Abstract
To improve production of fuel ethanol from renewable raw materials, laccase from the white rot fungus Trametes versicolor was expressed under control of the PGK1 promoter in Saccharomyces cerevisiae to increase its resistance to phenolic inhibitors in lignocellulose hydrolysates. It was found that the laccase activity could be enhanced twofold by simultaneous overexpression of the homologous t-SNARE Sso2p. The factors affecting the level of active laccase obtained, besides the cultivation temperature, included pH and aeration. Laccase-expressing and Sso2p-overexpressing S. cerevisiae was cultivated in the presence of coniferyl aldehyde to examine resistance to lignocellulose-derived phenolic fermentation inhibitors. The laccase-producing transformant had the ability to convert coniferyl aldehyde at a faster rate than a control transformant not expressing laccase, which enabled faster growth and ethanol formation. The laccase-producing transformant was also able to ferment a dilute acid spruce hydrolysate at a faster rate than the control transformant. A decrease in the content of low-molecular-mass aromatic compounds, accompanied by an increase in the content of high-molecular-mass compounds, was observed during fermentation with the laccase-expressing strain, illustrating that laccase was active even at the very low levels of oxygen supplied. Our results demonstrate the importance of phenolic compounds as fermentation inhibitors and the advantage of using laccase-expressing yeast strains for producing ethanol from lignocellulose.
Lignocellulose is used as a renewable energy source worldwide. Lignocellulosic materials can be hydrolyzed to sugars, which then can be fermented to ethanol by microorganisms, such as the yeast Saccharomyces cerevisiae (16). Ethanol obtained from lignocellulosic materials can be used as an environmentally friendly liquid fuel. A problem associated with efficient conversion of cellulose and hemicellulose sugars to ethanol is that during dilute acid hydrolysis a broad range of compounds which inhibit the fermenting microorganism are liberated or formed along with the sugars. Inhibitors of fermentation include furan derivatives, such as furfural and 5-hydroxymethyl-furfural (5-HMF) (12, 17, 25, 26); low-molecular-mass aliphatic acids, such as acetic acid, formic acid, and levulinic acid (12); and phenolic compounds (2, 9, 14). Detoxification is often needed to increase fermentability, which increases the final cost of the ethanol (31).
In recent investigations, laccase from the filamentous fungus Trametes versicolor was used to treat hardwood and softwood hydrolysates, which resulted in increased fermentability by baker's yeast, S. cerevisiae (9, 14). The results indicated that phenolic compounds significantly contribute to inhibition of the yeast by the hydrolysates. Analyses of the concentrations of putative inhibitors demonstrated that a hydrolysate could be efficiently fermented if only the phenolic inhibitors were removed from the hydrolysate by laccase treatment, leaving the concentrations of the aliphatic acids and the furan derivatives unchanged (14).
The copper-containing blue oxidase laccase (p-diphenol oxidase; EC 1.10.3.2), an extracellular enzyme produced by a variety of plants and fungi (18), catalyzes the reduction of molecular oxygen to water and the one-electron oxidation of organic substrates, primarily phenolic compounds, to radicals. These radicals are unstable and subsequently form high-molecular-mass polymerization products, which leads to removal of low-molecular-mass phenolic compounds from the hydrolysate (9, 14). An alternative approach to adding laccase to the hydrolysate would be to genetically engineer S. cerevisiae for production of active laccase. This would allow ethanolic fermentation and detoxification to proceed simultaneously, which would combine two steps in the process into one, thus reducing the cost and time associated with production or purchase of laccase and with performing the detoxification step.
Laccase-encoding cDNAs have been isolated previously from the wood-degrading white rot basidiomycete T. versicolor (10) and have been used for heterologous expression in the yeasts Pichia pastoris (10) and S. cerevisiae (5). Laccase is difficult to express in yeasts and has been obtained in only very low amounts (5, 10). P. pastoris has a number of advantages for production of heterologous proteins compared to S. cerevisiae, and a high level of protein secretion is one of them (20). P. pastoris, however, cannot efficiently produce ethanol from lignocellulose due to the high oxygen demand of the organism. Therefore, it is highly desirable to increase the activity of laccase produced by S. cerevisiae. In this work, we attempted to increase the production of active laccase by S. cerevisiae through enhancement of secretion by overexpression of Sso2p. The Sso2 protein, a t-SNARE, is a membrane protein involved in the protein secretion machinery (1), and it has been suggested that this protein is involved in a rate-limiting step in protein secretion by S. cerevisiae (21). Secretion of a homologous protein, invertase, and secretion of a heterologous protein, α-amylase from Bacillus sp., by S. cerevisiae have been enhanced by overexpression of Sso2p (21).
A laccase-expressing, Sso2p-overexpressing S. cerevisiae transformant was cultivated in the presence of coniferyl aldehyde, an inhibitory compound present in lignocellulose hydrolysates (13, 14), in order to examine resistance to phenolic fermentation inhibitors. The transformant was also cultivated in a dilute acid hydrolysate of spruce to investigate the possible advantage of using laccase-expressing S. cerevisiae to produce fuel ethanol from lignocellulose.
MATERIALS AND METHODS
Strains and vectors used for cloning and expression.
Escherichia coli DH5α (obtained from Life Technologies, Rockville, Md.) was used for subcloning procedures. S. cerevisiae INVSC1 (MATα his3-Δ1 leu2 trp1-289 ura3-52) (Invitrogen, Groningen, The Netherlands) was used for expression of laccase. The SSO2 gene was obtained from the vector YEpSSO2-M (21). A fragment of YEpSSO2-M, containing the SSO2 gene and its control elements, was inserted into plasmid pAJ401 (23), a derivative of plasmid pFL60 (15), and plasmid pAJ401/lcc2 (5). These plasmids contain the β-lactamase gene, the ColE1 replication origin, the replication origin of the 2μ plasmid, the URA3 selection marker, and the PGK1 expression cassette. The vector pAJ401/lcc2 also contains the T. versicolor lcc2 gene inserted between the PGK1 5′ and 3′ control regions.
