Abstract
Sex determination is a critical element of successful vertebrate development, suggesting that sex chromosome systems might be evolutionarily stable across lineages. For example, mammals and birds have maintained conserved sex chromosome systems over long evolutionary time periods. Other vertebrates, in contrast, have undergone frequent sex chromosome transitions, which is even more amazing considering we still know comparatively little across large swaths of their respective phylogenies. One reptile group in particular, the gecko lizards (infraorder Gekkota), shows an exceptional lability with regard to sex chromosome transitions and may possess the majority of transitions within squamates (lizards and snakes). However, detailed genomic and cytogenetic information about sex chromosomes is lacking for most gecko species, leaving large gaps in our understanding of the evolutionary processes at play. To address this, we assembled a chromosome-level genome for a gecko (Sphaerodactylidae: Sphaerodactylus) and used this assembly to search for sex chromosomes among six closely related species using a variety of genomic data, including whole-genome re-sequencing, RADseq, and RNAseq. Previous work has identified XY systems in two species of Sphaerodactylus geckos. We expand upon that work to identify between two and four sex chromosome cis-transitions (XY to a new XY) within the genus. Interestingly, we confirmed two different linkage groups as XY sex chromosome systems that were previously unknown to act as sex chromosomes in tetrapods (syntenic with Gallus chromosome 3 and Gallus chromosomes 18/30/33), further highlighting a unique and fascinating trend that most linkage groups have the potential to act as sex chromosomes in squamates.
Keywords: genome evolution, genomics, herpetology, sex chromosomes, sex determination
Sexual reproduction is ubiquitous in vertebrates but the ways in which species determine sex differs (Otto and Lenormand 2002; Graves 2008). Most vertebrate species determine sex using genetic cues inherited from one of their parents (i.e., sex chromosomes), either from the sperm (male heterogamety; XY) or the egg (female heterogamety; ZW). Traditionally, cytogeneticists identified sex chromosomes by karyotyping a male and female of a species and looking for morphological differences between the two karyotypes (Stevens 1905). Until recently, the majority of sex chromosome research was restricted to these species whose sex chromosomes were heteromorphic, or visibly different under a light microscope, such as mammals (XY) and birds (ZW). As a consequence, much of what we know about vertebrate sex chromosomes comes from studies in mammals and birds who possess ancient, degenerated sex chromosomes, where transitions in sex-determining systems are rare or non-existent (Ohno 1967; Bachtrog 2003; Graves 2008; Zhou et al. 2014). However, other vertebrate groups, such as fish, amphibians, and squamate reptiles, frequently possess homomorphic sex chromosomes, which appear identical under the light microscope, historically stifling investigations of sex chromosome evolution in these groups (Hillis and Green 1990; Ezaz et al. 2009; Schultheis et al. 2009; and reviewed in Gamble 2010; Adolfsson and Ellegran 2013; Furman et al. 2020; Kostmann et al. 2021).
Sex chromosomes evolve when one member of an autosomal pair acquires a sex determining allele (Muller 1914; Ohno 1967; Graves 2008). Through a multitude of mechanisms, recombination can be suppressed between the nascent X/Y or Z/W chromosomes (Ohno 1967; Charlesworth 1991; Ponnikas et al. 2018). After recombination is suppressed, the sex-limited chromosome (Y or W) begins to accumulate deleterious mutations and degenerate by losing functional copies of genes and accumulating segments of repetitive DNA (Muller 1918; 1964; Ohno 1967; Bull 1983; Charlesworth 1991; Charlesworth and Charlesworth 2000; Bachtrog 2013; Wright et al. 2016). In some cases, this non-recombining region can expand outward over time—reducing sequence identity across the sex-linked region of the sex chromosome pair until the sex-limited chromosome becomes heteromorphic (Stevens 1905; Charlesworth 1978; Lahn and Page 1999; García-Moreno and Mindell 2000; Handley et al. 2004; Graves 2008; Bachtrog 2013). The ability to observe heteromorphic sex chromosomes under a light microscope has led to numerous discoveries in sex chromosome evolution, but as most vertebrate species do not possess heteromorphic sex chromosomes, other technologies are needed to identify sex chromosomes in these species (Bachtrog et al. 2014). Thanks to recent advances in sequencing and cytogenetic methods, empiricists are now able to identify and characterize homomorphic sex chromosomes in diverse taxa (Gamble et al. 2015, 2017; Augstenová et al. 2018; Nielsen et al. 2018, 2019a, 2019b, 2020; Pan et al. 2019, 2021a, b; Rovatsos et al. 2019; Sidhom et al. 2020; Keating et al. 2020, 2021). The recent ability to characterize homomorphic sex chromosomes has allowed researchers to test existing hypotheses in new ways, transforming our understanding of sex chromosome evolution (Bull 1983; Ogata et al. 2007; Uno et al. 2008; Blaser et al. 2013; Gamble et al. 2015a; Augstenová et al. 2018; Jeffries et al. 2018; Hundt et al. 2019; Kottler et al. 2020).
An extremely common method for identifying sex chromosomes in species lacking heteromorphic sex chromosomes is the identification of sex-specific genetic markers from restriction-site associated DNA sequencing (RADseq) data. By sequencing multiple males and females for a species with this method, alleles can be identified that exist in one sex and not the other (Gamble and Zarkower 2014; Gamble et al. 2015a). However, this method alone may not identify both the sex chromosome system and linkage group (or syntenic genomic region) to which those sex-limited alleles belong. There are exceptions to this where, by chance, sex-linked genes can be identified and successfully mapped to a distant reference genome in order to identify the sex chromosome linkage group (e.g., Nielsen et al. 2019a, 2020; Keating et al. 2020). However, estimating the total number of transitions among sex chromosomes from changes in heterogamety alone likely underestimates the true number of turnovers by a large margin (Gamble et al. 2015a; Jeffries et al. 2018). Indeed, at shallow evolutionary scales, RADseq data alone are unable to distinguish between (1) sex chromosomes and their associated sex-determining systems inherited from a common ancestor and (2) cis-transitions that independently evolved to the same linkage group, i.e., homologous cis-transitions (Bachtrog et al. 2014; Blaser et al. 2014; Augstenová et al. 2018; Jeffries et al. 2018). High-quality genome assemblies can be used to supplement this linkage information to elucidate the presence of cryptic transitions, but for reptile groups with few or no high-quality reference genomes, such as chameleons and geckos, transitions have only been broadly estimated using changes in patterns in heterogamety in these groups, i.e., XY to ZW or vice versa (Gamble et al. 2015a; Nielsen et al. 2018). To successfully recognize these more difficult to identify transitions and test hypotheses regarding sex chromosome turnover, there should be a push to generate high-quality reference genomes within groups that possess homomorphic sex chromosomes (Stöck et al. 2021)—including groups such as geckos (infraorder Gekkota).
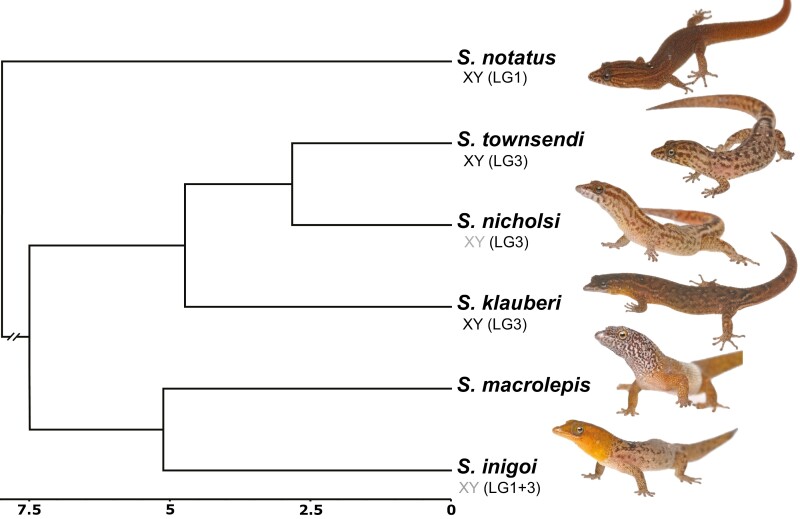
Geckos are a speciose clade of squamate reptiles, making up approximately 20% of all squamate species (Uetz et al. 2021). Impressively, geckos also account for more than 1/3 of all known transitions in sex determining systems across all reptiles (Gamble et al. 2015a; Gamble et al. 2018). Ancestrally, geckos possessed temperature-dependent sex determination (TSD) and have since undergone more than 25 transitions between TSD, XY, and ZW systems, with sex chromosomes that are generally homomorphic (Pokorna and Kratochvíl 2009; Gamble et al. 2015a; Rovatsos et al. 2019). Although there is an extremely useful model system to study sex chromosomes, geckos have not, until recently, had chromosome-level reference genomes available to estimate linkage information for sex chromosome turnovers in geckos (Liu et al. 2015; Xiong et al. 2016; Hara et al. 2018; Yamaguchi et al. 2021). Thus, previous work in most gecko groups has been restricted to characterizing the aforementioned broad changes in heterogametic systems without the ability to test hypotheses about sex chromosome conservation and turnover (although there are some exceptions, e.g., Nielsen et al. 2019a; Rovatsos et al. 2019, 2021; Keating et al. 2020). Indeed, until now this has also been the case for the charismatic Neotropical geckos of the genus Sphaerodactylus (Figure 1).
Figure 1.
Overview of the study system: Time-calibrated phylogenetic tree (Daza et al. 2019) for Sphaerodactylus geckos from within the Puerto Rican Bank and an outgroup (S. notatus; node ~20 mya) with previously identified sex chromosome sex systems in grey; new information identified here in black.
The gecko family Sphaerodactylidae comprises 12 genera distributed across 5 continents and a diversity of environments, yet only 4 genera have any information regarding sex chromosomes (reviewed in Gamble et al. 2018). Karyotypes of male and female Euleptes europaea suggest an XY system with an unknown linkage group (Gornung et al. 2013). A conserved ZW system was discovered across the Caribbean genus Aristelliger, syntenic with Gallus chromosome 2 (Keating et al. 2020). Gamble et al. (2018) found an XY system in the South American (Trinidad) Gonatodes ferrugineus, albeit with an unknown linkage group. Lastly, XY systems were discovered in Sphaerodactylus nicholsi and S. inigoi (both native to the Puerto Rican Bank), also with unknown linkage groups (Gamble et al. 2015a). Taken together, these results suggest a high diversity of sex chromosome systems within Sphaerodactylidae and likely many more will be uncovered. However, the glaring deficiency—not knowing the sex chromosome linkage groups in most taxa—hampers our development of a broader understanding of sex chromosome evolution in this group. Therefore, the logical next step in diagnosing the diversity of sex chromosomes across sphaerodactylids is to connect heterogamety (XY vs. ZW) in species with known sex chromosomes and their close relatives with linkage groups.
