Significance
Bacteriophages (phages) are viruses that infect bacteria. Phages are abundant in the gut and are compositionally unique across the human population. While phages are in constant evolutionary battle with bacteria, their potential evolution with the mammalian gut remains overlooked. Here, we test whether phages are capable of adapting directly to the mammalian “host.” Using a coculture of phages, bacteria, and a gut-like mucosa, we found that phage evolution was driven by de novo mutations and recombination. This contributed to a unique phage capsid mutation, lending enhanced phage persistence in the mucus layer. Our findings propose a potential coevolutionary mechanism between phages and the gut mucosa, which could contribute to the individuality of gut viromes.
Keywords: virus, evolution, symbiosis, lab-on-a-chip, mucus
Abstract
The majority of viruses within the gut are obligate bacterial viruses known as bacteriophages (phages). Their bacteriotropism underscores the study of phage ecology in the gut, where they modulate and coevolve with gut bacterial communities. Traditionally, these ecological and evolutionary questions were investigated empirically via in vitro experimental evolution and, more recently, in vivo models were adopted to account for physiologically relevant conditions of the gut. Here, we probed beyond conventional phage–bacteria coevolution to investigate potential tripartite evolutionary interactions between phages, their bacterial hosts, and the mammalian gut mucosa. To capture the role of the mammalian gut, we recapitulated a life-like gut mucosal layer using in vitro lab-on-a-chip devices (to wit, the gut-on-a-chip) and showed that the mucosal environment supports stable phage–bacteria coexistence. Next, we experimentally coevolved lytic phage populations within the gut-on-a-chip devices alongside their bacterial hosts. We found that while phages adapt to the mucosal environment via de novo mutations, genetic recombination was the key evolutionary force in driving mutational fitness. A single mutation in the phage capsid protein Hoc—known to facilitate phage adherence to mucus—caused altered phage binding to fucosylated mucin glycans. We demonstrated that the altered glycan-binding phenotype provided the evolved mutant phage a competitive fitness advantage over its ancestral wild-type phage in the gut-on-a-chip mucosal environment. Collectively, our findings revealed that phages—in addition to their evolutionary relationship with bacteria—are able to evolve in response to a mammalian-derived mucosal environment.
Bacteriophages (phages) are viruses that predate upon bacteria in order to replicate and are thus as ubiquitous as their bacterial counterparts. Their significance in shaping the underlying bacterial community in an ecosystem is reflected by the manifold studies on phage–bacteria antagonistic coevolution (1–4). The human gut is an exemplary ecosystem where phages are abundantly found alongside a rich bacterial community, known collectively as the gut microbiome. Here, antagonistic coevolutionary dynamics between gut phages and their bacterial hosts are key in maintaining long-term microbiome homeostasis and diversity (5–8). While the focus on phages and bacteria within traditional gut microbiome studies has been extremely insightful (9), there is a rapidly expanding interest in the role the mammalian host provides for these microbial communities. In particular, the interaction between the bacteria and the mammalian host remains a popular research paradigm where the complexity of bacterial–mammalian gut interactions is increasingly unfolding and disentangling (10). However, phage–mammalian interactions were often overlooked and, consequently, much less is known regarding these interactions and their coevolutionary potential.
Bacteriophages were previously demonstrated to adhere directly to mucus to form a non–host-derived immunity against bacterial invasion (11). Phage–mucosal adherence was shown to promote increased encounter rates with bacterial hosts resulting in phage enrichment within the mucosal layer (12). Subsequent studies have expanded on this phage–mucus interaction. A study by Almeida et al. (13) found that phages bind to primary mucus on rainbow trout and persisted for up to 7 d, protecting against the pathogenic bacterium Flavobacterium columnare. In addition, the mucosal environment altered the bacterial phenotype lending increased bacterial virulence but at the cost of augmented susceptibility to phage infection, resulting in an ecological tradeoff that maintains phages in metazoan mucosal surfaces. Other studies have further demonstrated the direct effect of the mammalian epithelial cell layer (14) and mammalian mucin glycans (15) on enhancing phage virulence toward their bacterial hosts within mucosal surfaces. At an ecological level, the gut mucosa segregates phage and bacterial populations, establishing spatial refuges, which further promote phage–bacteria coexistence (16). Collectively, these studies underscore the importance of the mammalian component—in this case, the mucosal epithelium—for phage persistence, ecology, and evolution within the gut.
Mucosal surfaces are critical layers that interface the mammalian host with its environment. They are both principal sites of defense and habitats for large and diverse microbial communities. The mucus layer itself is primarily formed by large, mammalian-derived mucin glycoproteins that are extensively glycosylated with diverse glycans (17). The adherence and enrichment of phages in mucus occur via binding interactions between these diverse glycan residues covering mucin glycoproteins and immunoglobulin (Ig)-like protein domains exposed on the phage capsids (11). Importantly, these Ig-like proteins can accommodate large sequence variations of >1013 potential alternatives, while still maintaining their structural fold integrity and function (18). As such, there is potential for phage Ig-like proteins to adapt increased or altered adherence to the mammalian host’s mucin glycosylation patterns. However, to our knowledge, there is no evidence demonstrating the evolutionary potential between phages and the mammalian “host.” To this end, we questioned whether phages are capable of evolving in response to the mammalian mucosal environmental within a tripartite setting.
Using an in vitro lab-on-a-chip device to simulate a mammalian mucosal layer colonized with a bacterial host population (19), we tested if phage coevolution would result in phenotypes that persist within the mammalian mucosal environment. We showed that both phage–bacteria populations and phage evolutionary dynamics were unique across replicate populations in the gut-on-a-chip, and that genetic recombination was a key evolutionary force in driving mutational fitness. Despite the interreplicate disparity, we were able to derive an evolved phage with a mutation affecting the phage Ig-like domain within the Hoc capsid protein, that was previously shown to mediate phage adherence to mucus (11). This mutation conferred a fitness advantage by altering phage binding affinity to fucosylated mucin glycans. In summary, we present empirical evidence on the capacity of phages to coevolve within a mammalian mucosal environment to adapt increased persistence and adherence to the mucosa.
Results
The Gut-on-a-Chip Supports Phage–Bacteria Coexistence within a Mucosal Layer.
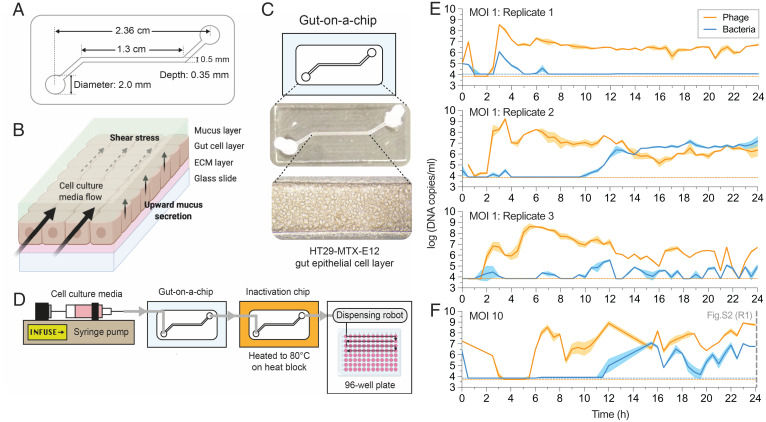
To investigate the capacity of phages to adapt to the mammalian mucosal environment, we fabricated a simple gut-on-a-chip microfluidic device that recapitulates key features of the mammalian gut mucosa (Fig. 1A). These devices are experimentally amenable, provide an accessible platform for biological replication, and recapitulate essential organ-level functions of the gut (19, 20) (Fig. 1B). Our gut-on-a-chip consisted of a single channel containing a confluent HT29-MTX-E12 colonic cell layer that produced a mucosal surface (Fig. 1C and SI Appendix, Fig. S1 A–C).
