Abstract
Cryptosporidium, an enteric parasite of humans and a wide range of other mammals, presents numerous challenges to the supply of safe drinking water. We performed a wildlife survey, focusing on white-tailed deer and small mammals, to assess whether they may serve as environmental sources of Cryptosporidium. A PCR-based approach that permitted genetic characterization via sequence analysis was applied to wildlife fecal samples (n = 111) collected from September 1996 to July 1998 from three areas in lower New York State. Southern analysis revealed 22 fecal samples containing Cryptosporidium small-subunit (SSU) ribosomal DNA; these included 10 of 91 white-tailed deer (Odocoileus virginianus) samples, 3 of 5 chipmunk (Tamias striatus) samples, 1 of 2 white-footed mouse (Peromyscus leucopus) samples, 1 of 2 striped skunk (Mephitis mephitis) samples, 1 of 5 racoon (Procyon lotor) samples, and 6 of 6 muskrat (Ondatra zibethicus) samples. All of the 15 SSU PCR products sequenced were characterized as Cryptosporidium parvum; two were identical to genotype 2 (bovine), whereas the remainder belonged to two novel SSU sequence groups, designated genotypes 3 and 4. Genotype 3 comprised four deer-derived sequences, whereas genotype 4 included nine sequences from deer, mouse, chipmunk, and muskrat samples. PCR analysis was performed on the SSU-positive fecal samples for three other Cryptosporidium loci (dihydrofolate reductase, polythreonine-rich protein, and beta-tubulin), and 8 of 10 cloned PCR products were consistent with C. parvum genotype 2. These data provide evidence that there is sylvatic transmission of C. parvum involving deer and other small mammals. This study affirmed the importance of wildlife as potential sources of Cryptosporidium in the catchments of public water supplies.
Cryptosporidium is a genus of protozoan parasites that causes gastrointestinal disease in humans and other vertebrates (6). Transmission occurs when infectious oocysts shed in the feces of an infected host are ingested by a new host. Numerous species of Cryptosporidium are currently recognized as affecting mammals; these species include C. parvum, C. muris, C. wrairi, and C. felis (6, 24). C. parvum has been reported to be present in approximately 70 species of mammals belonging to nine orders (11, 25, 37), whereas C. muris, C. wrairi, and C. felis appear to have more restricted host ranges (reviewed in reference 24). In addition, C. parvum variants or strains have been identified; although there is controversy regarding the phylogenetic status and nomenclature of these organisms, a convention for genotype designations using numbers and/or names has developed (49). Two C. parvum genotypes, genotype 1 (human) and genotype 2 (bovine), appear to be responsible for the majority of infections in humans (19, 20, 23; summarized in reference 48).
The natural ecology of C. parvum probably involves sylvatic cycles with transmission among wild mammals in which the infection may be largely asymptomatic (7, 37). Zoonotic cycles also occur in agricultural settings where high infection rates are seen in domestic animals, particularly dairy cattle (12, 21, 28, 40, 43). There is also an anthroponotic cycle that is either direct via person-to-person transmission or indirect (e.g, when human sewage contaminates the drinking water supply) (21). Water may serve as an important intersection for these cycles; effluents from sewage treatment plants and septic tanks and runoff from agricultural lands and forests can all have an impact on the surface water supply (17, 26, 34, 35). The observation that rainfall appears to have been an important factor in some drinking water outbreaks supports this premise (31).
Surface water supplies are vulnerable to Cryptosporidium contamination from a variety of sources. Watershed protection programs attempt to control such inputs from anthropogenic and agricultural sources (1, 34), but wildlife remains largely beyond the reach of management efforts. Numerous wild mammals have been shown to be infected with Cryptosporidium spp. (11, 25, 37). White-tailed deer (Odocoileus virginianus) are of particular concern in the eastern United States because of their high population densities in both rural and suburban areas, including the watersheds of New York City's Catskill-Delaware and Croton water supply systems. Cryptosporidium infection has been reported in both captive and wild white-tailed deer populations in the mid-Atlantic, southern, and western United States (11, 13, 29). We performed a survey of wildlife in lower New York State; focusing on white-tailed deer and small mammals, to assess whether they might be an environmental source of Cryptosporidium.
MATERIALS AND METHODS
Wildlife sample collection.
Wildlife fecal samples were collected from three sites in New York State: Black Rock Forest, a 1,500-ha research station located in Cornwall, N.Y., 50 miles north of New York City; Louis Calder Center of Fordham University, a biological field station located in Westchester County in Armonk, N.Y., within the local watershed of the Kensico reservoir; and forested lands buffering the New Croton Reservoir, adjacent to the Stanwood community in Westchester County (with permission from the New York City Department of Environmental Protection).
Deer fecal samples were collected on the basis of their morphology and relationship to tracks and visual observation. Fresh fecal material was collected from the ground. During June and July 1998, a total of 91 fecal specimens from white-tailed deer were collected at the three sites; 19, 54, and 18 samples were collected at the Calder Center, Stanwood, and Black Rock Forest sites, respectively.
Chipmunk (Tamias striatus) (n = 5), white-footed mouse (Peromyscus leucopus) (n = 2), striped skunk (Mephitis mephitis) (n = 2), and racoon (Procyon lotor) (n = 5) fecal samples were collected at the Calder Center in September and October 1996. These samples were provided by Thomas J. Daniels of Fordham University and were from animals that were trapped as part of an ongoing study of the role of small mammals in the transmission of Lyme disease (8). Muskrat (Ondatra zibethicus) fecal samples (n = 6) were collected at the Calder Center in July 1998.
Before analysis, fresh fecal material was either frozen and stored at −20°C (deer and muskrat specimens) or held in sodium acetate-acetic acid-formalin preservative for less than 30 days, washed in 1× phosphate-buffered saline (PBS), and refrigerated at 4°C (mice, chipmunk, racoon, and skunk specimens).
Molecular epidemiology.
Routine methods for detection and diagnosis of Cryptosporidium infection in feces are inadequate for the purposes of molecular epidemiology. Besides being relatively insensitive (42), they provide incomplete information with respect to the species of Cryptosporidium detected (i.e., there is limited specificity for C. parvum versus other species) and no means of differentiating among different genotypes, isolates, or strains (42). The diagnostic PCR protocols available at the time of this study typically involved either purified oocysts or diarrheal samples known a priori to contain Cryptosporidium (18). In contrast, we developed a diagnostic PCR assay (see below) to detect Cryptosporidium DNA when oocysts are present in formed stools at low concentrations. In order to increase both the specificity and the sensitivity of the PCR assay, Southern analysis was included. A positive result was defined as detection of a DNA product of the expected size following Southern analysis. This PCR-based approach also permitted genetic characterization of the isolates through cloning and sequencing of PCR products.
