Abstract
Motor skills learning is classically associated with brain regions including cerebral and cerebellar cortices and basal ganglia nuclei. Less is known about the role of the hippocampus in the acquisition and storage of motor skills. Here, we show that mice receiving a long-term training in the accelerating rotarod display marked hippocampal transcriptional changes and reduced pyramidal neurons activity in the CA1 region when compared with naive mice. Then, we use mice in which neural ensembles are permanently labeled in an Egr1 activity-dependent fashion. Using these mice, we identify a subpopulation of Egr1-expressing pyramidal neurons in CA1 activated in short-term (STT) and long-term (LTT) trained mice in the rotarod task. When Egr1 is downregulated in the CA1 or these neuronal ensembles are depleted, motor learning is improved whereas their chemogenetic stimulation impairs motor learning performance. Thus, Egr1 organizes specific CA1 neuronal ensembles during the accelerating rotarod task that limit motor learning. These evidences highlight the role of the hippocampus in the control of this type of learning and we provide a possible underlying mechanism.
SIGNIFICANCE STATEMENT It is a major topic in neurosciences the deciphering of the specific circuits underlying memory systems during the encoding of new information. However, the potential role of the hippocampus in the control of motor learning and the underlying mechanisms has been poorly addressed. In the present work we show how the hippocampus responds to motor learning and how the Egr1 molecule is one of the major responsible for such phenomenon controlling the rate of motor coordination performances.
Keywords: Egr1, engrams, hippocampus, learning and memory, motor coordination, neurons
Introduction
It is now well established that the hippocampus is involved in the formation of detailed cognitive “maps” of the context in which learning occurs (Spiers, 2020). However, in addition to its classical role in spatial maps formation, the hippocampus is also involved in tasks that primarily rely on other brain regions. For example, the hippocampus is activated during goal-directed behaviors and strategies (Fidalgo et al., 2012; Palombo et al., 2019) and modulates contextual associations during drug-of-abuse administration (Sjulson et al., 2018; Zhou et al., 2019) or appetitive conditioning (Ito et al., 2008). Despite these previous reports, the role of the hippocampus in motor learning and the underlying mechanisms remain poorly explored. Understanding the implications of the hippocampus in the regulation of motor learning could help to decipher the alterations in neural circuits of complex motor diseases such as Huntington's and/or Parkinson's disease in which a severe hippocampal atrophy and/or dysfunction have already been described (Spargo et al., 1993; Camicioli et al., 2003; Calabresi et al., 2013; Begeti et al., 2016; Harris et al., 2019).
Potential contributions of the hippocampus to motor learning and coordination have been proposed on the basis of human studies and imaging approaches (Albouy et al., 2013). Cooperative or competitive interactions between the human hippocampus and other brain regions such as the striatum or motor cortex seem to coordinate and synchronize during the acquisition of motor abilities (Albouy et al., 2008; Döhring et al., 2017; Boutin et al., 2018), but the underlying molecular mechanisms and the neural ensembles involved remain unknown. In this line, although initial animal studies with acute hippocampal lesions showed improvements in cued-learning (A.S. Lee et al., 2008) or object recognition memory (Oliveira et al., 2010), they did not support a role of the hippocampus in the regulation of motor skills (Curlik et al., 2013; Fouquet et al., 2013). In contrast, neuroimaging approaches have shown that the hippocampus displays higher rates of micro-structural changes in rotarod-trained mice compared with untrained controls and, accordingly, this brain region is larger in the best performers (Scholz et al., 2015). Furthermore, the accelerating rotarod task induces Fos expression (a marker of neural activation) labeling (Nagai et al., 2017), mTOR and cAMP-dependent protein kinase expression (Bergeron et al., 2014; Chagniel et al., 2014), and neurogenesis (DiFeo et al., 2015) in the hippocampus. These observations indicate that the hippocampus is recruited and likely to play a role in the modulation of motor skills learning.
In the present work, we show major hippocampal transcriptional changes in long-term trained (LTT) mice in the accelerating rotarod task when compared with untrained mice. Transcriptional profiling reveals the importance of changes in hippocampal synaptic genes during motor learning. Accordingly, the hippocampal CA1 progressively stabilizes and decreases its activity along trials. We then used transgenic mice to tag neuronal ensembles in CA1 in a Egr1-dependent fashion and show that depletion of such neural populations improves motor learning whereas their chemogenetic activation has a selective negative impact. Thus, our results reveal the existence of hippocampal neuronal ensembles that tightly modulate the rates of motor skills learning.
Materials and Methods
Animals
For this study we used adult (12-week-old) C57/BL6 males (MGI catalog #5657800, RRID:MGI:5657800) in experiments related with fiber photometry, RNAseq analyses and Egr1 downregulation. For the rest of the experiments, we used the Egr1-CreERT2 mice (Longueville et al., 2021). These mice carry a bacterial artificial chromosome (BAC) including the Egr1 gene in which the coding sequence was replaced by that of CreERT2 fusion protein. Egr1-CreERT2 mice were used as heterozygous in the chemogenetic experiments or they were crossed with R26RCE mice (Gt(ROSA)26Sortm1.1(CAG-EGFP)Fsh/Mmjax, Strain 004077, The Jackson Laboratory), which harbor the R26R CAG-boosted EGFP (RCE) reporter allele with a loxP-flanked STOP cassette upstream of the enhanced green fluorescent protein (EGFP) gene to create the double heterozygous mutant Egr1-CreERT2 x R26RCE mice for the experiments related with characterization of neural populations or depletion of neural ensembles. Genotypes were determined from an ear biopsy as described elsewhere (Martín-Ibáñez et al., 2012). For genotyping of the Cre and EGFP transgenes we used standard PCR assays following The Jackson Laboratory manufacturer's instructions. All mice were housed together in numerical birth order in groups of mixed genotypes (three to five mice per cage). The animals were housed with access to food and water ad libitum in a colony room kept at 19–22°C and 40–60% humidity, under an inverted 12/12 h light/dark cycle (from 8 A.M. to 8 P.M.). All animal procedures were approved by local committees [Universitat de Barcelona, CEEA (133/10); Generalitat de Catalunya (DAAM 5712); and Animal Care Committee of the University of Paris (APAFIS #15638)], in accordance with the European Communities Council Directive (86/609/EU).
Stereotaxic surgery and viral transduction in vivo
Animals were stereotaxically injected with one of the following adeno-associated viruses (AAVs): AAV-flex-taCasp3-TEVp (UNC vector core); AAV-U6-shRNA-Egr1-mCherry (#shAAV-258146, Vector Biolabs), AAV-U6-Scramble-mCherry (#1781, Vector Biolabs), AAV-CAG-FLEX-tdTomato (UNC vector core), and pAAV-hSyn-DIO-hM3D(Gq)-mCherry (RRID:Addgene_44361). Briefly, mice were anaesthetized with ketamine-xylazine (100 and 10 mg/kg, respectively), and bilaterally injected with AAVs (∼2.6 × 109 GS per injection) in the CA1 of the dorsal hippocampus, from the bregma (millimeters); antero-posterior, –2.0; lateral, ±1.5; and dorso-ventral, −1.3. AAV injection was conducted in 2 min. The needle was left in place for 7 min for complete virus diffusion before being slowly pulled out of the tissue. After 2 h of careful monitoring, mice were returned to their home cage for three weeks. All mice subjected to surgery that survived and were healthy without clinical problems (such as head inclination or >15% of body weight loss) were also behaviorally characterized. Once the behavioral characterization was done, half of the brain was used to verify the site of injection by immunofluorescence (see below, Tissue fixation, immunofluorescence, and confocal imaging). Mice that showed no correct viral transduction and location were excluded from the entire study.
Pharmacological treatments
We used single intraperitoneal injections of 4-hydroxytamoxifen (4-HT; Sigma, #H7904) 50 mg/kg or clozapine-N-oxide (CNO; Sigma, #C0832) 3 mg/kg. The 4-HT's vehicle was peanut oil (Sigma, #2144; with a previous dissolution by heating in 100% EtOH) and for CNO was distilled water. 4-HT was always administered 1 h before the behavioral testing and CNO was always administered 30 min before the behavioral testing or 2 h before the mice killing and brain tissue collection.
Fiber photometry
Male C57BL/6 mice were anaesthetized with isoflurane and received 10 mg kg−1 intraperitoneal injection of Buprécare (buprenorphine 0.3 mg) diluted 1:100 in NaCl 9 g l–1 and 10 mg kg−1 ketofen (ketoprofen 100 mg) diluted 1:100 in NaCl 9 g l–1, and placed on a stereotactic frame (Model 940, David Kopf Instruments); 1 µl of virus (AAV9.CamKII.GCaMP6f.WPRE.SV40, titer ≥ 1 × 101³ vg/ml, working dilution 1:10), was injected unilaterally into the CA1 (L = −1.25; AP = −2; V = −1.1–1.2, in mm) at a rate of 0.1 µl min−1. pENN.AAV.CamKII.GCaMP6f.WPRE.SV40 was a gift from James M. Wilson (Addgene viral prep #100834-AAV9).
A chronically implantable cannula (Doric Lenses) composed of a bare optical fiber (400-µm core, 0.48 N.A.) and a fiber ferrule was implanted at the location of the viral injection site. The fiber was fixed onto the skull using dental cement (Super-Bond C&B, Sun Medical). Real time fluorescence emitted from GCaMP6f-expressing neurons was recorded using fiber photometry as described (Berland et al., 2020). Fluorescence was collected using a single optical fiber for both delivery of excitation light streams and collection of emitted fluorescence. The fiber photometry setup used two light-emitting LEDs: 405-nm LED sinusoidally modulated at 330 Hz and a 465-nm LED sinusoidally modulated at 533 Hz (Doric Lenses) merged in a FMC4 MiniCube (Doric Lenses) that combines the two wavelengths excitation light streams and separate them from the emission light. The MiniCube was connected to a Fiberoptic rotary joint (Doric Lenses) connected to the cannula. A RZ5P lock-in digital processor controlled by the Synapse software (Tucker-Davis Technologies; TDT), commanded the voltage signal sent to the emitting LEDs via the LED driver (Doric Lenses). The light power before entering the implanted cannula was measured with a power meter (PM100USB, Thorlabs) before the beginning of each recording session. The irradiance was ∼9 mW/cm2. The fluorescence emitted by GCaMP6f in response to light excitation was collected by a femtowatt photoreceiver module (Doric Lenses) through the same fiber patch cord. The signal was received by the RZ5P processor (TDT). Real time fluorescence because of 405 nm and 465 nm excitations was demodulated online by the Synapse software (TDT). A camera was synchronized with the recording using the Synapse software. Signals were exported to MATLAB R2016b (MathWorks) and analyzed offline. After careful visual examination of all trials, they were clean of artifacts in these time intervals. The timing of events was extracted from the video.