Construction of expression plasmids and transformants.
All enzymes used for cloning and restriction analyses were obtained from Roche (Mannheim, Germany) unless stated otherwise. Standard techniques were used for cloning, transformation, and analysis (3).
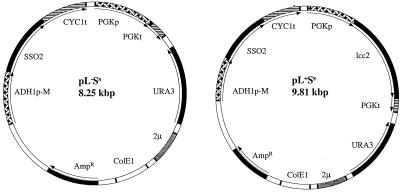
A 2.7-kb fragment containing the SSO2 gene between the ADH1-derived promoter and the CYC1 terminator of YEpSSO2-M was excised with SalI and HindIII, blunt ended with the Klenow fragment, and purified from an agarose gel by the QiaQuick procedure (Qiagen, Hilden, Germany). The pAJ401/lcc2 plasmid was linearized with PvuII, dephosphorylated with shrimp alkaline phosphatase, and purified from an agarose gel. The SSO2 gene was inserted into the pAJ/lcc2 plasmid by blunt-end ligation with T4 DNA ligase. The resulting construct was referred to as pL+Ss (pAJ401/lcc2/SSO2). Using the same procedure, the 2.7-kb SSO2 fragment was inserted into the pAJ401 plasmid without the lcc2 gene. The resulting construct was referred to as pL−Ss (pAJ401/SSO2). Both constructs are shown in Fig. 1.
FIG. 1.
Plasmids pL−Ss and pL+Ss.
S. cerevisiae INVSCI was transformed with pL+Ss and pL−Ss by electroporation (3). Transformants were selected on SC-Ura agar plates (24) containing 0.2 mM 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and 0.1 mM CuSO4. The integrity of transformant L+Ss was confirmed by PCR. Spheroplasts were prepared by using Zymolyase (ICN Biochemicals, Aurora, Ohio). The spheroplasts were then osmotically lysed, and plasmid DNA was isolated with a Quantum Prep plasmid miniprep kit (Bio-Rad, Hercules, Calif.). An upstream primer complementary to the PGK1 promoter (3′-CATCAAGGAAGTAATTATCTACT-5′) and a downstream primer complementary to the lcc2 gene (5′-CGGAATTCCATTTACTGGTCGCTCGGGTCG-3′) were used for PCR analysis. The concentrations of primers, nucleotides, Taq polymerase, and Mg2+ ions were those recommended by the supplier of the DNA polymerase (Roche), and the amplification conditions were as follows: 94°C for 1 min, 55°C for 1 min, and 70°C for 3 min for 30 cycles, followed by a final extension step at 72°C for 7 min. The PCR was performed by using GeneAmp PCR System 9700 (Perkin-Elmer Corp., Norwalk, Conn.). The PCR products were analyzed by agarose gel electrophoresis. The integrity of the yeast transformant harboring pL−Ss was confirmed by plasmid rescue, transformation of E. coli DH5α, and restriction analysis of plasmid DNA isolated from the bacterium. The vectors and yeast transformants used are shown in Table 1.
TABLE 1.
Plasmids and yeast transformants
| Plasmid | Reference(s) | Transformant | Description |
|---|---|---|---|
| pL− | 15, 23 | L− | S. cerevisiae INVSC1 harboring pL− (control for L+) |
| pL+ | 5 | L+ | S. cerevisiae INVSC1 harboring pL+ (laccase [lcc2] expressing) |
| pL−Ss | This study | L−Ss | S. cerevisiae INVSC1 harboring pL−Ss (Sso2p-overexpressing control for L+Ss) |
| pL+Ss | This study | L+Ss | S. cerevisiae INVSC1 harboring pL+Ss (laccase expressing and Sso2p overexpressing) |
Hydrolysate.
Dilute acid spruce hydrolysate was purchased from R. Eklund (Mitthögskolan, Örnsköldsvik, Sweden). The hydrolysate was prepared in two steps. Freshly chipped Norway spruce (Picea abies) was impregnated with sulfuric acid (0.5%, wt/vol) and then loaded into a 250-liter batch reactor. Steam was then added to a pressure of 12 × 105 Pa (190°C), and this pressure was kept constant for 10 min. The material from the first step was discharged into a collecting vessel, and the liquid and solid fractions were separated by filtration. The solid residue was reimpregnated with sulfuric acid and loaded into the reactor. Steam was added to a pressure of 21 × 105 Pa (215°C), and the pressure was kept constant for 10 min, after which the material was discharged. The liquid fraction from the second step was recovered by filtration. The liquid fractions from the two steps were pooled and are referred below to as the hydrolysate.
The hydrolysate contained (per liter) 20.5 g of glucose, 14.9 g of mannose, 7.0 g of xylose, 2.9 g of galactose, 1.4 g of arabinose, 2.8 g of acetic acid, 0.7 g of formic acid, 1.1 g of levulinic acid (N.-O. Nilvebrant, personal communication), 1.4 g of furfural, and 2.3 g of 5-HMF (measured by high-performance liquid chromatography [HPLC]), as well as 2.9 g of phenolic compounds (determined spectrophotometrically by using the Prussian blue method [7] and vanillin as the standard).
Cultivation of S. cerevisiae transformants.
All chemicals were analytical grade, unless stated otherwise. The cultivation pH was 5.5. All cultivations were done at 22°C since it has been reported previously that this temperature is within the optimal temperature range for heterologous expression of laccase in S. cerevisiae (5). Inocula were grown in two stages. Cells were obtained from SC-Ura agar plates and were grown overnight in 15-ml tubes containing 10 ml of SC-Ura medium. The cells were harvested by centrifugation at 1,200 × g and 4°C for 10 min and were washed with 0.9% (wt/vol) NaCl. The cells were then transferred to 1,000-ml baffled shake flasks containing 500 ml of SC-Ura medium. The cells were harvested in the late exponential phase and were used as an inoculum for shake flask or fermentor cultivation.
