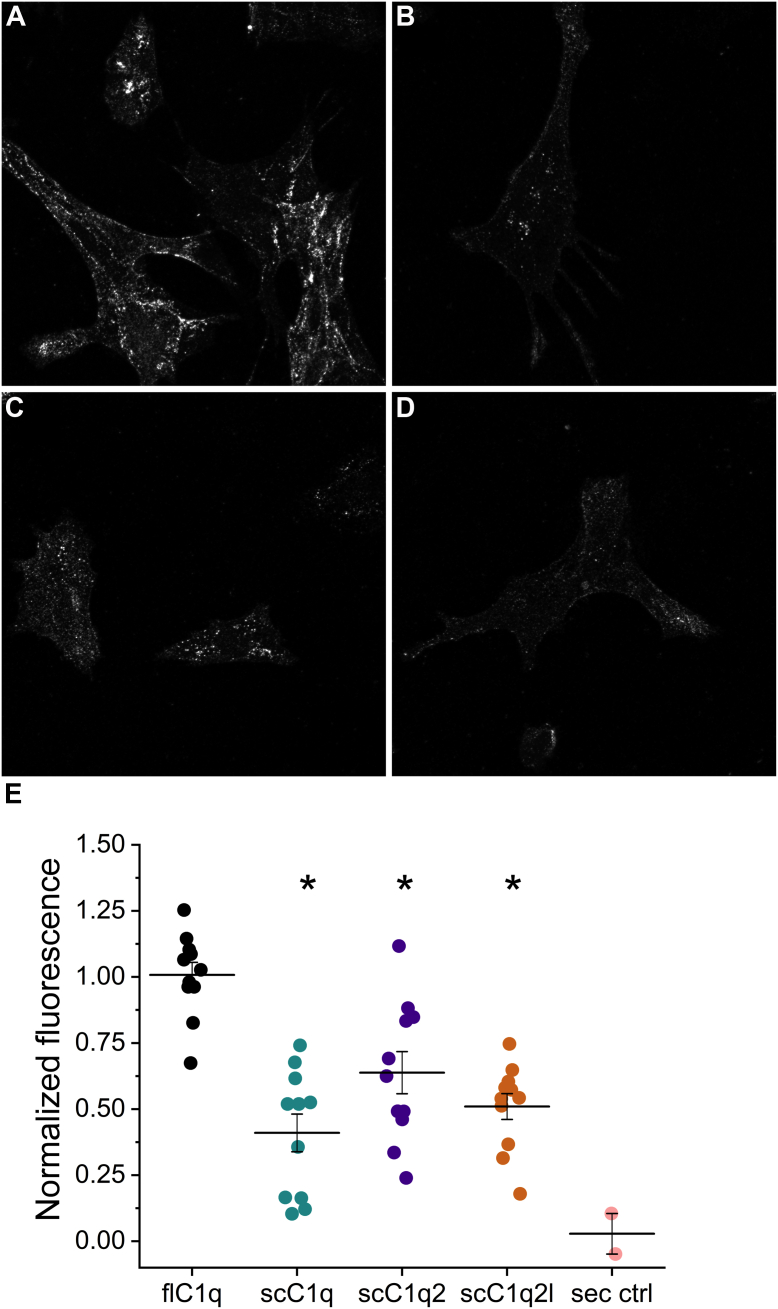
Finally, to test the inhibitory potential of the sc-gC1q constructs in a cellular environment, a competitive assay was carried out in cell culture. We incubated immortalized mouse hippocampal neurons (mHippoE-14 cell line; Cedarlane Corp) in a medium containing externally added full-length C1q at a concentration of 2 μg/ml. About 2% (v/v) serum contains approximately the same amount of C1q, which we applied in our previous experiments. Then, we treated the cells with different forms of sc-gC1q proteins in order to compete with the full-length C1q. Confocal fluorescence microscopy examination of neurons immunolabeled for CLR of C1q showed that all sc-gC1q constructs significantly lowered the cell surface–bound full-length C1q levels to a similar extent (Fig. 9). These findings support the idea that our sc-gC1q constructs can be utilized as tools to inhibit C1q binding.
Figure 9.
Competition of sc-gC1qs with full-length C1q binding to neuronal cell surface. Representative confocal fluorescence microscopy images are shown (A–D). After incubation with full-length C1q, cells were treated with sc-C1q-free medium (A), medium containing sc-gC1q (B), sc-gC1q2 (C), and sc-gC1q2l (D). Detection of full-length C1q and separation from sc-gC1q was based on applying primary antibody specific to C1q tail region. E, histograms of fluorescence intensities were determined by ImageJ program. Normalization was performed between solely secondary antibody–treated (sec. crtl) experiment and uninhibited measurement with full-length C1q treatment (flC1q). All three sc-gC1qs lowered significantly the full-length C1q signal in immortalized neural cell culture. One-way ANOVA was performed with post hoc Tukey’s test for mean comparison between flC1q and each sc-gC1q treatments. At the 0.05 level, the population means are different (F = 17). Sc-gC1q, sc-gC1q2, and sc-gC1q2l significantly lowered flC1q signal (p = 3 × 10−7, p = 0.001, and p = 1.16 × 10−5, respectively). Values represent mean ± SEM (from 11 images from uninhibited and inhibited samples and two images from secondary control; two individual experiments were performed from which five and six images were taken). gC1q, globular part of C1q; sc-gC1q, single-cell gC1q.
