Abstract
We have previously shown that the serine/threonine kinase PKCα triggers MAPK/ERK kinase (MEK)-dependent G1→S cell cycle arrest in intestinal epithelial cells, characterized by downregulation of cyclin D1 and inhibitor of DNA-binding protein 1 (Id1) and upregulation of the cyclin-dependent kinase inhibitor p21Cip1. Here, we use pharmacological inhibitors, genetic approaches, siRNA-mediated knockdown, and immunoprecipitation to further characterize antiproliferative ERK signaling in intestinal cells. We show that PKCα signaling intersects the Ras-Raf-MEK-ERK kinase cascade at the level of Ras small GTPases and that antiproliferative effects of PKCα require active Ras, Raf, MEK, and ERK, core ERK pathway components that are also essential for pro-proliferative ERK signaling induced by epidermal growth factor (EGF). However, PKCα-induced antiproliferative signaling differs from EGF signaling in that it is independent of the Ras guanine nucleotide exchange factors (Ras-GEFs), SOS1/2, and involves prolonged rather than transient ERK activation. PKCα forms complexes with A-Raf, B-Raf, and C-Raf that dissociate upon pathway activation, and all three Raf isoforms can mediate PKCα-induced antiproliferative effects. At least two PKCα–ERK pathways that collaborate to promote growth arrest were identified: one pathway requiring the Ras-GEF, RasGRP3, and H-Ras, leads to p21Cip1 upregulation, while additional pathway(s) mediate PKCα-induced cyclin D1 and Id1 downregulation. PKCα also induces ERK-dependent SOS1 phosphorylation, indicating possible negative crosstalk between antiproliferative and growth-promoting ERK signaling. Importantly, the spatiotemporal activation of PKCα and ERK in the intestinal epithelium in vivo supports the physiological relevance of these pathways and highlights the importance of antiproliferative ERK signaling to tissue homeostasis in the intestine.
Keywords: PKCα, RasGRP3, H-Ras protein, Raf kinase, ERK, p21Cip1, cyclin D1, Id1, growth arrest, EGF
Abbreviations: 4-OHT, 4-hydroxytamoxifen; BIM I, bisindolylmaleimide I; DAG, diacylglycerol; DiC8, 1,2-dioctanoyl-sn-glycerol; FBS, fetal bovine serum; mESC, mouse embryonic stem cell; PMA, phorbol 12-myristate 13-acetate
Self-renewal of the epithelial lining of the intestine requires precise coordination of rapid cell proliferation in the crypt with growth arrest at the crypt/villus junction (1, 2). Previous studies have determined that signaling downstream of extracellular signal-regulated kinase (ERK) 1 and 2 plays a central role in this process. Ligands such as epidermal growth factor (EGF), secreted by Paneth cells and mesenchymal cells, lead to activation of ERK in intestinal crypt cells (3). Multiple studies have established a growth stimulatory role for the ERK pathway in the intestinal epithelium. For example, inhibition of ERK signaling prevents cell cycle progression/proliferation in IEC-6 nontransformed intestinal epithelial cells, Caco-2 colon cancer cells (3), as well as Lgr5+ stem cells and transit amplifying cells in mouse intestinal organoids (4). However, evidence also points to important postmitotic roles of ERK signaling in the intestinal epithelium. The high levels of active ERK seen in proliferating cells of the crypt are maintained in nonproliferating cells of the villus in fetal and adult intestine (3, 5). In vitro, sustained activation of ERK correlates with G1 arrest and is required for differentiation in postconfluent Caco-2 cells (3, 6). In vivo, deletion of ERKs in the murine intestinal epithelium leads to loss of intestinal epithelial function, increased proliferation, and crypt elongation (7, 8), pointing to a defect in growth arrest at the crypt–villus junction. At a mechanistic level, our previous studies have determined that activation of protein kinase C α (PKCα) induces a program of cell-cycle withdrawal in intestinal epithelial cells that involves sustained activation of ERK (9, 10, 11). Thus, studies in the intestinal epithelium have determined that, in addition to the well-established role of ERK signaling in supporting cell proliferation, maintenance of intestinal homeostasis requires antiproliferative ERK signaling that can be triggered by PKCα.
ERK signaling plays a major role in transmission of extracellular signals that regulate fundamental cellular processes such as proliferation, differentiation, development, stress responses, and apoptosis (12). Canonical ERK activation involves a signaling cascade in which membrane anchored Ras small GTPases (H-Ras, K-Ras, and N-Ras) recruit and activate Raf kinases (A-Raf, B-Raf, and C-Raf), which then phosphorylate and activate MEKs (MEK1 and MEK2); MEKs, in turn, phosphorylate and activate ERKs (ERK1 and ERK2) (13, 14, 15, 16). Activity of this cascade is governed by cycling of the Ras GTPases between the inactive GDP-bound state and the active GTP-bound state. This process is predominantly regulated by Ras GTPase–activating proteins (Ras-GAPs) and Ras guanine nucleotide exchange factors (Ras-GEFs), with GEFs thought to play the major role in activation of Ras signaling (17, 18). Mammalian cells express three classes of Ras-GEFs: SOS proteins (SOS1, SOS2), the RasGRFs (RasGRF1 and RasGRF2), and the RasGRPs (RasGRP1, RasGRP3, and RasGRP4). Following pathway activation, ERK can phosphorylate multiple downstream targets in the cytoplasm and nucleus, including kinases such as RSK that further propagate the signal (19). This wide range of substrates allows ERKs to carry out their multiple roles in the cell. Several mechanisms have been implicated in directing ERK signaling to a particular physiological outcome, including signal strength and duration, scaffolding proteins, subcellar localization, and interaction with other signaling pathways (19). However, in most cases, these mechanisms remain poorly defined.
PKCα is a member of the classical subgroup (cPKCs) of the PKC family of serine/threonine kinases (20, 21, 22). Physiological activation of PKCα is calcium dependent and driven by membrane accumulation of diacylglycerol (DAG), generated by phospholipase C downstream of ligand binding to receptor tyrosine kinases (RTKs) or G protein–coupled receptors (20, 21, 22). DAG recruits PKCα to the membrane, leading to a conformational change that results in activation of the kinase (20, 21). In addition to physiological activation, membrane recruitment and activation of PKCα can be achieved experimentally using short chain DAGs, such as 1,2-dioctanoyl-sn-glycerol (DiC8), or pharmacological agents such as phorbol 12-myristate 13-acetate (PMA) (22, 23, 24).
Immunohistochemical analysis of intestinal tissue by our group demonstrated that PKCα is activated precisely at the point of growth arrest, as indicated by plasma membrane translocation of the enzyme at the crypt/villus junction (9, 25, 26). In vitro analysis using nontransformed intestinal epithelial cells confirmed that PKCα activation triggers a program of cell cycle withdrawal, involving downregulation of pro-proliferative cyclin D1 and inhibitor of DNA-binding 1 (Id1) and upregulation of the cyclin-dependent kinase (CDK) inhibitor p21Cip1 (9, 10, 26, 27, 28). Notably, these effects are dependent on MEK activation (11), indicating that PKCα initiates growth inhibitory ERK signaling in intestinal epithelial cells. While it has long been recognized that PKC signaling can activate the ERK pathway, there is controversy regarding the mechanism involved. While some studies have indicated that PKC activation of the pathway is Ras independent (29) and can be mediated by direct phosphorylation of C-Raf by PKCα (30), other studies indicate that Ras is required (31). Previous studies also support the ability of classical PKCs to activate ERK through a mechanism that does not require Raf phosphorylation (32) and that PKCα may directly activate MEK (33). Thus, mechanisms underlying PKCα-mediated activation of ERK remain to be elucidated.
In the present study, we define crosstalk between PKCα and the ERK activation cascade during growth suppressive signaling in intestinal epithelial cells. Our analysis has determined that PKCα intersects the canonical ERK activation pathway at the level of Ras and that PKCα triggers at least two antiproliferative ERK signaling pathways. One pathway involves activation of RasGRP3 and H-Ras for ERK-dependent induction of p21Cip1, while at least one other PKCα–Ras–ERK pathway triggers downregulation of the pro-proliferative proteins, cyclin D1 and Id1.
Results
Activation of PKCα by PMA induces ERK-dependent antiproliferative signaling in intestinal epithelial cells
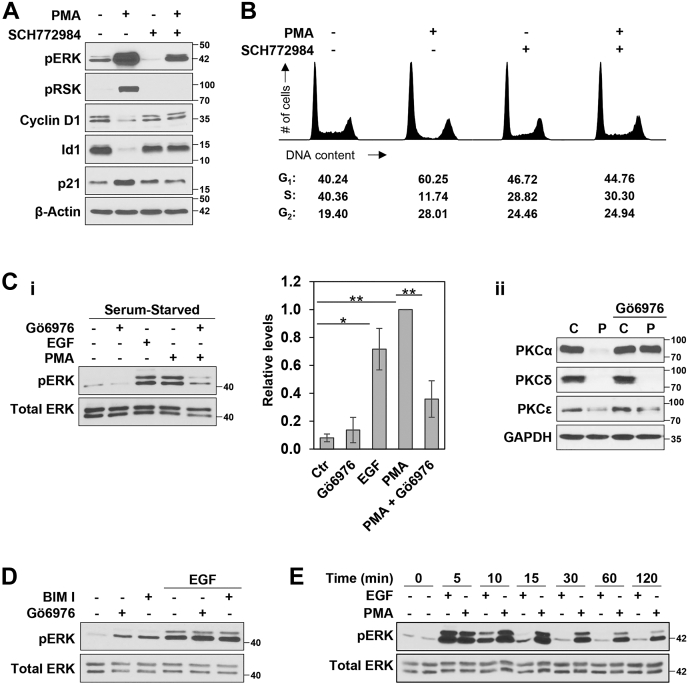
We have previously reported that pharmacological activation of PKCα in nontransformed IEC-18 intestinal crypt-like cells induces cell cycle arrest in association with activation of the ERK signaling pathway, as determined using antiphospho-ERK (pERK) immunoblotting (ref. (11) and Fig. 1A). Phosphorylation of the ERK substrate RSK in PMA-treated IEC-18 cells confirmed functional activation of ERK, an effect that was blocked by the selective ATP-competitive ERK inhibitor, SCH772984 (Fig. 1A). To determine the involvement of ERK in the antiproliferative activity of PKCα, we tested the effects of SCH772984 on PMA/PKCα-induced cell cycle arrest and downstream cell cycle regulatory targets. Treatment of IEC-18 cells with PMA for 6 h led to G1→S arrest, as indicated by a 50% increase in the percentage of cells in G1-phase and a >70% reduction in the percentage of cells in S-phase (Fig. 1B). Consistent with a role for pro-proliferative ERK signaling in intestinal epithelial cells, ERK inhibition with SCH772984 led to a modest decrease (∼30%) in cells in S-phase. However, the ERK inhibitor also abrogated PMA/PKCα-induced G1→S arrest in these cells (Fig. 1B, right panels). PKCα-induced growth inhibition is associated with downregulation of the pro-proliferative proteins, cyclin D1 and Id1, and upregulation of the CDK inhibitor p21Cip1 (refs. (9, 26) and Fig. 1A). Notably, SCH772984 also blocked the ability of PMA to modulate the expression of these proteins (Fig. 1A), confirming that the effects of PKCα on these downstream molecules are dependent on ERK activity.
Figure 1.
PMA induces antiproliferative PKCα-ERK signaling in intestinal epithelial cells.A, IEC-18 cells were pretreated with vehicle (−) or 1 μM SCH772984 (+) for 1 h, followed by addition of vehicle (−) or 100 nM PMA (+) for 2 h, and samples were subjected to immunoblot analysis for the expression/phosphorylation of the indicated proteins. Note that SCH772984 is an ATP-competitive inhibitor and does not prevent phosphorylation of ERK by MEK. B, IEC-18 cells were treated with 100 nM PMA for 6 h in the presence or absence of SCH772984 as indicated and DNA content/cell cycle distribution was determined by flow cytometric analysis. The percentage of cells in G1, S, and G2/M phase is shown below the DNA histograms. C, i, IEC-18 cells, cultured overnight in medium containing 0.5% FBS (serum-starved), were treated (10 min) with vehicle (−), 50 ng/ml EGF, or 100 nM PMA in the absence or presence of 4 μM Gö6976 as indicated, prior to Western blot analysis. The graph to the right of the blots shows densitometric analysis of relative levels of pERK normalized to loading control (±SD, n = 5, ∗p ≤ 0.05; ∗∗p ≤ 0.01); ii, IEC-18 cells were treated with PMA for 6 h in the absence or presence of 4 μM Gö6976 and analyzed for the expression of PKCα, δ, or ε by Western blotting. D, Serum-starved IEC-18 cells were pretreated for 1 h with 4 μM Gö6976 or 5 μM BIM I as indicated, before treatment with vehicle (−) or EGF for 10 min. E, serum-starved IEC-18 cells were treated with PMA or EGF for the indicated times and subjected to Western blot analysis for pERK and total ERK. All data are representative of at least three independent experiments. BIM I, bisindolylmaleimide I; PMA, phorbol 12-myristate 13-acetate.
To further characterize PKCα-triggered antiproliferative ERK signaling, we compared PMA/PKCα-induced ERK activation with ERK activation induced by a known growth stimulatory factor, EGF. To avoid confounding effects of serum growth factors in the medium, these experiments were performed under serum-starved conditions (0.5% serum). As shown in Figure 1Ci, EGF and PMA induced comparable levels of ERK phosphorylation in IEC-18 cells. Follow-up experiments tested the requirement for PKC activity in ERK activation by EGF and PMA using bisindolylmaleimide (BIM I), a pan-PKC inhibitor, or Gö6976, an inhibitor that selectively targets the cPKCs (PKCα, βI, βII, and γ). The only PMA-responsive PKC isozymes expressed in IEC-18 cells are PKCα, PKCδ, and PKCε (10, 11, 25); thus, Gö6976 selectively inhibits PKCα in these cells. The selectivity of Gö6976 for PKCα was confirmed by its ability to block PKC agonist (PMA) induced PKCα downregulation, which is dependent on PKCα activity, while not affecting the activity-dependent downregulation of PKCδ or PKCε (Fig. 1Cii). Neither BIM I nor Gö6976 affected the ability of EGF to activate ERK, indicating that PKC activity is not involved in this effect of EGF (Fig. 1D). In contrast, Gö6976 largely prevented the increase in phospho-ERK induced by PMA (Fig. 1Ci).