Shake flask cultivation was performed in 500-ml baffled flasks containing 50 or 400 ml of SC-Ura medium. The inoculum was added to a final optical density at 620 nm of 0.5, as measured spectrophotometrically (model U-2000 spectrophotometer; Hitachi, Tokyo, Japan), which corresponded to a concentration of 0.1 g (dry weight) per liter. The yeast cells were incubated with shaking (150 rpm). Samples (2 ml) were withdrawn every fourth hour for spectrophotometric determination of cell growth and for a laccase activity assay. The cells were cultivated for 48 h under aseptic conditions. Each cultivation was repeated at least twice.
Fermentor vessels (1-liter;-Belach AB, Stockholm, Sweden) containing 800 ml of medium were used for batch fermentation under oxygen-limited conditions. Nitrogen gas was flushed through the medium to obtain an anaerobic environment, after which a mixture of nitrogen gas (0.15 liter/min) and nitrogen gas supplemented with 0.5% oxygen (0.05 liter/min) was supplied. The exhaust gas was passed through a reflux cooler in order to minimize evaporation of ethanol. The pH in the fermentor was maintained at 5.5 by using 3 M NaOH and 3 M H2SO4. The stirring speed was 250 rpm. Samples used for laccase activity, substrate consumption, and product formation measurements were withdrawn, flash frozen in a dry ice-ethanol bath, and stored at −80°C until they were analyzed, whereas optical density measurement and gravimetric analysis of biomass were performed immediately after sample withdrawal.
The following media were used for the fermentor cultivations: (i) SC-Ura medium, (ii) SC-Ura medium containing coniferyl aldehyde at a concentration of 1.25 mM, and (iii) hydrolysate supplemented with yeast nitrogen base and amino acids at concentrations corresponding to those in SC-Ura medium. In all fermentor cultivations an ergosterol-Tween 80 solution was added so that the concentrations of these compounds were 10 and 420 mg/liter, respectively (30). Inocula for the fermentors were grown as described above, and the fermentors were inoculated to final optical densities of 0.5 (SC-Ura medium and SC-Ura medium containing coniferyl aldehyde) and 5 (hydrolysate), which corresponded to 0.1 and 1 g (dry weight) per liter, respectively. Each cultivation was repeated twice.
Analyses.
Biomass was determined gravimetrically (as dry weight) at the beginning and end of each fermentation. To determine dry weight, 1.5 ml of culture broth was filtered through predried nitrocellulose filters with a pore size of 0.45 μm (Gelman Sciences, Ann Arbor, Mich.). The filtered pellets were then washed with 4.5 ml of double-deionized water (Millipore, Bedford, Mass.) and dried in a microwave oven (Whirlpool, Benton Harbor, Mich.) at a power setting of 3.5 for 15 min.
Samples used for subsequent laccase activity assay and HPLC measurements were filtered through 0.2-μm-pore-size membrane filters (Advantech, Pleasanton, Calif.). The concentrations of glucose, mannose, ethanol, glycerol, acetic acid, and lactic acid were determined by using an HPLC system (Gilson Inc., Middleton, Wis.) equipped with an Aminex HPX-87H column (Bio-Rad) and a refractive index detector (RID-6A; Shimadzu, Kyoto, Japan) operating at 45°C; the mobile phase was 5 mM H2SO4 at a flow rate of 0.6 ml/min.
Consumption of coniferyl aldehyde was determined with an HPLC system (Waters, Milford, Mass.) equipped with a BioSil-C18 column (Bio-Rad) and a UV detector (model 2487; Waters) set at a wavelength of 254 nm and operating at the ambient temperature. The mobile phase consisted of 40% aqueous methanol adjusted to pH 3 with concentrated HCl and was supplied at a flow rate of 0.40 ml/min.
The total concentration of phenolic compounds in the hydrolysate during fermentation was monitored by the Prussian blue method, as described by Graham (8), using vanillin as the standard.
Formation of high-molecular-mass compounds in the cultivations with coniferyl aldehyde was investigated by using gel permeation chromatography (GPC) with a PD-10 column (Pharmacia, Uppsala, Sweden). A cell-free sample (1.5 ml) was mixed with 1 ml of 20 mM sodium acetate buffer (pH 5.2) and applied to the column. Elution was performed with the same buffer (20 mM sodium acetate, pH 5.2), and 1-ml fractions were collected after the entire 2.5-ml sample diluted with buffer had entered the column. The fractions were analyzed by measuring absorption at 280 nm. Aqueous solutions of coniferyl aldehyde (1.25 mM) and coniferyl alcohol (1.25 mM) were separately applied to the column as standards. The standards were diluted with buffer and eluted in the same way as the samples from the cultivations, except that sodium chloride was added at a final concentration of 200 mM to the 20 mM sodium acetate (pH 5.2) used as buffer.
Laccase stability test.
To determine the optimal pH for fermentation, the stability of a commercial laccase preparation from T. versicolor (synonym, Coriolus versicolor) (Jülich GmbH, Jülich, Germany) was assessed after incubation of 10.3 nkat of laccase for 24 h at approximately 22°C and at pH values ranging from 2.4 to 9. The following buffer solutions were used: glycine-HCl (pH 2.4, 3.0, and 3.4), sodium acetate (pH 4.0 and 5.0), phosphate (pH 6.0, 7.0, and 8.0), and glycine-NaOH (pH 9.0). The concentration of each of the buffers was 50 mM. After incubation, enzyme activity was measured spectrophotometrically at 414 nm as described elsewhere (5). The final pH in the cuvette was 5.2 for all the samples.
Laccase activity assay.
