Significance
Many cetacean species can dive to extraordinary depths on a single breath, but the evolutionary origins of deep-sea foraging in ancestral cetaceans remain unclear. We present a resurrected ancestral cetacean visual protein (rhodopsin) to investigate this critical innovation from a visual ecological perspective. Our results indicate that the ancestor of modern cetaceans was a deeper diver; we discovered both a deep-sea spectral shift and accelerated retinal kinetics over the terrestrial-to-aquatic transition. These findings suggest that ancient whales were active at mesopelagic depths and had evolved a faster dark adaptation rate, a trait that allows diving mammals to rapidly adjust to dimming light. As such, our study provides compelling evidence for deep-diving behavior before the divergence of toothed and baleen whales.
Keywords: cetacean evolution, ancestral sequence reconstruction, metarhodopsin II, retinal release, spectral tuning
Abstract
Cetaceans are fully aquatic mammals that descended from terrestrial ancestors, an iconic evolutionary transition characterized by adaptations for underwater foraging via breath-hold diving. Although the evolutionary history of this specialized behavior is challenging to reconstruct, coevolving sensory systems may offer valuable clues. The dim-light visual pigment, rhodopsin, which initiates phototransduction in the rod photoreceptors of the eye, has provided insight into the visual ecology of depth in several aquatic vertebrate lineages. Here, we use ancestral sequence reconstruction and protein resurrection experiments to quantify light-activation metrics in rhodopsin pigments from ancestors bracketing the cetacean terrestrial-to-aquatic transition. By comparing multiple reconstruction methods on a broadly sampled cetartiodactyl species tree, we generated highly robust ancestral sequence estimates. Our experimental results provide direct support for a blue-shift in spectral sensitivity along the branch separating cetaceans from terrestrial relatives. This blue-shift was 14 nm, resulting in a deep-sea signature (λmax = 486 nm) similar to many mesopelagic-dwelling fish. We also discovered that the decay rates of light-activated rhodopsin increased in ancestral cetaceans, which may indicate an accelerated dark adaptation response typical of deeper-diving mammals. Because slow decay rates are thought to help sequester cytotoxic photoproducts, this surprising result could reflect an ecological trade-off between rod photoprotection and dark adaptation. Taken together, these ancestral shifts in rhodopsin function suggest that some of the first fully aquatic cetaceans could dive into the mesopelagic zone (>200 m). Moreover, our reconstructions indicate that this behavior arose before the divergence of toothed and baleen whales.
The first cetaceans in the fossil record (archaeocetes) iconically document major morphological adaptations for increased underwater activity throughout the late Eocene (1). The closest living relatives to cetaceans are the hippopotamids, which together form the clade Whippomorpha within the order Cetartiodactyla (1, 2) (Fig. 1). Arguably, the most impressive adaptations in ancestral cetaceans arose from the need to support foraging with breath-hold diving. Nevertheless, the evolution of this specialization, particularly when cetaceans began to exploit the aphotic zones of the ocean (beyond ∼200 m), remains unclear (3). The common ancestor of crown cetaceans (Neoceti) likely resembled some of the first fully aquatic archaeocetes, which had a dolphin-like body, including tail flukes and vestigial hind limbs, but the diving behavior of these animals is uncertain (4, 5).
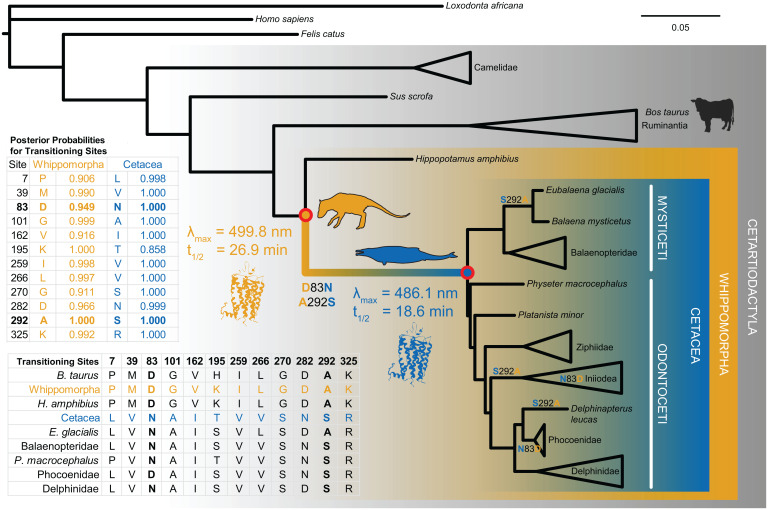
Fig. 1.
Phylogeny of cetartiodactyl rhodopsin coding sequences and reconstructed terrestrial-to-aquatic transitioning residues. We used codon-based likelihood models of evolution to infer the rhodopsin coding sequence at internal nodes of the tree (Cetacea and Whippomorpha). The reconstructed sequences had high posterior probabilities and revealed 12 transitioning residues. We then synthesized and expressed the ancestral genes in vitro via transfection into mammalian tissue culture cells for experimental tests of evolutionary hypotheses. We found significant shifts in both spectral tuning (λmax) and meta II decay (retinal release t1/2) along the transitional branch, caused by substitutions D83N and A292S. The tree topology shown is based on established species relationships, where branch lengths are proportional to the number of nucleotide substitutions per codon (see Materials and Methods and SI Appendix for further details).
Sensory systems can provide important evidence for investigating major environmental transitions because of their critical role in mediating the daily activities of an organism. The visual pigment rhodopsin is a model G protein–coupled receptor (GPCR) responsible for dim-light vision in most vertebrates (6). Found in rod photoreceptors, rhodopsin is a transmembrane apoprotein with a covalently bound vitamin A chromophore, 11-cis-retinal. Light absorption initiates the phototransduction cascade by isomerizing the chromophore to all-trans retinal, which triggers a conformational change in the rhodopsin apoprotein to the active photo-intermediate, metarhodopsin II (meta II) (7). Experimental studies across model and nonmodel systems have demonstrated adaptive shifts in rhodopsin light activation occurring with changes in the light environment over major evolutionary transitions (8–15).
Like other aquatic vertebrates, most cetacean rhodopsins have blue-shifted spectral sensitivity (λmax), presumably to better match the predominant light spectrum in ocean water (16–19). Several studies have also demonstrated that λmax decreases predictably for deeper-dwelling fish species (12, 20, 21), and this relationship appears to be true for diving mammals as well (18, 19, 22, 23). Comparative experimental work from our group and others has revealed that spectral blue-shifts between model cetacean rhodopsins and terrestrial outgroups arise from amino acid substitutions at positively selected sites near the chromophore (D83N and A292S) (11, 18, 24). Moreover, we found in a previous study that evolutionary rates in cetacean rhodopsin correlate with species foraging depth (11).
