Significance
Blue cone monochromacy (BCM) is an inherited retinal disorder characterized by low vision and poor color vision and caused by mutations in the multicopy gene cluster encoding the long- and middle-wavelength-sensitive cone photoreceptor visual pigments. We showed that structural genomic mutations at the gene cluster explain about one-third of those affected among 213 genetically confirmed BCM families. Our study expands the known spectrum of structural mutations causing BCM by a factor of 4 and provides a comprehensive landscape of their extent and fine structure as well as a deep insight into the underlying molecular mechanisms. We observed evidence that occurrence of BCM-linked structural mutations may be driven by inherent increased instability of individual gene clusters with large copy numbers.
Keywords: human visual pigment genes, BCM, opsin gene deletion, gene conversion, locus control region
Abstract
Blue cone monochromacy (BCM) is an X-linked retinal disorder characterized by low vision, photoaversion, and poor color discrimination. BCM is due to the lack of long-wavelength-sensitive and middle-wavelength-sensitive cone photoreceptor function and caused by mutations in the OPN1LW/OPN1MW gene cluster on Xq28. Here, we investigated the prevalence and the landscape of submicroscopic structural variants (SVs) at single-base resolution in BCM patients. We found that about one-third (n = 73) of the 213 molecularly confirmed BCM families carry an SV, most commonly deletions restricted to the OPN1LW/OPN1MW gene cluster. The structure and precise breakpoints of the SVs were resolved in all but one of the 73 families. Twenty-two families—all from the United States—showed the same SV, and we confirmed a common ancestry of this mutation. In total, 42 distinct SVs were identified, including 40 previously unreported SVs, thereby quadrupling the number of precisely mapped SVs underlying BCM. Notably, there was no “region of overlap” among these SVs. However, 90% of SVs encompass the upstream locus control region, an essential enhancer element. Its minimal functional extent based on deletion mapping in patients was refined to 358 bp. Breakpoint analyses suggest diverse mechanisms underlying SV formation as well as in one case the gene conversion-based exchange of a 142-bp deletion between opsin genes. Using parsimonious assumptions, we reconstructed the composition and copy number of the OPN1LW/OPN1MW gene cluster prior to the mutation event and found evidence that large gene arrays may be predisposed to the occurrence of SVs at this locus.
Blue cone monochromacy (BCM, Online Mendelian Inheritance in Man [OMIM] no. 303700) is a congenital retinal disorder characterized by low vision, severe color vision abnormality, photoaversion, and a frequent occurrence of sensory defect nystagmus. Its prevalence has been estimated to about one in 100,000 in Western populations (1).
BCM is inherited as an X-linked disorder and caused by mutations in the OPN1LW/OPN1MW gene cluster encoding the genes for the long-wavelength-sensitive (LWS) and the middle-wavelength-sensitive (MWS) cone opsins on Xq28 (2, 3).
In healthy individuals the OPN1LW/OPN1MW gene cluster comprises a single OPN1LW gene followed by one or multiple OPN1MW gene copies organized in a tandem repeat structure. OPN1LW and OPN1MW sequences are more than 98% identical, including introns and the intergenic sequence between gene copies (4). A model of nonallelic homologous recombination (NAHR) between these conserved sequences has been proposed to explain the variability in copy number and the frequent occurrence of OPN1LW•OPN1MW hybrid genes (5). Expression of the gene copies within the OPN1LW/OPN1MW gene cluster is governed by a locus control region (LCR) upstream of the gene cluster. Through physical interaction of the LCR with the proximal promoter of the OPN1LW or the OPN1MW gene, the mutually exclusive expression of a single gene copy in an individual cone photoreceptor is determined and maintained (6–8). Yet, the probability of activation of gene expression decays with distance and only the two copies closest to the LCR are therefore expressed to an extent that influences color vision (9).
BCM is due to a simultaneous loss of functional LWS and MWS opsins. Three main mutation mechanisms have been described to underlie BCM: 1) deletions or other structural variants (SVs) at the OPN1LW/OPN1MW gene cluster, 2) single or multiple gene copies carrying inactivating point mutations, and 3) single or multiple gene copies carrying rare exon 3 variant haplotypes inducing a splicing defect (2, 3, 10, 11).
Deletions and SVs represent the category of mutations that has been studied least intensively. Except for the initial work by Nathans et al. (2), only a few additional SVs have been mapped at the nucleotide level (12–18), most likely due to the tandem repeat structure of the OPN1LW/OPN1MW gene cluster which hampers mapping of breakpoints located within the repeat sequence.
In this study we investigated the frequency, extent, and precise breakpoint location of SVs underlying BCM in a cohort of 213 genetically confirmed BCM families collected over a period of 25 y. Seventy-three families were shown to carry an SV, and we determined its structure and breakpoints at single-base resolution in all but one family. Forty-two distinct SVs were observed, of which 40 are not reported previously. With this, we quadrupled the number of precisely mapped distinct SVs associated with BCM. This uniquely large collection of BCM-linked SVs not only includes a plethora of unique cases (such as the intrachromosomal gene conversion of an intragenic deletion or an SV undergoing a subsequent loss of a residual OPN1MW gene copy) but also allows us to extract some general features of the structural composition and insights into the mechanisms of nonrecurrent SVs at the OPN1LW/OPN1MW gene cluster. Moreover, our data suggest that such SVs often originate from subjects carrying high-number multicopy gene arrays which are predisposed to undergo genomic rearrangements.
Results
SVs at the OPN1LW/OPN1MW Gene Cluster Are a Common Cause of BCM.
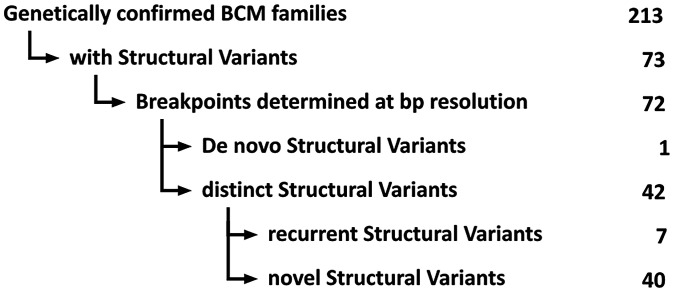
Among our cohort of genetically confirmed BCM families (n = 213) which were collected and genetically investigated over a period of more than 25 y, we identified 73 families (34%) carrying an SV as a cause of the disease (Fig. 1). X-linked inheritance of the condition was commonly reported and documented in the families’ pedigree (Dataset S1). The cosegregation of the SV with the disease was tested and confirmed where affected family members were available. We only observed a single case for which we could establish a de novo event in the lineage of an individual family (BCM 17/SVar10, Table 1) (19).
Fig. 1.
SVs in BCM families: Study overview. Root diagram of the study population and the subcategorization of SVs. Numbers on the right indicate numbers of BCM families in the respective subcategory.
Table 1.