Samples from the cultivations in SC-Ura medium and SC-Ura medium containing coniferyl aldehyde were filtered through 0.2-μm-pore-size membrane filters (Advantech) and dialyzed prior to a laccase activity assay in order to remove interfering compounds. For dialysis, Spectra/Por membrane tubing (Spectrum Laboratories, Rancho Dominguez, Calif.) with a molecular weight cutoff of 12,000 to 14,000 was used. The samples were dialyzed against 50 mM sodium acetate buffer (pH 5.2) supplemented with 0.1 mM CuSO4. The laccase activities in samples from the cultivations in hydrolysate were determined after GPC with a PD-10 column (Pharmacia). Each cell-free sample (1.5 ml) was mixed with 1 ml of 20 mM sodium acetate buffer (pH 5.2), and the column was eluted with the same buffer. Fractions (1 ml) were collected, and fractions 2 to 4 were assayed for laccase activity. The activity was determined by using ABTS as the reducing substrate (5). The change in absorption at 414 nm was recorded with a model U-2000 instrument (Hitachi) after incubation for 24 h at approximately 22°C, and 3.6 × 104 M−1 cm−1 (6) was used as the extinction coefficient for calculating the activity.
Calculations.
Laccase activity was expressed in nanokatals per milliliter of sample. Specific activity was expressed either as laccase activity in the supernatant as a function of biomass (in nanokatals per gram [dry weight] of cells) or as laccase activity as a function of the total amount of protein in the supernatant (in nanokatals per gram of protein). The dilution was taken into account when activity after GPC was calculated. The ethanol yield on total sugar was calculated as the amount of ethanol produced divided by the total amount of fermentable sugar (glucose, mannose, and galactose). The ethanol yield on consumed sugars was calculated as the amount of ethanol produced divided by the consumed amount of fermentable sugars. The biomass yield was calculated as the amount of biomass formed (in grams [dry weight]) divided by the amount of consumed sugar. The maximum growth rate (μmax) was calculated from logarithmic optical density obtained values in the exponential phase.
RESULTS
The performance of S. cerevisiae expressing laccase from T. versicolor was investigated in defined medium (SC-Ura medium) under aerobic conditions and in SC-Ura medium, SC-Ura medium with 1.25 mM coniferyl aldehyde, and hydrolysate supplemented with nutrients under oxygen-limited conditions. Before fermentor cultivation was performed, the effects of pH, aeration, and Sso2p overexpression on laccase activity were examined. Aerobic fermentation was done in shake flasks (50 ml of medium in 500-ml flasks), whereas oxygen-limited fermentation was done in both shake flasks (400 ml of medium in 500-ml flasks) and a fermentor. The effects of coniferyl aldehyde and hydrolysate on cell growth and ethanol formation were studied in the fermentor. All fermentations were performed in duplicate, and the results presented in Table 2 and below are the mean values.
TABLE 2.
Product yields and growth rates in fermentor cultivations
| Fermentation medium |
YEtOH tot (g/g)a
|
YEtOH cons (g/g)b
|
YX (g/g)c
|
μmax (h−1)
|
||||
|---|---|---|---|---|---|---|---|---|
| L+Ss | L−Ss | L+Ss | L−Ss | L+Ss | L−Ss | L+Ss | L−Ss | |
| SC-Ura | 0.38 | 0.38 | 0.38 | 0.38 | 0.07 | 0.07 | 0.060 | 0.058 |
| SC-Ura + coniferyl aldehyde | 0.44 | 0.00 | 0.44 | 0.17 | 0.06 | 0.00 | 0.060 | 0.000 |
| Hydrolysate | 0.44 | 0.02 | 0.47 | 0.25 | 0.02 | 0.00 | 0.012 | 0.000 |
YEtOH tot, ethanol yield on total sugar.
YEtOH cons, ethanol yield on consumed sugars.
Yx, biomass yield.
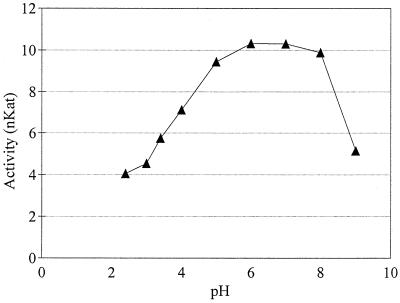
Laccase was most stable at pH 6.0 and 7.0 (Fig. 2); at these pH values 99% of the initial activity was retained after incubation for 24 h. The pH values determined at the end of shake flask cultivation in SC-Ura medium without pH control ranged from 2.8 to 3.5. The stability test indicated that at these low pH values only a fraction of the activity was retained (Fig. 2). We therefore concluded that pH control of fermentation was necessary. The pH of the medium in the fermentors was kept at 5.5.
FIG. 2.
Stability of native T. versicolor laccase at different pH values. Laccase was incubated for 24 h at 22°C in the following buffers (concentrations, 50 mM): glycine-HCl (pH 2.4, 3.0, and 3.4), sodium acetate (pH 4.0 and 5.0), phosphate (pH 6.0, 7.0, and 8.0), and glycine-NaOH (pH 9.0). After incubation, the activity at pH 5.2 was determined by using ABTS as the reducing substrate.
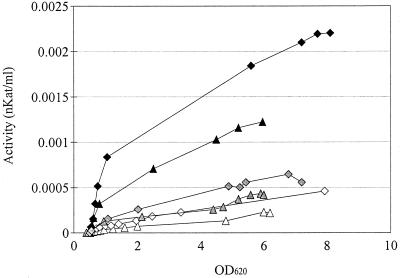
Lower laccase activity was observed when the cultivation conditions in shake flasks were aerobic (50 ml of medium in 500-ml flasks) than when the conditions were oxygen-limited (400 ml of medium in 500-ml flasks) (Fig. 3). After 36 h of incubation of the transformant L+ under aerobic and oxygen-limited conditions, the specific activities based on biomass were 0.18 and 0.36 nkat/g of cells, respectively, the specific activities based on total protein were 1.57 and 3.07 nkat/g of protein, respectively, and the total activities were 0.00022 and 0.00043 nkat/ml of culture supernatant, respectively. After 36 h of incubation of L+Ss under aerobic and oxygen-limited conditions, the specific activities based on biomass were 0.29 and 0.51 nkat/g of cells, respectively, the specific activities based on total protein were 2.56 and 3.72 nkat/g of protein, respectively, and the total activities were 0.00046 and 0.00067 nkat/ml of culture supernatant, respectively.
