Abstract
Photosystem II (PSII) catalyzes the oxidation of water at its active site that harbors a high-valent inorganic Mn4CaOx. cluster called the oxygen-evolving complex (OEC). Extrinsic subunits generally serve to protect the OEC from reductants and stabilize the structure, but diversity in the extrinsic subunits exists between phototrophs. Recent cryo-electron microscopy experiments have provided new molecular structures of PSII with varied extrinsic subunits. We focus on the extrinsic subunit PsbQ, that binds to the mature PSII complex, and on Psb27, an extrinsic subunit involved in PSII biogenesis. PsbQ and Psb27 share a similar binding site and have a four-helix bundle tertiary structure, suggesting they are related. Here, we use sequence alignments, structural analyses, and binding simulations to compare PsbQ and Psb27 from different organisms. We find no evidence that PsbQ and Psb27 are related despite their similar structures and binding sites. Evolutionary divergence within PsbQ homologs from different lineages is high, probably due to their interactions with other extrinsic subunits that themselves exhibit vast diversity between lineages. This may result in functional variation as exemplified by large differences in their calculated binding energies. Psb27 homologs generally exhibit less divergence, which may be due to stronger evolutionary selection for certain residues that maintain its function during PSII biogenesis and this is consistent with their more similar calculated binding energies between organisms. Previous experimental inconsistencies, low confidence binding simulations, and recent structural data suggest that Psb27 is likely to exhibit flexibility that may be an important characteristic of its activity. The analysis provides insight into the functions and evolution of PsbQ and Psb27, and an unusual example of proteins with similar tertiary structures and binding sites that probably serve different roles.
Keywords: Photosystem II, Extrinsic subunits, PsbQ, PsbQ’, Psb27
Introduction
Photosystem II (PSII) is a multisubunit, membrane intrinsic, pigment–protein complex that extracts electrons from water to provide reducing equivalents that fuel photosynthetic electron transport. In the mature enzyme, water oxidation is catalyzed by a high-valent inorganic Mn4CaOx cluster called the oxygen-evolving complex (OEC) that is bound on the lumenal side of PSII by the D1 and CP43 core subunits (Brudvig 2008; Vinyard et al. 2013; Shen 2015; Cox et al. 2020). Extrinsic subunits are bound on the lumenal side of the mature PSII complex that stabilize the structure and protect the active site from external reductants that would result in OEC damage (Roose and Pakrasi 2008; Bricker et al. 2012; Roose et al. 2016). In cyanobacteria, these extrinsic proteins are PsbO, PsbQ, PsbU, and PsbV; in red algae and diatoms, they are PsbO, PsbQ’, PsbU, and PsbV; and in plants and green algae, they are PsbO, PsbP, PsbQ, and PsbR (Enami et al. 2008; Roose and Pakrasi 2008; Bricker et al. 2012; Roose et al. 2016). All of these extrinsic subunits have been structurally resolved in complex with PSII, the most recent of which was PsbQ in cyanobacteria, in the cryo-EM structure of PSII from Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) (Gisriel et al. 2021b). We note that PsbQ in cyanobacteria has often been referred to as “CyanoQ” (e.g., as used in Fagerlund and Eaton-Rye 2011); however, the recent cryo-EM structure showing its binding site to be identical to that of PsbQ in plants and algae (Gisriel et al. 2021b) suggests that nomenclature to be inappropriate. We suggest that the “PsbQ” nomenclature for this cyanobacterial subunit be maintained by researchers in the future.
PSII has a high propensity for oxidative damage (Pospíšil 2009; Kale et al. 2017), causing fast turnover that requires a complex mechanism of repair and assembly (Komenda et al. 2012b; Nickelsen and Rengstl 2013; Heinz et al. 2016; Eaton-Rye and Sobotka 2017). During the PSII repair cycle, additional extrinsic subunits transiently bind and facilitate PSII maturation. Although most of the extrinsic subunits present in the mature PSII complex have been characterized and structurally resolved, assembly factors that transiently bind during PSII maturation have been challenging targets for study, and molecular structures of a few have only recently appeared in the literature (Huang et al. 2021; Xiao et al. 2021; Zabret et al. 2021). So far, the only lumenal assembly factor to be structurally resolved in complex with the PSII core is Psb27 from the thermophilic cyanobacteria Thermosynechococcus elongatus (Zabret et al. 2021) and Thermosynechococcus vulcanus (Huang et al. 2021).
Both PsbQ in the structure of the mature PSII complex from Synechocystis 6803 (Gisriel et al. 2021b) and Psb27 in the structures of PSII assembly intermediates (Huang et al. 2021; Zabret et al 2021) were determined to share binding sites analogous to the known PsbQ and PsbQ’ subunits (hereafter “PsbQ(′)”) in PSII structures from plants and algae (Ago et al. 2016; Wei et al. 2016; Su et al. 2017; Van Bezouwen et al. 2017; Pi et al. 2019; Sheng et al. 2019) (Fig. 1). This was somewhat surprising because (a) there is very low sequence identity between PsbQ(′) and Psb27 (Huang et al. 2021) (Supplementary Fig. 1 and Supplementary Table 1), (b) cross-linking experiments suggested that cyanobacterial PsbQ bound to the lumenal surface of PSII in a different site than PsbQ(′) in plants and algae (Liu et al. 2014), and (c) it is unlikely that there is functional similarity between Psb27 that is involved in PSII maturation and PsbQ(′) that are present in the mature PSII complex with other permanently bound lumenal subunits. In hindsight, however, their similar binding site makes sense because they ah exhibit a four-helix bundle tertiary structure. An additional similarity between the PsbQ(′) and Psb27 is that in most cyanobacteria, PsbQ and Psb27 both have an N-terminal lipid modification (Kashino et al. 2006; Fagerlund and Eaton-Rye 2011), but neither have this modification in plants or algae (Xingxing et al. 2018). The fact that both PsbQ(′) and Psb27 exhibit a four-helix bundle tertiary structure, that both bind to the same site on the CP43 subunit of PSII, and that both exhibit similar trends in the presence or absence of an N-terminal lipid modification between different species, suggests that these proteins are related in their function and possibly evolved from a common ancestor.
Fig. 1.
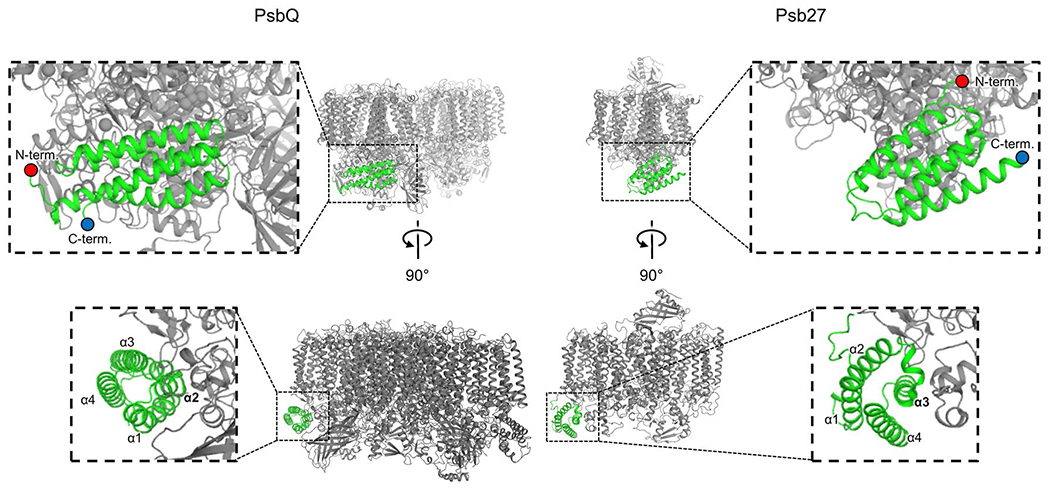
Overview of PsbQ(′) and Psb27 binding. Representative PSII structures are shown where PsbQ is bound (PDB 7N8O, left), which occupies the same binding site as PsbQ(′) in plants and algae, and where Psb27 is bound (PDB 7NHP, right). Protein ribbons are shown where the structures are colored grey except for PsbQ and Psb27, which are colored green. Two views are shown for each. In the top row, the N- and C-termini are labeled, and in the bottom row, the four α-helices are labeled from N- to C-terminus
A challenge in determining how PsbQ(′) and Psb27 are related is that their functions remain unclear, and experiments have produced a variety of results dependent upon the species studied. In the cyanobacterium Synechocystis 6803, PSII complexes containing PsbQ achieve higher rates of oxygen evolution than those that do not, leading to the hypothesis that PsbQ stabilizes the lumenal domain of PSII that harbors the active site (Roose et al. 2007). PsbQ from Synechocystis 6803 was also suggested to be involved in PSII assembly due to the isolation of a PSII complex lacking two of the extrinsic subunits typically found in mature PSII, PsbU and PsbV, and containing four PsbQ subunits per PSII dimer (Liu et al. 2015). Deletion of PsbQ in Arabidopsis thaliana has little effect except under low light conditions (Yi et al. 2006), but defects in PsbP that result in calcium and chloride sensitivity in spinach can be restored by the addition of PsbQ (Ifuku and Sato 2001, 2002). The function of PsbQ in plants is probably tightly intertwined with PsbP as is also exemplified by a recent cryo-EM structure of PSII from A. thaliana lacking PsbQ and PsbP but maintaining PsbO where the active site is found in an immature configuration lacking the OEC (Graça et al. 2021). Removal of PsbQ alone in A. thaliana resulted in less light-harvesting complexes and changes in state transitions, and so it could also be involved in the attachment of peripheral antenna subunits to the core (Allahverdiyeva et al. 2013). In halophytic plants, PsbQ genes were not expressed in varying NaCl concentrations and were thus considered non-essential (Pagliano et al. 2009; Trotta et al. 2012). This range of experimental results aiming to understand the role of PsbQ(′) subunits suggests that their functions are highly dependent upon other extrinsic subunits that vary between organisms.