Differences were also observed in the duration of ERK activation elicited by PMA/PKCα and EGF; while EGF-induced ERK activity returned to basal levels by 15 to 30 min, the ERK signal induced by PMA/PKCα was sustained for longer than 2 h (Fig. 1E). We have previously demonstrated that sustained activation of ERK is required for the antiproliferative effects of PKCα activation but not for pro-proliferative growth factor signaling (11). While 15 to 30 min of ERK activation is sufficient to promote proliferation in response to serum growth factors, we showed that 30 to 60 min of PKCα-ERK signaling is required for G1→S phase arrest at 6 h, and maintenance of PKCα-induced cell cycle blockade at 9 h requires continued PKCα-ERK signaling for 60 to 90 min (11). Together, the data indicate that PKCα activates antiproliferative ERK signaling that is qualitatively and temporally distinct from the canonical pro-proliferative ERK axis induced by growth factors.
PKCα-induced growth suppressive ERK signaling requires MEK, RAF, and Ras activity
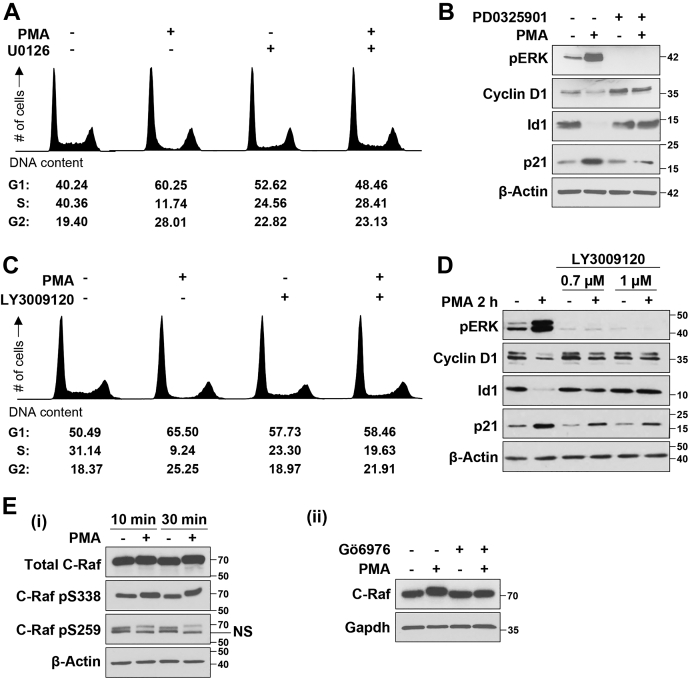
The canonical ERK signaling cascade involves sequential activation of Ras, Raf, and MEK (12), which stimulates ERK by phosphorylation. Subsequent experiments, therefore, investigated the requirement for canonical ERK pathway components in the antiproliferative effects of PKCα activation. As seen with ERK inhibition using SCH772984, blockade of ERK activation using the MEK inhibitor, U0126, led to a 30% to 35% reduction in cells in S-phase. Consistent with our previous findings (11), U0126 also prevented PMA/PKCα-induced G1→S-phase arrest (Fig. 2A, right panels). Further confirmation of the role of MEK in antiproliferative PKCα-ERK signaling (11, 26) was provided by the ability of the second generation MEK1/2 selective inhibitor, PD0325901, to block PMA-induced ERK activation, downregulation of cyclin D1 and Id1, and upregulation of p21Cip1 (Fig. 2B).
Figure 2.
PKCα requires MEK and RAF activity to induce cell cycle arrest in IEC-18 intestinal epithelial cells.A, flow cytometric analysis of IEC-18 cells pretreated with the MEK inhibitor U0126 (10 μM) for 1 h prior to addition of vehicle (−) or 100 nM PMA for 6 h. Note that the data are part of the same experiment shown in Figure 1B; vehicle and PMA-treated control panels on the left are therefore the same in both figures. B, Western blot analysis of IEC-18 cells pretreated with the MEK1/2 inhibitor PD0325901 (10 μM) for 1 h before addition of 100 nM PMA for 2 h. C, IEC-18 cells were pretreated with the pan-Raf inhibitor LY3009120 (3 μM) for 1 h, followed by 100 nM PMA for 6 h as indicated, and cell cycle distribution was analyzed by flow cytometry. D, control and LY3009120 pretreated IEC-18 cells were incubated with 100 nM PMA for 2 h and subjected to Western blot analysis for the indicated proteins. E, i, IEC-18 cells were treated with 100 nM PMA for the specified times before analysis of C-Raf phosphorylation by Western blotting. ii, IEC-18 cells were pretreated with 4 μM Gö6976 for 1 h prior to addition of PMA for 10 min as indicated and subjected to immunoblot analysis. All data are representative of at least three independent experiments.
To investigate if growth suppressive ERK signaling requires Raf activity, we used the pan-Raf inhibitor LY3009120, an ATP-competitive inhibitor that binds all Raf isoforms with the same affinity (34, 35). Inhibition of Raf activity by LY3009120 abrogated PMA/PKCα-induced G1→S phase arrest (Fig. 2C), activation of ERK, and alterations in the expression of cyclin D1, Id1, and p21Cip1 (Fig. 2D). PMA treatment also led to a mobility shift in C-Raf on SDS-PAGE, suggestive of effects on C-Raf phosphorylation (Fig. 2E). Changes in activating and inhibitory phosphorylation of C-Raf (36) were, therefore, analyzed using phospho-specific antibodies, which detected a PMA-induced increase in activating phosphorylation at S338 and a reduction in inhibitory phosphorylation at S259 (Fig. 2Ei). Importantly, the change in C-Raf mobility was inhibited by Gö6976 (Fig. 2Eii), indicating that PMA-induced Raf activation is part of the same antiproliferative, PKCα-dependent pathway identified previously. Thus, as with growth promoting ERK pathways, PKCα-triggered growth suppressive ERK signaling requires MEK activity downstream of Raf activation.
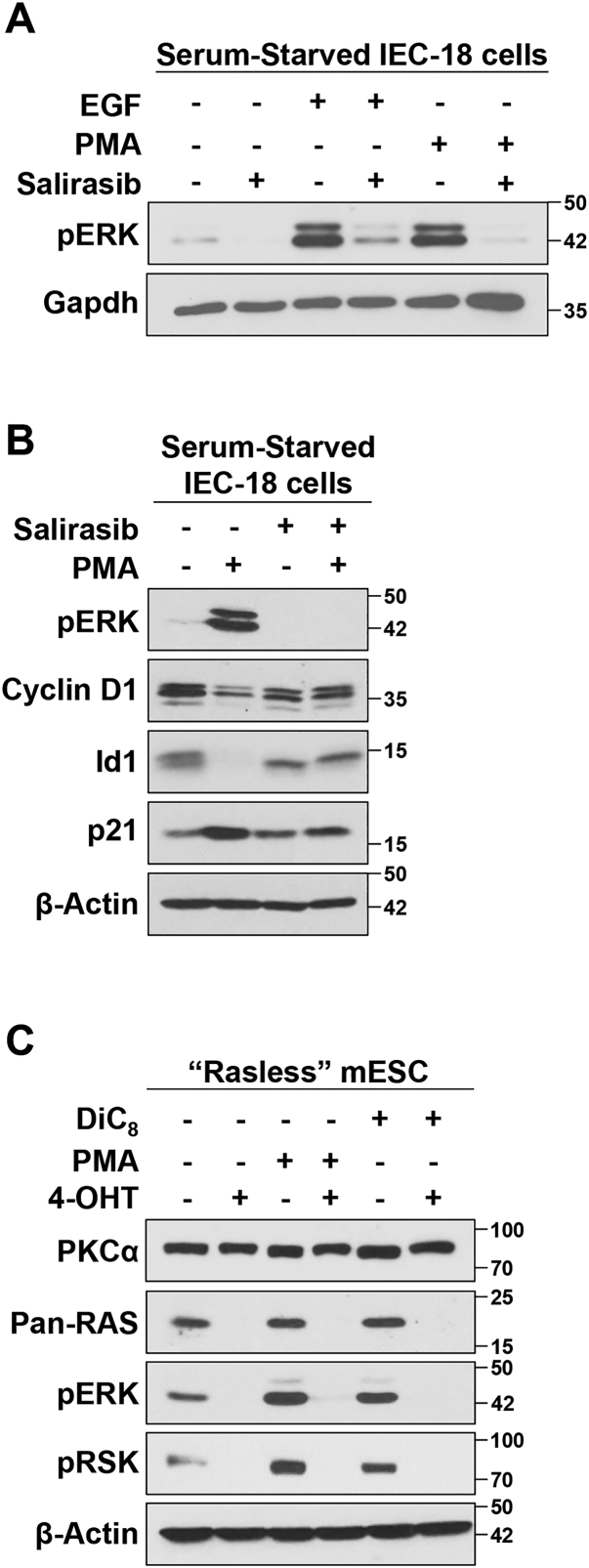
To assess the role of Ras proteins in PKCα-induced activation of ERK, we tested the effects of salirasib, a farnesylcysteine mimetic that blocks membrane association of activated Ras proteins (37). As also shown in Figure 1C, both EGF and PMA/PKCα strongly activated ERK in serum-starved IEC-18 cells (Fig. 3A). As expected, salirasib abrogated EGF-induced activation of ERK, which is known to be dependent on Ras activity (38). Salirasib also blocked the ability of PMA to activate ERK (Fig. 3, A and B) and to modulate the expression of cyclin D1, Id1, and p21Cip1 (Fig. 3B), pointing to a requirement for Ras activity in PKCα-induced growth suppressive ERK signaling. Since salirasib can have off target effects (39, 40, 41, 42), the requirement for Ras in PKC agonist-induced activation of ERK was further tested using “Rasless” N-Ras−/−; H-Ras−/−; K-Rasf/f; and Ubiq-CreERT2 mouse embryonic stem cells (mESCs). Additional validation was particularly important because there has been considerable confusion in the field regarding the role of Ras in transducing signals from PKC to ERK, with some studies indicating that PKC-mediated activation of Rafs is Ras independent (29, 30, 33, 43). These mESCs can be used for this analysis because they express PKCα at equal levels before and after KO of K-Ras (Fig. 3C). N-Ras−/−; H-Ras−/−; K-Rasf/f; and Ubiq-CreERT2 mESCs are knocked out for H-Ras and N-Ras and allow for conditional KO of floxed K-Ras following activation of CreERT2; thus, they lack expression of all three Ras isoforms following treatment with 4-hydroxytamoxifen (4-OHT) (ref. (44) and Fig. 3C). As expected based on the established role of Ras in growth factor–induced ERK activation, complete loss of Ras in these cells is reflected in loss of basal ERK and downstream RSK phosphorylation (Fig. 3C). Prior to KO of K-Ras, PMA treatment readily increased ERK activation in these mESCs. A similar effect was seen with the short chain DAG, DiC8, a more physiological PKC/PKCα agonist that mimics endogenous DAG signaling (45) and elicits the same physiological responses as phorbol esters in IEC-18 cells (9, 11, 26). However, following KO of K-Ras to generate “Rasless” cells, neither PMA nor DiC8 was able to induce ERK phosphorylation (Fig. 3C), confirming a requirement for Ras in PKC agonist-induced ERK signaling. Collectively, the data indicate that PKCα-activated growth-inhibitory ERK signaling, like growth-promoting ERK signaling, is mediated by activation of Ras in addition to Raf and MEK.
Figure 3.
PKCα-induced ERK activation and cell cycle arrest requires the activity of Ras.A, serum-starved IEC-18 cells were pretreated with vehicle (−) or 50 μM salirasib for 2 h, followed by 50 ng/ml EGF or 100 nM PMA for 10 min as indicated, and analyzed by Western blotting. B, cells were treated with PMA for 2 h in the absence or presence of salirasib and subjected to immunoblot analysis for the indicated proteins. C, N-Ras−/−; H-Ras−/−; K-Rasf/f; Ubiq-CreERT2 mESCs were treated with vehicle (−) or 1 μM 4-hydroxytamoxifen (4-OHT) for 7 days to knock out K-Ras and generate “Rasless” mESCs. The cells were then treated with 100 nM PMA or 20 μg/μl DiC8 for 15 min as indicated, prior to Western blot analysis for PKCα, Ras, pERK, and pRSK. The data are representative of three independent experiments. mESC, mouse embryonic stem cell; PMA, phorbol 12-myristate 13-acetate.
The scaffold proteins KSR1 and KSR2 are not required for PKCα-mediated antiproliferative ERK signaling
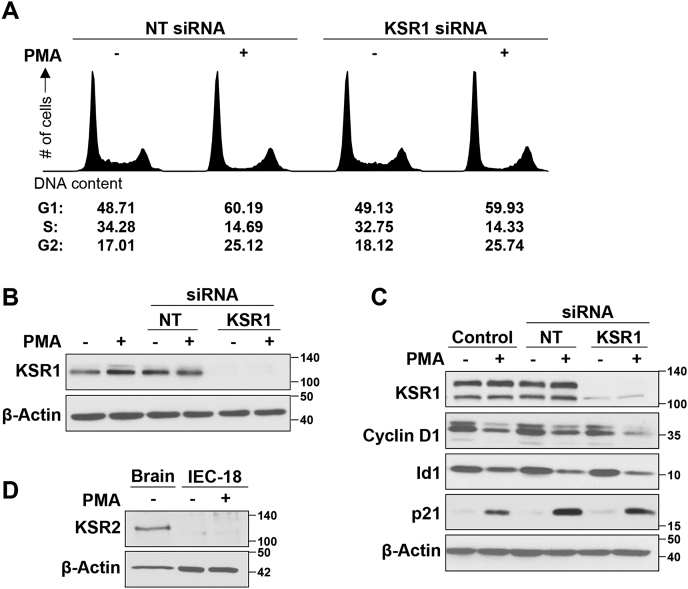
The ERK scaffolding proteins, KSR1 and KSR2, play an important role in modulation of growth promoting and oncogenic ERK signaling and have been implicated in promoting ERK-dependent differentiation (46, 47). Therefore, the role of these scaffolding proteins in PKCα-induced growth inhibitory signaling was examined. Knockdown of KSR1 in IEC-18 cells did not affect the ability of PMA to induce a reduction of cells in S-phase (Fig. 4, A and B), indicating that KSR1 is not required for PKCα/ERK-mediated G1→S arrest in these cells. Similarly, KSR1 knockdown did not prevent PMA-induced downregulation of cyclin D1 and Id1 or upregulation of p21Cip1 (Fig. 4C). Since KSR2 is not expressed in IEC-18 cells (Fig. 4D), these data indicate that PKCα-induced growth inhibitory Ras-ERK signaling is not regulated by KSR scaffolding proteins.
Figure 4.
ERK pathway scaffold proteins KSR1 and KSR2 do not regulate PKCα-induced growth arrest in IEC-18 cells. IEC-18 cells were transfected with the indicated siRNAs 96 h prior to treatment with PMA for 6 h, followed by flow cytometric analysis of DNA content/cell cycle distribution (A) or Western blot analysis for confirmation of KSR1 knockdown (B). C, IEC-18 cells were transfected with the indicated siRNAs 72 h prior to treatment with PMA for 2 h and Western blot analysis for the indicated proteins. D, Western blot analysis of KSR2 expression in IEC-18 cells, with rat brain lysate included as a positive control. Data are representative of three independent experiments. PMA, phorbol 12-myristate 13-acetate.