Compilation of SVs at the OPN1LW/OPN1MW gene cluster in BCM families in this study
| Variant | Variant (nomenclature)* | Deletion size | OPN1 copy no.† |
No. of fams‡ | Breakpoint analysis |
Likely SV mechanism§ | Ref. |
|---|---|---|---|---|---|---|---|
| Intragenic deletions | |||||||
| SVar1 | g.[154156356_154156497del; 154193470_154193611del] |
142 bp + 142 bp | 1 x LW 1 x MW |
1 | No homology | NHEJ + gene conversion | This study |
| LCR deletions | |||||||
| SVar2 | g.154140509_154140919del | 411 bp | 1 x LW 2 x MW |
1 | 4bp homology (GGGC) | MMEJ | This study |
| SVar3 | g.154139421_154142962 delins19 |
3,542 bp | 1 x LW 3 x MW |
1 | No homology | Replicative | This study |
| Deletions of the LCR and parts of the OPN1LW/OPN1MW gene cluster | |||||||
| SVar4 | g.154101548_154180938del | 79,391 bp | 3 x MW | 1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar5 | g.154106268_154173412delins AAAC |
67,145 bp | 2 x Hyb 2 x MW |
1 | No homology | NHEJ | This study |
| SVar6 | g.154107176_154212164del | 104,989 bp | 2 x MW | 1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar7 | g.154109499_154145822del | 36,324 bp | 1 x pLW 1 x Hyb 1 x MW |
1 | 3-bp homology (ATC) |
MMEJ | This study |
| SVar8 | g.154109808_154237456del | 127,649 bp | 1 x MW | 1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar9 | g.154112841_154214363del | 101,523 bp | 2 x MW | 2 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar10 | g.154118184_154266255del (de novo) |
148,071 bp | 1 x pMW | 1 | Homeology, Alu elements |
Alu-Alu | 19 |
| SVar11 | g.154120645_154184227 delinsTAGCAGAG |
63,583 bp | 1 x Hyb 3 x MW |
1 | No homology | NHEJ | This study |
| SVar12 | g.154124706_154153479del | 28,774 bp | 1 x pLW 1 x MW |
1 | 2-bp homology (GC) |
NHEJ | This study |
| SVar13 | g.[154125822_154125825del CAGC;154126040_154180538 delinsG] |
4 bp, 54,498 bp |
1 x Hyb 2 x MW |
1 | No homology | Replicative/NHEJ | This study |
| SVar14 | g.154127568_154170730del | 43,163 bp | 1 x Hyb 6 x MW |
1 | No homology | NHEJ | This study |
| SVar15 | g.154127629_154239784del | 112,156 bp | 1 x MW | 1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar16 | g.154130298_154239957del | 109,660 bp | 1 x Hyb 2 x MW |
1 | No homology | NHEJ | This study |
| SVar17 | g.154133406_154186565del | 53,160 bp | 1 x pMW 3 x MW |
1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar18 | g.154134412_154175618del | 41,207 bp | 2 x MW | 1 | 2bp homology (CT) |
NHEJ | This study |
| SVar19 | g.[154135035_154135036ins 26;154135114_154187762del] |
52,649 bp | 1 x pHyb 1 x Hyb 1 x MW |
4 | Insertion homology | Replicative | 14 |
| SVar20 | g.154135236_154235350del | 100,115 bp | 1 x MW | 1 | Homeology, Alu elements |
Alu-Alu | this study |
| SVar21 | g.154136252_154225156del | 88,904 bp | 1 x pMW 1 x MW |
2 | 4-bp homology (GTGC) | MMEJ | 14 |
| SVar22 | g.[154136509_154200716delins CT;154200800_154200814del) |
64,208 bp, 15 bp |
2 x MW | 2 | No homology | Replicative | This study |
| SVar23 | g.154136764_154145987del | 9,224 bp | 1 x pLW 2 x MW |
2 | 3-bp homology (ATC) |
MMEJ | This study |
| SVar24 | g.[154136800_154218125delins 51;154219480_154228735delins GCC] |
81,326 bp, 9,256 bp |
1 x pMW 1 x Hyb |
1 | No homology | Replicative | This study |
| SVar25 | g.154136950_154167537del | 30,588 bp | 1 x Hyb 3 x MW |
1 | 4-bp homology (CCAC) | MMEJ | This study |
| SVar26 | g.154138410_154178979del | 40,570 bp | 3 x Hyb 2 x MW |
22 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar27 | g.154139254_154194420del | 55,167 bp | 1 x pLW 1 x Hyb 1 x MW |
1 | 2-bp homology (CC) |
NHEJ | This study |
| SVar28 | g.154139374_154169145delins 139 |
29,772 bp | 3 x Hyb 10 x MW |
3 | No homology | Replicative | This study |
| SVar29 | g.154139739_154160981del | 21,242 bp | 1 x Hyb 4 x MW |
1 | 4-bp homology (GCTC) |
MMEJ | This study |
| Deletions of parts of the OPN1LW/OPN1MW gene cluster (LCR intact) | |||||||
| SVar30 | g.154143960_154258401del | 114,442 bp | 1 x pMW | 1 | 2-bp homology (AA) |
NHEJ | This study |
| SVar31 | g.154150103_154276841del | 126,739 bp | 1 x pLW | 1 | 3-bp homology (CAT) |
MMEJ | This study |
| SVar32 | g.154150323_154284093del | 133,771 bp | 1 x pLW | 1 | 3-bp homology (GAG) |
MMEJ | This study |
| SVar33 | g.154155559_154271811del | 116,253 bp | 1 x pLW | 1 | No homology | NHEJ | This study |
| Complete OPN1LW/OPN1MW locus deletions | |||||||
| SVar34 | g.154106985_154314620del | 207,636 bp | None | 1 | 3-bp homology (GGA), flanking Alu | MMEJ/ Alu-Alu |
This study |
| SVar35 | g.154113628_154285080del | 171,453 bp | None | 1 | No homology | NHEJ | This study |
| SVar36 | g.154120448_154281284delins 180 |
160,836 bp | None | 1 | Homeology with insertion | Replicative | This study |
| SVar37 | g.[154129722_154137642delins GCACT;154140507_ 154292032del] |
7,921 bp, 151,526 bp |
None | 1 | 4-bp homology (TGGG) | Replicative | This study |
| SVar38 | g.154135236_154273154del | 137,919 bp | None | 1 | Homeology, Alu elements |
Alu-Alu | This study |
| SVar39 | g.154136998_154279848del | 142,851 bp | None | 1 | 2bp homology (GC) |
NHEJ | This study |
| Complex SVs | |||||||
| SVar40 | g.154111506_(154233181_ 154257596)delins [NC_000020.11: g.(pter_2875532)_2895538inv] |
121–146 kb | 1 x MW | 1 | 3-bp homology (CTG), flanking Alu |
MMEJ/ Alu-Alu |
This study |
| SVar41 | g.154128542_154164019delins [g.154150904_154156875inv; AGTGCGG] |
35,377 bp | 1 x pLW 3 x MW |
1 | 5-bp homology (ACTCC) | Replicative | This study |
| SVar42 | g.154143928_154338057delins [394;g.154394091_154480236inv] |
Del: 194 kb, Dup-Inv: 86 kb |
None | 1 | No homology | Replicative | This study |
*Reference sequence: NC_000023.11 if not otherwise stated.
†Number and structure of remaining OPN1LW and OPNMW gene copies: LW, OPN1LW; MW, OPN1MW; Hyb, OPN1MW•OPN1LW hybrid; pLW/pMW, incomplete OPN1LW or OPNMW gene copy.