In ancestral cetaceans, however, the precise nature and extent of adaptive change in rhodopsin function remains unclear. Because D83N and A292S have blue-shifting effects in extant rhodopsin models, a spectral blue-shift of uncertain magnitude was long assumed to characterize the terrestrial-to-aquatic transition (25–27). Nevertheless, this hypothesis has not been tested experimentally in the context of the ancestral rhodopsins in which these substitutions are thought to have occurred. More recent studies also highlight the influence of retinal release and light-activated rhodopsin (meta II) decay on dark adaptation in the retina (15, 28, 29), a physiological response of critical importance to diving mammals (30). However, the only study of these mechanisms in a cetacean rhodopsin to date showed that they are affected by intramolecular epistasis (24); therefore, their relevance to ancestral cetaceans is unknown.
Here, we determine functional shifts in rhodopsin over the transitional branch of Cetacea using ancestral sequence reconstruction (ASR) and protein synthesis (sometimes called “resurrection”) to discuss implications for the evolution of visual ecology and diving. This interdisciplinary approach merges computational analyses with laboratory experiments to investigate potentially adaptive molecular evolutionary scenarios. Specifically, ancestral genetic backgrounds allow hypotheses about protein functional evolution to be directly tested in vitro through mutation experiments that replicate substitutions along an ancestral lineage (31, 32) (Fig. 1). Rhodopsin is one of many proteins for which considerable evolutionary insight has been gained through ASR in other vertebrate systems (8, 15, 33–35). The cetacean terrestrial-to-aquatic transition is widely recognized as a promising model for investigating the molecular foundations of macroevolutionary change, yet ancestral protein resurrection remains an underutilized approach in cetacean evolutionary research (36).
We investigate whether the cetacean visual system evolved adaptations for aphotic zone foraging before the divergence of the two modern sister clades, Odontoceti (toothed whales) and Mysticeti (baleen whales). To test this hypothesis, we use several ASR methods to infer the nucleotide and amino acid sequences of rhodopsin in both the Cetacea and Whippomorpha common ancestors (Fig. 1). Because no genetic material has as yet been recovered from transitional cetacean fossil specimens, these two nodes represent the narrowest time span (∼20 My) for bracketing the terrestrial-to-aquatic transition, which occurred between ∼55 and 35 Mya (1, 2). We then express and mutate the ancestral pigments to determine whether spectral tuning and meta II decay profiles (evaluated with absorbance and fluorescence spectroscopy, respectively) shift toward deep-sea signatures in the Cetacea ancestor.
Results
Ancestral Rhodopsin Expression Reveals Functional Shifts in Spectral Tuning and Meta II Decay during the Terrestrial-to-Aquatic Transition.
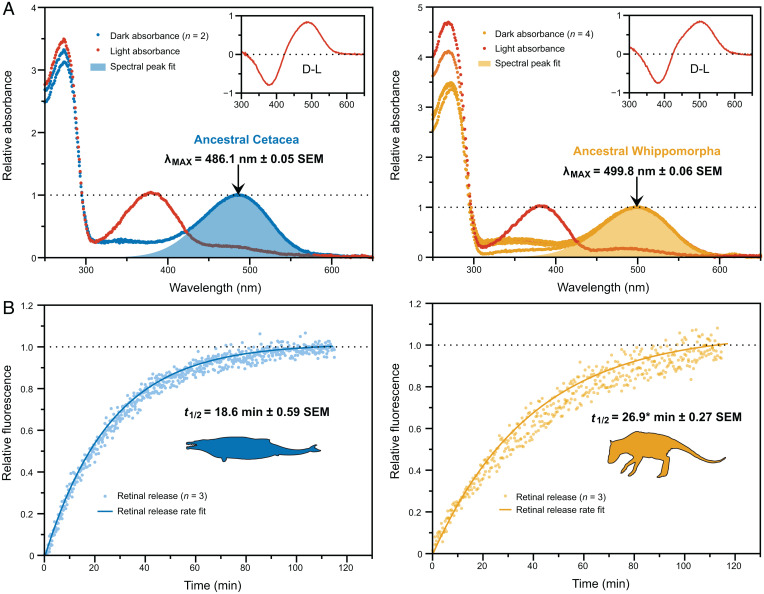
We used codon-based likelihood methods to infer the nucleotide and amino acid sequences of both the Cetacea and Whippomorpha ancestral rhodopsins (Fig. 1). Through in vitro expression in mammalian cell culture and protein purification, we generated properly folded rhodopsin pigment samples from both synthesized ancestral coding sequences. We successfully regenerated these pigments with 11-cis-retinal chromophore and obtained dark-state absorbance spectra typical for vertebrate rhodopsin (as compared with a bovine rhodopsin positive control, SI Appendix, Fig. S1). Both ancestral visual pigments responded to light exposure with a spectral shift to 380 nm (Fig. 2A), which is characteristic of the light-activated intermediate, meta II (37).
Fig. 2.
(A) Dark and light-activated absorption spectra. The indicated λmax values are the means (±SEs) of estimates calculated for separately eluted samples of each pigment (where n is the number of elutions per pigment). Light-activated spectral peaks are 380 nm, which is the characteristic signature of the light-activated intermediate, and insets show dark-light difference spectra. (B) Light-activation fluorescence time series. The indicated t1/2 values for retinal release rates are the means (±SEs) of estimates calculated for separate fluorescence time series (where n is the number of time series). *This value excludes an outlier (t1/2 = 29.1 ± 2.20 with the outlier).
Although the Whippomorpha rhodopsin had a spectral peak typical of terrestrial vertebrates at ∼500 nm (38, 39) (Figs. 1 and 2A), the Cetacea rhodopsin was significantly blue-shifted relative to Whippomorpha by 14 nm (t = 170.79, df = 3.64, P = 3.125E−8; Welch’s two-tailed t test; Fig. 2A). Computational evolutionary analyses of rhodopsin coding sequences have thus far implied that the ancestral crown cetacean had either a moderately blue-shifted spectral peak similar to modern delphinids (∼489 nm) (27) or an extreme shift similar to that of the bathypelagic-diving beaked whales (<485 nm) (26). However, we found that the ancestral Cetacea spectral peak was intermediate between these estimates at λmax = 486 nm (Figs. 1 and 2A), which matches a deep-diving pinniped, the elephant seal (40), and various teleost fish that inhabit the mesopelagic zone (∼200 to 1,000 m) (20, 21). Considering that baleen whales have spectral peaks ranging between 490 and 495 nm (19, 23, 26, 41), the ancestral Cetacea rhodopsin is on the lower end of the cetacean spectral range overall.