Association of PKCα with ERK signaling pathway components
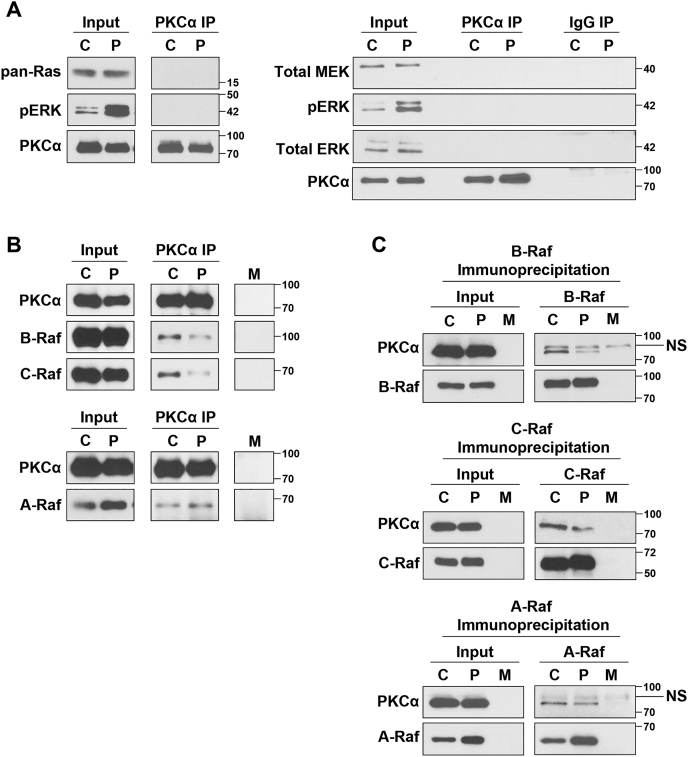
Having excluded a role for KSR scaffolds in regulation of growth suppressive ERK signaling, we examined the ability of PKCα to form complexes with components of the ERK pathway. Western blot analysis of PKCα immunoprecipitates using an antibody that recognizes all Ras isoforms failed to detect interaction between PKCα and Ras, either before or after activation of growth-inhibitory signaling with PMA (Fig. 5A). Similar analysis also failed to detect interaction between PKCα and MEK, total ERK, or pERK in unstimulated or PMA-treated cells (Fig. 5A).
Figure 5.
Interaction of PKCα with components of the RAS-ERK pathway.A and B, IEC-18 cells were treated with vehicle (C) or 100 nM PMA (P) for 10 min prior to lysis and immunoprecipitation (IP) of protein complexes using a monoclonal anti-PKCα antibody (PKCα IP). Immunoprecipitates were analyzed for the presence of the indicated proteins by Western blotting. Input represents 10% of the lysate used for immunoprecipitation. C, as in (A) except that complexes were immunoprecipitated with antibodies against A-Raf, B-Raf, or C-Raf as indicated. M: Mock immunoprecipitation using lysis buffer instead of cell extracts to detect antibody-derived nonspecific bands. NS: Nonspecific band. Data are representative of at least three independent experiments.
Mammalian cells express three Raf isoforms, A-Raf, B-Raf, and C-Raf (48, 49). In contrast to the lack of interaction between PKCα and Ras, MEK, or ERK, immunoprecipitation experiments clearly detected association of both B-Raf and C-Raf with PKCα in unstimulated cells (Fig. 5B). These interactions were confirmed in reciprocal experiments in which B-Raf or C-Raf immunoprecipitates were probed for the presence of PKCα (Fig. 5C). Notably, PMA treatment reduced the interaction of PKCα with B-Raf and C-Raf by 10 min (Fig. 5, B and C), indicating that activation of growth-inhibitory ERK signaling is associated with dissociation of PKCα–B-Raf and PKCα–C-Raf complexes.
Reciprocal immunoprecipitation experiments also consistently detected interaction between A-Raf and PKCα in unstimulated cells, although the signal was weaker than for the other Rafs (Fig. 5, B and C). It was not possible to determine the effects of PKCα activation on the strength of this interaction because of the ability of PMA to increase the levels of A-Raf in the lysates used for immunoprecipitation (Fig. 5, B and C, input). This increase, which occurred by 10 min of PMA treatment, was not seen in whole-cell lysates obtained using SDS solubilization buffer (e.g., Fig. 6) and, therefore, does not reflect an increase in A-Raf expression. Instead, the effect reflects increased solubility of PKCα-stimulated A-Raf in the NP-40/Igepal-630 containing buffer used for immunoprecipitation. While the mechanism(s) underlying the increased solubility of A-Raf remain to be determined, these data demonstrate that PKCα forms a complex with all three Raf isoforms and that, at least for B-Raf and C-Raf, these complexes dissociate during activation of growth-suppressive Ras-ERK signaling.
Figure 6.
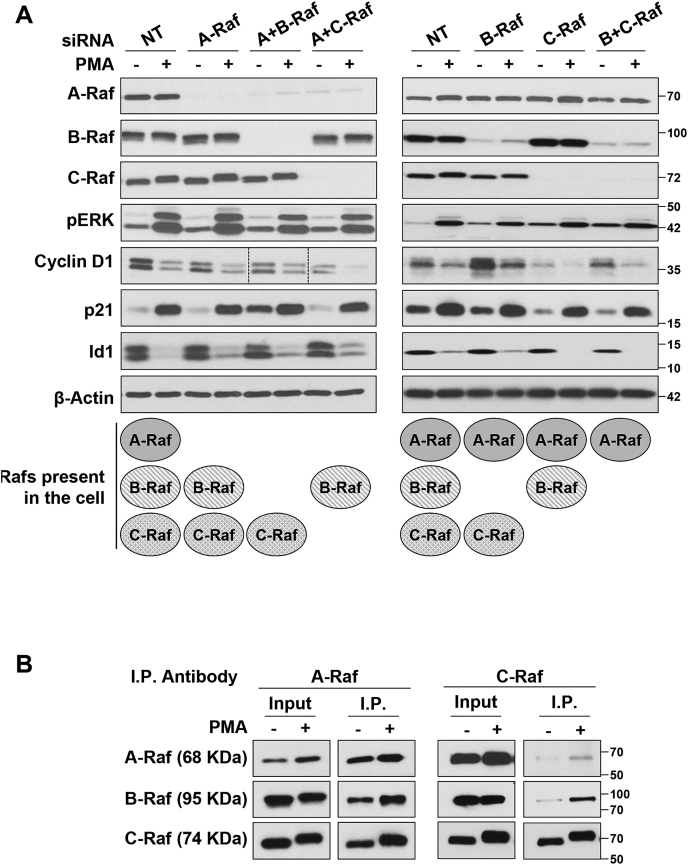
Raf proteins have redundant functions in PKCα-induced cell growth arrest in IEC-18 intestinal epithelial cells.A, IEC-18 cells were transfected with nontargeting siRNA (NT) or siRNA targeting the indicated Raf proteins 48 h prior to treatment with 100 nM PMA for 2 h. Expression of the indicated proteins was then assessed by Western blotting. The data are from single immunoblots; lanes between the vertical lines in the cyclin D1 blot (left panel) show a shorter exposure of the blot for clarity. B, IEC-18 cells were treated with vehicle (−) or 100 nM PMA for 30 min, followed by immunoprecipitation using A-Raf or C-Raf antibodies. Immunoprecipitated samples (I.P.) and 10% if the original lysate (Input) were subjected to Western blot analysis for the presence of A-Raf, B-Raf, and C-Raf. Data are representative of at least three independent experiments. PMA, phorbol 12-myristate 13-acetate.
Raf proteins have redundant functions in PKCα-induced growth inhibitory ERK signaling
The three Raf isoforms function as homodimers and/or heterodimers to regulate downstream signaling (49). To determine if PKCα-induced antiproliferative ERK signaling is specified by differential activation of individual Raf isoforms and/or formation of specific Raf homodimers or heterodimers, siRNA knockdown experiments were performed. Single knockdown of A-Raf, B-Raf, or C-Raf did not affect the ability of PMA to activate ERK, downregulate cyclin D1 or Id1, or upregulate p21Cip1 (Fig. 6A), excluding a requirement for a specific Raf isoform or heterodimer for these effects. To test if specific Raf homodimer(s) mediate the effect, simultaneous knockdown of all possible Raf pairs was performed, effectively leaving the cells with only A-Raf, B-Raf, or C-Raf (Fig. 6A) (note that simultaneous knockdown of all three Raf isoforms was not possible due to toxicity). Interestingly, dual Raf knockdown also failed to block the effects of PMA treatment on cyclin D1, Id1, or p21Cip1 (Fig. 6A). Together, the data point to (a) redundancy in the ability of Raf isoforms to mediate growth suppressive ERK activation and (b) the ability of Raf homodimers to mediate PKCα-induced growth-suppressive signaling.
The ability of all Raf isoforms to mediate PMA/PKCα-induced antiproliferative ERK signaling was further supported by analysis of Raf heterodimer formation in immunoprecipitation experiments. Analysis of C-Raf immunoprecipitates revealed that PMA/PKCα promotes the association of C-Raf with both A-Raf and B-Raf (Fig. 6B, C-Raf IP right panel). A similar increase in C-Raf–A-Raf and B-Raf–A-RAF complexes following PMA treatment was detected in A-Raf immunoprecipitates (Fig. 6B, A-Raf IP left panel). Interpretation of the A-Raf immunoprecipitation results is complicated by the ability of PKCα/PMA to increase the levels of A-Raf in lysates used for immunoprecipitation (see previous text). Nonetheless, these data indicate that PMA/PKCα increases the levels of A-Raf containing heterodimers in Igepal 630-soluble compartments. Although technical difficulties precluded analysis of B-Raf immunoprecipitates by Western blotting, the data collectively indicate that PMA/PKCα induces the formation of A-Raf/B-Raf, A-Raf/C-Raf, and B-Raf/C-Raf heterodimers and that there is redundancy in the ability of Raf isoforms and dimers to mediate PKCα-induced growth-suppressive ERK signaling. Thus, selective Raf activation does not appear to specify growth-suppressive ERK signaling pathways.
Role of Ras guanine exchange factors in PKCα-induced growth inhibitory ERK signaling
SOS1 and SOS2 are not required for PKCα-induced activation of the Ras-ERK pathway
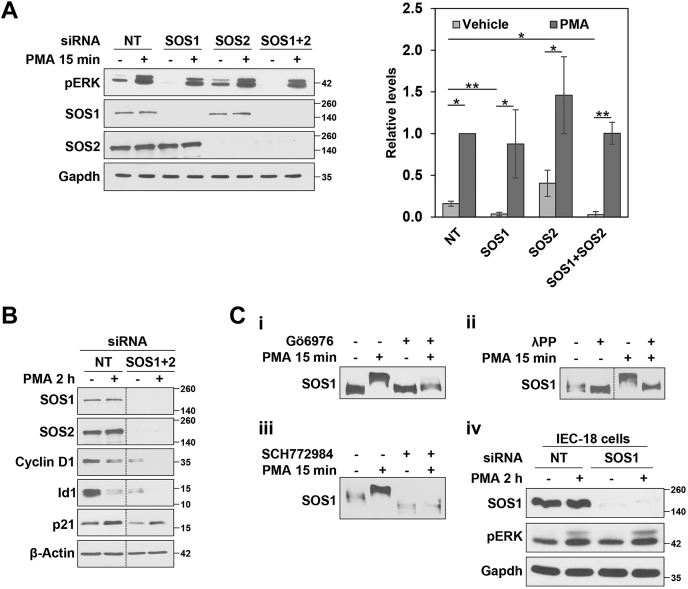
Having established that PKCα signaling intersects the ERK pathway at Ras, we investigated the role of Ras-GEFs in PKCα-induced growth-inhibitory ERK activation. Since RNA-Seq analysis indicated that IEC-18 cells do not express RasGRFs (Table 1), these studies focused on SOS and RasGRP Ras-GEFs (50). Consistent with the involvement of SOS1 and SOS2 in growth-promoting signaling (51), combined knockdown of SOS1 and SOS2 markedly reduced the expression of cyclin D1 and Id1 (Fig. 7, A and B). However, single or double knockdown of SOS1 and SOS2 failed to prevent PMA-induced ERK activation (Fig. 7A) and did not affect the ability of PMA/PKCα to further downregulate cyclin D1 and Id1 or to upregulate p21Cip1 (Fig. 7B), indicating that PKCα induces growth-inhibitory Ras-ERK signaling independently of SOS Ras-GEFs.
Table 1.
mRNA expression of Ras-GEFs and Ras isoforms in IEC-18 cells (revealed by RNA-Seq analysis)
| Gene Name | Expression relative to Gapdh (×103) |
|---|---|
| Sos1 | 3.27 |
| Sos2 | 3.20 |
| Rasgrp1 | 0.04 |
| Rasgrp2 | 0.53 |
| Rasgrp3 | 4.13 |
| Rasgrp4 | 0.00 |
| Rasgrf1 | 0.00 |
| Rasgrf2 | 0.00 |
| Hras | 18.03 |
| Kras | 11.73 |
| Nras | 27.24 |
Figure 7.
Role of SOS1/2 RasGEFs in PKCα-induced growth inhibitory ERK signaling.A and B, IEC-18 cells were transfected with nontargeting (NT) siRNA or siRNA targeting SOS1 and/or SOS2 as indicated. After 48 h, cells were treated with 100 nM PMA for 15 min (A) or 2 h (B) and analyzed for the indicated proteins by Western blotting. The graph to the right of the blots in (A) shows densitometric analysis of relative levels of pERK normalized to loading control (±SD, n = 3. ∗p ≤ 0.05; ∗∗p ≤ 0.01). All data in (B) are from single immunoblots; the vertical lines indicate rearrangement of the blot for clarity. C, i, IEC-18 cells were pretreated with vehicle (−) or 4 μM Gö6976 to inhibit PKCα activity, followed by 15 min treatment with vehicle (−) or 100 nM PMA as indicated. Lysates were run on Phos-tag gels and analyzed for SOS1 by immunoblotting. ii, IEC-18 cells were treated with 100 nM PMA for 15 min. Cells were then lysed, and half of each lysate was treated with lambda phosphatase (λPP) as indicated before being subjected to Phos-tag gel analysis. iii, Phos-tag gel analysis of cells treated with PMA in the absence or presence of the ERK inhibitor SCH772984 as indicated. iv, as in (A), except that (NT) siRNA or SOS1 siRNA transfected cells were treated as indicated for 2 h. Data are representative of at least three independent experiments, except C(ii) and C(iii) which are representative of two independent experiments. PMA, phorbol 12-myristate 13-acetate.