DNA extraction from wildlife feces.
Fecal samples were processed in a designated area separate from the PCR setup and post-PCR zones. Reagents and equipment, such as pipettors, were dedicated to each of the three stages (sample preparation, PCR setup, and post-PCR).
Fecal material (1 to 1.5 g) was mixed with 4 to 6 ml of ice-cold PBS. The resulting slurry was washed through a funnel screening device (Meridien Contrate) with additional PBS into a 50-ml tube, shaken vigorously, and centrifuged for 5 min at 500 × g (42). The supernatant was discarded, and the sediment (∼0.5 g) was transferred to a 2-ml tube (with a screw top and an Oring; Biospec Corp.) and centrifuged for 1 min at 16,000 × g (maximum speed). The supernatant was discarded, and 1.1 ml of DNAzol (MRC Corp.) plus ∼2.5 g of 0.1-mm-diameter zirconium-silica beads (Biospec Corp.) were added (50). Samples were homogenized at 4,200 rpm for 5 min in a Mini-BeadBeater (Biospec Corp.) and then incubated in a water bath for 10 min at 90°C (9, 50). The tubes were pierced with a 26G1/2 needle, and each tube was piggybacked on a new microcentrifuge tube inside a 15-ml tube and then centrifuged for 2 min at 2,000 × g. The lysate recovered (∼1 ml) was purified by phenol-chloroform-isoamyl alcohol (25:24:1) extraction, and the DNA was precipitated with 100% ethanol in the presence of 5 μl of polyacryl carrier (MRC Corp.). The DNA pellets were washed twice with 95% ethanol, air dried for 10 min, eluted with 100 μl of 8 mM NaOH, incubated at 65°C for 5 min, and centrifuged for 10 min at 16,000 × g to remove insoluble material. Each supernatant containing DNA was transferred to a new tube containing 10 μl of 0.1 M HEPES (final pH ∼8). The process controls used were GCHIP, an unprocessed fecal sample from an experimentally infected calf at the Tufts School of Veterinary Medicine (provided by G. Widmer); and a deer fecal sample spiked with 105 oocysts (previously purified from isolate GCH1) per g.
PCR analysis.
Formed stools contain an enormous array of microorganisms, and stools also often contain PCR inhibitors (10). The adequacy of DNA isolation and purification was assessed for each isolate by first performing PCR amplification with a nonspecific primer set (LSU primer set) targeting large-subunit rRNA genes (Table 1) that could prime from a broad range of templates. The template quantities were adjusted or additional phenol-chloroform extractions were performed as needed for the subset of isolates that did not initially produce a detectable (∼280-bp) PCR product with the LSU primer set. PCR amplification was then performed with the SSU primer set to amplify a ∼435-bp region of the small-subunit (SSU) rRNA gene (Table 1) that spans the hypervariable region. The SSU primers anneal to all known Cryptosporidium SSU rRNA genes (10). The presence of five copies of the ribosomal DNA (rDNA) unit in C. parvum (16) enhanced the assay's ability to detect very-low-level infections. PCR amplifications in which control DNA (Cryptosporidium DNA previously extracted from isolate GCH1 or KSU-1) was used with the diagnostic SSU primers consistently generated positive products (as determined by visual detection following gel electrophoresis and UV transillumination) at template levels of 1.0 pg of DNA (equivalent to approximately 20 oocysts).
TABLE 1.
Primer sets used in this study to identify and characterize Cryptosporidium
| Primer set | Target gene | Primer | Sequence | Expected product size (bp) | Annealing temp (°C) | Reference(s) |
|---|---|---|---|---|---|---|
| LSU | Large-subunit rRNA | LSU-F | 5′-GTAACTGCGAGTGAACAGGAA-3′ | 280 | 60 | This study |
| LSU-R | 5′-CCTCACGGTACTTGTTTGCT-3′ | |||||
| SSU | SSU rRNA | CPB-DIAGF | 5′-AAGCTCGTAGTTGGATTTCTG-3′ | 435a | 60 | 15 |
| CPB-DIAGR | 5′-TAAGGTGCTGAAGGAGTAAGG-3′ | |||||
| DHFR | Dihydrofolate reductase-thymidylate synthase | DHFR1 | 5′-TTGTGGCAGCTTCTGTTTTGA-3′ | 232 | 68 | This study |
| DHFR2 | 5′-GGATCGGCTTCATCTTGTGG-3′ | 39 | ||||
| POLY(T) | Polythreonine-rich glycoprotein | CRY44 | 5′-CTCTTAATCCAATCATTACAAC-3′ | 318 | 52 | 3, 5 |
| CRY39 | 5′-GAGTCTAATAATAAACCACTG-3′ | |||||
| B-TUB | β-Tubulin | BTUB5 | 5′-GATTGGTGCTAAATTCTGGG-3′ | 478 | 60 | 4, 45 |
| BTUB4 | 5′-CCTGATCCTGTACCACCTCC-3′ |
Corresponding to position 601 to position 1035 of the C. parvum SSU rRNA sequence (GenBank accession number L16996).
Multilocus analysis was performed with selected samples by PCR amplification of regions of the β-tubulin gene (B-TUB), a polythreonine-rich gene [POLY(T)], and the dihydrofolate reductase gene (DHFR) (3–5, 36, 39, 45) (Table 1). The target sequences have been analyzed for C. parvum genotypes 1 and 2 (the human and bovine genotypes, respectively), and the corresponding sequence polymorphisms have been documented; furthermore, each of these genes is present as a single copy (30, 36, 39, 45).