Light-activated retinal isomerization hydrolyzes the Schiff base linkage between all-trans retinal and the rhodopsin apoprotein, releasing the molecule from the binding pocket. In all vertebrate visual opsins, this retinal must then be replaced with a new 11-cis molecule before the visual cycle can repeat (42). The release of all-trans retinal can be measured in vitro by monitoring an intrinsic increase in fluorescence as the molecule migrates out of the protein, providing an estimate of the retinal release rate (t1/2), which varies among species (43, 44). The t1/2 for Cetacea rhodopsin at 18.6 min was significantly faster than the t1/2 for Whippomorpha rhodopsin at 26.9 min (t = 11.91, df = 2.96, P = 0.001; Welch’s two-tailed t test; Figs. 1 and 2B).
The ancestral Cetacea retinal release rate is intermediate between some terrestrial mammals such as human and bovine (12–15 min (39)) and other aquatic mammals such as the killer whale and hippopotamus (∼25 min (24)). This result is interesting given that the Cetacea pigment has high sequence similarity to killer whale rhodopsin, which is also blue-shifted, yet has a slower retinal release rate (24). Such a case exemplifies the potential risks associated with using an extant model system to make inferences about ancestral function. However, the spectral tuning and retinal release results for the ancestral Whippomorpha rhodopsin are comparable to hippopotamus rhodopsin; both have a typical terrestrial spectral peak (∼500 nm) and a long retinal release rate [t1/2 ≥ 25 min (24)]. The latter may be the result of increased aquatic/dimlight activity, a hypothesis that was similarly proposed to explain longer retinal release rates in the killer whale, hippopotamus, bats, and ancestral mammals, all of which have become increasingly nocturnal and/or aquatic relative to their ancestors (8, 14, 24).
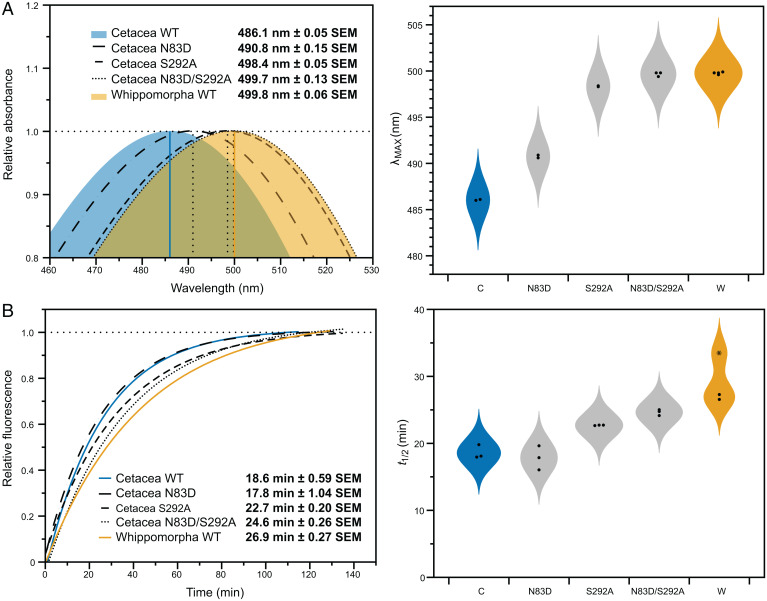
Sites 83 and 292 Mediate Functional Shifts in Ancestral Cetacean Rhodopsin.
Previous mutation experiments that we conducted in killer whale and hippopotamus rhodopsin (24) led us to hypothesize that sites 83 and 292, which are also under positive selection in cetaceans (11), may account for the majority of the functional shifts between the two ancestral states. As such, we created backward mutations at these sites in the ancestral Cetacea rhodopsin to determine whether they were sufficient to produce the functional phenotype of the Whippomorpha pigment. Single mutations at these sites significantly red-shifted spectral tuning (N83D: t = 29.73, df = 1.22, P = 0.011; S292A: t = 173.95, df = 2.00, P = 3.305E−5; Welch’s two-tailed t tests), although the effect from 292 was considerably greater (Fig. 3A). For retinal release, the shifts were minor although still significant for site 292 (t = 6.87, df = 2.01, P = 0.020; Welch’s two-tailed t test; Fig. 3B). When combined in a double mutant, sites 83 and 292 were sufficient to shift the spectral peak of Cetacea to within 1 nm from Whippomorpha (Fig. 3A), with no significant difference between the pigments (t = 0.73, df = 2.89, P = 0.518; Welch’s two-tailed t test). For retinal release, the double mutant and Whippomorpha pigments had similar values (Fig. 3B) yet were still significantly different (t = 5.09, df = 1.99, P = 0.037; Welch’s two-tailed t test), which suggests other residues may be contributing to the difference between the ancestral rhodopsins.
Fig. 3.
(A) The absorption spectra of the ancestral Cetacea rhodopsin mutants shift toward the ancestral Whippomorpha spectral peak. The double mutant matches the ancestral phenotype, with no significant difference between them (t = 0.73, df = 2.89, P = 0.518; Welch’s two-tailed t test). (B) The light-activation fluorescence time series of the ancestral Cetacea rhodopsin mutants also shift toward the ancestral Whippomorpha retinal release rate. The double mutant does not match the retinal release rate of the more ancestral phenotype (t = 5.09, df = 1.99, P = 0.037; Welch’s two-tailed t test). The data points comprising λmax and t1/2 estimates are shown on violin probability plots next to the spectrophotometric readings (*This value is an outlier for the Whippomorpha retinal release rate).
These results are consistent with growing evidence that the functional effects of these substitutions, especially at site 83, are variable across species. Although D83 is highly conserved in vertebrates (45), D83N is associated with slowed retinal release and increased meta II stability in dim-light-adapted species (9, 13, 14, 46, 47). However, N83D negligibly shifts retinal release in the killer whale due to the presence of serine at site 299, a case of intramolecular epistasis (24). The ancestral Cetacea rhodopsin also has S299 (SI Appendix, Fig. S2), and the effect of N83D on retinal release was also insignificant (t = 0.65, df = 3.18, P = 0.562; Welch’s two-tailed t test; Fig. 3B). As such, even though we found that the retinal release rate of this pigment was faster than killer whale rhodopsin (24), the minimal shifts resulting from our N83D mutant suggest that some epistatic masking was already established in the ancestral crown cetacean. In addition, the contribution of site 292 to metarhodopsin kinetics has received minimal attention relative to spectral tuning, yet here we found it significantly affected Cetacea retinal release, just as it did in the killer whale (24).