The function of Psb27 has also been challenging to determine. It is well documented to be involved in PSII assembly (Nowaczyk et al. 2006; Bentley et al. 2008; Liu et al. 2011b; Grasse et al. 2011; Komenda et al. 2012a; Bečková et al. 2017; Weisz et al. 2019), but the details of its structural influence are unclear. Psb27 has been suggested to block premature binding of the extrinsic subunits found in the mature PSII complex (Nowaczyk et al. 2006) so that the OEC can be efficiently assembled during a process called photoactivation (Bao and Burnap 2016). In a more complicated view, cyanobacterial Psb27 is also thought to modulate photoactivation by influencing loop E of CP43 that in turn alters the OEC-binding site (Roose and Pakrasi 2008; Liu et al. 2011a, 2013; Avramov et al. 2020; Gisriel et al. 2021a); however, a superposition of an apo-PSII structure (Gisriel et al. 2020), two assembly intermediate PSII structures that lack lumenal extrinsic subunits other than Psb27 (Huang et al. 2021; Zabret et al. 2021), and a mature PSII structure lacking only PsbQ (Kato et al. 2021), show surprisingly little difference in the structure of CP43 loop E (Supplementary Fig. 2). Thus, the influence of Psb27 on the lumenal domain of PSII is thought to be subtle (Zabret et al. 2021). In those structures that lack the OEC (all except the mature PSII structure), the C-terminus of the D1 subunit was found in a unique orientation relative to mature PSII. Because Psb27 was determined not to directly interact with the D1 C-terminus in either of the cyanobacterial structures where Psb27 is bound, it is unclear whether Psb27 stabilizes the lumenal domain of PSII in a way that alters the D1 C-terminus. It was also determined that when Psb27 is bound to the PSII core, its position only slightly overlaps with the position where PsbO is bound in mature PSII structures (Zabret et al. 2021); however, cyanobacterial PSII complexes containing both Psb27 and PsbO have been observed (Liu et al. 2011b). Cyanobacterial Psb27 binding has been observed prior to the insertion of the CP43 module during biogenesis (Komenda et al. 2012a; Heinz et al. 2016) so its function may vary dependent upon the PSII assembly state. Much less is known about Psb27 in higher plants. A. thaliana contains two Psb27 homologs that are also involved in PSII repair, but appear to have distinct functions (Chen et al. 2006; Wei et al. 2010; Hou et al. 2015) that may be unique from cyanobacterial Psb27 (Xingxing et al. 2018).
Now that a variety of molecular structures of these subunits in complex with PSII are available, a comparison of their similarities and differences is important to help determine their functions and variation between species. Here, we use structural and sequence analyses, homology modeling, and in silico docking simulations to compare and predict the differences between PsbQ(′) and Psb27 from a variety of photosynthetic organisms. Although the PsbQ(′) subunits are likely to have diverged from a common ancestor, which is consistent with previous observations (Kashino et al. 2002; Thornton et al. 2004; Balsera et al. 2005; De Las Rivas and Roman 2005; Jackson et al. 2010; Michoux et al. 2014), there is no evidence that Psb27 is related despite sharing a four-helix bundle tertiary structure and a very similar binding site. PsbQ(′) subunits have diverged strongly between lineages which is exemplified by low sequence identity, variable characteristics of the N-terminus, and a wide range of calculated binding energies. In contrast, Psb27 subunits generally exhibit less divergence between lineages, more similar N-terminal characteristics, and similar calculated binding energies between organisms. These observations suggest that the lumenal domain of the CP43 subunit has evolved to bind these four-helix bundle proteins in the same site for completely different functions dependent upon the assembly state of PSII. Furthermore, the evolutionary divergence of PsbQ homologs has probably been influenced by interactions with other lumenal extrinsic subunits. The greater identity observed in Psb27 sequences suggests selective pressure to maintain its function in PSII biogenesis between organisms.
Materials and methods
Sequence alignments
Multiple sequence alignments were performed using Clustal Omega (Sievers et al. 2011). Where structure-based sequence alignments were used, they were performed using PROMALS3D (Pei et al. 2008). The PsbQ(′) structures used for the PROMALS3D alignments were chain Q of PDB 7N8O (Synechocystis 6803), a homology model (see the next section on homology modeling) of PsbQ from T. elongatus, a homology model of PsbQ from T. vulcanus, chain Q of PDB 6KAC (Chlamydomonas reinhardtii), chain Q2 of PDB 4YUU (Cyanidium caldarium), chain Q of PDB 6JLU (Chaetoceros gracilis), chain Q of PDB 5XNL (Pisum sativum), chain Q of PDB 3JCU (Spinacea oleracea), and a homology model of PsbQ from A. thaliana. The Psb27 structures used for the PROMALS3D alignments were PDB 2KND (Synechocystis 6803), PDB 2Y6X (T. elongatus), chain 1 of PDB 7NHP (T. vulcanus, note that the sequences of Psb27 from T. elongatus and T. vulcanus are identical), a homology model of Psb27 from C. reinhardtii, a homology model of Psb27 from S. oleracea, and PDB 5X56 (A. thaliana). Standard multiple sequence alignments were performed with and without signal sequences but produced the same outcomes as described below.
Homology modeling
Due to their lack of experimentally determined molecular structures, homology models of PsbQ from A. thaliana, Psb27 from C. reinhardtii, and Psb27 from S. oleracea were created using SwissModel (Guex et al. 2009). These used input sequences from the National Center for Biotechnology Information (NCBI) with accession codes NP_001319873.1, XP_001700736.1, and XP_021864834.1, respectively. The structural template for A. thaliana PsbQ was chain Q of PDB 3JCU (PsbQ from S. oleracea). The structural template for Psb27 from both C. reinhardtii and S. oleracea was PDB 5X56 (Psb27 from A. thaliana).
Molecular docking
To estimate binding energies, each PsbQ(′) or Psb27 model was docked in silico to its respective PSII core using ClusPro (Kozakov et al. 2017). For the PsbQ(′) subunits, the following molecular structures were used. Synechocystis 6803: Chain Q of PDB 7N8O was docked to PDB 7N8O with chain Q removed. T. elongatus: PDB 3ZSU was docked to 6W1O. T. vulcanus: PDB 3ZSU was docked to PDB 7D1T. C. reinhardtii: Chain Q of PDB 6KAC was docked to PDB 6KAC with chain Q removed. C. caldarium: Chain Q2 of PDB 4YUU was docked to PDB 4YUU with chain Q2 removed. C. gracilis: Chain Q of PDB 6JLU was docked to PDB 6JLU with chain Q removed. P. sativum: Chain Q of PDB 5XNL was docked to PDB 5XNL with chain Q removed. S. oleracea: Chain Q of PDB 3JCU was docked to PDB 3JCU with chain Q removed. A. thaliana: The homology model of A. thaliana PsbQ was docked to PDB 5MDX. The restraint was applied to maintain > 1 Å and < 4 Å between any atom in the highly conserved PsbQ(′) binding interaction described below.
For Psb27, the following molecular structures were used. Synechocystis 6803: PDB 2KND was docked to PDB 7N8O with extrinsic subunits removed. T. elongatus: 2Y6X was docked to PDB 6W1O with extrinsic subunits removed. T. vulcanus: Chain 1 of PDB 7NHP was docked to PDB 7D1T with extrinsic subunits removed. C. reinhardtii: The homology model of C. reinhardtii Psb27 was docked to PDB 6KAC with extrinsic subunits removed. S. oleracea: The homology model of S. oleracea Psb27 was docked to PDB 3JCU with extrinsic subunits removed. A. thaliana: PDB 5X56 was docked to PDB 5MDX with extrinsic subunits removed. The restraint was applied to maintain > 1 Å and < 4 Å between any atom in the highly conserved Psb27-subunit binding interaction described below.
Phylogenetic trees
Phylogenetic trees were created using MEGA7 (Kumar et al. 2016) from the sequences in the sections, “Sequence alignments” and “Homology modeling” above. The evolutionary history for the PsbQ(′) and Psb27 homologs were inferred using the Maximum Likelihood method based on the Le Gascuel model (Le and Gascuel 2008). The bootstrap consensus tree (100 replicates) was taken to represent the evolutionary history of the taxa analyzed (Felsenstein 1985). Initial tree(s) for the heuristic searches were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (3 categories).
Results
Lack of evidence for common ancestry between PsbQ(′) and Psb27
A multiple sequence alignment of nine PsbQ(′) sequences and six Psb27 sequences was performed using Clustal Omega (Sievers et al. 2011) (Supplementary Fig. 1). The resulting sequence identity matrix shows that sequence identity between PsbQ(′) and Psb27 sequences is never greater than ~ 20% (Supplementary Table 1) which is below the “twilight zone” of ~ 25% where sequences can no longer be used to infer common ancestry (Doolittle 1986; Rost 1999); however, it may be possible that small regions of high sequence similar could still imply relatedness. To assess this, we analyzed the Clustal Omega sequence alignment but no regions of obvious sequence similarity are observed between PsbQ(′) sequences and Psb27 sequences (Supplementary Fig. 1). This is consistent with the hypothesis that despite their similar tertiary structure and binding sites (Fig. 1), Psb27 and PsbQ(′) do not function similarly and do not share a common ancestor, their similar characteristics instead arising due to convergent evolution.