Discussion
C1q is a well-known molecule of the immune system, the first element of the classical pathway of the complement cascade. The role of C1q in CP activation is well characterized: it mostly binds to immune complexes and PTXs triggering the complement cascade and generating inflammation, phagocytosis, and cell lysis. Recently, C1q has also been implicated in numerous diseases of the CNS, such as Alzheimer’s disease, epilepsy, and frontotemporal dementia (19, 20, 21, 22). C1q serves as a signal on synapses that need to be eliminated; however, the detailed molecular mechanisms are still unexplored (13, 19). To investigate this enigmatic function of C1q, the need for potent C1q inhibitors has emerged.
In this study, we created potential inhibitors of C1q function, which comprise the recognition part of C1q, without its effector domain to circumvent potentially confounding effects of the latter. We designed a single-chain form of the globular head of mouse C1q, consisting of the globular domains of C1qA, C1qC, and C1qB chains similar to the work of Moreau et al. (32). First, we carried out an in silico study to model the structure of the designed sc-gC1q molecule and verified that its domains can organize in the known jelly-roll β-sandwich structure (5) consisting of antiparallel β-sheets (Figs. 1 and S2). The structural model showed a similarity to the X-ray structure of the human single-chain C1q head. MD simulations indicated that the structure is stable and revealed its dynamic properties (Fig. S2). Although full-length C1q is an extracellular protein with several disulfide bonds, we managed to express properly folded and functional sc-gC1q molecules in the cytoplasm of prokaryotic cells in a cost-effective manner. Note that mouse gC1qB and gC1qC contain only one surface-exposed disulfide bond, which is presumably formed after cell lysis and revealed by MS experiments (Fig. S4). Molecular weight study by analytical ultracentrifugation sedimentation velocity experiments indicated that the single-chain C1q head exhibits a homogeneous solution with a well-packed globular fold and an apparent molecular mass close to the theoretical 47.8 kDa (Fig. 2B).
Secondary structure of sc-gC1q was examined by CD spectroscopy and was compared with the secondary structure content of the in silico model and that of the crystal structure of the similar human sc-gC1q protein (PDB ID: 5HKJ) (32) (Table 1 and Fig. 3). Fold estimation by BeStSel resulted in a jelly-roll topology, which is in agreement with our model structure and the literature (5) (Table 1). In order to make our system similar to the original C1q, we designed multimer constructs, dimers and trimers with shorter (5 amino acids) and longer (13 amino acids) linkers between sc-gC1q and the multimerizing coiled-coil sequence. The GCN4 leucine zipper and the SpD neck region served as dimerizing or trimerizing regions, respectively (40, 41).
Analytical gel filtration and analytical ultracentrifugation experiments clarified that the dimers assembled, whereas trimers probably remained monomers (Fig. 2). Note that Kishore et al. (41) successfully trimerized the globular head domain of the human C1q B chain (corresponding to one globular head of C1q) using the helical neck region of an immune molecule, SpD. Our construct, aiming the assembly of three complete heads, could not trimerize even with the longer linker. We suppose that the stabilizing interactions in SpD coiled-coil are insufficient to hold together our larger complex. Usage of other trimeric coiled-coils may help overcome these problems, that is, certain types of modified GCN4 peptides that are able to multimerize proteins even with higher molecular weight (42, 43) or T4 foldon, which was also successfully used to trimerize severe acute respiratory syndrome coronavirus 2 S protein (44). Although our trimer constructs were shown to be monomers, several functional measurements were carried out with them to see if the nearly 50 residue-long C-terminal extensions interfere with the function of the single-chain head. The similar behavior of the sc-gC1q, sc-gC1g3, and sc-gC1q3l suggest that C-terminal fusions do not affect the binding properties of the head and can be used to address specific functions in the future.
In order to compare dissociation constants and characterize the binding of sc-gC1q variants to Igs and NPTXs, we performed SPR experiments. In this work, we claimed that the globular head region of C1q interacts with NPTXs. The affinities of sc-gC1q variants to various ligands were highly similar. The dimers could be characterized with a lower apparent dissociation rate and Kd than the monomer (Fig. 6), which is supposed to originate in its bivalent nature causing avidity. To assess the competitive potency of the properly folded sc-gC1qs in vitro, we carried out C4 ELISA using the Igs and NPTXs as CP activators. All the sc-gC1qs inhibited CP activation in a concentration-dependent manner; however, without the clear trend of the dimers being more effective. In contrast to the SPR results, here only the sc-gC1q2l seemed to be more potent than the monomeric constructs, which could be explained by the difference in ligand immobilization (chemistry and ligand density).
As in vivo surfaces are typically not covered exclusively by one ligand as in the case of SPR and ELISA experiments, ex vivo functional tests were conducted. We investigated the inhibitory effect of our molecules on SRBC lysis (Fig. 8) and C1q deposition on mHippoE-14 mouse embryonal hippocampal cells (Fig. 9). These experiments confirmed that sc-gC1qs are valuable CP inhibitors in biological environments, as well. In the hemolysis assay, the sc-gC1q2l proved to be the most potent inhibitor again, and all variants were similarly capable of inhibiting C1q binding to cell surfaces, thereby arresting classical complement activation at the very first step. Applying sc-gC1qs could help us identify in vivo in animal models the key steps of functions of C1q in the CNS and determine to what extent they are complement dependent and associated to the classical complement cascade.