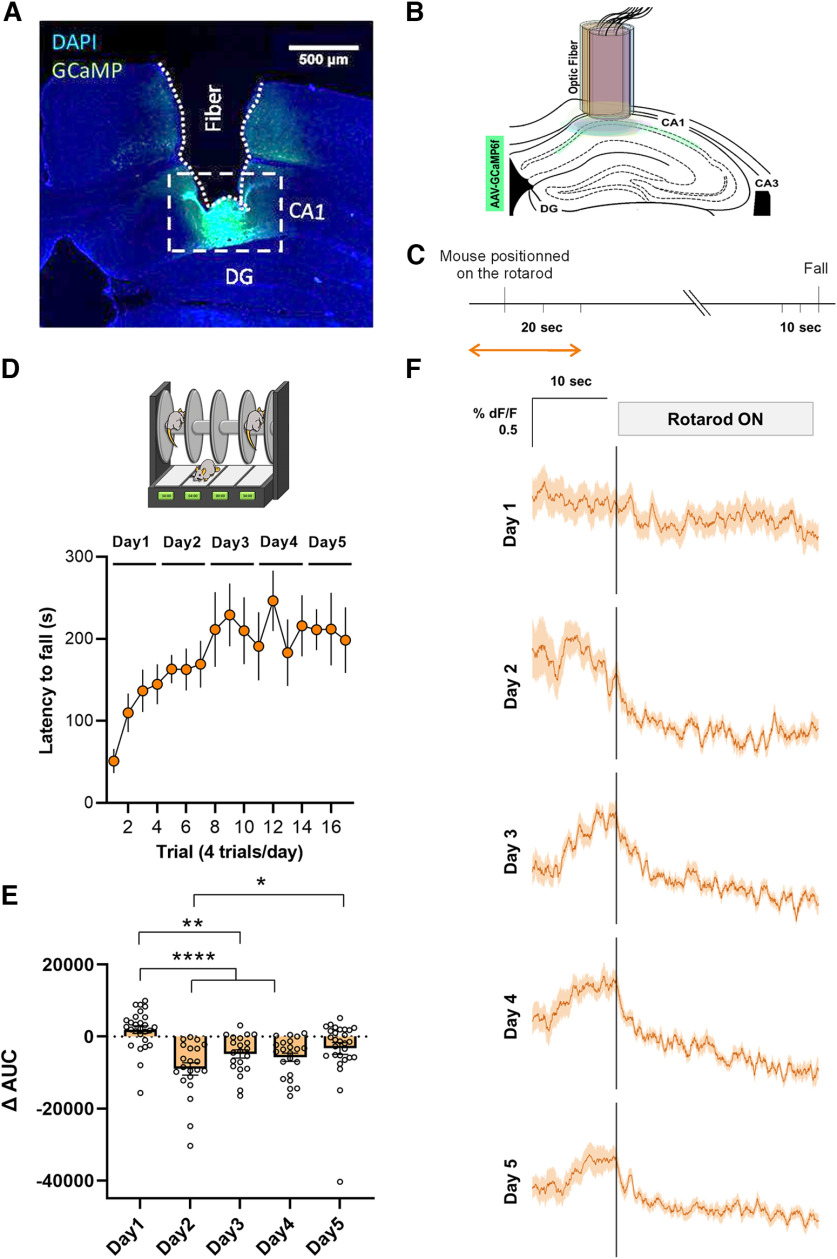
To calculate ΔF/F, a linear least-squares fit was applied to the 405-nm signal to align it to the 465-nm signal, producing a fitted 405-nm signal. This was then used to normalize the 465-nm signal as follows: ΔF/F = (465-nm signal – fitted 405-nm signal)/fitted 405-nm signal (Lerner et al., 2015). For each trial, signal analysis was performed from −10 to +20 s around the moment the mouse is positioned on the rotarod. The percentage of change of the area under the curve (AUC) was calculated between [−10 0] and [0 10] s relative to the moment when animals were placed in the from rotarod.
Accelerating rotarod
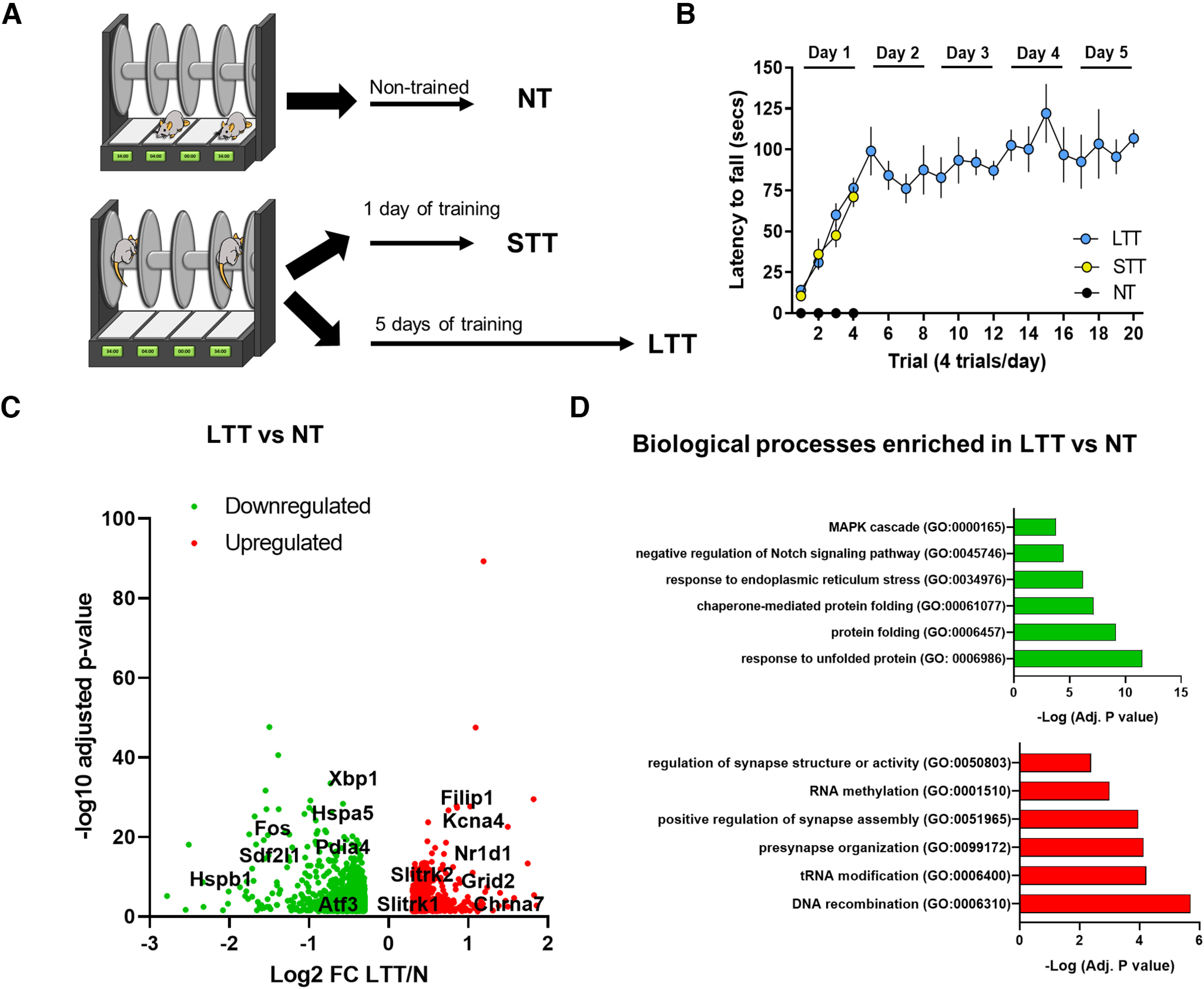
As previously described (Giralt et al., 2013), animals were placed on a motorized rod (30-mm diameter, Panlab). The rotation speed was gradually increased from 4 to 40 r.p.m. over the course of 5 min. The fall latency time was recorded when the animal was unable to keep up with the increasing speed and fell. Rotarod training/testing was performed four times per day with 30 min as intertrial time interval. The results show the average of fall latencies per trial during the 5 d of training. In experiments depicted in Figures 1, 3 and 5, we used three groups, a nontrained (NT) mice but exposed to the rotarod for 1 d, the short-term trained mice (STT) exposed to only 1 d of training in the rotarod; and the LTT mice exposed to 5 d of training in the rotarod.
Figure 1.
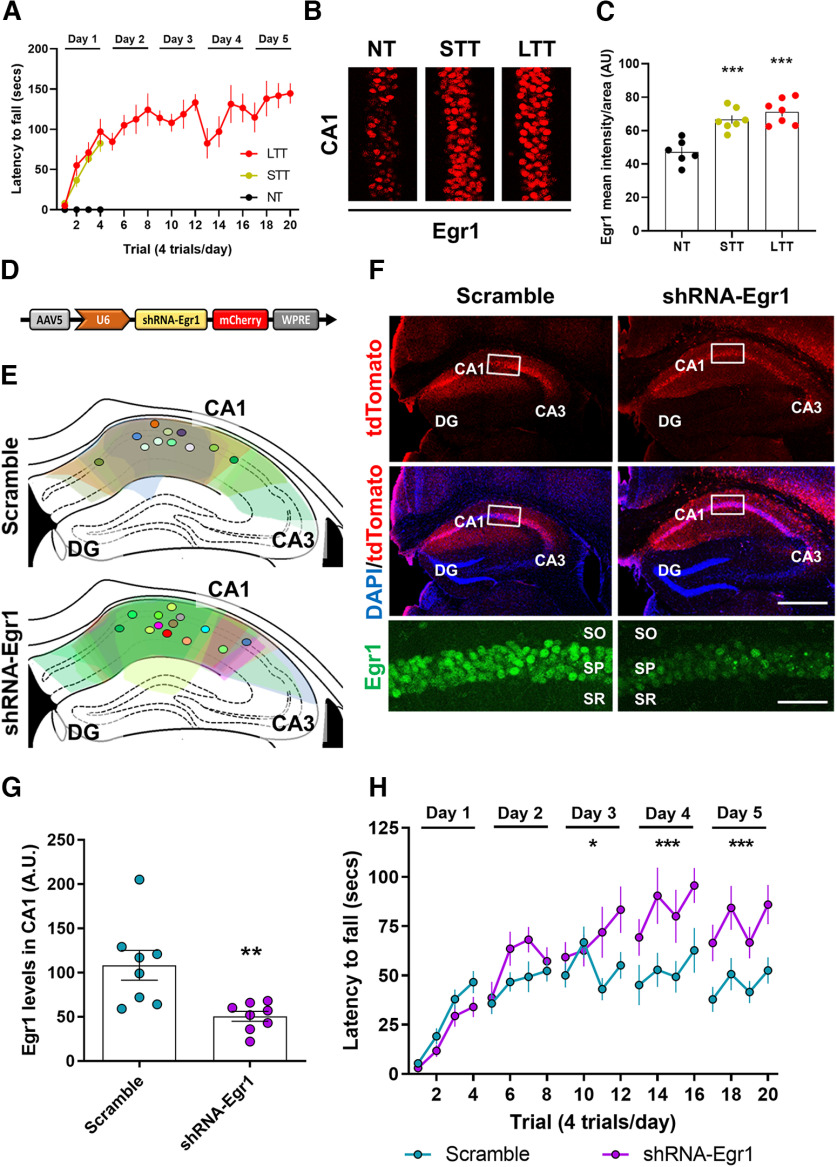
Hippocampal gene expression profile following the accelerating rotarod task. A, Schematic of experimental design. WT mice were trained on a rotarod. Three groups were evaluated (n = 5 per group): NT, STT, and LTT mice. The transcriptome of the five performers in each group was analyzed by RNA sequencing 24 h after the last trial. B, Latency to fall in the accelerating rotarod task in NT, STT, and LTT mice. Data are mean ± SEM. C, Volcano plots showing significant mRNA differential expression genes between LTT and NT samples (red: upregulated genes; green: downregulated genes; Adj p-value < 0.05; log2FC > 0.3 or < −0.3, n = 5 per group). The name of representative mRNAs is indicated. For full data, see Extended Data Table 1-1. D, Gene ontology analysis for transcriptomic enrichment of upregulated genes (upper panel), and downregulated genes in LTT (lower panel), by using Metascape analysis including all significant DEGs (FDR < 0.05) in LTT.
Figure 3.