Fig. 1.
Gut-on-a-chip supports phage–bacteria coculture within a mucus environment. (A) Schematic and channel dimensions of the gut-on-a-chip. (B) Mucus turnover dynamics from the device is driven by shear stress from fluid flow and upward mucus secretion from the epithelial layer. ECM, extracellular matrix. (C) The HT29-MTX-E12 cell line grows and differentiates within the device channel environment to produce a mucus layer at ∼72 h postseeding. (D) Schematic for the overall gut-on-a-chip setup for chip perfusion, continuous sample inactivation via heat, and automated sample collection for qPCR. (E) qPCR-quantified phage–bacteria population in three replicate devices (n = 3) initiated at MOI 1 at 30-min intervals over 24 h. (F) qPCR-quantified phage–bacteria population in one device (n = 1) initiated at MOI 10 at 30-min intervals over 24 h. The plotted lines and shaded regions in E and F represent mean ± SEM of three qPCR technical replicates (n = 3) per experimental replicate. Horizontal orange and blue dotted lines in E and F represent the qPCR LOD threshold for phage and bacteria, respectively, in each experimental replicate (refer to Dataset S1 for standard curves and LOD thresholds). The vertical gray dashed line in F at 24 h corresponds to end-point population density measurement in replicate 1 (R1) of SI Appendix, Fig. S2.
We seeded 104 colony-forming units (CFUs) of Escherichia coli bacteria and 104 plaque-forming units (PFUs) of T4 phages—resulting in an inoculation with multiplicity of infection (MOI) 1—into three replicate devices. The coculture was maintained for 24 h under constant perfusion with sterile media at 120 µL/h, equating to 12 turnover cycles of the channel environment per hour. This flow rate within the device’s channel dimensions resulted in a calculated shear stress of 0.025 dyne⋅cm−2, which is within physiological range reported in the human intestine (21, 22). An automated sample collection system was developed where egressing samples from the device were heat-inactivated, followed by collection at 30-min intervals, with phages and bacteria subsequently quantified via qPCR (Fig. 1E, SI Appendix, Fig. S1D, and Dataset S1). Despite inoculating each replicate device with similar phage–bacteria densities, we observed immediate differences within the first hour across the replicates. Hereafter, both phages and bacteria remained under the limit of detection (LOD) until an initial growth phase between 2 and 4 h where phages in particular achieved high densities accompanied by a small bacterial population recovery above the LOD. Subsequently, we saw a second phage growth phase between 5 and 6 h in tandem with bacterial population decrease below the LOD, suggesting lytic phage suppression during this window. Thereafter, we saw divergent ecological dynamics occurring between replicate gut-on-a-chip devices. In replicate 1, bacteria remained under the LOD while phage populations were consistently high. Given the turnover rate of the channel environment of 12 times per hour, the stable phage persistence suggests that the phages are maintained by a nonextinct but undetectable bacterial population. In replicate 2, the bacterial population overtook the phage population potentially through the emergence of phage resistance, but both phages and bacteria nonetheless remained in coexistence. Finally, in replicate 3, phage–bacteria populations fluctuated above and below the LOD, exhibiting classical prey–predator dynamics.
Given the disparities in phage–bacteria dynamics across the replicate devices, we also tested if initiating the population at a higher MOI would lend similar stochastic outcomes in phage–bacteria populations. High temporal resolution of population densities initiated at MOI 10 demonstrated a similar initial phage and bacterial crash under the LOD, likely from robust phage predation on the bacterial population (Fig. 1F). However, phage population recovery at MOI 10 occurred at 6 h postinoculation, which was delayed in comparison with devices at MOI 1, where phage recovery typically occurs 2 to 3 h postinoculation. This suggests that the higher phage presence at MOI 10 contributed to a more drastic initial crash of the bacterial population compared with MOI 1. The bacterial population then recovered at ∼12 h postinoculation and subsequently engaged in a prey–predator fluctuation, indicating sustained predation by phages thereafter. We further initiated an additional five replicate gut-on-a-chip devices at MOI 10 that were measured only for their population densities at the 24-h end point (SI Appendix, Fig. S2). Despite the higher initial MOI, both phages and bacteria were recovered from all five replicates, but phage–bacterial densities were disparate at the experimental end point.
In summary, we demonstrated that the gut-on-a-chip was able to support phage–bacteria coexistence for up to 24 h, albeit with considerable variation between replicate devices in terms of population abundances and dynamics. Our results suggest that MOI impacts the initial establishing dynamics of phage–bacterial populations within the mucosal environment but, following this, each device was unique and delineated by inherent fluctuations and ecological stochasticity. Overall, the gut-on-a-chip provided a tripartite model system that supported mammalian, bacterial, and phage coculture.
The Mammalian Mucus Layer Influences Phage Evolution.
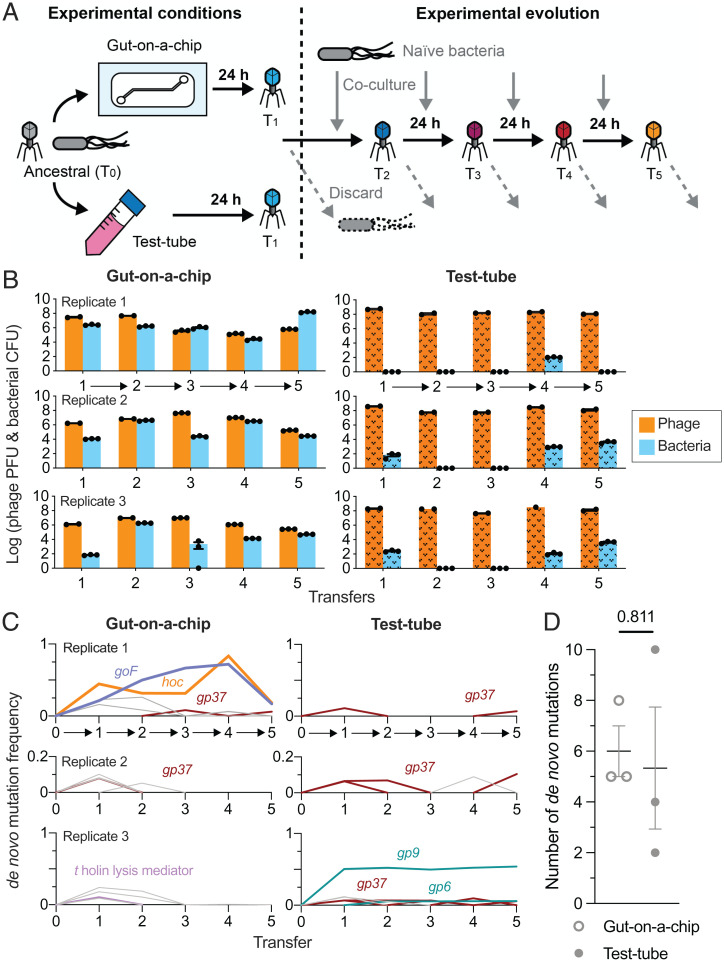
Based on experimental evidence that our gut-on-a-chip supports a tripartite coculture (Fig. 1E), we next experimentally evolved independent phage populations in either the tripartite gut-on-a-chip system or a test-tube control, which lacked a mammalian mucus layer (Fig. 2A). Here, both systems were inoculated with populations of T4 phages and E. coli bacteria at MOI 1 (the founding phage population herein is referred to as the “ancestral” phage), which were maintained for 24 h as previously described. At the end of the 24-h period, we collected the entire population from both the gut-on-a-chip and test-tube environments, followed by phage and bacterial quantification via traditional plating. We then conducted successive transfers of the “evolved” phage populations into either fresh gut-on-a-chip devices grown from naïve gut cells and seeded with naïve bacterial populations (“naïve” referring to entities that had no prior exposure to phages), or new test-tube controls with naïve bacterial populations. Both gut cells and bacterial populations were kept naïve in order to capture phage evolution within the two differing environmental contexts (i.e., gut-on-a-chip and test tube). In total, we performed five successive transfers of evolved phages across three biological replicates in gut-on-a-chip devices and in test tubes, respectively (Fig. 2B).