To begin addressing sex chromosome evolution in Sphaerodactylus geckos, we sequenced and assembled a chromosome-scale genome for Townsend’s least gecko (Sphaerodactylus townsendi) from Puerto Rico and examined patterns of sex chromosome conservation and turnover among a small number of related Sphaerodactylus species. We chose to focus on the Puerto Rican Sphaerodactylus because we know more about their sex determining systems than most other genera in the family (Gamble et al. 2015a, 2018) and recent phylogenetic analyses provide a robust evolutionary framework for examining traits within this system (Daza et al. 2019; Pinto et al. 2019a). We set out to determine whether or not Sphaerodactylus possesses an ancestral XY system that has been conserved across sampled Sphaerodactylus species. To test this, we collected a patchwork of genomic data from six Sphaerodactylus species (five from the Puerto Rican species radiation and one outgroup, S. notatus, native to southern Florida and the northern Caribbean) to accompany the new S. townsendi reference genome. This sampling represents ~5% of described Sphaerodactylus species (6 of 107 species—Uetz et al. 2021). The data included in this study were restriction-site associated DNA sequencing (RADseq), RNA sequencing (RNAseq), and whole-genome re-sequencing (WGS). We used these data to identify and confirm the sex chromosome linkage group in a subset of these species (S. townsendi, S. nicholsi, S. inigoi, and S. notatus). Then, we used a preliminary dataset generated from additional taxa (S. klauberi and S. macrolepis) to extrapolate from these more well-substantiated species to detect conserved patterns on the sex chromosomes. We identified multiple cis-transitions within Sphaerodactylus XY systems and report that these transitions are utilizing two linkage groups whose syntenic regions in chicken (Gallus gallus) were previously unknown to act as sex chromosomes in other tetrapods (Figure 1). With these results, we begin to gauge the dynamic nature of sex chromosome evolution in Sphaerodactylus, which in turn may provide insight into the sex chromosome evolution of other underrepresented taxa with frequent sex chromosome transitions across the tree of life.
Methods
Data Generation
We generated a high-quality reference genome for a male S. townsendi (indiv. TG3544 [male]) collected in Playa de Ponce, Puerto Rico (17.96439, −66.61387). Importantly, S. townsendi is one of the smallest terrestrial vertebrate species, weighing < 0.5 g and an average snout-vent length (SVL) of 24.6 mm (Thomas and Schwartz 1966), and as a result, there is no voucher of TG3544 because the entire specimen was used for HMW DNA extraction. Genome assembly combined linked-read sequencing (10X genomics), chromatin-contact sequencing (Hi-C), nanopore long-read sequencing, and whole-genome re-sequencing (WGS) using paired Illumina reads. For linked-read and nanopore long read sequencing, we extracted high molecular-weight (HMW) DNA from blood and liver tissue of one S. townsendi (TG3544 [male]) using a published DNA extraction protocol designed for low input (Pinto et al. 2021). For re-sequencing and all other DNA-related experiments described herein, we used Qiagen DNeasy DNA extractions of tail or liver tissues.
For the reference genome sequencing, we generated and sequenced a single 10X Chromium library (S. townsendi TG3544 [male]) across 2 lanes of Illumina HiSeqX (HudsonAlpha Institute for Biotechnology, Huntsville, AL), proximally ligated input DNA from blood and liver tissue in-house (S. townsendi TG3718 [male]) using the Arima-HiC kit (Arima Genomics, San Diego, CA) and sequenced it as a 400-600bp insert Illumina library using the NEBNext Ultra II Library Preparation kit (New England Biolabs [NEB], Ipswich, MA) on an Illumina NovaSeq lane (Novogene, Davis, CA); we generated 2 nanopore sequencing libraries (Oxford Nanopore Technologies [ONT], Oxford, UK) using the Ligation Sequencing Kit (SQK-LSK109) of HMW DNA (indiv. TG3544) and sequenced each library on its own flowcell (FLO-MINSP6) to completion (~60 h) on a single MinION device (MIN-101B); lastly, we made and sequenced a 400–600 bp insert re-sequencing library on an Illumina HiSeqX (Psomagen, Rockville, MD).
For reference genome annotation, we conducted additional RNA sequencing in S. townsendi. RNA sequencing (RNAseq) methods are described thoroughly by Pinto et al. (2019b); briefly, we extracted RNA from flash-frozen tissues stored at −80°C in Trizol reagent and generated sequencing libraries using the KAPA Stranded mRNA-Seq Kit for Illumina Platforms (KR0960 [v5.17]). We deep-sequenced RNAseq libraries from a whole head from a male (TG3467) and a whole embryo, 11 days post-oviposition (dpo) of unknown sex (TG3715), which were sequenced on an Illumina HiSeqX (Psomagen, Rockville, MD). For downstream sex chromosome analyses (see below), we sequenced additional RNAseq libraries from whole heads (males and females) of S. macrolepis and S. inigoi preserved in RNAlater. These libraries were sequenced using paired-end reads (125-bp) on an Illumina HiSeq2500 (Medical College of Wisconsin, Milwaukee, WI).
To identify and explore the sex-linked regions of the genome, we generated whole-genome re-sequencing data for 1M/1F of S. townsendi, S. nicholsi, S. klauberi, and S. notatus. Additionally, we acquired population-level RADseq data for multiple males and females of S. townsendi, S. nicholsi, S. inigoi, and S. notatus. For whole genome re-sequencing data, we generated Illumina libraries for each individual using the NEBNext Ultra II kit (New England Biolabs). For RADseq data, we followed a modified protocol from Etter et al. (2011) as outlined in Gamble et al. (2015a) (Rohland and Reich 2012). Libraries were pooled and sequenced using paired-end 100-bp or 150-bp reads on an Illumina HiSeq2000 at the University of Minnesota Genomics Center (Minneapolis, MN) or an Illumina HiSeqX at Psomagen. In sum, our final dataset for assessing sex chromosome dynamics in Sphaerodactylus included re-sequencing [1M, 1F] of S. townsendi, S. nicholsi, S. klauberi, S. notatus, and a single S. macrolepis male; RADseq data for S. townsendi [7M, 7F], S. nicholsi [6M, 6F], S. inigoi [7M, 9F] (from Gamble et al. 2015a), S. notatus [8M, 7F]; and RNAseq data from S. macrolepis [2M, 2F] and S. inigoi [2M, 2F]. The four RADseq species contained representative samples from across their known range. These data sources are summarized in Table 1.
Table 1.
Table tracking the available data for each species used in this study
| Species | Reference genome | Re-sequencing | RADseq | RNAseq |
|---|---|---|---|---|
| S. townsendi | Yes | 1.1 | 7.7 | 1.0 |
| S. nicholsi | — | 1.1 | 6.6 | — |
| S. klauberi | — | 1.1 | — | — |
| S. inigoi | — | — | 7.9 | 2.2 |
| S. macrolepis | — | 1.0 | — | 2.2 |
| S. notatus | — | 1.1 | 8.7 | — |
Notation in each cell refers to males and females (M.F).
Transcriptome Assembly
We quality and adapter trimmed our RNAseq reads using Trim Galore!, filtered PCR duplicates using bbmap, and subsampled 50,000,000 PE reads for each tissue using seqtk. In an isolated docker computing environment (Merkel 2014), we normalized cleaned reads and assembled de novo transcriptomes for each tissue using Trinity [v2.8.4] (Grabherr et al. 2011) in the De novo RNAseq Assembly Pipeline (DRAP) [v1.92] (Cabau et al. 2017). For S. townsendi, we generated both a “head” and “embryo” de novo assembly and combined them using the runMeta function in DRAP. An in-depth description of the utility of DRAP in the production of high-quality transcriptome assemblies can be found elsewhere (Cabau et al. 2017; Pinto et al. 2019b).
Reference Assembly, Annotation, and Characterization
We used a 6-part, iterative assembly approach to integrate the five different sequencing experiments (outlined in Table 2). In an effort to make these genome assembly efforts reproducible across platforms, all genome assembly steps—except for the initial SuperNova assembly (conducted at HudsonAlpha) and three steps conducted in docker environments (details below)—were conducted in conda virtual environments that contained the following versions of these programs (in alphabetical order): ARCS [v1.1.1] (Yeo et al. 2018), assembly-stats [v1.0.1], bamtools [v2.5.1] (Barnett et al. 2011), BBmap [v38.79] (Bushnell, 2014), bcftools [v1.9] (Li, 2011), bedtools [v2.29.2] (Quinlan and Hall, 2010), diamond [v0.9.14] (Buchfink et al. 2015), FastQC (Andrews, 2010), freebayes [v1.3.2] (Garrison and Marth, 2012), HiSat2 [v2.1] (Kim et al. 2019), merqury [v1.3.0] (Rhie et al. 2020), minimap2 [v2.17] (Li, 2018), mosdepth [v0.2.6] (Pedersen and Quinlan, 2018), parallel [v20200322] (Tange, 2018), picard tools [v2.22], pixy [v1.1.1] (Korunes and Samuk, 2021), sambamba [v0.7.1] (Tarasov et al. 2015), samtools [v1.6] (Li and Durbin 2009), seqkit [v0.12] (Shen et al. 2016), seqtk [v1.3] (https://github.com/lh3/seqtk), STACKS [v2.3] (Catchen et al. 2013), Tigmint [v1.1.2] (Jackman et al. 2018), TGS-GapCloser [v1.0.1] (Xu et al. 2020), Trim Galore! [v0.5] (Martin, 2011; https://doi.org/10.5281/zenodo.5127899), and vcftools [v0.1.15] (Danecek et al. 2011).
Table 2.
Tracking contiguity of the genome assembly across versions using 4 common metrics: Scaffold N50, size of the smallest scaffold comprising the largest 50% of the assembly; Scaffold L50 number of scaffolds comprising the largest 50% of the genome; Scaffolds, total number of scaffolds comprising the full assembly; Size, the approximate number of base pairs in the assembly. BUSCO—percent complete Core Vertebrate Genes (CVG)
| Assembly | Step | N50 | L50 | Scaffolds | Size | BUSCO |
|---|---|---|---|---|---|---|
| v1.1 | SuperNova | 12,629,056 | 37 | 58,149 | 2.0 Gb | 85.5% |
| v1.2 | Tigmint | 6,460,730 | 69 | 59,469 | 2.0 Gb | 85.5% |
| v1.3 | ARCS | 7,457,274 | 57 | 58,603 | 2.0 Gb | 85.5% |
| v1.4 | TGS-GapCloser | 7,468,733 | 57 | 58,603 | 2.0 Gb | 88.0% |
| v1.5 | NextPolish | 7,605,248 | 57 | 58,603 | 2.0 Gb | 88.8% |
| v1.6 | 3D-DNA | 126,215,344 | 7 | 56,114 | 2.0 Gb | 88.9% |
| v1.7 | Redundancy-filter | 134,006,883 | 6 | 32,127 | 1.9 Gb | 88.7% |
| v1.8-v2.1 | +10kb cutoff | 134,006,883 | 6 | 1,823 | 1.8 Gb | 88.3% |
To assemble the reference genome from sequence data, we generated an initial assembly using SuperNova [v2.1.1] (Weisenfeld et al. 2017) using ~80% of our total 10X sequencing reads [assembly v1.1]. To improve this assembly, we broke potential misassemblies accumulated during the assembly process using Tigmint [assembly v1.2] and re-scaffolded with 100% of our 10X reads using ARCS [assembly v1.3]. Next, we incorporated the quality-filtered ONT reads (total reads = 435,394; total bp = 6,565,554,881; mean read length = 15,079.6; largest/smallest read = 162,107/1,001) to fill gaps in the genome using TGS-GapCloser [assembly v1.4]. Then, we combined Illumina data with ONT data to polish the genome using NextPolish [v1.3.1] (Hu et al. 2020) [assembly v1.5]. We broke and re-scaffolded the polished assembly using 2 iterations of 3D-DNA [v201008] (Dudchenko et al. 2017), which yielded 17 chromosome-scale scaffolds with no apparent large-scale misassemblies [assembly v1.6]. We visualized the final HiC contact map for misassemblies and with no large-scale misassemblies visible, we removed only small “blemishes” from the contact map using Juicebox Assembly Tools [v1.11] (Durand et al. 2016). We removed duplicate assembled regions by mapping smaller assembled regions to the 17 chromosome-level scaffolds using RaGOO [v1.11] (Alonge et al. 2019) and removing scaffolds with high grouping confidence scores (i.e., 1.0) [assembly v1.7]. Lastly, to facilitate genome annotation, we removed scaffolds to a minimum length of 10Kb [assembly v1.8].