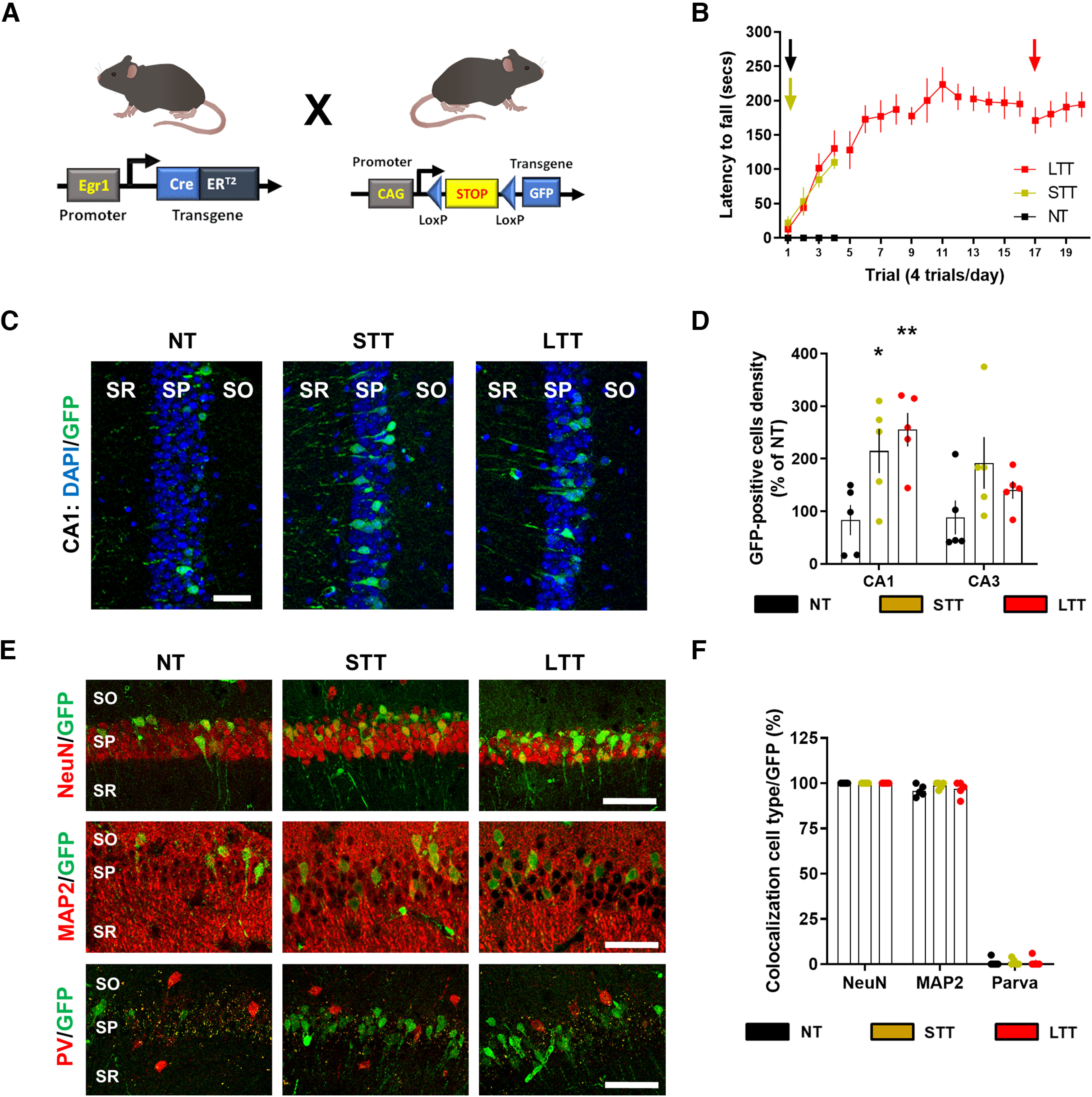
Characterization of Egr1-dependent activated neural cells during different phases of the rotarod task training. A, Schematic representation of the mutant mice used. B, Mice were subjected to three different conditions in an accelerating rotarod task (as in Fig. 1A): NT, STT, and LTT. All three groups of mice (n = 5 mice per group) received an injection of 4-HT 1 h before the rotarod training session (arrows) on the last day of training. C, Representative images of Egr1-dependent activation of neural cells (GFP-positive, green) co-stained with DAPI (blue) in dorsal CA1. D, Quantification of GFP-positive neural cells density per area in NT, STT, and LTT groups of mice. Scatter plot with mean ± SEM. Two-way ANOVA identified general significant changes between groups (F(2,88) = 16.9; p < 0.0001). Dunnett's post hoc analysis (*p < 0.05, **p < 0.01) compared with the NT group. E, Representative images of Egr1-dependent activation of neural cells (GFP-positive, green) co-stained with NeuN or MAP2 or parvalbumin (all in red) in CA1. F, Quantification of the percentage of GFP-positive neural cells that co-localizes with various neural markers (in red) in the CA1. PV: parvalbumin, CA1–CA3: cornu ammonis. SO: stratum oriens, SP: stratum pyramidale, SR: stratum radiatum. Scale bar: 40 µm (C, E).
Figure 5.
Effects of Egr1 downregulation in CA1 on accelerating rotarod training. A, WT mice were subjected to three different conditions in an accelerating rotarod task as in Figure 1 NT, STT, and LTT (4 trials per day). N = 6–7 per group. B, Egr1 staining (in red) for all three groups 2 h after the last trial with the rotarod. C, Quantification of Egr1 optical density (arbitrary units) in the pyramidal cell layer of CA1. Data are scatter plots (one point per mouse, average from two slices per mouse) and mean ± SEM. One-way ANOVA (F(2,17) = 20,12, p = 0.0001), Dunnett's post hoc: ***p < 0.001 compared with NT group. D, Schematic illustration of the transduced AAV (AAV-U6-shRNA-Egr1-mCherry) in a second and new cohort of WT mice. E, Schematic location and distribution of the injection sites in dorsal CA1 for all mice used in this experiment (only left hemisphere is shown). F, Representative images of hippocampi transduced with control AAV (AAV-U6-Scramble-mCherry, Scramble, left) or with the experimental AAV (AAV-U6-shRNA-Egr1-mCherry, shRNA-Egr1, right). Triple staining showing all cells (DAPI, blue), transduced cells (mCherry, red), and Egr1-positive cells (inset, green) for each group of mice. Scale bars: 300 µm, 60 µm (inset). G, Quantification of Egr1 optical density (arbitrary units) in the pyramidal cell layer of CA1. Data are scatter plots (one point per mouse, average from two slices per mouse) and mean ± SEM. Mann–Whitney t test, sum of ranks A, 95, B, 41, Mann–Whitney U, 5, **p = 0.003. N = 8 mice per group. H, Three weeks after viral transduction mice were subjected to the accelerating rotarod task for 5 d (4 trials per day). N = 11 per group. Data are mean ± SEM and analyzed by two-way ANOVA. A significant difference was detected between groups on day 3, F(1,80) = 6.30, p = 0.0141), day 4, F(1,80) = 16.91, p < 0.0001), and day 5, F(1,80) = 26.03, p < 0.0001. DG: dentate gyrus, CA1: cornu ammonis 1, CA3: cornu ammonis 3, SO: stratum oriens, SP: stratum pyramidale, SR: stratum radiatum.
Open field and novel object location (NOL) test
For the NOL test, an open-top arena (45 × 45 × 45 cm) with visual cues surrounding the apparatus was used. Mice were first habituated to the arena (1 d, 30 min). We considered this first exposition to the open arena as an open field paradigm. We monitored total traveled distance, time spent in the center of the arena and parallel index as measures of locomotor activity, anxiogenic behavior and spatial navigation strategies, respectively. On day 2, two identical objects (A1 and A2) were placed in the arena and explored for 10 min. Twenty-four hours later (day 3), one object was moved from its original location to the diagonally opposite corner and mice were allowed to explore the arena for 5 min. The object preference was measured as the time exploring each object × 100/time exploring both objects. Behavioral data were processed and analyzed using the Smart Junior software (Panlab).
RNA sequencing analysis
RNA extraction and quality control
Hippocampal were homogenized, and RNA extracted using RNeasy Lipid Tissue Mini kit (QIAGEN) according to manufacturer's recommendations. RNA purity and quantity were determined with a UV/V spectrophotometer (Nanodrop 1000), while RNA integrity was assessed with a 2100 Bioanalyzer (Agilent Technologies Inc.), according to manufacturers' protocols. The average RIN value for our samples was 9.5, and the RIN cutoff for sample inclusion was 8.0.
RNA sequencing and differential gene expression analysis
Libraries were prepared using the TruSeq Stranded mRNA Sample Prep kit v2 (ref. RS-122-2101/2) according to the manufacturer's protocol. Briefly, 500 ng of total RNA were used for poly(A)-mRNA selection using streptavidin-coated magnetic beads and were subsequently fragmented to ∼300 bp. cDNA was synthesized using reverse transcriptase (SuperScript II, ref. 18064-014, Invitrogen) and random primers. The second strand of the cDNA incorporated dUTP in place of dTTP. Double-stranded DNA was further used for library preparation. dsDNA was subjected to A-tailing and ligation of the barcoded Truseq adapters. Library amplification was performed by PCR using the primer cocktail supplied in the kit. All purification steps were performed using AMPure XP beads. Final libraries were analyzed using Fragment Analyzer to estimate the quantity and check size distribution and were then quantified by qPCR using the KAPA Library Quantification kit (ref. KK4835, KapaBiosystems) before amplification with Illumina's cBot. Sequencing was done using the HiSeq2500 equipment (Illumina), Single Read, 50 bp, using the v4 chemistry.
The quality of the sequencing data was checked using the FastQC software v0.11.5. Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data (available online at http://www.bioinformatics.babraham.ac.uk/projects/fastqc). An estimation of ribosomal RNA in the raw data were obtained using riboPicker version 0.4.3 (Schmieder et al., 2012). Reads were aligned to the GENCODE version of the Mus musculus genome, release M20 (GRMm38/mm10 assembly) using the STAR mapper (version 2.5.3a; Dobin et al., 2013). The raw read counts per gene was also obtained using STAR (–quantMode TranscriptomeSAM GeneCounts option) and the GENCODE release M20 annotation (ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M20/gencode.vM20.annotation.gtf.gz). The R/Bioconductor package DESeq2 version 1.22.2 (R version 3.5.0) was used to assess the differentially expressed genes (DEGs) between experimental groups, using the Wald statistical test and the False Discovery Rate for the p-value correction. Before the differential expression analysis, genes with the sum of raw counts across all samples below 10 were discarded, the library sizes were normalized using the default DeSeq2 method, and the read counts were log2 transformed. To exclude false positive genes, genes with low expression levels (baseMean <10) were excluded from the list of DEGs. Sequencing data has been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number (accession number pending).
Gene functional enrichment analysis
Metascape pathway enrichment analysis was performed to explore the functional roles of DEGs in the paired comparison of LTT, STT, and NT mice. To discover further possible connections between DEGs and transcription factors we used EnrichR. Functional annotations for modules of interest were generated using the web server SynGO (https://www.syngoportal.org/), which provides an expert-curated resource for synapse function and gene enrichment analysis (Koopmans et al., 2019).
Quantitative RT-PCR
The cDNA synthesis was performed at 37°C for 15 min and a final step at 85°C for 5 s in a final volume of 20 μl according to the manufacturer's instructions. The cDNA was then analyzed by quantitative RT-PCR using the following PrimeTime qPCR Assays (Integrated DNA Technologies): Arc (Mm.PT.58.5865502.g), Mdga1 (Mm.PT.58.32475502), Egr-1 (Mm.PT.58.29064929), Mdga2 (Mm.PT.58.10917976), Tnik1 (Mm.PT.58.32835672), Gsgl1 (Mm.PT.58.28965348), and Actinβ (Mm.PT.39a.22214843.g). Quantitative PCR was performed in 12 μl of final volume on 96-well plates using the Premix Ex Taq (Probe qPCR). Reactions included Segment 1: 1 cycle of 30 s at 95°C and Segment 2: 40 cycles of 5 s at 95°C and 20 s at 60°C. All quantitative PCR assays were performed in duplicate. To provide negative controls and exclude contamination by genomic DNA, the PrimeScript RTEnzyme was omitted in the cDNA synthesis step. To analyze the relative changes in gene expression the 2(–δδ C(T)) method was used.
Tissue fixation, immunofluorescence, and confocal imaging
Animals were deeply anaesthetized and subsequently intracardially perfused with 4% (weight/vol) paraformaldehyde in 0.1 m phosphate buffer. The brains were dissected out and kept 48 h in 4% paraformaldehyde. Sagittal sections (40 μm) were obtained using a vibratome (Leica VT1000). For immunofluorescence, after blocking/permeabilization [1 h in PBS containing 3 ml/l Triton X-100 and 10 g/l bovine serum albumin (BSA)], sections were incubated overnight with specific antibodies against MAP2 (1:500; Sigma-Aldrich catalog #M1406, RRID:AB_477171), NeuN (1:500, Millipore catalog #MAB377, RRID:AB_2298772), parvalbumin (1:1000, Swant catalog #PV27, RRID:AB_2631173), GFP FITC-conjugated (1:500, Abcam catalog #ab6662, RRID:AB_305635), Egr1 (1:1000, Cell Signaling Technology catalog #4154, RRID:AB_2097035), and cFos (1:150, Santa Cruz Biotechnology catalog #sc-52, RRID:AB_2106783). After incubation (2 h) with appropriate fluorescent secondary antibodies (Cy3-coupled or Cy2-coupled fluorescent secondary antibodies, 1:200; Jackson ImmunoResearch catalog #715-165-150, RRID:AB_2340813 and catalog #715-545-150, RRID:AB_2340846, respectively), nuclei were stained (10 min) with 4′,6-diamidino-2-phenylindole (DAPI; catalog #D9542, Sigma-Aldrich). The sections were mounted onto gelatinized slides and cover-slipped with Mowiol.