Fig. 2.
Phages evolve in response to the mammalian mucus layer in the gut-on-a-chip. (A) Ancestral (zeroth transfer; T0) T4 phage and E. coli bacterial hosts were inoculated into three gut-on-a-chip and test-tube setups, respectively. The cocultures were incubated for 24 h with phages subsequently harvested for the first transfer (T1). Phages from T1 were transferred to either fresh chips or test tubes seeded with naïve E. coli B hosts and the process was repeated until the fifth transfer (T5). (B) Density of phages and bacteria from the mucus layer collected from the gut-on-a-chip (plain colored bars) and test-tube (textured colored bars) replicates at the end of each 24-h passage. The arrows along the x axis represent the experimental evolution phage transfer in-line with A. (C) Frequency of de novo mutations emerging from the phage population over five transfers from the gut-on-a-chip and test-tube setups. Colored lines represent the mutations: D246N hoc mutation in orange, Δ21bp goF mutation in purple, gp37 (distal subunit phage long-tail fiber) in brown, t holin lysis mediator in pink, and gp6 and gp9 (phage baseplate subunits) in teal. Gray lines represent other transient and low-frequency de novo mutations (Dataset S3). The arrows along the x axis represent the experimental evolution phage transfer in-line with A. (D) Average number of de novo mutations from phage populations evolved in gut-on-a-chip and test-tube conditions. Data points in B were plotted as a bar chart with error bars representing mean ± SEM of technical replicates (n = 2 or 3, except test-tube replicate 3, transfer 4 for phage, where n = 1). Data points in D were mutational counts from independent experimental replicates as specified in Dataset S3, with lines and error bars plotted as mean ± SEM across three experimental replicates (n = 3). The P value in D was derived from an unpaired two-tailed t test from three experimental replicates (n = 3).
Both phages and bacteria were consistently recovered from the gut-on-a-chip (Fig. 2B). In line with our previous findings, phage–bacteria population densities varied across all replicates between experimental transfers which again reflects on the stochasticity inherent to the gut-on-a-chip system (Fig. 1 E and F and SI Appendix, Fig. S2). This contrasted with test-tube controls where bacterial populations frequently went extinct (Fig. 2B, test-tube). We posit that the absence of spatial heterogeneity within a test-tube setting lends a high likelihood for productive phage–bacteria encounters. In contrast, the mucosal layer is spatially heterogeneous, largely dictated by the mucin gradient along the polarized mucosal epithelial layer (23, 24). This heterogeneity provides spatial refuge for bacterial micropopulations which consequently tempers phage predation pressures within the mucosal environment, resulting in stable tripartite coculture of phages, bacteria, and the mammalian mucosa (16).
Next, we sought to determine the evolutionary changes that occurred in the phage populations between gut-on-a-chip devices and test tubes using whole-genome sequencing, followed by read alignment and mutational calling. First, we screened our ancestral T4 phage population and discovered a number of background mutations comprising single-nucleotide polymorphisms (SNPs) and single-nucleotide insertions, reflective of their long-term laboratory storage and genetic drift (25) (Dataset S2). We subtracted these background SNPs and insertions from our mutational readouts in order to highlight de novo mutations arising in our gut-on-a-chip and test-tube evolved phage populations.
In the case of our gut-on-a-chip populations, de novo mutations were found in genes encoding nucleotide-binding and metabolism proteins, structural proteins, and hypothetical proteins, most of which were transient and at low abundance (Dataset S3). While we did not observe parallel evolution across our chip-evolved populations, two mutations attained high abundance within the first gut-on-a-chip replicate. The first was a nonsynonymous SNP within the hoc (highly immunogenic outer capsid) gene, which encodes for an accessory outer capsid protein containing three Ig-like domains that has been demonstrated to facilitate phage adherence to mucus (11). This SNP resulted in an amino acid change at position 246 from aspartic acid to asparagine (henceforth referred to as D246N Hoc). The second mutation was an in-frame 21-bp deletion (Δ21bp) of the goF gene which encodes for a transcription antitermination factor that antagonizes the bacterial ρ (Rho) termination factor from prematurely degrading phage messenger RNA transcripts (SI Appendix, Fig. S3) (26). At their peak frequencies in the fourth transfer, D246N Hoc and Δ21bp goF mutations achieved 83.3 and 72% of the population, which was subsequently followed by a decrease to 18.3 and 16.9%, respectively. While the exact cause for the unexpected decrease in mutational frequencies is not known, we speculate that this is likely due to the strict population bottleneck imposed by our phage transfer. The stochastic nature of the gut-on-a-chip mucosal setting may also render the phage population vulnerable to sudden population loss, especially under bottleneck constraints. In combination, these factors likely contributed to the sudden decrease in D246N Hoc and Δ21bp goF mutational frequencies in the fifth transfer. Despite this, their rise to high frequencies during the initial four transfers implies a selective advantage for these mutations within the gut-on-a-chip (Fig. 2C, gut-on-a-chip replicate 1).
Unlike with replicate 1, we did not observe significant evolutionary events in gut-on-a-chip replicates 2 and 3. Here, mutations were transient and chiefly involved hypothetical genes and intergenic regions except the gp37 long-tail fiber subunit in replicate 2 and the t holin lysis mediator in replicate 3. Collectively, the varied mutational profiles between the replicate gut-on-a-chip phage populations further reflect on the stochastic nature of phage–bacterial populations within the tripartite system.