It is possible, however, PsbQ(′) and Psb27 have diverged so strongly that standard alignment strategies are insufficient to properly align sequences, but that alignments based on structural models may be useful for proper sequence alignment and thus inferring common ancestry (Sadekar et al. 2006; Orf et al. 2018). Because both Psb27 and the PsbQ(′) maintain a similar tertiary structure of a four-helix bundle, we additionally performed a structure-based sequence alignment of the same sequences and their accompanying molecular structures using PROMALS3D (Pei et al. 2008) (Supplementary Fig. 3). Of the nine sequences used from PsbQ(′) subunits, the one from A. thaliana did not have an accompanying molecular structure, and of the six sequences used for Psb27, the two from C. reinhardtii and S. oleracea did not have accompanying molecular structures; therefore, we created their homology models using SwissModel (Guex et al. 2009) (see “Materials and methods”). The sequence identity comparing Psb27 and the PsbQ(′) calculated from the structure-based sequence alignments is always worse than the corresponding sequence identity using the standard sequence alignment (Supplementary Table 2), which suggests that this is an inappropriate alignment strategy for comparing sequences. Like the standard multiple sequence alignment, an analysis of the structure-based sequence alignment showed no regions of homology (Supplementary Fig. 3), providing additional support for the hypothesis that Psb27 and PsbQ(′) are unrelated.
Diversity in PsbQ(′) subunits
Sequence and structural similarity in PsbQ(′)
The PsbQ(′) sequences share sequence identity with a minimum of ~ 12%, a maximum of ~ 70% (Supplementary Table 3), and a mean of ~ 26%. Despite this low identity, an analysis of the Clustal Omega (Sievers et al. 2011) sequence alignment including only PsbQ(′) sequences shows some homology, aligns the sequences corresponding to the known α-helices correctly, and aligns a fully conserved Trp residue as discussed below (Supplementary Fig. 4). This suggests that PsbQ(′) subunits from vastly different organisms (e.g., cyanobacteria and plants) share a common origin; therefore, we used this sequence alignment to generate a phylogenetic tree of the PsbQ(′) sequences (Supplementary Fig. 5). The tree shows that PsbQ(′) from higher plants form one clade and those from cyanobacteria form another clade. PsbQ of C. reinhardtii is more closely related to PsbQ from higher plants compared to the other algae, which could suggest that PsbQ functions similarly between plants and green algae. The location of PsbQ′ in the phylogenetic tree, between cyanobacterial PsbQ and plant/algal PsbQ, suggests that PsbQ’ exhibits intermediate traits of both the green lineage and cyanobacteria. This makes sense because the other extrinsic subunits of PSII from red algae and diatoms are more closely related to cyanobacterial extrinsic subunits than those from the green lineage (Enami et al. 1998, 2005).
To visualize the structural regions of sequence similarity, we mapped the sequence similarity to a representative structure of PsbQ, that from Synechocystis 6803, color coding as shown in Fig. 2a. Compared to other regions, the sequence similarity is greater in the second α-helix, or α2, which is the closest α-helix to the PSII core (Fig. 1 and Fig. 2a). This conservation of the interacting α-helix suggests that PsbQ(′) subunits that do not have an accompanying molecular structure in complex with the PSII core, such as PsbQ from thermophilic cyanobacteria and the higher plant A. thaliana, also bind in the same way as those whose molecular structures have been solved. Thus, regardless of the organism, at least the four-helix bundle domain of PsbQ(′) appears likely to bind similarly to a peripheral lumenal domain of CP43 between PsbO and either PsbV in cyanobacteria, red algae, and diatoms, or PsbP in green algae and plants (Fig. 1).
Fig. 2.
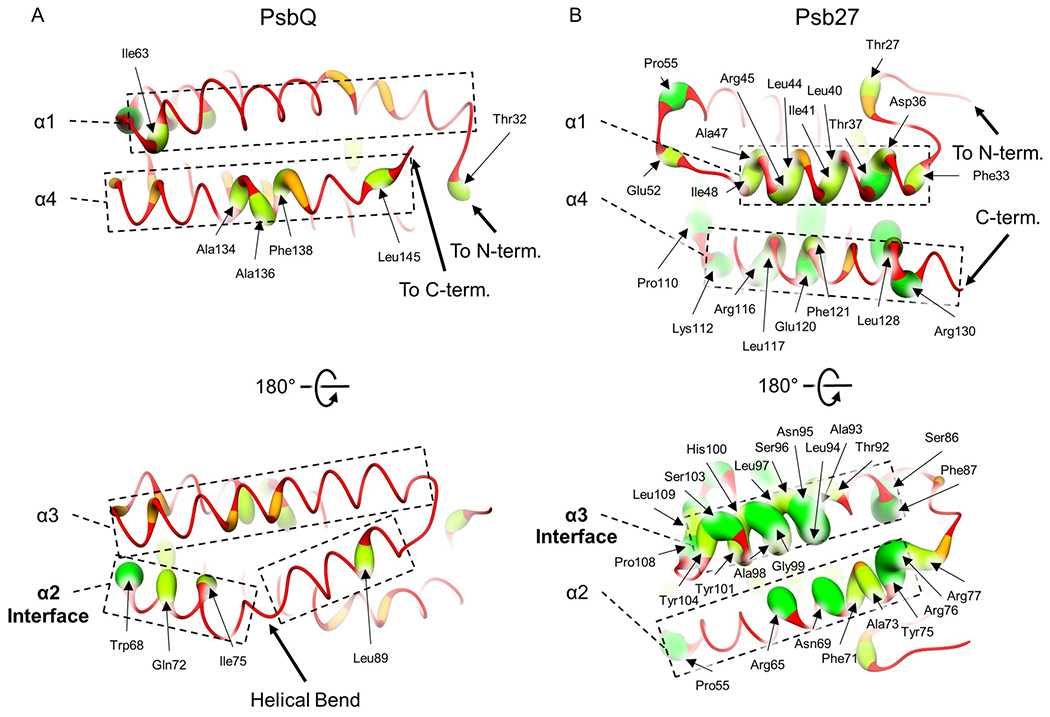
Local sequence similarity of PsbQ(′) and Psb27. In both panels, sequence alignments were performed using Clustal Omega (Sievers et al. 2011) and a representative structure of PsbQ(′) or Psb27 was visualized using UCSF Chimera (Pettersen et al. 2004), color coding sequence similarity from large radii and green ribbons (high similarity) to small radii and red ribbons (low similarity). Two views of each are shown and residues exhibiting ≥ 50% sequence similarity are labeled, α-helices are labeled from N- to C-terminus, and the one labeled in bold font is that which interfaces with the PSII core. The last modeled residues closest to the N- and C-termini, or the terminal residue if it is modeled in the corresponding structure, are additionally labeled. A Structure of PsbQ from Synechocystis 6803 (PDB 7N8O). The helical bend conserved between PsbQ(′) structures is labeled. B Structure of Psb27 from T. elongatus (PDB 7NHP)
Most of the conserved residues from the PsbQ(′) sequences are hydrophobic and their sidechains face the interior of the four-helix bundle (Fig. 2), suggesting they are conserved to maintain the four-helix bundle tertiary structure that requires a hydrophobic interior. Consistent with previous results (Thornton et al. 2004; Kashino et al. 2006), the cleavage site of a signaling sequence was detected by the SigP server (Almagro Armenteros et al. 2019) in the cyanobacterial sequences (highlighted red in Supplementary Fig. 4) just before a Cys residue that forms the thiol linkage to an N-terminal lipid modification that tethers PsbQ to the membrane (Kashino et al. 2006). Notably, the lipid modification is not present in algae and plants (Xingxing et al. 2018). The only fully conserved residue in the PsbQ(′) sequence alignment is a Trp found at the beginning of α3 within the Ncap motif as noted previously (Jackson et al. 2010; Michoux et al. 2014), Trp68 in the sequence from Synechocystis 6803 (Fig. 2 and Supplementary Fig. 4). Interestingly, this Trp residue is among the closest residues to the only part of PsbQ that interacts with PsbO (Gisriel et al. 2021b). It may be that the Trp sidechain is involved, directly or indirectly, in stabilizing interactions between PsbQ(′) and PsbO. Consistent with this hypothesis, PsbQ-Trp68 in the Synechocystis 6803 PSII structure is directly adjacent to PsbQ-Arg122, which was thought to participate in an important electrostatic interaction for PsbQ binding (Gisriel et al. 2021b).
The most highly conserved of the PsbQ(′) binding interactions to the PSII core is a H-bond from the indole N of a conserved Trp residue in loop E of CP43 to the backbone carbonyl in a helical bend of α2 in PsbQ(′) (Fig. 3) (Gisriel et al. 2021b). This interaction is not obvious by the sequence alignments of PsbQ(′) (Supplementary Fig. 4) because only the backbone carbonyl of a PsbQ(′) residue is involved. The H-bond may be important for the bent conformation of α2, which is likely an integral part of the four-helix bundle structure for binding to PSII. Furthermore, the various hydrophobic residues on the interior of the bundle probably contribute to the helical bend motif, which is consistent with the observation that the majority of conserved residues in PsbQ(′) are hydrophobic and directed toward the center of the bundle as noted above.
Fig. 3.
