Significance
Heme (iron-protoporphyrin IX complex) is extremely cytotoxic when in excess, although it plays an important role in numerous biological processes. Due to its lipophilicity, free heme is concentrated in lipid membranes, generating harmful reactive oxygen species. Pathogenic bacteria overcome heme toxicity via degradation, sequestration, and extrusion, while they assimilate heme derived from the host heme-containing proteins as an iron nutrient. To eliminate heme toxicity, Corynebacterium diphtheriae and many gram-positive bacteria possess a heme-dedicated efflux pump, HrtBA, in their cytoplasmic membranes. Our functional and structural data reveal how HrtBA expels heme from the membrane using ATP, elucidating a mechanism of bacterial heme detoxification in host blood.
Keywords: heme, ABC transporter, membrane, pathogenic bacteria, heme toxicity
Abstract
Bacterial pathogens acquire heme from the host hemoglobin as an iron nutrient for their virulence and proliferation in blood. Concurrently, they encounter cytotoxic-free heme that escapes the heme-acquisition process. To overcome this toxicity, many gram-positive bacteria employ an ATP-binding cassette heme-dedicated efflux pump, HrtBA in the cytoplasmic membranes. Although genetic analyses have suggested that HrtBA expels heme from the bacterial membranes, the molecular mechanism of heme efflux remains elusive due to the lack of protein studies. Here, we show the biochemical properties and crystal structures of Corynebacterium diphtheriae HrtBA, alone and in complex with heme or an ATP analog, and we reveal how HrtBA extracts heme from the membrane and releases it. HrtBA consists of two cytoplasmic HrtA ATPase subunits and two transmembrane HrtB permease subunits. A heme-binding site is formed in the HrtB dimer and is laterally accessible to heme in the outer leaflet of the membrane. The heme-binding site captures heme from the membrane using a glutamate residue of either subunit as an axial ligand and sequesters the heme within the rearranged transmembrane helix bundle. By ATP-driven HrtA dimerization, the heme-binding site is squeezed to extrude the bound heme. The mechanism sheds light on the detoxification of membrane-bound heme in this bacterium.
Heme (iron-protoporphyrin [PP] IX complex) is one of the most pivotal metal cofactors across the kingdoms of life and is utilized in numerous biological processes, including oxygen metabolism. For their survival and virulence in hosts, pathogenic bacteria can break down the host erythrocytes and acquire the free heme from hemoglobin via a bacterial heme-acquisition system that includes heme-binding proteins and an ATP-binding cassette (ABC) importer (1). The bacteria utilize the internalized heme intact as a component of hemoproteins, although they can also endogenously synthesize it or enzymatically extract the iron from the internalized heme for use as a nonheme iron (1). However, because of its lipophilicity and unruly chemical reactivity, unassimilated heme can have deleterious effects on cells, such as causing membrane disorder and generating reactive oxygen species (ROS) that oxidatively damage proteins, lipids, and DNA (2).
The cytoplasmic membranes of many gram-positive bacteria are exposed to high concentrations of heme from lysed erythrocytes because these bacteria lack outer-membrane barriers. To tolerate heme toxicity, these bacteria have evolved heme-dedicated efflux pumps in the cytoplasmic membranes (1). The heme-regulated transporter genes hrtB and hrtA (hrtBA), encoding an ABC transporter, were discovered among genes that were highly expressed in Staphylococcus aureus in response to exogenous heme (3) and were found to confer heme resistance (4–7). Intriguingly, Corynebacteriales, gram-positive diderms possessing atypical outer membranes (8, 9), also carry these genes (10) (SI Appendix, Fig. S1). While HrtA contains a well-conserved nucleotide-binding domain (NBD), HrtB is predicted to possess a transmembrane (TM) domain (TMD) with four TM helices and an extracytoplasmic domain (ECD) protruding between TM1 and TM2 (11) (SI Appendix, Fig. S1). Although heme detoxification strategies can entail sequestration in proteins, degradation, and extrusion of heme (in addition to ROS dismutation), genetic analyses revealing that heme generates ROS in cytoplasmic membranes (11, 12) suggest that HrtBA extrudes heme from membranes (11).
The molecular mechanism of heme efflux, however, remains unclear because to the best of our knowledge, biochemical and structural studies have not been performed. In the present study, we characterized the functional properties of HrtBA from a gram-positive diderm—Corynebacterium diphtheriae, the causative agent of diphtheria—and determined the crystal structures of its unliganded (substrate- and nucleotide-free), heme-bound, and nucleotide-bound states. Based on these data, we reveal a heme detoxification mechanism in which HrtBA extracts heme from the membrane and releases it using ATP.
Results
Functional Characterization of HrtBA.
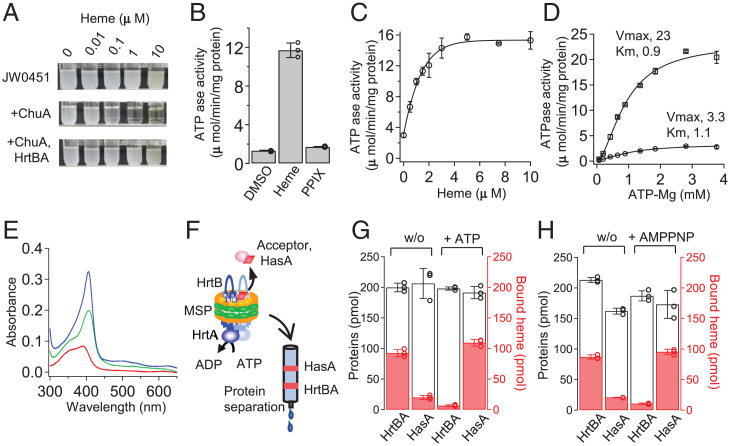
We first examined heme detoxification by HrtBA using a recombinant heme-sensitive Escherichia coli K-12 strain. The parental E. coli strain, JW0451, is naturally resistant to exogenous heme, with growth doubling times of 1.1 h at 1 and 10 μM heme (Fig. 1 A, Top and SI Appendix, Fig. S2); its outer membrane is impermeable to heme because of the lack of a heme receptor protein. However, the expression of ChuA protein, which is responsible for heme internalization across the outer membrane, from the pathogenic bacterium E. coli O157 (13) led to growth retardation at 1 and 10 μM heme with prolonged doubling times (2.4 and 2.7 h, respectively) (Fig. 1 A, Middle and SI Appendix, Fig. S2). These observations indicate that heme that passes through the outer membrane is cytotoxic to the bacterium. The expression of HrtBA in the heme-sensitive cells rescued growth at 1 and 10 μM heme (doubling times of 1.4 and 1.5 h, respectively), showing that HrtBA expressed in the cytoplasmic membrane detoxifies exogenous heme (Fig. 1 A, Bottom and SI Appendix, Fig. S2).