FIG. 3.
Laccase activities in cultures of S. cerevisiae transformants L+ (triangles) and L+Ss (diamonds) in shake flasks under aerobic conditions (50 ml of medium in 500-ml shake flasks) (open symbols) and oxygen-limited conditions (400 ml of medium in 500-ml shake flasks) (shaded symbols) and in a fermentor under oxygen-limited conditions (solid symbols). The initial pH of each of the fermentations was 5.5. The pH was maintained at pH 5.5 in the fermentor but not in the shake flasks. OD620 optical density at 620 nm.
Laccase activity increased remarkably when oxygen-limited conditions were combined with pH control in the fermentor cultivations (Fig. 3). After 36 h of incubation of L+, the specific activity based on biomass was 1.02 nkat/g of cells and the total activity was 0.0012 nkat/ml of culture supernatant. After 36 h of incubation of L+Ss, the specific activity based on biomass was 1.38 nkat/g of cells and the total activity was 0.0022 nkat/ml of culture supernatant. Overexpression of Sso2p enhanced the activity of laccase under all cultivation conditions used (Fig. 3).
There was no difference either in growth or in product formation in SC-Ura medium between L+Ss and L−Ss (Table 2). Transformants L+ and L+Ss showed similar product formation patterns (data not shown), and the final ethanol yield on consumed sugar was the same for both transformants (0.38 g/g). However, there was a slight difference in biomass formation between L+Ss and L+. Transformant L+Ss had a higher μmax value than L+ (0.06 h−1 compared to 0.04 h−1), and it was able to reach a higher final cell mass (1.5 g [dry weight]/liter compared to 1.2 g/liter). Since both better growth and higher laccase activity were obtained with the transformant overexpressing Sso2p (L+Ss), further experiments with coniferyl aldehyde added to the medium or with hydrolysate were performed only with L+Ss and L−Ss.
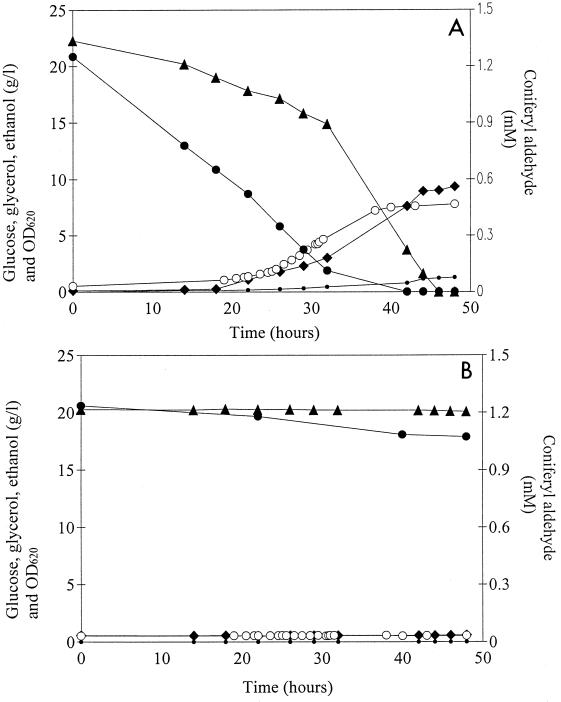
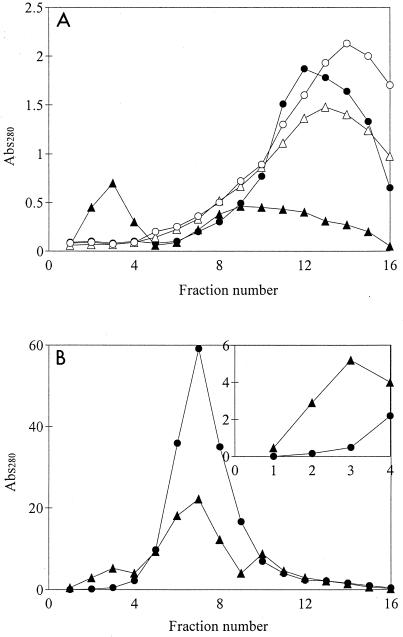
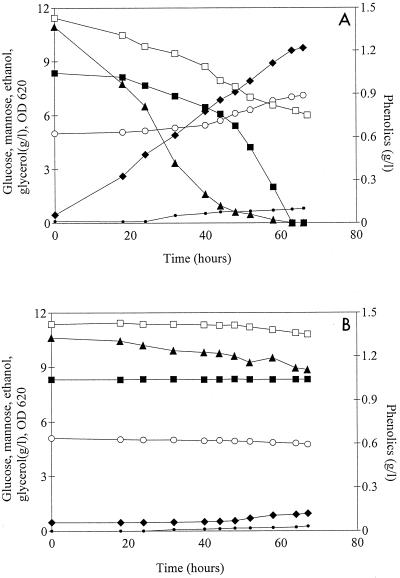
When coniferyl aldehyde was added to the cultivation medium, cell growth and ethanol production were observed only with L+Ss, the laccase-expressing strain, whereas L−Ss did not grow at all during the experiment (Fig. 4). Transformant L+Ss grew at a rate comparable to the rate in cultivations without coniferyl aldehyde added (Table 2), although the lag phase lasted longer (20 h compared to 9 h in the absence of coniferyl aldehyde). When the concentration of coniferyl aldehyde in the growth medium had decreased to approximately 0.6 mM, cell growth commenced and ethanol formation started to proceed at a higher rate. The yield of ethanol was higher than that in SC-Ura medium without coniferyl aldehyde, whereas the biomass yield was slightly lower (Table 2). A gel permeation chromatogram (Fig. 5) revealed the presence of high-molecular-mass compounds eluting in fractions 2 to 4 after cultivation with L+Ss, as well as a decrease in the amount of low-molecular-mass compounds eluting in fractions 8 to 16. Absorption of purified laccase at 280 nm was tested, and we concluded that the increase in absorption in fractions 2 to 4 could not be a consequence of absorption by the laccase molecules themselves. Most of the absorption in fractions 2 to 4 was most likely due to polymerization products. Coniferyl alcohol, which was used as a standard and was applied at a concentration which corresponded to the concentration after a tentative equimolar conversion of the aldehyde to alcohol, eluted with a peak in fraction 13, and the absorption was much greater than that in the L+Ss fermentation sample. The broad peak obtained with the L+Ss sample, stretching between fractions 8 and 16, indicated that several compounds were present. The HPLC analyses of the first sample (zero time) from the L+Ss fermentation revealed the presence of only one major peak, which was identified as coniferyl aldehyde. The disappearance of coniferyl aldehyde in subsequent samples (14 to 48 h) coincided with the formation of several other peaks, indicating that there was heterogeneity in the product pattern.