In therapeutics, mAbs are cutting-edge tools, as they can directly target proteins or cells and they can act as powerful inhibitors. Moreover, they promote immune responses in case of a variety of diseases (45, 46, 47). Despite their numerous benefits, mAbs also have some fundamental limitations. Most importantly, mAbs are produced by mammalian cell lines with extremely high production costs. Proteins containing less disulfide bonds and less glycosylation are suitable to be expressed in prokaryotic cells. Most mAbs are IgGs with approximately 150 kDa molecular weight. This size decelerates diffusion and penetration into deeper tissue layers when compared with smaller proteins. For this purpose, engineering of mAbs becomes more popular, employing Fab or single-chain variable fragments with 50 or 30 kDa molecular weights, respectively (48). Our sc-gC1q molecule is of moderate size, and its mechanism of inhibition differs from that of mAbs being competitors of full-length C1q instead of binding to it. Similarly, Hebecker et al. (49) produced the functional parts of a complement regulator protein, factor H, calling it mini factor H that efficiently inhibited SRBC lysis and C3 deposition. Moreover, there are naturally occurring complement inhibitors lacking an effector region, like the alternative splice variants of the MASP1 and MASP2 genes, MAp44 (50) and MAp19 (51), respectively.
In summary, we produced mouse single-chain C1q globular head constructs, which were proven to be properly folded, functional, and capable of binding to various partners, such as IgG, IgM, and NPTXs. We demonstrated that sc-gC1qs are potent inhibitors of C1q function by hemolysis assay and C4 ELISA. Moreover, they compete with C1q in binding to the neuronal cell membrane. SPR experiments corroborate that dimer constructs bind to CP activators with a one order of magnitude lower apparent dissociation constant than the monomer. The application of sc-gC1qs can reveal neuronal localization and functions of C1q in assays in vivo and might serve as a basis for engineering inhibitors acting at the initiation of the cascade for therapeutic purposes.
Experimental procedures
In silico molecular modeling
An initial homology model of sc-gC1q was generated by the Phyre2 server (33) using the highest scoring template of the human single-chain globular C1q head structure (PDB ID: 5HKJ). The model was successfully created on 94% of the sequence with 100% confidence. The N-terminal sequence was built into the model manually using the SwissPDBViewer program. This model was subjected to MD simulations as implemented in GROMACS (35), using the AMBER-ff99SB∗-ILDNP force field (52). The system was solvated by water molecules with TIP4P parametrization (53). The total charge of the system was neutralized, and the physiological salt concentration was set by placing Na+ and Cl− ions. Energy minimization of the starting structures was followed by sequential relaxation of constraints on protein atoms in three steps and an additional NVT step (all of 200 ps) to stabilize pressure. One microsecond trajectories of NPT simulation at 300 K were recorded (collecting snapshots at every 20 ps). Secondary structure compositions of the frames of MD trajectories were determined by DSSP algorithm (54). Molecular graphics was performed with the UCSF CHIMERA package (55). Hydrophilic and hydrophobic surfaces were calculated using GETAREA (http://curie.utmb.edu/getarea.html) with water probe of a size of 1.4 Å (56).
Vector constructs
Single-chain C1q head (sc-gC1q) coding DNA sequence was generated from complementary DNA encoding the head domain regions of mouse C1qA (Arg110–Ala245; UniProt ID: P98086), C1qC (Lys116–Asp246; UniProt ID: Q02105), and C1qB (Gly115–Ala253; UniProt ID: P14106) chains fused sequentially with 4-residue-long GSGA or GSGG linkers. Dimers and trimers were created using the GCN4 leucine-zipper coding fragment (Leu-253–Arg281; UniProt ID: P03069) and the α-helical neck region of SpD (Asp222–Phe253; UniProt ID: P50404), respectively (Fig. 1, A and B) (40, 41). DNA sequences coding dimerizing or trimerizing peptide sequences were fused to the C terminus of the sc-gC1q applying NEBuilder (New England BioLabs, Inc). Two types of linker lengths were designed to connect globular head domains to the multimerizing part. Shorter and longer linkers contain 5 and 13 residues, respectively (Fig. 1A). All the sc-gC1q DNA constructs were cloned into a modified pET vector, called pEW encoding N-terminal His6 tag followed by SplB protease cleavage site.
Protein expression and purification
The E. coli BL21∗(DE3) cells transformed with the expression plasmids were grown at 37 °C in LB/Amp medium. When the absorbance reached 0.7 at 600 nm, the cultures were cooled to 18 °C and the protein expression was induced with 100 μM IPTG overnight. The cells were spun down, resuspended in 25 mM Tris (pH 7.4), 500 mM NaCl (immobilized metal ion affinity chromatography [IMAC] buffer), and then disintegrated by ultrasonication on ice. The cell debris was removed by centrifugation, and the His6-tagged proteins were purified by Ni2+-affinity chromatography (Profinity IMAC resin; Bio-Rad Laboratories). After excessive wash with IMAC buffer, proteins were eluted with a 300 mM imidazole-containing IMAC buffer. Samples were diluted with 25 mM Tris (pH 7.4) to decrease the conductivity to approximately 5 mS/cm, then loaded onto a cation exchange column (HiTrap SP HP; GE Lifesciences) connected to an ÄKTA pure system (GE Healthcare). After washing with 25 mM Tris (pH 7.4) and 50 mM NaCl, the elution was carried out with a linear gradient of 25 mM Tris (pH 7.4) and 1 M NaCl. The proteins were eluted at a conductance of 25 mS/cm. The purity of the preparations was analyzed by SDS-PAGE and size-exclusion chromatography (described later).
MS
LC–MS analysis
Mass spectrometric measurements were performed on a high-resolution hybrid quadrupole-time-of-flight mass spectrometer (Waters Select Series Cyclic IMS; Waters Corp). The mass spectrometer operated in positive V mode. Leucine enkephalin was used as lock mass standard. Chromatographic separations were performed on a Waters Acquity I-Class UPLC system, coupled directly to the mass spectrometer.