Fig. 1.
ATP-dependent heme-efflux functions of HrtBA. (A) Growth rescue of heme-sensitive E. coli with HrtBA. Photographs of the culture test tubes after a 24-h incubation. JW0451, parental strain. (B) ATPase activity of HrtBA reconstituted in nanodiscs. Heme and PPIX dissolved in dimethyl sulfoxide (DMSO) were added at a final concentration of 10 μM in the presence of 2 mM ATP. n = 3. (C) Heme-dependent ATPase activity of HrtBA in the presence of 2 mM ATP. n = 3. (D) Kinetics of ATPase activity in the presence (square) or absence (circle) of 5 μM heme. n = 3. (E) UV-visible spectra of heme. Two micromolar of heme in buffer (red), empty nanodiscs (turquoise), and HrtBA-nanodiscs (blue). The nanodisc solutions were used at 4 μM for full binding to the added heme. (F) Schematic of heme transfer assay using a heme-binding protein. Heme-loaded HrtBA nanodisc was incubated with HasA in the presence or absence of ATP and subjected to gel filtration column chromatography. In total, 200 pmol of the proteins with 100 pmol heme was analyzed. (G) Heme transfer in the presence (+) or absence (w/o) of ATP. (H) Heme transfer in the presence (+) or absence (w/o) of AMPPNP.
For biochemical analyses, we next solubilized and purified HrtBA from the membrane fractions of recombinant E. coli in the presence of n-dodecyl-β-D-maltoside (DDM) (SI Appendix, Fig. S3). The HrtBA transporter, reconstituted into lipid nanodiscs (SI Appendix, Fig. S3), displayed heme-dependent ATPase activity (Fig. 1 B–D). The half-maximal effective concentration (EC50) of heme was estimated to be 0.9 μM (Fig. 1C). In terms of the physiological conditions in which the transporter drives heme efflux by ATP hydrolysis, this value is consistent with the free heme concentration that affects the growth of the HrtBA-lacking C. diphtheriae strain (10). Fitting the ATP-dependent kinetic data to the Hill equation indicated that the maximum velocity was increased by heme, but the Km for ATP was not affected, with a Hill coefficient of 1.7 (Fig. 1D).
Measurement of the ultraviolet (UV)-visible absorption spectrum for heme, exogenously added to the nanodisc-embedded HrtBA solution, confirmed heme binding to HrtBA. Heme in the aqueous buffer exhibited a broad Soret band at ∼400 nm, which is attributed to its dimeric state (14) (Fig. 1E, red). In the presence of empty nanodiscs, the band became sharp at 407 nm (Fig. 1E, turquoise), indicating that heme preferentially migrates to the lipid bilayers and resides in the monomeric state (15) because of its lipophilicity (16, 17). Heme binding to the nanodisc-embedded HrtBA generated a sharper Soret peak at 405 nm, which was blue-shifted compared to that of heme in the empty nanodiscs (Fig. 1E, blue vs. turquoise). These spectral data strongly suggest that HrtBA can bind heme that migrates from the buffer to lipids, although we cannot exclude the possibility that HrtBA could receive heme directly from the buffer.
To examine the role of ATP in heme binding to HrtBA, we carried out a heme transfer assay in the presence or absence of ATP, in which a secretory high-affinity heme-binding protein, HasA from Serratia marcescens (18), was used as an acceptor (the experimental design is illustrated in Fig. 1F). Nucleotide-free HrtBA predominantly retained heme even in the presence of HasA (Fig. 1 G, Left). In contrast, HrtBA transferred most heme to HasA in the presence of ATP (Fig. 1 G, Right). The use of a nonhydrolyzable ATP analog, adenylyl-imidodiphosphate (AMPPNP), produced a similar result (Fig. 1H).
We further studied the role of ATP hydrolysis in heme binding and detoxification. HrtA contains conserved motifs (Walker A, Q-loop, signature, Walker B, D-loop, and H-loop) that play essential roles in ATP binding and hydrolysis (SI Appendix, Fig. S1). Mutations in Walker A (K49A), signature (G143A), and Walker B (E165Q) motifs of HrtA failed to rescue growth in the presence of heme, with doubling times of ∼3 h (SI Appendix, Fig. S2 A, Middle and B, Left). Although ATPase activities were lost (SI Appendix, Fig. S4A), consistent with the findings of a previous study on equivalent mutations in the purified HrtA subunit of S. aureus (19), heme transfer assays indicated that these mutants possessed varying degrees of ATP-dependent heme release activities (SI Appendix, Fig. S4B). Specifically, although G143A presented a similar profile as the wild-type protein (SI Appendix, Fig. S4 B, Middle), K49A and E165Q transferred significant amounts of heme to HasA in the absence of ATP, indicating their low heme-binding affinities (SI Appendix, Fig. S4 B, Left and Right). Meanwhile, addition of ATP to K49A resulted in some retention of heme in the mutant despite its low heme-binding affinity, indicating that K49A may also reduce ATP-binding affinity. The partial defect in ATP binding of K49A is consistent with a previous nucleotide-titration study (20). ATP binding and concomitant heme release are unaffected in E165Q, which agrees with the structure determinations of other ABC transporter Gln mutants in the ATP-bound, substrate-free state (21, 22). Thus, to varying degrees, the HrtBA catalytic mutants bind ATP and modulate the TMDs to release bound heme, while impairing ATPase activity.