By contrast, phages evolving in test tubes largely possessed mutations in coding regions that were directly involved with bacterial infection. This was anticipated since bacteria were the only other biological entity alongside phages in test-tube settings. In particular, mutations in gp37—encoding the phage long-tail fiber distal subunit responsible for phage adsorption onto bacterial hosts—were found across all test-tube replicate populations. However, these gp37 mutations were observed transiently and at low frequency in all test-tube replicate populations (Fig. 2C, test-tube replicates and Dataset S3). We also observed other mutations affecting phage baseplate-associated genes (gp6 and gp9), whose gene products facilitate phage genome injection into the bacterial host during infection. We note that these baseplate mutations were only present in test-tube replicate 3, and only one mutation affecting gp9 which resulted in a synonymous SNP at residue 141 persisted throughout transfers 1 to 5 (Fig. 2C, test-tube replicate 3). Despite the disparate de novo mutation profiles between gut-on-a-chip and test-tube populations, we did not observe significant differences in total number of de novo mutations across the five transfers between the gut-on-a-chip (6.3 ± 0.9 mutations) and test-tube phage populations (7.0 ± 2.1 mutations) (Fig. 2D and Dataset S3; unpaired two-tailed t test: t = 0.2561, degrees of freedom [d.f.] = 4, n = 3 [three experimental replicates], P = 0.8105). Nonetheless, the contrasting mutational profiles between phages evolving in the gut-on-a-chip and test tube suggest the potential role of the mucosal layer as an active environmental contributor in driving phage evolution. This reflects upon the increasing need to conduct phage evolution studies in more representative environmental conditions over the classical, “gold standard” test-tube approach.
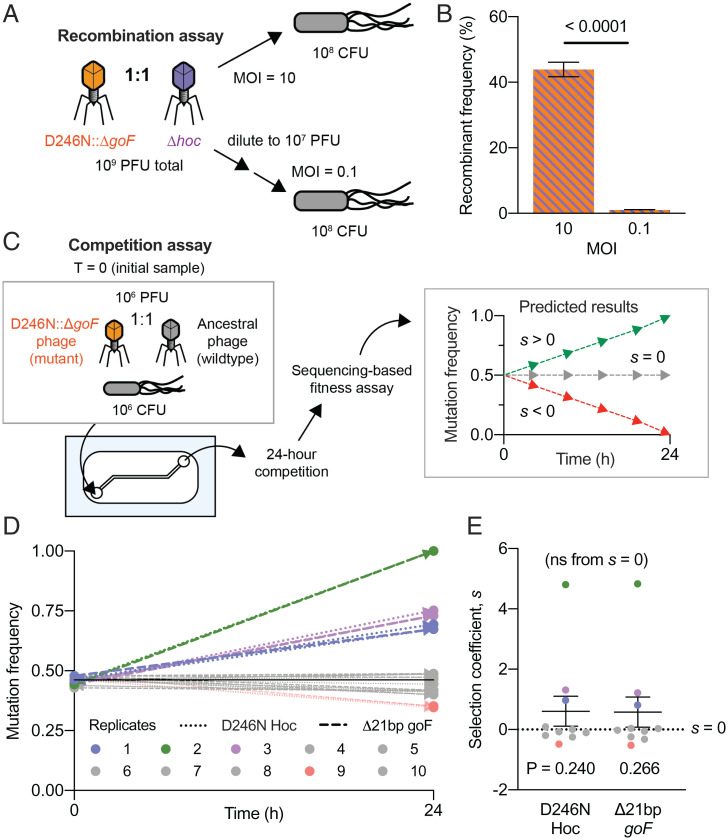
High MOI Is a Driver for Phage Recombination.
In asexual populations, such as phages, genetic recombination is key to enhancing fitness by alleviating clonal interference and genetic hitchhiking (27, 28). For lytic phages like T4, recombination occurs when multiple phage genotypes coinfect the same bacterial host, allowing for allelic exchange between the phage genomes (29). Since coinfections drive recombination, higher MOIs typically render higher recombination rates (29). Crucially, high MOIs were sustained in our gut-on-a-chip devices, where elevated phage-to-bacteria ratios were observed (Figs. 1E and 2B). In our first gut-on-a-chip replicate, we saw the D246N Hoc and Δ21bp goF mutations emerged from the first transfer and progressed with independent frequencies until the third transfer. Hereafter, both mutations followed an intertwined frequency trajectory, increasing beyond 50% of the population in the fourth transfer (Fig. 2C, test-tube replicate 1). Their intertwined trajectories surpassing 50% frequencies indicate that the mutations had recombined onto a shared genetic background to overcome clonal interference. We sought to verify whether high phage-to-bacteria ratios—and thus high MOI—were drivers for recombination in lytic phage populations. To this end, we screened the phage populations from transfer 4 of our experimental evolution and were able to successfully plaque-isolate the D246N Hoc::Δ21bp goF double-mutant phage (herein referred to as the D246N::ΔgoF phage). Next, we initiated one-step phage growth experiments at high and low MOIs (i.e., 10 and 0.1, respectively) with a 1:1 mix of two phage mutants: 1) experimentally derived D246N::ΔgoF phage, and 2) laboratory-stock hoc-deletion mutant (Δhoc) with a wild-type goF gene (Fig. 3A). We also performed one-step growths of the two phage mutants in isolation to confirm that there were no significant differences in latent phase and burst size between the phages (SI Appendix, Fig. S4). Using these two independent gene-deletion phage mutants, namely the Δ21bp goF (gene position: 5842 to 6267) and the Δhoc (gene position: 110187 to 111317), allowed for PCR screening of recombinant phages. By limiting the phages to a single growth step, we limit phage recombination within a single replicative cycle. Following PCR screening of individual plaques, we found that 44% of phage progeny were recombinants at high MOI conditions, with a bias toward wild-type recombinants (43/98 phages screened were recombinants; 31/43 of wild-type recombinants; Fig. 3B; unpaired two-tailed t test, t = 19.59, d.f. = 194, n = 98 [plaques screened], P < 0.0001; SI Appendix, Fig. S5). Meanwhile, only 1 wild-type recombinant phage was detected from 98 isolates screened from low-MOI conditions, namely ∼1% recombinant frequency (Fig. 3B and SI Appendix, Fig. S5). Collectively, this demonstrates that a high phage-to-bacteria ratio facilitates genetic recombination in lytic phages. Furthermore, the rapid emergence of recombinants within a single phage replication cycle suggests that recombination is a key driving force for phage evolution, particularly within the context of sustained high phage-to-bacteria ratios as seen in the gut-on-a-chip devices. This in turn alleviates clonal interference and potentially promotes the selection of beneficial mutations.
Fig. 3.