PKCα signaling may promote ERK-dependent feedback inhibition of SOS1
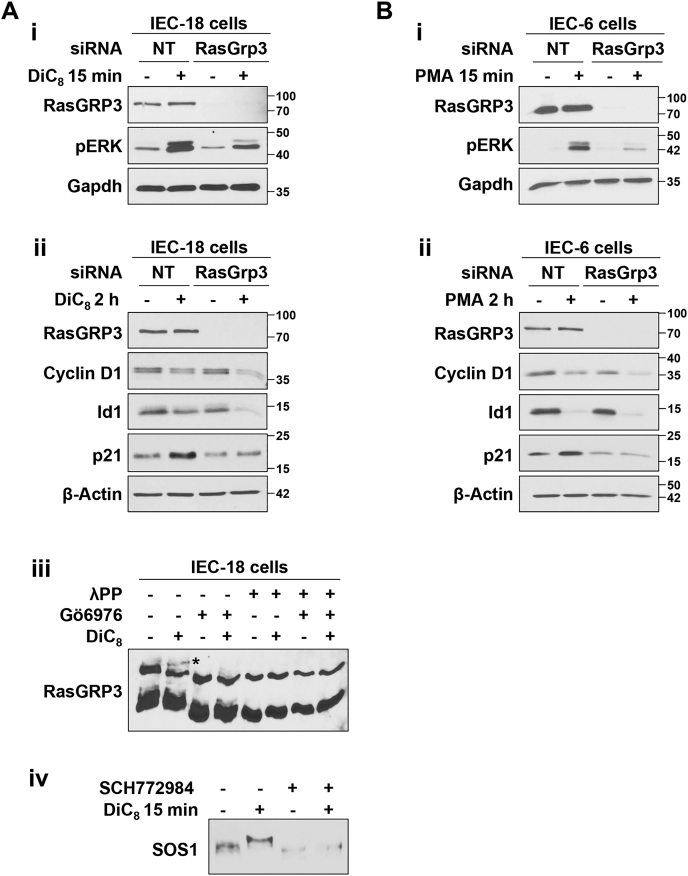
While our analysis identified SOS1/2-independent growth suppressive PKCα-ERK signaling, the data did not exclude the possibility that antiproliferative PKCα-ERK signaling may also intersect growth-promoting ERK pathways through inhibition of SOS proteins. ERK-dependent phosphorylation of SOS1 is a major mechanism for negative feedback of growth factor–induced Ras-ERK signaling. ERK phosphorylates SOS1 on Ser1132, Ser1167, Ser1178, and Ser1193 in the C-terminal domain, which inhibits SOS1 function by reducing its association with Grb2, Shc, and the EGF receptor (51, 52, 53). Negative feedback of Ras stimulation can also result from ERK→Rsk-mediated phosphorylation of SOS1 (e.g., on Ser1134 and Ser1161) (54, 55). Follow-up studies, therefore, examined the ability of PMA/PKCα to induce ERK-dependent inhibitory phosphorylation of SOS1/2 (51). Since antibodies targeting these phosphosites are not commercially available, we used Phos-tag gels for the analysis. These SDS-polyacrylamide gels contain phosphate-binding metal chelate complexes that specifically retard migration of phosphorylated proteins during electrophoresis (56). While PMA/PKCα had no effect on SOS2 migration in Phos-tag gels (data not shown), PMA treatment induced a PKCα-dependent change in SOS1 phosphorylation, indicated by a mobility shift that was prevented by the PKCα selective inhibitor Gö6976 (Fig. 7Ci) and abrogated by λ-phosphatase treatment (Fig. 7Cii). Notably, PMA-induced SOS1 phosphorylation was also prevented by inhibition of ERK with SCH772984, indicating that the effect is mediated by ERK (Fig. 7Ciii). Since ERK-induced phosphorylation of SOS leads to downregulation of growth factor signaling (57), these data point to the possibility that PKCα can dampen pro-proliferative signaling in intestinal cells by promoting ERK-mediated inhibitory phosphorylation of SOS1. However, it is clear that the activation of antiproliferative Ras-ERK by PKCα is SOS1 independent, as further supported by the ability of PMA to induce prolonged (>2 h) phosphorylation of ERK in cells with SOS1 knockdown (Fig. 7Civ).
PKCα-induced activation of the Ras-ERK pathway is partially dependent on RasGRP3
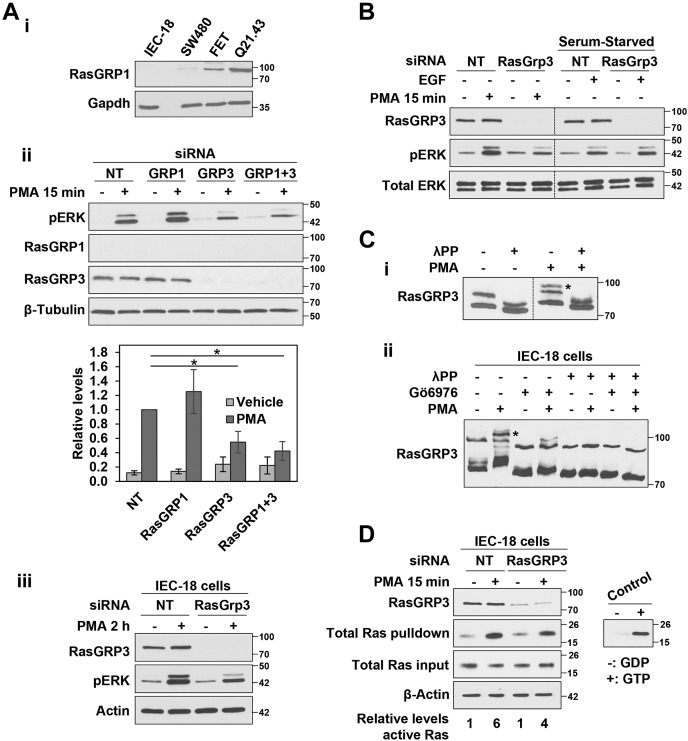
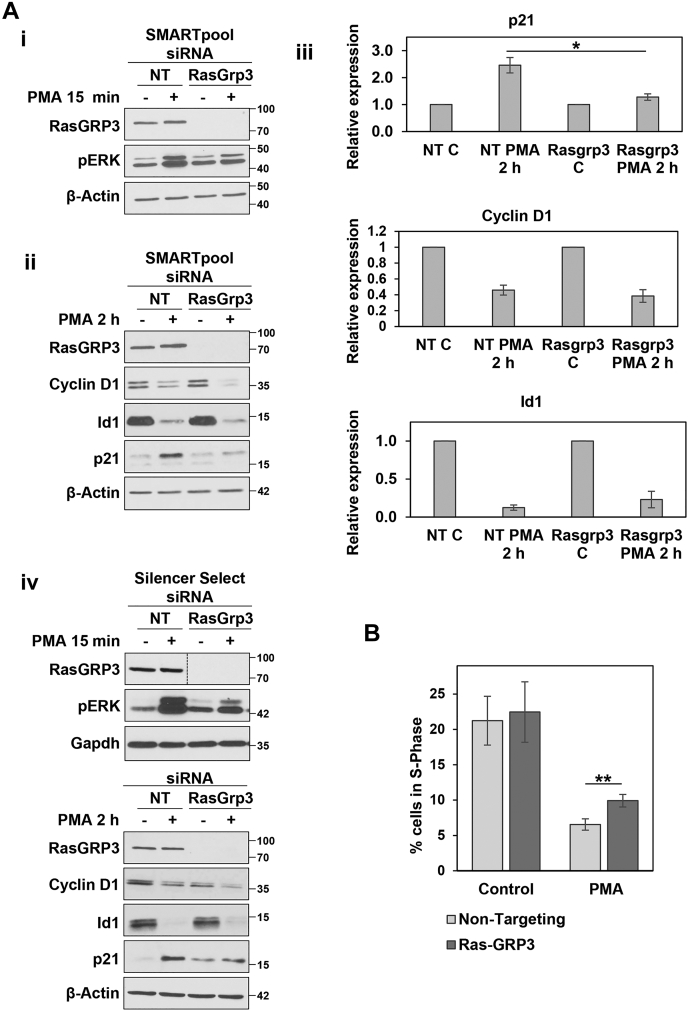
IEC-18 cells do not express RasGRP1 or RasGRP4 (Table 1 and Fig. 8Ai), but RasGRP2 and RasGRP3 were detected in these cells (Table 1 and Fig. 8Aii). Since RasGRP2 does not activate Ras (58), we focused our analysis on RasGRP3. In contrast to SOS1/2, siRNA-mediated knockdown of RasGRP3 consistently reduced the level of ERK activation seen following PMA treatment (Figs. 8, Aii, iii and B and 9Ai), indicating that this GEF is partially responsible for mediating PKCα activation of ERK. RasGRP3 deficiency reduced the ability of PKCα to induce ERK activation at early times (15 min) (Fig. 8Aii) and to sustain ERK activity at 2 h (Fig. 8Aiii), although ERK activity was still elevated relative to control cells at this later time. While RasGRP3 knockdown inhibited ERK activation by PMA/PKCα, the absence of this GEF did not affect ERK activation by EGF (Fig. 8B), supporting a role for RasGRP3 in antiproliferative rather than growth-promoting ERK signaling in intestinal epithelial cells. As mentioned previously, RasGRP1 is not expressed in control IEC-18 cells and a compensatory role for RasGRP1 that has been suggested in other systems (59, 60) was excluded since (a) Western blot analysis failed to detect RasGRP1 in control or RasGRP3 knockdown cells and (b) RasGRP1 siRNA, either alone or in combination with RasGRP3 knockdown, did not affect the ability of PMA to activate ERK (Fig. 8Aii).
Figure 8.
RasGRP3 is involved in PKCα-induced growth inhibitory ERK signaling.A, i, Western blot analysis confirms that IEC-18 cells do not express RasGRP1. SW480 and FET colon cancer cells and Q21.43 patient-derived colon cancer organoids are used as positive controls for expression of RasGRP1. ii, IEC-18 cells were transfected with NT siRNA or ON-TARGETplus SMARTpool siRNA targeting RasGRP1 and/or RasGRP3 as indicated 24 h prior to treatment with PMA for 15 min. The graph under the blots shows densitometric analysis of relative levels of pERK normalized to loading control (±s.d., n = 3. ∗p ≤ 0.05). iii, as in (ii), except that cells were treated with PMA for 2 h. B, IEC-18 cells were transfected with NT or ON-TARGETplus SMARTpool RasGRP3-targeted siRNA and treated for 15 min with PMA (in full serum), or with EGF (after serum-starvation), before Western blot analysis for the indicated proteins. The data are from single immunoblots; vertical lines are included for clarity. C, i, IEC-18 cells were treated with 100 nM PMA for 15 min before lysis and treatment with lambda phosphatase (λPP) as indicated. Lysates were run on Phos-tag gels and analyzed by immunoblotting. ∗ Slower mobility RasGRP3 species induced by PMA treatment. Data are from a single membrane; the vertical line indicates rearrangement of the blot for clarity. ii, IEC-18 cells were pretreated with 4 μM Gö6976 to inhibit PKCα activity, followed by 15 min treatment with 100 nM PMA as indicated. Cells were then lysed and lysates were divided and treated with lambda phosphatase as indicated prior to Phos-tag gel/immunoblot analysis. ∗ As in C(i). D, nontargeting (NT) and RasGRP3 siRNA transfected cells were treated with vehicle or PMA as in (A) before lysis and isolation of active GTP-bound Ras by GST-Raf1-RBD pulldown. Levels of the indicated proteins in the original lysates and Raf1-RBD pulldowns (Total Ras pulldown) were detected by Western blotting. Detection of Total Ras used a pan-Ras antibody that targets all three Ras proteins. The panel to the right shows the signal obtained following pulldown of GDP- and GTP-loaded Ras as negative and positive controls, respectively. All data are representative of at least three independent experiments. PMA, phorbol 12-myristate 13-acetate.
Figure 9.
RasGRP3 partially mediates PKCα-induced antiproliferative signaling.A, loss of RasGRP3 significantly inhibits PKCα-induced activation of ERK and upregulation of p21Cip1. IEC-18 cells were transfected with nontargeting siRNA (NT) or ON-TARGETplus SMARTpool siRNA targeting RasGRP3 (Dharmacon) as indicated. After 48 h, cells were treated with 100 nM PMA for 15 min (i) or 2 h (ii) and analyzed for the indicated proteins by Western blotting. iii, densitometric quantification of p21Cip1, cyclin D1, and Id1 expression (normalized to β-actin) from three independent experiments. ∗p = 0.03. iv, cells were treated as in (A(i)) except that the siRNA targeting RasGRP3 was from Invitrogen (Silencer Select predesigned siRNA Rasgrp3). Data are from single immunoblots; the vertical line indicates rearrangement of the blot for clarity. B, IEC-18 cells were transfected with nontargeting siRNA (NT) or siRNA targeting RasGRP3 (ON-TARGETplus SMARTpool, Dharmacon). After 24 h, cells were treated with 100 nM PMA for 6 h, as indicated, and the percentage of cells in S-phase was assessed by flow cytometry. ∗∗p = 0.003, n = 4. PMA, phorbol 12-myristate 13-acetate.
Phosphorylation of RasGRP3 at Thr133 by conventional PKCs has been shown to be important for RasGRP3 activation in B cells (61, 62). Since a reliable reagent for detection of phospho-Thr133-RasGRP3 in rat cells is not available, we conducted Phos-tag gel analysis to determine if PMA/PKCα alters the phosphorylation of RasGRP3 in intestinal cells. Treatment of IEC-18 cells with PMA led to the appearance of a slower migrating RasGRP3 immunoreactive band following Phos-tag electrophoresis (Fig. 8C, asterisk). This shift was inhibited in cells treated with Gö6976 and was not seen when the lysates were treated with λ-phosphatase; thus, these data indicate that activation of antiproliferative ERK signaling is associated with PKCα-induced phosphorylation of RasGRP3 (Fig. 8C).
As we have previously reported (11), PKCα signaling activates Ras in IEC-18 cells, as determined by Ras activity assays using GST-Raf1-RBD fusion protein to pull down the activated GTP-bound form of Ras (Fig. 8D). Importantly, active Ras pull-down assays confirmed a role for RasGRP3 in Ras activation, since knockdown of RasGRP3 reduced the level of activated Ras seen in PMA-treated cells (Fig. 8D).
RasGRP3 is partially responsible for PKCα-induced antiproliferative signaling in intestinal epithelial cells
Analysis of the effects of RasGRP3 knockdown revealed that distinct ERK-dependent mechanisms underlie the ability of PKCα to regulate pro-proliferative and antiproliferative proteins. While RasGRP3 knockdown had no effect on PMA-induced downregulation of cyclin D1 or Id1, knockdown of this Ras-GEF with two different siRNA reagents (OnTarget SMARTpool or Silencer Select) markedly impaired the ability of PMA to upregulate of p21Cip1 (Fig. 9Ai–iv). These data indicate that induction of p21Cip1 by PMA/PKCα is mediated by a RasGRP3-dependent ERK signaling pathway.