Intact reverse-phase chromatography–MS
Reverse-phase LC–MS analysis of the intact proteins were performed on a Waters Acquity BEH300 C4 UPLC column (2.1 × 150 mm, 1.7 μm) under the following parameters: mobile phase “A”: 0.1% trifluoroacetic acid in water, mobile phase “B”: 0.1% trifluoroacetic acid in acetonitrile; flow rate: 400 μl/min; column temperature: 80 °C; gradient: 1 min: 5% B, 12 min: 50% B, and 12.5 min: 90% B. UV detection was performed at 220 and 280 nm. The m/z range was 400 to 2000. Deconvolution was performed by the MaxEnt 1 software (Waters Corporation).
Peptide mapping
Sequences of the proteins were analyzed after proteolysis by trypsin (Promega Corporation) using 1:50 enzyme:protein ratio for 4 h at 37 °C. Digestion was stopped by adding formic acid in a final concentration of 0.2% (v/v). Gradient elution was performed on a Waters Acquity CSH Peptide C18 UPLC column (2.1 × 150 mm, 1.7 μm) under the following parameters: mobile phase “A”: 0.1% formic acid in water, mobile phase “B”: 0.1% formic acid in acetonitrile; flow rate: 300 μl/min; column temperature: 60 °C; gradient: 2 min: 2% B, 80 min: 5% B, and 81 min: 85% B. MSE experiments were performed using fragmentation in the trap cell with collision voltage ramping. MS data acquisition was performed under the following parameters: m/z 50 to 2000, scan time: 0.3 s, single lock mass: leucine enkephalin; low energy: 6 V, high energy: ramping 19 to 45 V. BiopharmaLynx 1.3.5 software (Waters Corp) was used to for data analysis.
Gel filtration
Purified sc-gC1q proteins (approximately 100–200 μg) were applied to Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with a buffer containing 25 mM Tris (pH 7.4) and 400 mM NaCl at room temperature using ÄKTA pure system. Eluted proteins were monitored at 280 nm. The column was calibrated using gel filtration protein standard (Bio-Rad Laboratories; catalog no.: 1511901).
Analytical ultracentrifugation experiments
To verify the correct folding and determine the molecular size distribution of sc-gC1q, sc-gC1q2, and sc-gC1q3 samples, analytical ultracentrifugation was used. Samples were dialyzed against a buffer of 50 mM sodium phosphate, 200 mM NaCl, pH 7.5 overnight before the measurements. Sedimentation velocity measurements were performed on a Beckman–Coulter Optima XL-1 analytical ultracentrifuge equipped with absorbance optics. Samples were first centrifuged at 3000 rpm (700g) for 20 min to stabilize the temperature, and after precentrifugation, the rotor speed was increased to 45,000 rpm (156,000g) and absorbance data at 237 nm were collected at intervals of 10 min. All measurements were carried out at a constant temperature of 25 °C with a radial increment of 0.003 cm in the continuous scanning mode. Buffer density and viscosity were calculated from the buffer composition using SEDNTERP program (http://jphilo.mailway.com/sednterp.htm) (57). Data analysis was carried out with continuous c(s) distribution using the SEDFIT program (http://jphilo.mailway.com/sedfit.htm) (58).
CD spectroscopy
Far-UV CD experiments of the sc-gC1q proteins were performed using a Jasco J-810 spectropolarimeter (Japan Spectroscopic Co) and a 0.1 cm path-length quartz cuvette in 25 mM Tris (pH 7.5), 250 mM NaCl, and 10% glycerol at 25 °C. Three spectra were accumulated in the wavelength range of 200 to 250 nm with a bandwidth of 1 nm, scanning speed of 10 nm/min, and response time of 8 s. After subtraction of the buffer reference, the spectra were normalized and analyzed for secondary structure composition using the BeStSel software (http://bestsel.elte.hu/) (36). Average of three spectra with BeStSel fitting is presented. SRCD spectra were recorded at the DISCO beamline of SOLEIL French Synchrotron Facility. Samples at ∼1 mg/ml were measured in CaF2 cells with a path length of 50 μm. Twelve scans were accumulated in the 190 to 270 nm wavelength range. One nanometer data steps with a lock-in time constant of 300 ms and integration time of 1200 ms were used. After baseline subtraction, the spectrum was corrected with the d-10-camphorsulfonic acid calibration (59). Protein concentration was determined by directly measuring the absorbance of the CD sample and buffer reference at 205 and 214 nm (60, 61).