Image analysis and stereological counting
Images (at 1024 × 1024 pixel resolution) in a mosaic format were acquired with a Leica Confocal SP5 with a × 40 oil-immersion or x20 normal objectives and standard (1 Airy disk) pinhole (1 AU) and frame averaging (three frames per z step) were held constant throughout the study. For pseudo-stereological counting, we analyzed three sagittal sections, from 1.4 to 2.0 mm relative to bregma, spaced 300 μm apart. The areas of analysis were, dorsal CA1 and dorsal CA3. Unbiased blind counting of GFP-positive or MAP2-positive or parvalbumin-positive or NeuN-positive neural cells relative to genotype and condition was performed and normalized to the area of counting.
Statistics
Analyses were done using Prism version 8.0.2 for Windows (GraphPad Software). Data are expressed as mean ± SEM. Normal distribution was tested with the D'Agostino and Pearson omnibus test. If no difference from normality was detected, statistical analysis was performed using two-tailed Student's t test or ANOVA and Tukey's or Dunnett's post hoc tests. If distribution was not normal, nonparametric two-tailed Mann–Whitney test was used. The p < 0.05 was considered as significant.
Results
Differential regulation of gene expression in hippocampus during motor learning
Although previous studies implicated individual genes or genetic pathways in learning and memory in the hippocampus, they did not investigate gene expression patterns during motor skill learning. Therefore, to analyze potential changes in hippocampal gene expression during the acquisition of a motor skill we used a transcriptome-scale screening. We subjected two groups of mice to the accelerating rotarod task (Fig. 1A,B). The first group (STT) was trained just 1 d in the task to assess the initial phase of motor learning, and a second group was trained for 5 d to acquire well-learned motor skill (LTT). Both groups were compared with mice that were not trained but were placed in the apparatus as a control (NT). Twenty-four hours after the last day of training or exposure to the rotarod, the dissected dorsal hippocampus of the mice from the three groups was subjected to deep sequencing analysis (RNAseq).
Supplementary. Download Table 1-1, XLSX file (84.6KB, xlsx) .
We assessed the overall transcriptional changes in response to training comparing STT versus NT and LTT versus NT (Adj p-value < 0.05; log2fold change > 0.3 or < −0.3; Extended Data Table 1-1). In the comparison of the transcriptional profile between STT and NT we found only two genes that were significantly downregulated in STT compared with NT, Gsgl1 [an AMPA receptor (AMPAR) auxiliary subunit] and Dusp1, an inactivator of mitogen-activated protein kinase (MAPK). In contrast, the comparison between LTT and NT revealed that a large number of genes were downregulated and upregulated (797 and 540, respectively; Fig. 1C). To assess the functional profile of DEGs in the LTT, we used enrichment analysis by mapping to Metascape database. We found that downregulated DEGs were significantly enriched in processes related to response to unfolded protein, NOTCH and MAPK pathways (Fig. 1D), whereas upregulated DEGs were DNA recombination, RNA modifications and synaptic organization, structure and activity, particularly in presynaptic processes (Fig. 1D).
Ca2+ activity decreases in the CA1 pyramidal cells at the end of the motor learning task
Hippocampal cell populations are activated during simple locomotion (Bocchio et al., 2020) and during a motor learning task (Albouy et al., 2013). To better understand the hippocampal dynamics during the acquisition of a motor skill, we transduced dorsal CA1 pyramidal cells with AAV-GCaMP6f and we placed a fiber-optic probe to monitor pyramidal neurons activity in CA1 during the accelerating rotarod task (Fig. 2A–C). First, we observed that mice performed well and progressively learned the accelerating rotarod task (Fig. 2D). Interestingly, we observed that during the initial trials of the accelerating rotarod task the Ca2+-dependent signal dynamics were irregular and heterogenous whereas at the last trials of the task the signal progressively decreased and stabilized (Fig. 2E,F). Altogether, these results suggest that the pyramidal cells of the CA1 reduce their activity associated with a progressive consolidation of motor learning.
Figure 2.
Accelerating rotarod task induced-functional plasticity in the hippocampal CA1 neurons. A, Representative viral expression of AAV-GCaMP6f and (B) placement of the fiber-optic probe in CA1 for each mouse depicted in different colors accordingly. DG, dentate gyrus. C, Experimental design of fiber photometry. Analysis was performed during the time windows indicated by the orange arrow. D, Mice implanted with the fiber-optic probe were subjected to the accelerating rotarod task. E, Quantification of the change in the AUC between [−10, 0 s] and [0, 10 s] relative to the placement of the animal in the rotarod. F, Averaged traces of GCaMP6f signal expressed as ΔF/F% for 10 s before and 20 s after the mouse was placed on the rotarod. All the trials (n = 7 mice) were averaged on each training sessions (days 1–5).
Egr1-dependent neuronal subpopulations are permanently activated in CA1 during motor learning
We showed that acquisition and consolidation of motor learning is accompanied by changes in gene expression as well as in Ca2+ dynamics. To identify which neural cells are activated during different phases of motor learning, we used a novel mouse line that allows a permanent tagging of neurons activated by experience: the Egr1-CreERT2 transgenic mice (Longueville et al., 2021). We chose Egr1 as promoter because it is a crucial IEG significantly induced in the hippocampus as well as in the cortico-striatal network during initial learning of a motor skill (Hernandez et al., 2006), and it is tightly linked to physiological firing activity of neurons (Vaccarino et al., 1992; Wang et al., 1994). Concretely, these mice express the Cre recombinase fused to modified estrogen receptor (ERT2) under the control of the Egr1 promoter. Egr1 drives the expression of the CreERT2 recombinase that is only active in the presence of tamoxifen metabolite, 4-HT. Egr1-CreERT2 mice were crossed with R26RCE mice, a reporter line in which EGFP expression requires recombination by Cre (Fig. 3A).
In double transgenic Egr1-CreERT2 x R26RCE mice, cells in which Egr1 is induced by neuronal activity and in the presence of 4-HT become permanently labeled with EGFP. We then subjected Egr1-CreERT2 x R26RCE mice to the accelerating rotarod task using the same three groups (NT, STT, and LTT; Fig. 3B). Each group received a single injection of 4-HT 1 h before the last session of training in the task (Fig. 3B). To ensure detectable recombination, 3 d (72 h) after the 4-HT injection, their brains were processed. We then counted the density of GFP-positive neural cells in the CA1 and CA3 of the dorsal hippocampus (Fig. 3C,D). The density of GFP-positive neural cells was increased in CA1 in both, STT and LTT groups compared with NT group whereas they remained unchanged in CA3 (Fig. 3C,D). These results indicated a more sustained Egr1 activation in a specific group of neuronal cells in CA1 compared with CA3. We then characterized the GFP-positive neural cells in CA1. Co-localization studies revealed that most GFP-positive cells in CA1 were NeuN-positive and MAP2-positive (and parvalbumin-negative; Fig. 3E,F), indicating they were pyramidal neurons. Finally, we evaluated whether endogenous Egr1 levels are induced in the dorsal CA1 after STT and LTT. This result was in contrast with Egr1 mRNA levels in our RNAseq. Specifically, 24 h after the last trial with the rotating rod, Egr1 expression was reduced only in LTT mice (Gene: Egr1, log2 FC: −1,30, Adj p-value: 6,50E-22; Extended Data Table 1-1). Altogether suggest a complex and time-dependent regulation of Egr1 on motor learning in the rotarod.
Egr1 downstream targets in the hippocampus during motor performance
To evaluate the potential Egr1-dependent gene expression, we analyzed in our transcriptome data the DEGs with Egr1-binding motifs in their promoter region (Extended Data Table 4-1). We identified 1 DEG and 94 DEGs (7% of DEGs in LTT) in STT and LTT, respectively, which can be regulated by Egr1. In particular, Gsg1l was a common DEG in STT and LTT compared with NT. Fold-change heatmap (Fig. 4A) shows the genes downregulated and upregulated in LTT with no significant changes observed in STT (Fig. 4A). Interestingly, using the SynGo platform, we identified in this gene dataset, 11 genes mapping with processes in synapse assembly, organization and function, and regulation of postsynaptic membrane neurotransmitter receptor levels (Fig. 4A). To further validate these results on a larger number of samples (n = 8 for the NT group, n = 7 for the STT group and n = 7 for the LTT group), we performed qPCR analysis on genes Mdga1, Mdga2, Gsgl1, Arc, and Tnik (Fig. 4B–F). We confirmed that the expression of Mdga1, Arc and Gsgl1 is reduced in LTT compared with NT group (Fig. 4B–D). Meanwhile, the expression levels of the other two selected genes (Mdga2 and Tnik) showed a trend in increase but these differences were not statistically significant (Fig. 4E,F) which might be explained by the difference in sensitivity of the methods being used. Taken together these data indicate that during the progressive learning of a motor skill, long-term transcriptional changes associated with Egr1 activity occur in the hippocampus associated with its acquisition and/or maintenance.
Figure 4.
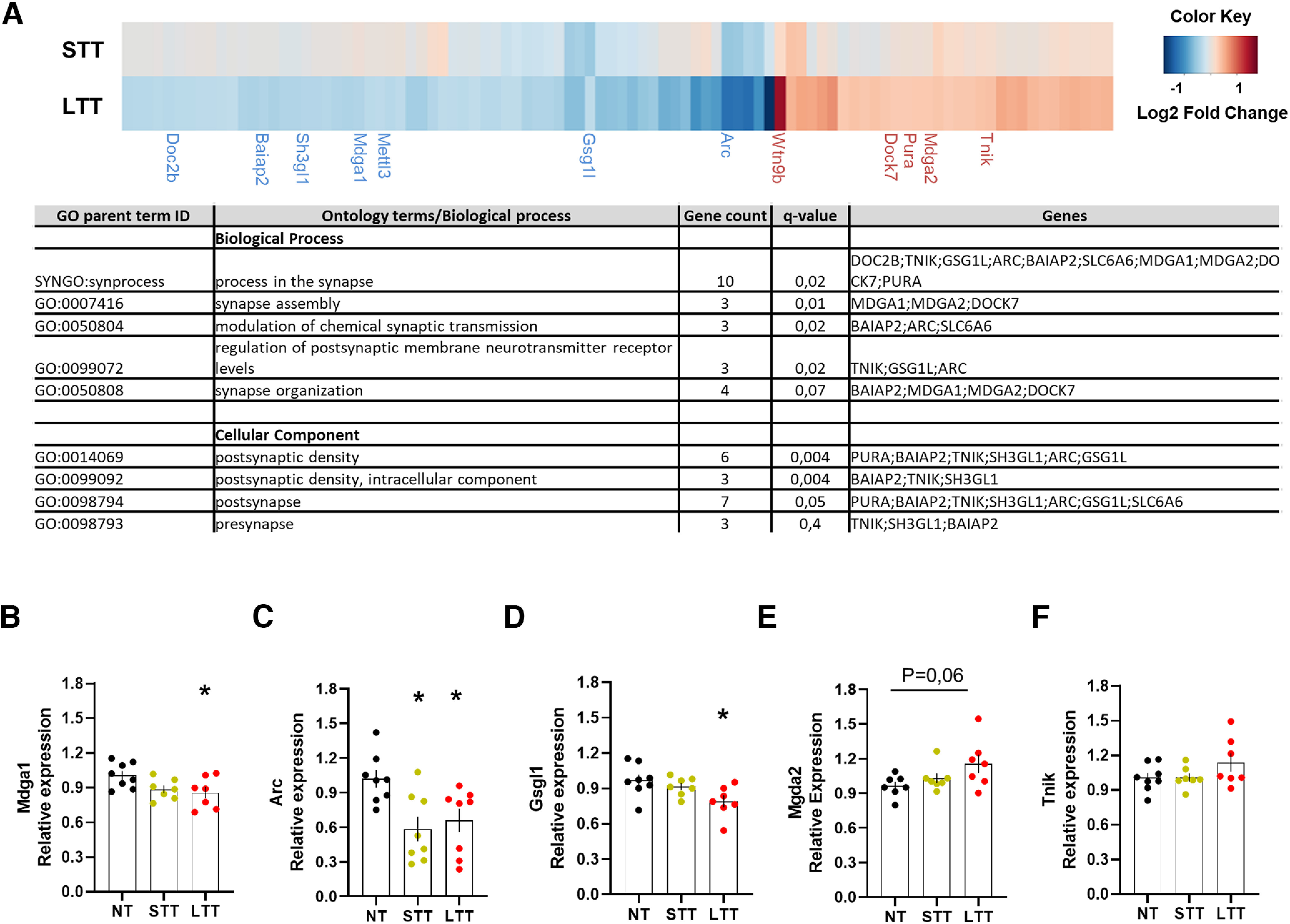
Transcriptional changes associated with Egr1 promoter activity during the rotarod task training. A, Heatmap (upper panel) showing log2FC of DEGs with putative Egr1 binding sites in LTT and STT groups relative to the NT group. The results were considered statistically significant at Adj p-value < 0.05 and log2FC > 0.3 or < −0.3 in LTT. DEGs in STT are not statistically significant except for Gsgl1. Only synaptic genes are named. A table (lower panel) is also depicted to show the name of all the putative Egr1 binding sites in the LTT group. For full data, see Extended Data Table 4-1. Validation of RNA-seq data by quantitative real-time PCR (qRT-PCR) in NT, STT, and LTT groups of mice. Results from qRT-PCR performed on a few select genes: B, Mdga1; C, Arc; D, Gsgl1; E, Mdga2; F, Tnik, enriched in synapse processes. The error bars represent the range of the fold change values and data are analyzed by one-way ANOVA. A significant difference was detected between groups for Mdga1 (F(2,19) = 3762; p < 0.05), Arc (F(2,21) = 5957; p < 0.05), and Gsgl1 (F(2,19) = 4323; p < 0.05). Tukey's post hoc analysis indicated the significant differences (p < 0.05) compared with the NT group.