To functionally annotate the genome assembly, we used the Funannotate pipeline [v1.5.0] (Palmer 2018) in an isolated docker computing environment (Merkel 2014). Briefly, Funannotate provides a pipeline to soft-mask the assembly (https://github.com/Dfam-consortium/RepeatModeler) and predict gene models using both curated databases (Simão et al. 2015) and custom transcriptomic data (Haas et al. 2008; Keller et al. 2011; Hoff et al. 2015). To facilitate genome annotation, we provided transcriptomic data in the form of our aforementioned de novo meta transcriptome assembly. These files were then incorporated directly into the funannotate pipeline to inform the annotation process. The final annotated genome assembly was recoded to be submitted to GenBank as “MPM_Stown_v2.2.” (https://www.ncbi.nlm.nih.gov/assembly/GCA_021028975.1). This annotation was sufficient to inform the questions asked in this study; however, genome annotation is a challenging process and re-annotation using the RefSeq pipeline at NCBI, which has been shown to improve annotations, is currently underway.
To assess the completeness and quality of the reference genome and de novo transcriptome, we employed metrics that query the assemblies for highly-conserved orthologous proteins and kmers. First, we used Benchmarking Universal Single-Copy Orthologs (BUSCO) [v5.1.2] (Simão et al. 2015), implemented on the gVolante web server [v2.0.0] (Nishimura et al. 2017), to query multiple databases of conserved orthologs: Core Vertebrate Genes (CVG) and tetrapoda_odb10. We calculated these metrics at each stage of genome assembly [assembly v1.1-v1.8] in its completeness as Supplementary Table 1 and present a subset of this information in Table 2. Second, we calculated completeness and quality metrics using merqury, which compares kmers from the genome assembly with the unassembled Illumina WGS reads.
Sex Chromosome Identification and Comparative Genomics
WGS + RNAseq
We mapped WGS data to the genome using minimap2 and RNAseq data using hisat2. For WGS, we quantified per-individual read depth in 500Kb windows using mosdepth. We normalized each sample by its median read depth before calculating the male/female read depth in R [v3.6.2] (R Core Team 2016). Importantly, for all species with WGS data, we identified no differences in read depth between males and females, which suggested that analyses examining sequence differences in this region would be successful. We called SNPs for WGS using freebayes to generate an “all-sites” vcf file and calculated pi in 500Kb windowed using pixy. For RNAseq data, we called SNPs separately using freebayes to include only variable sites and calculated pi in 500Kb windowed using vcftools.
RADseq
Restriction site-associated DNA sequencing (RADseq) has been shown to be an essential tool for the identification of sex chromosome systems in species lacking heteromorphic sex chromosomes (e.g., Gamble et al. 2015a, 2018; Nielsen et al. 2019, 2020; Keating et al. 2020; Pan et al. 2021a, b). The expectation is that the only genomic region that should contain sex-specific RADtags are the non-recombining regions of the Y/W chromosomes (Gamble and Zarkower 2014; Gamble 2016). We can interpret areas with an abundance of mapped sex-specific RADtags as regions within the non-recombining region of the sex chromosomes. When analyzed alone, RADseq can identify sex chromosome systems but can say nothing of sex chromosome linkage or the size of the non-recombining region in the focal taxon (Gamble et al. 2015a, 2017, 2018; Fowler and Buonaccorsi 2016; Hundt et al. 2019; Nielsen et al. 2019a, 2020). However, when analyzed in conjunction with a reference genome, we can both map these sex-specific RADtags to identify the linkage group and analyze the sequences in this region by calling SNPs from the raw data (Gamble 2016; Pan et al. 2019). Thus, we can use both methods to confirm a region is sex-specific by looking for coincident locations of male/female differences across species.
Reference-Free Analyses
We identified sex-specific RAD loci and their gametologous counterparts using the published RADtools pipeline plus a custom perl script (Gamble et al. 2015a; Nielsen et al. 2019b). We validated a subset of S. townsendi RADtags as Y-linked via PCR; primer pairs S70_8.05_F1/R1 [[5ʹ-CTTGTCACTTTTAGTGGGCACTG-3ʹ/5ʹ-GGATGCACGTTGTTGAACAAAAC-3ʹ]] and S272_192_F2/R1 [5ʹ-TTCAAAGCAAGAGATGTTCAGCG-3ʹ/5ʹ-GATCCTGGAATACGGMACCATGA-3ʹ], whereas those in S. nicholsi and S. inigoi were validated previously (Gamble et al. 2015a,b).
Reference-Assisted Analyses
We mapped RADseq reads to the genome using minimap2 and used refmap.pl pipeline in STACKS to call SNPs separately for each species. We calculated male/female FST across the genome in 500Kb windows using vcftools and mapped the sex-specific RAD loci identified using the RADtools pipeline to the genome (Weir and Cockerham 1984). We expected that each dataset would converge on specific areas in the genome (high M/F FST and many sex-specific markers added).
Genome Synteny and Characterization
As this is one of the only two chromosome-scale gecko genomes currently available, to investigate synteny among a gecko and other reptiles, we conducted a few analyses to characterize it relative to other reptile genomes. Specifically, we identified the syntenic regions of the S. townsendi genome across four high-quality genomes available on Ensembl for reptiles: green anole (Anolis carolinensis, Acar2.0; Alföldi et al. 2011), Indian cobra (Naja naja, Nanav5; Suryamohan et al. 2020), common wall lizard (Podarcis muralis, PodMur_1.0; Andrade et al. 2019), and domestic chicken (Gallus gallus, GRCg6a). We identified syntenic regions in S. townsendi with these other reptile taxa using MCScanX (Wang et al. 2012). To visualize MCScanX synteny results, we generated synteny plots using SynVisio software (https://github.com/kiranbandi/synvisio). In addition to synteny, GC content is thought to be an important characteristic of the genome with potential implications for the recombination landscape and life history strategies across taxa and has not been extensively characterized across squamates (Eyre-Walker et al. 2001; Charlesworth et al. 2020). Thus, we comparatively explored GC content within Podarcis, Anolis, and Naja in 500Kb windows using python script (slidingwindow_gc_content.py) from Schield et al. (2019).
Results
Genome Characterization
The best a priori estimate of haploid chromosome number in Sphaerodactylus townsendi is n = 17 as identified from the karyotypes of the closely related species S. ariasae, S. plummeri, and S. streptophorus (presented here in Supplementary Figure 1). For the new S. townsendi reference genome, 97.3% of the de novo assembly was anchored onto the 17 chromosome-length scaffolds using HiC. We estimated the genome size using kmers from the raw illumina reads at 1.87 Gb, which is close to our final assembly size of approximately 1.82 Gb. The total GC content was 46.0% (±11.1%) and we soft-masked 44.47% of the genome modeled as repetitive DNA. Our BUSCO score calculated against 5310 conserved tetrapod orthologs (tetrapoda_odb10) was 88.3% complete. The assembly contained 87.6% single-copy orthologs, 0.7% duplicated ortholog copies, 3.9% fragmented copies, and 7.8% missing gene copies. When examining a subset of core vertebrate genes (CVG) with BUSCO, which may be a more reliable subset of genes for BUSCO to identify when present than other ortholog datasets (Yamaguchi et al. 2021), our assembly maintained a score of 95.7%. However, given the overall limitations that constrain evaluations of genomic completeness using BUSCO (Botero-Castro et al. 2017; Peona et al. 2020), we accompanied these measures using a kmer-based method with merqury. Similar to BUSCO, we calculated a completeness value of 89.2% using our S. townsendi re-sequencing data with merqury.
We compared synteny maps with three other reptile species: chicken (Gallus gallus), green anole (Anolis carolinensis), and wall lizard (Podarcis muralis) and the information from the physically mapped Hokou gecko (Gekko hokouensis) genome (Srikulnath et al. 2015). The Indian cobra (Naja naja) was omitted from the table due to its collinearity with Anolis macrochromosomes. Most linkage groups (chromosome-scale scaffolds) maintained a one-to-one relationship with Podarcis chromosomes and the known syntenic configurations in G. hokouensis (Table 3).
Table 3.
A key to navigate synteny across largest fragments of the reference genome assembly relative to Anolis, Podarcis, and Gallus, according to the Gekko hokouensis (Gekko) physical mapping
| Sphaerodactylus townsendi | Anolis carolinensis | Podarcis muralis |
Gallus
gallus |
Gekko hokouensis |
|---|---|---|---|---|
| LG1 | 1q | 3 | 3 | 1p |
| LG2 | 1p | 1 | 5,7 | 2 |
| LG3 (XY) | 2q | 2 | 12,13,16,18,30,33 | 1q |
| LG4 | 3q | 4 | 1q,14 | 13 |
| LG5 | 4q | 6,18 | 8,26,28 | 3 |
| LG6 | 5p | 10 | 1p,23 | 14 |
| LG7 | 2p | 11,17 | ZW | ZW |
| LG8 | 3p | 5,14 | 6,9 | 15 |
| LG9 | 4p | 7 | 2q | unplaced |
| LG10 | micro | 9 | 4q | 7 |
| LG11 | 6q | 12 | 2p,27 | 8 |
| LG12 | micro | 15,ZW | 17,22,24*** | 9 |
| LG13 | micro | 16,ZW | 4p,15 | 11 |
| LG14 | 4 | 8 | 11 | unplaced |
| LG15 | 6p | 13 | 27 | 12 |
| LG16 | micro | 8 | 21 | unplaced |
| LG17 | micro | 14 | 10 | unplaced |
Scaffolds were called if linkage groups described by Srikulnath et al. (2015) were corroborated by syntenic mapping to Anolis, Podarcis, and/or Gallus. Note that the snake (Naja) was omitted due to its collinearity with Anolis genome.