These functional data unambiguously indicate that the HrtBA wild type binds heme with a high affinity in the absence of nucleotides, and it releases the bound heme upon ATP binding via reducing heme affinity. Therefore, heme detoxification in vivo necessitates ATP hydrolysis for binding and release of the heme and is not fulfilled by sequestration of the heme. To elucidate the structural basis of the heme efflux mechanism, we next performed X-ray crystallography on HrtBA (Fig. 2).
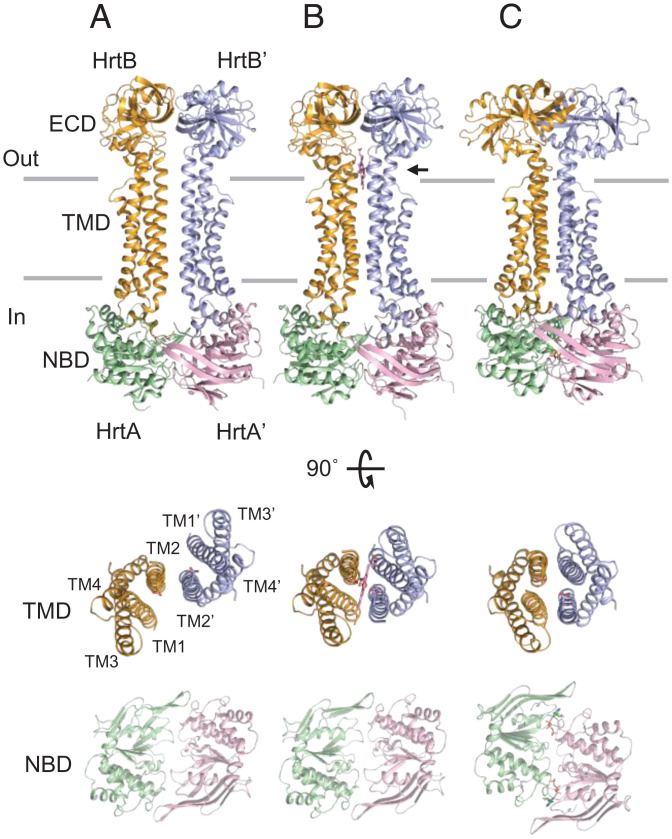
Fig. 2.
Crystal structures of HrtBA. (Top) (A) Unliganded form. (B) Heme-bound form. (C) AMPPNP-bound form. (Middle) Top views of the TM helices of the HrtB dimer. The aa (1 to 12, 249 to 263, 332 to 343) are omitted. (Bottom) Top views of the HrtA subunits. Glu219 of HrtB, heme, and AMPPNP are indicated by stick models. Horizontal lines indicate the hydrophobic layer of the membrane. An arrow indicates heme.
Overall Structure of HrtBA.
We determined the crystal structures of HrtBA in the unliganded, heme-bound, and AMPPNP-bound states at 2.65, 3.27, and 2.88 Å, respectively (summarized in SI Appendix, Table S1). HrtBA comprises two copies of the HrtB-HrtA pair (Fig. 2). The overall architecture is strikingly reminiscent of MacB (23–26), the membrane component of the macrolide efflux transporter, and the lipoprotein translocator LolCDE (27, 28), which are both type VII members of the ABC transporter family (29, 30) (SI Appendix, Fig. S5A).
HrtB is a membrane-embedded subunit consisting of the TMD (amino acids [aa] 1 to 42 and 214 to 344) and ECD (aa 52 to 204) (SI Appendix, Fig. S5 B and C). The TMD contains four TM helices (TM1 to TM4), an N-terminal elbow helix, a coupling helix between TM2 and TM3, and a C-terminal helix. TM1 and TM2 are bundled together and extend to the extramembrane space, peripherally accompanied by TM3 and TM4 (SI Appendix, Fig. S5B). The two linker helices 1 (aa 43 to 51) and 2 (aa 205 to 213) connect the ECD to TM1 and TM2, respectively. The intracellular components (the elbow, coupling, and C-terminal helices) contact the HrtA subunit mainly through hydrophobic interactions. The two linker helices are perpendicularly bent in HrtB (SI Appendix, Fig. S5 B and D), while the corresponding regions are straight and fused to TM1 and TM2, respectively, in MacB and LolCDE (SI Appendix, Fig. S5E).
The HrtA subunit exhibits the ATPase structure typically conserved in the ABC transporter superfamily (30). The NBD consists of a well-conserved RecA-like domain and a structurally diverse helical domain (SI Appendix, Fig. S5B). The HrtA subunit forms a groove on the NBD to accommodate the coupling and C-terminal helices of HrtB.
Structure of the Heme-Binding Site.