Ancestral Rhodopsin 3D Structure Suggests Multiple Mechanisms for Transitioning Residues.
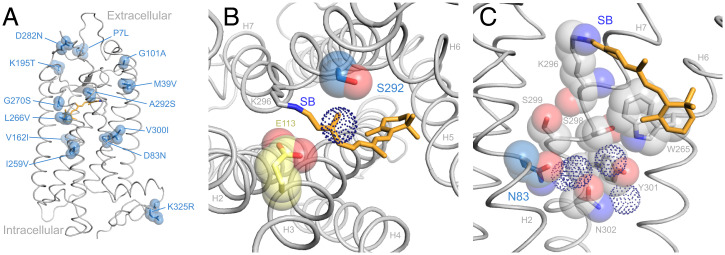
The ancestral sequences we reconstructed in PAML4.9 (48) show 12 substitutions between the Whippomorpha and Cetacea nodes (Fig. 1, SI Appendix, Fig. S2). We constructed a homology model of the ancestral Cetacea rhodopsin to understand the structural significance of these transitioning residues. Most of them (7 sites) are in transmembrane helical regions, 3 of which (sites 83, 292, and 300) are near the Schiff base of the retinal chromophore (Fig. 4A). The remaining sites are located near other functional regions, including hydrogen-bond networks, potential dimerization interfaces, glycosylation sites, transducin/arrestin-binding regions, and visual disease sites (45).
Fig. 4.
(A) Amino acid substitutions that occurred over the terrestrial-to-aquatic transition (including uncertain site 300) are highlighted with van der Waals spheres. (B) Top-down view showing the proximity of S292 to the chromophore (orange), Schiff base (SB), and counterion (E113). The serine side chain may influence chromophore activation energy indirectly through a nearby water molecule (dotted sphere). (C) Side view highlighting residues (including N83) and water molecules (dotted spheres) participating in a hydrogen bond network that facilitates packing and light-activated rotation among helices H2, H6, and H7 (45, 50). The ancestral Cetacea rhodopsin is a homology model based on the bovine rhodopsin crystal structure (PDB: 1U19).
Our mutation results and ancestral rhodopsin 3D structure are consistent with recent studies that emphasize the contrasting structural-functional roles of sites 83 and 292. In vertebrate rhodopsins overall, D83 and A292 are highly conserved (45). While the A292S substitution correlates strongly with aquatic spectral tuning (20, 24, 25), D83N has varied effects with respect to both spectral tuning and metarhodopsin kinetics in the context of different pigment backgrounds (14, 24, 46, 47). Spectral tuning mechanisms that are associated with D83N and A292S have been extensively investigated in multiple vertebrate rhodopsin systems using both computational simulations and mutation experiments. These investigations generally agree that the large blue-shift associated with A292S is due to the close proximity of the side chain to the Schiff base of the chromophore (24, 45, 49) (Fig. 4B). In contrast, the mechanism of D83N appears to be through its participation in a protein-wide hydrogen-bond network that mediates the conformational shift of rhodopsin through its light-activated metarhodopsin states (13, 24, 45, 50) (Fig. 4C).
Discussion
We report here the experimental characterization of reconstructed ancestral cetacean visual pigments and provide estimates for the genotypic and phenotypic states of rhodopsin at the phylogenetic nodes separating cetaceans from their terrestrial relatives. Our spectral tuning measurements have confirmed the long-standing hypothesis of a spectral blue-shift on the ancestral cetacean branch. The magnitude of this shift places the ancestor of crown cetaceans closer to the deep-sea spectral profiles of beaked whales and mesopelagic teleosts than to the more intermediate spectral range of most modern cetaceans (19, 25–27, 41). Our retinal release assays also showed a decrease in active-state (meta II) stability over the terrestrial-to-aquatic transition, which has since been recovered in some modern descendants such as the killer whale (24). We propose that decreased meta II stability may correspond to increased rates of retinal regeneration, and subsequently dark adaptation, a trait that allows for faster rod acclimation to dimming light levels in extant deep-diving marine mammals (30). Taken together, our results suggest that some of the first fully aquatic cetaceans were mesopelagic divers.
The ancestor of crown cetaceans (stem Neoceti) is thought to have derived from the earliest fully aquatic archaeocetes, particularly dorudontine members of the Basilosauridae in the Middle to Late Eocene (5). These ancient whales likely possessed tail muscles that were suitable for powered diving strokes (1, 4) and had reduced thickness in periosteal compact bone similar to modern cetaceans (51). Reconstructions of net surface charge on ancestral cetacean myoglobin also indicate the evolution of increased muscle oxygen storage capacity on the root branch of Cetacea before the divergence of Odontoceti and Mysticeti, in addition to further increases on descendant branches (52). Exposure to the vast prey resources provided by deep-sea cephalopods, many of which are bioluminescent and emit in the blue part of the spectrum, is thought to have influenced the evolution of deep diving and echolocation in early odontocetes (3), but analyses of tooth microwear and ancestral state reconstructions of cetacean feeding strategies indicate that cephalopods were among basilosaurid prey as well (53, 54).
In addition, extant cetaceans have at most two functioning visual pigments (rhodopsin and long-wave cone opsin), and several lineages are rod monochromats (27). The timing of short- and long-wave cone opsin loss events in cetacean evolutionary history is uncertain, but predominant hypotheses agree that pseudogenization occurred multiple times following the divergence of Odontoceti and Mysticeti, presumably due to advanced auditory adaptations (e.g., echolocation) reducing sensory reliance on vision (27). As such, the ancestral crown cetacean likely had the full complement of mammalian visual pigments, but was not specialized for either ultra- or infrasonic hearing (5). Consequently, these animals depended on the visual system (more than their descendants) for foraging activities. Given that mesopelagic prey were likely a critical food source for oceanic cetaceans before drastic increases in ocean primary productivity during the Oligocene (54), the resulting selection pressure on dim-light visual function would have included rhodopsin light-activation mechanisms.