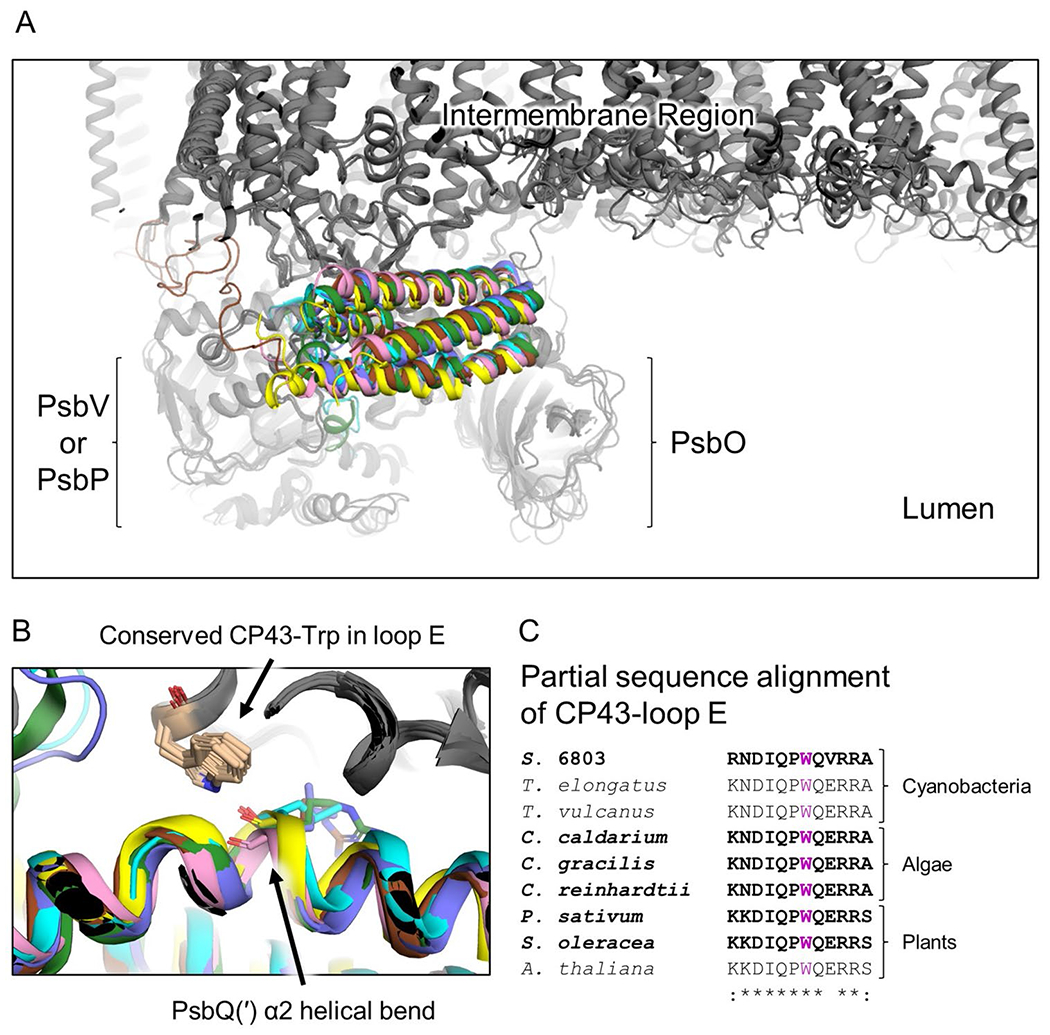
Binding site of PsbQ(′) and conserved CP43-Trp interaction. A Superimposed PsbQ(′)-containing PSII structures. Structures are colored grey except the PsbQ(′) subunit where that from Synechocystis 6803 is yellow, C. caldarium is pink, C. gracilis is brown, C. reinhardtii is green, P. sativum is blue, and S. oleracea is cyan. The regions of PsbO and PsbV/PsbP, the intermembrane region of the protein complexes, and the lumenal space, are labeled. B Focus on the PsbQ(′)-CP43 interface where a conserved CP43-Trp sidechain donates a H-bond to the backbone of α2 in its helical bend of each PsbQ(′) subunit. PsbQ(′) is colored as in A. C Partial sequence alignment of the CP43-loop E showing the conservation of the Trp residue (purple) in B. Bold sequences correspond to those structures shown in A and B, which are those PSII structures with associated PsbQ(′) subunits. The Clustal Omega (Sievers et al. 2011) sequence similarity identifiers are shown below the sequence alignment. Synechocystis 6803 is abbreviated S. 6803
PsbQ(′) calculated binding energies
The recent cryo-EM structure of PSII from Synechocystis 6803 containing PsbQ showed that electrostatic interactions stabilized PsbQ-binding to PSII from the mesophilic cyanobacterium to a greater extent than for two thermophilic cyanobacteria, T. elongatus and T. vulcanus (Gisriel et al. 2021b). To test whether PsbQ(′) exhibits major differences in binding energies between a broader range of organisms, we performed in silico docking of those subunits to their respective PSII cores using ClusPro (Kozakov et al. 2017) to calculate their PIPER binding energies (Table 1 and Supplementary Fig. 6). In short, ClusPro docks two models, iteratively performing energy minimizations based on coefficient sets that weigh molecular interactions differently (Kozakov et al. 2017). The results are consensus models that represent low-energy docked structures for a given coefficient set, the highest scoring and lowest energy of which we refer to as the “top model”. We note that the PIPER binding energies reported should not be taken as valid binding free energies because they do not include entropic contributions, and the coefficient weights are approximations optimized to yield reasonable cluster populations in docking (Kozakov et al. 2017); however, comparison of the PIPER energies provides insight into which cluster population is more favorable. A single constraint was applied to maintain the interaction of CP43-Trp with the PsbQ(′) subunit’s α2 helical bend (see “Materials and methods”) due to this interaction’s full conservation between organisms (Fig. 3). Nine docking simulations were performed in total using structures from the cyanobacteria Synechocystis 6803, T. elongatus, and T. vulcanus, the algae C. reinhardtii, C. caldarium, and C. gracilis, and the plants P. sativum, S. olercea, and A. thaliana. No structure of PsbQ was available from A. thaliana. Therefore, we created its homology model from the structure of PsbQ from S. oleracea.
Table 1.
Lowest energy coefficient set and calculated PIPER binding energy of the top model associated with each docking simulation of PsbQ(′) binding
| Type | Organism | Lowest energy coefficient set | Top model’s energy (kcal/mol)a |
|---|---|---|---|
| Cyanobacteria | S. 6803b | Electrostatic-favored | −978 |
| T. elongatus | Electrostatic-favored | −737 | |
| T. vulcanus | Electrostatic-favored | −790 | |
| Algae | C. reinhardtii | Electrostatic-favored | −2907 |
| C. caldarium | Hydrophobic-favored | −970 | |
| C. gracilis | Electrostatic-favored | −2092 | |
| Plants | P. sativum | Electrostatic-favored | −2003 |
| A. thaliana | Electrostatic-favored | −1363 |
The S. oleracea simulation is excluded from this list because no docking cluster was consistent with the experimentally derived molecular structure. Top model energies likely to be overestimated are in bold
The energies reported in this table are the PIPER energies calculated by ClusPro and do not correspond to valid binding free energies, but can be used for a general comparison of binding interactions from the docking algorithm (Kozakov et al. 2017)
Synechocystis 6803 is abbreviated as S. 6803 in the table
Every docking simulation resulted in the top model being consistent with the experimentally derived molecular structure except that from S. oleracea (Supplementary Fig. 6). This suggests that, aside from the S. oleracea simulation, the calculated PIPER binding energies can be used to assess the relative binding energies for different models. We list the top model’s PIPER binding energy for each simulation and its corresponding coefficient set except from S. oleracea in Table 1. The calculated PIPER binding energies vary widely, with an average energy of − 1480 kcal/mol and standard deviation of 778 kcal/mol. An electrostatic-favored coefficient set resulted in the lowest PIPER binding energy for every simulation except that from C. caldarium which gave the lowest energy for a hydrophobic-favored coefficient set. These results suggest that electrostatic interactions are the primary force that drive PsbQ(′)-binding in the majority of cases. The top model’s PIPER binding energy for thermophilic cyanobacteria was ~ 200 kcal/mol greater than for the mesophilic cyanobacterium, Synechocystis 6803, which is consistent with previous results and may explain why PsbQ(′) is partially maintained in the molecular structure of PSII from Synechocystis 6803 but not in structures from thermophilic cyanobacteria: it does not bind as tightly (Gisriel et al. 2021b).
Generally, cyanobacterial PsbQ-binding is observed to be looser than PsbQ(′) binding in plants and algae, except maybe the red algae C. caldarium. We note that all of the models used in the docking simulations are incomplete to some extent at their N-terminus (Supplementary Table 4). In cyanobacteria, the N-terminus is tethered to the membrane and thus the missing residues probably do not strongly influence the calculated PIPER binding energy. In plants and algae, the much longer N-terminus interacts with other regions on the lumenal surface of the PSII complex (an example is shown in Supplementary Fig. 7); therefore, missing residues are more likely to influence the docking simulation. Thus, the PIPER binding energies calculated for plants and algae are probably overestimates, especially for C. caldarium that is missing the greatest number of C-terminal residues from its model. For C. caldarium, this may also make the determination of the lowest energy coefficient set unreliable which strengthens the argument that electrostatic-favored interactions are the primary force involved in PsbQ-binding. These results suggest that the PsbQ(′) subunits are bound more tightly in plants and algae than in cyanobacteria which is consistent with the fact that only one cyanobacterial PSII structure maintains PsbQ, and only at partial occupancy (Gisriel et al. 2021b), whereas many molecular structures of PSII from plants and algae are available that maintain PsbQ(′). Lower PsbQ-binding affinity in cyanobacteria makes sense because it is kept in close proximity to the core by the N-terminal lipid anchor, whereas tighter PsbQ(′)-binding in algae and plants is required in the absence of a membrane tether.