Standard 50-μl PCR mixtures consisted of 5 to 50 ng of DNA template and the following reagents (final concentrations): 2.5 mM MgCl2, each deoxyribonucleoside triphosphate at a concentration of 0.2 mM, 1× PCR buffer, 250 μg of bovine serum albumin per ml, 0.4 mM forward primer, 0.4 mM reverse primer, and 1.25 U of Taq polymerase (AmpliTaq Gold; PE Applied Biosystems). Positive and negative controls containing 1 pg of C. parvum DNA and no template, respectively, were included in all experiments. A hot start PCR was performed (model 480 thermal cycler; PE Applied Biosystems) by using the following conditions: 95°C for 9 min (Taq activation), followed by 40 cycles of 94°C for 45 s, 60°C (or another temperature as specified in Table 1) for 45 s, and 72°C for 45 s, and a final extension at 72°C for 7 min. A 10-μl aliquot of each PCR mixture was examined by gel electrophoresis on 3.5% agarose (NuSieve; FMC Corp) that was stained with 1% GelStar DNA stain (FMC Corp.) and visualized with a UV transilluminator. DNA was transferred from the gels via Southern blotting onto nylon filters by standard methods (2).
Southern analysis.
Southern analysis of the products from the PCR amplifications was performed by standard methods (2). All of the DNA probes used were PCR products amplified from C. parvum genomic DNA (GCH1 or KSU-1) that had been cloned into pCRII. The following probes were used: SSU rRNA gene p17-16 (16); β-tubulin gene B-TUB3/4 (45); polythreonine-rich gene CRY44/37 (5); and dihydrofolate reductase gene DHFR201. The SSU rRNA, β-tubulin, and polythreonine-rich gene probes were all kindly provided by G. Widmer (Tufts University). The dihydrofolate reductase gene probe was generated in this study. Standard protocols for the manipulation of plasmids (2) were used throughout this study.
DNA probes were prepared from gel-purified plasmid fragments by using random priming with [α-32P]dATP (NEN Research Products). Overnight hybridization was performed at 65°C, and the stringency of the final posthybridization wash was 0.2× SSC at 65°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Records of the hybridization signals were made by autoradiography. Longer exposure times were required during autoradiography to detect weak signals from certain samples.
Sequence analysis.
PCR products were cloned into the pCRII-TOPO plasmid vector by using a TOPO cloning kit (Invitrogen). The nucleotide sequences of the clones were determined by the dideoxy chain termination method, and the PCR primers were trimmed from the final sequence. The sequences of both strands of the SSU and B-TUB PCR products were determined. Searches of the DNA databases were performed by using the BLAST server service (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were aligned by using the BLAST 2 tool (http://www.ncbi.nlm.nih.gov/gorf/b12.html) and CLUSTALW (14) with default parameters; this analysis included using the identity matrix for scoring alignments and generating distances and using the neighbor-joining algorithm (32) for generating trees. Slightly negative branches were set to zero. Trees were displayed by using NJplot, as described by M. Guoy, University of Lyon, Lyon, France (ftp.epi.ac.uk/pub/software/mac/NJplot.sea.hqx). Because the simplest measurement of distance in which gaps are ignored was used, the distances in the tree for single sequences with large insertions, such as C. felis SSU rDNA (accession number AF112575), may have been underestimated.
Nucleotide sequence accession numbers.
The GenBank accession numbers for PCR products from fecal samples are as follows: AF297511 to AF297515 and AF297517 Cryptosporidium SSU rRNA gene sequences from white-tailed deer fecal samples 524, 563, 569, 570, 586, and 598, respectively; AF297502 to AF297507 for SSU rRNA gene sequences from muskrat fecal samples 589, 590, 591, 592, 593, and 603, respectively; AF297508 for SSU rRNA gene sequence from white-footed mouse fecal sample 4227; AF297509 and AF297510 for SSU rRNA gene sequences from chipmunk fecal samples 4237 and 4247, respectively; AF303051 and AF303052 for Cryptosporidium DHFR sequences from white-footed mouse fecal samples 4252 and 4227, respectively; AF303047, AF303049, and AF303050 for DHFR sequences from chipmunk fecal samples 4237, 4247, and 4234, respectively; AF303048 and AF303053 for DHFR sequences from muskrat fecal samples 591 and 603, respectively; AF303054 for Cryptosporidium POLY(T) sequences from chipmunk fecal sample 4237; AF303055 for POLY(T) sequence from muskrat fecal sample 603; and AF303056 for Cryptosporidium B-TUB sequence from muskrat fecal sample 603.
The accession numbers of the SSU rRNA gene reference sequences used in the alignments are as follows: AF093489 and AF093492 (46) for C. parvum genotype 1; AF093490 (46) and AF015772 (16) for C. parvum genotype 2 type A; AF308600 for C. parvum genotype 2 type B; AF093494 for C. parvum deer isolate; AF093495 for C. baileyi; AF093496 and AF093498 for C. muris; AF093499 and AF093500 (46) for C. serpentis; AF115377, AF112576, AF112570, AF112571, and AF112572 for C. parvum pig, dog, kangaroo, mouse, and ferret isolates, respectively; AF115378 for C. wrairi; AF112573 for Cryptosporidium sp.; AF112574 for C. meleagridis; and AF112575 (49) for C. felis. The accession numbers of the dihydrofolate reductase reference sequences are U41366 and U41365 for C. parvum genotype 1 and C. parvum genotype 2, respectively (39); the accession numbers of the β-tubulin reference sequences are AF115399 and AF115398 for C. parvum genotype 1 and C. parvum genotype 2, respectively (30); and the accession number of the polythreonine-rich gene reference sequence of C. parvum genotype 2 is U83169 (5).
RESULTS
Diagnostic SSU PCR of wildlife samples.
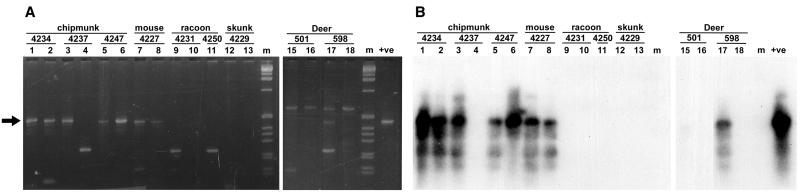
Twenty-two Cryptosporidium-positive wildlife samples were detected among the 111 fecal samples tested by using the diagnostic SSU rDNA PCR assay (Fig. 1; Table 2). Four additional wildlife fecal samples were scored as provisionally positive samples (Table 2). Positive controls resulted in strong gel bands and intense hybridization in all instances, and no products were detected in lanes containing negative controls (Fig. 1; data not shown). In general, the hybridization signal was consistent with the intensity of the band visualized on a stained gel in the expected size range, ∼435 bp (Fig. 1). Southern analysis revealed four positive samples that were not apparent from the stained gels. For certain samples, amplification was inconsistent, reflecting variable levels of target DNA and/or inhibiting substances in the template preparations (e.g., chipmunk sample 4237 and deer sample 598) (Fig. 1). Nonspecific gel bands outside the expected size range did not hybridize with the SSU probe and could represent products from organisms other than Cryptosporidium spp. (Fig. 1).