ELISA binding assays
Maxisorp 96-well ELISA plate (Nunc, Invitrogen) was coated with either 40 nM NPTX1 (human NPTX1; UniProtID: Q15818; catalog no.: 7707-NP-050; R&D Systems), NPTX2 (human NPTX2; UniProtID: P47972; catalog no.: 7816-NP-050; R&D Systems), sc-gC1q, sc-gC1q2, or sc-gC1q2l in PBS overnight at 4 °C. Surfaces were blocked with 2% (w/v) bovine serum albumin (BSA) (Sigma–Aldrich) dissolved in PBS for 1 h at room temperature. For binding assays with PTXs, sc-gC1q, sc-gC1q2, and sc-gC1q2l were used in a serial dilution starting at 3 μM, whereas in case of the interaction analysis with sc-gC1qs, mouse IgG (Sigma–Aldrich; catalog no.: I5381) and hIgG1 (kind gift of Péter Závodszky, Research Center for Natural Sciences) were added to wells from 0.5 μM concentration, human IgM (Sigma–Aldrich; catalog no.: I8260) was used from 0.1 μM concentration. Sc-gC1qs, IgGs, and IgMs were diluted in 0.05% (v/v) Tween-20 containing PBS (PBS-T) and were incubated with the wells for 1 h at room temperature. After washing with PBS-T, PTX‒sc-gC1q complex–containing surfaces were treated with anti-C1qA antibody diluted in PBS-T (catalog no.: PAD207Mu01; Cloud Clone Corp; 1:500 dilution) for 1 h at room temperature. After washing, wells were treated with anti-rabbit horseradish peroxidase (HRP)–conjugated (catalog no.: A1949; Sigma–Aldrich; 1:5000 dilution), antimouse HRP-conjugated (catalog no.: 715-035-150; Jackson ImmunoResearch; 1:8000 dilution), antihuman-IgM-HRP-conjugated (catalog no.: A0420; Sigma–Aldrich; 1:1500 dilution), or antihuman-IgG-HRP-conjugated (catalog no.: A6029; Sigma–Aldrich; 1:1500 dilution) secondary antibodies diluted in PBS-T for 45 min at room temperature. 3,3′,5,5′-Tetramethylbenzidine substrate was added to the surfaces and after developing, reactions were stopped with 1 M HCl. Absorbance at 450 nm was measured using Synergy H4 plate reader (BioTek Instruments, Inc). Hill equation was fitted using Origin8 software (OriginLab).
SPR measurements
Binding affinities between sc-gC1q proteins (monomers and dimers) and IgG, IgM, or NPTXs were measured using a ProteOn XPR36 (Bio-Rad Laboratories) instrument equipped with a GLC sensor chip (Bio-Rad Laboratories). Five ligand channels were activated for 5 min with a solution containing 5.5 mM 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide HCl and 1.5 mM sulfo-N-hydroxysuccinimide. IgG (30 μg/ml), IgM (30 μg/ml), NPTX1 (10 μg/ml), NPTX2 (10 μg/ml), and BSA (30 μg/ml) were immobilized by amine coupling in 10 mM sodium acetate buffer (pH 4.5). Before surface deactivation, 0.15 mg/ml BSA was used to covalently block the unreacted N-hydroxysuccinimide groups. The remaining reactive sites were subsequently deactivated using 1 M ethanolamine-HCl (pH 8.5; Bio-Rad Laboratories). Then, 150, 75, 37.5, 18.75, 9.375, and 0 nM sc-gC1qs were injected simultaneously onto the sensor chip for 200 s in SPR buffer (20 mM Hepes, 150 mM NaCl, 0.005% [v/v] Tween-20 [pH 7.5]). Dissociation of sc-gC1qs was monitored for 420 s. Responses generated on the control channel were subtracted from those measured on the specific ligand channels. Apparent kinetic constants of dimer proteins were obtained from fitting association and dissociation phases with the following equations by Origin8 software:
where R is response in response unit; Req is response at equilibrium in response unit, Rt→∞ is response offset, kd is apparent dissociation rate constant (s−1), and ka is apparent association rate constant (M−1 s−1). Apparent dissociation rate constants and association rate constants of monomer protein were received by fitting two-phase exponential decays and two-phase association equations using Origin8 software:
ELISA inhibition assays
Forty nanomolar hIgG, IgM, NPTX1, or NPTX2 were immobilized on Maxisorp 96-well ELISA plate in PBS (Thermo Fisher Scientific) for 3 h at room temperature. Surfaces were blocked with 2% (w/v) BSA (Sigma–Aldrich) dissolved in PBS for 1 h at room temperature. After washing with 0.05% (v/v) Tween-20 containing Dulbecco’s PBS supplemented with Ca2+, Mg2+ (Ca2+/Mg2+ PBS-T; catalog no.: 14040091; Thermo Fisher Scientific), wells were treated with a mixture that consists of 2% (v/v) normal human serum with a serial dilution of sc-gC1q, sc-gC1q2, or sc-gC1q2l (initial concentration was 2 μM) in Ca2+/Mg2+ PBS-T for 30 min at 37 °C. After washing with Ca2+/Mg2+ PBS-T, anti-C4 primary antibody was added to the wells (Quidel Corporation; catalog no.: A305; 1:1000 dilution; diluted in Ca2+/Mg2+ PBS-T) for 1 h at room temperature. Next, surfaces were washed with 0.05% (v/v) PBS-T, then antigoat HRP-conjugated (Agilent Dako; catalog no.: P0449; 1:2000 dilution; diluted in PBS-T) secondary antibody was added to the wells for 45 min at room temperature. 3,3′,5,5′-Tetramethylbenzidine substrate was added to the surfaces, and reactions were stopped with 1 M HCl. Absorbance at 450 nm was measured using Epoch microplate reader (BioTek). To investigate the extent of autoactivation without immobilized activator, control measurement was performed completely the same way as activator-induced experiments, except to the immobilization. Normalization was performed between uninhibited and autoactivated values. Hill equation was fitted using Origin8 software.
Hemolysis studies
Hemolytic assay was performed using SRBCs (Culex Gp) coated with rabbit anti-SRBC antibody (hemolysin; Sigma–Aldrich; catalog no.: S1389-1VL).