Ion-binding sites in PsbQ(′)
Zn2+-binding sites have been observed in each of two X-ray crystal structures of PsbQ from S. oleracea (Calderone et al. 2003; Balsera et al. 2005) and one X-ray crystal structure of PsbQ from Synechocystis 6803 (Jackson et al. 2010), and a sulfate-binding site was assigned in another X-ray crystal structure of PsbQ from T. elongatus (Michoux et al. 2014), all of which are structures of the subunit alone not bound to the PSII core. None of these cation-binding sites are similar between the structures, and in all the structures the cations were present in the crystallization buffer, so it is challenging to determine their physiological relevance. Physiological roles for a Zn2+-binding site have been suggested for cyanobacterial PsbQ (Jackson et al. 2010) and plant PsbQ (Balsera et al. 2005), but all of the cation-binding sites in the X-ray crystal structures are found near crystallographic symmetry axes and most are partially coordinated by an adjacent symmetry mate. This suggests that the cation-binding sites found in these structures are a result of crystallization and may not be present in vivo. This notion is supported by the fact that none of the molecular structures of PsbQ(′) bound to the PSII core contain bound cations. We cannot rule out that cations bind to any PsbQ(′) subunits in vivo, but we currently see no strong evidence for this based on observations from structural biology approaches.
Diversity in Psb27
Sequence and structural similarity in Psb27
The sequence identity for Psb27 sequences from different organisms is generally higher than for the PsbQ(′) sequences, with a minimum of ~ 30%, a maximum of ~ 56% (Supplementary Table 5), and a mean of ~ 40%. Obvious regions of sequence homology are observed in the alignment (Supplementary Fig. 8), consistent with the hypothesis that Psb27 proteins from all organisms share a common origin. A phylogenetic tree constructed from the sequence alignment follows the same topology as that of PsbQ(′) (Supplementary Fig. 5); however, Psb27 sequences could not be found using the blastp tool (Goujon et al. 2010) from the NCBI (query sequence of Psb27 from T. elongatus, NCBI code WP_011058295.1) for C. caldarium, C. gracilis, and P. sativum; therefore, the phylogenetic analysis is not ideally robust. The inability to identify these sequences could be due either to the absence of psb27 in their genomes or high divergence of Psb27 relatives that function similarly but contain too little sequence similarity to detect. We are unaware of any other extrinsic subunits transiently bound during PSII assembly, except possibly cyanobacterial PsbQ as discussed below, so Psb27 evolution may not be intertwined with that of other extrinsic subunits unlike the case for PsbQ(′). The higher degree of similarity between Psb27 sequences may suggest that some residues are under strong evolutionary selection to maintain Psb27’s function, resulting in slower evolutionary divergence.
Sequence similarity was mapped onto the structure of Psb27 from T. elongatus (Zabret et al. 2021) (Fig. 2) which shows that the highest sequence conservation is present in α3, the closest α-helix to the PSII core (Fig. 1), suggesting similar binding between organisms. As with PsbQ(′), many hydrophobic residues within the α-helices facing the center of the four-helix bundle are conserved (Fig. 2), probably exemplifying the importance of maintaining the tertiary structure of the bundle. Also like PsbQ(′), the SigP server (Almagro Armenteros et al. 2019) predicts a cleavage site in the cyanobacterial sequences (highlighted red in Supplementary Fig. 8) just before the Cys that is thiol-linked to Psb27’s lipid modification in vivo which agrees with previous results (Nowaczyk et al. 2006). The most highly conserved region in α3 of Psb27 corresponds to an interaction where the backbone carbonyl of a Pro residue in CP43’s loop C appears to form a H-bond to a highly conserved Asn sidechain from Psb27 (Fig. 4). Although this interaction was discussed in one of the publications presenting a structure of PSII with Psb27 bound (Huang et al. 2021), in both models, the sidechain of the Asn has its O atom point toward the O atom of the Pro’s backbone carbonyl moiety which is an unlikely configuration. Since the measured distance between those two O atoms is ~ 3.0 Å in the structures, we think it much more likely that the correct orientation of the Asn sidechain is flipped where the Asn N atom would donate a H-bond to the O atom of the Pro’s backbone carbonyl (yellow-highlighted arrow in Fig. 4a). Interestingly, the CP43-Trp residue important for PsbQ(′)-binding (Fig. 3) is very close to the CP43-Pro residue that is important for Psb27-binding (Fig. 4); the distance between the Cα atoms of those residues in the T. elongatus and T. vulcanus Psb27-PSII structures are 10.7 Å and 12.8 Å, respectively.
Fig. 4.
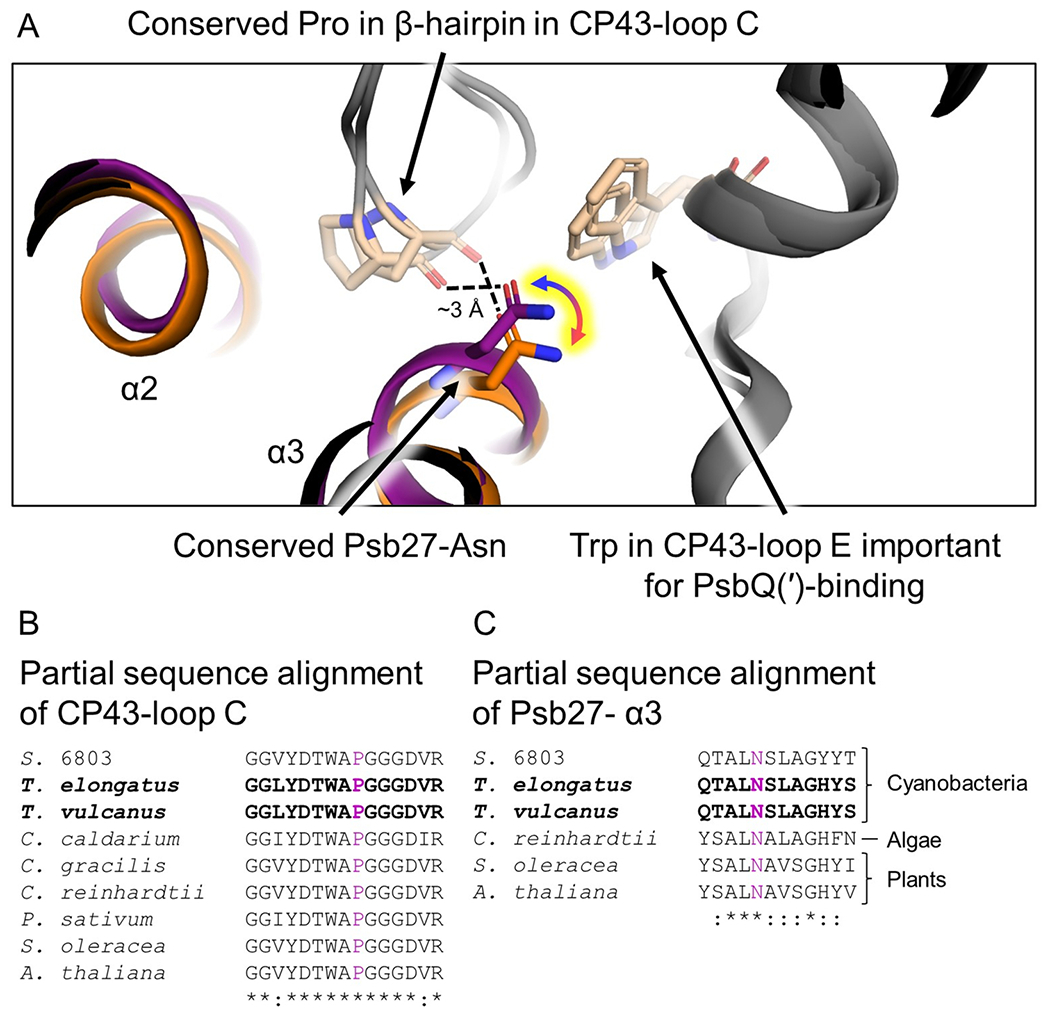
Binding site of Psb27 and conserved CP43-Pro interaction. A Superimposed Psb27-containing PSII structures. Structures are colored grey except the Psb27 subunits where that from T. elongatus (Zabret et al. 2021) is orange and that from T. vulcanus (Huang et al. 2021) is purple. Additionally, the CP43-Pro residue that we suggest forms a H-bond with a Psb27-Asn sidechain important for Psb27-binding, and the CP43-Trp residue important for PsbQ(′)-binding, are colored tan. A yellow-highlighted arrow signifies that the Asn sidechain in both models are likely modeled incorrectly where the O and N atoms of the sidechain should be reversed. Applicable α-helices from Psb27 are labeled. B Partial sequence alignment of the CP43-loop C showing the conservation of the β-hairpin containing an important Pro residue (purple) in A. C Partial sequence alignment of the Psb27-α3 showing the conservation of the Asn residue (purple) in A. For B and C, bold sequences correspond to those structures shown in A, which are those PSII structures containing Psb27, and the Clustal Omega (Sievers et al. 2011) sequence similarity identifiers are shown below the sequences. Synechocystis 6803 is abbreviated S. 6803
Psb27-binding energies
To gain insight into the binding energies of Psb27 subunits to PSII, we additionally performed molecular docking simulations of Psb27 models to CP43 models in the absence of other extrinsic subunits (Supplementary Fig. 9). Each simulation was restrained to maintain proximity of the CP43-Pro to the conserved Psb27-Asn due to this interaction’s high conservation (Fig. 3). Experimental models of Psb27 for C. reinhardtii and S. oleracea are not available, so we created their homology models using the A. thaliana Psb27 structure as a template (see “Materials and methods”). Because Psb27 sequences could not be identified for P. sativum, C. caldarium, and C. gracilis, they were neglected. It is important to consider that the calculation of the PIPER binding energy should only be considered reliable if the associated top model resembles the experimentally determined orientation; however, there are only two structures of Psb27 in complex with the PSII core and the two structures are nearly identical in how they bind Psb27, which makes sense because they are from highly similar thermophilic cyanobacteria (Huang et al. 2021; Zabret et al. 2021). Therefore, the experimental data for comparison of docking simulations is much less robust for Psb27 compared to what is currently available for the PsbQ(′) subunits. More structures of Psb27 bound to PSII from other organisms, like plants and algae, are desirable for more confident comparisons in the future.