‡Number of BCM families.
§NHEJ, nonhomologous end joining; MMEJ, microhomology-mediated end joining; Replicative, replication-based mechanism of CNV formation (e.g., MMBIR); Alu-Alu, involving pairs of Alu repeat elements (e.g., Alu-mediated NAHR).
Landscape of SVs at the OPN1LW/MW Gene Cluster in BCM Patients.
Of the 73 independent BCM families that tested positive for an SV in the initial PCR-based screening, we were able to define the exact breakpoint(s) at single-base resolution in all but one family. For breakpoint junction sequences see SI Appendix, Figs. S1–S4. In total, we identified 42 different SVs, of which 40 are not reported previously (Table 1). Seven of the SVs were observed recurrently in more than one family, including one very common deletion observed in 22 families (SVar26; see below).
The 42 distinct SVs can be subclassified into 1) intragenic SVs (n = 1), 2) SVs restricted to the LCR (n = 2), 3) SVs encompassing the LCR and parts of the OPN1LW/OPN1MW gene cluster (n = 27), 4) SVs encompassing parts of the OPN1LW/OPN1MW gene cluster but excluding the LCR (n = 4), 5) SVs encompassing the LCR and the entire OPN1LW/OPN1MW gene cluster (n = 5), and 6) complex SVs (n = 3) (Table 1 and Fig. 2). All SVs involve deletions. Their size ranges from 142 bp to 207 kb with a majority between 20 and 100 kb in size and thus may escape detection by commercial CGH arrays. In fact, a benchmark experiment with DNA samples from three patients with SVs (SVar26, SVar41, and SVar42) using the Cytoscan HD probe array did only correctly call an SV at the OPN1LW/OPN1MW gene cluster for SVar42 (SI Appendix, Table S1).
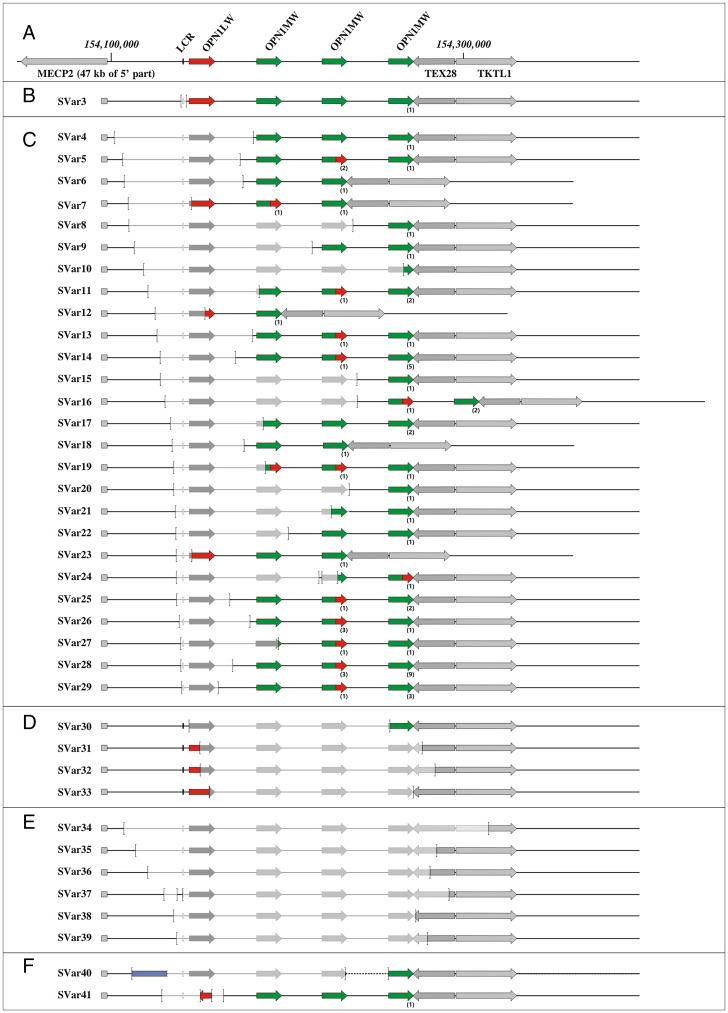
Fig. 2.
Structure, extent, and composition of BCM-linked SVs observed in this study. (A) Map of the OPN1LW/OPN1MW gene array with a single OPN1LW and three downstream OPN1MW gene copies (according to the GRCh38/hg38 genome assembly). The OPN1LW and OPN1MW gene(s) are depicted by red and green arrows, respectively. The LCR is shown as a rectangle upstream of the OPN1LW gene. Flanking genes (MECP2, TEX28, and TKTL1) are shown by gray arrows. (B–F) Categories of BCM-linked SVs including deletions restricted to the LCR (B), SVs covering the LCR and parts of the OPN1LW/OPN1MW gene cluster (C), SVs covering OPN1LW/OPN1MW gene cluster but intact LCR (D), deletions of the OPN1LW/OPN1MW gene cluster (including the LCR) and extending into the downstream TEX28 and TEKTL1 genes (E), and complex structural rearrangements (F). The SV breakpoints are marked by brackets and deleted parts are indicated by lighter gray color. The presence of OPN1MW•OPN1LW hybrid genes is indicated by arrows half-colored in green and red. The blue box in SVar40 represents an interstitial insertion of chromosome 20 sequences. Additional copies of OPN1MW or OPN1MW•OPN1LW hybrid genes are indicated by the number in parentheses below the arrows. Note that the structure of SVar1, SVar2, and SVar42, which cannot be properly displayed at this scale, is displayed in SI Appendix, Figs. S5, S8, and S2, respectively.
The centromeric breakpoints of all BCM-linked SVs are located between MECP2 and the OPN1LW/OPN1MW gene cluster or within the gene cluster, while nine SVs extend further into and disrupt downstream genes, either TEX28 or both TEX28 and TKTL1. Two SVs (SVar24 and SVar37) had single long, contiguous, and correctly oriented segments of OPN1LW/OPN1MW sequence of 1,353 bp and 2,863 bp, respectively, inserted between the proximal and distal breakpoints (SI Appendix, Fig. S1) and were thus formally considered as two consecutive deletions in cis (Table 1). More complex SVs were seen in SVar40, SVar41, and SVar42. SVar40 comprises a deletion of the LCR and large parts of the gene cluster combined with an interstitial insertion of >20 kb of chromosome 20 sequences, SVar41 is a deletion of the LCR and the OPN1LW gene combined with an insertion of a large segment of OPN1LW (exons 2 to 5) in reverse orientation, and SVar42 represents a deletion of the entire OPN1LW/OPN1MW gene cluster and the downstream genes TEX28 and TKTL1 combined with an inverted duplication of >80 kb of downstream sequence including eight genes (RPL10, DNASE1L1, TAZ, ATP6AP1, GDI1, FAM50A, PLXNA, and LAGE3) (SI Appendix, Fig. S2). Notably, the inverted duplication is inserted about 60 kb upstream of its native copy and separated by the nonduplicated EMD and FLNA genes and their flanking low copy inverted sequence repeat which drives the frequent inversion of EMD and FLNA (20).
Breakpoint Sequence Analysis and Molecular Mechanism Underlying SVs.