*** indicates changes in annotated chicken chromosomes making up the linkage group from that reported by Srikulnath et al. (2015) from “21 and 25” to “22 and 24.”
Sex Chromosome Identification and Description
Across species with whole-genome re-sequencing data (WGS) for both a male and female, we observed no differences in read depth between the sexes in any species (Supplementary Figure 2). Since read mapping did not differ between the sexes, we could successfully call SNPs and analyze sequence differences between the sexes. Thus, we called and analyzed SNPs for each of our datasets: WGS, RNAseq, and RADseq.
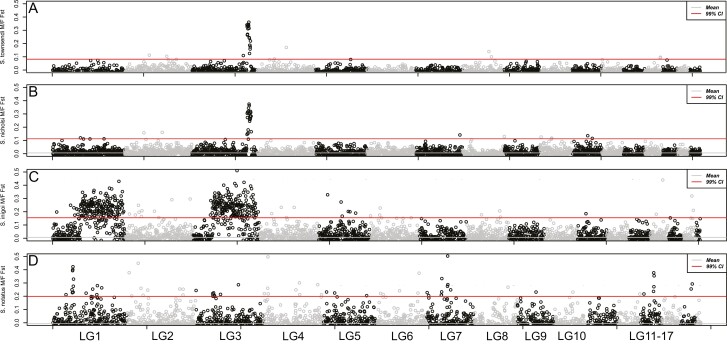
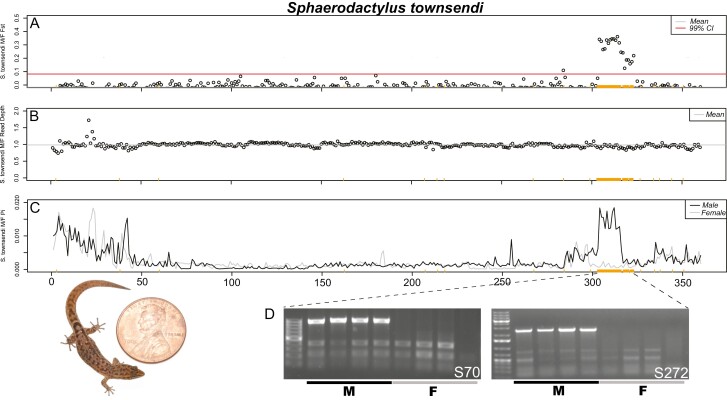
For species with RADseq data from multiple males and females, we identified a list of sex-specific RADtags using the Gamble et al. (2015a, b) pipeline. For all species, we identified an excess of confirmed male-specific RADtags: S. townsendi (M = 431/F = 0), S. nicholsi (M = 186/F = 11), S. inigoi (M = 157/F = 0), and S. notatus (M = 21/F = 2). Previous work had validated a subset of these male-specific markers as Y-linked in S. nicholsi and S. inigoi using PCR (Gamble et al. 2015a). The majority of male-specific RADtags identified in each species, mapped to a small number of linkage groups in the S. townsendi genome: S. townsendi (LG3—87%), S. nicholsi (LG3—86%), S. inigoi (LG1—46%; LG3—51%), and S. notatus (LG1—62%). Importantly, the eight-remaining male-specific RADtags for each species mapped randomly throughout the rest of the genome. When examining male/female FST, we observed a single, solitary peak of elevated FST in the same location for both S. townsendi (LG3) and S. nicholsi (LG3) (Figures 2–4), whereas S. inigoi presented two regions of elevated FST spanning both LG1 and LG3 (Figures 2, 4, and 5). The FST scan for S. notatus included more noise than the other three taxa, likely due to its phylogenetic distance from the reference taxon (diverged ~20 mya; Figure 1). However, we identified a credible peak on LG1 that coincided with a majority (48%) of the mapped male-specific RADtags (Figure 5). Notably, in S. notatus, two male-specific RADtags mapped coincidentally to the FST peak on LG4; however, the remaining spuriously mapped RADtags did not co-locate with any of the alternative FST peaks. Thus, the genomic regions where the sex-specific RADtags mapped overlap with regions of elevated M/F FST values (LG3, Figure 4; LG1, Figure 5).
Figure 2.
Whole-genome M/F FST scan in 500 Kb windows using RADseq data for 4 taxa: (A) S. townsendi, (B) S. nicholsi, (C) S. inigoi, and (D) S. notatus.
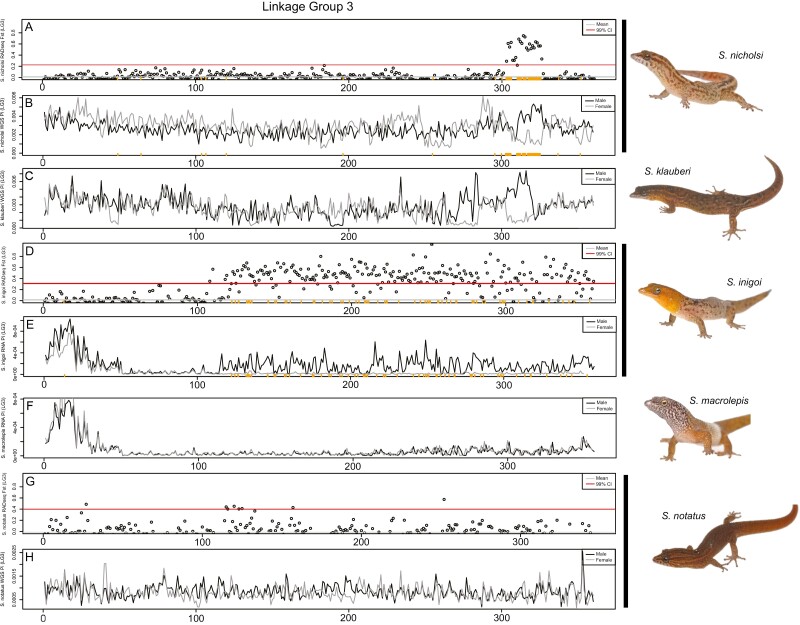
Figure 4.
Comparative genomics of the S. townsendi sex chromosome (LG3) across multiple Sphaerodactylus species in 500 kb windows. (A and B) S. nicholsi M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively; (C) S. klauberi M and F nucleotide diversity (WGS); (D and E) S. inigoi M/F FST values (RADseq) and M and F nucleotide diversity (RNAseq), respectively; (F) S. macrolepis M and F nucleotide diversity (RNAseq); (G and H) S. notatus M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively. Sex-specific RADtags mapped to S. nicholsi (A and B) and S. inigoi (D and E) along the X axis (orange ticks). Note: slight shifts on the X-axis are due to the differences in programs used to calculate values, i.e., WGS used pixy, while RADseq and RNAseq used vcftools.
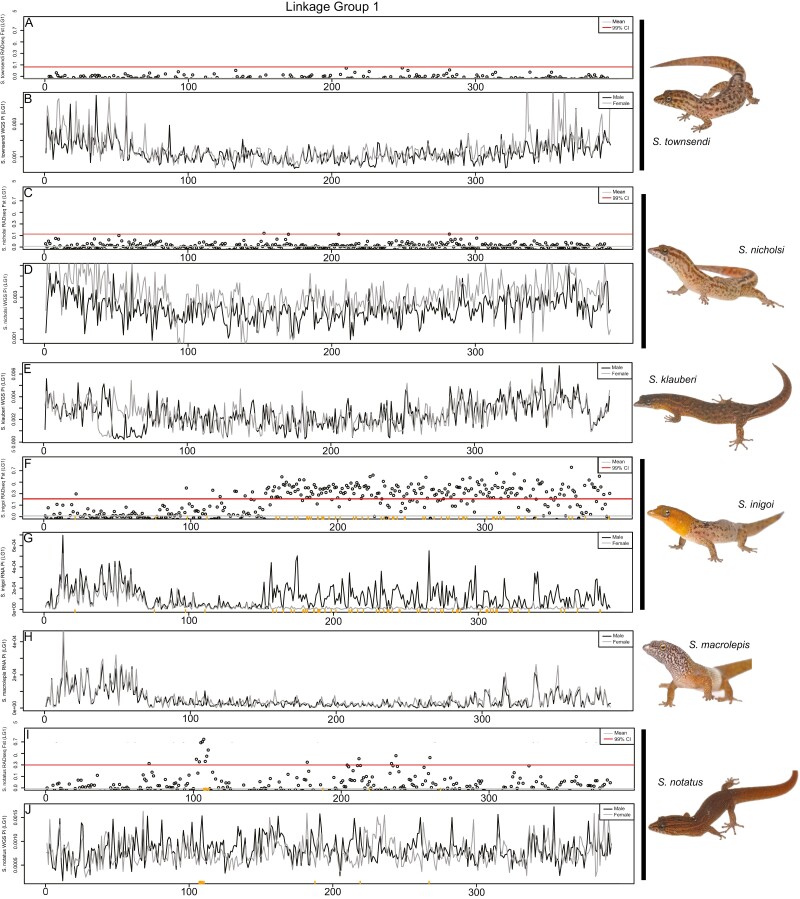
Figure 5.
Comparative genomics of the S. notatus sex chromosome (LG1) across multiple Sphaerodactylus species in 500kb windows. (A and B) S. townsendi M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively; (C and D) S. nicholsi M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively; (E) S. klauberi M and F nucleotide diversity (WGS); (F and G) S. inigoi M/F FST values (RADseq) and M and F nucleotide diversity (RNAseq), respectively; (H) S. macrolepis M and F nucleotide diversity (RNAseq); (I and J) S. notatus M/F FST values (RADseq) and M and F nucleotide diversity (WGS), respectively. Sex-specific RADtags mapped to S. inigoi (F and G) and S. notatus (I and J) along the X axis (orange ticks). Note: slight shifts on the X-axis are due to the differences in programs used to calculate values, i.e., WGS used pixy, while RADseq and RNAseq used vcftools.
After identifying the non-recombining region of the sex chromosomes in each species with RADseq, we used the WGS and RNAseq datasets to further characterize and corroborate these regions. We calculated nucleotide diversity (π) across the sex chromosomes. In recently evolved non-recombining regions where both X and Y reads map, we expect increased nucleotide diversity in males due to the increase in heterozygosity in this region relative to the rest of the genome (Schield et al. 2019)—note that we did not phase the X and Y haplotypes so both X and Y reads can potentially map to the collapsed X scaffold. We confirmed that this was indeed the case in species where we had already identified the sex chromosomes using RADseq, i.e., S. townsendi, S. nicholsi, S. inigoi, and S. notatus (Figures 3–5; Supplementary Figures 4 and 5). Next, we looked to species without available RADseq data, i.e., S. klauberi and S. macrolepis. We observed an increase in male π in S. klauberi WGS data at the same location in the sister species (S. townsendi and S. nicholsi), suggesting a conserved XY system in this clade (Figure 4C). However, we saw no such elevation in π, nor FST, in S. macrolepis RNAseq data, either indicating that these data are too sparse to locate the non-recombining region or that it is not located on this linkage group (Figures 4F and 5H; Supplementary Figure 6).