Mucus layer supports phage recombination and selection of beneficial mutants. (A) A gut-on-a-chip–evolved D246N::ΔgoF phage mutant was mixed at a 1:1 ratio with a laboratory-derived Δhoc mutant. Naïve E. coli were infected with the 1:1 phage mixture at MOIs 10 and 0.1 following a one-step growth protocol to ensure that only a single round of viral replication could occur. (B) Percentage frequency of phage recombinants from PCR screening for Δ21bp goF-Δhoc or wild type–reconstituted recombinants. Forty-nine phage isolates (n = 49) were screened per experimental replicate. There were two experimental replicates (n = 2) leading to a total of 98 phage isolates (n = 98) screened per experimental condition (MOI 10 or 0.1) (SI Appendix, Fig. S5). (C) Competition experiment between the D246N::ΔgoF phage mutant with ancestral phage T4 in the gut-on-a-chip. A gut-on-a-chip seeded with naïve E. coli was inoculated with equal proportions of the respective phage genotypes. Chip effluents were collected for 1 h at 24 h of competition and the samples were subjected to whole-genome sequencing to track D246N Hoc and Δ21bp goF mutations after 24 h of competition. Estimated selection coefficients could be positive (s > 0), neutral (s = 0), or negative (s < 0). (D) D246N Hoc and Δ21bp goF mutational frequencies measured from 10 independent gut-on-a-chip replicates (n = 10) between the initial inoculum, namely T = 0 and 24 h. (E) Plot of the estimated mean selection coefficient for D246N Hoc and Δ21bp goF mutations across 10 experimental replicates (n = 10). The black solid line in D represents the initial (T = 0) average frequency of D246N Hoc mutation at 45.8% and Δ21bp goF mutation at 46.7%, across 10 experimental replicates (n = 10). Error bars in B and the line with error bars in E represent mean ± SEM across experimental replicates (B: n = 2, i.e., two experimental replicates, n = 98 screened per experimental replicate; E: n = 10, i.e., ten experimental replicates). P values in B were derived from an unpaired two-tailed t test between treatment conditions (MOI 10 and 0.1). P values in E were derived from an unpaired two-tailed t test between coefficients of D246N Hoc and Δ21bp goF mutations against s = 0 (no significance; ns), respectively.
Phage Mutant Outcompetes the Ancestor Phage in the Gut-on-a-Chip.
To assess the fitness of the evolved D246N::ΔgoF phage, we competed the evolved phage against its ancestral counterpart in the gut-on-a-chip mucus environment. Competition between the D246N::ΔgoF phage and ancestral phage was initiated by inoculating both phages at a 1:1 ratio into 10 gut-on-a-chip replicate devices seeded with a naïve bacterial host. Each device was maintained at a 120 µL/h flow rate for 24 h as previously described, with the effluent collected for 1 h at the 24-h end point of the competition assay. The sampled effluent was subsequently whole genome–sequenced to track the D246N Hoc and Δ21bp-goF mutation frequencies over 24 h of competition (Fig. 3C). We verified that our devices were accurately seeded with roughly equal proportions of mutant and wild-type phages as reflected by ∼46% frequency of both the D246N Hoc and Δ21bp goF mutations at the initial experimental time point (t = 0) (Fig. 3D). We observed the D246N::ΔgoF phage outcompeted the wild-type phage in 3 out of 10 replicate devices, eventually fixing in one of the replicate populations. Meanwhile, six replicates showed no change from the initial frequency whereas the one remaining replicate showed a decline of the mutant phage to 35% (Fig. 3D). To ascertain the strength of selection, we quantified the selection coefficient (s) across the replicate populations with coefficients being either positive (s > 0), neutral (s = 0), or negative (s < 0) (Fig. 3C and Dataset S4). On average, we found a positive selection coefficient with s = 0.6 for both D246N Hoc and Δ21bp goF, although significance from null selection, namely s = 0, was not attained due to significant variability between experimental replicates (Fig. 3E; unpaired two-tailed t test of each mutation against s = 0, tD246N Hoc = 1.216, tΔ21bp goF = 1.149, d.f.D246N Hoc = 18, d.f.Δ21bp goF = 18, PD246N Hoc = 0.2398, PΔ21bp goF = 0.2657; Dataset S4). In summary, the D246N and ΔgoF mutations confer a mild fitness benefit to the phage within the mucosal environment.
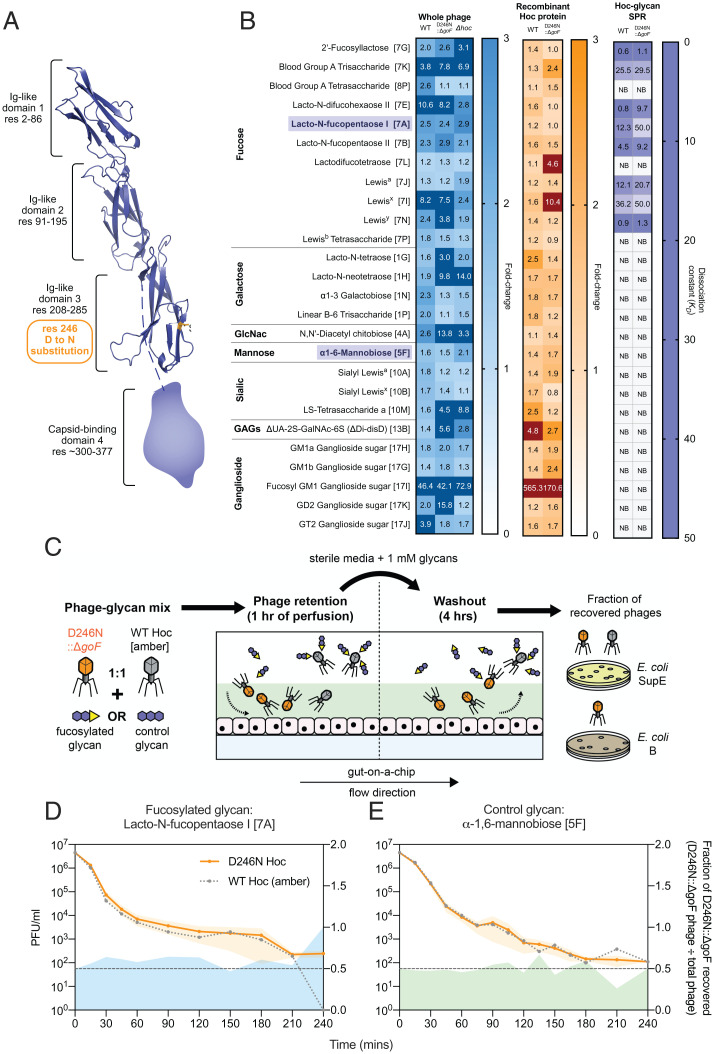
Hoc Mutation Alters the Phage Mucus-Adherence Phenotype.