Mammalian rhodopsins are characterized by a rate of meta II decay that is 50 times slower than the cone opsins (29). As such, the trait is thought to be a dim-light adaptation in vertebrate evolution (35, 55, 56). For example, mutation-induced destabilization of meta II reduced dim-light photosensitivity in transgenic mouse models (57), and there is evidence to suggest that the occurrence of N83, which shifts the equilibrium of meta states to favor meta II, may be a dim-light adaptation in bats and deep-dwelling cichlids (13). However, the evolutionary reasons for the slowed meta II decay remain uncertain. Early studies suggested that slowed retinal release rates may be associated with greater signal amplification and photosensitivity (58), but this is likely not the case considering the speed and efficiency of arrestin binding (57).
Alternatively, increased meta II stability combined with arrestin binding can serve a photoprotective role. Meta II helps to sequester all-trans retinal in bright conditions, as excessive all-trans retinal photoproducts (such as from a full bleach of the retina) are known to be cytotoxic (59). This relationship may explain the prevalence of counterintuitive substitutions that increase rhodopsin thermal stability at an unprecedented cost to meta II stability (which becomes cone-like) in some deep-dwelling teleost fish that are never exposed to bright light (58, 60). However, dim-light-adapted mammals tend to have stronger pupillary responses, which help protect the retina from full bleaching (30), but also have a greater proportion and density of rod cells (61). Therefore, the characteristically stable meta II of rhodopsin appears to be constrained in most tetrapods (60) and is exaggerated in several dim-light-adapted mammals such as bats (14).
Our results suggest that the common ancestor of cetaceans and hippos (Whippomorpha) had a very slow meta II decay rate. This ancestor likely resembled members of the fossil genus Indohyus, herbivorous rodent-like artiodactyls that were semiaquatic in that they used small streams and rivers to escape from predators (62). If, like hippos, this ancestor was also nocturnally active, then the slow meta II decay rate may reflect a greater photoprotective need arising from a strongly rod-dominated retina. In contrast, our results demonstrate a significant shift toward faster meta II decay rates in the ancestor of crown cetaceans that has since been reversed in some extant species such as the killer whale (24). As such, there must be an alternative function associated with meta II decay rates that is critical to the terrestrial-to-aquatic transition even at the cost of rod photoprotection.
Aside from rod photoprotection, meta II decay rates may have important implications for retinal regeneration and dark adaptation. In the phototransduction pathway in vivo, phosphorylation of meta II by rhodopsin kinase and binding by rod arrestin halts G protein (transducin) signaling; this activity culminates in the release of both retinal and arrestin to allow the light-responsive (inactive) state of rhodopsin to regenerate (42). Natural rhodopsin variants that favor a less stable meta II may thus experience accelerated regeneration of the inactive state in cases where the retina is only partially bleached (e.g., in dimmer light environments, with a strong pupillary response). Indeed, there is some evidence from transgenic studies that the decay of light-activated rhodopsin can affect rates of dark adaptation (28, 29, 63). At the molecular level, Hauser and colleagues discovered that N83D occurred in three lineages of neotropical cichlids that have transitioned from turbid water habitats to clear water, and proposed a trade-off between dim-light photosensitivity and retinal regeneration to explain the accelerated retinal release rate caused by the substitution (9).
A similar trade-off was recently proposed to explain increased meta II decay rates and a spectral red-shift over a marine-to-freshwater transition in croakers. Specifically, rapid retinal regeneration may be a benefit for vertically navigating the water column in freshwater habitats where light attenuates quickly with depth and tends to be longer wavelength (15). The cetacean terrestrial-to-aquatic transition is a similar scenario, except that the spectral change is for a shorter wavelength (blue-shifted) environment. Interestingly, the increased meta II decay rates in the croakers are due in part to the substitution E122I, which also causes a spectral blue-shift (the croakers have several other substitutions to generate the spectral red-shift) (15). In several oceanic fish lineages, transitions to blue-shifted waters coincide with E122I and several other substitutions to recover the concurrent reduction in meta II stability (60). However, E122 is constrained in tetrapods (60), so the double D83N/A292S substitution in ancestral cetaceans may reflect an alternative evolutionary pathway toward both blue-shifted spectral tuning and rapid retinal regeneration. Furthermore, the background residue S299 may facilitate this pathway by epistatically negating the meta II-stabilizing effect of D83N (24).
Following on these works, we propose an ecological trade-off between rod photoprotection and dark adaptation in how they are affected by light-activated meta II stability. For mesopelagic (and deeper) foraging marine mammals, faster regeneration rates facilitated by rapid retinal release and meta II decay may accelerate rod dark adaptation during dives. Although rates of dark adaptation have not as yet been measured in a cetacean, the deep-diving elephant seal can adjust to dim light at a rate three to five times faster than shallow diving pinnipeds and terrestrial mammals (30). Given that rhodopsin regeneration is thought to be limited by meta II decay when the retina is only partially bleached (29, 59), the ancestral cetacean may have had a dark adaptation rate that was faster than in some extant cetaceans such as the killer whale (which is not a deep diver), but perhaps more comparable to deep-diving mammals such as the elephant seal. This hypothesis could be evaluated further by comparing rhodopsin retinal release rates across a variety of marine mammal species with different diving abilities.
Taken together, the deep-sea spectral signature and accelerated retinal release of ancestral cetacean rhodopsin suggest that the progenitor of modern cetaceans was exploiting the mesopelagic zone, diving to depths where light is both more limited and blue-shifted. The evolution of diving behavior has received much attention in certain lineages, such as the bathypelagic-diving sperm and beaked whales (5), but there has been comparatively little paleontological insight into the diving abilities of ancestral crown cetaceans. Through our study of ancestral cetacean rhodopsin, the question of when aphotic diving first evolved in cetaceans now has an experimentally supported hypothesis, one that invites further corroboration from future paleontological discoveries and molecular evolutionary data from additional physiological systems. Evolutionary insight into the ecology and behavior of ancient organisms is notoriously elusive, even with a plentiful fossil record, but laboratory re-creation of ancestral proteins provides an invaluable context for testing structural-functional hypotheses with a direct bearing on organismal physiology and lifestyle.
Materials and Methods
Ancestral Rhodopsin Expression and Site-Directed Mutagenesis.