Figure 3.
Confirmation of the Sphaerodactylus townsendi sex chromosome on LG3. (A) RADseq M/F FST scan in 500 kb windows (zoomed in on LG3 from Figure 2); (B) M/F read depth differences across the length of LG3; (C) male and female nucleotide diversity (π) along LG3. The same set of male-specific RADtags mapped to LG3 are denoted by orange ticks along the bottom of each graph (same in each panel). (D) Gel images from a subset of these markers illustrate that they are located on the Y chromosome. Picture of an adult male S. townsendi scaled with a penny, USA currency (diameter = 19.05 mm).
Lastly, we used the RNAseq data in S. townsendi and S. inigoi to explore whether both X and Y alleles are being expressed, essentially using these data as another reduced-representation genomic dataset (conceptually similar to RADseq). Indeed, for S. townsendi and S. inigoi, we scanned each for genomic signatures identified in RADseq and WGS data (e.g., Figure 2). In S. townsendi, we identified a peak in male nucleotide diversity that coincides with the identified SDR on LG3 (Supplementary Figure 3). In S. inigoi, we observed the same patterns in the RNAseq data as seen in RADseq data for both LG1 and LG3 calculating both FST (Supplementary Figures 3 and 4) and nucleotide diversity (Figures 4D and E and 5F and G). However, in S. macrolepis, for whom we also had RNAseq data, we saw no differences between males and females on either linkage group coinciding with the SDRs identified in this study when examining FST (Supplementary Figure 5) or nucleotide diversity (Figures 4F and 5H), nor did we see any elevation in nucleotide diversity in the single male WGS data (not shown). Thus, we are as of yet unable to identify the sex chromosome linkage group in S. macrolepis.
We examined synteny across the genome to construct a quick-reference synteny table correlating each S. townsendi linkage group with their syntenic regions in Podarcis, Anolis, and Gallus (Table 3). Of note, Naja was omitted from the table as its macrochromosomes were collinear with Anolis. We used these correlations to approximate the locations of these linkage groups in the physically mapped Gekko genome (Srikulnath et al. 2015). More specifically, for the sex chromosomes, we present a fine-scale synteny analysis comparing the sex chromosome linkage groups identified here with their counterparts in Podarcis, Anolis, and Gallus (Supplementary Figure 7). We identified that most Sphaerodactylus linkage groups are represented in other species as a single syntenic block (e.g., Podarcis and Gallus macrochromosomes), whereas others are whole chromosome arms (Anolis macrochromosomes) or made up of many smaller linkage groups in other more distantly related lineages (i.e., Gallus microchromosomes). This information provides a simple reference for future work investigating genome synteny in geckos.
We examined the annotated S. townsendi genes present within the identified non-recombining region (or sex-determining region; SDR) for each species. The number of annotated genes varied by nearly two orders of magnitude (smallest to largest): S. notatus (23 LG1 genes), S. townsendi (236 LG3 genes), S. nicholsi (283 LG3 genes), and S. inigoi (3,225 LG1 genes + 2,330 LG3 genes = 5,555 total). A full list of genes is available in Supplementary Appendix. Differences in the number of annotated genes may indicate the relative stage of sex chromosome degeneration in each species or the approximate time since fixing an ancestral population in each lineage. Among the ~250 and ~25 annotated genes in the S. townsendi group and S. notatus SDRs, respectively, we searched for putative sex-determining genes from a relatively short list of known or likely sex-determining genes (i.e., the “usual suspects”; Herpin and Schartl 2015; Dor et al. 2019) and none were apparent.
Discussion
Reference Genome Description
The final genome assembly of Sphaerodactylus townsendi achieved chromosome-level status (Table 3). This is the second such assembly in a gecko and one of only a handful of high-quality assemblies in squamate reptiles (Yamaguchi et al. 2021). Other publicly available chromosome-level squamate assemblies include those from the Indian cobra (Naja naja;Suryamohan et al. 2020) and prairie rattlesnake (Crotalus viridis; Schield et al. 2019), as well as the physically-mapped green anole (Anolis carolinsensis) genome (Alfoldi et al. 2011), and the common wall lizard (Podarcis muralis; Andrade et al. 2019), with more being sequenced, assembled, and published on a regular basis. The first chromosome-level genome assembly for a gecko (Paroedura picta) was published while this manuscript was in review, and we were unable to include it in our analyses (Yamaguchi et al. 2021). A non-exhaustive list of publicly available Lepidosaur reference genomes is provided in Supplementary Table 2.
Sex Chromosome Evolution in Sphaerodactylus
Across our sampled taxa, we found that five out of six Sphaerodactylus species have XY sex chromosomes while the sixth, S. macrolepis, remains unknown. Among the taxa with an identified sex chromosome system, three maintain a conserved XY system encompassing (presumably) a single stratum of the sex-determining region (SDR) on LG3 (S. townsendi, S. nicholsi, and S. klauberi). Our outgroup, S. notatus, possesses a distinct sex chromosome system from the other taxa located on LG1, which rejects a hypothesis of a conserved XY system across all sampled Sphaerodactylus species. Sphaerodactylus inigoi maintains a sex chromosome system that includes both LG1 and LG3, likely due to chromosomal fusion. The S. inigoi sex-linked region is extremely large and encompasses most of LG1 and LG3, including the SDR of S. townsendi on LG3 but excluding the SDR of S. notatus on LG1, which is found on a different region of the same linkage group. Thus, we cannot reject the hypothesis that S. townsendi and S. inigoi inherited a sex chromosome system on LG3 from their most-recent common ancestor (MRCA). Notably, the sex chromosome system in S. macrolepis remains unknown. As S. inigoi, a close relative to S. macrolepis, has a clear pattern of sex linkage, we might predict that there has been a transition within this lineage. Alternatively, S. inigoi and S. macrolepis could indeed share a homologous sex chromosome system, but the evidence for sex-linkage was not captured in our data, possibly due to the sex-linked region being comparatively small in S. macrolepis. While both hypotheses open up intriguing lines of investigation, we do not have the data to address them here. The rest of this section further explores the current lines of evidence on the evolution of LG3 as a sex chromosome within the sampled species of Puerto Rican Sphaerodactylus.
Following speciation, the non-recombining regions of the species’ respective sex chromosomes can diverge rapidly from one another, both in genomic location (e.g., addition of evolutionary strata or expansion of non-recombining region) and level of sequence degeneration (Lahn and Page 1999; Bachtrog 2006; Graves 2008). These factors can make confirming, or rejecting, homologous sex chromosomes (single origin) from those derived from a homologous cis transition (multiple origins) difficult. In the case of Sphaerodactylus from Puerto Rico—S. townsendi, S. nicholsi, S. klauberi (herein the “S. townsendi group”), and S. inigoi—all possess a sex-linked LG3. However, the non-recombining region of the Y in S. inigoi encompasses the entirety of S. townsendi group SDR. This pattern could be generated by either a single origin of LG3 as a sex chromosome in the MRCA of S. townsendi and S. inigoi, and then a subsequent fusion of LG3 to LG1 in S. inigoi, or multiple origins of LG3 as a sex chromosome. Indeed, if the SDR identified in the S. townsendi group was present in the MRCA of S. townsendi and S. inigoi—having remained largely static in the S. townsendi group but expanded greatly in S. inigoi—we might expect to see an overall increase of sex-specific markers or FST values located in this region in S. inigoi (indicating an older stratum, followed by addition of a secondary stratum/strata), or conserved male-specific RAD markers on the Y chromosomes of each species. However, we see none of these lines of evidence, presenting the possibility that this SDR may not have been present in the MRCA of S. townsendi and S. inigoi and LG3 was recruited as a sex chromosome multiple times independently (similarly to LG1 in S. inigoi and S. notatus). Additional data will be required to definitively distinguish between these two hypotheses. These scenarios, for example, could be distinguished by assembling haplotype-resolved genomes for at least two of these species, e.g., S. townsendi or S. nicholsi and S. inigoi, and examining gene trees from windowed regions within the S. townsendi SDR (García-Moreno and Mindell 2000; Natri et al. 2013; Sardell et al. 2021). More elaborately, this region could also be targeted using advanced cytogenetic techniques in a comparative context. Either experiment would necessitate collection of new samples and generation of additional data beyond the scope of the present study.
Although there are several published examples for recent cis- (e.g., XY to XY) and trans- (e.g., XY to ZW) transitions in sex chromosomes to different linkage groups at shallow scales (e.g., Jeffries et al. 2018; Tao et al. 2021), there are far fewer confirmed examples of homologous cis-transitions. Empirical examples of trans-transitions to the same linkage group in other systems have emerged in recent literature. The Japanese wrinkled frog (Glandirana rugosa) possesses independently derived XY and ZW systems on the same linkage group with two independent derivations of the ZW system accompanied by lineage-specific W-degradation (Ogata et al. 2003; 2007; Miura et al. 2012) and the XY system in the southern platyfish (Xiphophorus maculatus) chromosome 21 (Xma21) has been recruited multiple times within the genus Xiphophorus (i.e., as a ZW system in X. helleri; Franchini et al. 2018). However, cis-transitions to the same linkage group have only been identified within ranid frogs (Jeffries et al. 2018), stickleback fishes (independent derivations of an XY system on LG12; Ross et al. 2009), and possibly also multiple times within Xiphophorus fishes (M. Schartl pers. comm.).
Along the same vein, recent research has found that certain linkage groups have been recruited as sex chromosomes multiple times, while others have remained unutilized (Graves and Peichel 2010; O’Meally et al. 2012; Jeffries et al. 2018; Kratochvíl et al. 2021). For example, the syntenic regions of the bird ZW system have been independently recruited as a sex chromosome in both a turtle (Staurotypus triporcatus; XY) and two geckos (Gekko hokouensis; ZW and Phyllodactylus wirshingi; ZW) (summarized in Nielsen et al. 2019a). In sphaerodactylids, the only linkage group previously identified as a sex chromosome linkage group was the ZW system in Aristelliger (Gallus 2; Keating et al. 2020). Within Sphaerodactylus, this is the first identified use of S. townsendi LG1 (syntenic with Gallus 3) or LG3 (specifically, regions of the chromosome syntenic with Gallus 18/30/33; Table 3). To the best of our knowledge, this is the first time this region has been recruited into a sex determining role in geckos (Augstenová et al. 2021), as well as the first time the syntenic regions of Gallus 3 and 30/33 have been recruited as a sex chromosome in tetrapods (Kratochvíl et al. 2021). Within S. townsendi, LG3 has only been found as a partial component (i.e., Gallus chromosome 18, not including Gallus 30/33) of the sex chromosome linkage group in one other species—the ZW system of the night lizard Xantusia henshawi (Nielsen et al. 2020). Thus, no other tetrapod group is currently known to have recruited either of these linkage groups as a sex chromosome—lending support to the hypothesis that any linkage group may act a sex chromosome (e.g., Hodgkin, 2002).