FIG. 1.
PCR analysis of wildlife fecal sample DNA amplified with the SSU diagnostic primer set. PCR amplification products of DNA extracted from wildlife fecal samples and amplified with the SSU diagnostic primer set were visualized directly on agarose gels (A) and by Southern analysis with the SSU rDNA probe (B). Two template concentrations (1× and 2×) were used for each sample except sample 4250. The left and right lanes of each pair of lanes contained the high and low concentrations, respectively Lane +ve contained a PCR-positive control consisting of 2 pg of C. parvum GCH1 DNA. Lanes m contained size markers. The arrow indicates the expected 435-bp product.
TABLE 2.
Results of PCR amplification of DNA from wildlife fecal samples followed by Southern analysisa
| Host | Siteb | Sample | SSU rDNA
|
DHFR
|
POLY(T)
|
B-TUB
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| PCRc | Sequenced | PCRc | Sequenced | PCRc | Sequenced | PCRc | Sequenced | |||
| Calfe | GCH1P | + | 2 | + | 2 | + | 2 | + | 2 | |
| Deer | S | 506 | + | − | + | + | ||||
| Deer | S | 524 | + | 3 | − | − | + | |||
| Deer | S | 540 | ∼ | − | − | ∼ | ||||
| Deer | B | 553 | + | − | − | − | ||||
| Deer | S | 563 | + | 3 | − | − | + | |||
| Deer | S | 569 | + | 3 | − | − | − | |||
| Deer | S | 570 | + | 3 | − | − | − | |||
| Deer | S | 571 | + | − | − | − | ||||
| Deer | C | 576 | + | − | − | − | ||||
| Deer | S | 578 | ∼ | − | − | ∼ | ||||
| Deer | S | 586 | + | 4 | − | − | ∼ | |||
| Deer | C | 598 | + | 2 | − | − | − | |||
| Skunk | C | 4229 | + | − | + | + | ||||
| Raccoon | C | 4250 | + | − | − | − | ||||
| Mouse | C | 4227 | + | 4 | + | 2 | + | + | ||
| Mouse | C | 4252 | ∼ | + | 2 | ∼ | − | |||
| Chipmunk | C | 4234 | + | + | 2f | − | + | |||
| Chipmunk | C | 4237 | + | 2 | + | 2 | + | 2 | + | |
| Chipmunk | C | 4244 | ∼ | + | − | − | ||||
| Chipmunk | C | 4247 | + | 4 | + | 2 | + | − | ||
| Muskrat | C | 589 | + | 4 | − | ∼ | + | |||
| Muskrat | C | 590 | + | 4 | − | − | + | |||
| Muskrat | C | 591 | + | 4 | + | 2 | ∼ | + | ||
| Muskrat | C | 592 | + | 4 | − | − | + | |||
| Muskrat | C | 593 | + | 4 | − | − | + | |||
| Muskrat | C | 603 | + | 4 | + | 2 | + | —g | + | —g |
A total of 10 of 91 deer samples, 1 of 2 skunk samples, 1 of 5 raccoon samples, 1 of 2 mouse samples, 3 of 5 chipmunk samples, and 6 of 6 muskrat samples were positive with the SSU primer set. Two deer samples, one mouse sample, and one chipmunk sample were provisionally positive with the SSU primer set. Samples that were negative for the SSU PCR products are not shown.
S, Stanwood; B, Black Rock Forest; C, Calder Center.
The results of Southern analysis of PCR products from amplifications with primer pairs are indicated as follows: +, positive (signal corresponding to product within 5% of the expected size) ∼, provisional (signal very weak or corresponding to a product between 5 and 10% larger or smaller than expected); −, negative (no signal or signal corresponding to a product either more than 10% larger or more than 10% smaller than expected).
The genotypes of the cloned and sequenced PCR products were as follows: for SSU rDNA, C. parvum genotype 2 (16, 46) or C. parvum genotype 3 or 4 (this study) (Table 3; Fig. 2); for DHFR, C. parvum genotype 2 (39); for POLY(T) C. parvum genotype 2 (5); and for B-TUB, C. parvum genotype 2 (30).
C. parvum genotype 2 reference positive control.
Allelic variant.
Alleles that are not attributed to a Cryptosporidium species or genotype.
Sequence analysis of SSU PCR products.
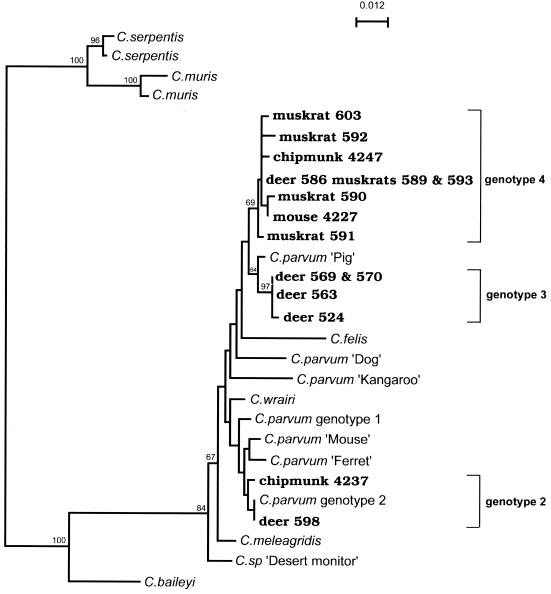
Fifteen SSU PCR products from wildlife samples were sequenced; these products included six products from deer samples, one product from mouse samples, two products from chipmunk samples, and six products from muskrat samples (Table 2). BLAST analysis of the GenBank databases indicated that the most significant alignment for all of these sequences individually was the alignment with Cryptosporidium SSU rDNA sequences. Multiple sequence alignment with reference Cryptosporidium SSU rDNA sequences showed that the wildlife-derived SSU sequences were most similar to C. parvum sequences (Table 3; data not shown). The distance relationships among the wildlife-derived SSU rDNA sequences and the reference sequences are illustrated in Fig. 2.
TABLE 3.