Phage adherence to mucus has been described as a mechanism that facilitates phage enrichment and persistence within the mammalian mucosal layers (11). For the T4 phage, this adherence phenotype is facilitated by the outer capsid protein Hoc, which has three externally displayed Ig-like domains and a highly conserved fourth C-terminal capsid-binding domain (30, 31). The D246N Hoc mutation removes an acidic residue (aspartic acid) and replaces it with a neutral residue (asparagine). This mutation is located within the third Ig-like domain, potentially altering Hoc binding affinity to mucin glycans (Fig. 4A). To test for altered glycan adherence, we fluorescently labeled whole-phage particles of wild-type Hoc, D246N::ΔgoF, and Δhoc genotypes, and assayed for glycan binding on a microarray printed with 153 unique glycan structures (Dataset S5). Binding was measured as fold changes relative to the array background signal and verified for P value significance. Overall, we were able to observe binding of whole phages across seven glycan families. D246N::ΔgoF phages generally exhibited altered glycan binding compared with wild-type phage, while Δhoc phages had lower overall fold-change intensities relative to wild-type and D246N::ΔgoF phages (Fig. 4B, whole phage). To further investigate the specificity of Hoc–glycan interactions, we recombinantly expressed wild-type and D246N Hoc proteins (SI Appendix, Fig. S6) and tested the proteins on the glycan array. We showed that the Hoc protein–glycan binding largely matched whole-phage binding results (Fig. 4B, recombinant Hoc protein). Next, surface plasmon resonance (SPR) was adopted to quantify the binding strength between glycans and surface-immobilized Hoc proteins. We focused on a subset of 26 glycans that were amenable for SPR measurements taken in solution under flow (see Dataset S5 for full glycan array analysis). The SPR data demonstrated that both wild-type and D246N Hoc–glycan binding were specific for interactions with the same subset of fucosylated glycans (Fig. 4B, Hoc-glycan SPR). Furthermore, the D246N Hoc protein had higher dissociation values (KDs), indicating weaker binding to a number of fucosylated glycans than the wild-type Hoc (Fig. 4B, Hoc-glycan SPR and S7). Fucosylated mucin glycans are ubiquitous along the human gastrointestinal tract in individuals possessing a functional copy of the α-1,2-fucosyltransferase (FUT2) gene (known as “secretors”) (32). Our gut-on-a-chip HT29-MTX-E12 cell line possesses FUT2 and is capable of producing a fucosylated mucus layer in-line with the secretor phenotype (33).
Fig. 4.
Phages evolved in the mammalian mucus layer exhibit altered the mucus-adherence phenotype. (A) T4 Hoc protein structure model demonstrating the position of the D246N mutation within the third Ig-like domain, highlighted in orange. The capsid-binding fourth domain was not modeled due to the lack of structural homologs in the Protein Data Bank. (B) Normalized fold-change fluorescence intensities of the 26 top glycan array hits (glycan ID corresponding to Dataset S5 as indicated in square brackets) of labeled, ultrapurified whole phages: wild-type (WT), D246N::ΔgoF phage, and Δhoc: blue heatmap; recombinantly expressed Hoc proteins: wild-type and D246N: orange heatmap; followed by SPR assessing glycan-to-Hoc protein binding strength: purple heatmap. Numerical values in glycan array heatmaps represent fold-change magnitude normalized against background fluorescence where dark-color panels indicate high fold-change values that were out-of-bounds from the heatmap gradient. Numerical values in the SPR heatmap represent KD values where higher KD values indicate lower binding affinity. “NB” in the SPR heatmap indicates no binding event. Bolded and purple-highlighted glycans 7A (lacto-N-fucopentaose I) and 5F (α-1,6-mannobiose) represent the glycans selected for phage retention and washout experiments in D and E. (C) Experimental setup for phage retention and washout from the gut-on-a-chip, where equal proportions of wild-type Hoc (with am43−/44− mutation) and D246N::ΔgoF phages in 1 mM glycan solutions were perfused in the gut-on-a-chip for 1 h during the retention phase. Subsequently, sterile media supplemented with 1 mM glycan were perfused for 4 h to initiate phage washout from the mucus layer. Washouts were collected at set time intervals and phages were quantified via selective plating on E. coli SupE (permissive for both wild-type Hoc [am43−/44−] and D246N::ΔgoF phage) and E. coli B (only permissive for D246N::ΔgoF phage). (D) Washout of wild-type Hoc and D246N::ΔgoF phages from the gut-on-a-chip under flow with 1 mM fucosylated glycan 7A (lacto-N-fucopentaose I) or (E) control glycan 5F (α-1,6-mannobiose) over 4 h. Lines with shaded regions in D and E were plotted along the left y axis as mean ± SEM of three experimental replicates (n = 3) per time point. The plotted gray dotted line with data points represents the expected number of wild-type Hoc phages recovered by subtracting D246N::ΔgoF phages from the total phage number recovered. Shaded regions extending along the x axis and right y axis represent the D246N::ΔgoF phage fraction over the total phage number recovered (D246N::ΔgoF phage ÷ total phage recovered) from the gut-on-a-chip across time. The black dotted line along 0.5 of the right y axis represents the fraction expected if D246N Hoc and wild-type Hoc phages were recovered at equal proportions.
With this knowledge, we proceeded to validate the glycan-binding phenotype of the D246N::ΔgoF phage within the gut-on-a-chip mucosal environment. We tested this by competing the D246N::ΔgoF phage against the wild-type Hoc phage in a phage retention and washout assay within the gut-on-a-chip, in the presence of either a fucosylated (lacto-N-fucopentaose I [7A]) or nonfucosylated (α-1,6-mannobiose [5F]) glycan. Importantly, the fucosylated glycan chosen (lacto-N-fucopentaose I [7A]) showed an ∼4-fold reduction in KDs between the wild-type and D246N::ΔgoF phages, while the nonfucosylated glycan showed no interaction with either phage. We initiated the experiment by infusing three replicate devices, each with a 1:1 ratio of wild-type Hoc and D246N::ΔgoF phages suspended in either fucosylated or nonfucosylated glycan solutions, followed by washout of the phages from the devices using the same glycan solutions (Fig. 4C and SI Appendix, Fig. S8). We posit that the wild-type Hoc phage, possessing higher affinity to dissolved fucosylated glycans, will be competitively sequestered from the mucus layer during the initial infusion, while the D246N::ΔgoF phage, with its lower affinity to fucosylated glycans, will be selectively retained in the mucus. Consequently, during the washout, we expect higher recovery of the D246N::ΔgoF phage for extended periods over the wild-type Hoc phage. To determine the fraction of the D246N::ΔgoF phage recovered from the total phage infused, we utilized a wild-type Hoc phage possessing amber mutations on genes 43 and 44 (herein known as T4 am43−/44−) that was permissive only on E. coli strain SupE, while the D246N::ΔgoF phage was permissive on both E. coli strains B and SupE. Our results showed that in the presence of the fucosylated glycan lacto-N-fucopentaose I (7A), the D246N::ΔgoF phage was consistently recovered at higher levels in the first 2 h of washout and remained detectable up to 4 h, whereas the wild-type Hoc phage was eliminated by 4 h (Fig. 4D and SI Appendix, Fig. S8B). Conversely, in the control setting with the nonfucosylated glycan α-1,6-mannobiose (5F), fewer D246N::ΔgoF phages were recovered compared with wild-type Hoc phages in the first 1.5 h, followed by recovery at approximately equal proportions up to 4 h of washout (Fig. 4E and SI Appendix, Fig. S8B). Collectively, this indicates that more D246N::ΔgoF phages were retained and recovered from devices perfused with fucosylated glycan over nonfucosylated glycan, as predicted from our SPR results (SI Appendix, Fig. S8C). In summary, we demonstrate that the T4 phage outer capsid protein Hoc binds specifically to fucosylated glycans and exhibited changes in glycan binding affinity through mutation in the protein’s Ig-like binding domain. On a broader perspective, our results revealed the capacity of phages to mutate phage display proteins in order to adapt to the tripartite conditions imparted by the gut-on-a-chip mucosal environment.