Supplementary. Download Table 4-1, XLSX file (24.3KB, xlsx) .
Egr1 knock-down in CA1 potentiates motor performance
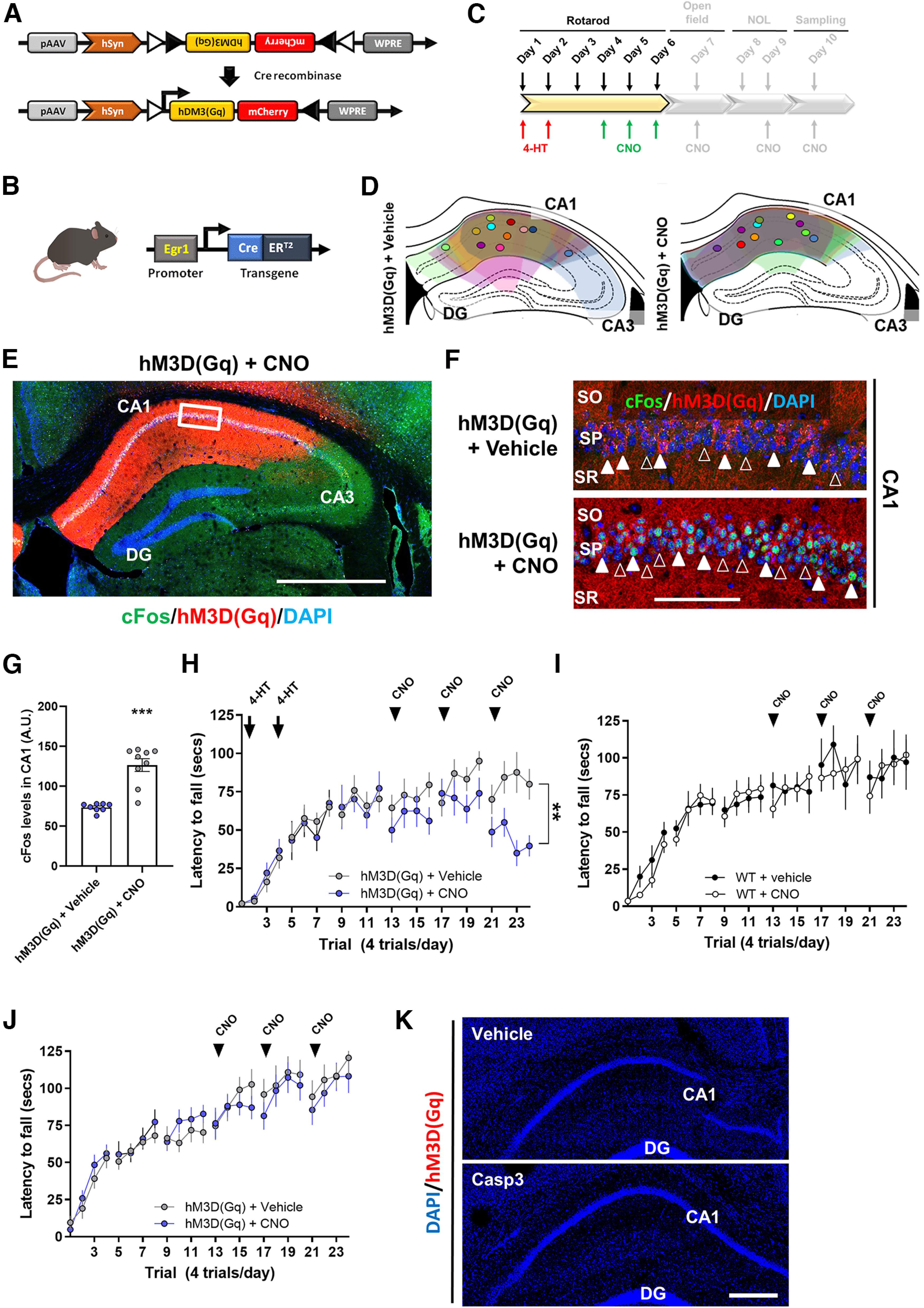
To test whether Egr1 levels in the dorsal CA1 control the acquisition and/or maintenance of the motor skills required for the accelerating rotarod task, we first evaluated the endogenous levels of Egr1 during the accelerating rotarod task. We observed that 2 h after the training with the rotarod, Egr1 expression was increased in the pyramidal cell layer of the CA1 in both, STT and LTT mice compared with NT mice (Fig. 5A–C). This is in agreement with our ensembles' experiment (Fig. 3) which suggests and early upregulation of Egr1 just after rotarod training, but it is in contrast with the RNAseq 24 h after the rotarod (Extended Data Table 1-1) which shows an Egr1 mRNA levels reduction 24 h later only in the LTT group. Thus, to prove the relevance of the acute Egr1 increase in the CA1 during motor learning (Figs. 3, 5A–C), we transduced the CA1 of wild-type (WT) mice with an AAV expressing a shRNA against the Egr1 transcript (shRNA-Egr1 group) or a control shRNA (scramble group; Fig. 5D,E). After three weeks of viral transduction, we observed a significant reduction of Egr1 immuno-reactivity in the pyramidal cells of the dorsal CA1 in shRNA-Egr1 mice compared with the scramble group (Fig. 5F,G). To test the effects of downregulating Egr1 in CA1 we subjected the shRNA-Egr1 and scramble groups of mice to the accelerating rotarod task (Fig. 5H). Scramble mice progressively learned the task and reached a plateau of performance from the second day of training on, whereas, in contrast, the shRNA-Egr1 mice displayed, from day 3 of training, significant increased latencies to fall from the rotarod compared with the scramble mice (Fig. 5H). These results suggest that Egr1 downregulation in the pyramidal neurons of the CA1 improves the accelerating rotarod performance.
Depletion of CA1 Egr1-dependent neuronal subpopulations enhances motor performance
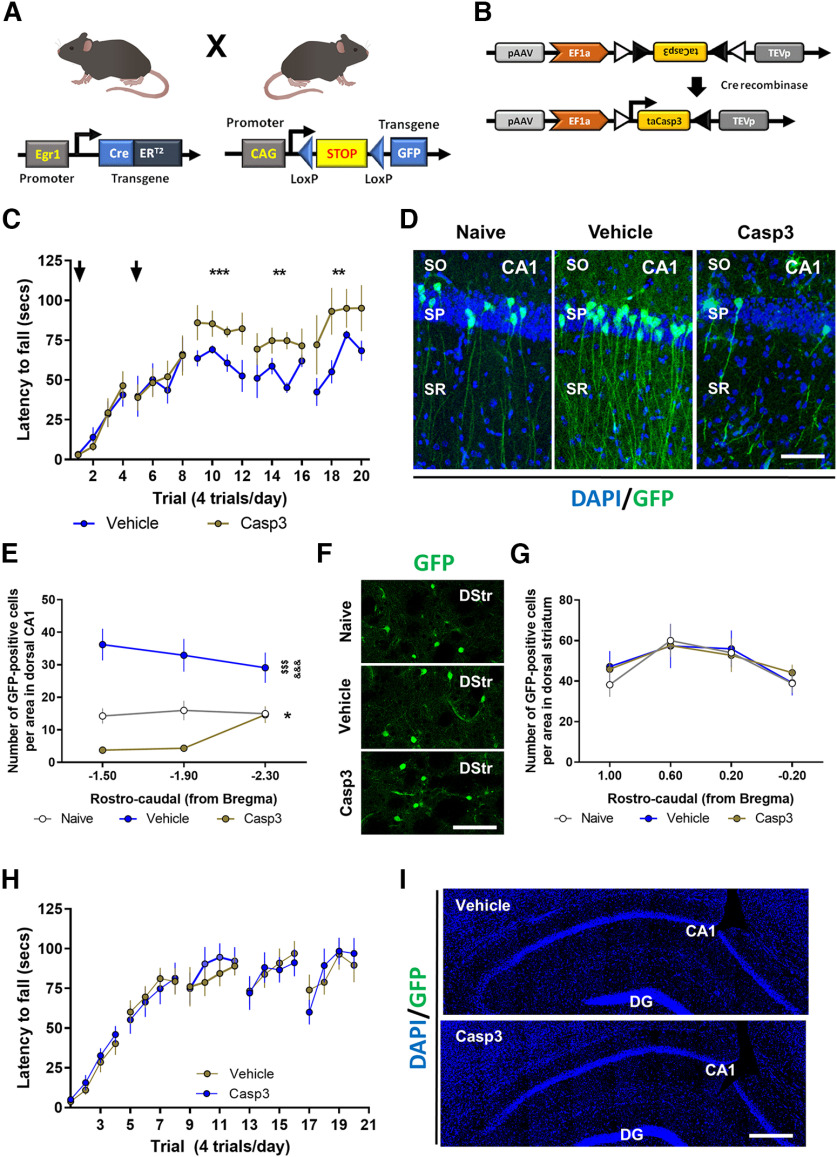
We observed the activation of Egr1-dependent neuronal subpopulations in CA1 during the acquisition of motor skills in the accelerating rotarod task and we also showed that a global downregulation of Egr1 in the dorsal CA1 enhances the performance in this task. We therefore tested the consequences of depleting these CA1 Egr1-dependent activated neuronal subpopulations on motor learning to evaluate their contribution. We used the double mutant Egr1-CreERT2 x R26RCE mice (Fig. 6A). These mice were transduced bilaterally in the dorsal CA1 with vehicle or AAV-flex-taCasp3-TEVp (Fig. 6B). Three weeks later, all mice were subjected to the accelerating rotarod task (Fig. 6C) and received an intraperitoneal injection of 4-HT 1 h before the training session. This injection was administered at days 1 and 2 of the task. With this design, specific Egr1-dependent CreERT2 induction in activated CA1 pyramidal cells of the CA1 would induce Caspase-3 expression in the presence of 4-HT resulting in cell death of the hippocampal neuronal cells activated in a Egr1-dependent fashion. Thus, mice transduced with AAV-flex-taCasp3-TEVp (Casp3 group) in CA1 displayed higher latencies to fall on days 3, 4, and 5 compared with control mice (Vehicle group; Fig. 6C). Brains from these mice were examined to verify the depletion of the Egr1-dependent neuronal ensembles (Fig. 6D,E). As expected, mice infused with vehicle in CA1 showed an increase in GFP-positive cells in the pyramidal layer of CA1 after rotarod training when compared with NT mice. In contrast, the density of GFP-positive cells in the dorsal CA1 was dramatically reduced in rotarod-trained mice that were transduced with Casp3 (Fig. 6D,E). This latter result confirmed a depletion of Egr1-dependent activated neuronal subpopulations.
Figure 6.
