Abstract
In order to understand the role of the mar locus in Salmonella with regard to multiple antibiotic resistance, cyclohexane resistance, and outer membrane protein F (OmpF) regulation, a marA::gfp reporter mutant was constructed in an antibiotic-sensitive Salmonella enterica serovar Typhimurium DT104 background. Salicylate induced marA, whereas a number of antibiotics, disinfectants, and various growth conditions did not. Increased antibiotic resistance was observed upon salicylate induction, although this was shown to be by both mar-dependent and mar-independent pathways. Cyclohexane resistance, however, was induced by salicylate by a mar-dependent pathway. Complementation studies with a plasmid that constitutively expressed marA confirmed the involvement of mar in Salmonella with low-level antibiotic resistance and cyclohexane resistance, although the involvement of mar in down regulation of OmpF was unclear. However, marA overexpression did increase the expression of a ca. 50-kDa protein, but its identity remains to be elucidated. Passage of the marA::gfp reporter mutant with increasing levels of tetracycline, a method reported to select for mar mutants in Escherichia coli, led to both multiple-antibiotic and cyclohexane resistance. Collectively, these data indicate that low-level antibiotic resistance, cyclohexane resistance, and modulation of OMPs in Salmonella, as in E. coli, can occur in both a mar-dependent and mar-independent manner.
The multiple antibiotic resistance locus (mar) is reported to be present in Salmonella, Shigella, Klebsiella, Citrobacter, Hafnia, Enterobacter, and Escherichia coli (13). In recent years, much work has been carried out to elucidate the structure, function, and clinical significance of the mar locus, mainly in E. coli but also in Salmonella (1, 18, 29, 30, 42, 43). The marRAB operon, which is located at min 34.8 of the chromosomal map of E. coli K-12 (20), controls multiple antibiotic resistance in E. coli by production of MarA (12, 17, 27), a 127-amino-acid protein of the XylS/AraC family of transcriptional activator proteins, which alters the expression of several target genes (1). For example, in E. coli MarA positively regulates acrAB (25), which encodes a stress-induced efflux system, and micF (12, 38), which encodes an antisense RNA involved in the regulation of the porin outer membrane protein F (OmpF), through which hydrophilic substances enter the cell (35). The repressor MarR (43), encoded by marR, binds to the marO operator region (39) to negatively regulate expression of marRAB. Neither the function of MarB nor the natural inducer of the marRAB operon is known. A number of unrelated substances, including tetracycline, chloramphenicol, dinitrophenol, menadione, paraquat, plumbagin, benzoate, and sodium salicylate and related compounds, have been shown to induce the operon in E. coli (10, 38, 39), of which salicylate is the most potent inducer (10). In addition to the involvement of mar in multiple antibiotic resistance, it has also been shown to be involved in organic solvent tolerance (7, 47), resistance to disinfectants such as pine oil (31), and resistance to weak acids (5).
E. coli passaged on low levels of tetracycline or chloramphenicol produced mar mutants at a rate of about 10−8 per cell division, and these mutants had increased resistance to the unrelated antibiotics penicillin G, ampicillin, cephalothin, puromycin, rifampin, nalidixic acid, and fluoroquinolones (11, 19). Furthermore, E. coli strains resistant to pine oil, which is used in household disinfectants, showed resistance to multiple antibiotics (tetracycline, ampicillin, chloramphenicol, and nalidixic acid) that was associated with increased expression of marA (31). Continued passage of first-step mar mutants on media with increasing levels of tetracycline or chloramphenicol resulted in increased levels of antibiotic resistance (19). However, the genetic basis for second-step high-level resistant mutants was only partially attributed to mar, because transduction of the locus from high- or low-level mar mutants produced only a low level of multiple antibiotic resistance (1).
E. coli induced for mar has been demonstrated to have resistance to several unrelated antibiotics, which is in part associated with reduced levels of OmpF (11, 12). However, cyclohexane resistance in E. coli has been shown to be independent of OmpF but dependent on the acrAB efflux pump (6). For Salmonella, the involvement of reduced levels of OmpF in antibiotic and cyclohexane resistance is less well characterized. A Salmonella enterica serovar Typhi isolate resistant to chloramphenicol, carbenicillin, and ampicillin that lacked OmpF and did not encode a chloramphenicol acetyltransferase has been described (45). However, a lack of correlation between reduced levels of OmpF and quinolone resistance in clinical isolates of serovar Typhimurium from two patients that failed ciprofloxacin therapy has been shown (37).
There is an increasing concern regarding the veterinary use of antibiotics, which prompts a closer examination of the mechanisms of resistance in zoonotic pathogens. While there has been considerable work done in relation to the role of the mar locus of E. coli, the role of the mar locus in the biology of Salmonella has not been investigated in such depth. It cannot be assumed that the mar locus will be isofunctional in E. coli and Salmonella. Thus, the aim of this work was to investigate the role of mar in Salmonella in antibiotic resistance, cyclohexane resistance, and modulation of OmpF.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used are listed in Tables 1 and 2, respectively. Additionally, 44 serotypes of Salmonella were used. These are not listed in Table 1. All Salmonella strains were obtained from the collection of strains at the Veterinary Laboratories Agency, Weybridge, United Kingdom, and were originally isolated from animals or their environment. As negative controls for marA DNA hybridization studies, the following strains (not listed in Table 1) were used: E. coli NCTC 10418, Enterobacter aerogenes NCTC 10006, Klebsiella aerogenes NCTC 418, Pseudomonas diminuta NCTC 8545, Proteus vulgaris NCTC 4175, Streptococcus mutans NCTC 10920, Streptococcus agalactiae NCTC 8181, and Staphylococcus aureus NCTC 1803.