The majority of BCM-linked SVs were deletions with or without few nucleotides (range 1 bp to 51 bp) inserted at the deletion breakpoint. Besides the more complex SVs with larger contiguous insertions (see above), we also observed five SVs where single (SVar3, SVar24, and SVar36) or multiple short discontiguous fragments (SVar28 and SVar42) of OPN1LW/OPN1MW gene cluster-specific sequences were inserted at the breakpoint junctions in direct or inverted orientation (SI Appendix, Fig. S3). Moreover, we observed three SVs (SVar13, SVar19, and SVar22) with additional small deletions ranging from 4 bp to 26 bp in close vicinity upstream or downstream of the principal SV breakpoint (SI Appendix, Fig. S1). Such small insertions of remnant sequences and close-by indels are typical features of replicative processes underlying SV formation (21, 22).
We also investigated a 300-bp window of sequence upstream and downstream of the breakpoints for sequence homology and the presence of repetitive sequences which may provide clues for the underlying molecular mechanism. From this analysis we inferred about equal proportions of SVs most likely caused by nonhomologous end joining (NHEJ, n = 12), by microhomology-mediated end joining (MMEJ, n = 10), by inter-Alu deletion events (e.g., Alu-based NAHR, n = 10), and by replication-based structural rearrangements such as microhomology-mediated break-induced replication (MMBIR, n = 10) (Table 1) (23, 24).
All but six of the SVs have their centromeric breakpoint located in a 39-kb region between MECP2 and the LCR. We observed some clustering of breakpoints, notably a cluster of six breakpoints in a narrow 750-bp region 7.2 to 8 kb upstream of OPN1LW. This sequence is characterized by a marked increase in GC content from below 25% to higher than 75% (SI Appendix, Fig. S5) and the presence of some predicted distal enhancer elements.
Evidence for the “Spread” of an Intragenic Deletion by Gene Conversion.
In family BCM 262 we detected an intraexonic deletion of 142 bp (c.807_948del) which is deduced to result in a truncated cone opsin lacking important functional domains of the polypeptide. Notably, this 142-bp deletion was present in the proximal OPN1LW gene as well as in the single distal OPN1MW gene copy (Table 1 and SI Appendix, Fig. S4). Such sequence homogenization of mutations in multiple OPN1LW/OPN1MW gene copies in the same gene array has been observed in several families with point mutations (11, 25, 26) and is explained by intrachromosomal gene conversion. The introduction of deleterious variants through gene conversion between paralogs (e.g., a pseudogene and a functional gene copy) usually involves base changes and small indels (27). To the best of our knowledge, the 142-bp deletion observed in this study is the largest reported disease-associated deletion most likely spread through intrachromosomal gene conversion. Mechanistically gene conversion relies on sequence homology between the donor and the recipient sequence. Therefore, gene conversion of a donor sequence containing a deletion in comparison to the recipient is sterically hindering and may involve a larger proportion of flanking homologous sequences.
SVar26 Is a Founder Mutation in BCM Families from the United States.
SVar26 was found recurrently in a total of 22 families, all originating from the United States. We employed haplotype marker analysis using 11 microsatellites encompassing a region of 3.9 Mb in the vicinity of the OPN1LW/OPN1MW gene cluster and demonstrated that all the 22 tested families share a common haplotype covering a physical region of 0.47 Mb (SI Appendix, Fig. S6). Similarly, we observed common haplotypes in families sharing SVar19 (three families investigated, all from the United States) and SVar28 (all three families investigated, all from France), respectively (SI Appendix, Fig. S7).
Do SVs Still Undergo Unequal Homologous Recombination?
NAHR between the highly homologous OPN1LW and OPN1MW gene sequences has been attributed to underlie the variability in gene copy number at this locus as well as the frequent occurrence of OPN1LW•OPN1MW and OPN1MW•OPN1LW hybrid genes (5). Since NAHR relies on the homology between the recombining sequences, we asked whether NAHR can still occur on chromosomes rearranged by SVs. The occurrence of the common SVar26 deletion with multiple remaining OPN1LW/OPN1MW gene copies downstream of the deletion in 22 seemingly independent families provided the opportunity to address this question at a semipopulation level. We determined the copy number of OPN1LW and OPN1MW gene copies in all families carrying SVar26 and observed that there were five gene copies (three OPN1MW•OPN1LW hybrid gene copies and two OPN1MW gene copies as deduced from the multiplex ligation-dependent probe amplification [MLPA] results) in all tested subjects. Given the considerable age of this founder mutation—as suggested by the microsatellite marker data (SI Appendix, Fig. S6)—this finding may be taken as evidence that intrachromosomal NAHR at the OPN1LW/OPN1MW gene locus is suppressed in subjects carrying SVar26.
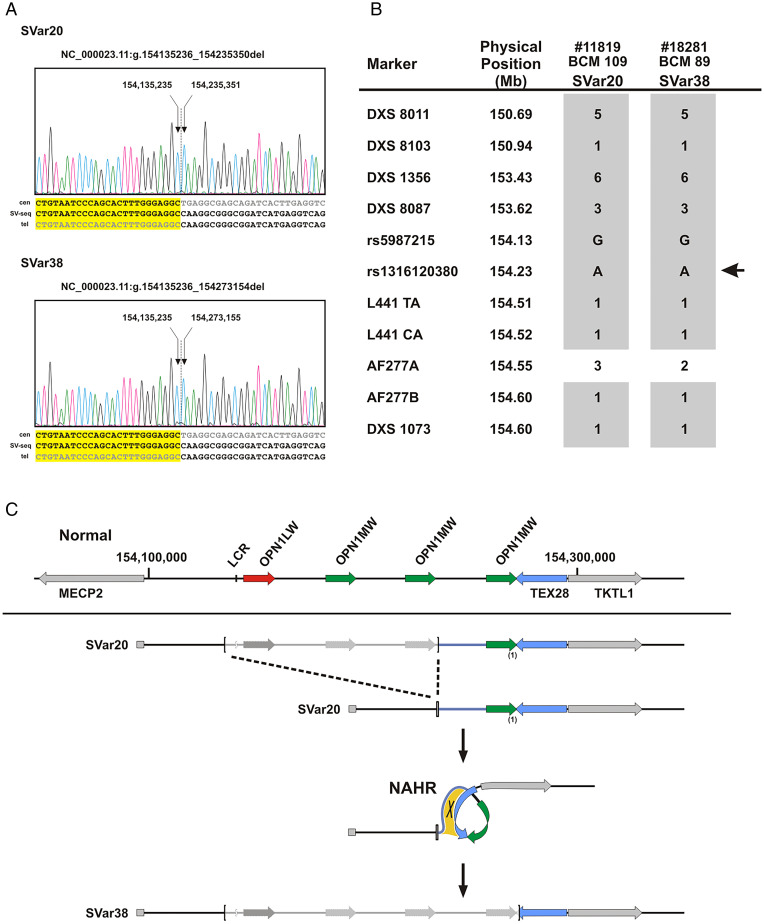
In contrast, we also noted that the centromeric breakpoints of SVar20 and SVar38 were identical while the telomeric breakpoints were fully conserved in sequence but just differ by the presence (in SVar20) or absence (in SVar38) of a single OPN1MW gene copy. Since the independent occurrence of these unique SVs at the homologous nucleotide positions is rather unlikely, we hypothesize that SVar38 derives from SVar20, or vice versa, by a subsequent intrachromosomal or interchromosomal NAHR event. In fact, we observed in these two families, both of German origin, a common haplotype including microsatellite and additional SNP markers strongly supporting our hypothesis that SVar20 and SVar38 belong to one lineage and that SVar20 underwent an intrachromosomal NAHR to give rise to SVar38 (Fig. 3).