Discussion
Phages are largely considered inert with respect to the mammalian host and chiefly respond to antagonistic selection from their immediate replicative bacterial hosts. However, the mammalian milieu—in this case, the gut mucosa—is also a complex environment that can impose additional selection pressures such as mucus turnover dynamics and glycosylation that act on both bacterial and viral entities (32, 34, 35). Under these mucosal selection pressures, phage variants arising from propagation on resident bacterial populations that display competitive advantage will be retained and further propagated. Our gut-on-a-chip system recapitulates these selection pressures and hence favors phage phenotypes that persist within the mucosal layer. Here, we adopt the model phage T4 which has previously been demonstrated to adhere to mucus (11). By experimentally evolving the phage within the gut-on-a-chip mucosa inoculated with bacterial hosts, we revealed the capacity of the phage to evolve its mucus-binding domains within a tripartite setting, resulting in enhanced persistence within the mucosal environment. While our findings are specific to the T4 phage, a significant utility of the gut-on-a-chip—and more broadly, the organ-on-a-chip system—is its amenability to also extend evolutionary and ecological forays into alternative phage–bacteria pairs within various tripartite contexts. The tripartite setting could also be built upon by culturing multiphage and microbial communities [e.g., microbiome-on-a-chip (20)]. Given the growing importance of environmental and community contexts in phage evolution (36, 37), our study provides a proof of concept for leveraging the organ-on-a-chip platform to examine phage evolution within the mammalian mucosal context.
Phage–bacteria population dynamics was disparate between independent gut-on-a-chip populations. We collectively demonstrated this through high temporal resolution (Fig. 1E; n = 3) and 24-h end-point measurements (Fig. 2B; n = 15), both of which amounted to 18 independent measurements, each reporting unique phage and bacteria densities in the gut-on-a-chip environment. Similarly, disparity was also observed in mutations emerging from independently evolving phage populations in the gut-on-a-chip (Fig. 2C). This led us to speculate that stochastic ecological effects arising from demographic noise and mucosal spatial complexity (38) could be key factors in determining mucosal selection within independent gut environments. In a broader perspective, these observations may partly reflect on the interpersonal variations seen in gut viral community dynamics (5, 39), although more nuanced empirical approaches are warranted to test these speculations. Despite the ecological variation and mutational disparity between gut-on-a-chip phage populations, we acquired a genetically recombined phage mutant exhibiting altered affinity toward fucosylated glycans (Fig. 4C) via a mutation in the phage Hoc mucus-adhering domain (Fig. 4A).
We demonstrated that the acquired phage Hoc mutant conferred a fitness advantage within the mucus layer by altering phage Hoc affinity to mucin glycans, specifically by decreasing affinity to fucosylated glycan structures (Figs. 3E and 4B). While diminished phage–glycan binding may appear counterintuitive as a fitness advantage for persistence in the mucosal environment, we note that 1) the exact glycosylation profile and glycan abundance of the gut-on-a-chip mucus layer were unknown, and 2) our SPR screen was limited to a small subset of fucosylated glycans. Nonetheless, we were able to show that the Hoc mutation lent a detectable phenotypic response within the mucosal environment when titrated against fucosylated and nonfucosylated glycans (Fig. 4 D and E). Meanwhile, the Δ21bp goF mutation—also found within the D246N Hoc mutant genetic background—resulted in the loss of seven amino acids within an acidic region of the protein previously reported in other RNA-binding proteins (SI Appendix, Fig. S3) (40). Given that GoF functions as an intracellular antitermination factor, we hypothesized that this deletion likely results in a loss-of-function phenotype, affecting the phage replicative lifecycle. However, we reported no significant differences in one-step growth curves between the wild-type phage and the phage harboring the Δ21bp goF mutation (SI Appendix, Fig. S4). To date, the phenotype conferred by the Δ21bp goF mutation remains elusive and selection benefits may be conferred through a mechanism that is yet to be known. Collectively, our findings provide empirical support for the capacity of phages to adapt to mucin glycosylation, which subsequently enhanced phage persistence and propagation within the gut-on-a-chip mucosal environment.
Mucin fucosylation is widespread along the gastrointestinal tract of functional FUT2 human genotypes (known as secretors), especially within the proximal and distal colon (35). This suggests that the human host genotype and glycosylation demography directly influence gut phage biogeography at the inter- and intraindividual level, respectively. Moreover, the majority of the gut phageome possesses open reading frames for variable glycan-binding superfamily domains (41, 42). The model T4 phage adopted in this study is one of such phages with glycan-binding domains and is also a natural component of the human gut phageome in both healthy and diseased states (43–45). While not all phages possess capsid display Ig-like domains, these domains are found within approximately a quarter of sequenced Caudovirales phages to date (18). However, since most gut phages remain unannotated despite recent sequencing and genome assembly efforts (46, 47), there exist an immense abundance, diversity, and prevalence of phage structural domains that could play similar roles in associating with the mammalian entity, such as proteoglycan-binding domains (15), C-type lectin folds (41, 48), and the Ig folds of Bacteroides-associated carbohydrate-binding often N-terminal (BACON) domains (49). This suggests that gut phages have immense adaptive freedom to respond and coevolve with an individual’s unique mucosal glycosylation patterns to foster persistence (11).
Given the potential diversity of gut phages with capsid display proteins—which can accommodate large sequence variations—coupled with the individuality of the mammalian gut, the evolutionary potential between phages and their mammalian environment remains largely untapped. Our study represents a foray into this evolutionary potential between phages and their mammalian host. We posit that the mammalian gut has a significant role in selecting successful phage variants that emerge and persist in the gut, which will then propagate and potentially recombine to promote the fixation of beneficial mutations within the population. This subsequently dictates the phage populations that will reside and further engage in evolution with both the individual’s gut microbiome and gut environment. In a broader view, this tripartite evolutionary interplay might lend stable, long-term, and highly personalized viromes and microbiomes, often recapitulated in human metagenomic cohort studies (5, 46). To this end, we also envisage future directions toward human host-centric intelligent phage design in synergy with host-directed phage evolution for highly personalized medicine and refined in vivo phage applications.
Methods and Materials
Methods Summary.
Gut-on-a-chip devices were manufactured using soft lithography techniques. Each device was seeded with 3.0 × 105 cells of an HT29-MTX-E12 tumorigenic goblet cell line, maintained in antibiotic-free complete Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) under flow conditions; a 120 µL/h flow rate was used for all experiments unless stated otherwise. Phage–bacteria dynamics in the gut-on-a-chip was assessed by inoculating each replicate device with phages and bacteria at an MOI of 1 or 10, followed by 24-h incubation under flow and quantification using qPCR for high temporal resolution tracking, or traditional agar plating for 24-h end-point densities. For the phage experimental evolution, five experimental evolution transfers were performed per experimental replicate, with each transfer initiated on a fresh gut-on-a-chip device (i.e., naïve HT29-MTX-E12 mucosal layer and 104 CFUs E. coli B), with 104 phage PFUs recovered from the previous device at the 24-h end point. Recovered phages from each passage including the ancestral population were whole genome–sequenced and analyzed for mutations using the breseq polymorphism mixed population pipeline (50). A phage recombination assay was initiated at MOI 10 and MOI 0.1 between the laboratory-stock Δhoc phage and D246N::ΔgoF mutant phage (double plaque isolated from experimental evolution transfer 4), where recombinants were screened for deletions using PCR. A sequencing-based competition assay was performed by competing equal proportions of 106 PFUs wild-type T4 phage and the double-plaque isolated D246N::ΔgoF mutant phage in the gut-on-a-chip for 24 h under flow. Meanwhile, binding assays to a printed glycan array were performed by assessing fluorescence intensities of bound whole phages and recombinantly expressed Hoc protein on the array. For SPR, Hoc proteins were immobilized on HisCap biosensors (Sartorius) followed by glycan perfusion and elution steps to quantify glycan–Hoc binding affinity. Finally, phage retention and washout assays were performed by inoculating each replicate gut-on-a-chip device with equal proportions of 107 PFUs per milliliter wild-type Hoc T4 phage (am43−/44−) and D246N::ΔgoF mutant phage followed by phage washout for 4 h under flow, where the phages recovered over time were selectively plated and quantified on E. coli B and E. coli SupE lawns.