The reconstructed coding sequences for the ancestral Cetacea and Whippomorpha rhodopsins (see below) were synthesized with BamHI and EcoRI restriction sites on their 5′ and 3′ ends, respectively, for insertion into the p1D4-hGFP II expression vector (64) using the services of GeneArt (Thermo Fisher Scientific). We introduced mutations to each pigment (N83D, S292A, N83D/S292A in Cetacea; V300I in Whippomorpha; SI Appendix, Fig. S3) using the QuikChange II site-directed mutagenesis protocol (Agilent). Before heterologous expression, we confirmed all of the mutant-vector constructs by Sanger sequencing. As described in previous work from our group (11, 64), we transiently transfected maxi-preps (Lipofectamine 2000, Invitrogen; 8 μg DNA/10 cm plate) of each rhodopsin-vector construct into cultured HEK293T cells (ATCC CRL-11268). Each transfection also included standard bovine rhodopsin constructs as a positive control. After 48 h, we harvested the cells, regenerated them with 11-cis-retinal in the dark, and solubilized them in 1% dodecylmaltoside detergent. We then purified the rhodopsin samples by immunoaffinity nutation with the 1D4 monoclonal antibody (University of British Columbia 95062, lot 1017) coupled to a hydrazide resin, followed by washing and sample elution in sodium phosphate buffers.
We determined the spectral tuning of eluted rhodopsin samples from UV-visible absorption spectra as recorded by a Cary 4000 double-beam spectrophotometer (Agilent) in the dark (25 °C). We then fit template curves for opsins bound to 11-cis-retinal (65) to the normalized absorption spectra to estimate the spectral peak (λmax). To confirm light-activation ability, we also recorded the absorption spectrum for a single elution per pigment after 60 s of white-light bleaching (fiberoptic lamp, Dolan-Jenner), which allowed us to calculate the dark-light difference spectrum. For the remaining unbleached samples, we recorded the release rate of all-trans retinal after a 30-s white-light bleach (20 °C) using a Cary Eclipse fluorescence spectrophotometer (Agilent) (24). Where necessary, we diluted elution samples to a maximum concentration of roughly 0.3 μM for the assay, which allowed us to run additional time courses for higher yield samples. The spectrophotometer detects the intrinsic increase in fluorescence over time as migration of all-trans retinal from the binding pocket unquenches nearby tryptophan residues in the protein. The retinal release half-life (t1/2) can then be estimated by fitting a first-order exponential curve (y = y0 + a(1 – e−kx), where t1/2 = ln (2)/k) to the fluorescence time course (43).
Ancestral Rhodopsin Sequence Reconstruction.
We obtained all available complete rhodopsin nucleotide coding sequences for cetartiodactyl species (22 cetaceans, 11 noncetacean cetartiodactyls; SI Appendix, Table S3) from GenBank and aligned them by PRANK (66) within the BLASTPhyME program (67). We included rhodopsin coding sequences from cat (representing Laurasiatheria), human (representing Euarchontoglires), and elephant (representing Afrotheria) as outgroups. For inference of ancestral sequences, we constructed a species tree topology using cetartiodactyl relationships established in previous studies (2, 68).
We used codon-based likelihood models to infer the ancestral rhodopsin coding sequences at the cetartiodactyl nodes representing Whippomorpha and Cetacea, which span the terrestrial-to-aquatic transition (Fig. 1). We implemented analyses in the codeml program of PAML 4.9 (48) using a variety of random sites and clade models (all with default F3 × 4 estimated codon equilibrium frequencies) (SI Appendix, Tables S1 and S2). Codon-based likelihood models, by making use of both the nucleotide sequence and the codon structure encoding the translated amino acid sequence, offer several advantages over commonly used amino acid- and nucleotide-based reconstruction methods, including higher accuracy and the incorporation of rate heterogeneity arising from varying selection pressure across the coding sequence and/or phylogenetic tree (SI Appendix). The latter is achieved via modeling of nonsynonymous versus synonymous substitution rates, often expressed as the ratio dN/dS = ω . This allowance may be relevant for ASRs, where varying selection pressure exists in the study system, a case that we previously demonstrated for cetacean rhodopsin (11).
The few studies that have used codon-based methods for ASR tend to rely on random-sites models (8, 15, 33, 35). Consequently, there has been little exploration of whether more complex models that incorporate rate variation over different parts of the phylogeny generate results that are consistent with them. In our previous work (11), the dataset of cetacean rhodopsin sequences was fit significantly better by a model that allowed evolutionary rates to vary differently on clade partitions corresponding to different diving profiles. As such, we included clade models for these diving class partitions in our analyses, and they also gave the best fits to our dataset according to the Akaike information criterion (SI Appendix, Table S2). The only other study we know of that used clade models for ASR also found that they provided the best fit to data with rate heterogeneity that varied over the tree (15).
Even so, for both ancestral pigments, the resulting amino acid sequences had high posterior probabilities (Fig. 1) and were either identical (Cetacea) or nearly identical (Whippomorpha), regardless of codon model complexity (SI Appendix). For the Whippomorpha sequence, the codon models were inconsistent at a single site (similar probabilities for either V300 or I300), but the alternative residues did not have a significant effect on either spectral tuning or retinal release (SI Appendix, Fig. S3). Nevertheless, to further assess the robustness of our results, we sampled the posterior distribution of the best-fitting codon model and repeated sequence reconstruction using less appropriate but more commonly used nucleotide- and amino acid–based methods (SI Appendix). However, the amino acid models resulted in three substitutions that had highly improbable underlying nucleotide substitutions according to the codon models (SI Appendix, Figs. S4–S6). Apart from these three sites, these methods estimated ancestral sequences at the Cetacea and Whippomorpha nodes that were consistent with the codon models.
Homology Modeling.
To visualize the structural locations of terrestrial-to-aquatic transitioning residues and to better understand the structural-functional context of sites 83 and 292, we modeled the three-dimensional structure of the reconstructed ancestral Cetacea rhodopsin amino acid sequence using the dark-state bovine rhodopsin crystal structure (PDB: 1U19 (50)). Using the Modeler package (69), we generated 100 models of Cetacea rhodopsin (optimized 10 times per model by the objective function) and ranked them according to discrete optimized protein energy (DOPE) score (70). We selected the best model (lowest DOPE score) for examination in MacPyMol (Delano Scientific). The Cetacea rhodopsin had a similar total energy to the bovine rhodopsin crystal structure [comparable Z scores in ProSA-web (71))], and highly probable stereochemical bond conformations among amino acid residues [ProCheck (72)].
Statistical Tests.