Depletion of the CA1 Egr1-dependent activated neurons during the rotarod task. A, Schematic representation of the double mutant mice used in this experiment. B, Schematic representation of the AAV-flex-taCasp3-TEVp vector. Mice were bilaterally injected in CA1 with AAV-flex-taCasp3-TEVp (Casp3, n = 8) or vehicle (Vehicle, n = 7). A group of NT mice (Naive, n = 5) was also used. C, All three groups received a single injection 4-HT 1 h before the rotarod training (arrows) on days 1 and 2 of training. Data are mean ± SEM, analyzed by two-way ANOVA. A significant difference was detected between groups on day 3, F(1,52) = 15.78, p = 0.0002; day 4, F(1,52) = 9.9588, p = 0.0027; and day 5, F(1,52) = 10.05, p = 0.0025. D, Representative images of Egr1-dependent activation of neural cells (GFP-positive, green) co-stained with DAPI (blue) in the dorsal CA1 of the hippocampus in the three groups. Scale bar: 60 µm. E, Quantification of hippocampal GFP-positive neural cells density per area in the three groups. Data are mean ± SEM. Two-way ANOVA identified general significant changes between groups, F(2,51) = 49.27, p < 0.0001. Tukey's post hoc analysis indicated that both, Naive (p < 0.05) and Casp3 (p < 0.001) showed significantly different GFP-positive cell density compared with Vehicle mice. *: Naive versus Vehicle groups; $: Casp3 versus Naive; &: Casp3 versus Vehicle. F, Representative images of Egr1-dependent activation of neural cells (GFP-positive, green) in the dorsal striatum (DStr) from the three groups. Scale bar: 120 µm. G, Quantification of striatal GFP-positive neural cells density per area in the three groups. H, A new cohort of double mutant mice as in A, were subjected to the same experimental design as in B, C but without 4-HT administration to rule out potential leakiness of the system. Data are mean ± SEM, analyzed by two-way ANOVA (no significant differences were detected between groups). I, Postmortem hippocampal examination in mice from H shows no recombination because of the absence of 4-HT administration. SO: stratum oriens, SP: stratum pyramidale, SR: stratum radiatum.
We also evaluated whether the improvement of Casp3 mice in the rotarod task could be accompanied by a higher activation of neuronal ensembles in the striatum as a compensatory consequence/mechanism of the depletion of Egr1-dependent activated neuronal subpopulations. To do so, in the same mice, we analyzed the density of GFP-positive cells in the dorsal striatum (Fig. 6F,G). There was no change in the number of Egr1-dependent activated neurons in the dorsal striatum (Fig. 6F,G). Finally, to rule out potential leakiness of the Egr1-CreERT2 x R26RCE mice we repeated the entire experiment but without 4-HT administration. No changes were observed in the rotarod performance in any group as well as no recombination was detected in brains from Egr1-CreERT2 x R26RCE mice treated with vehicle or AAV-flex-taCasp3-TEVp (Fig. 6H,I). Taken together, our results reinforce the idea of an important role of CA1 Egr1-activated pyramidal cells in the modulation of motor skills during an accelerating rotarod task.
Chemogenetic activation of CA1 Egr1-dependent activated neuronal subpopulations impairs motor performance
To further characterize the role of this Egr1-dependent neuronal ensemble in CA1 induced by motor learning, we used an opposite strategy, aiming to activate it using designer receptors exclusively activated by designer drugs (DREADDs) technology. We transduced the dorsal CA1 of Egr1-CreERT2 mice with an AAV expressing the activator DREADD hM3D(Gq) using a FLEX switch vector (Fig. 7A,B). Three weeks after AAV injection, the mice were first subjected to the accelerating rotarod task (Fig. 7C). On days 1 and 2 of rotarod training, all mice received an injection of 4-HT to induce Cre-mediated recombination in the Egr1-expressing neuronal subpopulations. On the last 3 d of the rotarod task these transduced Egr1-dependent cells were activated using CNO. The same mice were also tested in the open field (day 7) and NOL (days 8 and 9, see below). At the end of the study, we tested the efficacy of hM3D(Gq) receptor stimulation by measuring cFos induction in mice treated with CNO or vehicle 2 h before killing and histologic analysis (Fig. 7C). Postmortem histology indicated that CA1 was well targeted with the vector expressing the hM3D(Gq) receptor (Fig. 7D,E). Moreover, CNO induced a robust upregulation of cFos immunoreactivity in the transduced pyramidal cells in CA1 as compared with nontransduced cells or transduced cells from mice treated with vehicle (Fig. 7F,G).
Figure 7.
Effects of chemogenetic activation of the CA1 Egr1-dependent activated neurons during the rotarod task. A, Design of the AAV vector to express hM3D(Gq) only in cells with Cre recombinase activity. B, Egr1-CreERT2 mutant mice used. C, Scheme depicting the behavioral characterization in the present figure (colored). D, Schematic location of the injection site for each mouse (only left hemisphere is shown). E, Representative image of a transduced hippocampus from a mouse treated with CNO. F, Representative images of CA1 in mice from E. Empty arrowheads designate nontransduced cells, white arrowheads designate transduced cells. G, Quantification of cFos immunofluorescence intensity in the CA1 pyramidal cell layer of transduced mice and treated with 4-HT and Vehicle (n = 8) or CNO (n = 9). Mean ± SEM. Mann–Whitney t test (sum of ranks A, 36; B, 117, U = 0, p < 0.0001). H, Accelerating rotarod task in transduced mice. Arrows indicate treatment with 4-HT and arrowheads treatment with CNO. Data are mean ± SEM. Two-way ANOVA identified significant differences between groups on last day of training (F(1,76) = 26,41, p < 0.0001). I, Two independent groups of WT mice treated with vehicle (n = 10) or CNO (3 mg/kg, n = 11), respectively, were subjected to the accelerating rotarod task with the same design as in H. J, A new cohort of mutant mice as in B, were subjected to the same experimental design as in A–C but without 4-HT administration to rule out potential leakiness of the system. Data are mean ± SEM, analyzed by two-way ANOVA (no significant differences were detected between groups). K, Postmortem hippocampal examination in mice from J shows no recombination because of the absence of 4-HT administration. Data are mean ± SEM. Scale bars: 300 µm (E), 60 μm (F), and 100 μm (K). DG: dentate gyrus, CA1: cornu amonis 1, CA3: cornu amonis 3, SO: stratum oriens, SP: stratum pyramidale, SR: stratum radiatum.
Concerning the rotarod performance, Egr1-CreERT2 mice transduced with hM3D(Gq) and injected with 4-HT on days 1 and 2, progressively learned the task (Fig. 7H). On days 4, 5, and 6, we treated one group of mice with CNO and the other with vehicle. Egr1-CreERT2 mice treated with vehicle showed significantly better scores than those treated with CNO in the rotarod performance. The effect became more pronounced at the last day of training (Fig. 7H). To rule out the possibility of unspecific or off-target effects of CNO, we repeated the same experiment as in Figure 7H using a new cohort of WT mice only treated with CNO or vehicle (Fig. 7I). CNO per se did no induce any effect on the accelerating rotarod performance at any time or session. This result ruled out unspecific off-target effects induced by CNO in motor learning per se. Furthermore, to rule out potential leakiness of the Egr1-CreERT2 mice we repeated in parallel the entire experiment (as in Fig. 7A–H) in a new cohort of mice, but without 4-HT administration. No changes were observed in the rotarod performance in any group as well as no recombination was detected in brains from Egr1-CreERT2 mice transduced with hM3D(Gq) and treated with vehicle or CNO (Fig. 7J,K).
Next, we aimed to assess whether the manipulation of the Egr1-dependent activated neuronal subpopulations induced during the accelerating rotarod task could affect unspecific and/or general hippocampal-dependent functions such as navigation, anxiety or spatial learning. Thus, we subjected the same Egr1-CreERT2 mice transduced with hM3D(Gq) receptor (from Fig. 7) to the open field and NOL tasks (Fig. 8A). Mice were injected 30 min before the open field test with vehicle or CNO (Fig. 8B). Mice treated with CNO did not display differences in terms of traveled distance (Fig. 8C), time spent in the center (Fig. 8D), and parallel index (Fig. 8E) during the 30-min session in the arena. In the NOL test, mice treated with CNO showed no modification in new object location preference (Fig. 8F,G). These results show that the alteration of rotarod performance caused by CNO because of the activation of the Egr1-dependent neuronal ensemble induced during the accelerating rotarod task was specific.
Figure 8.
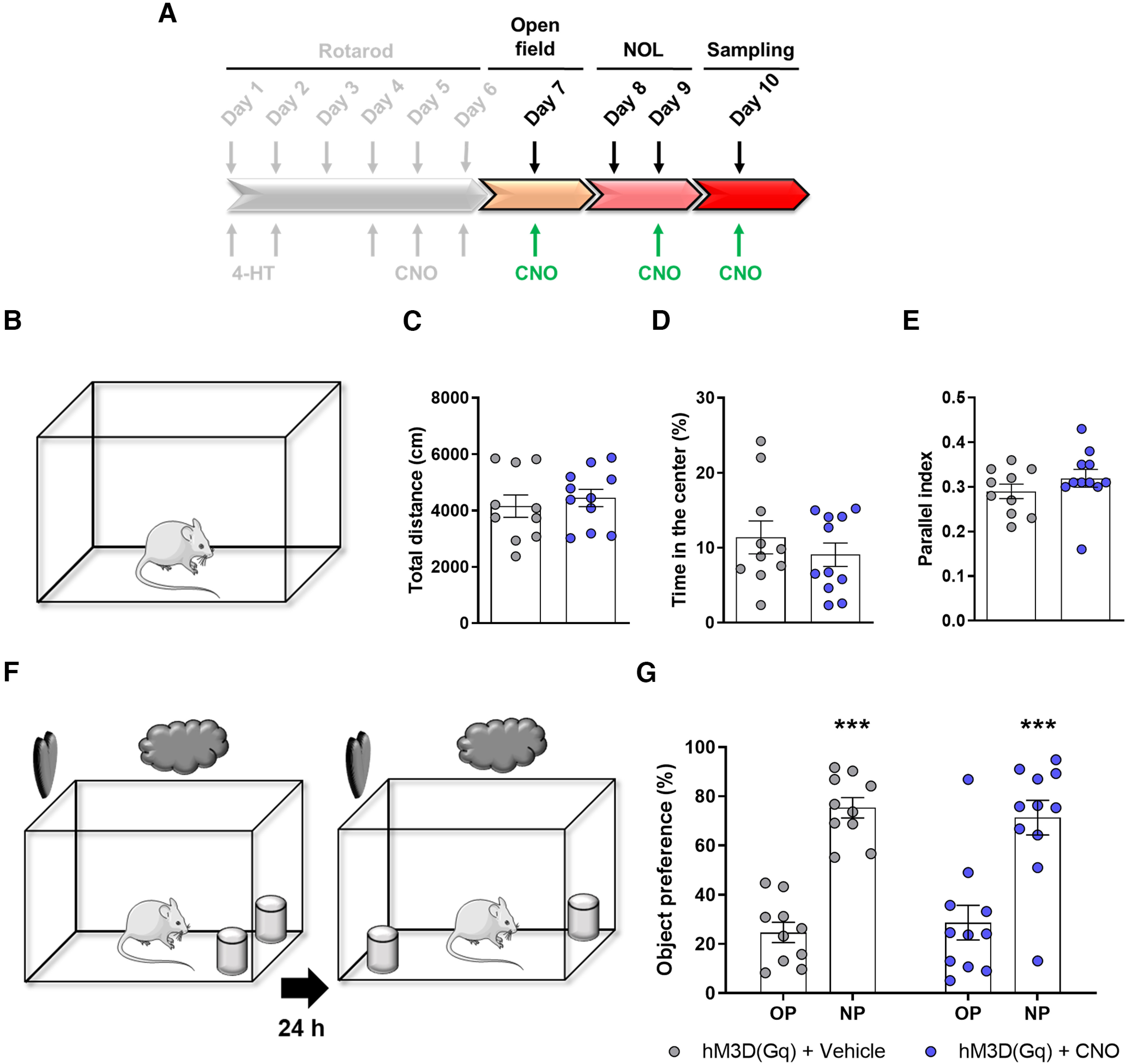
Chemogenetic modulation of the CA1 Egr1-dependent activated neurons does not alter performance in other hippocampal-related tasks. A, After the accelerating rotarod task showed in Figure 7A–H, gray tone scheme, the Egr1-CreERT2 mutant mice transduced with AAV-hSYN-DIO-hM3D(Gq)-mCherry construct and treated with 4-HT at the beginning of the rotarod training (Fig. 7) were subjected to hippocampal-related tasks; 30 min before open field task and before the testing session in the NOL task, all mice received a single intraperitoneal injection of vehicle (n = 10) or CNO (3 mg/kg, n = 11). In the open field task (B), CNO injection effects on the total distance traveled (C), the time spent in the center of the arena (D), and the parallel index (E) are shown. Unpaired t test in D, E or Mann–Whitney t test in G did not detect significant differences between groups in any parameter. F, In the NOL test, CNO injection effects on the NOL preference are shown G. Two-way ANOVA identified significant and equal NOL preference (NOL preference effect: F(1,38) = 62,11, p < 0.0001) but no differences were found between groups (group effect: F(1,38) = 1.9 10−14, p = 0.99). *: NP versus OP. OP: old position, NP: new position. In C–E, G, data are individual values for every mouse and mean ± SEM are shown.