Of the six docking simulations performed, only one resulted in top models where the Psb27 orientation was similar to the experimentally determined Psb27-PSII structures: that from the plant A. thaliana (Supplementary Fig. 9). Two other simulations resulted in docking orientations close to the experimentally determined structures, those from T. vulcanus and S. oleracea. Unlike PsbQ(′), the calculated PIPER energies associated with Psb27 binding are relatively similar, with an average energy of −752 kcal/mol and a standard deviation of only 83 kcal/mol. All of the top models were achieved using an electrostatic-favored coefficient set, suggesting that like PsbQ(′), binding of Psb27 to the PSII core is also facilitated mostly by electrostatic interactions, at least for the docking simulations that produced top models that are close to experimental structures.
Ion-binding sites in Psb27
Like PsbQ(′), we see no structural evidence that Psb27 binds ions in vivo. There are two available X-ray crystal structures of Psb27 not in complex with PSII: one from T. elongatus (Michoux et al. 2012) and another from A. thaliana (Xingxing et al. 2018). The latter does not contain any ion-binding site but the X-ray crystal structure of Psb27 from T. elongatus contains a Cl− ion that is coordinated only by atoms within the asymmetric unit, possibly implying physiological relevance. The Psb27 structure in complex with PSII and Psb28, another assembly factor bound to the stromal side of PSII, does not contain an ion-binding site, even though the buffer used for cryo-EM sample preparation contained Cl− (Zabret et al. 2021), suggesting against physiological relevance for the Cl− site bound in the X-ray crystal structure (Michoux et al. 2012). On the other hand, the same Cl−-binding site found in the X-ray crystal of Psb27 from T. elongatus (Michoux et al. 2012) is modeled in the other cryo-EM structure from T. vulcanus where Psb27 is bound to PSII (Huang et al. 2021). To investigate this possible Cl−-binding site, we analyzed the accompanying cryo-EM map (Huang et al. 2021) but found no corresponding electrostatic potential (Supplementary Fig. 10). The authors reported that the starting model for Psb27 in the cryo-EM structure was the X-ray crystal structure of Psb27 from T. elongatus (Huang et al. 2021); therefore, the Cl− ion may have been accidentally left in the coordinates. Although these observations do not rule out the possibility of ion-binding sites in Psb27 in vivo, like PsbQ(′), there is presently no strong evidence of such.
Discussion
A protein’s molecular structure is a valuable tool for understanding its function (Berman et al. 2000; Lee et al. 2007). Here, we show that PsbQ(′) and Psb27 exhibit very different properties even though their four-helix bundle tertiary structure, binding site, and molecular interactions with PSII are highly similar. The four-helix bundle is a common structural motif for globular proteins and fulfills a range of different functions in nature (Presnell and Cohen 1989), which is one reason it is an attractive motif for use in de novo protein design (Schafmeister et al. 1997). Protein multimerization has been shown to protect against deleterious mutations by forming hydrophobic interfaces (Hochberg et al. 2020). In evolutionary time, it is possible that the initial pressure to bind lumenal subunits such as PsbQ(′) and Psb27 to the PSII core was advantageous to select against mutations to the core subunits that would deactivate water oxidation. This is consistent with the observation that peripheral and extrinsic PSII subunits exhibit less sequence identity between cyanobacterial species compared to the core subunits (Gisriel et al. 2021b). As the lumenal subunits have themselves accumulated mutations, they have likely been further functionalized. The differing functional roles for PsbQ(′) and Psb27 probably highlights the complex mechanism of assembly and repair that the cell requires for maintaining PSII’s function (Heinz et al. 2016). We also note that, while PsbQ(′) in mature PSII appear to all maintain the same binding site, the observation that up to four PsbQ subunits bind per cyanobacterial PSII dimer (Liu et al. 2015) suggests that the function of PsbQ(′) could vary, dependent upon the assembly state, and that PsbQ(′) could fill one or more other binding sites on PSII. Furthermore, another four-helix bundle extrinsic subunit found in diatoms, Psb31, was observed in the cryo-EM structure of the PSII supercomplex from C. gracilis. Psb31 binds to the CP47 soluble domain (Pi et al. 2019) and shares very low sequence identity with PsbQ(′) (Okumura et al. 2008) but some structural similarity (Nagao et al. 2013). Coupled with our results, these observations imply that four-helix bundle accessory subunits are commonly found during PSII maturation, both in immature assembly states and the mature complex, and exhibit much diversity between organisms.
Although PsbQ(′) from different organisms are clearly derived from a common ancestor and the binding of most or all appear to be driven by electrostatic interactions, their variable levels of divergence suggests an increased rate of evolution resulting in functional variation between organisms. This is a reasonable hypothesis because (a) experiments aiming to understand the role of PsbQ(′) have produced a broad range of organism-dependent results, (b) PsbQ(′) functions in complex with other extrinsic subunits that vary between organisms, and (c) our calculated PIPER binding energies vary drastically between organisms. An important factor that likely influences PsbQ(′) evolution and thus function between organisms is the variety of N-terminal interactions, which may be linked to the presence of peripheral antenna proteins with PSII or lack thereof. No peripheral transmembrane antenna proteins are known to associate with cyanobacterial PSII; therefore, there may be nothing to hinder the membrane-tethered PsbQ(′) from maintaining proximity with the lumenal domain of PSII for delivery of the subunit to the PSII core during assembly. On the other hand, PSII from higher plants and algae have transmembrane peripheral light-harvesting antenna systems that surround the PSII core that could block, to some extent, the diffusion of lipid-tethered PsbQ(′). If the cyanobacterial PsbQ state is ancestral, it would suggest that plants and algae lost the PsbQ(′) membrane tether to accommodate peripheral antenna complexes. Consistent with this hypothesis, the cyanobacteria Prochlorococcus have evolved to use transmembrane Chl a/b-binding proteins as antenna rather than phycobilisomes (Ting et al. 2002), the PsbQ subunit is not found (Thornton et al. 2004; Fagerlund and Eaton-Rye 2011), and Psb27 lacks a lipid modification (Fagerlund and Eaton-Rye 2011). Coupled with functional variation due to interaction of the N-terminal extension with other extrinsic subunits, the loss of a membrane tether in plants and algae may also explain why a higher binding affinity is important compared to the lower binding affinity in most cyanobacteria where PsbQ is tethered. This hypothesis is also consistent with the fact that removal of the N-terminal lipid linkage from cyanobacterial PsbQ results in its depletion from Synechocystis 6803 thylakoid membrane preparations (Juneau et al. 2016).
For Psb27, previous cross-linking results from two different groups led to the proposal of conflicting Psb27-binding sites (Liu et al. 2011a; Cormann et al. 2016), and both were inconsistent with the recent experimental structures of Psb27 bound to the PSII core (Huang et al. 2021; Zabret et al. 2021). Few of our Psb27 docking simulations are consistent with the experimental Psb27-PSII structures, but we reiterate that Psb27 generally exhibits less divergence compared to PsbQ(′). Coupled with the high conservation of the CP43 subunits between organisms to which Psb27 binds, and lack of strong evidence suggesting that Psb27 binds in the presence of other extrinsic subunits that would result in drastic functional variation, we think it likely that Psb27 binds similarly in all organisms. Furthermore, it seems evident that a major factor in PsbQ(′) divergence, both functionally and evolutionarily, is the sequence divergence found in N-terminal extensions in plants and algae or its absence in cyanobacteria; however, for Psb27, there probably are no N-terminal extensions. Whereas it is well established that most cyanobacterial Psb27 have an N-terminal lipid modification (Nowaczyk et al. 2006), Psb27 in plants and algae instead have a bacterial peptidase cleavage site found in their sequence at a similar location (blue highlights in Supplementary Fig. 8). In A. thaliana, it is assumed that cleavage occurs at this site (Chen et al. 2006), but we could not find any direct evidence for this; however, it is a reasonable suggestion because the chloroplast in eukaryotes arose from a cyanobacterium in an endosymbiotic event (Gokøyr 1967; Martin et al. 2002). If Psb27 is cleaved by a bacterial peptidase in green plant lineage organisms, it would make the N-terminus a similar length to Psb27 from cyanobacteria, suggesting similar functions between organisms, consistent with our observation of relatively low sequence divergence (Supplementary Fig. 8 and Supplementary Table 5). Future studies that characterize the N-termini of Psb27 from members of the green plant lineage may provide insight into functional diversity in Psb27.