SRBCs (stored in Alsever’s solution) were washed three times with veronal buffer (veronal-buffered saline [VBS]; Lonza Group AG; catalog no.: 12-624E) supplemented with 0.3 mM CaCl2, 0.5 mM MgCl2 (Ca2+/Mg2+ VBS), each washing step was followed by sedimentation of the SRBCs (800g, 5 min, 4 °C). SRBC concentration is set to 107 cells/reaction by Bürker chamber counting. Hemolysin is added to SRBCs diluted 1:4000 for 20 min at room temperature. Next, mixture of 0.5% (v/v) normal human serum, SRBCs, and a serial dilution of sc-gC1qs (2 μM initial concentration), anti-C1q (Abcam; catalog no.: ab76425; 1:200 dilution) or anti-C4 (Quidel; catalog no.: A305; 1:400 dilution) antibodies in Ca2+/Mg2+ VBS were incubated with 300 rpm shaking for 1 h at 37 °C. After sedimentation (800g, 5 min, 4 °C), absorbance at 414 nm was detected using Epoch microplate reader to measure hemoglobin content of the supernatant. Incubation with heat-inactivated normal human serum was used as negative control, and we used a maximum hemolytic value given by 1% normal human serum. Hill equation was fitted using Origin8 software.
Cell culture
Embryonic mouse hippocampal cell line mHippoE-14 was cultured in Dulbecco’s modified Eagle’s medium (Lonza) containing l-glutamine, phenol red, supplemented with 10% (v/v) fetal bovine serum (Euroclone SpA) and penicillin/streptomycin/amphotericin B (Lonza) in a humidified incubator with 5% CO2/95% air.
Immunofluorescence microscopy
MHippoE-14 cells were seeded in a 24-well plate (Corning Incorporated) containing coverslips (Assistent). Next, cells were incubated with 2 μM or 0 μM sc-gC1qs for 20 min at 37 °C after treating with 4 nM full-length C1q for 20 min. After extensive washing with PBS, cells were fixed with 4% (w/v) formaldehyde for 15 min at room temperature. After washing with PBS three times, cells were blocked and permeabilized with PBS containing 5% (w/v) BSA (Sigma–Aldrich) and 0.3% (v/v) Triton-X (Sigma–Aldrich) for 1 h at room temperature. Full-length C1q bound to cell surfaces was detected with anti-C1q antibody (GeneTex; catalog no.: GTX54404; 1:200 dilution) diluted in PBS containing 1% (w/v) BSA and 0.1% (v/v) Triton-X. Cells were incubated with primary antibody for 1 h at room temperature. Coverslips were washed with PBS, cells were treated with antimouse Alexa Fluor 488–conjugated secondary antibody (Jackson ImmunoResearch; catalog no.: 7 15-545-151; 1:400 dilution) diluted in PBS. Cells were washed with PBS, and then coverslips were mounted to microscopy slides using Mowiol 4-88 (Sigma–Aldrich) embedding medium. Imaging was performed by a Zeiss LSM800 confocal laser scanning microscope. The procedure was performed twice; five and six images were recorded of each treatment as z-stacks. Maximal intensity z-projects and histograms were obtained using ImageJ software (http://imagej.nih.gov/ij/; National Institutes of Health). All pixel intensities were summed. Data were normalized between the mean of the solely full-length C1q-treated controls and solely secondary antibody–treated controls. Statistical analysis was carried out using one-way ANOVA with Tukey’s test for multiple comparisons using a significance level of 0.05.
Data availability
All experimental data for this article are available upon e-mail request to: József Kardos (kardos@elte.hu).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr J. Michael Klopf for the English proofreading, Mihály Józsi (Eötvös Loránd University) for valuable discussions and providing antibodies for C4 ELISA experiments, and Keiichi Yamaguchi (Osaka University) for his advices in analytical ultracentrifuge experiments. This study was supported by the National Research, Development and Innovation Office of Hungary (grants 2017-1.2.1-NKP-2017-00002, 2018-1.2.1-NKP-2018-00005, FIEK_16-1-2016-0005, K120391, K138937, PD135510, 2019-2.1.11-TÉT-2019-00079, 2019-2.1.11-TÉT-2020-00101, KMOP-4.2.1/B-10-20, and VEKOP-2.3.3-15-2016-00007) and by the Hungarian Ministry for Innovation and Technology (ELTE Thematic Excellence Programme). SRCD measurements were supported by SOLEIL Synchrotron (proposal no.: 20191810).
Author contributions
H. V., B. K., and J. K. conceptualization; H. V., A. M., G. S., R. Á. K., B. A. G., Y. G., B. U., K. L., and J. K. methodology; H. V., A. M., G. S., R. Á. K., B. A. G., Y. G., B. U., K. L., and J. K. investigation; H. V., B. K., and J. K. writing–original draft; B. K. and J. K. writing–review & editing; H. V., A. M., G. S., and T. S. visualization; K. A. K. and J. K. supervision; Y. G. and J. K. funding acquisition.
Funding and additional information
This study was supported by the Japan Society for the Promotion of Science, Core-to-Core Program A (Advance Research Networks) to Y. G.