Fig. 3.
Identical centromeric breakpoints and sequence-conserved telomeric breakpoints in SVar20 and SVar38 support a single lineage intrachromosomal NAHR event. (A) SVar20 and SVar38 share identical centromeric breakpoints while telomeric breakpoints share the same breakpoint sequence but differ by the presence/absence of a single OPN1MW gene copy. (B) Patient #11819/BCM 109 carrying SVar20 and patient #18281/BCM 89 carrying SVar38 share the same marker haplotype at Xq28. Markers including nine microsatellites and two SNPs are ordered according to their physical position (top to bottom). The localization of the OPN1LW/OPN1MW gene cluster is indicated by the arrow on the right. Shared alleles (microsatellite alleles coded in numbers) are depicted as gray squares. (C) Proposed sequence of events linking SVar20 and SVar38. Deletion of the LCR and parts of the OPN1LW/OPN1MW gene cluster results in SVar20, which retains a single OPN1MW gene copy. Subsequently, SVar20 undergoes intrachromosomal NAHR through homologous sequences downstream of the OPN1MW gene copies (relevant area of sequence homology indicated by the yellow patch; note that the intergenic sequence between OPN1MW gene copies is homologous to large parts of the TEX28 gene) which results in the loss of the terminal OPN1MW gene copy as seen in SVar38. In comparison with the structure of the normal gene cluster (Top), the SV breakpoints are marked by brackets and deleted parts are indicated by lighter gray color.
High OPN1LW/OPN1MW Gene Copy Number May Predispose to the Occurrence of SVs.
Given the high prevalence of SVs in BCM we asked whether certain features of the OPN1LW/OPN1MW gene cluster predispose to the occurrence of SVs. A special characteristic of the OPN1LW/OPN1MW gene cluster is its repetitive nature with multiple copies of genes and intergenic sequences with very high sequence homology, a feature which is known to induce nonallelic recombination events. We therefore investigated whether a large copy number (prior to the SV event) may predispose to the occurrence of nonrecurrent SVs. For this purpose, we determined the copy number of remaining OPN1LW and OPN1MW genes in subjects with SVs utilizing qPCR and MLPA. We used these data to deduce—by parsimonious assumptions—the minimal number of copies of the individual ancestral array prior to the occurrence of the SV event. For instance, for a subject with a single full or partially intact OPN1MW copy we conservatively assumed an ancestral copy number of two (one OPN1LW and one OPN1MW gene copy). We excluded SVar1, the intragenic 142-bp deletion which implicates gene conversion, as an underlying mechanism as well as all SVs involving deletions of the entire gene locus or all OPN1MW genes which prohibits ancestral copy number reconstruction.
We compared the data from the BCM-SV group (n = 33; i.e., distinct SVs) with those from a group of color vision-normal observers of German descent (CVNO, n = 35) (Table 2).
Table 2.
Deduced total OPN1LW/OPN1MW copy number prior to SV in comparison with healthy controls
| ∑ OPN1LW and OPN1MW copies | No. of BCM (prior to SV)* | No. of controls† | ||
|---|---|---|---|---|
| n = 2 | 7 | 16 | 5 | 26 |
| n = 3 | 9 | 21 | ||
| n = 4 | 7 | 17 | 3 | 9 |
| n = 5 | 6 | 5 | ||
| n = 6 | 2 | 1 | ||
| n = 7 | 0 | 0 | ||
| n = 8 | 1 | 0 | ||
| .. | 0 | 0 | ||
| n = 14 | 1 | 0 | ||
| Sum | 33 | 35 | ||
*Number of deduced ancestral gene arrays (prior to the SV formation) with a given total OPN1LW and OPN1MW copy number among BCM-linked SVs.
†Number of subjects with experimentally determined total OPN1LW and OPN1MW copy number in healthy controls. Shadings distinguish groups of subjects with three or fewer and more than three OPN1LW/MW gene copies, respectively.
Mean total copy number was 3.31 for the CVNO cohort and 4.03 for the BCM-SV group. However, this difference in mean total copy number is in parts inflated due to the very large gene arrays (reconstructed with n = 8 and n = 14 copies) in SVar14 and SVar28 in the BCM-SV group. Irrespective of these outliers, the distribution of the total copy number was shifted to higher copy numbers in the BCM-SV group with a statistically significant increase in the fraction of subjects with three or more gene copies in the BCM-SV group (chi-squared test, P = 0.028) and a reduced fraction of OPN1LW/OPN1MW arrays with three gene copies in the BCM-SV group (chi-squares test, P = 0.0065), with this statistical approach being rather robust against outliers. Moreover, we noted that a significant higher proportion of arrays in the BCM-SV group carry OPN1MW•OPN1LW hybrid genes (15/33, 44%) compared to the CVNO group (3/35, 8.5%). We thus hypothesize that large OPN1LW/OPN1MW gene arrays are less stable, i.e., prone to NAHR and the formation of OPN1MW•OPN1LW hybrid genes, and predisposed for the occurrence of deleterious nonrecurrent SVs.
Discussion
SVs account for 34% of the BCM index cases in our cohort of 213 molecularly confirmed families. There are only a few other genes or loci underlying inherited retinal dystrophies with a similar or exceeding fraction of SVs in their mutation spectrum, such as CHM, EYS, PRPF31, and CLN3, the latter due to the high prevalence of a founder mutation (see also ref. 28) or in a few retinal dystrophies in which SVs are the primary disease mechanisms due to chromatin topology disorganization (MCDR3, ref. 29; RP17-linked Retinitis pigmentosa, ref. 30; autosomal dominant cone dystrophy with early tritan color vision defect, ref. 31).
SVs associated with BCM have been reported in a number of prior studies (2, 3, 12–18, 32–41). However, to the best of our knowledge, exact breakpoints of the SVs were only reported in a rather small subset of only 14 BCM families, all carrying different SVs except for an SV shared in two families (17). This scarce information on exact breakpoints is likely due to the complexity and repetitive structure of the OPN1LW/OPN1MW gene cluster that complicates and hampers molecular analysis. In the present study, we determined the exact breakpoints in 72 additional independent BCM families. We identified 42 different SVs of which 40 are not reported previously. Our study thereby increases the number of BCM families with precisely mapped SVs sixfold (from 14 to 86) and the number of different SVs by a factor of 4 (from 13 to 53). The knowledge of the precise SV breakpoints enabled us to design diagnostic PCR assays (primer sequences in SI Appendix, Table S2), which now allow easy and reliable female carrier testing in families at risk. This obviates the need for qPCR-based copy number analysis of LCR sequences or—for SVs with intact LCR—indirect marker-based segregation analysis in BCM families which are typically used for this purpose until now.