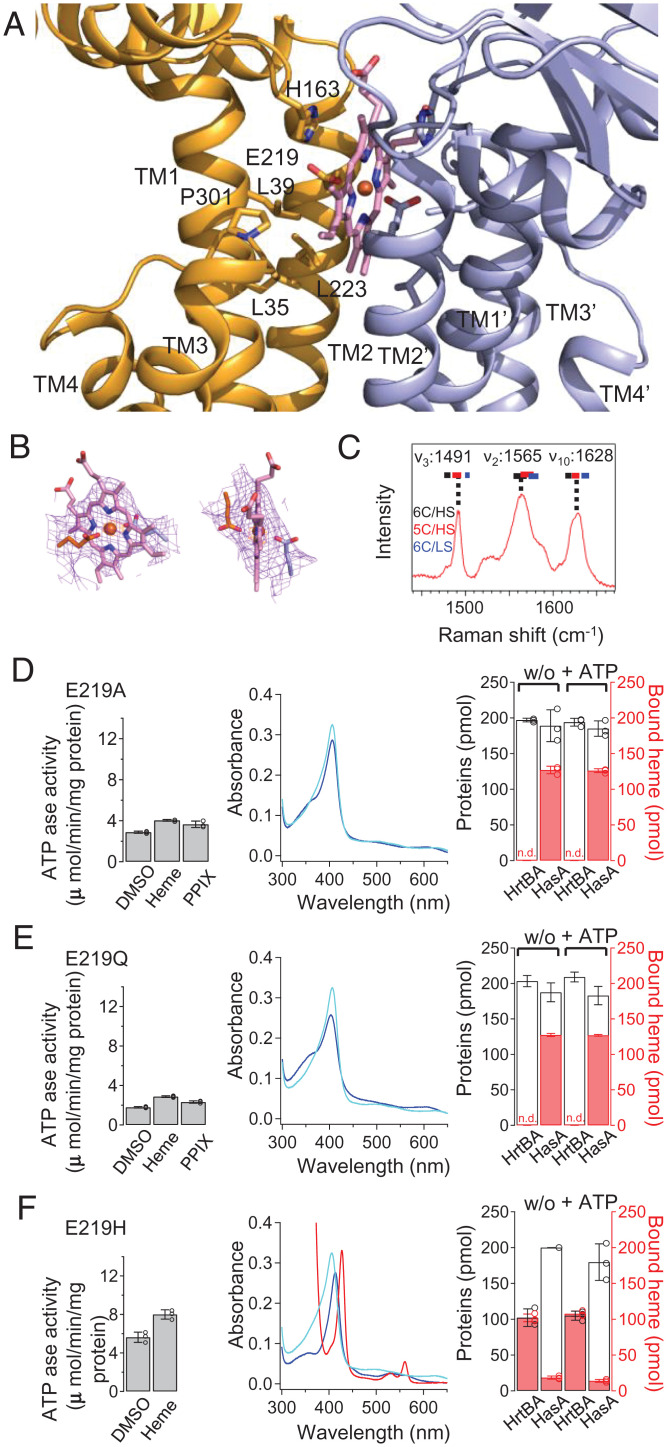
The structure of the heme-bound form revealed that the substrate-binding site containing one heme molecule is located within the TM helices beneath the ECDs. The position is at the same height as the boundary between the outer leaflet of the membrane and the peptidoglycan layer (Fig. 2B). Heme is enclosed by the kinked TM1 and straight TM2 from each HrtB subunit (Figs. 2B and 3A). The lower half of the protoporphyrin ring is surrounded by the well-conserved hydrophobic residues of TM1 (L35, L39), TM2 (L223), and Pro301 of the shoulder loop between TM3 and TM4 (Fig. 3A). The upper half is stacked by the imidazole group of His163 from each ECD. Glu219 residues are located close to the heme iron. The electron densities of the Glu219 side chains suggest ligation to the heme iron (Fig. 3B). However, the coordination state of the heme iron (penta- or hexa-coordination) was not determined from the densities because they represent an average of the molecules in the crystal. Instead, by measuring the UV-visible absorption spectrum (Fig. 1E) and the resonance Raman spectrum assignable to the five-coordination state (Fig. 3C), we concluded that one Glu residue serves as a fifth-axial ligand for the heme iron.
Fig. 3.
Structure and properties of the heme-binding site. (A) Structure of the heme-binding site in the heme-bound state. Well-conserved aa residues are indicated by stick models. (B) 2Fo-Fc electron density map of the bound heme and Glu219 contoured at 1.0 σ (pink) with an Fe anomalous peak (contoured at 9.5 σ, orange). (C) Resonance Raman spectrum of heme-bound HrtBA. (D–F) Mutational analyses of Glu219 mutants E219A, E219Q, and E219H, respectively. ATPase (Left), UV-visible spectrum (Middle), and heme transfer (Right). (Left) ATPase was measured at 10 μM heme or PPIX. (Middle) To fully bind to 2 μM heme, the transporter nanodiscs were used at twofold concentrations in E219A and E219Q (blue). In H219H, 2-μM HrtBA nanodiscs containing 2 μM heme were analyzed, and dithionite-reduced spectrum is shown as a red line. Lines of the wild type are overlaid (cyan). (Right) In E219A and E219Q, 200-pmol nanodiscs were added to 100 pmol heme and analyzed by gel filtration column chromatography. In E219H, 100-pmol nanodiscs containing 100 pmol heme were used.
Glu coordination is rare for heme-binding proteins (31–36), although it is present in cytochrome b595 of the cytochrome bd oxidase complex from Geobacillus thermodenitrificans (37) and E. coli (38, 39). We assessed the role of Glu219 of HrtB in heme binding using Glu219-mutated proteins. The mutation of Glu219 to unligandable Ala or Gln almost abolished the heme-dependent activation of the ATPase, although the mutants retained basal activity (Fig. 3 D, Left and E, Left). This is consistent with the observation that metal-free PPIX did not increase the activity (Fig. 1B). Specific heme binding to the mutants was impaired significantly but not completely, judging from the heme absorption spectra of these mutants (Fig. 3 D, Middle and E, Middle). A heme transfer assay verified that the heme-binding affinities were eliminated even in the absence of ATP (Fig. 3 D, Right and E, Right). Nevertheless, expression of the mutants was able to rescue growth, with slightly prolonged doubling times (1.5 to 1.8 h) in the presence of heme (SI Appendix, Fig. S2 A, Bottom). These data indicate that the heme-binding site can accommodate heme by hydrophobic interactions with the protoporphyrin moiety, and Glu219 contributes to high-affinity heme binding. The basal level of ATPase activity is capable of heme extrusion from the membrane, and the axial ligation of heme enables HrtBA to detoxify higher concentrations of heme by increasing the ATPase turnover rate.