Edited by Roger Colbran
Supporting information
References
- 1.Reid K.B., Porter R.R. Subunit composition and structure of subcomponent C1q of the first component of human complement. Biochem. J. 1976;155:19–23. doi: 10.1042/bj1550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ying S.C., Gewurz A.T., Jiang H., Gewurz H. Human serum amyloid P component oligomers bind and activate the classical complement pathway via residues 14-26 and 76-92 of the A chain collagen-like region of C1q. J. Immunol. 1993;150:169–176. [PubMed] [Google Scholar]
- 3.McGrath F.D.G., Brouwer M.C., Arlaud G.J., Daha M.R., Hack C.E., Roos A. Evidence that complement protein C1q interacts with C-reactive protein through its globular head region. J. Immunol. 2006;176:2950–2957. doi: 10.4049/jimmunol.176.5.2950. [DOI] [PubMed] [Google Scholar]
- 4.Bottazzi B., Vouret-Craviari V., Bastone A., De Gioia L., Matteucci C., Peri G., et al. Multimer formation and ligand recognition by the long pentraxin PTX3. J. Biol. Chem. 1997;272:32817–32823. doi: 10.1074/jbc.272.52.32817. [DOI] [PubMed] [Google Scholar]
- 5.Gaboriaud C., Juanhuix J., Gruez A., Lacroix M., Darnault C., Pignol D., et al. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 6.Eggleton P., Reid K.B.M., Tenner A.J. C1q – how many functions? How many receptors? Trends Cell Biol. 1998;8:4. doi: 10.1016/s0962-8924(98)01373-7. [DOI] [PubMed] [Google Scholar]
- 7.Steinø A., Jørgensen C.S., Laursen I., Houen G. Interaction of C1q with the receptor calreticulin requires a conformational change in C1q. Scand. J. Immunol. 2004;59:485–495. doi: 10.1111/j.0300-9475.2004.01425.x. [DOI] [PubMed] [Google Scholar]
- 8.Ghebrehiwet B., Peerschke E.I. Structure and function of gC1q-R: a multiligand binding cellular protein. Immunobiology. 1998;199:225–238. doi: 10.1016/S0171-2985(98)80029-6. [DOI] [PubMed] [Google Scholar]
- 9.Ghebrehiwet B., Geisbrecht B.V., Xu X., Savitt A.G., Peerschke E.I.B. The C1q Receptors: focus on gC1qR/p33 (C1qBP, p32, HABP-1) Semin. Immunol. 2019;45 doi: 10.1016/j.smim.2019.101338. [DOI] [PubMed] [Google Scholar]
- 10.Kandov E., Kaur A., Kishore U., Ji P., Williams J., Peerschke E.I.B., et al. C1q and C1q receptors (gC1qR and cC1qR) as potential novel targets for therapy against breast cancer. Curr. Trends Immunol. 2018;19:59–76. [Google Scholar]
- 11.Kim K.-B., Yi J.-S., Nguyen N., Lee J.-H., Kwon Y.-C., Ahn B.-Y., et al. Cell-surface receptor for complement component C1q (gC1qR) is a key regulator for lamellipodia formation and cancer metastasis. J. Biol. Chem. 2011;286:23093–23101. doi: 10.1074/jbc.M111.233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peerschke E.I.B., Ghebrehiwet B. cC1qR/CR and gC1qR/p33: observations in cancer. Mol. Immunol. 2014;61:100–109. doi: 10.1016/j.molimm.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Stevens B., Allen N.J., Vazquez L.E., Howell G.R., Christopherson K.S., Nouri N., et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 14.Györffy B.A., Kun J., Török G., Bulyáki É., Borhegyi Z., Gulyássy P., et al. Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6303–6308. doi: 10.1073/pnas.1722613115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Schaarenburg R.A., Magro-Checa C., Bakker J.A., Teng Y.K.O., Bajema I.M., Huizinga T.W., et al. C1q deficiency and neuropsychiatric systemic lupus erythematosus. Front. Immunol. 2016 doi: 10.3389/fimmu.2016.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stegert M., Bock M., Trendelenburg M. Clinical presentation of human C1q deficiency: how much of a lupus? Mol. Immunol. 2015;67:3–11. doi: 10.1016/j.molimm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Botto M., Dell’ Agnola C., Bygrave A.E., Thompson E.M., Cook H.T., Petry F., et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 18.Roos A., Nauta A.J., Broers D., Faber-Krol M.C., Trouw L.A., Drijfhout J.W., et al. Specific inhibition of the classical complement pathway by C1q-binding peptides. J. Immunol. 2001;167:7052–7059. doi: 10.4049/jimmunol.167.12.7052. [DOI] [PubMed] [Google Scholar]
- 19.Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajishengallis G., Reis E.S., Mastellos D.C., Ricklin D., Lambris J.D. Novel mechanisms and functions of complement. Nat. Immunol. 2017;18:1288–1298. doi: 10.1038/ni.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronica E., Boer K., van Vliet E.A., Redeker S., Baayen J.C., Spliet W.G.M., et al. Complement activation in experimental and human temporal lobe epilepsy. Neurobiol. Dis. 2007;26:497–511. doi: 10.1016/j.nbd.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.-Y., et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott-Hewitt N., Perrucci F., Morini R., Erreni M., Mahoney M., Witkowska A., et al. Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia. EMBO J. 2020;39 doi: 10.15252/embj.2020105380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovács R.Á., Vadászi H., Bulyáki É., Török G., Tóth V., Mátyás D., et al. Identification of neuronal pentraxins as synaptic binding partners of C1q and the involvement of NP1 in synaptic pruning in adult mice. Front. Immunol. 2021 doi: 10.3389/fimmu.2020.599771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D., Hopf C., Reddy R., Cho R.W., Guo L., Lanahan A., et al. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- 26.Rogers J., Cooper N.R., Webster S., Schultz J., McGeer P.L., Styren S.D., et al. Complement activation by beta-amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10016–10020. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tacnet-Delorme P., Chevallier S., Arlaud G.J. β-Amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for aβ on the C1q globular regions. J. Immunol. 2001;167:6374–6381. doi: 10.4049/jimmunol.167.11.6374. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H., Burdick D., Glabe C.G., Cotman C.W., Tenner A.J. beta-Amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J. Immunol. 1994;152:5050–5059. [PubMed] [Google Scholar]