Discussion
The role of specific brain regions including the striatum, motor cortex and cerebellum in the control of motor learning is well established (De Zeeuw and Ten Brinke, 2015; Giordano et al., 2018; Papale and Hooks, 2018). Less explored is the role of the hippocampus. Potentially, the hippocampus is capable of playing either cooperative or competitive roles during the acquisition of motor skills as suggested by human studies (Poldrack and Packard, 2003; Ghiglieri et al., 2011). However, how the hippocampus is playing such a complex role is puzzling and the underlying cellular and molecular mechanisms remain completely unknown (Albouy et al., 2013). Here, we provide evidence for a negative regulation of motor learning implying Egr1 as a gene controlling a specific subset of pyramidal neurons in the CA1.
To our knowledge, our study is the first to profile the transcriptome in the hippocampus following a motor learning task. Our in-depth genome-wide regional comparative study of mRNAs showed that the acquisition of a motor skill involves changes in gene expression particularly during the last phases of the motor learning. This is in line with previous reports showing that the hippocampus shows major structural changes after a long-term training procedure in the accelerating rotarod task (Scholz et al., 2015). Indeed, our cluster and pathway analysis of hippocampal transcriptomic data in response to LTT identified genes related with synapse assembly and organization, regulation of signaling transduction, response to unfolded protein, RNA methylation and DNA recombination which are in turn important processes for formation of hippocampal memories (MacDonald et al., 2006; Svitkina et al., 2010). We observed that the progressive downregulation of the Ca2+ signaling in the CA1 pyramidal cells correlated with a progressive improvement of motor learning and with global and changes in gene expression such as a downregulation of genes like Gsg1l, Arc, and Mdga1. Gsg1l is a novel gene with some roles previously described in the regulation of synaptic activity and plasticity via modulation of AMPARs (Coombs et al., 2019). Another couple of interesting genes are Mdga1 and Mdga2 which are downregulated and upregulated, respectively, in LTT mice compared with NT mice. Mdga1 is enriched in the hippocampus and its loss has been related with impairments in cognitive skills (Connor et al., 2017). Furthermore, Mdga1 is a negative regulator of inhibitory synapses (K. Lee et al., 2013). Therefore, in our trained mice, reduced Mdga1 levels could be related with an increase of inhibitory activity as observed by our in vivo calcium imaging recordings. In contrast, deficiencies in Mdga2 increases the presence of AMPARs in the synaptic membrane and increases excitatory transmission (Connor et al., 2016). In our trained mice, Mdga2 trends to increase which could result in a decrease of AMPARs in the synaptic membrane and a consequent decrease on hippocampal excitatory activity. Notably, among others, Gsgl1, Mdga1, and Mdga2 are potentially regulated at a transcriptional level by Egr1.
Here, we also demonstrate that, a rapid and early increase of Egr1-positive ensembles as well as a fast (at 2 h after training) but transient increase on Egr1 levels in the CA1 is induced on motor leaning. However, as shown by gene expression analysis, Egr1 decreases 24 h after LTT suggesting a complex expression pattern depending on the time point on which Egr1 expression is analyzed after the rotarod training. Similar decrease was observed with Arc. Such reduction in the abundance of IEG transcripts to levels below the baseline may reflect an active mechanism for transcriptional shut down (Link et al., 1995; Waltereit et al., 2001). Importantly, the subset of hippocampal pyramidal neurons activated by Egr1 exert a negative regulation on motor learning. This interpretation is indicated by improved learning when these neurons are selectively depleted by Casp3 activation and, on the contrary, impaired rotarod performance when they are chemogenetically stimulated. Moreover, the downregulation of Egr1 in the hippocampus with shRNA also improved the accelerating rotarod learning performance, indicating a causal role for Egr1 in these regulations. Thus, our results give support to a role of this CA1 neuronal ensemble as a negative regulator of motor systems giving a molecular/cellular explanation of the phenomena previously suspected in human subjects (Poldrack and Packard, 2003; Ghiglieri et al., 2011). As far as we know, to date there are just a few indirect observations involving Egr1 with motor-like skills. For example, Egr1 is induced in the thalamus in motor-driven behaviors induced by singing in vocal learning birds (Horita et al., 2012) or in operant conditioning-based tasks in the cortico-striatal pathway (Hernandez et al., 2006), but there is no information about the role of Egr1 in the procedural motor learning in the rotarod. It is possible that the induced Egr1 activation in the hippocampus may oppose to the same pathway when activated in other brain regions such as in the striatum. In this line, cocaine-induced hyperlocomotion or extensive instrumental training induced both of them an Egr1 increase in the striatum (Hernandez et al., 2006; Xu and Kang, 2014). Also, we previously reported in a mouse model of Huntington's disease with severe motor coordination alterations that overexpression of Rsk1 (a transcription factor) in their striata rescued their motor deficits with a concomitant rescue of their aberrantly reduced Egr1 levels (Anglada-Huguet et al., 2016).
A remarkable phenomenon in our study is that all the manipulations exerted their effects only at the last days of the rotarod training whereas the induction of the Egr1-positive engrams was observed in both, the first and the last days of training. This apparent mismatch could be provoked by distinct mechanisms related with short-term and long-term memories (Izquierdo et al., 2002; Cowan, 2008) in which Egr1-dependent actions would be only affected by long-term or consolidation or re-activation (Buzsáki, 2015; Giri et al., 2019; Grosmark et al., 2021) mechanisms. Another interpretation is that other systems (striatum, motor cortex, cerebellum) highly involved in the acquisition of motor skills (Turner and Desmurget, 2010; De Zeeuw and Ten Brinke, 2015; Kawai et al., 2015) could compensate for any hippocampal manipulation performed, at least, in the early phases of motor learning.
Another intriguing point is that Egr1-induction has classically been involved with neuronal activity (Duclot and Kabbaj, 2017), but we observed a decreased hippocampal activity concomitant with Egr1 upregulation 2 h after motor learning. This apparent discrepancy could be associated to different phenomena. First, Egr1 is not only involved with enhancement but also with repression of gene expression (Trizzino et al., 2021). Thus, we strongly believe that the underlying transcriptional changes mediated by upregulated Egr1 activity in the particular ensemble would be involved with a long-term and long-lasting inhibition of hippocampal neuronal circuits as observed in Figure 2. Second, there exist internal loops implying superficial pyramidal cells, deep pyramidal cells, and parvalbumin interneurons in the CA1 that could be autoinhibitory (Valero and de la Prida, 2018). Then, it is conceivable that our Egr1-dependent ensemble is labeling such loop although further studies should we performed to demonstrate such statements.
Another question raised by this study is the type of information coded by Egr1-dependent neuronal ensembles during motor learning in the hippocampus. The hippocampus could play a role in the modulation of anxiety levels related to the task (Cha et al., 2016), or encoding contextual information (Smith and Bulkin, 2014) or in the regulation of goal-directed actions in synchrony with the striatum (Albouy et al., 2013; Palombo et al., 2019) and even in the regulation of basal locomotion per se (Arriaga and Han, 2017). Thereby, the depletion of the Egr1-dependent neuronal ensemble could eliminate contextual or emotional “distracters” and, therefore, enhance the task performance. Yet it is important to underlie that we did not observe compensatory changes in the striatal activation or major changes in anxiety, locomotion, navigation or spatial learning processes when we manipulated the ensembles induced by the accelerating rotarod task. Alternatively, the activated neurons identified here could code for specific contextual information of the task. However, the hippocampus is capable to create multiple compensatory representations of the same spatial context (Sheintuch et al., 2020). Our study reveals that the molecular mechanism could be driven by Egr1 in a particular group of pyramidal cells of the CA1, which opens the way for further investigation of procedural memory formation.
Footnotes
A.G. is a Ramón y Cajal Fellow (RYC-2016-19466). This work was supported by Ministerio de Ciencia, Innovación y Universidades Grants RTI2018-094678-A-I00 (to A.G.), RTI2018-094374-B-I00 (to S.G.), PID2020-119386RB-100 (to J.A.), and PID2020-116474RB-I00 (to V.B.). We thank Ana López (María de Maeztu Unit of Excellence, Institute of Neurosciences, University of Barcelona, MDM-2017-0729, Ministry of Science, Innovation and Universities) for technical support. We also thank María Calvo from the Advanced Microscopy Service (Centres Científics i Tecnològics Universitat de Barcelona) for her help in the acquisition, analysis, and interpretation of the confocal images and Daniel del Toro and Eulàlia Martí, from the Institut de Neurociències, for their insightful comments and advice.
The authors declare no competing financial interests.