In contrast, substitution with a canonical axial ligand residue, His (31), resulted in a purified protein containing one heme molecule of E. coli–derived heme within the heterotetramer in the six-coordination state, judging from the heme and protein analyses of the protein solution (40) (Fig. 3 F, Middle), indicating that the position of aa 219 allows ligation to the heme iron. Probably due to its high-affinity heme binding, the His mutant possessed relatively high basal activity without a significant heme-dependent elevation (Fig. 3 F, Left), and this mutation abolished heme transfer to HasA (Fig. 3 F, Right) and detoxification in E. coli, with doubling times of ∼3 h (SI Appendix, Fig. S2 A, Bottom).
The glutamate residue at position 219 seems to provide moderate heme-binding affinity (EC50 = 0.9 μM in Fig. 1C) that can be regulated by ATP binding for the association/dissociation of heme. The use of a glutamate residue is in contrast to the stringent heme binding of the heme-acquisition system, in which heme-binding proteins use His and Tyr as axial ligands, although specific heme binding in the TMD of the importer has not been characterized (1, 41, 42).
The Heme-Binding Site Is Laterally Accessible to Membrane-Buried Heme in the Unliganded State and Collapses in the Nucleotide-Bound State.
The structure of the unliganded form revealed how the heme-binding site extracts the heme from the membrane. The TMD monomers contact each other along TM2 (Fig. 2A). In comparison with the heme-bound state, TM2 pivots at the terminus of the coupling helix, generating a lateral shift of the TM helices with a 5.4-Å distance for Glu219, while the NBD subunits are nearly overlaid (Fig. 2 A and B and SI Appendix, Fig. S6 A and B). No inward-facing substrate-binding cavity is present inside the TMD dimer. Instead, the Glu219 of each HrtB is exposed to the surface of the TMD dimer on the boundary of the outer leaflet surface of the membrane (Fig. 2A and SI Appendix, Fig. S6C). The location of the heme-binding site in the TM helices is consistent with the intercalation of heme in the lipid layer (43). Thus, the unliganded state is competent to associate the heme that laterally accesses one of the two Glu21 residues from the membrane outer leaflet. The bound heme is autonomously housed in the rearranged four-TM helix bundle in the heme-bound conformation.
In contrast, the HrtBA-AMPPNP complex adopts a four-helix bundle structure of tightly associated TM1s and TM2s. The upper half of the TM1 straightens (Fig. 2C), rendering the intervening space incapable of accommodating heme. This is consistent with the functional data that nucleotide binding to the NBD transmits allosteric signals to the heme-binding site in the TMD to abolish its binding capability (Fig. 1 G and H).
ATP-Mediated NBD Dimerization Squeezes TM Helices for Heme Extrusion.
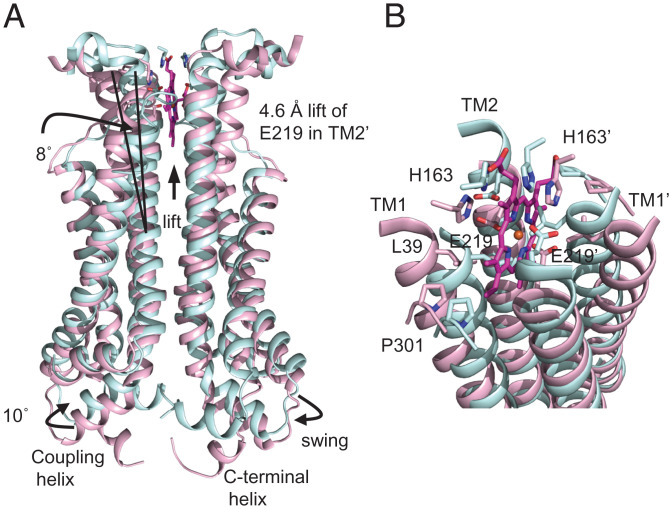
The structural difference between the heme-bound and AMPPNP-bound states evokes an ATP-dependent allosteric mechanism for heme extrusion. In the AMPPNP-bound state, the HrtA subunits adopt a nucleotide-mediated dimer structure (Fig. 2 C, Bottom). Gln90 of each subunit coordinates the nucleotide-associated Mg2+ in the nucleotide-binding site, and the signature motif of the other subunit interacts with the nucleotide (SI Appendix, Fig. S7A), similar to that observed with other known ABC transporters (22). The nucleotide-bound structure is generated from the unliganded state with the induced-fit rotation of the A-loop and the helical domain containing the Q-loop and signature motif (SI Appendix, Fig. S7B, magenta to blue) and the shift of the signature motif of the other subunit (SI Appendix, Fig. S7B, isolated fragments, red to turquoise). These structural changes lift the coupling helix concomitant with a swing of ∼10° (SI Appendix, Fig. S7C). Collectively, the nucleotide-mediated NBD dimerization elevates the TM2s (E219 lift of ∼4.6 Å) (Fig. 4A). In concert with the vertical shift, a horizontal rotation of the ECDs and linker helices (Fig. 2, Top B vs. C and SI Appendix, Fig. S7C) may cause an inward movement of the upper part of TM1 to TM2 of the other subunit, straightening the TM helices (Fig. 4B). Formation of the tightly associated four-helix TM bundle is accompanied by a migration of Leu39 and His163 into the heme-binding pocket to collapse the binding site (Fig. 4B).
Fig. 4.
Conformational changes in the heme-binding site upon nucleotide binding. (A) Helical shift in the heme-bound TMD dimer upon AMPPNP binding based on superimposition of the TMDs. The heme-bound and AMPPNP-bound states are shown in pink and cyan, respectively. (B) Close-up of the heme-binding site.
Discussion
Heme is a beneficial but deleterious nutrient for organisms. To manage this contradiction, many gram-positive bacteria possess a heme-dedicated ABC-type efflux pump, HrtBA, in the cytoplasmic membranes in addition to a heme importer. HrtBA rescues the cells from heme toxicity since the cytoplasmic membranes are exposed to high concentrations of heme in the host blood (SI Appendix, Fig. S8 A and B).