The LCR upstream of the OPN1LW/OPN1MW gene cluster governs the expression of the downstream cone opsin genes (6, 7). Its absence (e.g., due to deletions) strongly impairs OPN1LW and OPN1MW gene expression and results in BCM. This offers the opportunity to indirectly map the extent of functionally relevant sequences of the human LCR in BCM patients with SVs. In the present patient series, we identified two deletions that were restricted to the LCR: SVar3 (in family BCM 215) with a deletion of 3,542 bp and SVar2 (in family BCM 74) with a deletion of 411 bp. The latter is the smallest LCR deletion ever reported in BCM. Together with an upstream breakpoint (HS102) reported by Nathans et al. (2), the present data refine the minimal crucial sequence of the LCR to 358 bp which covers the evolutionarily highest conserved sequence element in this region (SI Appendix, Fig. S8). Given the utility of the LCR/OPN1LW promoter (6, 42), our refinement may be important for the further development of more compact promoters to drive transgene expression in gene therapy applications aiming for strong cone photoreceptor-specific expression.
The vast majority of SVs in the present study were unique and only observed in single families. However, we also identified one SV, SVar26, which was rather common and accounted for about 30% of all BCM families with SVs in the present study. Remarkably, SVar26 was found exclusively in families from the United States and our marker analysis confirmed the presence of a founder mutation.
The size of the deletions and the localization of breakpoints of the SVs vary considerably. While the centromeric breakpoint is always located downstream of MECP2, the extent of deletion at the telomeric side can include parts of or the entire TEX28 and TKTL1 genes downstream of the OPN1LW/OPN1MW gene cluster. Notably, there is not a single region of overlap shared by all SVs. Although the majority of SVs did involve deletions of the LCR, it is still intact in five of the SVs. Therefore, multiple probes targeting different parts of the OPN1LW/OPNMW gene cluster are required to be tested for reliable detection or exclusion of SVs linked to BCM.
Thirty-two of the SVs have at least one of the breakpoints located within the OPN1LW/OPN1MW gene cluster which represents a tandem arrangement of highly homologous sequences of 37- to 38-kb unit size composed of genic (OPN1LW or OPN1MW or OPN1LW•OPN1MW hybrid genes) and intergenic sequences. Such low copy repeat arrangements are frequently involved in the formation of recurrent copy number variations but also favor the occurrence of nonrecurrent SVs such as those reported in this study (24). In line with this, telomeric breakpoints of nonrecurrent duplications and complex SVs observed in patients with MECP2 duplication syndrome are frequently located in the OPN1LW/OPN1MW gene cluster (43–45). Such a predisposition to genomic rearrangements may further be enhanced by an increased number of repeats. Therefore, a major finding of this study is the evidence that BCM-linked SVs may occur most frequently in subjects with larger than average OPN1LW/OPN1MW gene arrays. Or, conversely, subjects with large arrays are at higher risk for the formation of a deleterious SV. While we were unable to determine the ancestral copy number (prior to the occurrence of the SV) experimentally, we applied very conservative and parsimonious assumptions for the “reconstruction” of the composition of the ancestral gene array. Thus, on average, the true OPN1LW/OPN1MW copy number on the ancestral chromosomes likely had been even higher.
There is some prior evidence from human disease that a higher copy number of low copy repeat sequences predisposes to genomic rearrangements. For instance, Liu et al. observed that triplication at the CMT1A locus emerges from duplications (three copies of the CMT1A low copy repeat) at a much higher rate than the rate of de novo duplications (46). Moreover, duplication and subsequent triplication of the SCNA gene in different branches of the same family with autosomal dominant parkinsonism (47) and the unique presence of PRSS1 duplications and triplications in French families with hereditary pancreatitis that originate from a single founder allele (48) argue for an increased susceptibility for genomic rearrangement. Notably, in all three instances these changes in copy number involve intrachromosomal (i.e., interchromatidal recombination) rather than interchromosomal NAHR events suggesting some inherent instability of a chromosome carrying multiple copies of a low copy repeat sequence. All these studies involve a further gain in copy number. Yet, this likely represents an ascertainment bias since reversion events (e.g., a duplication reverted to normal single copy) will hardly be detected in such patient studies. In contrast, a predominance of copy losses over copy gains has already been noted in the rates of mutations and revertants at the Drosophila bar locus (e.g., bar to ultrabar, and vice versa) (49, 50), a prototypical locus for dosage effects due to changes in copy number (51, 52), and also observed in humans at several loci for genomic disorders upon sperm genotyping (53) as well as in the frequency of reciprocal recurrent deletions/duplication in the allelic Smith–Magenis syndrome and Potocki–Lupski syndrome (54). This predominance of deletions is at least in part due to intrachromosomal (i.e., intrachromatidal or interchromatidal) NAHR which is equal or higher than interchromosomal NAHR at all tested loci (53). Other than in these instances, our data suggest that also nonrecurrent SVs involving deletions are driven by an inherent increased instability of large, high-copy gene arrays at the OPN1LW/OPN1MW gene cluster.
Although further studies are recommended for validation, such an increased instability of large arrays may explain the high proportion of SVs in subjects with BCM.
Subjects and Materials and Methods
Patient Recruitment and Clinical Evaluation.
The study was conducted pro- and retrospectively in accordance with the tenets of the World Medical Association Declaration of Helsinki and approval was obtained from the respective local research and ethical boards or dependent on the local regulatory bodies at the time the patients were recruited as part of local clinical studies or ad hoc at different centers specialized in inherited retinal diseases during routine clinical diagnostics. A clinical diagnosis of BCM was based on ophthalmologic examination according to local protocols or local clinical standards of the recruiting centers. Venous blood was taken from patients and family members after informed consent and sent to the Tuebingen group for genetic analysis. Specifically, this study was approved by the Ethics Board of the Medical Faculty, Eberhard Karls University Tuebingen under the study no. 349/2003V and 116/2015BO2.
Genotyping of the OPN1LW/OPN1MW Gene Cluster.
Genomic DNA was isolated from blood samples according to standard procedures. All patients underwent a routine screening protocol to test for the structure and integrity of the OPN1LW/OPN1MW gene cluster which includes PCR-based sequence-tagged site (STS) content mapping for amplicons covering the LCR (amplicon A), the proximal promoter and exon 1 of the OPN1LW gene and the OPN1MW gene, respectively (amplicons B and C), and amplicons D and E covering exon 4 and exon 5 not distinguishing between OPN1LW and OPN1MW (32). Each PCR assay for amplicons A to C was performed as duplex PCR including an additional primer pair for an autosomal locus as amplification control. The specificity of the screening protocol was validated in 50 males of European origin with normal color vision.
Subjects with evidence for deletions or rearrangements from the initial screen were further investigated to delineate the structure, extent, and the breakpoints of the SVs. As a first step, we applied STS content mapping in unique sequences outside the OPN1LW/OPN1MW sequence repeat to refine centromeric breakpoints between MECP2 and OPN1LW, and where possible also the telomeric breakpoint downstream of the gene cluster. To cover breakpoint junctions, we used multiple approaches including 1) long-distance PCR with primers as defined by refined breakpoint mapping, 2) long-distance PCRs with a forward primer as defined by refined breakpoint mapping and a panel of reverse primers located at distinct sites within the OPN1LW/OPN1MW gene cluster, 3) genome walking applying the APAgene Gold Genome Walking Kit (Bio S&T), and 4) inverse PCR (55).