For detecting differences in λmax and t1/2 among our ancestral and mutated pigments, we used Welch’s two-sample t tests (two-tailed) to accommodate unequal variances. To assess normality, we tested the distribution of data from our bovine rhodopsin control pigment (λmax: W = 0.90, P = 0.34, n = 10; t1/2: W = 0.97, P = 0.88, n = 9; Shapiro-Wilk tests). We determined minimum viable sample sizes for pigment expression (β ≤ 0.2) by conducting a power analysis for detecting shifts of at least 2 nm in λmax and 5 min in t1/2 (SI Appendix, Table S4), differences that are thought to be biologically significant (34, 35, 38, 60).
Supplementary Material
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) post-graduate scholarship (S.Z.D.), a University of Toronto Vision Science Research Program fellowship (S.Z.D.), and an NSERC Discovery Grant (B.S.W.C.). The 11-cis-retinal chromophore used in visual pigment experiments was generously provided by Dr. Rosalie Crouch (Medical University of South Carolina), and the posterior distribution sampling algorithm was kindly shared by Dr. Eric Gaucher (Georgia Institute of Technology). Silhouette images were obtained from PhyloPic (phylopic.org), with credit to Nobu Tamura (https://creativecommons.org/licenses/by/3.0/). We also thank two anonymous reviewers for helpful feedback and suggestions to improve the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118145119/-/DCSupplemental.
Data Availability
Dataset files include: Alignment of rhodopsin (Rh1) sequences formatted for PAML, corresponding species tree in Newick format for PAML, and ancestral Rh1 amino acid sequences (for Cetacean and Whippomorpha nodes) estimated with PAML (random sites, clade, and amino acid models), Datamonkey, and ProtASR. Data have been deposited in Zenodo, https://zenodo.org/record/5797505#.Yei6h_hyaUl (73).
References
- 1.Uhen M. D., The origin(s) of whales. Annu. Rev. Earth Planet. Sci. 38, 189–219 (2010). [Google Scholar]
- 2.Gatesy J., et al. , A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 66, 479–506 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Lindberg D. R., Pyenson N. D., Things that go bump in the night: Evolutionary interactions between cephalopods and cetaceans in the tertiary. Lethaia 40, 335–343 (2007). [Google Scholar]
- 4.Uhen M. D., Form, function, and anatomy of Dorudon atrox (Mammalia, Cetacea): An archaeocete from the Middle to Late Eocene of Egypt. Pap. Paleontol. 34 (2004). [Google Scholar]
- 5.Pyenson N. D., The ecological rise of whales chroniPcled by the fossil record. Curr. Biol. 27, R558–R564 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K., et al. , Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Pugh E. N., Lamb T. D., Amplification and kinetics of the activation steps in phototransduction. Biochim. Biophys. Acta 1141, 111–149 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Bickelmann C., et al. , The molecular origin and evolution of dim-light vision in mammals. Evolution 69, 2995–3003 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Hauser F. E., et al. , Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol. Biol. Evol. 34, 2650–2664 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Castiglione G. M., et al. , Evolution of nonspectral rhodopsin function at high altitudes. Proc. Natl. Acad. Sci. U.S.A. 114, 7385–7390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dungan S. Z., Kosyakov A., Chang B. S. W., Spectral tuning of killer whale (Orcinus orca) rhodopsin: Evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323–336 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Hunt D. M., Fitzgibbon J., Slobodyanyuk S. J., Bowmaker J. K., Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Res. 36, 1217–1224 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Sugawara T., Imai H., Nikaido M., Imamoto Y., Okada N., Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol. Biol. Evol. 27, 506–519 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez E. A., et al. , Functional shifts in bat dim-light visual pigment are associated with differing echolocation abilities and reveal molecular adaptation to photic-limited environments. Mol. Biol. Evol. 35, 2422–2434 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Van Nynatten A., Castiglione G. M., de A Gutierrez E., Lovejoy N. R., Chang B. S. W., Recreated ancestral opsin associated with marine to freshwater croaker invasion reveals kinetic and spectral adaptation. Mol. Biol. Evol. 38, 2076–2087 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warrant E. J., Locket N. A., Vision in the deep sea. Biol. Rev. Camb. Philos. Soc. 79, 671–712 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Fasick J. I., Cronin T. W., Hunt D. M., Robinson P. R., The visual pigments of the bottlenose dolphin (Tursiops truncatus). Vis. Neurosci. 15, 643–651 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Fasick J. I., Robinson P. R., Spectral-tuning mechanisms of marine mammal rhodopsins and correlations with foraging depth. Vis. Neurosci. 17, 781–788 (2000). [DOI] [PubMed] [Google Scholar]
- 19.McFarland W. N., Cetacean visual pigments. Vision Res. 11, 1065–1076 (1971). [DOI] [PubMed] [Google Scholar]
- 20.Hunt D. M., Dulai K. S., Partridge J. C., Cottrill P., Bowmaker J. K., The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J. Exp. Biol. 204, 3333–3344 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Hope A. J., Partridge J. C., Dulai K. S., Hunt D. M., Mechanisms of wavelength tuning in the rod opsins of deep-sea fishes. Proc. Biol. Sci. 264, 155–163 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavigne D. M., Ronald K., Pinniped visual pigments. Comp. Biochem. Physiol. 52, 325–329 (1975). [DOI] [PubMed] [Google Scholar]
- 23.Southall K. D., Oliver G. W., Lewis J. W., Le Boeuf B. J., Levenson D. H., Visual pigment sensitivity in three deep diving marine mammals. Mar. Mammal Sci. 18, 275–281 (2002). [Google Scholar]
- 24.Dungan S. Z., Chang B. S. W., Epistatic interactions influence terrestrial-marine functional shifts in cetacean rhodopsin. Proc. Biol. Sci. 284, 20162743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasick J. I., Robsinson P. R., Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry 37, 433–438 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Bischoff N., Nickle B., Cronin T. W., Velasquez S., Fasick J. I., Deep-sea and pelagic rod visual pigments identified in the mysticete whales. Vis. Neurosci. 29, 95–103 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Meredith R. W., Gatesy J., Emerling C. A., York V. M., Springer M. S., Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9, e1003432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederiksen R., et al. , Rhodopsin kinase and arrestin binding control the decay of photoactivated rhodopsin and dark adaptation of mouse rods. J. Gen. Physiol. 148, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss E., Shedding light on dark adaptation. Biochem (Lond.) 42, 44–50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levenson D. H., Schusterman R. J., Dark adaptation and visual sensitivity in shallow and deep-diving pinnipeds. Mar. Mammal Sci. 15, 1303–1313 (1999). [Google Scholar]