References
- Albouy G, Sterpenich V, Balteau E, Vandewalle G, Desseilles M, Dang-Vu T, Darsaud A, Ruby P, Luppi PH, Degueldre C, Peigneux P, Luxen A, Maquet P (2008) Both the hippocampus and striatum are involved in consolidation of motor sequence memory. Neuron 58:261–272. 10.1016/j.neuron.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Albouy G, King BR, Maquet P, Doyon J (2013) Hippocampus and striatum: dynamics and interaction during acquisition and sleep-related motor sequence memory consolidation. Hippocampus 23:985–1004. 10.1002/hipo.22183 [DOI] [PubMed] [Google Scholar]
- Anglada-Huguet M, Giralt A, Rué L, Alberch J, Xifró X (2016) Loss of striatal 90-kDa ribosomal S6 kinase (Rsk) is a key factor for motor, synaptic and transcription dysfunction in Huntington's disease. Biochim Biophys Acta 1862:1255–1266. 10.1016/j.bbadis.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Arriaga M, Han EB (2017) Dedicated hippocampal inhibitory networks for locomotion and immobility. J Neurosci 37:9222–9238. 10.1523/JNEUROSCI.1076-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begeti F, Schwab LC, Mason SL, Barker RA (2016) Hippocampal dysfunction defines disease onset in Huntington's disease. J Neurol Neurosurg Psychiatry 87:975–981. 10.1136/jnnp-2015-312413 [DOI] [PubMed] [Google Scholar]
- Bergeron Y, Chagniel L, Bureau G, Massicotte G, Cyr M (2014) mTOR signaling contributes to motor skill learning in mice. Front Mol Neurosci 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland C, et al. (2020) Circulating triglycerides gate dopamine-associated behaviors through DRD2-expressing neurons. Cell Metab 31:773–790.e11. 10.1016/j.cmet.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchio M, Gouny C, Angulo-Garcia D, Toulat T, Tressard T, Quiroli E, Baude A, Cossart R (2020) Hippocampal hub neurons maintain distinct connectivity throughout their lifetime. Nat Commun 11:4559. 10.1038/s41467-020-18432-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin A, Pinsard B, Boré A, Carrier J, Fogel SM, Doyon J (2018) Transient synchronization of hippocampo-striato-thalamo-cortical networks during sleep spindle oscillations induces motor memory consolidation. Neuroimage 169:419–430. 10.1016/j.neuroimage.2017.12.066 [DOI] [PubMed] [Google Scholar]
- Buzsáki G (2015) Hippocampal sharp wave-ripple: a cognitive biomarker for episodic memory and planning. Hippocampus 25:1073–1188. 10.1002/hipo.22488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi P, Castrioto A, Di Filippo M, Picconi B (2013) New experimental and clinical links between the hippocampus and the dopaminergic system in Parkinson's disease. Lancet Neurol 12:811–821. 10.1016/S1474-4422(13)70118-2 [DOI] [PubMed] [Google Scholar]
- Camicioli R, Moore MM, Kinney A, Corbridge E, Glassberg K, Kaye JA (2003) Parkinson's disease is associated with hippocampal atrophy. Mov Disord 18:784–790. 10.1002/mds.10444 [DOI] [PubMed] [Google Scholar]
- Cha J, Greenberg T, Song I, Blair Simpson H, Posner J, Mujica-Parodi LR (2016) Abnormal hippocampal structure and function in clinical anxiety and comorbid depression. Hippocampus 26:545–553. 10.1002/hipo.22566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagniel L, Bergeron Y, Bureau G, Massicotte G, Cyr M (2014) Regulation of tyrosine phosphatase STEP61 by protein kinase A during motor skill learning in mice. PLoS One 9:e86988. 10.1371/journal.pone.0086988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor SA, Ammendrup-Johnsen I, Chan AW, Kishimoto Y, Murayama C, Kurihara N, Tada A, Ge Y, Lu H, Yan R, LeDue JM, Matsumoto H, Kiyonari H, Kirino Y, Matsuzaki F, Suzuki T, Murphy TH, Wang YT, Yamamoto T, Craig AM (2016) Altered cortical dynamics and cognitive function upon haploinsufficiency of the autism-linked excitatory synaptic suppressor MDGA2. Neuron 91:1052–1068. 10.1016/j.neuron.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Connor SA, Ammendrup-Johnsen I, Kishimoto Y, Karimi Tari P, Cvetkovska V, Harada T, Ojima D, Yamamoto T, Wang YT, Craig AM (2017) Loss of synapse repressor MDGA1 enhances perisomatic inhibition, confers resistance to network excitation, and impairs cognitive function. Cell Rep 21:3637–3645. 10.1016/j.celrep.2017.11.109 [DOI] [PubMed] [Google Scholar]
- Coombs ID, Soto D, McGee TP, Gold MG, Farrant M, Cull-Candy SG (2019) Homomeric GluA2(R) AMPA receptors can conduct when desensitized. Nat Commun 10:4312. 10.1038/s41467-019-12280-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan N (2008) What are the differences between long-term, short-term, and working memory? Prog Brain Res 169:323–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curlik DM, Maeng LY, Agarwal PR, Shors TJ (2013) Physical skill training increases the number of surviving new cells in the adult hippocampus. PLoS One 8:e55850. 10.1371/journal.pone.0055850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zeeuw CI, Ten Brinke MM (2015) Motor learning and the cerebellum. Cold Spring Harb Perspect Biol 7:a021683. 10.1101/cshperspect.a021683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFeo G, Curlik DM, Shors TJ (2015) The motirod: a novel physical skill task that enhances motivation to learn and thereby increases neurogenesis especially in the female hippocampus. Brain Res 1621:187–196. 10.1016/j.brainres.2014.11.045 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhring J, Stoldt A, Witt K, Schönfeld R, Deuschl G, Born J, Bartsch T (2017) Motor skill learning and offline-changes in TGA patients with acute hippocampal CA1 lesions. Cortex 89:156–168. 10.1016/j.cortex.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Duclot F, Kabbaj M (2017) The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front Behav Neurosci 11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo C, Conejo NM, González-Pardo H, Lazo PS, Arias JL (2012) A role for dorsal and ventral hippocampus in response learning. Neurosci Res 73:218–223. 10.1016/j.neures.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Fouquet C, Babayan BM, Watilliaux A, Bontempi B, Tobin C, Rondi-Reig L (2013) Complementary roles of the hippocampus and the dorsomedial striatum during spatial and sequence-based navigation behavior. PLoS One 8:e67232. 10.1371/journal.pone.0067232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglieri V, Sgobio C, Costa C, Picconi B, Calabresi P (2011) Striatum–hippocampus balance: from physiological behavior to interneuronal pathology. Prog Neurobiol 94:102–114. 10.1016/j.pneurobio.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Giordano N, Iemolo A, Mancini M, Cacace F, De Risi M, Latagliata EC, Ghiglieri V, Bellenchi GC, Puglisi-Allegra S, Calabresi P, Picconi B, De Leonibus E (2018) Motor learning and metaplasticity in striatal neurons: relevance for Parkinson's disease. Brain 141:505–520. 10.1093/brain/awx351 [DOI] [PubMed] [Google Scholar]
- Giralt A, Sanchis D, Cherubini M, Ginés S, Cañas X, Comella JX, Alberch J (2013) Neurobehavioral characterization of Endonuclease G knockout mice reveals a new putative molecular player in the regulation of anxiety. Exp Neurol 247:122–129. 10.1016/j.expneurol.2013.03.028 [DOI] [PubMed] [Google Scholar]
- Giri B, Miyawaki H, Mizuseki K, Cheng S, Diba K (2019) Hippocampal reactivation extends for several hours following novel experience. J Neurosci 39:866–875. 10.1523/JNEUROSCI.1950-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmark AD, Sparks FT, Davis MJ, Losonczy A (2021) Reactivation predicts the consolidation of unbiased long-term cognitive maps. Nat Neurosci 24:1574–1585. 10.1038/s41593-021-00920-7 [DOI] [PubMed] [Google Scholar]
- Harris KL, Armstrong M, Swain R, Erzinclioglu S, Das T, Burgess N, Barker RA, Mason SL (2019) Huntington's disease patients display progressive deficits in hippocampal-dependent cognition during a task of spatial memory. Cortex 119:417–427. 10.1016/j.cortex.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Hernandez PJ, Schiltz CA, Kelley AE (2006) Dynamic shifts in corticostriatal expression patterns of the immediate early genes Homer 1a and Zif268 during early and late phases of instrumental training. Learn Mem 13:599–608. 10.1101/lm.335006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita H, Kobayashi M, Liu W, Oka K, Jarvis ED, Wada K (2012) Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLoS One 7:e42173. 10.1371/journal.pone.0042173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R, Robbins TW, Pennartz CM, Everitt BJ (2008) Functional interaction between the hippocampus and nucleus accumbens shell is necessary for the acquisition of appetitive spatial context conditioning. J Neurosci 28:6950–6959. 10.1523/JNEUROSCI.1615-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo LA, Barros DM, Vianna MRM, Coitinho A, deDavid e Silva T, Choi H, Moletta B, Medina JH, Izquierdo I (2002) Molecular pharmacological dissection of short- and long-term memory. Cell Mol Neurobiol 22:269–287. 10.1023/A:1020715800956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai R, Markman T, Poddar R, Ko R, Fantana AL, Dhawale AK, Kampff AR, Ölveczky BP (2015) Motor cortex is required for learning but not for executing a motor skill. Neuron 86:800–812. 10.1016/j.neuron.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans F, et al. (2019) SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103:217–234.e4. 10.1016/j.neuron.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Duman RS, Pittenger C (2008) A double dissociation revealing bidirectional competition between striatum and hippocampus during learning. Proc Natl Acad Sci U S A 105:17163–17168. 10.1073/pnas.0807749105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kim Y, Lee SJ, Qiang Y, Lee D, Lee HW, Kim H, Je HS, Südhof TC, Ko J (2013) MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc Natl Acad Sci U S A 110:336–341. 10.1073/pnas.1219987110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K (2015) Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162:635–647. 10.1016/j.cell.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D (1995) Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A 92:5734–5738. 10.1073/pnas.92.12.5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longueville S, Nakamura Y, Brami -Cherrier K, Coura R, Hervé D, Girault J (2021) Long-lasting tagging of neurons activated by seizures or cocaine administration in Egr1-CreER T2 transgenic mice. Eur J Neurosci 53:1450–1472. 10.1111/ejn.15060 [DOI] [PubMed] [Google Scholar]
- MacDonald JF, Jackson MF, Beazely MA (2006) Hippocampal long-term synaptic plasticity and signal amplification of NMDA receptors. Crit Rev Neurobiol 18:71–84. 10.1615/critrevneurobiol.v18.i1-2.80 [DOI] [PubMed] [Google Scholar]
- Martín-Ibáñez R, Crespo E, Esgleas M, Urban N, Wang B, Waclaw R, Georgopoulos K, Martínez S, Campbell K, Vicario-Abejón C, Alberch J, Chan S, Kastner P, Rubenstein JL, Canals JM (2012) Helios transcription factor expression depends on Gsx2 and Dlx1&2 function in developing striatal matrix neurons. Stem Cells Dev 21:2239–2251. 10.1089/scd.2011.0607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, de Vivo L, Bellesi M, Ghilardi MF, Tononi G, Cirelli C (2017) Sleep consolidates motor learning of complex movement sequences in mice. Sleep 40:zsw059. 10.1093/sleep/zsw059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AMM, Hawk JD, Abel T, Havekes R (2010) Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learn Mem 17:155–160. 10.1101/lm.1625310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo DJ, Hayes SM, Reid AG, Verfaellie M (2019) Hippocampal contributions to value-based learning: converging evidence from fMRI and amnesia. Cogn Affect Behav Neurosci 19:523–536. 10.3758/s13415-018-00687-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papale AE, Hooks BM (2018) Circuit changes in motor cortex during motor skill learning. Neuroscience 368:283–297. 10.1016/j.neuroscience.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Packard MG (2003) Competition among multiple memory systems: converging evidence from animal and human brain studies. Neuropsychologia 41:245–251. 10.1016/s0028-3932(02)00157-4 [DOI] [PubMed] [Google Scholar]
- Schmieder R, Lim YW, Edwards R (2012) Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics 28:433–435. 10.1093/bioinformatics/btr669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Niibori Y, Frankland PW, Lerch JP (2015) Rotarod training in mice is associated with changes in brain structure observable with multimodal MRI. Neuroimage 107:182–189. 10.1016/j.neuroimage.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Sheintuch L, Geva N, Baumer H, Rechavi Y, Rubin A, Ziv Y (2020) Multiple maps of the same spatial context can stably coexist in the mouse hippocampus. Curr Biol 30:1467–1476.e6. 10.1016/j.cub.2020.02.018 [DOI] [PubMed] [Google Scholar]
- Sjulson L, Peyrache A, Cumpelik A, Cassataro D, Buzsáki G (2018) Cocaine place conditioning strengthens location-specific hippocampal coupling to the nucleus accumbens. Neuron 98:926–934.e5. 10.1016/j.neuron.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Bulkin DA (2014) The form and function of hippocampal context representations. Neurosci Biobehav Rev 40:52–61. 10.1016/j.neubiorev.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spargo E, Everall IP, Lantos PL (1993) Neuronal loss in the hippocampus in Huntington's disease: a comparison with HIV infection. J Neurol Neurosurg Psychiatry 56:487–491. 10.1136/jnnp.56.5.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers HJ (2020) The hippocampal cognitive map: one space or many? Trends Cogn Sci 24:168–170. 10.1016/j.tics.2019.12.013 [DOI] [PubMed] [Google Scholar]
- Svitkina T, Lin WH, Webb DJ, Yasuda R, Wayman GA, Van Aelst L, Soderling SH (2010) Regulation of the postsynaptic cytoskeleton: roles in development, plasticity, and disorders. J Neurosci 30:14937–14942. 10.1523/JNEUROSCI.4276-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Zucco A, Deliard S, Wang F, Barbieri E, Veglia F, Gabrilovich D, Gardini A (2021) EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci Adv 7:eaaz8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RS, Desmurget M (2010) Basal ganglia contributions to motor control: a vigorous tutor. Curr Opin Neurobiol 20:704–716. 10.1016/j.conb.2010.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino FM, Hayward MD, Nestler EJ, Duman RS, Tallman JF (1992) Differential induction of immediate early genes by excitatory amino acid receptor types in primary cultures of cortical and striatal neurons. Brain Res Mol Brain Res 12:233–241. 10.1016/0169-328X(92)90089-T [DOI] [PubMed] [Google Scholar]
- Valero M, de la Prida LM (2018) The hippocampus in depth: a sublayer-specific perspective of entorhinal–hippocampal function. Curr Opin Neurobiol 52:107–114. 10.1016/j.conb.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D (2001) Arg3.1/Arc mRNA induction by Ca 2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci 21:5484–5493. 10.1523/JNEUROSCI.21-15-05484.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Daunais JB, McGinty JF (1994) Role of kainate/AMPA receptors in induction of striatal zif/268 and preprodynorphin mRNA by a single injection of amphetamine. Brain Res Mol Brain Res 27:118–126. 10.1016/0169-328X(94)90192-9 [DOI] [PubMed] [Google Scholar]
- Xu S, Kang UG (2014) Cocaine induces ubiquitination of Egr-1 in the rat dorsal striatum. Neuroreport 25:1362–1367. 10.1097/WNR.0000000000000273 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu H, Liu Z, Chen X, Su X, Ma C, Tian Z, Huang B, Yan E, Liu X, Ma L (2019) A ventral CA1 to nucleus accumbens core engram circuit mediates conditioned place preference for cocaine. Nat Neurosci 22:1986–1999. 10.1038/s41593-019-0524-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary. Download Table 1-1, XLSX file (84.6KB, xlsx) .
Supplementary. Download Table 4-1, XLSX file (24.3KB, xlsx) .