- 29.Pisalyaput K., Tenner A.J. Complement component C1q inhibits β-amyloid- and serum amyloid P-induced neurotoxicity via caspase- and calpain-independent mechanisms. J. Neurochem. 2008;104:696–707. doi: 10.1111/j.1471-4159.2007.05012.x. [DOI] [PubMed] [Google Scholar]
- 30.Benoit M.E., Hernandez M.X., Dinh M.L., Benavente F., Vasquez O., Tenner A.J. C1q-induced LRP1B and GPR6 proteins expressed early in alzheimer disease mouse models, are essential for the C1q-mediated protection against amyloid-β neurotoxicity. J. Biol. Chem. 2013;288:654–665. doi: 10.1074/jbc.M112.400168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelek W.M., Xie L., Morgan B.P., Harris C.L. Compendium of current complement therapeutics. Mol. Immunol. 2019;114:341–352. doi: 10.1016/j.molimm.2019.07.030. [DOI] [PubMed] [Google Scholar]
- 32.Moreau C., Bally I., Chouquet A., Bottazzi B., Ghebrehiwet B., Gaboriaud C., et al. Structural and functional characterization of a single-chain form of the recognition domain of complement protein C1q. Front. Immunol. 2016 doi: 10.3389/fimmu.2016.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 35.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., et al. Gromacs 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Micsonai A., Wien F., Kernya L., Lee Y.-H., Goto Y., Réfrégiers M., et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nauta A.J., Bottazzi B., Mantovani A., Salvatori G., Kishore U., Schwaeble W.J., et al. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur. J. Immunol. 2003;33:465–473. doi: 10.1002/immu.200310022. [DOI] [PubMed] [Google Scholar]
- 38.Pillemer L., Seifter S., Chu F., Ecker E.E. Function of components of complement in immune hemolysis. J. Exp. Med. 1942;76:93–101. doi: 10.1084/jem.76.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper N.R. The classical complement pathway: activation and regulation of the first complement component. Adv. Immunol. 1985;37:151–216. doi: 10.1016/s0065-2776(08)60340-5. [DOI] [PubMed] [Google Scholar]
- 40.Rapali P., Radnai L., Süveges D., Harmat V., Tölgyesi F., Wahlgren W.Y., et al. Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishore U., Strong P., Perdikoulis M.V., Reid K.B.M. A recombinant homotrimer, composed of the α helical neck region of human surfactant protein D and C1q B chain globular domain, is an inhibitor of the classical complement pathway. J. Immunol. 2001;166:559–565. doi: 10.4049/jimmunol.166.1.559. [DOI] [PubMed] [Google Scholar]
- 42.Eckert D.M., Malashkevich V.N., Kim P.S. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J. Mol. Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Farzan M., Wyatt R., Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2000;74:5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zahavi D., Weiner L. Monoclonal antibodies in cancer therapy. Antibodies. 2020;9:34. doi: 10.3390/antib9030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nahta R., Esteva F.J. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26:3637–3643. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 47.Bayry J., Lacroix-Desmazes S., Kazatchkine M.D., Kaveri S.V. Monoclonal antibody and intravenous immunoglobulin therapy for rheumatic diseases: rationale and mechanisms of action. Nat. Clin. Pract. Rheumatol. 2007;3:262–272. doi: 10.1038/ncprheum0481. [DOI] [PubMed] [Google Scholar]
- 48.Cruz E., Kayser V. Monoclonal antibody therapy of solid tumors: clinical limitations and novel strategies to enhance treatment efficacy. Biol. Targets Ther. 2019;13:33–51. doi: 10.2147/BTT.S166310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hebecker M., Alba-Domínguez M., Roumenina L.T., Reuter S., Hyvärinen S., Dragon-Durey M.-A., et al. An engineered construct combining complement regulatory and surface-recognition domains represents a minimal-size functional factor H. J. Immunol. 2013;191:912–921. doi: 10.4049/jimmunol.1300269. [DOI] [PubMed] [Google Scholar]
- 50.Degn S.E., Hansen A.G., Steffensen R., Jacobsen C., Jensenius J.C., Thiel S. MAp44, a human protein associated with pattern recognition molecules of the complement system and regulating the lectin pathway of complement activation. J. Immunol. 2009;183:7371–7378. doi: 10.4049/jimmunol.0902388. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi M., Endo Y., Fujita T., Matsushita M. A truncated form of mannose-binding lectin-associated serine protease (MASP)-2 expressed by alternative polyadenylation is a component of the lectin complement pathway. Int. Immunol. 1999;11:859–863. doi: 10.1093/intimm/11.5.859. [DOI] [PubMed] [Google Scholar]
- 52.Aliev A.E., Kulke M., Khaneja H.S., Chudasama V., Sheppard T.D., Lanigan R.M. Motional timescale predictions by molecular dynamics simulations: case study using proline and hydroxyproline sidechain dynamics. Proteins. 2014;82:195–215. doi: 10.1002/prot.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 54.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 57.Laue T., Shah B., Ridgeway T., Pelletier S. Computer-aided Interpretation of Sedimentation Data for Proteins. Royal Society of Chemistry; Cambridge, U.K: 1992. [Google Scholar]
- 58.Brown P.H., Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen G.C., Yang J.T. Two-point calibration of circular dichrometer with d-10-camphorsulfonic acid. Anal. Lett. 1977;10:1195–1207. [Google Scholar]
- 60.Anthis N.J., Clore G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuipers B.J.H., Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data for this article are available upon e-mail request to: József Kardos (kardos@elte.hu).