Alignment of the hypervariable regions of the Cryptosporidium SSU rDNA PCR products from wildlife samples and reference sequencesa
| Source (sample) | Genotypeb | Sequence at positions 632 to 667c | Sequence at positions 678 to 698c |
|---|---|---|---|
| 598 | C. parvum genotype 2d | ATATAAAATATTTTGAT--GAATATT--TATATAATATTA | TATT-------ACTA--TATATTT----TAGTAT |
| 4237 | C. parvum genotype 2e | ATATAAAATATTTT-----GAATATT--TATATAACATTA | TATT-------ACTA--TATTTTT----TAGTAT |
| C. parvum ferret | ATATAAAATATTTTGAT--TAATATT--TATATAATATTA | TATT-------ACTA--AATTTTTGTT-TGGTAT | |
| C. parvum mouse | ATATAAAATATTTTAAT--TAATATT--TATATAATATTA | TATT-------ACTAT-AATTATTTTT-TAGTAT | |
| C. parvum genotype 1 | ATATAAAATATTTTGAT--GAATATT--TATATAATATTA | TATT-------ACTA--TTTTTTTTTT-TAGTAT | |
| C. meleagridis | ATATATAATATTT-GAT--TAATATT--TATATAATATTA | TATT-------ACTA--AATTTAT----TAGTAT | |
| C. wrairi | ATATATAATATTTTGA---AAATATT--TATATAATATTA | TATT-------ACTA--TATATTTT---TAGTAT | |
| 586 | Genotype 4 | ATATATAATATTTTA-T---GATATT--TATATAATATTA | TATT-------ACTAT---TATTATT---AGTAT |
| 591 | Genotype 4 | ATATATAATATTTTA-C---GATATT--TATATAATATTA | TATT-------ACTAT---TAT---T---AGTAT |
| 524 | Genotype 3 | ATATATAATATTTTATT---AATATT--TATATAGTATTA | TATT-------ACTATA---TTTTAT---AGTAT |
| 563 | Genotype 3 | ATATATAATATTTTATT---AATATT--TATATAGTATTA | TATT-------ACTAT----TTTAT----AGTAT |
| C. parvum pig | ATATATAATATTTT--T---AATATT--TATATAATATTA | TATT-------ACTATAATTTTTATT---AGTAT | |
| C. felis | ATATATAATATTTTTTTTTAAATATTATTATGTAAGATTA | TATTTTTTAAGACTGAATTTTTAGTTTTGATAAT | |
| C. parvum dog | ATATATAATATTTAA-----CATATT--TATATAATATTA | TATT-------ACTAT--TTAT-------AGTAT | |
| C. parvum kangaroo | ATATATTATACTTTTT----AAGGTGTTTATATAATATTA | TATT-------ACTATATTTTTTT-----AGTAT | |
| Cryptosporidium sp. | ATATATAATATTACG------GTATT--TATATAATATTA | TATT-------ACTTTATTTTTAG-----AGTAT | |
| ∗∗∗∗∗ ∗∗∗ ∗ ∗ ∗∗∗ ∗∗ ∗∗∗∗ | ∗∗∗∗ ∗∗∗ ∗∗ |
The sequences from wildlife fecal samples 586, 590, 4227, 4247, 603, 592, 589, and 593 are identical in the hypervariable region, as are the sequences from samples 524, 569, and 570. The accession numbers for the PCR products from wildlife fecal samples are listed in Materials and Methods, as are the accession numbers for reference sequences.
The genotypes of reference sequences are listed by number or name. Genotypes 3 and 4 are new genotypes (this study).
Nucleotide positions in the C. parvum SSU rRNA gene sequence (accession number L16996). Asterisks indicate nucleotide identity.
Type A SSU rDNA sequence (16).
Type B SSU rDNA sequence (16).
FIG. 2.
Distance relationships among SSU rDNA PCR products from wildlife samples and reference Cryptosporidium sequences. The tree illustrates the sequence dissimiliarity among the SSU rDNA PCR products from wildlife samples and reference sequences (corresponding to positions 622 to 1014 of the C. parvum SSU rRNA gene sequence [accession number L16996]). The region examined comprises 22.5% of the SSU rRNA gene and includes the main hypervariable regions. Bootstrap values greater than 50% are shown at the nodes. Wildlife-derived sequences are indicated by boldface type along with the host, sample number, and genotype. Bar = 0.012 substitution per site.
As expected, most of the heterogeneity among the wildlife-derived SSU sequences occurred in the hypervariable region, and the sequences could be divided into three groups based on the sequence differences in this region (Table 3). The sequences from deer sample 598 and chipmunk sample 4237 were identical to C. parvum genotype 2 type A and type B SSU rDNA sequences, respectively (16). The two other groups were designated genotypes 3 and 4. To name the different alleles, numerical designations were used rather than the host animals, because common sequences were derived from different host species and some host species contained more than one genotype (Table 2). Genotype 3 comprised four deer-derived sequences (>99% identical); three of these sequences were identical in the hypervariable region (Table 3). Genotype 4 included nine sequences from deer, mouse, chipmunk, and muskrat samples (98% identical); eight of these sequences were identical in the hypervariable region (Table 3). As noted above, intraorganism sequence polymorphisms occur in the hypervariable region of the C. parvum SSU rRNA gene (i.e., between the type A and type B rDNA units) (16, 47). This implies, for example, that among the genotype 4 sequences, the sequence from muskrat sample 591 could be the companion type B unit for type A sequences amplified from other samples. Apart from sequences from muskrat sample 591 and deer sample 563, the differences among the genotype 3 and 4 alleles lay outside the hypervariable region and were limited to one or two nucleotides; it is possible that some of these differences may have been due to PCR artifacts resulting from Taq misincorporation. Genotypes 3 and 4 were most similar to the C. parvum pig genotype reference sequence (49) (98% identical) (Table 3; Fig. 2).
DHFR, POLY(T), and B-TUB PCR products.