Heme predominantly accumulates in the cytoplasmic membrane when it escapes from the heme-binding proteins of the acquisition system in the peptidoglycan layer. The cytotoxicity of membrane-accumulated heme has been ascribed to ROS, despite the multifaced mechanisms of heme toxicity (2). ROS, which cause oxidative damage to membrane proteins, are generated during heme redox cycle with menaquinones, respiratory components in cytoplasmic membranes (11, 12). The membrane localization of heme and menaquinones suggests that HrtBA detoxifies heme by expelling it from the cytoplasmic membrane rather than by exporting it from the cytoplasm across the membrane (11). It is noteworthy that HrtBA-driven extraction of heme from the cytoplasmic membrane accompanies decrease of heme in the cytoplasm according to the partition equilibrium of heme between the cytoplasm and membrane (16, 17), which can account for the accumulation of the cytoplasmic heme in the HrtBA-lacking mutant (44).
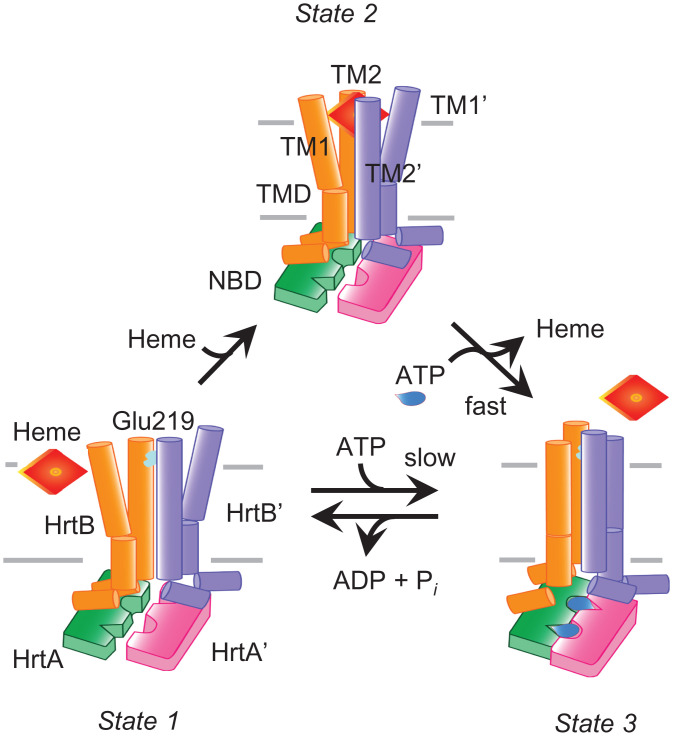
Our functional and structural data strongly suggest that HrtBA expels heme from the membrane. Heme predominantly binds to lipid membranes (Fig. 1E), and HrtBA can extract the membrane-embedded heme and release it upon ATP binding (Fig. 1 E and G). Furthermore, the heme-binding site containing heme-ligandable Glu219 residues in the HrtB dimer is located on the surface of the outer leaflet of the membrane (Fig. 2). The lack of a substrate-translocating cavity across the membrane in the TMDs (Fig. 2) also indicates that HrtBA does not directly receive heme from the cytoplasm. Thus, we propose a plausible model of the ATP-dependent heme efflux cycle (Fig. 5). In HrtBA, the membrane-localized heme accesses one of the two Glu219 residues of the TMDs exposed to the protein-lipid interface at the extracytoplasmic side and is sequestered into the four-helix TM bundle (Fig. 5, State 1 to State 2). Upon ATP binding, heme-bound HrtBA transitions into the ATP hydrolysis-competent state (ATP-bound state), releasing the bound heme (Fig. 5, State 2 to State 3). The subsequent hydrolysis of ATP resets HrtBA to the unliganded state (Fig. 5, State 3 to State 1). Note that unliganded HrtBA less frequently transitions into the ATP hydrolysis-competent state (Fig. 5, State 1 to State 3), giving basal activity because of the high transition energy (45).
Fig. 5.
Mechanism of heme efflux in the membrane by HrtBA. (State 1) Unliganded state, laterally accessible heme embedded in the outer leaflet of the membrane. (State 2) Heme-bound state. (State 3) ATP-bound state. TM1, TM2, and NBD are depicted. The heme ligand residue Glu219 of TM2 in HrtB is colored cyan. Heme and ATP are colored red and blue, respectively. See text for a description of the proposed heme-translocation cycle.
Our functional and structural study consequently strengthens the position that type VII ABC transporters may share lateral access to substrates at the extracytoplasmic side and nucleotide-dependent four-helix bundle formation for substrate release. Consistently, in MacB and LolCDE, their substrate vestibules are also located at the interspace between the outer leaflet of the cytoplasmic membrane and the peptidoglycan layer, and substrate binding is abolished upon nucleotide binding (24, 27, 28, 46).
Gram-positive commensal and opportunistic pathogens can cause septicemia and meningitis. Such fatal diseases are initiated by bacterial propagation in the bloodstream. Inhibitors for HrtBA functions may prevent bacterial proliferation in the host blood. The structure-function studies presented herein will, thus, be helpful in development of antibiotics.
Materials and Methods
Cloning and Expression of HrtBA.
The hrtBA gene was amplified by PCR from C. diphtheriae NCTC13129 (ATCC700971D) genomic DNA. The hrtBA DNA fragment and a synthetic DNA duplex of Strep-tag II were ligated to pMAL-p2G and pRSET-C generating pTac-Cd hrtBAstr and pT7-Cd hrtBAstr, respectively, in which the Strep-tag II DNA is fused in frame to the 3′ end of the hrtA gene (SI Appendix, Fig. S9). The gene encoding the outer-membrane heme receptor ChuA was amplified by PCR from the E. coli O157:H7 strain Sakai genomic DNA. The promoter for the ampicillin-resistance gene (PAp) of pACYC177 and chuA DNA fragment was subcloned into pACYC184, generating pCm-PAp-chuA (SI Appendix, Fig. S7). The E. coli K12 strain JW0451(acrB::Kmr) carrying pCm-PAp-chuA and pTac-Cd hrtBA was used in the growth rescue experiments in the presence of heme. The E. coli BL21(DE3) Star strain was used for protein production. Details are provided in SI Appendix, Detailed Methods.