Though Psb27 probably binds similarly between organisms, all the following observations suggest that the nature of bound Psb27 is relatively flexible: (a) there are conflicting cross-linking results (Liu et al. 2011a; Cormann et al. 2016), (b) molecular structures of Psb27 in complex with PSII show that Psb27 overlaps with the PsbO-binding site (Huang et al. 2021; Zabret et al. 2021) yet PSII complexes with both Psb27 and PsbO bound have been observed (Liu et al. 2011b), (c) the region of Psb27 in cryo-EM maps of Psb27-PSII exhibits the lowest local resolutions in the entire map for both structures (Huang et al. 2021; Zabret et al. 2021), (d) there is high variation and low confidence in docking simulations performed here and reported previously (Cormann et al. 2016), and (e) the calculated binding energies that do seem reliable reported here suggest weak molecular interactions (Supplementary Fig. 9). In consideration of these observations, we suggest that the conserved interaction of the Pro in the CP43 loop C acts as a hinge from which Psb27 exhibits high mobility. This is probably be an important characteristic of Psb27 function so as to decrease the binding affinity of PsbO as suggested previously (Zabret et al. 2021) prior to photoactivation, while also avoiding a strong influence on the CP43 loop E that would interrupt photoactivation. This is consistent with the observation that the soluble domain of CP43 exhibits little variation whether Psb27 is bound (Huang et al. 2021; Zabret et al. 2021) or unbound (Gisriel et al. 2020) (Supplementary Fig. 2). We note that multiple groups have suggested that flexibility in the CP43 soluble domain itself is an important characteristic of PSII during photoactivation (Gisriel et al. 2020; Tokano et al. 2020; Avramov et al. 2020); therefore, contrary to previous suggestions that Psb27 stabilizes PSII, we suggest that Psb27’s inherent mobility when bound may be an important influencer of cation entry into the OEC-binding site during photoactivation.
Conclusions
There are remarkable similarities between PsbQ(′) and Psb27. Namely, they all exhibit four-helix bundle tertiary structures, they bind to the same site on the soluble domain of CP43, electrostatic interactions appear to be the primary force that drives their binding, and both PsbQ(′) and Psb27 exhibit an N-terminal lipid modification in prokaryotes whereas the eukaryotic proteins do not. However, they also exhibit many differences that are not obvious at first glance: PsbQ(′) exhibits high divergence between organisms which probably results from variation of the N-terminus, while Psb27 shows much less divergence. It also exhibits a range of binding energies between organisms whereas Psb27 probably do not, and the flexibility of bound Psb27 may contribute to its unique function unlike PsbQ(′) that are important in stabilization of the PSII lumenal domain. Further investigation is required to better understand the roles of these subunits between organisms, especially Psb27 about which little is understood.
Supplementary Material
Acknowledgements
This work was supported by Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences Grant DE-FG02-05ER15646 to G.W.B. Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number K99GM140174 to C.J.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Figure 2 was generated using UCSF Chimera which was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH Grant P41-GM103311.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s11120-021-00888-2.
Conflict of interest The authors declare that they have no conflict of interest.
References
- Ago H, Adachi H, Umena Y et al. (2016) Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J Biol Chem 291:5676–5687. 10.1074/jbc.M115.711689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Suorsa M, Rossi F et al. (2013) Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J 75:671–684. 10.1111/tpj.12230 [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK et al. (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Avramov AP, Hwang HJ, Burnap RL (2020) The role of Ca2+ and protein scaffolding in the formation of nature’s water oxidizing complex. Proc Natl Acad Sci USA 117:28036–28045. 10.1073/pnas.2011315117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M, Arellano JB, Revuelta JL et al. (2005) The 1.49 Å resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J Mol Biol 350:1051–1060. 10.1016/j.jmb.2005.05.044 [DOI] [PubMed] [Google Scholar]
- Bao H, Burnap RL (2016) Photoactivation: the light-driven assembly of the water oxidation complex of photosystem II. Front Plant Sci 7:578. 10.3389/fpls.2016.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bečková M, Gardian Z, Yu J et al. (2017) Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol Plant 10:62–72. 10.1016/j.molp.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Bentley FK, Luo H, Dilbeck P et al. (2008) Effects of inactivating psbM and psbT on photodamage and assembly of photosystem II in Synechocystis sp. PCC 6803. Biochemistry 47:11637–11646. 10.1021/bi800804h [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Zukang F et al. (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker TM, Roose JL, Fagerlund RD et al. (2012) The extrinsic proteins of photosystem II. Biochim Biophys Acta - Bioenerg 1817:121–142. 10.1016/j.bbabio.2011.07.006 [DOI] [PubMed] [Google Scholar]
- Brudvig GW (2008) Water oxidation chemistry of photosystem II. Philos Trans R Soc B Biol Sci 363:1211–1219. 10.1098/rstb.2007.2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone V, Trabucco M, Vujicić A et al. (2003) Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep 4:900–905. 10.1038/sj.embor.embor923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang D, Guo J et al. (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61:567–575. 10.1007/s11103-006-0031-x [DOI] [PubMed] [Google Scholar]
- Cormann KU, Möller M, Nowaczyk MM (2016) Critical assessment of protein cross-linking and molecular docking: an updated model for the interaction between photosystem II and Psb27. Front Plant Sci 7:157. 10.3389/fpls.2016.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N, Pantazis DA, Lubitz W (2020) Current understanding of the mechanism of water oxidation in photosystem II and its relation to XFEL data. Annu Rev Biochem 89:795–820. 10.1146/annurev-biochem-011520-104801 [DOI] [PubMed] [Google Scholar]
- De Las RJ, Roman A (2005) Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photochem Photobiol Sci 4:1003–1010. 10.1039/b506874f [DOI] [PubMed] [Google Scholar]
- Doolittle RF (1986) Of urfs and orfs: a primer on how to analyze derived amino acid sequences. University Science Books, Mill Valley [Google Scholar]
- Eaton-Rye JJ, Sobotka R (2017) Editorial: Assembly of the photosystem II membrane-protein complex of oxygenic photosynthesis. Front Plant Sci 8:884. 10.3389/fpls.2017.00884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami I, Kikuchi S, Fukuda T et al. (1998) Binding and functional properties of four extrinsic proteins of photosystem II from a red alga, Cyanidium caldarium, as studied by release–reconstitution experiments. Biochemistry 37:2787–2793. 10.1021/bi9724624 [DOI] [PubMed] [Google Scholar]
- Enami I, Suzuki T, Tada O et al. (2005) Distribution of the extrinsic proteins as a potential marker for the evolution of photosynthetic oxygen-evolving photosystem II. FEBS J 272:5020–5030. 10.1111/j.1742-4658.2005.04912.x [DOI] [PubMed] [Google Scholar]
- Enami I, Okumura A, Nagao R et al. (2008) Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynth Res 98:349–363. 10.1007/s11120-008-9343-9 [DOI] [PubMed] [Google Scholar]
- Fagerlund RD, Eaton-Rye JJ (2011) The lipoproteins of cyanobacterial photosystem II. J Photochem Photobiol B: Biol 104:191–203. 10.1016/j.jphotobiol.2011.01.022 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidential limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- Gisriel CJ, Zhou K, Huang H-L et al. (2020) Cryo-EM structure of monomeric photosystem II from Synechocystis sp. PCC 6803 lacking the water-oxidation complex. Joule 4:2131–2148 [Google Scholar]
- Gisriel CJ, Shen G, Ho M-Y et al. (2021a) Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f. J Biol Chem 297:101424. 10.1016/jobc.2021.101424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisriel CJ, Wang J, Liu J et al. (2021b) High-resolution cryo-EM structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803 Proc Natl Acad Sci USA. 10.1073/pnas.2116765118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksøyr J (1967) Evolution of eucaryotic cells. Nature 214:1161. 10.1038/2141161a0 [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça AT, Hall M, Persson K, Schröder WP (2021) High-resolution model of Arabidopsis photosystem II reveals the structural consequences of digitonin-extraction. Sci Rep 11:15534. 10.1101/2021.01.15.426676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasse N, Mamedov F, Becker K et al. (2011) Role of novel dimeric photosystem II (PSII)-Psb27 protein complex in PSII repair. J Biol Chem 286:29548–29555. 10.1074/jbc.M111.238394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-Pdb-Viewer: a historical perspective. Electrophoresis 30:S162–S173. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- Heinz S, Liauw P, Nickelsen J, Nowaczyk M (2016) Analysis of photosystem II biogenesis in cyanobacteria. Biochim Biophys Acta - Bioenerg 1857:274–287. 10.1016/j.bbabio.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Hochberg GKA, Liu Y, Marklund EG et al. (2020) A hydrophobic ratchet entrenches molecular complexes. Nature 588:503–508. 10.1038/s41586-020-3021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Fu A, Garcia VJ et al. (2015) PSB27: A thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc Natl Acad Sci USA 112:1613–1618. 10.1073/pnas.1424040112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Xiao Y, Pi X et al. (2021) Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus. Proc Natl Acad Sci USA 118:e2018053118. 10.1073/pnas.2018053118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Sato F (2001) Importance of the N-terminal sequence of the extrinsic 23 kDa polypeptide in photosystem II in ion retention in oxygen evolution. Biochim Biophys Acta 1546:196–204. 10.1016/S0167-4838(01)00139-X [DOI] [PubMed] [Google Scholar]
- Ifuku K, Sato F (2002) A truncated mutant of the extrinsic 23-kDa protein that absolutely requires the extrinsic 17-kDa protein for Ca2+ retention in photosystem II. Plant Cell Physiol 43:1244–1249. 10.1093/pcp/pcf136 [DOI] [PubMed] [Google Scholar]