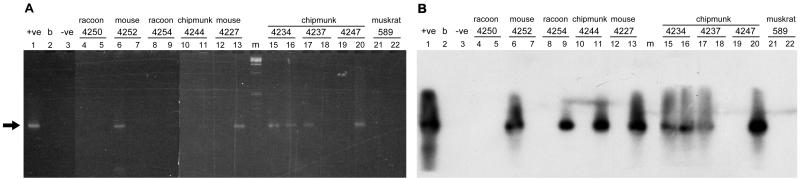
The 22 Cryptosporidium SSU-positive wildlife samples and four provisionally positive wildlife samples were subjected to PCR amplification with three additional primer sets for multilocus analysis, DHFR, POLY(T), and B-TUB (Fig. 3; Tables 1 and 2). Positive controls resulted in strong gel bands and intense hybridization in all instances, and no products were detected in lanes containing negative controls (Table 2; Fig. 3). There were 8, 6, and 13 positive PCR with the DHFR, POLY(T), and B-TUB primers, respectively, among the 26 samples. Three wildlife fecal samples (mouse sample 4227, chipmunk sample 4237, and muskrat sample 603) gave positive PCR products with all three primer pairs; five wildlife fecal samples (one deer sample, one skunk sample, two chipmunk samples, and one muskrat sample) were positive with two primer pairs (Table 2). Six wildlife samples were positive with one primer pair, and eight were negative for all three primer pairs (Table 2). Many of the samples failed to amplify; this may have been due to suboptimal reaction conditions or lower sensitivity with these primers than with the SSU set. Two of the eight rodent fecal samples (mouse sample 4252 and chipmunk sample 4244) that produced the expected DHFR product were samples that were scored as provisionally positive with the diagnostic SSU PCR assay, suggesting that they were indeed positive (Fig. 3; Table 2).
FIG. 3.
PCR analysis of wildlife fecal samples amplified with the DHFR primer set. PCR amplification products of DNA extracted from wildlife fecal samples and amplified with the DHFR typing primer set were visualized directly on agarose gels (A) and by Southern analysis with the DHFR probe (B). Two template concentrations (1× and 0.1×) were used for each sample. The left and right lanes of each pair of lanes contained the high and low concentrations, respectively. Lane +ve contained a PCR positive control consisting of 2 pg of C. parvum GCH1 DNA. Lane −ve contained a no-template control. Lane m contained size markers, and lane b was empty. The arrow indicates the expected 232-bp product.
Multilocus genotypic analysis.
The eight DHFR-positive PCR products were obtained from rodent host species, and seven of these (two products from mouse samples, three products from chipmunk samples, and two products from muskrat samples) were cloned and sequenced (Table 2). Six of the sequences were identical to the DHFR sequence of C. parvum genotype 2, while the sequence from chipmunk sample 4234 differed from the genotype 2 sequence by three nucleotides (98% identical) (data not shown). The chipmunk sample 4234 sequence was more similar to the C. parvum genotype 2 sequence than to any of the other previously reported Cryptosporidium DHFR sequences (i.e., the sequences of genotype 1 and the mouse and marsupial DHFR genotypes) (22, 39), and therefore it was considered an allele of C. parvum genotype 2.
Two of the six positive POLY(T) PCR products were cloned and sequenced. The sequence from chipmunk sample 4237 was identical to the C. parvum genotype 2 reference sequence (5). The sequence from muskrat sample 603 was only 84% identical to the C. parvum genotype 2 sequence at the nucleotide level (data not shown). For a 143-bp segment of the POLY(T) product, 10 base pair differences were reported between the sequences of C. parvum genotypes 1 and 2 (44), whereas 24 differences were detected between the muskrat sample 603 and genotype 2 sequences. BLAST analysis of the nucleotide sequence from muskrat sample 603 showed significant alignment for the entire sequence only with the C. parvum sequence. However, in the absence of more extensive information on sequence heterogeneity among polythreonine-rich genes of Cryptosporidium species and strains, it is not possible to assign the mouse sample 603 POLY(T) allele to a genotype.
Of the 13 B-TUB-positive PCR products, sequence information was limited to the product from muskrat sample 603. The sequence was 20 bp longer than expected, and the size discrepancy was due primarily to differences in the size of the intron. BLAST analysis showed that the most significant alignment with sequences in the GenBank database was the alignment with the C. parvum β-tubulin gene, followed by alignments with β-tubulin sequences from other microorganisms. When it was compared to the C. parvum genotype 1 and 2 reference sequences (30), the muskrat sample 603 sequence had 86 and 89% identity, respectively, compared with 98% identity between the sequences of the established genotypes. There were 33 polymorphic nucleotides in the exon sequences of the 458-bp PCR product, but there was only one amino acid difference between the muskrat sample 603 putative open reading frame and the C. parvum reference sequences (data not shown). The muskrat sample 603 B-TUB allele may have originated from an unknown organism, or it may be bona fide, perhaps reflecting a Cryptosporidium species or strain difference.
DISCUSSION
Molecular tools for detection and characterization of Cryptosporidium from wildlife fecal samples were successfully developed and applied in this study. These tools were used to obtain evidence of sylvatic C. parvum transmission cycles involving deer and other mammalian hosts in lower New York State, affirming the potential role of wildlife species as sources of Cryptosporidium in the catchments of public water supplies. The sylvatic cycles involve C. parvum genotype 2 strains, as well as two additional novel strains that closely resemble but are distinct from the established C. parvum genotypes in terms of their SSU rDNA alleles. With the exception of genotype 2, the previously reported genotypes have been closely associated with a single host species (24, 48), whereas in this study we detected a unique strain in four different mammalian hosts (deer, mouse, muskrat, and chipmunk).
The fecal samples in which Cryptosporidium DNA was detected were recovered from asymptomatic hosts (i.e., formed stools) and apparently were associated with relatively low levels of oocyst excretion. The extent to which oocysts present in wildlife feces enter water is not known. The survival and movement of oocysts in agricultural and sylvatic settings have received only limited study (41). In this regard, finding Cryptosporidium infections in muskrats may be of some significance as these animals are aquatic rodents that spend much of their time in water. There have been no previous reports of Cryptosporidium infection in either muskrats or skunks, except for a recent report of infection in muskrats in Poland (33). Also, the present study represents the first time that C. parvum isolates from chipmunks or muskrats have been characterized genetically.
Larger wild mammals, such as white-tailed deer, may provide a significant reservoir of infection because of the substantial quantity of droppings which they produce (37). In the present study 11% (10 of 91) of the deer samples tested were positive as determined by the diagnostic SSU PCR assay. Although this study was not designed to accurately characterize the prevalence of infection, one can appreciate the role that deer may play in terms of transmission among sylvatic mammals and water quality impact. Also, regardless of the prevalence or concentration of Cryptosporidium oocysts in wildlife species, it is clear that there is a contribution to the environmental load (37). Even in the complete absence of anthroponotic or agricultural sources, watersheds preserved in a pristine state may not be adequately protected in this regard.