Purification of HrtBA.
The E. coli membranes containing HrtBA were solubilized with 1% (wt/vol) DDM at 4 °C for 30 min. The HrtBA protein was purified by Strep-Tactin Superflow column chromatography and Superdex 200 pg column chromatography in the presence of DDM or lauryl maltose neopentyl glycol. Details are provided in SI Appendix, Detailed Methods.
Incorporation of HrtBA into Lipid Nanodiscs.
HrtBA was incorporated into lipid nanodiscs formed by a membrane scaffold protein, MSP1D1 (47). Details are provided in SI Appendix, Detailed Methods.
Measurement of ATPase Activity.
The ATPase activity of HrtBA was determined using a reduced nicotinamide adenine dinucleotide (NADH) oxidation-coupled enzyme assay (48). The reaction was initiated by the addition of nanodisc-embedded HrtBA in the presence of heme or PPIX at 37 °C. The oxidation of NADH was monitored by measuring the absorbance at 340 nm. Details are provided in SI Appendix, Detailed Methods.
Spectroscopic Measurements.
Visible spectra were recorded at 25 °C using a Shimadzu UV-2500 spectrophotometer equipped with a micro cuvette holder. Resonance Raman spectra were obtained using a single polychromator, Jovin Yvon SPEX750M, equipped with a liquid nitrogen-cooled charge-coupled device detector. Details are provided in SI Appendix, Detailed Methods.
Heme Transfer Assay.
The secretory heme-binding protein, HasA, was purified from the culture medium of E. coli BL21 (DE3) expressing the PCR-amplified hasADE genes from S. marcescens strain CDC3100-71 as previously described (18) with slight modifications. HrtBA nanodiscs containing a half equivalent of heme were mixed with HasA in the presence or absence of nucleotides and were subjected to Superdex 200 Increase column chromatography. The eluted proteins and heme were monitored by measuring A280 nm and A405 nm, respectively, and their contents were determined from the area of the peaks in the elution profile. Details are provided in SI Appendix, Detailed Methods.
Crystallization, Data Collection, Phase Calculation, and Structure Refinement.
Crystallization of HrtBA-Mn•AMPPNP, HrtBA-Mg•AMPPNP, HrtBA-manganese PPIX (MnPP), HrtBA-heme, and unliganded HrtBA proteins was performed using the sitting-drop vapor-diffusion technique. For the first screening, MemGold HT-96 and MemGold2 HT-96 were used with a mosquito robot automated system. The crystals obtained were subjected to cryoprotectant treatment. X-ray diffraction data were collected under cryogenic conditions using beamlines BL26B1/B2, BL41XU, and BL32XU at SPring-8, Harima, Japan. The diffraction data were integrated and scaled using HKL2000 (49) or XDS (50) software. Structural analysis for the Mn•AMPPNP dataset was conducted using the single anomalous dispersion technique, and the other datasets were analyzed using the molecular replacement technique in Coot (51) and Phenix software package (52). The quality of the structural models was assessed using MolProbity (53). Details are provided in SI Appendix, Detailed Methods. The data and refinement statistics are summarized in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This study was supported by RIKEN Incentive Research Projects (H14-54600) and MEXT, Grants-in-Aid for Science Research (C) (16K07309) to H.N.; the RIKEN Pioneering Project “Dynamic Structural Biology” to M.S.; and the RIKEN Pioneering Project “Molecular System” and MEXT, Grants-in-Aid for Science Research (S) (26220807) to Y.S. We thank K. Takatsu for growing the bacterial cultures and preparing the membrane fractions. We thank T. Kurachi and M. Tanzawa for crystallization of the proteins in the initial phase of the project. We are grateful to M. Inoue and K. Hanada for DNA sequencing and N. Dohmae for N-terminal protein sequence analysis. We thank H. Sugimoto for technical support in the structural analyses of the unliganded form and for critical discussion. We appreciate T. Someya for critically reading the manuscript. We thank M. Hirai for access to the UV2500 spectrophotometer and M. Kubo for the use of the Raman equipment. We also thank the staff of beamlines BL26B1/B2, BL32XU, and BL41XU at SPring-8 (Proposal No. 2018A2580) for their assistance with data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123385119/-/DCSupplemental.
Data Availability
Coordinates and structure factors have been deposited in the Protein Data Bank, under the accession codes 7W78 (54) for HrtBA-Mg•AMPPNP, 7W79 (55) for HrtBA-Mn•AMPPNP, 7W7A (56) for MnPP-bound crystal data (anomalous), 7W7B (57) for MnPP-bound crystal data (high resolution), 7W7C (58) for unliganded HrtBA, and 7W7D (59) for heme-bound crystal data. All other study data are included in the article and/or SI Appendix.
Note.
Heme refers to ferrous iron PPIX, whereas hemin refers to ferric iron PPIX. In this article, the term “heme” is used as a generic expression irrespective of the iron valence state. The term “free heme” means the exchangeable heme that reversibly binds to proteins, lipids, and small ligand molecules, distinct from the heme tightly bound to hemoproteins (60). Ideally, free heme exists in very small quantities due to its insolubility and lipophilicity (60).