To confirm a role for RasGRP3 in growth-inhibitory PKCα-ERK signaling, we examined whether RasGRP3 knockdown inhibited the ability of PMA/PKCα to induce G1→S phase arrest, as determined by changes in the percentage of cells in S-Phase (Fig. 9B). Knockdown of RasGRP3 did not affect the level of S-phase in untreated cells and did not prevent the ability of PMA treatment to reduce the number of cells in S-phase, consistent with the presence of antiproliferative RasGRP3-independent PKCα signaling that channels through cyclin D1 and Id1. However, the growth-inhibitory effects of PMA/PKCα were significantly reduced by RasGRP3 deficiency, as indicated by a highly consistent increase in the proportion of S-phase cells in PMA-treated RasGRP3 knockdown cells compared with PMA-treated control cells (p = 0.003, n = 4; Fig. 9B). Thus, inhibition of the PKCα–RasGRP3–ERK–p21Cip1 axis dampened the ability of PKCα signaling to induce G1→S phase arrest in intestinal cells. Together, the data indicate that there are at least two ERK signaling pathways that contribute to the growth-suppressive effects of PKCα in intestinal epithelial cells: a PKCα–RasGRP3–ERK signaling axis that induces upregulation of p21Cip1 and additional RasGRP3-independent PKCα-ERK signaling module(s) that lead to downregulation of cyclin D1 and Id1. Consistent with the requirement for sustained ERK activity in PKCα-induced growth arrest, the RasGRP3-independent PKCα-ERK signaling module can also sustain enhanced ERK signaling for more than 2 h (Fig. 8Aiii).
H-Ras mediates PMA/PKCα-induced upregulation of p21Cip1
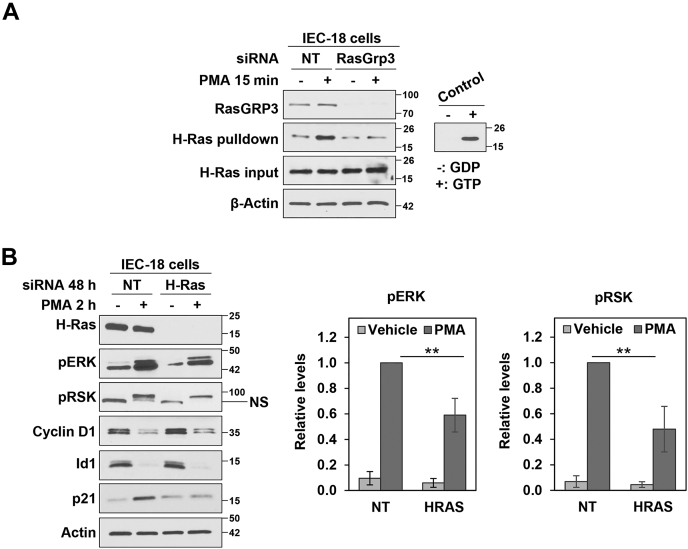
To further characterize growth-suppressive ERK signaling, we next addressed the involvement of individual members of the Ras family. These studies focused on H-Ras based on evidence that this isoform is highly expressed in postmitotic/mature intestinal cells (63) and that H-Ras signaling reduces proliferation and induces the expression of differentiation markers (e.g., brush border hydrolases) in CaCo-2 colon cancer cells (3, 6). As shown in Figure 10A, active Ras pull-down assays confirmed that PMA/PKCα promotes GTP loading/activation of H-Ras in IEC-18 cells. siRNA-mediated knockdown of H-Ras in IEC-18 cells reduced ERK activation in response to PMA, confirmed by a reduction in phosphorylation of RSK (Fig. 10B). Thus, H-Ras appears to be partially responsible for activation of ERK by PKCα. Notably, PMA-induced H-Ras activation was dependent on RasGRP3, as indicated by the inability of PMA to activate H-Ras in RasGRP3 knockdown cells (Fig. 10A), pointing to a role for H-Ras in PKCα-induced p21Cip1 upregulation. Remarkably, the effects of H-Ras knockdown on downstream proteins phenocopied those of loss of RasGRP3 on all growth regulatory targets analyzed: while no effect was seen on PMA-induced downregulation of cyclin D1 or Id1, PMA-induced upregulation of p21Cip1 was abrogated in H-Ras knockdown cells (Fig. 10B). Together, these data further delineate growth-suppressive PKCα-ERK signaling by (a) indicating that upregulation of p21Cip1 is mediated by RasGRP3-induced H-Ras activation and (b) determining that K-Ras and/or N-Ras can mediate downregulation of cyclin D1 and Id1.
Figure 10.
H-Ras is required for PKCα-induced p21Cip1upregulation.A, IEC-18 cells were transfected with nontargeting siRNA (NT) or siRNA targeting RasGRP3 for 24 h and subjected to GST-Raf1-RBD pulldown analysis of levels of active GTP-bound H-Ras. Levels of the indicated proteins in the original lysates and GST-Raf-RBD pulldowns (H-Ras pulldown) were detected by Western blotting. The panel to the right shows negative and positive controls. B, IEC-18 cells were transfected with nontargeting (NT) siRNA or siRNA targeting H-Ras. About 48 h later, cells were treated with 100 nM PMA for 2 h and lysates were analyzed for expression of the indicated proteins by Western blotting. NS: nonspecific band. The graphs to right of the blots show densitometric analysis of relative levels of pERK and pRSK normalized to loading control (±s.d., n = 4. ∗p ≤ 0.05). Data are representative of two (A) or at least three (B) independent experiments.
The growth inhibitory PKCα-RasGRP3-H-Ras-ERK-p21Cip1 signaling module is also triggered by the PKCα agonist, DiC8, and is seen in other intestinal epithelial cells
To exclude the possibility that the differential effects of RasGRP3 on downstream targets of growth-inhibitory PKCα-ERK signaling are restricted to phorbol ester treatment, we tested the effects of the PKC agonist, DiC8, a short chain DAG that, as discussed previously, mimics the effects of endogenous DAG and promotes PKCα-induced cell cycle arrest in IEC-18 cells (9, 10, 45). RasGRP3 knockdown reduced ERK activation and inhibited p21Cip1 upregulation following DiC8 treatment of IEC-18 cells, without affecting downregulation of cyclin D1 or Id1 (Fig. 11Ai, ii). DiC8 also induced a slower migrating RasGRP3 band in Phos-tag gels (Fig. 11Aiii, asterisk) that was prevented by the PKCα-selective inhibitor Gö6976 and by λ-phosphatase, indicating that DiC8 also recapitulates the effects of PMA on RasGRP3 phosphorylation. Finally, this short chain DAG recapitulated the ability of PMA to induce ERK-dependent phosphorylation of SOS1 (Fig. 11Aiv; cf. Fig. 7Ciii), demonstrating that none of the observed effects of PKCα activation are phorbol ester–specific but can also be elicited by this “physiological” agonist.
Figure 11.
The PKCα–RasGRP3–p21Cip1signaling axis is induced by the short chain DAG, DiC8, and is also present in IEC-6 intestinal epithelial cells.A, i and ii, IEC-18 cells were transfected with nontargeting siRNA (NT) or Silencer Select siRNA targeting RasGRP3 (Invitrogen) 24 h prior to treatment with 20 μg/ml DiC8 for 15 min (i) or 2 h (with DiC8 replenished at 1 h) (ii) and analysis of the indicated proteins by Western blotting. iii, cells were treated with DiC8 for 15 min in the absence or presence of 4 μM Gö6976 prior to lysis. Lysates were treated with lambda phosphatase as indicated, run on Phos-tag gels, and analyzed by immunoblotting. The slower mobility RasGRP3 species seen in Figure 8 is also induced by DiC8 (∗) and is inhibited by the PKCα-selective inhibitor, Gö6976. iv, IEC-18 cells were treated with DiC8 for 15 min in the absence or presence of SCH772984 as indicated and phosphorylation of SOS1 was analyzed by Phos-tag gel electrophoresis and immunoblotting. B, IEC-6 cells were transfected as in (A) and treated with 100 nM PMA for 15 min (i) or 2 h (ii) prior to Western blot analysis. All data are representative of three independent experiments except (A(iv)) which is representative of two independent experiments. DAG, diacylglycerol; DiC8, 1,2-dioctanoyl-sn-glycerol; PMA, phorbol 12-myristate 13-acetate.
Importantly, the RasGRP3-dependent pathway that is required for upregulation of p21Cip1, but not downregulation of cyclin D1 or Id1, was also observed in PMA-treated IEC-6 cells (Fig. 11B), an independently derived nontransformed intestinal epithelial cell line (64, 65) that was previously used to demonstrate a role for growth-promoting ERK signaling in intestinal epithelial cells (3). Thus, the presence of a PKCα-induced RasGRP3–H-Ras–dependent growth-inhibitory ERK signaling axis that leads to upregulation of p21Cip1, as well as RasGRP3–H-Ras–independent ERK signaling module(s) that mediate PKCα-induced downregulation of cyclin D1 and Id1, appears to be a general characteristic of intestinal epithelial cells.
Discussion
In the present study, we have defined antiproliferative ERK signaling pathways downstream of PKCα using intestinal epithelial cells as a model. The finding that PKCα signaling intersects the ERK pathway by inducing the activation of Ras addresses longstanding confusion in the field regarding the requirement for Ras activity in PKC-mediated signaling to ERK. The dependence on Ras activity is consistent with findings from the Marshall group (31) showing that C-Raf activation by PKC requires the formation of Ras-GTP/C-Raf complexes. Our data further exclude a role for direct activation of Raf (30) or MEK (33) in ERK activation by PKCα in the models used in this study, while shedding new light on the involvement of individual Ras proteins in PKCα signaling. We demonstrate for the first time that PKCα specifically activates H-Ras for upregulation of the negative cell cycle regulator p21Cip1 and partial cell cycle arrest. An antiproliferative function of H-Ras in intestinal epithelial cells is consistent with high expression of this isoform in postmitotic/mature intestinal epithelial cells (63) and the ability of H-Ras to reduce proliferation and induce differentiation markers in Caco-2 colon cancer cells (3, 6). Interestingly, H-Ras is the least frequently mutated Ras isoform in human cancer (4% versus 85% for K-Ras) and activating mutations in H-Ras are not observed in colorectal cancer (66), perhaps reflecting the growth-inhibitory function of this isoform in intestinal epithelial tissue.
The finding that H-Ras knockdown does not affect PKCα-induced downregulation of cyclin D1 and Id1 points to the existence of at least one additional growth-suppressive ERK signaling axis downstream of PKCα in intestinal cells. Data showing that PKCα-induced cyclin D1 and Id1 downregulation is Ras-ERK dependent support the conclusion that PKCα is also able to activate ERK through K-Ras and/or N-Ras. Furthermore, since loss of H-Ras only partially blocked the ability of PKCα to induce cell cycle arrest, these distinct ERK pathways appear to collaborate in mediating PKCα-induced cell cycle withdrawal in the intestinal epithelium. Engagement of multiple downstream pathways is a common feature of growth regulatory signaling. For example, antiproliferative effects of TGFβ are mediated by multiple factors downstream of both canonical SMAD-mediated signaling and noncanonical signaling involving Ras-ERK, p38, and JNK cascades, among others (67). Thus, the identification of multiple signaling nodes downstream of PKCα supports the importance of PKCα in regulation of intestinal homeostasis, while pointing to its potential role as a signaling nexus for coordination of growth-suppressive pathways in this tissue.
The finding that the Ras-GEF, RasGRP3, is required for H-Ras activation and p21Cip1 induction in intestinal epithelial cells provides the first evidence for a PKCα–RasGRP3–H-Ras–Raf–MEK–ERK signaling axis. RasGRP3 has been shown to activate H-Ras in other systems (e.g., 293T cells), and in vitro studies with the RasGRP3 catalytic domain showed that RasGRP3 can promote GDP release from H-Ras (68, 69, 70). Like PKCs, the RasGRPs have a DAG/phorbol ester–binding C1 domain and DAG/PMA redistributes RasGRP3 to the plasma membrane to induce Ras-GTP loading (71, 72). However, studies in lymphocytes have revealed that PKCs can phosphorylate RasGRP3 at Thr133 (62, 73) and that PKC-mediated phosphorylation is required for RasGRP3 activation and downstream signaling (61). These published findings, combined with our demonstration of PKCα-dependent phosphorylation of RasGRP3 in intestinal epithelial cells (Figs. 8 and 11), suggest a model in which DAG/phorbol ester treatment leads to membrane concentration of PKCα and RasGRP3, facilitating RasGRP3 phosphorylation and activation by the kinase. Interestingly, RasGRP family members have been implicated in negative regulation of cell proliferation in intestinal epithelial cells, with RasGRP1 opposing EGFR–SOS1–Ras signals in crypt cells through a negative feedback loop (74, 75). Ras-GRP1 also restricts proliferation of colorectal cancer cells and low levels of RasGRP1 predict poor clinical outcome in colorectal cancer patients (74, 75). Since RasGRP1 is not expressed in IEC-18 intestinal crypt-like cells (Fig. 8), the data collectively indicate that both RasGRP1 and RasGRP3 have anti-proliferative functions in the intestinal epithelium, functions that are consistent with frequent loss of both proteins in colorectal cancer cells (refs. (74, 75), and TCGA data).
Although details of the ERK pathway(s) that mediate downregulation of cyclin D1 and Id1 remain to be elucidated, a potential contributing mechanism involves the ability of PMA/PKCα to induce ERK-dependent phosphorylation of SOS1. SOS1 is subject to feedback phosphorylation by ERK or ERK→Rsk at multiple sites, and ERK-regulated phosphorylation negatively modulates SOS1 interaction with Grb2, Shc, and the EGF receptor to dampen growth factor signaling (57). Since SOS proteins are known to play a key role in maintaining cyclin D1 and Id1 levels in proliferating cells, as indicated by a reduction in expression of these proteins in IEC-18 cells following knockdown of SOS1 and SOS2 (Fig. 7), inhibition of SOS1 function through PKCα-ERK “feedback” phosphorylation is a potential mechanism for the loss of cyclin D1 and Id1 seen following PKCα activation (Fig. 12B). However, the ability of PKCα to further downregulate cyclin D1 and Id1 in SOS1/SOS2 double-knockdown cells (Fig. 7) indicates that other SOS-independent mechanism(s) are also involved.
Figure 12.