Genome Architecture and Synteny Across Squamates
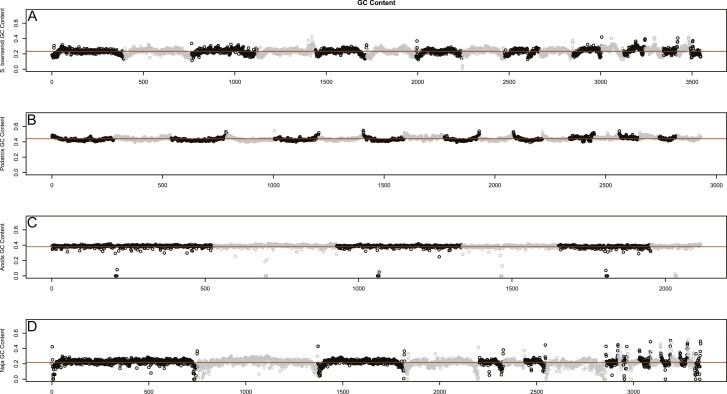
Most scaffolds in S. townsendi maintain a one-to-one relationship with Podarcis chromosomes (Table 3). This is interesting because geckos and wall lizards—unlike most other squamate reptiles—lack microchromosomes (loosely defined as chromosomes ~30 Mb in size; Perry et al. 2020). Instead, they possess a series of graded acrocentric chromosomes (Olmo et al. 1990; Srikulnath 2013). The current interpretation suggests multiple origins of this genomic architecture in squamates (i.e., independently evolved in geckos and wall lizards), however additional data are still needed. As a quick assessment of similarities between Sphaerodactylus and Podarcis, we compared genome-wide GC content between four representative squamates (the aforementioned two taxa lacking microchromosomes, with two that possess them, Anolis and Naja). Qualitative patterns of windowed GC content were most similar between Sphaerodactylus and Naja despite being less closely related to each other than other sampled taxa (Figure 6). Interestingly, Anolis and Podarcis are diurnal, while geckos and snakes are both ancestrally nocturnal (Gamble et al. 2015b; Simões et al. 2016; Pinto et al. 2019c), and it is plausible the genome-wide decrease in per-window GC content resulted in independent losses of highly thermo-stable DNA in both lineages (Fullerton et al. 2001). Alternatively, this could also be lineage-specific to Sphaerodactylus (i.e., not a property of geckos as a whole; Scantlebury et al. 2011). Indeed, contrasting patterns of genome-wide patterns of GC content (and potentially other indicators of genome organization) could be explained by two independent origins of macrochromosome-only karyotypes. Alternatively, these patterns could be explained by changes in recombination landscape between taxa (Charlesworth 1994; Charlesworth et al. 2020) or related to the presence/absence of isochores (Eyre-Walker et al. 2001). These ideas can and should be further tested with multiple chromosome-level genome assemblies across geckos, snakes, and additional squamates.
Figure 6.
Genome-wide patterns of GC content across representative squamate taxa, orange line representing the genomic mean. Broadly, pattern of GC content appears most similar, both in chromosome patterns and mean per-window GC content (~0.2), between (A) Sphaerodactylus and (D) Naja. Both (B) Podarcis and (C) Anolis have a considerably higher mean per-window GC (~0.4), and Podarcis shows an inverse pattern to Sphaerodactylus and Naja in that GC goes up at the tips of chromosomes instead of down. We believe that the Anolis patterns here are less informative in this regard as the sequencing method employed is not directly comparable to the other 3 genomes.
Future Directions
In recent years, much has been learned about vertebrate sex chromosome evolution. Just within geckos, our knowledge of sex chromosome evolution has expanded exponentially (see Gamble et al. 2015a and Augstenová et al. 2021). In sphaerodactylids, we have discovered three distinct sex chromosome linkage groups within two genera (Keating et al. 2020; this study) and at least two other XY systems that currently lack linkage information (Gornung et al. 2013; Gamble et al. 2018). The identification of sex chromosome linkage groups is fast becoming more feasible as new reference genomes become available, as well as new tools that permit the functional analysis of mechanisms of sex determination and sexual differentiation, both practically and financially (Rasys et al. 2019; Stöck et al. 2021). Thus, the sprightly sphaerodactyls are poised to become a potent model system for genomic research. We here point out two potentially worthwhile research avenues.
First, future work focusing on unsampled species both nested within our focal taxa (e.g., S. macrolepis and others), closely related outgroup taxa (e.g., S. roosevelti), as well as more-distant relatives, could help develop a clear hypothesis for when and how these newly identified sex chromosome linkage groups were recruited within this genus. For example, a closer look at the sister species, S. inigoi and S. grandisquamis, may provide insight into the timing of the putative chromosomal fusion we hypothesize here in S. inigoi may help better estimate the total number of sex chromosome transitions in other groups (Daza et al. 2019). Research including S. roosevelti and other more distantly related species will illuminate whether the LG3 sex-linked region was inherited from a common ancestor or independently derived between the two Puerto Rican clades (the clades containing S. inigoi + S. grandisquamis and the S. townsendi + S. klauberi clade; see Daza et al. 2019).
Second, there are many examples of sexual dimorphisms, especially sexual dichromatism in fishes, linked to sex chromosomes (Kallman 1970; Kottler and Schartl 2018). Sphaerodactylids also display an impressive phenotypic diversity, such as body size and sexual dichromatism (Griffing et al. 2018; Daza et al. 2019). Indeed, it has been posited that sexual dichromatism has evolved repeatedly within Sphaerodactylus (Regalado, 2015; Daza et al. 2019). Coincidentally, one such loss of dichromatism is hypothesized between the sister clades containing the dichromatic S. inigoi + S. macrolepis and the monochromatic S. townsendi + S. klauberi. As we are just now becoming privy to the sex-linked regions in sphaerodactylids, it remains to be seen if any sexually dimorphic traits are linked to the non-recombining region of the Y chromosome. If dichromaticism is connected to sex chromosomes in S. inigoi (encompassing almost 2 entire chromosomes)—and that degenerated system were ancestral—the loss of the S. inigoi system in the S. townsendi clade could have been selected for to relieve predation pressures, or to resolve sexual conflict (van Doorn and Kirkpatrick 2007; Stöck et al. 2011).
Conclusions
We presented data and analyses of the sex chromosomes for a small percentage of the known taxonomic diversity within Sphaerodactylus geckos. Within this small subset of species, our analyses reject the hypothesis that there are conserved sex chromosomes maintained across Sphaerodactylus geckos. We identified and characterized at least two cis-transitions between species with XY sex chromosome systems. These newly identified sex chromosome linkage groups are syntenic with regions that have not previously been characterized as sex chromosomes in an amniote: LG1 (syntenic with Gallus 3) and LG3 (syntenic regions with Gallus 18/30/33). We posit that the recruitment of LG3 (S. townsendi) as a sex chromosome in S. townsendi and S. inigoi may be independent although additional data are required to validate this. We reviewed the data for and against multiple recruitments of this chromosome between these taxa and suggest that a putative sex chromosome fusion in S. inigoi may correspond with a cis-transition specific to this lineage. Overall, our results highlight that contemporary estimates of sex chromosome transitions within gecko lizards are overly conservative and that more transitions will likely be uncovered in the future, further emphasizing that gekkotan sex chromosome evolution is far more dynamic than previously hypothesized (Gamble et al. 2015a, 2018; Nielsen et al. 2019a; Rovatsos et al. 2019). The foundation is laid for this group to serve as an essential model to study sex chromosome evolution.
Supplementary Material
Acknowledgments
We would like to thank J. Bernstein, M. “Toño” García, E. Glynne, N. Holovacs, J. Titus-McQuillan, C. Rivera, V. Rodriguez, D. Zarkower, E. Blumenthal, and D. DeFilippis for their respective contributions to the completion of this work. Animals were collected with permission of Departamento de Recursos Naturales y Ambientales (DRNA) Del Gobierno de Puerto Rico; under permits 2014-IC-042, 2013-IC-006, and 2016-IC-091). All experiments were carried out in accordance with Institutional Animal Care and Use Committee (IACUC) protocols at Marquette University (AR279 and AR288) and the University of Minnesota: 0810A50001 and 1108A03545. Specimens from Florida were collected by the permission of the Florida Fish and Wildlife Conservation Commission given to the Florida Museum of Natural History. Sociedad Ornitológica de la Hispaniola for assisting with logistics and the Ministerio de Medio Ambiente y Recursos Naturales for providing them with permits necessary for the collection and exportation of specimens in the Dominican Republic (0512-0515). The authors would also like to thank three anonymous reviewers and the corresponding editor (A. Suh) for their valuable feedback in constructing the final version of this manuscript.
Contributor Information
Brendan J Pinto, Milwaukee Public Museum, Milwaukee, WI 53233, USA; School of Life Sciences, Arizona State University, Tempe, AZ 85281, USA; Center for Evolution and Medicine, Arizona State University, Tempe, AZ 85281, USA.
Shannon E Keating, Department of Biological Sciences, Marquette University, Milwaukee, WI 53233, USA.
Stuart V Nielsen, Department of Biological Sciences, Louisiana State University in Shreveport, Shreveport, LA 71115, USA; Division of Herpetology, Florida Museum of Natural History, Gainesville, FL 32611, USA.
Daniel P Scantlebury, Washington, DC, USA.
Juan D Daza, Department of Biological Sciences, Sam Houston State University, Huntsville, TX 77340, USA.
Tony Gamble, Milwaukee Public Museum, Milwaukee, WI 53233, USA; Department of Biological Sciences, Marquette University, Milwaukee, WI 53233, USA; Bell Museum of Natural History, University of Minnesota, St Paul, MN 55455, USA.
Funding
Helen T. and Frederick M. Gaige fund [2018] (American Society of Ichthyologists and Herpetologists: ASIH) to BJP, the Dean’s Research Enhancement Award [2018] from Marquette University (MU) to BJP, the Denis J. O’Brien Summer Research Fellowship [2018] (MU) to BJP, and the American Genetic Association (AGA) – Ecological, Evolutionary, and Conservation Genomics (EECG) Research Award to BJP, MU laboratory startup funds to TG, National Science Foundation (NSF) DEB1110605 and DEB0920892 (to R. Glor), NSF IOS1146820 (to D. Zarkower), and NSF DEB1657662 (to T.G.). B.J.P. was funded by the Department of Biological Sciences Graduate Research Fellowship (MU; 2018–2020), Catherine Grotelueschen Scholarship (MU; 2019), and by National Institutes of Health (NIH) project number 2R01GM116853-05 to M. Kirkpatrick (2020–2021).