References
- 1.Choby J. E., Skaar E. P., Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 428, 3408–3428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S., Bandyopadhyay U., Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157, 175–188 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Friedman D. B., et al. , Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2, e87 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres V. J., et al. , A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1, 109–119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauff D. L., Skaar E. P., Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 72, 763–778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechardeur D., et al. , Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB-HrtA transporter in Lactococcus lactis. J. Biol. Chem. 287, 4752–4758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joubert L., et al. , Visualization of the role of host heme on the virulence of the heme auxotroph Streptococcus agalactiae. Sci. Rep. 7, 40435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H., Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber B., et al. , Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibb L. A., Schmitt M. P., The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 192, 4606–4617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert L., Derré-Bobillot A., Gaudu P., Gruss A., Lechardeur D., HrtBA and menaquinones control haem homeostasis in Lactococcus lactis. Mol. Microbiol. 93, 823–833 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Wakeman C. A., et al. , Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86, 1376–1392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres A. G., Payne S. M., Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23, 825–833 (1997). [DOI] [PubMed] [Google Scholar]
- 14.de Villiers K. A., Kaschula C. H., Egan T. J., Marques H. M., Speciation and structure of ferriprotoporphyrin IX in aqueous solution: Spectroscopic and diffusion measurements demonstrate dimerization, but not mu-oxo dimer formation. J. Biol. Inorg. Chem. 12, 101–117 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Tipping E., Ketterer B., Christodoulides L., Interactions of small molecules with phospholipid bilayers. Binding to egg phosphatidylcholine of some organic anions (bromosulphophthalein, oestrone sulphate, haem and bilirubin) that bind to ligandin and aminoazo-dye-binding protein A. Biochem. J. 180, 327–337 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon J. B., Kuo F. S., Pasternack R. F., Wong N. M., Muller-Eberhard U., Kinetics of the interaction of hemin liposomes with heme binding proteins. Biochemistry 23, 3715–3721 (1984). [DOI] [PubMed] [Google Scholar]
- 17.Light W. R. III, Olson J. S., The effects of lipid composition on the rate and extent of heme binding to membranes. J. Biol. Chem. 265, 15632–15637 (1990). [PubMed] [Google Scholar]
- 18.Izadi N., et al. , Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry 36, 7050–7057 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Stauff D. L., et al. , Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 190, 3588–3596 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerner-Marmarosh N., Gimi K., Urbatsch I. L., Gros P., Senior A. E., Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of wild-type, Walker A, and Walker B mutant proteins. J. Biol. Chem. 274, 34711–34718 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Johnson Z. L., Chen J., ATP binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Manolaridis I., et al. , Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563, 426–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick A. W. P., et al. , Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2, 17070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crow A., Greene N. P., Kaplan E., Koronakis V., Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, 12572–12577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada U., et al. , Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 8, 1336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H. B., et al. , Structure of a MacAB-like efflux pump from Streptococcus pneumoniae. Nat. Commun. 9, 196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X., et al. , Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nat. Struct. Mol. Biol. 28, 347–355 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Sharma S., et al. , Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat. Commun. 12, 4687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas C., et al. , Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas C., Tampé R., Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Andreini C., Putignano V., Rosato A., Banci L., The human iron-proteome. Metallomics 10, 1223–1231 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Kakar S., Hoffman F. G., Storz J. F., Fabian M., Hargrove M. S., Structure and reactivity of hexacoordinate hemoglobins. Biophys. Chem. 152, 1–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulos T. L., Cytochrome P450: Molecular architecture, mechanism, and prospects for rational inhibitor design. Pharm. Res. 5, 67–75 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Zhong L., et al. , Heme axial methionine fluxionality in Hydrogenobacter thermophilus cytochrome c552. Proc. Natl. Acad. Sci. U.S.A. 101, 8637–8642 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma K. D., Andersson L. A., Loehr T. M., Terner J., Goff H. M., Comparative spectral analysis of mammalian, fungal, and bacterial catalases. Resonance Raman evidence for iron-tyrosinate coordination. J. Biol. Chem. 264, 12772–12779 (1989). [PubMed] [Google Scholar]
- 36.Johnson E. A., Russo M. M., Nye D. B., Schlessman J. L., Lecomte J. T. J., Lysine as a heme iron ligand: A property common to three truncated hemoglobins from Chlamydomonas reinhardtii. Biochim. Biophys. Acta, Gen. Subj. 1862, 2660–2673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safarian S., et al. , Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 352, 583–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safarian S., et al. , Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 366, 100–104 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Theßeling A., et al. , Homologous bd oxidases share the same architecture but differ in mechanism. Nat. Commun. 10, 5138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isogai Y., et al. , Design and synthesis of a globin fold. Biochemistry 38, 7431–7443 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Woo J. S., Zeltina A., Goetz B. A., Locher K. P., X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat. Struct. Mol. Biol. 19, 1310–1315 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Naoe Y., et al. , Crystal structure of bacterial haem importer complex in the inward-facing conformation. Nat. Commun. 7, 13411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giri R. P., et al. , Continuous uptake or saturation-investigation of concentration and surface-packing-specific hemin interaction with lipid membranes. J. Phys. Chem. B 122, 7547–7554 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Wakeman C. A., Stauff D. L., Zhang Y., Skaar E. P., Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J. Bacteriol. 196, 1335–1342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikant S., Gaudet R., Mechanics and pharmacology of substrate selection and transport by eukaryotic ABC exporters. Nat. Struct. Mol. Biol. 26, 792–801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greene N. P., Kaplan E., Crow A., Koronakis V., Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front. Microbiol. 9, 950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie T. K., et al. , Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karasawa A., et al. , Physicochemical factors controlling the activity and energy coupling of an ionic strength-gated ATP-binding cassette (ABC) transporter. J. Biol. Chem. 288, 29862–29871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W78/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 55.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W79/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 56.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W7A/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 57.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W7B/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 58.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W7C/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 59.T. Hisano et al., Data from “Heme exporter HrtBA in complex with Mg-AMPPNP.” Protein Data Bank. 10.2210/pdb7W7D/pdb. Deposited 4 December 2021. [DOI] [Google Scholar]
- 60.Gallio A. E., Fung S. S., Cammack-Najera A., Hudson A. J., Raven E. L., Understanding the logistics for the distribution of heme in cells. JACS Au 1, 1541–1555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank, under the accession codes 7W78 (54) for HrtBA-Mg•AMPPNP, 7W79 (55) for HrtBA-Mn•AMPPNP, 7W7A (56) for MnPP-bound crystal data (anomalous), 7W7B (57) for MnPP-bound crystal data (high resolution), 7W7C (58) for unliganded HrtBA, and 7W7D (59) for heme-bound crystal data. All other study data are included in the article and/or SI Appendix.