- Jackson SA, Fagerlund RD, Wilbanks SM, Eaton-Rye JJ (2010) Crystal structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 Å: implications for binding and function in cyanobacterial photosystem II. Biochemistry 49:2765–2767. 10.1021/bi100217h [DOI] [PubMed] [Google Scholar]
- Juneau AD, Frankel LK, Bricker TM, Roose JL (2016) N-terminal lipid modification is required for the stable accumulation of CyanoQ in Synechocystis sp. PCC 6803. PLoS ONE 11:e0163646–e0163646. 10.1371/journal.pone.0163646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale R, Hebert AE, Frankel LK et al. (2017) Amino acid oxidation of the D1 and D2 proteins by oxygen radicals during photoinhibition of photosystem II. Proc Natl Acad Sci USA 114:2988–2993. 10.1073/pnas.1618922114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA et al. (2002) Proteomic analysis of a highly active Photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41:8004–8012. 10.1021/bi026012+ [DOI] [PubMed] [Google Scholar]
- Kashino Y, Inoue-Kashino N, Roose JL, Pakrasi HB (2006) Absence of the PsbQ protein results in destabilization of the PsbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. J Biol Chem 281:20834–20841. 10.1074/jbc.m603188200 [DOI] [PubMed] [Google Scholar]
- Kato K, Miyazaki N, Hamaguchi T et al. (2021) High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams. Commun Biol 4:382. 10.1038/s42003-021-01919-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Knoppová J, Kopečná J et al. (2012a) The Psb27 assembly factor binds to the CP43 complex of Photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 158:476–486. 10.1104/pp.111.184184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012b) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15:245–251. 10.1016/j.pbi.2012.01.017 [DOI] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Xia B et al. (2017) The ClusPro web server for protein–protein docking. Nat Protoc 12:255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lee D, Redfern O, Orengo C (2007) Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol 8:995–1005. 10.1038/nrm2281 [DOI] [PubMed] [Google Scholar]
- Liu H, Huang RY-C, Chen J et al. (2011a) Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc Natl Acad Sci USA 108:18536–18541. 10.1073/pnas.1111597108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Roose JL, Cameron JC, Pakrasi HB (2011b) A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J Biol Chem 286:24865–24871. 10.1074/jbc.M111.246231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen J, Huang RY-C et al. (2013) Mass spectrometry-based footprinting reveals structural dynamics of loop E of the chlorophyll-binding protein CP43 during photosystem II assembly in the cyanobacterium Synechocystis 6803. J Biol Chem 288:14212–14220. 10.1074/jbc.M113.467613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Weisz DA et al. (2014) MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc Natl Acad Sci USA 111:4638–4643. 10.1073/pnas.1323063111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Weisz DA, Pakrasi HB (2015) Multiple copies of the PsbQ protein in a cyanobacterial photosystem II assembly intermediate complex. Photosynth Res 126:375–383. 10.1007/s11120-015-0123-z [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E et al. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99:12246–12251. 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michoux F, Takasaka K, Boehm M et al. (2012) Crystal structure of the Psb27 assembly factor at 1.6 Å: implications for binding to photosystem II. Photosynth Res 110:169–175. 10.1007/s11120-011-9712-7 [DOI] [PubMed] [Google Scholar]
- Michoux F, Boehm M, Bialek W et al. (2014) Crystal structure of CyanoQ from the thermophilic cyanobacterium Thermosynechococcus elongatus and detection in isolated photosystem II complexes. Photosynth Res 122:57–67. 10.1007/s11120-014-0010-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao R, Suga M, Niikura A et al. (2013) Crystal structure of Psb31, a novel extrinsic protein of photosystem II from a marine centric diatom and implications for its binding and function. Biochemistry 52:6646–6652. 10.1021/bi400770d [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64:609–635. 10.1146/annurev-arplant-050312-120124 [DOI] [PubMed] [Google Scholar]
- Nowaczyk MM, Hebeler R, Schlodder E et al. (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of Photosystem II. Plant Cell 18:3121–3131. 10.1105/tpc.106.042671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Nagao R, Suzuki T et al. (2008) A novel protein in photosystem II of a diatom Chaetoceros gracilis is one of the extrinsic proteins located on lumenal side and directly associates with PSII core components. Biochim Biophys Acta 1797:160–166. 10.1016/j.bbabio.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Orf GS, Gisriel C, Redding KE (2018) Evolution of photosynthetic reaction centers: insights from the structure of the heliobacterial reaction center. Photosynth Res 138:11–37. 10.1007/s11120-018-0503-2 [DOI] [PubMed] [Google Scholar]
- Pagliano C, La Rocca N, Andreucci F et al. (2009) The extreme halophyte Salicornia veneta is depleted of the extrinsic PsbQ and PsbP proteins of the oxygen-evolving complex without loss of functional activity. Ann Bot 103:505–515. 10.1093/aob/mcn234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-HH, Grishin NV (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC et al. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pi X, Zhao S, Wang W et al. (2019) The pigment–protein network of a diatom photosystem II-light-harvesting antenna supercomplex. Science 365:eaax4406. 10.1126/science.aax4406 [DOI] [PubMed] [Google Scholar]
- Pospíšil P (2009) Production of reactive oxygen species by photosystem II. Biochim Biophys Acta 1787:1151–1160. 10.1016/j.bbabio.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Presnell SR, Cohen FE (1989) Topological distribution of four-alpha-helix bundles. Proc Natl Acad Sci USA 86:6592–6596. 10.1073/pnas.86.17.6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB (2008) The Psb27 protein facilitates manganese cluster assembly in photosystem II. J Biol Chem 283:4044–4050. 10.1074/jbc.M708960200 [DOI] [PubMed] [Google Scholar]
- Roose JL, Kashino Y, Pakrasi HB (2007) The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proc Natl Acad Sci USA 104:2548–2553. 10.1073/pnas.0609337104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Frankel LK, Mummadisetti MP, Bricker TM (2016) The extrinsic proteins of photosystem II: update. Planta 243:889–908. 10.1007/s00425-015-2462-6 [DOI] [PubMed] [Google Scholar]
- Rost B (1999) Twilight zone of protein sequence alignments. Protein Eng Des Sel 12:85–94. 10.1093/protein/12.2.85 [DOI] [PubMed] [Google Scholar]
- Sadekar S, Raymond J, Blankenship RE (2006) Conservation of distantly related membrane proteins: photosynthetic reaction centers share a common structural core. Mol Biol Evol 23:2001–2007. 10.1093/molbev/msl079 [DOI] [PubMed] [Google Scholar]
- Schafmeister CE, LaPorte SL, Miercke LJW, Stroud RM (1997) A designed four helix bundle protein with native-like structure. Nat Struct Biol 4:1039–1046. 10.1038/nsb1297-1039 [DOI] [PubMed] [Google Scholar]
- Shen J-R (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66:23–48. 10.1146/annurev-arplant-050312-120129 [DOI] [PubMed] [Google Scholar]
- Sheng X, Watanabe A, Li A et al. (2019) Structural insight into light harvesting for photosystem II in green algae. Nat Plants 5:1320–1330. 10.1038/s41477-019-0543-4 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D et al. (2011) Fast, scalable generation of high quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ma J, Wei X et al. (2017) Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357:815–820. 10.1126/science.aan0327 [DOI] [PubMed] [Google Scholar]
- Thornton LE, Ohkawa H, Roose JL et al. (2004) Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16:2164–2175. 10.1105/tpc.104.023515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CS, Rocap G, King J et al. (2002) Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol 10:134–142. 10.1016/s0966-842x(02)02319-3 [DOI] [PubMed] [Google Scholar]
- Tokano T, Kato Y, Sugiyama S et al. (2020) Structural dynamics of a protein domain relevant to the water-oxidizing complex in photosystem II as visualized by high-speed atomic force microscopy. J Phys Chem B 124:5847–5857. 10.1021/acs.jpcb.0c03892 [DOI] [PubMed] [Google Scholar]
- Trotta A, Redondo-Gómez S, Pagliano C et al. (2012) Chloroplast ultrastructure and thylakoid polypeptide composition are affected by different salt concentrations in the halophytic plant Arthrocnemum macrostachyum. J Plant Physiol 169:111–116. 10.1016/j.jplph.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Van Bezouwen LS, Caffarri S, Kale R et al. (2017) Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat Plants 3:17080. 10.1038/nplants.2017.80 [DOI] [PubMed] [Google Scholar]
- Vinyard DJ, Ananyev GM, Charles Dismukes G (2013) Photosystem II: the reaction center of oxygenic photosynthesis. Annu Rev Biochem 82:577–606. 10.1146/annurev-biochem-070511-100425 [DOI] [PubMed] [Google Scholar]
- Wei L, Guo J, Ouyang M et al. (2010) LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem 285:21391–21398. 10.1074/jbc.M110.105064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Su X, Cao P et al. (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534:69–74. 10.1038/nature18020 [DOI] [PubMed] [Google Scholar]
- Weisz DA, Johnson VM, Niedzwiedzki DM et al. (2019) A novel chlorophyll protein complex in the repair cycle of photosystem II. Proc Natl Acad Sci USA 116:21907. 10.1073/pnas.1909644116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Huang G, You X et al. (2021) Structural insights into cyanobacterial photosystem II intermediates associated with Psb28 and Tsl0063. Nat Plants 7:1132–1142. 10.1038/s41477-021-00961-7 [DOI] [PubMed] [Google Scholar]
- Xingxing C, Jiuyang L, Huan Z et al. (2018) Crystal structure of Psb27 from Arabidopsis thaliana determined at a resolution of 1.85 Å. Photosynth Res 136:139–146. 10.1007/s11120-017-0450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Hargett SR, Frankel LK, Bricker TM (2006) The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J Biol Chem 281:26260–26267. 10.1074/jbc.M603582200 [DOI] [PubMed] [Google Scholar]
- Zabret J, Bohn S, Schuller SK et al. (2021) Structural insights into photosystem II assembly. Nat Plants 7:524–538. 10.1038/s41477-021-00895-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