FIG. 4.
Consumption of the substrates glucose (▴) and coniferyl aldehyde (●) and formation of ethanol (⧫), glycerol (•), and biomass (○) (measured as optical density at 620 nm [OD620]) by L+Ss (A) and L−Ss (B). The fermentations were performed in a 1-liter fermentor by using SC-Ura medium supplemented with 1.25 mM coniferyl aldehyde.
FIG. 5.
Effect of laccase on the size distribution of compounds with absorption at 280 nm (Abs280) during fermentation with S. cerevisiae transformants L+Ss (▴) and L−Ss (●). (A) GPC of fermentation medium after cultivation with 1.25 mM coniferyl aldehyde. Elution of the standards, coniferyl aldehyde (○) and coniferyl alcohol (Δ), is also shown. (B) GPC of a dilute acid hydrolysate of spruce at the end of fermentation. The inset shows a close-up for fractions 1 to 4.
In contrast, GPC of the sample from the cultivation with L−Ss resulted in an elution pattern very similar to the elution pattern of the coniferyl aldehyde standard, which had only a slightly higher absorption value. This result was in agreement with the results of the HPLC analyses, which showed that there was only a slight decrease in the coniferyl aldehyde concentration after cultivation with L−Ss.
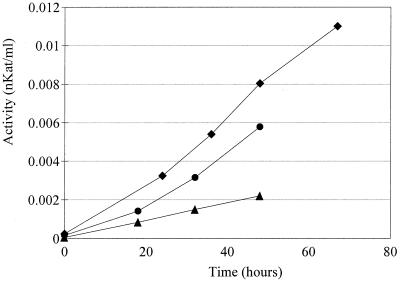
The enzyme activity of L+Ss was significantly higher in SC-Ura medium with coniferyl aldehyde than in SC-Ura medium alone (Fig. 6). Laccase specific activity was greater as well. In the absence of coniferyl aldehyde, the specific activity based on biomass after 48 h of fermentation was 1.41 nkat/g (dry weight) of cells, whereas in the presence of coniferyl aldehyde the specific activity increased to 3.72 nkat/g (dry weight) of cells.
FIG. 6.
Laccase activity of L+Ss during cultivation as a function of time. L+Ss was cultivated in SC-Ura medium (▴), in SC-Ura medium containing 1.25 mM coniferyl aldehyde (●), and in hydrolysate (⧫).
The highest total activity value (Fig. 6) and highest laccase specific activity value (7.75 nkat/g [dry weight] of cells after 48 h) were obtained with the hydrolysate cultivations. Laccase activity could not be measured readily after dialysis of cell-free hydrolysate samples. This was most likely due to interference with the ABTS assay by high-molecular-mass compounds present in the hydrolysate, which were retained within the dialysis membrane together with laccase. However, it was possible to measure the laccase activity after GPC. High-molecular-mass compounds to some extent coeluted with the enzyme, as indicated by the brown color present in fractions 2 to 4, where the laccase activity was found. Activity values for fraction 2 are shown in Fig. 6, since this fraction had the highest laccase activity. The transformant which did not express laccase, L−Ss, was not able to grow in the hydrolysate and produced only 0.9 g of ethanol per liter in 67 h (Fig. 7B). The laccase-expressing strain, L+Ss, on the other hand, produced ethanol with a yield of 0.44 g/g on the total fermentable sugars present (Table 2). All glucose and mannose were consumed (Fig. 7A). Cell growth, on the other hand, was quite limited. Both the biomass yield and the growth rate were significantly lower than the values obtained for the cultivations in which only coniferyl aldehyde was added (Table 2). The total amount of phenolic compounds decreased during the fermentation (Fig. 7A), whereas the amount of high-molecular-mass compounds increased (Fig. 5). There was also a minor decrease in the concentration of phenolic compounds during the fermentation with L−Ss (Fig. 7B); however, it was not at all comparable to the decrease observed with the laccase-expressing transformant.
FIG. 7.
Fermentation of a dilute acid spruce hydrolysate by L+Ss (A) and L−Ss (B) at a high cell density. The consumption of glucose (▴) and mannose (■), the concentration of phenolic compounds (□), and the production of ethanol (⧫), glycerol (•), and biomass (○) (measured as optical density at 620 nm [OD 620]) were determined.
DISCUSSION
Before the performance of laccase-expressing S. cerevisiae in fermentor cultivations with phenolic inhibitors was tested, the effects of pH, aeration, and overexpression of Sso2p were examined. A problem obtaining heterologous laccase activity when the pH of the cultivation medium was low was observed previously with P. pastoris (10). However, the reason for this was not known. Expression of laccase in S. cerevisiae was affected by the same problem, which could be explained by the finding that laccase exhibits poor stability at low pH values.