Refer to SI Appendix, Methods and Materials for details of culture, experimental procedures, and materials along with additional data.
Supplementary Material
Acknowledgments
This work, including the efforts of J.J.B., was funded by an Australian Research Council Discovery Early Career Researcher Award (DE170100525). We thank the Monash Ramaciotti cryoelectron microscopy platform for the use of their facilities and A. de Marco for granting access to the scanning electron microscope. This work was performed in part at the Melbourne Centre for Nanofabrication in the Victorian Node of the Australian National Fabrication Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116197119/-/DCSupplemental.
Data Availability
Raw sequence read data reported in this article have been deposited in the National Center for Biotechnology Information, Sequence Read Archive under the accession no. PRJNA737295 (51) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA737295/). All other study data are included in the article and/or supporting information.
References
- 1.Buckling A., Rainey P. B., Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269, 931–936 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall A. R., Scanlan P. D., Buckling A., Bacteria-phage coevolution and the emergence of generalist pathogens. Am. Nat. 177, 44–53 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Betts A., Kaltz O., Hochberg M. E., Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 111, 11109–11114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson S., et al. , Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes A., et al. , Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes A., Wu M., McNulty N. P., Rohwer F. L., Gordon J. I., Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. U.S.A. 110, 20236–20241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Sordi L., Khanna V., Debarbieux L., The gut microbiota facilitates drifts in the genetic diversity and infectivity of bacterial viruses. Cell Host Microbe 22, 801–808.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 8.De Sordi L., Lourenço M., Debarbieux L., “I will survive”: A tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 10, 92–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirzaei M. K., Maurice C. F., Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I., Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Barr J. J., et al. , Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr J. J., et al. , Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. U.S.A. 112, 13675–13680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida G. M. F., Laanto E., Ashrafi R., Sundberg L. R., Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. mBio 10, e01984-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan J., et al. , Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 8, 5091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green S. I., et al. , Targeting of mammalian glycans enhances phage predation in the gastrointestinal tract. mBio 12, e03474-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lourenço M., et al. , The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hansson G. C., Mucins and the microbiome. Annu. Rev. Biochem. 89, 769–793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser J. S., Yu Z., Maxwell K. L., Davidson A. R., Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J. Mol. Biol. 359, 496–507 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Kim H. J., Li H., Collins J. J., Ingber D. E., Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U.S.A. 113, E7–E15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalili-Firoozinezhad S., et al. , A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son Y., Determination of shear viscosity and shear rate from pressure drop and flow rate relationship in a rectangular channel. Polymer (Guildf.) 48, 632–637 (2007). [Google Scholar]
- 22.Kim H. J., Huh D., Hamilton G., Ingber D. E., Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Pelaseyed T., et al. , The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tropini C., Earle K. A., Huang K. C., Sonnenburg J. L., The gut microbiome: Connecting spatial organization to function. Cell Host Microbe 21, 433–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subedi D., Barr J. J., Temporal stability and genetic diversity of 48-year-old T-series phages. mSystems 6, e00990-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanson B., Uzan M., Sequence and characterization of the bacteriophage T4 comC alpha gene product, a possible transcription antitermination factor. J. Bacteriol. 174, 6539–6547 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang G. I., et al. , Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald M. J., Rice D. P., Desai M. M., Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531, 233–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosig G., The effect of multiplicity of infection on recombination values in bacteriophage T4D. Z. Vererbungsl. 93, 280–286 (1962). [DOI] [PubMed] [Google Scholar]
- 30.Sathaliyawala T., et al. , Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 77, 444–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fokine A., et al. , Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J. Virol. 85, 8141–8148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickard J. M., Chervonsky A. V., Intestinal fucose as a mediator of host-microbe symbiosis. J. Immunol. 194, 5588–5593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns M. T., et al. , Glycosylation-related gene expression in HT29-MTX-E12 cells upon infection by Helicobacter pylori. World J. Gastroenterol. 23, 6817–6832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson J. K., et al. , Dynamic changes in mucus thickness and ion secretion during Citrobacter rodentium infection and clearance. PLoS One 8, e84430 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashyap P. C., et al. , Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 110, 17059–17064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevallereau A., Pons B. J., van Houte S., Westra E. R., Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol. 20, 49–62 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Blazanin M., Turner P. E., Community context matters for bacteria-phage ecology and evolution. ISME J. 15, 3119–3128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vellend M., Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Lim E. S., et al. , Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller E. S., et al. , Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67, 86–156 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minot S., Grunberg S., Wu G. D., Lewis J. D., Bushman F. D., Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 109, 3962–3966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutilh B. E., et al. , A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhillon T. S., Dhillon E. K. S., Chau H. C., Li W. K., Tsang A. H., Studies on bacteriophage distribution: Virulent and temperate bacteriophage content of mammalian feces. Appl. Environ. Microbiol. 32, 68–74 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackermann H. W., Nguyen T. M., Sewage coliphages studied by electron microscopy. Appl. Environ. Microbiol. 45, 1049–1059 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furuse K., et al. , Bacteriophage distribution in human faeces: Continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol. 64, 2039–2043 (1983). [DOI] [PubMed] [Google Scholar]
- 46.Shkoporov A. N., et al. , The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Camarillo-Guerrero L. F., Almeida A., Rangel-Pineros G., Finn R. D., Lawley T. D., Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMahon S. A., et al. , The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 12, 886–892 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Jonge P. A., Meijenfeldt F. A. B. V., Rooijen L. E. V., Brouns S. J. J., Dutilh B. E., Evolution of BACON domain tandem repeats in crAssphage and novel gut bacteriophage lineages. Viruses 11, 1085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deatherage D. E., Traverse C. C., Wolf L. N., Barrick J. E., Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front. Genet. 5, 468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin W. H., et al. , Data files pertaining to “Bacteriophages evolve enhanced persistence to a mucosal surface.” https://www.ncbi.nlm.nih.gov/bioproject/PRJNA737295. Deposited 14 June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence read data reported in this article have been deposited in the National Center for Biotechnology Information, Sequence Read Archive under the accession no. PRJNA737295 (51) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA737295/). All other study data are included in the article and/or supporting information.