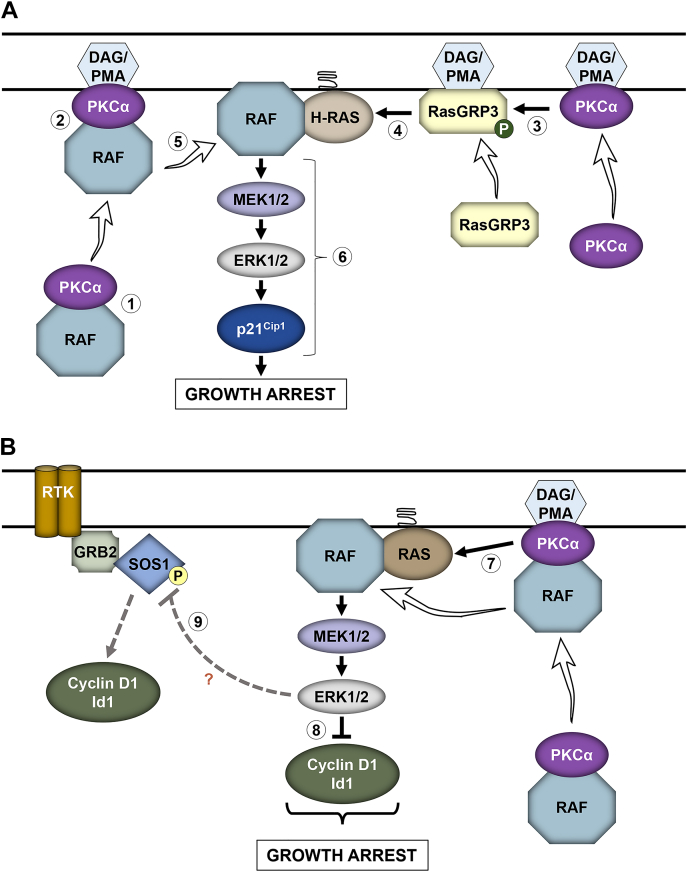
Model for PKCα-induced ERK-dependent cell cycle arrest in the intestinal epithelium.A, PKCα-induced RasGRP3–H-Ras–ERK–p21Cip1 signaling. In the resting state, inactive PKCα associates with inactive Raf proteins in the cytoplasm (1); PKC agonists, such as PMA or DAG (generated by ligand induced hydrolysis of membrane phospholipids), recruit PKCα and associated Raf proteins to the plasma membrane (2); PMA/DAG also recruit RasGRP3 to the plasma membrane, where it is phosphorylated (P) and activated by membrane-localized PKCα (3); activated RasGRP3 promotes GTP-loading and activation of H-Ras (4); PKCα dissociates from Raf proteins, which can then interact with nearby activated Ras (5) for efficient activation of the ERK signaling pathway and induction of the cell cycle inhibitory molecule p21Cip1 (6) and growth arrest. B, PKCα-induced Ras–ERK–cyclin D1/Id1 signaling. PKCα also activates K- and/or N-Ras through at least one other undefined mechanism (7), leading to ERK-dependent downregulation of cyclin D1 and Id1 (8). PKCα also promotes ERK-dependent phosphorylation of SOS1 (9), which may negatively modulate SOS1 interaction with Grb2, Shc, or the EGF receptor to dampen growth factor signaling, thereby downregulating growth factor–induced cyclin D1 and Id1 and inhibiting proliferation. DAG, diacylglycerol; PMA, phorbol 12-myristate 13-acetate.
While our studies revealed selectivity in the effects of Ras isoforms in mediating PKCα antiproliferative signaling, they revealed redundancy at the level of Raf (Fig. 6). Such redundancy has also been noted in pro-proliferative Raf signaling, although B-Raf/C-Raf heterodimers have the highest catalytic activity and appear to predominate in Ras-mediated signaling in many systems (49, 76, 77). While extensive evidence supports a role for Rafs in promoting cell proliferation (78, 79), antiproliferative roles have also been reported, with several studies linking high activity of Rafs with cell cycle arrest rather than proliferation (80, 81). Analysis of the Raf-interactome has identified PKCs among the proteins that form a complex with Raf proteins (78). To our knowledge, our studies provide the first evidence that PKCα forms complexes with all three Raf proteins in unstimulated cells that dissociate with PKCα activation (Fig. 5). While PKCα/Raf interactions are not sufficient to induce ERK activation, as indicated by the requirement for Ras in the pathway, they likely contribute to growth-inhibitory signaling. In unstimulated cells, both PKCα and Raf proteins reside in the cytoplasm in an inactive conformation and are activated by recruitment to the plasma membrane (22, 36). Thus, the interaction of PKCα with Rafs suggests the following model (Fig. 12, A and B): (a) in the resting state, inactive PKCα associates with inactive Raf proteins in the cytoplasm; (b) PKC agonists, such an PMA or DAG, recruit PKCα and associated Raf proteins to the plasma membrane; (c) PMA/DAG also recruit RasGRP3 to the plasma membrane, where it is phosphorylated and activated by membrane-localized PKCα; (d) activated RasGRP3 promotes GTP loading and activation of H-Ras and PKCα activates K- and/or N-Ras through at least one other mechanism; and (e) PKCα dissociates from Raf proteins, which can then interact with nearby activated Ras for efficient activation of growth-inhibitory ERK signaling. Our previous studies have also determined that PMA and DAG recruit PKCα to membrane subdomains such as lipid rafts (24, 82, 83); thus, the PKCα/Raf interaction may also ensure spatially restricted activation of ERK within a signaling milieu that supports growth-inhibitory signaling.
Although PKCα-induced growth inhibitory ERK signaling parallels growth-promoting ERK signaling in its requirement for canonical ERK pathway components, we show that these pathways differ in several respects. At the level of Ras activation, in contrast to the established role of SOS proteins in activation of Ras downstream of RTKs, SOS1/2 are dispensable for PKCα-induced ERK activation (Fig. 7). Conversely, while RasGRP3 contributed to ERK activation by PKCα, depletion of this Ras-GEF did not affect ERK activation by EGF (Fig. 8B). The duration of ERK activation following stimulation by PKCα agonists or EGF also differentiates these pathways: PKCα signaling induces prolonged ERK activation (>2 h) while ERK activation induced by EGF is transient (ref. (11) and Fig. 1E). Interestingly, both RasGRP3-mediated and RasGRP3-independent pathways stimulated by PKCα induce prolonged activation of ERK (>2 h, Figs. 7Civ and 8Aiii). Sustained ERK activation is a hallmark of antiproliferative signaling mediated by the Ras–Raf–MEK–ERK cascade (11, 80, 84, 85), and our previous studies have confirmed that prolonged ERK pathway activation, which is dependent on sustained activation of PKCα, is required for PKCα-induced intestinal epithelial cell cycle arrest (11). The finding that PKCα-ERK signaling was not affected by knockdown of KSR1 also points to the possibility that differential utilization of scaffolding proteins may direct the ERK signaling pathway to growth inhibition.
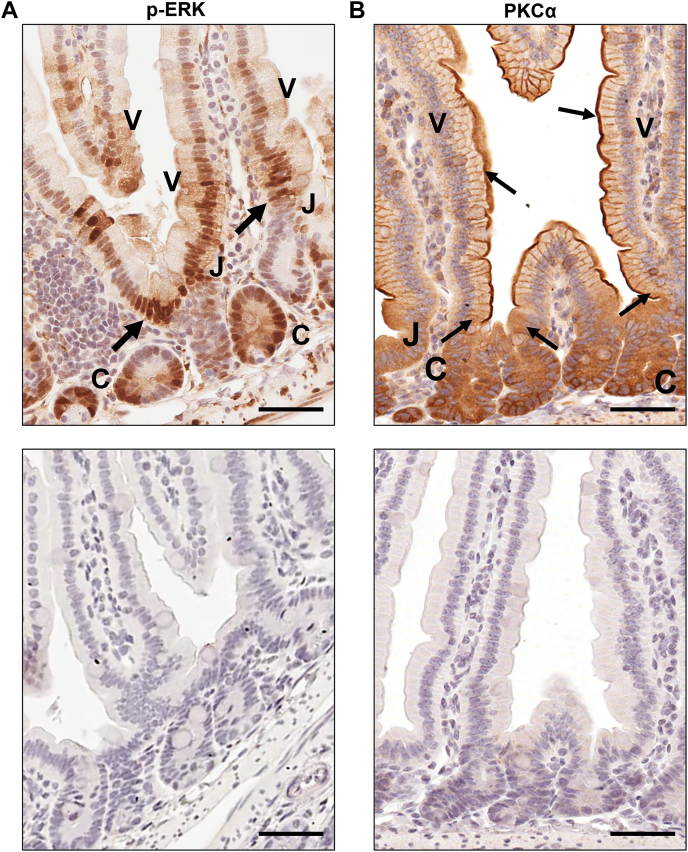
While the current study defines the pathway by which PKCα activates ERK for growth inhibition in intestinal epithelial cells, the factor(s) regulating PKCα activation in this tissue remain to be identified. We have shown that PKCα is cytosolic and inactive in proliferating cells of the intestinal crypt and that the enzyme translocates to the plasma membrane for activation coincident with growth arrest near the crypt–villus junction, remaining membrane associated/active along the length of the villus (refs. (9, 24, 25, 27) and Fig. 13B). A systematic analysis of canonical and noncanonical Wnt pathway components in the murine gastrointestinal tract (86) points to several potential upstream regulators of PKCα including Wnt-2b, Wnt-4, Wnt-5a, Frizzled 4, Frizzled 6, and Dickkopf (DKK) family members, all of which are expressed in epithelial or mesenchymal cells of the villus/colon surface. Notably, several of these candidates (Wnt-4 (87), Wnt-5a (88), Frizzled 6 (88), and DKK1 (89)) are also PKC/PKCα agonists in other systems. Like PKCα (90), these candidates have been reported to inhibit canonical Wnt signaling, which is required for intestinal epithelial cell proliferation and/or promote cell growth arrest/differentiation. Studies of the major signaling pathways regulating intestinal epithelial self-renewal also support a potential role for growth-suppressive members of the bone morphogenetic protein (BMP) and transforming growth factor β (TGFβ) families (91, 92). Although the role of these factors in upstream activation of PKCα remain to be defined, analysis of the self-renewing intestinal epithelium in vivo supports a role for growth-suppressive ERK signaling downstream of PKCα in this tissue. Activation of PKCα at the crypt/villus junction coincides with cyclin D1 and Id1 downregulation and p21Cip1 upregulation in the intestinal epithelium (9, 26). While phosphorylated active ERK is readily detected in the crypt epithelium (Fig. 13A), consistent with its established role in supporting proliferation within this compartment, a postmitotic role for the kinase is also indicated by the presence of phospho-ERK in the nonproliferating cells of the villus in both adult and fetal intestine (ref. (3, 5) and Fig. 13A). Notably, live imaging of the small intestine of transgenic mice ubiquitously expressing a FRET biosensor for ERK activity also detected active ERK in crypt and villus epithelial cells (93), with particularly strong ERK activity observed in the region of growth arrest at the crypt–villus transition (93). A role for ERK signaling in precise regulation of intestinal epithelial cytostasis is further supported by recent in vivo studies showing crypt cell hyperproliferation and crypt elongation following targeted deletion of ERK1 and ERK2 in the murine small intestinal epithelium (7, 8). Since strong ERK activity coincides with membrane translocation/activation of PKCα (24, 27), the spatiotemporal distribution of these molecules supports a model in which PKCα activates growth-suppressive ERK signaling to (a) drive cell cycle arrest at the crypt–villus junction and (b) maintain the postmitotic state on the villus (Fig. 13, A and B).
Figure 13.
PKCα membrane translocation/activation coincides with strong ERK activity at the crypt/villus junction in the mouse intestinal epithelium.A, upper panel, phospho-ERK immunostaining of formaldehyde-fixed paraffin-embedded murine intestinal epithelial tissue shows that ERK is active in both proliferating crypt cells (C) and postmitotic villus cells (V). Strong pERK staining is detected at the crypt/villus junction (J) where cells undergo growth arrest (arrows). Lower panel, negative control in which primary antibody was omitted demonstrates specificity of staining. B, upper panel, immunohistochemical analysis shows that PKCα is diffusely distributed in the cytoplasm and inactive in proliferating crypt cells (C) and is recruited to the plasma membrane, a hallmark of PKC activation, coincident with growth arrest at the crypt/villus junction (J) (arrows). PKCα remains membrane-associated/activated in postmitotic cells of the villus (V) (arrows). Lower panel, negative control demonstrates specificity of staining. Magnification Bar: 50 μm.
Experimental procedures
Cell culture
IEC-18 (ATCC CRL-1589) and IEC-6 (ATTC CRL-1592) rat intestinal crypt-like cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 4 mM L-glutamine, 1 mM sodium pyruvate, and 5 μg/ml insulin at 37 °C and in 5% CO2. For serum starvation, cells were cultured for 24 h in the same medium containing 0.5% FBS and lacking insulin. N-Ras−/−; H-Ras−/−; K-Rasf/f; and Ubiq-CreERT2 mESCs (“Rasless” mESCs) were cultured at optimum density (2.5–3 × 106 in 10 cm dish) on 0.1% gelatin (Millipore Sigma)-coated plates in DMEM supplemented with 15% FBS, 1000 U/ml leukemia inhibitory factor (Millipore Sigma), 1% Glutamax (Invitrogen), 0.1 mM nonessential amino acids, 1% penicillin/streptomycin, and 55 mM β-mercaptoethanol (44). For KO of K-Ras, fresh medium containing 1 μM 4-OHT (Sigma) was added daily for 7 days. Medium was changed every day, and cells were subcultured every second or third day depending on confluency.
Drug treatments and reagents
For PKC/PKCα activation, cells were treated with 100 nM PMA (Sigma) dissolved in ethanol or 20 μg/ml DiC8 (Cayman Chemicals) dissolved in acetonitrile. DiC8 was added repeatedly (every hour) in fresh medium to compensate for its rapid metabolism in cells as we have described (24). EGF was dissolved in PBS and was added to the culture medium at 50 to 100 ng/ml for the indicated times. For inhibitor treatments, cells were pretreated for 1 h with the indicated concentrations of BIM I (Calbiochem), Gö6976 (Calbiochem), SCH772984 (SelleckChem), LY3009120 (Med Chem BioExpress), PD0325901 (SelleckChem), or salirasib (SelleckChem) dissolved in dimethyl sufoxide before addition of PKCα agonist, unless otherwise stated. 4-OHT was dissolved in ethanol prior to addition to the culture medium. The relevant vehicle was added to controls and the final concentration of solvents was ≤0.2%.