However, the pH effect did not explain the difference in activity between the shake flasks with different volumes, since in neither case was the pH controlled. The laccase activity increased when the culture volume increased and aeration decreased. If the increase in activity was caused by decreased aeration, this phenomenon is highly desirable for ethanol production from lignocellulose hydrolysates, since the best conditions for obtaining high laccase activity would then coincide with the preferred conditions for performing ethanolic fermentation. Oxygen-limited conditions, particularly anaerobic conditions, also lead to a lower biomass yield. An increase in the specific activity of an enzyme is thus counteracted by a decrease in the formation of protein-producing biomass. It should be mentioned that a decrease in biomass formation is beneficial during production of ethanol from lignocellulose, since more carbon is then converted to ethanol instead of being incorporated into biomass. One possibility is that the increase in laccase activity was a consequence of an effect of decreased oxygenation on regulation of transcription. Laccase was expressed in S. cerevisiae under control of the PGK1 (phosphoglycerate kinase gene) promoter, which is known to be induced by glucose (28). More recently, a slight increase (ratio, 1.3) in expression of PGK1 under anaerobic conditions compared to aerobic conditions was found by using glucose-limited chemostat cultures of S. cerevisiae (27). Moreover, the gene encoding phosphoglycerate kinase in Penicillium chrysogenum has previously been reported to be induced by decreased oxygen levels (19), and the same effect has been observed in mammalian cells (11, 22). This possible explanation deserves to be investigated further, considering that heterologous proteins are often expressed in S. cerevisiae under control of the PGK1 promoter and under aerobic conditions. Oxygen-limited conditions were superior to well-aerated conditions for obtaining higher levels of laccase activity, but additional studies of other proteins expressed in S. cerevisiae under control of the PGK1 promoter are required to conclusively establish the influence of the oxygen supply on the expression level.
Overexpression of Sso2p increased laccase activity, regardless of the cultivation conditions. The two Sso2p-overexpressing transformants used (with and without lcc2) had similar growth characteristics. This is in agreement with a study on enhancement of production of heterologous α-amylase by S. cerevisiae (21). In contrast, Vad et al. found that even if overexpression of Sso2p in S. cerevisiae enhanced secretion, the total amount of the heterologous protein, human parathyroid hormone, was not increased due to impaired growth of the cells (29).
Besides pH, oxygenation level, and Sso2p overexpression, the presence or absence of inhibitory phenolic compounds affected laccase activity in S. cerevisiae. Higher laccase activity (both specific activity and total activity) was observed when coniferyl aldehyde and various phenolic compounds present in the spruce hydrolysate were included in the medium than when the organism was cultivated in SC-Ura medium alone. Since laccase catalyzes conversion of inhibitory phenolic compounds to less inhibitory polymerization products (9), heterologous expression of laccase by yeast cells growing under these conditions is a metabolic asset rather than a burden.
Laccase-expressing and Sso2p-overexpressing S. cerevisiae was able to grow in medium containing coniferyl aldehyde at a concentration that completely inhibited the transformant not expressing laccase. This ability was attributed to laccase-mediated transformation of phenolic compounds into radicals, which was indicated by the formation of polymerization products detected by GPC. HPLC chromatograms showed the presence of several peaks after the disappearance of coniferyl aldehyde. Previous results showed that coniferyl aldehyde is converted to coniferyl alcohol and dihydroconiferyl alcohol by baker's yeast (13). The complex product pattern observed could therefore be explained by combined action on coniferyl aldehyde by laccase and by as-yet-unidentified enzymes from S. cerevisiae that are able to catalyze reduction of coniferyl aldehyde. Reduced products of coniferyl aldehyde, such as coniferyl alcohol, may also serve as substrates for laccase, giving rise to further heterogeneity in product formation.
Transformant L+Ss also efficiently fermented the spruce hydrolysate to ethanol, strengthening the conclusion that low-molecular-mass phenolic compounds are important inhibitors of ethanolic fermentation in spruce lignocellulose hydrolysates (14). Growth was still significantly inhibited in the hydrolysate cultivations, most likely due to the presence of furan derivatives, which have previously been shown to have this effect (4, 12, 17, 25, 26). The higher initial dry weight, which may have contributed to higher total laccase activity at the beginning of the hydrolysate cultivation, was deliberately chosen due to the presence in the hydrolysate of inhibitors other than phenolic compounds, such as furfural and 5-HMF, since the inhibitory effect of furan derivatives can be reduced by using a large inoculum (7). It is probable that the laccase activity in the GPC fractions was even higher than that reported here due to the remaining negative influence of high-molecular-mass compounds in the hydrolysate on the assay.
In conclusion, the laccase activity of S. cerevisiae was increased by overexpressing Sso2p and by modifying the cultivation conditions (pH and aeration). It is possible that the level of expression of laccase in S. cerevisiae could be enhanced further by improving important steps in the protein expression machinery other than secretion, such as folding. The activity obtained was, however, sufficient for achieving very good fermentability of an inhibitory lignocellulose hydrolysate without any prior detoxification step. Our results demonstrate the importance of low-molecular-mass phenolic compounds as inhibitors and the potential of using genetic engineering for the development of microbial strains with enhanced resistance to fermentation inhibitors.
ACKNOWLEDGMENTS
The Swedish National Energy Administration and the Carl Tryggers Foundation are acknowledged for financial support.
Sirkka Keränen (VTT, Espoo, Finland) is gratefully acknowledged for providing plasmid YEpSSO2-M. Nils-Olof Nilvebrant and Anders Reimann (STFI, Stockholm, Sweden) are gratefully acknowledged for performing the analysis of sugars and aliphatic acids in the hydrolysate. Christer Larsson is gratefully acknowledged for performing the analysis of furan derivatives in the hydrolysate.