Breakpoint junction PCR and long-distance PCR fragments were sequenced by primer walking using BigDye Terminator Cycle Sequencing 1.1 chemistry (Thermo Fisher Scientific GmbH) and sequencing products electrophoretically separated on an ABI 3130XL capillary sequencer instrument. Breakpoint junction sequences were used to design primers for SV-specific diagnostic PCR assays which were used for segregation analysis and carriership detection (SI Appendix, Table S2).
The copy number of inserted chromosome 20 sequences in SVar40, and duplicated-inverted X-chromosomal sequences in SVar42 was determined by custom designed real-time qPCR assays using SYBR Green chemistry (Quantitect SYBR Green PCR Kit; Qiagen) on an ABI 7500 instrument. Prior to the copy number assay, DNA samples of patients and controls were adjusted and normalized based on Ct values obtained with a custom copy number control assay targeting a sequence at the SDC4 locus on chromosome 20q.
The total number of OPN1LW/OPN1MW gene copies was determined by means of qPCR or MLPA with genomic DNA as template. For qPCR, the amount of input DNA from probands and controls was adjusted based on the result obtained with a RNaseP TaqMan Copy Number Reference Assay (Life Technologies). We used two different TaqMan assays that target different parts of the OPN1LW/OPN1MW gene cluster: the HS_01912094 assay (Thermo Fisher/Life Technologies) targeting a common sequence in exon 6 of OPN1LW and OPN1MW and a custom-designed TaqMan assay (employing RGCP_TQF [5′-CCCAACAGAAAGCTGAAAGC-3′] as forward and RGCP_TQR [5′-GTGCAAAACTTTCGGATTGG-3′] as reverse primers, respectively, and RGCP_TQP [5′-CAGCCCGAGTCCTGCCATTGG-3′] with 5′-FAM and 3′-BHQ1 modifications as probe primer) targeting a common segment of intron 1. The two qPCR assays targeting OPN1LW and OPN1MW sequences and another RNaseP TaqMan Copy Number Reference Assay—performed as triplicates for each sample—were done in parallel on the same instrument run.
For MLPA we used a premarketing release version of the SALSA X080-B1 Opsin probe-mix (MRC Holland) targeting sequences of the LCR, and individual exons of OPN1LW and OPN1MW specifically. MLPA was performed according to the manufacturer’s recommendations and amplification products separated by capillary electrophoresis on an ABI 3130XL instrument. Data analysis was done using the Coffalyser.Net software (MRC Holland) which calculates ratios in comparison to reference samples coprocessed with the test samples. For both qPCR and MLPA, we used a series of male controls with defined OPN1LW/OPN1MW copy number (ranging from one to six copies) as determined by Southern blot of NotI-digested and pulsed-field electrophoretic separated genomic DNA and/or Fiber-FISH (56), to generate copy number-dependent probe/control ratio and ΔCt calibration curves, respectively.
In Silico Analysis of SV Breakpoints.
We performed in silico sequence analysis of 600 bp in the vicinity of the SV breakpoints (300 bp upstream and 300 bp downstream of the actual breakpoint). We used RepeatMasker for the identification of repetitive sequences and performed a BLAST2seq analysis of the upstream and downstream breakpoint sequences for the detection of sequence homologies. Local G/C content was calculated with a sliding window of 50 bp.
Microsatellite Markers.
Eleven microsatellite markers (including five markers from public databases and six novel designed CA- and TA-repeat markers) encompassing a 3.9-Mb region on chromosome Xq28 were used for haplotyping. Microsatellite markers were PCR-amplified from genomic DNA using AmpliTaq Gold (Thermo Fisher) or Multiplex PCR Master Kit (Qiagen) reagents and—owing to fluorescence labeling of primers—separated and detected on an ABI3130-XL capillary electrophoresis instrument.
Array-CGH Analysis.
Array-CGH analysis applying the Affymetrix Cytoscan HD array was done at Atlas Biolabs GmbH, Berlin, Germany.
Web Resources.
Resources used in this study include BLAST2Seq (https://blast.ncbi.nlm.nih.gov/Blast.cgi), GenBank (https://www.ncbi.nlm.nih.gov/genbank/), HGMD (www.hgmd.cf.ac.uk/), OMIM (https://omim.org/), and RepeatMasker (https://www.repeatmasker.org/).
Supplementary Material
Acknowledgments
We thank all families for participation in this study and Miss Sabine Tippmann and the late Miss Monika Papke for technical assistance in the initial phase of this project. We also want to thank the late Christian Hamel for the excellent and amicable cooperation before his much-too-early passing. The work was supported by a grant of the German Research Council (Wi1189/12-1 to B.W.), the Blue Cone Monochromacy Family Foundation, and Dr. Renata Sarno as well as in parts by funds of the UK Medical Research Council (to A.J.H.) and a grant from the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology (to M.M.). Patient clinical care and recruitment in Tuebingen was supported by the Tistou and Charlotte Kerstan Foundation. The Parisian group (I.A.) was supported by LabEx LifeSenses, IHU FOReSIGHT, and a Fondation Fighting Blindness grant (BR-GE-0619-0761-INSERM). DNA samples collected by the Parisian group originate from NeuroSensCol, a biobank for research in neuroscience (Principal Investigator: J. A. Sahel; co-Principal Investigator: I.A., partner with Centre Hospitalier National d’Ophtalmologie des Quinze-Vingts, INSERM and CNRS). Katarina Stingl, Isabelle Audo, Isabelle Meunier, Michel Michaelides, Helene Dollfus, Birgit Lorenz, Markus Preising, Eberhart Zrenner, and Susanne Kohl are members of ERN-EYE (European Reference Network for Rare Eye Diseases).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115538119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Aboshiha J., Dubis A. M., Carroll J., Hardcastle A. J., Michaelides M., The cone dysfunction syndromes. Br. J. Ophthalmol. 100, 115–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathans J., et al. , Molecular genetics of human blue cone monochromacy. Science 245, 831–838 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Nathans J., et al. , Genetic heterogeneity among blue-cone monochromats. Am. J. Hum. Genet. 53, 987–1000 (1993). [PMC free article] [PubMed] [Google Scholar]
- 4.Nathans J., Thomas D., Hogness D. S., Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science 232, 193–202 (1986a). [DOI] [PubMed] [Google Scholar]
- 5.Nathans J., Piantanida T. P., Eddy R. L., Shows T. B., Hogness D. S., Molecular genetics of inherited variation in human color vision. Science 232, 203–210 (1986b). [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., et al. , A locus control region adjacent to the human red and green visual pigment genes. Neuron 9, 429–440 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Smallwood P. M., Wang Y., Nathans J., Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc. Natl. Acad. Sci. U.S.A. 99, 1008–1011 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng G. H., Chen S., Active opsin loci adopt intrachromosomal loops that depend on the photoreceptor transcription factor network. Proc. Natl. Acad. Sci. U.S.A. 108, 17821–17826 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winderickx J., Battisti L., Motulsky A. G., Deeb S. S., Selective expression of human X chromosome-linked green opsin genes. Proc. Natl. Acad. Sci. U.S.A. 89, 9710–9714 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner J. C., et al. , Three different cone opsin gene array mutational mechanisms with genotype-phenotype correlation and functional investigation of cone opsin variants. Hum. Mutat. 35, 1354–1362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buena-Atienza E., et al. , De novo intrachromosomal gene conversion from OPN1MW to OPN1LW in the male germline results in Blue Cone Monochromacy. Sci. Rep. 6, 28253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladekjaer-Mikkelsen A. S., Rosenberg T., Jørgensen A. L., A new mechanism in blue cone monochromatism. Hum. Genet. 