Cryptosporidium infection was first reported in white-tailed deer in a captive herd in Georgia in a small sample of neonatal fawns and their mothers; most infections were asymptomatic (13). Recently, the first study of Cryptosporidium infection in wild white-tailed deer was described (29); the prevalence of infection was determined by microscopic methods. Among fawns (age, <6 months) at a wildlife center in Virginia, 9% (3 of 34 fawns) were infected. Deer that were 6 months to more than 7 years old were surveyed throughout Mississippi and were found to have an overall prevalence of Cryptosporidium infection of 5% (18 of 360 deer). From these data and our findings in New York, it appears that Cryptosporidium infection in white-tailed deer may be common and widespread.
Based on morphometric analysis, in the previous reports of infections in deer cryptosporidia were designated C. parvum (13, 29). In addition, for one of the captive deer isolates, the SSU rRNA gene sequence was identical to the C. parvum genotype 2 sequence (GenBank accession number AF093494). In the present study, the sequences from two of the positive fecal samples, one from a deer and one from a chipmunk, were also identical to C. parvum genotype 2 alleles in the amplified regions of the SSU rRNA gene; multilocus genotypic analysis of the chipmunk fecal sample provided further evidence that the identified sample contained C. parvum genotype 2 oocysts.
The relevance of the novel strains of C. parvum detected in the present study to human health is not clear. It is not known whether the C. parvum strains with SSU rRNA genotype 3 or 4 alleles are phenotypically equivalent to genotype 2 strains and whether they are capable of infecting a similar range of hosts. Interestingly, C. parvum genotype 4 SSU rDNA sequences were detected in fecal samples from four different mammalian hosts, suggesting that this genotype could have a broad host range. The novel strains were not detected in fecal samples from human cryptosporidiosis cases in New York City (unpublished data).
C. parvum genotypes 1 and 2 appear to account for the majority of cryptosporidiosis cases in humans (19; reviewed in reference 48). However, C. felis, C. meleagridis, and the C. parvum Dog genotype have been identified in human immunodeficiency virus-infected persons with diarrhea (23, 27). Thus, the risks from species or strains of Cryptosporidium that do not conform to C. parvum genotypes 1 and 2 may not be dismissed as irrelevant. Instead, the emergence of zoonotic strains in humans deserves close attention (38). There is a clear need to identify the factors responsible for human infectivity in C. parvum and to find correlative genetic markers that can be used to assess the phenotypes of sylvatic strains.
ACKNOWLEDGMENTS
This work was supported by an Environmental Protection Agency National Center for Environmental Research STAR graduate fellowship to J.F.P. and by the Columbia University Center for Environmental Research and Conservation.
The help of many persons was crucial to this study. Our thanks go to Bill Schuster and John Brady of Black Rock Forest, Tom Daniels (Fordham University), the New York City Department of Environmental Protection, Miguel Gelpi (Clinical Microbiology Service, Columbia Presbyterian Medical Center), Giovanni Widmer (Tufts University), Richard Friedman (Columbia University Computing Facility), and Terri Wu.
ADDENDUM IN PROOF
The genotype 3 Cryptosporidium SSU rRNA gene sequence from white-tailed deer fecal sample 563 (accession no. AF297512) is identical to the W4 genotype sequence obtained from storm water samples in lower New York State (accession no. AF262328) reported by Xiao et al. (Appl. Environ. Microbiol. 66:5492–5498, 2000).
REFERENCES
- 1.Ashendorff A, Princippe M A, Seeley A, DaLuca J, Beckhardt L, Faber W J, Mantus J. Watershed protection for New York City's water supply. J Am Water Works Assoc. 1997;89:75–82. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 3.Barnes D A, Bonnin A, Huang J X, Gousset L, Wu J, Gut J, Doyle P, Dubremetz J F, Ward H, Petersen C. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol. 1998;96:93–110. doi: 10.1016/s0166-6851(98)00119-4. [DOI] [PubMed] [Google Scholar]
- 4.Caccio S, La Rosa G, Pozio E. The beta-tubulin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1997;89:307–311. doi: 10.1016/s0166-6851(97)00122-9. [DOI] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Current W L, Garcia L S. Cryptosporidiosis. Clin Microbiol Rev. 1991;4:325–358. doi: 10.1128/cmr.4.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Current W L, Reese N C. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J Protozool. 1986;33:98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 8.Daniels T J, Boccia T M, Varde S, Marcus J, Le J, Bucher D J, Falco R C, Schwartz I. Geographic risk for lyme disease and human granulocytic ehrlichiosis in southern New York state. Appl Environ Microbiol. 1998;64:4663–4669. doi: 10.1128/aem.64.12.4663-4669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva A J, Bornay-Llinares F J, del Aguila de la Puente C del A, Moura H, Peralta J M, Sobottka I, Schwartz D A, Visvesvara G S, Slemenda S B, Pieniazek N J. Diagnosis of Enterocytozoon bieneusi (microsporidia) infections by polymerase chain reaction in stool samples using primers based on the region coding for small-subunit ribosomal RNA. Arch Pathol Lab Med. 1997;121:874–879. [PubMed] [Google Scholar]
- 10.da Silva A J, Bornay-Llinares F J, Moura I N, Slemenda S B, Tuttle J L, Pieniazek N J. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol Diagn. 1999;4:57–64. doi: 10.1016/s1084-8592(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 11.Deng M Q, Cliver D O. Improved immunofluorescence assay for detection of Giardia and Cryptosporidium from asymptomatic adult cervine animals. Parasitol Res. 1999;85:733–736. doi: 10.1007/s004360050623. [DOI] [PubMed] [Google Scholar]
- 12.Faubert G M, Litvinsky Y. Natural transmission of Cryptosporidium parvum between dams and calves on a dairy farm. J Parasitol. 2000;86:495–500. doi: 10.1645/0022-3395(2000)086[0495:NTOCPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Fayer R, Fischer J R, Sewell C T, Kavanaugh D M, Osborn D A. Spontaneous cryptosporidiosis in captive white-tailed deer (Odocoileus virginianus) J Wildl Dis. 1996;32:619–622. doi: 10.7589/0090-3558-32.4.619. [DOI] [PubMed] [Google Scholar]