Antibodies
Anti-KSR1 (ab68483) and anti-PKCα (ab32376 used for immunoblot analysis; ab221611 used for immunoprecipitation) antibodies were from Abcam; anti-H-Ras (A19619), anti–pan-Ras (A4735), anti-RasGRP1 (A10495), and anti-PKCε (A2110) antibodies were from Abclonal; anti-KSR2 (H00283455-M08) antibody was from Abnova; anti-A-Raf (75804S), anti-B-Raf (14814S), anti–phospho-Ser259 C-Raf (9421), anti–phospho-Ser338 C-Raf (9427), anti-cyclin D1 (2978), anti-Gapdh (5174), anti–pan-Ras (3339S), anti-pERK (9106 and 4370), anti-pMEK (9121), anti–total ERK (9102), anti–total MEK (9122), anti-SOS1 (D3T7T), and anti-pRSK(S380) (9341) antibodies were from Cell Signaling Technology; anti-C-Raf (610151) and anti-p21Cip1 (556430) antibodies were from BD Pharma; anti-Id1 (BCH-1/195-14-50) antibody was from BioCheck; anti-pRSK(S380) (04-418) was from Millipore; anti-p21Cip1 (NBP2-29463) antibody was from Novus Biologicals; anti-RasGRP3 (sc-271068), anti-SOS2 (sc-393667), and anti-PKCδ (sc-8402) antibodies were from Santa Cruz Biotechnology; and anti-actin (A2066) antibody was from Sigma–Aldrich. Secondary antibodies were horseradish peroxidase–conjugated goat anti-rabbit IgG (AP132P, Millipore), horseradish peroxidase–conjugated goat anti-rabbit IgG (111-035-144, Jackson Immunoresearch Laboratories), and horseradish peroxidase–conjugated goat antimouse IgG (170-6516, Bio-Rad).
Western blot analysis
Cells were washed twice with ice cold PBS and lysed in 1% SDS lysis buffer (1% SDS, 10 mM Tris–HCl, pH 7.4). Equal amounts of protein were subjected to SDS-PAGE and transferred to nitrocellulose membrane as described previously (9). Membranes were stained with 0.1% fast green (Sigma–Aldrich) to confirm equal loading and even transfer. Following blocking with 5% milk or 5% bovine serum albumin (Sigma) in Tris-buffered saline containing 0.1% Tween-20 (TBS-T), membranes were incubated with primary antibody (in blocking buffer) overnight at 4 °C, washed (3 × 10 min in TBS-T), and incubated with horseradish peroxidase–conjugated anti-rabbit IgG or anti-mouse IgG secondary antibody for 2 h at room temperature. All antibodies were used at a dilution of 1:1000 except anti-actin, anti-Gapdh, and anti-PKCα antibodies (1:20,000); anti-C-Raf, anti-p-ERK, and anti-total ERK antibodies (1:2000); and the anti-RasGRP3, anti-SOS2, anti-H-Ras, and anti-p21Cip1 antibodies (1:500). Signal detection on washed membranes was performed using SuperSignal West (Pierce).
siRNA transfection
IEC-18 or IEC-6 cells were transfected with 100 pmol siRNA using Lipofectamine RNAiMAX reagent (Invitrogen). Analysis was performed 48 h after transfection, unless otherwise stated. siRNAs were as follows: ON-TARGETplus nontargeting siRNA #1 (D-001810-01-05; Dharmacon); ON-TARGETplus rat A-Raf SMARTpool siRNA (L-088540-02-0005; Dharmacon); ON-TARGETplus rat B-Raf SMARTpool siRNA (L-094802-02-0005; Dharmacon); ON-TARGETplus rat C-Raf SMARTpool siRNA (L-087699-02-0005; Dharmacon); ON-TARGETplus rat H-Ras SMARTpool siRNA (L-084782-01-0005;Dharmacon); ON-TARGETplus rat SMARTpool KSR1 siRNA (L-085072-02-0005; Dharmacon); ON-TARGETplus rat RasGRP1 SMARTpool siRNA; Dharmacon (L-100584-02-0005); ON-TARGETplus Rat RasGRP3 SMARTpool siRNA (L-098911-02-0005; Dharmacon); Silencer select predesigned siRNA RasGRP3, GGAAGUAGCUAGUCAACUATT (4392420; Invitrogen), Silencer select predesigned siRNA SOS1, CAAGCACGCUUUCGAAAUATT (Invitrogen); Silencer select predesigned siRNA SOS2, GCUUUUGAAUUAGUAUCCATT (4390771; Invitrogen). ON-TARGETplus SMARTpool siRNAs are a pool of four siRNAs designed and modified to increase specificity and reduce off-target effects.
Flow cytometry
Subconfluent IEC-18 cells were briefly washed with PBS and treated with trypsin (Gibco) to obtain single cell suspensions as described (10). About 9 × 105 cells were fixed in 70% ethanol and stained overnight at 4 °C with Telford’s reagent (90 mM EDTA, 0.1% (v/v) Triton X-100, 50 μg/ml propidium iodide in PBS). DNA content was determined by flow cytometric analysis in the Flow Cytometry Research Facility at UNMC.
Immunoprecipitation
IEC-18 cells were treated with PMA or vehicle for 10 min, rinsed twice with ice-cold PBS, and lysed for 15 to 30 min on ice in immunoprecipitation buffer (1% NP-40/Igepal CA-630, 137 mM NaCl, 20 mM Tris–HCl, pH 8.0 with freshly added protease inhibitor cocktail and phosphatase inhibitor cocktails I and II [Sigma]). Lysates were centrifuged at 13,000 rpm for 15 min, and protein concentration of the supernatants was quantified using the bicinchoninic acid assay kit (Pierce). About 500 to 1000 μg of protein were incubated in immunoprecipitation buffer containing the specified antibody (diluted as recommended by the manufacturer) or isotype matched IgG (control) overnight with rocking at 4 °C. Antibody-protein complexes were incubated with protein A/G-agarose beads (Santa Cruz) for 2 h at 4 °C with rocking. Beads were collected by centrifugation, and washed three times with ice-cold immunoprecipitation buffer. Bound proteins were extracted with 2× SDS buffer (containing freshly added DTT, 350 mM final concentration) and subjected to Western blotting.
Active Ras pull-down assay
Active Ras pull-down assays were performed using an Active Ras Detection Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Briefly, nontargeting siRNA or RasGRP3 siRNA transfected IEC-18 cells were treated with vehicle or 100 nM PMA for 15 min and lysed in 1× lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (Sigma). Lysates were cleared by centrifugation and protein concentration in the supernatant was quantified using the bicinchoninic acid assay kit (Pierce). Active Ras in lysates (containing equal amounts of protein) was pulled down with GST-Raf1-RBD (Ras-binding domain of Raf) fusion protein and glutathione resin, and levels of total (pan) Ras or H-Ras in the pulldowns and original lysates were determined by Western blotting. Positive and negative controls for the pull-down procedures were generated by incubating lysates with GTPγS or GDP, respectively.
Phos-tag gel analysis
Cells were lysed in immunoprecipitation buffer or in immunoprecipitation buffer lacking phosphatase inhibitors, and lysates were cleared by centrifugation. For λ-phosphatase treatment, supernatants lacking phosphatase inhibitors were adjusted to 1 mM MnCl2, and 800 units of λ-phosphatase (P0753, New England Biolabs) were added for 30 min at 30 °C. Samples were subjected to SDS-PAGE/Western blotting using a 6% acrylamide resolving gel containing 20 μg Phos-tag reagent (Wako Pure Chemical Industries; AAL-107) in the presence of 0.1 mmol/l MnCl2. Following electrophoresis at 60 to 90 V, gels were equilibrated in transfer buffer containing 10 mM EDTA (2 × 10 min). The gels were then soaked in regular transfer buffer for another 10 min followed by immunoblotting.
Immunohistochemistry
Deparaffinized sections of formaldehyde-fixed, paraffin-embedded mouse intestinal epithelium were rehydrated with ddH2O for 5 min. For antigen retrieval, slides were heated in preheated 1× target retrieval solution (DAKO) for 30 to 70 min at 90 °C, followed by cooling for 20 min. Endogenous peroxidase was inactivated with 3% H2O2. For pERK staining, sections were blocked with 5% normal goat serum in TBS-T. Sections were then incubated with primary anti-pERK antibody (Cell Signaling Technology antibody 4370, diluted 1:400) overnight at 4 °C in a humidified chamber, followed by SignalStain boost anti-rabbit IgG polymer (Cell Signaling Technologies) and diaminobenzidine (DAKO) as recommended by the manufacturer. Immunostaining for PKCα used anti-PKCα antibody from Abcam (ab32376) as we have described (26). Controls for staining specificity involved omission of primary antibody or replacement of the primary antibody with normal rabbit serum. Sections were counterstained with hematoxylin and dehydrated before mounting with coverslips.
Software and statistical analysis
Cell cycle analysis of flow cytometric data and generation of histograms were performed using FlowJo (FlowJo LLC) and Modfit (Verity Software) software. Densitometric analysis of scanned Western blot data was performed with ImageJ Software (NIH) using multiple exposures of each blot. Levels of proteins of interest were normalized to the loading control and presented as relative expression levels. Due to very low levels of active ERK in many control samples, quantification of pERK and pRSK is expressed as relative to the levels in PMA-treated samples for consistency. Contrast and brightness of scanned images were adjusted using GMU Image Manipulation Program (GIMP), Adobe Photoshop, or Microsoft PowerPoint software. All adjustments to contrast and brightness were made equally across the entire blot and no individual lanes were treated differently than the rest of the blot. In some figures, dashed lines indicate where lanes have been rearranged for clarity: in all cases lanes in each panel are from the same exposure of a single blot except as indicated in Figure 6A. Graphs were generated using Microsoft Excel software, and figures were assembled and annotated in Microsoft PowerPoint software. Statistical analysis was performed using Microsoft Excel software. Statistical significance of differences was assessed using Student’s t-tests with p-values ≤0.05 considered significant.
Data availability
All data described in the article are contained within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Drs Mariano Barbacid and Sergio Ruiz Macias for providing “Rasless” mESCs for this study; Dr Moorthy Ponnusamy, Dr A. Angie Rizzino, and Phillip J. Wilder for help with mESC experiments; Dr Jixin Dong and his laboratory for guidance on Phos-tag gel analysis, and Dr Keith Johnson and members of the Black laboratory for helpful discussions. This work was supported in part by National Institutes of Health Grants DK060632, CA054807, CA191894, CA036727, P20GM121316, and a pilot award from P50 CA127297. Support was also received from DOD award W81XWH-20-1-0590.
Author contributions
N. K., R. E. L., A. R. B., and J. D. B. conceptualization; N. K., A. R. B., and J. D. B. formal analysis; N. K. and M. A. L. investigation; N. K. and A. R. B. data curation; N. K., A. R. B., and J. D. B. writing–original draft; A. R. B. and J. D. B. writing–review & editing; A. R. B. and J. D. B. visualization; A. R. B. and J. D. B. supervision; J. D. B. funding acquisition.
Funding and additional information
N. K. was supported by a UNMC Program of Excellence Assistantship (Nancy Lee Smith-Bequest Trust Fund Assistantship). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Alex Toker
References
- 1.Pinto D., Clevers H. Wnt, stem cells and cancer in the intestine. Biol. Cell. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 2.Karmakar S., Deng L., He X.C., Li L. Intestinal epithelial regeneration: active versus reserve stem cells and plasticity mechanisms. Am. J. Physiol. Gastrointest. Liver Physiol. 2020;318:G796–G802. doi: 10.1152/ajpgi.00126.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aliaga J.C., Deschênes C., Beaulieu J.-F., Calvo E.L., Rivard N. Requirement of the MAP kinase cascade for cell cycle progression and differentiation of human intestinal cells. Am. J. Physiol. Gastrointest. Liver Physiol. 1999;277:G631–G641. doi: 10.1152/ajpgi.1999.277.3.G631. [DOI] [PubMed] [Google Scholar]
- 4.Basak O., Beumer J., Wiebrands K., Seno H., van Oudenaarden A., Clevers H. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20:177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Heuberger J., Kosel F., Qi J., Grossmann K.S., Rajewsky K., Birchmeier W. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celano P., Berchtold C.M., Mabry M., Carroll M., Sidransky D., Casero R.A., Jr., et al. Induction of markers of normal differentiation in human colon carcinoma cells by the v-rasH oncogene. Cell Growth Differ. 1993;4:341–347. [PubMed] [Google Scholar]
- 7.de Jong P.R., Taniguchi K., Harris A.R., Bertin S., Takahashi N., Duong J., et al. ERK5 signalling rescues intestinal epithelial turnover and tumour cell proliferation upon ERK1/2 abrogation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei G., Gao N., Chen J., Fan L., Zeng Z., Gao G., et al. Erk and MAPK signaling is essential for intestinal development through Wnt pathway modulation. Development. 2020;147 doi: 10.1242/dev.185678. [DOI] [PubMed] [Google Scholar]
- 9.Frey M.R., Clark J.A., Leontieva O., Uronis J.M., Black A.R., Black J.D. Protein kinase C signaling mediates a program of cell cycle withdrawal in the intestinal epithelium. J. Cell Biol. 2000;151:763–778. doi: 10.1083/jcb.151.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey M.R., Saxon M.L., Zhao X., Rollins A., Evans S.S., Black J.D. Protein kinase C isozyme-mediated cell cycle arrest involves induction of p21(waf1/cip1) and p27(kip1) and hypophosphorylation of the retinoblastoma protein in intestinal epithelial cells. J. Biol. Chem. 1997;272:9424–9435. doi: 10.1074/jbc.272.14.9424. [DOI] [PubMed] [Google Scholar]
- 11.Clark J.A., Black A.R., Leontieva O.V., Frey M.R., Pysz M.A., Kunneva L., et al. Involvement of the ERK signaling cascade in protein kinase C-mediated cell cycle arrest in intestinal epithelial cells. J. Biol. Chem. 2004;279:9233–9247. doi: 10.1074/jbc.M312268200. [DOI] [PubMed] [Google Scholar]
- 12.Rubinfeld H., Seger R. The ERK cascade: a prototype of MAPK signaling. Mol. Biotechnol. 2005;31:151–174. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- 13.Cox A.D., Der C.J. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 15.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 16.McKay M.M., Morrison D.K. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 17.Hennig A., Markwart R., Esparza-Franco M.A., Ladds G., Rubio I. Ras activation revisited: role of GEF and GAP systems. Biol. Chem. 2015;396:831–848. doi: 10.1515/hsz-2014-0257. [DOI] [PubMed] [Google Scholar]