Authors’ Contributions
B.J.P. designed study, directed fieldwork, conducted genome sequencing and later RADseq experiments, acquired funding, assembled and analyzed data, and wrote the manuscript; S.E.K. constructed RNAseq libraries; S.V.N. conducted fieldwork and constructed RADseq and RNAseq libraries; D.P.S. conducted fieldwork; J.D.D. conducted fieldwork and provided organismal expertise; T.G. assisted in design of study, conducted fieldwork and initial RADseq experiments, performed karyotyping, acquired funding, and oversaw the project. All authors read and approved the final manuscript.
Data Availability
Sequencing reads generated for this study: 10X Chromium sequencing, long-read sequencing, Illumina re-sequencing, RNAseq, and RADseq are available on NCBI’s Sequence Read Archive (SRA) under project number PRJNA746057 and a full list of individual metadata is available in Supplementary Table 3. Assembled and annotated genome and transcriptomes are available on Figshare (https://doi.org/10.6084/m9.figshare.12291236) and is available on NCBI at https://www.ncbi.nlm.nih.gov/nuccore/JAHVXK010000000. All genomic computation took place on a custom-built, 24-core Intel Xeon and 128Gb RAM system running Ubuntu 16.04.
References
- Adolfsson S, Ellegren H.. 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol. 30:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 477:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC.. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade P, Pinho C, i de Lanuza GP, Afonso S, BrejchaJ, Rubin CJ, Wallerman O, Pereira P, Sabatino SJ, Bellati A, et al. 2019. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. PNAS. 116:5633–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Augstenová B, Pensabene E, Veselý M, Kratochvíl L, Rovatsos M.. 2021. Are geckos special in sex determination? Independently evolved differentiated ZZ/ZW sex chromosomes in carphodactylid geckos. Genome Biol Evol. 13:evab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augstenová B, Pokorná MJ, Altmanová M, Frynta D, Rovatsos M, Kratochvíl L.. 2018. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution. 72:1701–1707. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2003. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in Drosophila. Nat Genet. 34:215–219. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2006. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 16:578–585. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, Perrin N, Ross L, Valenzuela N, Vamosi JC.. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT.. 2011. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 27:1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N.. 2013. Sex-chromosome turnovers induced by deleterious mutation load. Evolution. 67:635–645. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N.. 2014. Sex-chromosome turnovers: the hot-potato model. Am Nat. 183:140–146. [DOI] [PubMed] [Google Scholar]
- Botero-Castro F, Figuet E, Tilak MK, Nabholz B, Galtier N.. 2017. Avian genomes revisited: hidden genes uncovered and the rates versus traits paradox in birds. Mol Biol Evol. 34:3123–3131. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Bull JJ. 1983. Evolution of sex determining mechanisms. San Francisco (CA):Benjamin/Cummings Publishing Company, Inc. [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner (No. LBNL-7065E). Berkeley (CA):Lawrence Berkeley National Lab (LBNL). [Google Scholar]
- Cabau C, Escudié F, Djari A, Guiguen Y, Bobe J, Klopp C.. 2017. Compacting and correcting Trinity and Oases RNA-Seq de novo assemblies. PeerJ. 5:e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA.. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22:3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1978. Model for evolution of Y chromosomes and dosage compensation. PNAS. 75:5618–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science. 251:1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1994. Patterns in the genome. Curr Biol. 4:182–184. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 2000. The degeneration of Y chromosomes. Phil Trans Roy Soc Lond B. 355:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Zhang Y, Bergero R, Graham C, Gardner J, Yong L.. 2020. Using GC content to compare recombination patterns on the sex chromosomes and autosomes of the Guppy, Poecilia reticulata, and its close outgroup species. Mol Biol Evol. 37:3550–3562. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza JD, Pinto BJ, Thomas R, Herrera-Martinez A, García LF, Scantlebury D, Perry G, Balaraman RP, Gamble T.. 2019. The sprightly little sphaerodactyl: systematics and biogeography of the Puerto Rican dwarf geckos Sphaerodactylus (Gekkota, Sphaerodactylidae). Zootaxa. 4712:151–201. [DOI] [PubMed] [Google Scholar]
- Dor L, Shirak A, Kohn YY, Gur T, Weller JI, Zilberg D, Seroussi E, Ron M.. 2019. Mapping of the sex determining region on linkage group 12 of guppy (Poecilia reticulata). G3. 9:3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. . 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL.. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter P, Bassham S, Hohenlohe P, Johnson E, Cresko W.. 2011. SNP discovery and genotyping for evolutionary genetics using RAD sequencing. In: Orgogozo V, Rockman M, editors. Molecular methods for evolutionary genetics. New York: Springer. p. 157–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD.. 2001. The evolution of isochores. Nat Rev Genet. 2:549–555. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Sarre SD, O’Meally D, Graves JA, Georges A.. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res. 127:249–260. [DOI] [PubMed] [Google Scholar]
- Fowler BL, Buonaccorsi VP.. 2016. Genomic characterization of sex-identification markers in Sebastes carnatus and Sebastes chrysomelas rockfishes. Mol Ecol. 25:2165–2175. [DOI] [PubMed] [Google Scholar]
- Franchini P, Jones JC, Xiong P, Kneitz S, Gompert Z, Warren WC, Walter RB, Meyer A, Schartl M.. 2018. Long-term experimental hybridisation results in the evolution of a new sex chromosome in swordtail fish. Nat Comm. 9:5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG.. 2001. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 18:1139–1142. [DOI] [PubMed] [Google Scholar]
- Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, Mank JE.. 2020. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol. 12:750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T. 2010. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex Dev. 4:88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T. 2016. Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Mol Ecol. 25:2114–2116. [DOI] [PubMed] [Google Scholar]
- Gamble T, Castoe TA, Nielsen SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W.. 2017. The discovery of XY sex chromosomes in a boa and python. Curr Biol. 27:2148–2153.e4. [DOI] [PubMed] [Google Scholar]
- Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D.. 2015a. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among Gecko sex-determining systems. Mol Biol Evol. 32:1296–1309. [DOI] [PubMed] [Google Scholar]
- Gamble T, Greenbaum E, Jackman TR, Bauer AM.. 2015b. Into the light: diurnality has evolved multiple times in geckos. Biol J Linn Soc. 115: 896–910. [Google Scholar]
- Gamble T, McKenna E, Meyer W, Nielsen SV, Pinto BJ, Scantlebury DP, Higham TE.. 2018. XX/XY sex chromosomes in the South American dwarf gecko (Gonatodes humeralis). J Hered. 109:462–468. [DOI] [PubMed] [Google Scholar]
- Gamble T, Zarkower D.. 2014. Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol Ecol Resour. 14:902–913. [DOI] [PubMed] [Google Scholar]
- García-Moreno J, Mindell DP.. 2000. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): a case study using avian CHD. Mol Biol Evol. 17:1826–1832. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv. 1207.3907. [Google Scholar]
- Gornung E, Mosconi F, Annesi F, Castiglia R.. 2013. The first cytogenetic description of Euleptes europaea (Gené, 1839) from Northern Sardinia reveals the highest diploid chromosome number among sphaerodactylid geckos (Sphaerodactylidae, Squamata). Comp Cytogenet. 7:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. . 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Ann Rev Genet. 42: 565–586. [DOI] [PubMed] [Google Scholar]
- Graves J, Peichel C.. 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing AH, Daza JD, DeBoer JC, Bauer AM.. 2018. Developmental osteology of the parafrontal bones of the Sphaerodactylidae. The Anatomical Record, 301: 581–606. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell R, Wortman JR.. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley LJ, Ceplitis H, Ellegren H.. 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 167:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Takeuchi M, Kageyama Y, Tatsumi K, Hibi M, Kiyonari H, Kuraku S.. 2018. Madagascar ground gecko genome analysis characterizes asymmetric fates of duplicated genes. BMC Biology. 16: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M.. 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16:1260–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Green DM.. 1990. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J Evol Biol. 3: 49–64. [Google Scholar]
- Hodgkin J. 2002. Exploring the envelope. Systematic alteration in the sex-determination system of the nematode caenorhabditis elegans. Genetics. 162:767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2015. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Fan J, Sun Z, Liu S.. 2020. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 36:2253–2255. [DOI] [PubMed] [Google Scholar]
- Hundt PJ, Liddle EB, Nielsen SV, Pinto BJ, Gamble T.. 2019. Sex chromosome evolution in combtooth blennies (Blenniidae, Istiblennius). Mar Ecol Prog. 627:195–200. [Google Scholar]
- Jackman SD, Coombe L, Chu J, Warren RL, Vandervalk, BP, Yeo S, Xue Z, Mohamadi H, Bohlmann J, Jones S, Birol I.. 2018. Tigmint: correcting assembly errors using linked reads from large molecules. BMC Bioinformatics. 19:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet P-A, Dufresnes C, et al. . 2018. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Comm. 9:4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallman KD. 1970. Different genetic basis of identical pigment patterns in two populations of platyfish, Xiphophorus maculatus. Copeia. 3:472–487. [Google Scholar]
- Keating SE, Blumer M, Grismer LL, Lin A, Nielsen SV, Thura MK, Wood PL, Quah ES, Gamble T.. 2021. Sex chromosome turnover in bent-toed geckos (Cyrtodactylus). Genes. 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SE, Griffing AH, Nielsen SV, Scantlebury DP, Gamble T.. 2020. Conserved ZZ/ZW sex chromosomes in Caribbean croaking geckos (Aristelliger: Sphaerodactylidae). J Evol Biol. 33:1316–1326. [DOI] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S.. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 27:757–763. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korunes KL, Samuk K.. 2021. pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 21:1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostmann A, Kratochvíl L, Rovatsos M.. 2021. Poorly differentiated XX/XY sex chromosomes are widely shared across skink radiation. Proc Roy Soc B. 288:20202139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler VA, Feron R, Nanda I, Klopp C, Du K, Kneitz S, Helmprobst F, Lamatsch DK, Lopez-Roques C, Lluch J, et al. . 2020. Independent Origin of XY and ZW Sex Determination Mechanisms in Mosquitofish Sister Species. Genetics. 214:193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler V, Schartl M.. 2018. The colorful sex chromosomes of teleost fish. Genes. 9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochvíl L, Gamble T, Rovatsos M.. 2021. Sex chromosome evolution among amniotes: is the origin of sex chromosomes non-random? Phil Trans Roy Soc Lond B. 376:20200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC.. 1999. Four evolutionary strata on the human X chromosome. Science. 286:964–967. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou Q, Wang Y, Luo L, Yang J, Yang L, Liu M, Li Y, Qian T, et al. . 2015. Gekko japonicus genome reveals evolution of adhesive toe pads and tail regeneration. Nat Comm. 6:10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main H, Scantlebury DP, Zarkower D, Gamble T.. 2012. Karyotypes of two species of Malagasy ground gecko (Paroedura: Gekkonidae). African J Herpetol. 61:81–90. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 17:10–12. [Google Scholar]