- 14.Higgins D G, Thompson J D, Gibson T J. Using Clustal for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 15.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 17.LeChevallier M W, Norton W D. Examining relationships between particle count and Giardia, Cryptosporidium and turbidity. J Am Water Works Assoc. 1992;84:54–60. [Google Scholar]
- 18.Leng X, Mosier D A, Oberst R D. Simplified method of recovery and PCR detection of Cryptosporidium DNA from bovine feces. Appl Environ Microbiol. 1996;62:643–647. doi: 10.1128/aem.62.2.643-647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLauchlin J, Amar C, Pedraza-Diaz S, Nichols G L. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–3990. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLauchlin J, Pedraza-Diaz S, Amar-Hoetzeneder C, Nichols G L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinhardt P L, Casemore D P, Miller K B. Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol Rev. 1996;18:118–136. doi: 10.1093/oxfordjournals.epirev.a017920. [DOI] [PubMed] [Google Scholar]
- 22.Morgan U M, Monis P T, Fayer R, Deplazes P, Thompson R C A. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J Parasitol. 1999;85:1126–1133. [PubMed] [Google Scholar]
- 23.Morgan U M, Weber R, Xiao L, Sulaiman I, Thompson R C A, Ndiritu W, Lal A, Moore A, Deplazes P. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–1183. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 25.O'Donoghue P J. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- 26.Ongerth J E, Stibbs H H. Identification of Cryptosporidium oocysts in river water. Appl Environ Microbiol. 1987;53:672–676. doi: 10.1128/aem.53.4.672-676.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N, Arrowood M J, Ditrich O, Addiss D G. New Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quy R J, Cowan D P, Haynes P J, Sturdee A P, Chalmers R M, Bodley-Tickell A T, Bull S A. The Norway rat as a reservoir host of Cryptosporidium parvum. J Wildl Dis. 1999;35:660–670. doi: 10.7589/0090-3558-35.4.660. [DOI] [PubMed] [Google Scholar]
- 29.Rickard L G, Siefker C, Boyle C R, Gentz E J. The prevalence of Cryptosporidium and Giardia spp. in fecal samples from free-ranging white-tailed deer (Odocoileus virginianus) in the southeastern United States. J Vet Diagn Invest. 1999;11:65–72. doi: 10.1177/104063879901100111. [DOI] [PubMed] [Google Scholar]
- 30.Rochelle P A, Jutras E M, Atwill E R, De Leon R, Stewart M H. Polymorphisms in the β-tubulin gene of Cryptosporidium parvum differentiate between isolates based on animal host but not geographic origin. J Parasitol. 1999;85:986–989. [PubMed] [Google Scholar]
- 31.Rose J B. Environmental ecology of Cryptosporidium and public health implications. Annu Rev Public Health. 1997;18:135–161. doi: 10.1146/annurev.publhealth.18.1.135. [DOI] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Sinski E, Bednarska M, Bajer A. The role of wild rodents in ecology of cryptosporidiosis in Poland. Folia Parasitol (Prague) 1998;45:173–174. [PubMed] [Google Scholar]
- 34.Sischo W M, Atwill E R, Lanyon L E, George J. Cryptosporidia on dairy farms and the role these farms may have in contaminating surface water supplies in the northeastern United States. Prev Vet Med. 2000;43:253–267. doi: 10.1016/s0167-5877(99)00107-5. [DOI] [PubMed] [Google Scholar]
- 35.Smith H V, Rose J B. Waterborne cryptosporidiosis. Parasitol Today. 1990;6:8–12. doi: 10.1016/0169-4758(90)90378-h. [DOI] [PubMed] [Google Scholar]
- 36.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturdee A P, Chalmers R M, Bull S A. Detection of Cryptosporidium oocysts in wild animals of mainland Britain. Vet Parasitol. 1999;80:273–280. doi: 10.1016/s0304-4017(98)00226-x. [DOI] [PubMed] [Google Scholar]
- 38.Tzipori S, Griffiths J K. Natural history and biology of Cryptosporidium parvum. Adv Parasitol. 1998;40:5–36. doi: 10.1016/s0065-308x(08)60116-5. [DOI] [PubMed] [Google Scholar]
- 39.Vasquez J R, Gooze L, Kim K, Gut J, Petersen C, Nelson R G. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–165. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 40.Wade S E, Mohammed H O, Schaaf S L. Prevalence of Giardia sp., Cryptosporidium parvum and Cryptosporidium muris (C. andersoni) in 109 dairy herds in five counties of southeastern New York. Vet Parasitol. 2000;93:1–11. doi: 10.1016/s0304-4017(00)00337-x. [DOI] [PubMed] [Google Scholar]
- 41.Walker M J, Montemagno C D, Jenkins M B. Source water assessment and nonpoint sources of acutely toxic contaminants: a review of research related to survival and transport of Cryptosporidium parvum. Water Resour Res. 1998;34:3383–3392. [Google Scholar]
- 42.Weber R, Bryan R, Bishop H S, Wahlquist S, Sullivan J, Juranek D. Threshold of detection of Cryptosporidium oocysts in human stool specimens: evidence for low sensitivity of current diagnostic methods. J Clin Microbiol. 1991;29:1323–1327. doi: 10.1128/jcm.29.7.1323-1327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webster J P, Macdonald D W. Parasites of wild brown rats (Rattus norvegicus) on UK farms. Parasitology. 1995;11:247–255. doi: 10.1017/s0031182000081804. [DOI] [PubMed] [Google Scholar]
- 44.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:223–239. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 45.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the β-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Xiao L, Alderisio K, Limor J, Royer M, Lal A A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol. 2000;66:5492–5498. doi: 10.1128/aem.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao L, Limor J R, Li L, Morgan U, Thompson R C A, Lal A A. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J Eukaryot Microbiol. 1999;46:44S–45S. [PubMed] [Google Scholar]
- 48.Xiao L, Morgan U M, Fayer R, Thompson R C A, Lal A A. Cryptosporidium systematics and implications for public health. Parasitol Today. 2000;16:287–292. doi: 10.1016/s0169-4758(00)01699-9. [DOI] [PubMed] [Google Scholar]
- 49.Xiao L, Morgan U M, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu G, Marchewka M J, Ennis J G, Keithly J S. Direct isolation of DNA from patient stools for polymerase chain reaction detection of Cryptosporidium parvum. J Infect Dis. 1998;177:1443–1446. doi: 10.1086/517834. [DOI] [PubMed] [Google Scholar]