- 18.Vigil D., Cherfils J., Rossman K.L., Der C.J. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muta Y., Matsuda M., Imajo M. Divergent dynamics and functions of ERK MAP kinase signaling in development, homeostasis and cancer: lessons from fluorescent bioimaging. Cancers. 2019;11:513. doi: 10.3390/cancers11040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newton A.C. Protein kinase C: perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 2018;53:208–230. doi: 10.1080/10409238.2018.1442408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antal C.E., Newton A.C. Tuning the signalling output of protein kinase C. Biochem. Soc. Transact. 2014;42:1477–1483. doi: 10.1042/BST20140172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black A.R., Black J.D. The complexities of PKCα signaling in cancer. Adv. Biol. Regul. 2021;80 doi: 10.1016/j.jbior.2020.100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooke M., Magimaidas A., Casado-Medrano V., Kazanietz M.G. Protein kinase C in cancer: the top five unanswered questions. Mol. Carcinog. 2017;56:1531–1542. doi: 10.1002/mc.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lum M.A., Barger C.J., Hsu A.H., Leontieva O.V., Black A.R., Black J.D. Protein kinase Calpha (PKCalpha) is resistant to long term desensitization/down-regulation by prolonged diacylglycerol stimulation. J. Biol. Chem. 2016;291:6331–6346. doi: 10.1074/jbc.M115.696211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxon M.L., Zhao X., Black J.D. Activation of protein kinase C isozymes is associated with post-mitotic events in intestinal epithelial cells in situ. J. Cell Biol. 1994;126:747–763. doi: 10.1083/jcb.126.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao F., Pysz M.A., Curry K.J., Haas K.N., Seedhouse S.J., Black A.R., et al. Protein kinase Cα signaling regulates inhibitor of DNA binding 1 in the intestinal epithelium. J. Biol. Chem. 2011;286:18104–18117. doi: 10.1074/jbc.M110.208488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hizli A.A., Black A.R., Pysz M.A., Black J.D. Protein kinase C alpha signaling inhibits cyclin D1 translation in intestinal epithelial cells. J. Biol. Chem. 2006;281:14596–14603. doi: 10.1074/jbc.M601959200. [DOI] [PubMed] [Google Scholar]
- 28.Guan L., Song K., Pysz M.A., Curry K.J., Hizli A.A., Danielpour D., et al. Protein kinase C-mediated down-regulation of cyclin D1 involves activation of the translational repressor 4E-BP1 via a phosphoinositide 3-kinase/Akt-independent, protein phosphatase 2A-dependent mechanism in intestinal epithelial cells. J. Biol. Chem. 2007;282:14213–14225. doi: 10.1074/jbc.M610513200. [DOI] [PubMed] [Google Scholar]
- 29.Ueda Y., Hirai S., Osada S., Suzuki A., Mizuno K., Ohno S. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J. Biol. Chem. 1996;271:23512–23519. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 30.Kolch W., Heidecker G., Kochs G., Hummel R., Vahidi H., Mischak H., et al. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature. 1993;364:249–252. doi: 10.1038/364249a0. [DOI] [PubMed] [Google Scholar]
- 31.Marais R., Light Y., Mason C., Paterson H., Olson M.F., Marshall C.J. Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science. 1998;280:109–112. doi: 10.1126/science.280.5360.109. [DOI] [PubMed] [Google Scholar]
- 32.Schönwasser D.C., Marais R.M., Marshall C.J., Parker P.J. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol. Cell. Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen-Sheng W. Protein kinase C alpha trigger Ras and Raf-independent MEK/ERK activation for TPA-induced growth inhibition of human hepatoma cell HepG2. Cancer Lett. 2006;239:27–35. doi: 10.1016/j.canlet.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 34.Peng S.B., Henry J.R., Kaufman M.D., Lu W.P., Smith B.D., Vogeti S., et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell. 2015;28:384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Brummer T., McInnes C. RAF kinase dimerization: implications for drug discovery and clinical outcomes. Oncogene. 2020;39:4155–4169. doi: 10.1038/s41388-020-1263-y. [DOI] [PubMed] [Google Scholar]
- 36.Terrell E.M., Morrison D.K. Ras-mediated activation of the Raf family kinases. Cold Spring Harb. Perspect. Med. 2019;9 doi: 10.1101/cshperspect.a033746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotblat B., Ehrlich M., Haklai R., Kloog Y. The Ras inhibitor farnesylthiosalicylic acid (salirasib) disrupts the spatiotemporal localization of active Ras: a potential treatment for cancer. Methods Enzymol. 2008;439:467–489. doi: 10.1016/S0076-6879(07)00432-6. [DOI] [PubMed] [Google Scholar]
- 38.Oda K., Matsuoka Y., Funahashi A., Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baines A.T., Xu D., Der C.J. Inhibition of Ras for cancer treatment: the search continues. Future Med. Chem. 2011;3:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blum R., Jacob-Hirsch J., Amariglio N., Rechavi G., Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005;65:999–1006. [PubMed] [Google Scholar]
- 41.Goldberg L., Kloog Y. A Ras inhibitor tilts the balance between Rac and Rho and blocks phosphatidylinositol 3-kinase-dependent glioblastoma cell migration. Cancer Res. 2006;66:11709–11717. doi: 10.1158/0008-5472.CAN-06-1878. [DOI] [PubMed] [Google Scholar]
- 42.McMahon L.P., Yue W., Santen R.J., Lawrence J.C., Jr. Farnesylthiosalicylic acid inhibits mammalian target of rapamycin (mTOR) activity both in cells and in vitro by promoting dissociation of the mTOR-raptor complex. Mol. Endocrinol. 2005;19:175–183. doi: 10.1210/me.2004-0305. [DOI] [PubMed] [Google Scholar]
- 43.Arai H., Escobedo J.A. Angiotensin II type 1 receptor signals through Raf-1 by a protein kinase C-dependent, Ras-independent mechanism. Mol. Pharmacol. 1996;50:522–528. [PubMed] [Google Scholar]
- 44.Mayor-Ruiz C., Olbrich T., Drosten M., Lecona E., Vega-Sendino M., Ortega S., et al. ERF deletion rescues RAS deficiency in mouse embryonic stem cells. Genes Dev. 2018;32:568–576. doi: 10.1101/gad.310086.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebeling J.G., Vandenbark G.R., Kuhn L.J., Ganong B.R., Bell R.M., Niedel J.E. Diacylglycerols mimic phorbol diester induction of leukemic cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 1985;82:815–819. doi: 10.1073/pnas.82.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kortum R.L., Costanzo D.L., Haferbier J., Schreiner S.J., Razidlo G.L., Wu M.-H., et al. The molecular scaffold kinase suppressor of Ras 1 (KSR1) regulates adipogenesis. Mol. Cell. Biol. 2005;25:7592–7604. doi: 10.1128/MCB.25.17.7592-7604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frodyma D., Neilsen B., Costanzo-Garvey D., Fisher K., Lewis R. Coordinating ERK signaling via the molecular scaffold kinase suppressor of Ras. F1000Res. 2017;6:1621. doi: 10.12688/f1000research.11895.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freeman A.K., Ritt D.A., Morrison D.K. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol. Cell. 2013;49:751–758. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freeman A.K., Ritt D.A., Morrison D.K. The importance of Raf dimerization in cell signaling. Small GTPases. 2013;4:180–185. doi: 10.4161/sgtp.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 51.Baltanás F.C., Zarich N., Rojas-Cabañeros J.M., Santos E. SOS GEFs in health and disease. Biochim. Biophys. Acta Rev. Cancer. 2020;1874 doi: 10.1016/j.bbcan.2020.188445. [DOI] [PubMed] [Google Scholar]
- 52.Corbalan-Garcia S., Yang S.S., Degenhardt K.R., Bar-Sagi D. Identification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2. Mol. Cell. Biol. 1996;16:5674–5682. doi: 10.1128/mcb.16.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozakis-Adcock M., van der Geer P., Mbamalu G., Pawson T. MAP kinase phosphorylation of mSos1 promotes dissociation of mSos1-Shc and mSos1-EGF receptor complexes. Oncogene. 1995;11:1417–1426. [PubMed] [Google Scholar]
- 54.Douville E., Downward J. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 55.Saha M., Carriere A., Cheerathodi M., Zhang X., Lavoie G., Rush J., et al. RSK phosphorylates SOS1 creating 14-3-3-docking sites and negatively regulating MAPK activation. Biochem. J. 2012;447:159–166. doi: 10.1042/BJ20120938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinoshita E., Kinoshita-Kikuta E., Koike T. Phos-tag SDS-PAGE systems for phosphorylation profiling of proteins with a wide range of molecular masses under neutral pH conditions. Proteomics. 2012;12:192–202. doi: 10.1002/pmic.201100524. [DOI] [PubMed] [Google Scholar]
- 57.Eblen S.T. Extracellular-regulated kinases: signaling from Ras to ERK substrates to control biological outcomes. Adv. Cancer Res. 2018;138:99–142. doi: 10.1016/bs.acr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stone J.C. Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer. 2011;2:320–334. doi: 10.1177/1947601911408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golec D.P., Dower N.A., Stone J.C., Baldwin T.A. RasGRP1, but not RasGRP3, is required for efficient thymic β-selection and ERK activation downstream of CXCR4. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y., Zhu M., Nishida K., Hirano T., Zhang W. An essential role for RasGRP1 in mast cell function and IgE-mediated allergic response. J. Exp. Med. 2007;204:93–103. doi: 10.1084/jem.20061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aiba Y., Oh-hora M., Kiyonaka S., Kimura Y., Hijikata A., Mori Y., et al. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16612–16617. doi: 10.1073/pnas.0407468101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng Y., Liu H., Coughlin J., Zheng J., Li L., Stone J.C. Phosphorylation of RasGRP3 on threonine 133 provides a mechanistic link between PKC and Ras signaling systems in B cells. Blood. 2005;105:3648–3654. doi: 10.1182/blood-2004-10-3916. [DOI] [PubMed] [Google Scholar]
- 63.Furth M.E., Aldrich T.H., Cordon-Cardo C. Expression of ras proto-oncogene proteins in normal human tissues. Oncogene. 1987;1:47–58. [PubMed] [Google Scholar]
- 64.Quaroni A., Isselbacher K.J. Cytotoxic effects and metabolism of benzo[a]pyrene and 7,12-dimethylbenz[a]anthracene in duodenal and ileal epithelial cell cultures. J. Natl. Cancer Inst. 1981;67:1353–1362. [PubMed] [Google Scholar]
- 65.Quaroni A., Wands J., Trelstad R.L., Isselbacher K.J. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hobbs G.A., Der C.J., Rossman K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ikushima H., Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita S., Mochizuki N., Ohba Y., Tobiume M., Okada Y., Sawa H., et al. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J. Biol. Chem. 2000;275:25488–25493. doi: 10.1074/jbc.M003414200. [DOI] [PubMed] [Google Scholar]
- 69.Ebinu J.O., Bottorff D.A., Chan E.Y., Stang S.L., Dunn R.J., Stone J.C. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 70.Nagy Z., Kovács I., Török M., Tóth D., Vereb G., Buzás K., et al. Function of RasGRP3 in the formation and progression of human breast cancer. Mol. Cancer. 2014;13:96. doi: 10.1186/1476-4598-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lorenzo P.S., Kung J.W., Bottorff D.A., Garfield S.H., Stone J.C., Blumberg P.M. Phorbol esters modulate the Ras exchange factor RasGRP3. Cancer Res. 2001;61:943–949. [PubMed] [Google Scholar]
- 72.Ksionda O., Limnander A., Roose J.P. RasGRP Ras guanine nucleotide exchange factors in cancer. Front. Biol. (Beijing) 2013;8:508–532. doi: 10.1007/s11515-013-1276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teixeira C., Stang S.L., Zheng Y., Beswick N.S., Stone J.C. Integration of DAG signaling systems mediated by PKC-dependent phosphorylation of RasGRP3. Blood. 2003;102:1414–1420. doi: 10.1182/blood-2002-11-3621. [DOI] [PubMed] [Google Scholar]
- 74.Depeille P., Henricks L.M., van de Ven R.A.H., Lemmens E., Wang C.-Y., Matli M., et al. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat. Cell Biol. 2015;17:804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gbenedio O.M., Bonnans C., Grun D., Wang C.-Y., Hatch A.J., Mahoney M.R., et al. RasGRP1 is a potential biomarker for stratifying anti-EGFR therapy response in colorectal cancer. JCI Insight. 2019;5 doi: 10.1172/jci.insight.127552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weber C.K., Slupsky J.R., Kalmes H.A., Rapp U.R. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–3598. [PubMed] [Google Scholar]
- 77.Rushworth L.K., Hindley A.D., O'Neill E., Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol. Cell. Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An S., Yang Y., Ward R., Liu Y., Guo X.-X., Xu T.-R. Raf-interactome in tuning the complexity and diversity of Raf function. FEBS J. 2015;282:32–53. doi: 10.1111/febs.13113. [DOI] [PubMed] [Google Scholar]
- 79.Thiel G., Ekici M., Rössler O.G. Regulation of cellular proliferation, differentiation and cell death by activated Raf. Cell Commun. Signal. 2009;7:8. doi: 10.1186/1478-811X-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woods D., Parry D., Cherwinski H., Bosch E., Lees E., McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu J., Woods D., McMahon M., Bishop J.M. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lum M.A., Pundt K.E., Paluch B.E., Black A.R., Black J.D. Agonist-induced down-regulation of endogenous protein kinase c alpha through an endolysosomal mechanism. J. Biol. Chem. 2013;288:13093–13109. doi: 10.1074/jbc.M112.437061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lum M.A., Balaburski G.M., Murphy M.E., Black A.R., Black J.D. Heat shock proteins regulate activation-induced proteasomal degradation of the mature phosphorylated form of protein kinase C. J. Biol. Chem. 2013;288:27112–27127. doi: 10.1074/jbc.M112.437095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raggatt L.J., Evdokiou A., Findlay D.M. Sustained activation of Erk1/2 MAPK and cell growth suppression by the insert-negative, but not the insert-positive isoform of the human calcitonin receptor. J. Endocrinol. 2000;167:93–105. doi: 10.1677/joe.0.1670093. [DOI] [PubMed] [Google Scholar]
- 85.Kerkhoff E., Rapp U.R. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 1998;58:1636–1640. [PubMed] [Google Scholar]
- 86.Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 87.Nishizuka M., Koyanagi A., Osada S., Imagawa M. Wnt4 and Wnt5a promote adipocyte differentiation. FEBS Lett. 2008;582:3201–3205. doi: 10.1016/j.febslet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 88.Sheldahl L.C., Park M., Malbon C.C., Moon R.T. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 89.Yamaguchi Y., Passeron T., Watabe H., Yasumoto K., Rouzaud F., Hoashi T., et al. The effects of dickkopf 1 on gene expression and Wnt signaling by melanocytes: mechanisms underlying its suppression of melanocyte function and proliferation. J. Invest. Dermatol. 2007;127:1217–1225. doi: 10.1038/sj.jid.5700629. [DOI] [PubMed] [Google Scholar]
- 90.Lee J.M., Kim I.S., Kim H., Lee J.S., Kim K., Yim H.Y., et al. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol. Cell. 2010;37:183–195. doi: 10.1016/j.molcel.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 91.Vanuytsel T., Senger S., Fasano A., Shea-Donohue T. Major signaling pathways in intestinal stem cells. Biochim. Biophys. Acta. 2013;1830:2410–2426. doi: 10.1016/j.bbagen.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 93.Muta Y., Fujita Y., Sumiyama K., Sakurai A., Taketo M.M., Chiba T., et al. Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat. Commun. 2018;9:2174. doi: 10.1038/s41467-018-04527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in the article are contained within the article.