REFERENCES
- 1.Aalto M K, Rönne H, Keränen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando S, Arai I, Kiyoto K, Hanai S. Identification of aromatic monomers in steam-exploded poplar and their influences on ethanol fermentation by Saccharomyces cerevisiae. J Ferment Technol. 1986;64:567–570. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. [Google Scholar]
- 4.Boyer L J, Vega L J, Klasson K T, Clausen E C, Gaddy J L. The effects of furfural on ethanol production by Saccharomyces cerevisiae in batch culture. Biomass Bioenergy. 1992;3:41–48. [Google Scholar]
- 5.Cassland P, Jönsson L J. Characterization of a gene encoding Trametes versicolor laccase A and improved heterologous expression in Saccharomyces cerevisiae by decreased cultivation temperature. Appl Microbiol Biotechnol. 1999;52:393–400. doi: 10.1007/s002530051537. [DOI] [PubMed] [Google Scholar]
- 6.Childs R E, Bardsley W G. The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem J. 1975;145:93–103. doi: 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung I S, Lee Y Y. Ethanol fermentation of crude acid hydrolyzate of cellulose using high-level yeast inocula. Biotechnol Bioeng. 1984;27:308–315. doi: 10.1002/bit.260270315. [DOI] [PubMed] [Google Scholar]
- 8.Graham H D. Stabilization of the Prussian blue color in the determination of polyphenols. J Agric Food Chem. 1992;40:801–805. [Google Scholar]
- 9.Jönsson L J, Palmqvist E, Nilvebrant N-O, Hahn-Hägerdal B. Detoxification of wood hydrolysates with laccase and peroxidase from the white-rot fungus Trametes versicolor. Appl Microbiol Biotechnol. 1998;49:691–697. [Google Scholar]
- 10.Jönsson L J, Saloheimo M, Penttilä M. Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr Genet. 1997;32:425–430. doi: 10.1007/s002940050298. [DOI] [PubMed] [Google Scholar]
- 11.Kress S, Stein A, Maurer P, Weber B, Reichert J, Buchmann A, Huppert P, Schwarz M. Expression of hypoxia-inducible genes in tumor cells. J Cancer Res Clin Oncol. 1998;124:315–320. doi: 10.1007/s004320050175. [DOI] [PubMed] [Google Scholar]
- 12.Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant N-O. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol. 1999;24:151–159. [Google Scholar]
- 13.Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant N O, Jönsson L J. The influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2000;84–86:617–632. doi: 10.1385/abab:84-86:1-9:617. [DOI] [PubMed] [Google Scholar]
- 14.Larsson S, Reimann A, Nilvebrant N-O, Jönsson L J. Comparison of different methods for the detoxification of lignocellulose hydrolyzates of spruce. Appl Biochem Biotechnol. 1999;77–79:91–103. [Google Scholar]
- 15.Minet M, Lacroute F. Cloning and sequencing of a human cDNA coding for a multifunctional polypeptide of the purine pathway by complementation of the ade2–101 mutant in Saccharomyces cerevisiae. Curr Genet. 1990;18:287–291. doi: 10.1007/BF00318209. [DOI] [PubMed] [Google Scholar]
- 16.Olsson L, Hahn-Hägerdal B. Fermentation of lignocellulose hydrolysates for ethanol production. Enzyme Microb Technol. 1996;18:312–331. [Google Scholar]
- 17.Palmqvist E, Almeida J S, Hahn-Hägerdal B. Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol Bioeng. 1999;62:447–454. doi: 10.1002/(sici)1097-0290(19990220)62:4<447::aid-bit7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Reinhammar B. Kinetic studies on Polyporus and tree laccases. In: Messerschmidt A, editor. Multi-copper oxidases. Singapore: World Scientific; 1997. pp. 167–200. [Google Scholar]
- 19.Renno D V, Saunders G, Bull A T, Holt G. Transcript analysis of penicillin genes from Penicillium chrysogenum. Curr Genet. 1992;21:49–54. doi: 10.1007/BF00318654. [DOI] [PubMed] [Google Scholar]
- 20.Romanos M A, Scorer C A, Clare J J. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 21.Ruohonen L, Toikkanen J, Tieaho V, Outola M, Söderlund H, Keränen S. Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso2 protein, a late-acting component of the secretory machinery. Yeast. 1997;13:337–351. doi: 10.1002/(SICI)1097-0061(19970330)13:4<337::AID-YEA98>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Salceda S, Beck I, Caro J. Absolute requirement of aryl hydrocarbon receptor nuclear translocator protein for gene activation by hypoxia. Arch Biochem Biophys. 1996;334:389–394. doi: 10.1006/abbi.1996.0469. [DOI] [PubMed] [Google Scholar]
- 23.Saloheimo A, Henrissat B, Hoffren A M, Teleman O, Penttilä M. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol Microbiol. 1994;13:219–228. doi: 10.1111/j.1365-2958.1994.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 24.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 25.Taherzadeh M J, Eklund R, Gustafsson L, Niklasson C, Lidén G. Characterization and fermentation of dilute-acid hydrolyzates from wood. Ind Eng Chem Res. 1997;36:4659–4665. [Google Scholar]
- 26.Taherzadeh M J, Gustafsson L, Niklasson C, Lidén G. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2000;53:701–708. doi: 10.1007/s002530000328. [DOI] [PubMed] [Google Scholar]
- 27.ter Linde J J, Liang H, Davis R W, Steensma H Y, van Dijken J P, Pronk J T. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J Bacteriol. 1999;181:7409–7413. doi: 10.1128/jb.181.24.7409-7413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuite M F, Dobson M J, Roberts N A, King R M, Burke D C, Kingsman S M, Kingsman A J. Regulated high efficiency expression of human interferon-alpha in Saccharomyces cerevisiae. EMBO J. 1982;1:603–608. doi: 10.1002/j.1460-2075.1982.tb01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vad R, Moe E, Saga K, Kvinnsland A M, O/yen T B. High-level production of human parathyroid hormone (hPTH) by induced expression in Saccharomyces cerevisiae. Protein Expr Purif. 1998;13:396–402. doi: 10.1006/prep.1998.0912. [DOI] [PubMed] [Google Scholar]
- 30.Verdyn C, Postma E, Scheffers A W, van Dijken J P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 31.von Sivers M, Zacchi G, Olsson L, Hahn-Hägerdal B. Cost analysis of ethanol production from willow using recombinant Escherichia coli. Biotechnol Prog. 1994;10:555–560. doi: 10.1021/bp00029a017. [DOI] [PubMed] [Google Scholar]