- 31.Harms M. J., Thornton J. W., Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 20, 360–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang B. S. W., Donoghue M. J., Recreating ancestral proteins. Trends Ecol. Evol. 15, 109–114 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Chang B. S. W., Jönsson K., Kazmi M. A., Donoghue M. J., Sakmar T. P., Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19, 1483–1489 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama S, Tada T., Zhang H., Britt L., Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 105, 13480–13485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., et al. , Scotopic rod vision in tetrapods arose from multiple early adaptive shifts in the rate of retinal release. Proc. Natl. Acad. Sci. U.S.A. 116, 12627–12628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGowen M. R., Gatesy J., Wildman D. E., Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 29, 336–346 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Ryba N. J., Marsh D., Uhl R., The kinetics and thermodynamics of bleaching of rhodopsin in dimyristoylphosphatidylcholine. Identification of meta-I, meta-II, and meta-III intermediates. Biophys. J. 64, 1801–1812 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauser F. E., Chang B. S. W., Insights into visual pigment adaptation and diversity from model ecological and evolutionary systems. Curr. Opin. Genet. Dev. 47, 110–120 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Morrow J. M., et al. , An experimental comparison of human and bovine rhodopsin provides insight into the molecular basis of retinal disease. FEBS Lett. 591, 1720–1731 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Lythgoe J. N., Dartnall H. J. A., A “deep sea rhodopsin” in a mammal. Nature 227, 955–956 (1970). [DOI] [PubMed] [Google Scholar]
- 41.Fasick J. I., Bischoff N., Brennan S., Velasquez S., Andrade G., Estimated absorbance spectra of the visual pigments of the North Atlantic right whale (Eubalaena glacialis). Mar. Mammal Sci. 27, E321–E331 (2011). [Google Scholar]
- 42.Lamb T. D., Pugh E. N. Jr., Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 23, 307–380 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Farrens D. L., Khorana H. G., Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 270, 5073–5076 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Schafer C. T., Farrens D. L., Conformational selection and equilibrium governs the ability of retinals to bind opsin. J. Biol. Chem. 290, 4304–4318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pope A., Eilers M., Reeves P. J., Smith S. O., Amino acid conservation and interactions in rhodopsin: Probing receptor activation by NMR spectroscopy. Biochim. Biophys. Acta 1837, 683–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Hazel I., et al. , A comparative study of rhodopsin function in the great bowerbird (Ptilonorhynchus nuchalis): Spectral tuning and light-activated kinetics. Protein Sci. 25, 1308–1318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bickelmann C., Morrow J. M., Müller J., Chang B. S. W., Functional characterization of the rod visual pigment of the echidna (Tachyglossus aculeatus), a basal mammal. Vis. Neurosci. 29, 211–217 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Luk H. L., et al. , Modulation of thermal noise and spectral sensitivity in Lake Baikal cottoid fish rhodopsins. Sci. Rep. 6, 38425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okada T., et al. , Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 5982–5987 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooper L. N., Maas M. C., “Bones and Teeth, Histology of” in Encyclopedia of Marine Mammals, Wursig B., Thewissen J.G.M., Kovacs K., Eds. (Elsevier, 2018). [Google Scholar]
- 52.Mirceta S., et al. , Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340, 1234192 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Fahlke J. M., Bastl K. A., Semprebon G. M., Gingerich P. D., Paleoecology of archaeocete whales throughout the Eocene: Dietary adaptations revealed by microwear analysis. Palaeogeogr. Palaeoclimatol. Palaeoecol. 386, 690–701 (2013). [Google Scholar]
- 54.Berta A., Lanzetti A., Feeding in marine mammals: An integration of evolution and ecology through time. Palaeontol. Electron. 23, a40 (2020). [Google Scholar]
- 55.Imai H., et al. , Molecular properties of rod and cone visual pigments from purified chicken cone pigments to mouse rhodopsin in situ. Photochem. Photobiol. Sci. 4, 667–674 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Kojima K., Imamoto Y., Maeda R., Yamashita T., Shichida Y., Rod visual pigment optimizes active state to achieve efficient G protein activation as compared with cone visual pigments. J. Biol. Chem. 289, 5061–5073 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imai H., et al. , Molecular properties of rhodopsin and rod function. J. Biol. Chem. 282, 6677–6684 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai H., et al. , Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl. Acad. Sci. U.S.A. 94, 2322–2326 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sommer M. E., Hofmann K. P., Heck M., Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat. Commun. 3, 995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castiglione G. M., Chang B. S. W., Functional trade-offs and environmental variation shaped ancient trajectories in the evolution of dim-light vision. Elife 7, e35957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peichl L., Diversity of mammalian photoreceptor properties: Adaptations to habitat and lifestyle? Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 287, 1001–1012 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Thewissen J. G. M., Cooper L. N., Clementz M. T., Bajpai S., Tiwari B. N., Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Ala-Laurila P., et al. , Visual cycle: Dependence of retinol production and removal on photoproduct decay and cell morphology. J. Gen. Physiol. 128, 153–169 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrow J. M., Chang B. S. W., The p1D4-hrGFP II expression vector: A tool for expressing and purifying visual pigments and other G protein-coupled receptors. Plasmid 64, 162–169 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Govardovskii V. I., Fyhrquist N., Reuter T., Kuzmin D. G., Donner K., In search of the visual pigment template. Vis. Neurosci. 17, 509–528 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Löytynoja A., Goldman N., webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11, 579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schott R. K., Gow D., Chang B. S. W., BlastPhyMe: A toolkit for rapid generation and analysis of protein-coding sequence datasets. bioRxiv [Preprint] (2016). 10.1101/059881 (Accessed 1 October 2021). [DOI]
- 68.Foley N. M., Springer M. S., Teeling E. C., Mammal madness: Is the mammal tree of life not yet resolved? Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 54, 5–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen M. Y., Sali A., Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiederstein M., Sippl M. J., ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M., PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993). [Google Scholar]
- 73.S. Dungan, B. Chang, Dataset for “Ancient whale rhodopsin reconstructs dim-light vision over a major evolutionary transition: Implications for ancestral diving behaviour.” Zenodo. https://zenodo.org/record/5797505#.Yei6h_hyaUl. Deposited 22 December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dataset files include: Alignment of rhodopsin (Rh1) sequences formatted for PAML, corresponding species tree in Newick format for PAML, and ancestral Rh1 amino acid sequences (for Cetacean and Whippomorpha nodes) estimated with PAML (random sites, clade, and amino acid models), Datamonkey, and ProtASR. Data have been deposited in Zenodo, https://zenodo.org/record/5797505#.Yei6h_hyaUl (73).