- Merkel D. 2014. Docker: lightweight linux containers for consistent development and deployment. Linux Journal. 239:2. [Google Scholar]
- Miura I, Ohtani H, Ogata M.. 2012. Independent degeneration of W and Y sex chromosomes in frog Rana rugosa. Chromosome Res. 20:47–55. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1914. A gene for the fourth chromosome of Drosophila. J Exp Zool. 17: 325–336. [Google Scholar]
- Muller HJ. 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics. 3:422–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res. 106:2–9. [DOI] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merilä J.. 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 30:1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SV, Banks JL, Diaz RE Jr, Trainor PA, Gamble T.. 2018. Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae). J Evol Biol. 31:484–490. [DOI] [PubMed] [Google Scholar]
- Nielsen SV, Daza JD, Pinto BJ, Gamble T.. 2019a. ZZ/ZW sex chromosomes in the endemic Puerto Rican leaf-toed gecko (Phyllodactylus wirshingi). Cytogenet Genome Res. 157:89–97. [DOI] [PubMed] [Google Scholar]
- Nielsen SV, Guzmán-Mendez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M.. 2019b. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol Lett. 15:20190498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SV, Pinto BJ, Guzmán-Méndez IA, Gamble T.. 2020. First report of sex chromosomes in night lizards (Scincoidea: Xantusiidae). J Hered. 111:307–317. [DOI] [PubMed] [Google Scholar]
- Nishimura O, Hara Y, Kuraku S.. 2017. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics. 33:3635–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I.. 2007. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity (Edinb). 100:92–99. [DOI] [PubMed] [Google Scholar]
- Ogata M, Ohtani H, Igarashi T, Hasegawa Y, Ichikawa Y, Miura I.. 2003. Change of the heterogametic sex from male to female in the frog. Genetics. 164:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin, Germany:Springer Verlag. [Google Scholar]
- Olmo E, Odierna G, Capriglione T, Cardone A.. 1990. DNA and chromosome evolution in lacertid lizards. In: Olmo E, editor Cytogenetics of Amphibians and Reptiles. Berlin, Germany:Birkhauser Verlag. p. 181–204. [Google Scholar]
- O’Meally D, Ezaz T, Georges A, Sarre SD, Graves JA.. 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 20:7–19. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T.. 2002. Resolving the paradox of sex and recombination. Nat Rev Genet. 3:252–261. [DOI] [PubMed] [Google Scholar]
- Palmer J. 2018. Funannotate: Eukaryotic genome annotation pipeline. doi: 10.5281/zenodo.2604804. [DOI] [Google Scholar]
- Pan Q, Feron R, Jouanno E, Darras H, Herpin A, Koop B, Rondeau E, Goetz FW, Larson WA, et al. . 2021a. The rise and fall of the ancient northern pike master sex-determining gene. ELife. 10:e62858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Feron R, Yano A, Guyomard R, Jouanno E, Vigouroux E, Wen M, Busnel JM, Bobe J, Concordet JP, et al. 2019. Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genetics. 15:e1008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Kay T, Depincé A, Adolfi M, Schartl M, Guiguen Y, Herpin A.. 2021b. Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Phil Trans Roy Soc B. 376:20200091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 34:867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peona V, Blom MPK, Xu L, Burri R, Sullivan S, Bunikis I, Liachko I, Haryoko T, Jønsson KA, Zhou Q, Irestedt M, Suh A.. 2020. Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol Ecol Res. 21:263–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BW, Schield DR, Adams RH, Castoe TA.. 2020. Microchromosomes exhibit distinct features of vertebrate chromosome structure and function with underappreciated ramifications for genome evolution. Mol Biol Evol. 38:904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto BJ, Card DC, Castoe TA, Diaz RE Jr, Nielsen SV, Trainor PA, Gamble T.. 2019b. The transcriptome of the veiled chameleon (Chamaeleo calyptratus): a resource for studying the evolution and development of vertebrates. Dev Dyn. 248:702–708. [DOI] [PubMed] [Google Scholar]
- Pinto BJ, Nielsen SV, Gamble T.. 2019c. Transcriptomic data support a nocturnal bottleneck in the ancestor to gecko lizards. Mol Phylogenet Evol. 141: 1066392. [DOI] [PubMed] [Google Scholar]
- Pinto BJ, Titus-McQuillan J, Daza JD, Gamble T.. 2019a. Persistence of a geographically-stable hybrid zone in Puerto Rican dwarf geckos. J Hered. 110:523–534. [DOI] [PubMed] [Google Scholar]
- Pinto BJ, Weis JJ, Gamble T, Paul R, Ode P, Zaspel J.. 2021. A chromosome-level genome assembly of the parasitoid wasp, Cotesia glomerata (Hymenoptera: Braconidae). J Hered. 112:558–564. [DOI] [PubMed] [Google Scholar]
- Pokorna M, Kratochvíl L.. 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc. 156: 168–183. [Google Scholar]
- Ponnikas S, Sigeman H, Abbott JK, Hansson B.. 2018. Why do sex chromosomes stop recombining? Trends Genet. 34:492–503. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasys AM, Park S, Ball RE, Alcala AJ, Lauderdale JD, Menke DB.. 2019. CRISPR-Cas9 Gene Editing in lizards through microinjection of unfertilized oocytes. Cell Rep. 28:2288–2292.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Regalado R. 2015. Does dichromatism variation affect sex recognition in dwarf geckos? Ethol Ecol Evol. 27: 56–73. [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM.. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D.. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL.. 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Gamble T, Nielsen SV, Georges A, Ezaz T, Kratochvíl L.. 2021. Do male and female heterogamety really differ in expression regulation? Lack of global dosage balance in pygopodid geckos. Phil Trans Roy Soc Lond B. 376:20200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Farkačová K, Altmanová M, Johnson Pokorná M, Kratochvíl L.. 2019. The rise and fall of differentiated sex chromosomes in geckos. Mol Ecol. 28:3042–3052. [DOI] [PubMed] [Google Scholar]
- Sardell JM, Josephson MP, Dalziel AC, Peichel CL, Kirkpatrick M.. 2021. Heterogeneous histories of recombination suppression on stickleback sex chromosomes. Mol Biol Evol. 38:4403–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scantlebury DP, Ng J, Landestoy M, Geneva A, Glor RE.. 2011. Notes on activity patterns of five species of Sphaerodactylus (Squamata: Sphaerodactylidae) from the Dominican Republic. Reptiles Amphib. 18:12–17. [Google Scholar]
- Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Adams RH, Corbin AB, Smith CF, Ramesh B, Demuth JP, Betrán E, Tollis M, Meik JM, Mackessy SP, Castoe TA.. 2019. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis C, Böhne A, Schartl M, Volff JN, Galiana-Arnoux D.. 2009. Sex determination diversity and sex chromosome evolution in poeciliid fish. Sex Dev. 3:68–77. [DOI] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F.. 2016. SeqKit: A cross-platform and ultrafast toolkit for fasta/q file manipulation. PLoS One. 11:e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhom M, Said K, Chatti N, Guarino FM, Odierna G, Petraccioli A, Picariello O, Mezzasalma M.. 2020. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology. 141:125738. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simões BF, Sampaio FL, Loew ER, Sanders KL, Fisher RN, Hart NS, Hunt DM, Partridge JC, Gower DJ.. 2016. Multiple rod–cone and cone–rod photoreceptor transmutations in snakes: evidence from visual opsin gene expression. Proc Roy Soc B. 283:20152624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K. 2013. The dynamics of chromosome evolution in reptiles. Thai J Genet. 6:77–79. [Google Scholar]
- Srikulnath K, Uno Y, Nishida C, Ota H, Matsuda Y.. 2015. Karyotype reorganization in the Hokou Gecko (Gekko hokouensis, Gekkonidae): the process of microchromosome disappearance in Gekkota. PLoS One. 10:e0134829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NM. 1905. A study of the germ cells of Aphis rosae and Aphis oenotherae. J Exp Zool. 2:313–333. [Google Scholar]
- Stöck M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C, Dufresnes C, Bonjour E, Dumas Z, et al. . 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9:e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Kratochvíl L, Kuhl H, Rovatsos M, Evans BJ, Suh A, Valenzuela N, Veyrunes F, Zhou Q, et al. 2021. A brief review of vertebrate sex evolution with a pledge for integrative research: towards “sexomics”. Phil Trans Roy Soc B. 376:20200426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan K, Krishnankutty SP, Guillory J, Jevit M, Schröder MS, Wu M, Kuriakose B, Mathew OK, Perumal RC, Koludarov I, et al. . 2020. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet. 52:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. 2018. Gnu parallel 2018. pp. 112. doi: 10.5281/zenodo.1146014 [DOI] [Google Scholar]
- Tao W, Xu L, Zhao L, Zhu Z, Wu X, Min Q, Wang D, Zhou Q.. 2021. High-quality chromosome-level genomes of two tilapia species reveal their evolution of repeat sequences and sex chromosomes. Mol Ecol Resour. 21:543–560. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P.. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Schwartz A.. 1966. Sphaerodactylus (Gekkonidae) in the greater Puerto Rico region. Bull Florida State Mus, Biol Sci. 10:193–260. [Google Scholar]
- Uetz P, Freed P, Aguilar R, Hošek J. (eds.) 2021. The Reptile Database, http://www.reptile-database.org, accessed July 16, 2021.
- Uno Y, Nishida C, Oshima Y, Yokoyama S, Miura I, Matsuda Y, Nakamura M.. 2008. Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res. 16:637–647. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M.. 2007. Turnover of sex chromosomes induced by sexual conflict. Nature. 449:909–912. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. . 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB.. 2017. Direct determination of diploid genome sequences. Genome Res. 27:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Dean R, Zimmer F, Mank J.. 2016. How to make a sex chromosome. Nat Comm. 7:12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Li F, Li Q, Zhou L, Gamble T, Zheng J, Kui L, Li C, Li S, Yang H, et al. 2016. Draft genome of the leopard gecko, Eublepharis macularius. GigaScience. 5:s13742–016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Guo L, Gu S, Wang O, Zhang R, Peters BA, Fan G, Liu X, Xu X, Deng L, et al. 2020. TGS-GapCloser: a fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience. 9:giaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Kadota M, Nishimura O, Ohishi Y, Naito Y, Kuraku S.. 2021. Technical considerations in Hi-C scaffolding and evaluation of chromosome-scale genome assemblies. Mol Ecol. 30:5923–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S, Coombe L, Warren RL, Chu J, Birol I.. 2018. ARCS: scaffolding genome drafts with linked reads. Bioinformatics. 34:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MT, Zhang G.. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 346:1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads generated for this study: 10X Chromium sequencing, long-read sequencing, Illumina re-sequencing, RNAseq, and RADseq are available on NCBI’s Sequence Read Archive (SRA) under project number PRJNA746057 and a full list of individual metadata is available in Supplementary Table 3. Assembled and annotated genome and transcriptomes are available on Figshare (https://doi.org/10.6084/m9.figshare.12291236) and is available on NCBI at https://www.ncbi.nlm.nih.gov/nuccore/JAHVXK010000000. All genomic computation took place on a custom-built, 24-core Intel Xeon and 128Gb RAM system running Ubuntu 16.04.