98, 403–408 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Gardner J. C., et al. , Blue cone monochromacy: Causative mutations and associated phenotypes. Mol. Vis. 15, 876–884 (2009). [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll J., et al. , Deletion of the X-linked opsin gene array locus control region (LCR) results in disruption of the cone mosaic. Vision Res. 50, 1989–1999 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizrahi-Meissonnier L., Merin S., Banin E., Sharon D., Variable retinal phenotypes caused by mutations in the X-linked photopigment gene array. Invest. Ophthalmol. Vis. Sci. 51, 3884–3892 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Katagiri S., et al. , Genotype determination of the OPN1LW/OPN1MW genes: Novel disease-causing mechanisms in Japanese patients with blue cone monochromacy. Sci. Rep. 8, 11507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatsenko S. A., et al. , High-resolution microarray analysis unravels complex Xq28 aberrations in patients and carriers affected by X-linked blue cone monochromacy. Clin. Genet. 89, 82–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C., et al. , Novel OPN1LW/OPN1MW deletion mutations in 2 Japanese families with blue cone monochromacy. Hum. Genome Var. 3, 16011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buena-Atienza E., Nasser F., Kohl S., Wissinger B., A 73,128 bp de novo deletion encompassing the OPN1LW/OPN1MW gene cluster in sporadic Blue Cone Monochromacy: A case report. BMC Med. Genet. 19, 107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Small K., Iber J., Warren S. T., Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat. Genet. 16, 96–99 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Carvalho C. M., et al. , Replicative mechanisms for CNV formation are error prone. Nat. Genet. 45, 1319–1326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhokarh D., Abyzov A., Elevated variant density around SV breakpoints in germline lineage lends support to error-prone replication hypothesis. Genome Res. 26, 874–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G., Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho C. M., Lupski J. R., Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyniers E., et al. , Gene conversion between red and defective green opsin gene in blue cone monochromacy. Genomics 29, 323–328 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Gardner J. C., et al. , X-linked cone dystrophy caused by mutation of the red and green cone opsins. Am. J. Hum. Genet. 87, 26–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casola C., Zekonyte U., Phillips A. D., Cooper D. N., Hahn M. W., Interlocus gene conversion events introduce deleterious mutations into at least 1% of human genes associated with inherited disease. Genome Res. 22, 429–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Schil K., et al. ; CNV Study Group, Mapping the genomic landscape of inherited retinal disease genes prioritizes genes prone to coding and noncoding copy-number variations. Genet. Med. 20, 202–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cipriani V., et al. , Duplication events downstream of IRX1 cause North Carolina macular dystrophy at the MCDR3 locus. Sci. Rep. 7, 7512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Bruijn S. E., et al. , Structural variants create new topological-associated domains and ectopic retinal enhancer-gene contact in dominant retinitis pigmentosa. Am. J. Hum. Genet. 107, 802–814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohl S., et al. , A duplication on chromosome 16q12 affecting the IRXB gene cluster is associated with autosomal dominant cone dystrophy with early tritanopic color vision defect. Hum. Mol. Genet. 30, 1218–1229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cideciyan A. V., et al. , Human cone visual pigment deletions spare sufficient photoreceptors to warrant gene therapy. Hum. Gene Ther. 24, 993–1006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichel E., Bruce A. M., Sandberg M. A., Berson E. L., An electroretinographic and molecular genetic study of X-linked cone degeneration. Am. J. Ophthalmol. 108, 540–547 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Feil R., et al. , Adrenoleukodystrophy: A complex chromosomal rearrangement in the Xq28 red/green-color-pigment gene region indicates two possible gene localizations. Am. J. Hum. Genet. 49, 1361–1371 (1991). [PMC free article] [PubMed] [Google Scholar]
- 35.Ayyagari R., et al. , Bilateral macular atrophy in blue cone monochromacy (BCM) with loss of the locus control region (LCR) and part of the red pigment gene. Mol. Vis. 5, 13 (1999). [PubMed] [Google Scholar]
- 36.Ayyagari R., et al. , Spectrum of color gene deletions and phenotype in patients with blue cone monochromacy. Hum. Genet. 107, 75–82 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Eksandh L., Kohl S., Wissinger B., Clinical features of achromatopsia in Swedish patients with defined genotypes. Ophthalmic Genet. 23, 109–120 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Kellner U., et al. , Blue cone monochromatism: Clinical findings in patients with mutations in the red/green opsin gene cluster. Graefes Arch. Clin. Exp. Ophthalmol. 242, 729–735 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Michaelides M., et al. , Blue cone monochromatism: A phenotype and genotype assessment with evidence of progressive loss of cone function in older individuals. Eye (Lond.) 19, 2–10 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Carroll J., et al. , The effect of cone opsin mutations on retinal structure and the integrity of the photoreceptor mosaic. Invest. Ophthalmol. Vis. Sci. 53, 8006–8015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo X., et al. , Blue cone monochromacy: Visual function and efficacy outcome measures for clinical trials. PLoS One 10, e0125700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye G. J., et al. , Cone-specific promoters for gene therapy of achromatopsia and other retinal diseases. Hum. Gene Ther. 27, 72–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Esch H., et al. , Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 77, 442–453 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho C. M., et al. , Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 18, 2188–2203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho C. M., et al. , Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 43, 1074–1081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P., et al. , Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 94, 462–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs J., et al. , Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology 68, 916–922 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Chauvin A., et al. , Elucidation of the complex structure and origin of the human trypsinogen locus triplication. Hum. Mol. Genet. 18, 3605–3614 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Zeleny C., The direction and frequency of mutation in the bar-eye series of multiple allelomorphs of Drosophila. J. Exp. Zool. 34, 202–233 (1921). [Google Scholar]
- 50.Sturtevant A. H., Morgan T. H., Reverse mutation of the BAR gene correlated with crossing over. Science 57, 746–747 (1923). [DOI] [PubMed] [Google Scholar]
- 51.Bridges C. B., The BAR “gene” a duplication. Science 83, 210–211 (1936). [DOI] [PubMed] [Google Scholar]
- 52.Tsubota S. I., Rosenberg D., Szostak H., Rubin D., Schedl P., The cloning of the Bar region and the B breakpoint in Drosophila melanogaster: Evidence for a transposon-induced rearrangement. Genetics 122, 881–890 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner D. J., et al. , Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 40, 90–95 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P., et al. , Frequency of nonallelic homologous recombination is correlated with length of homology: Evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 89, 580–588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochman H., Gerber A. S., Hartl D. L., Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf S., et al. , Direct visual resolution of gene copy number in the human photopigment gene array. Invest. Ophthalmol. Vis. Sci. 40, 1585–1589 (1999). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.