TABLE 1.
Strains used
| Strain | Properties or use | Reference |
|---|---|---|
| E. coli DH5α | F′endA1 hsdR17(rK mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1Δ (lacIZYA-argF) U169 deoR (φ80 [dlacΔ(lacZ) M15]; general-purpose cloning strain | 48 |
| E. coli K-12 S-17(λpir) | pro res+mod+ RP4 2-Tc::Mu-Km::Tn7 λpir; recipient strain for suicide plasmid construct | 40 |
| E. coli AG100 | Wild-type K-12 used as cyclohexane-sensitive control | 19 |
| E. coli AG102 | marR1 mutant of AG100 used as cyclohexane-resistant control | 19 |
| Serovar Typhimurium DT104 S3530/96 | DNA template for construction of marA::gfp::cam | This study |
| Serovar Typhimurium DT104 S3992/96 | Recipient strain for marA::gfp::cam construct (referred to as parent strain) | This study |
| LR1 to LR5 | marA::gfp::cam reporter mutant clones of S3992/96 | This study |
| LR6 | S3992/96 + marA plasmid pLR1 | This study |
| LR7 | LR1 + marA plasmid pLR1 | This study |
TABLE 2.
Plasmids used
| Plasmid | Description | Reference or source |
|---|---|---|
| pCR-SCRIPT Am+ | Direct cloning vector | Commercially available from Stratagene (9) |
| pERFORM-Z | pERFORM-C with cam resistance gene replaced by zeocin resistance gene | 2 |
| pLR1 | pCR-SCRIPT with marRAB 1.3 kb in MCSa | This study |
| pLR2 | As pLR1 with cam cloned into XcmI site of marA | This study |
| pLR3 | As pLR2 with gfp cloned into AgeI site of marA | This study |
| pLR4 | pERFORM-Z suicide plasmid with marA::gfp::cam cloned into MCS | This study |
MCS, multiple cloning site.
Antibiotics, chemicals, bacterial growth, and media.
Sodium ampicillin, erythromycin, streptomycin sulphate, sodium cefoperazone, sodium sulfadiazine, chloramphenicol, sodium nalidixic acid, and tetracycline hydrochloride were obtained from Sigma-Aldrich Company, Ltd., as was hexane, cyclohexane, and salicylic acid. Zeocin was obtained from Invitrogen. Co-amoxiclav (amoxicillin and clavulanate, 2:1) and ciprofloxacin were obtained from SmithKline Beecham and Bayer, respectively.
All bacteria were grown in Luria-Bertani (LB) broth or agar except where stated. For MIC determinations, strains grown overnight at 37°C and diluted 1:10 in saline were inoculated onto Iso-Sensitest agar (Oxoid CM471) supplemented with antibiotics as required and also with 2.5 or 10 mM salicylate when measuring the effect of salicylate on antibiotic resistance. When measuring the effect of salicylate on MICs, strains were also grown in broth containing salicylate. For production of antibiotic-resistant (mar-like) mutants, LR1 and its isogenic parent (Table 1) were grown at 30°C for 48 to 72 h on Trypticase soy agar (Difco) supplemented with tetracycline, according to the method of George and Levy (19).
For OMP extraction, bacteria were grown for 48 h at 37°C on M9 minimal agar supplemented with glucose (0.2% [wt/vol]), thiamine (4 mg/liter), and nicotinamide (0.6 mg/liter). For acid tolerance assays, strains were grown overnight in buffered peptone water (BDH). Acid tolerance was determined in prewarmed buffered peptone water adjusted to specific pH values with hydrochloric acid.
To assess induction of marA, the serovar Typhimurium marA::gfp::cam reporter mutant and isogenic parent were grown at 30 and 37°C in the presence of salicylate ranging from 2.5 to 20 mM. Additionally, induction of marA at 37°C was monitored when strains were grown in the presence of the antibiotics and disinfectants at one-fourth or one-half of the MICs for those strains. The disinfectants tested were a synthetic phenol-xylenol mixture, a quaternary ammonium compound, a peroxygen compound (acetic acid in equilibrium with hydrogen peroxide), and a compound product of formaldehyde, glutaraldehyde, and the quaternary ammonium compound. To monitor the effects of some environmental stresses on induction of marA, strains were grown in LB broth supplemented with 15 and 30% (wt/vol) sucrose (high osmolarity), LB without NaCl (low osmolarity), or in M9 minimal medium supplemented with 0.025% (wt/vol) glucose (carbon limitation). Overnight LB cultures were held at 46°C for 2 h (heat shock).
PCR amplification.
The oligonucleotide primers used are listed in Table 3 and were designed from the published mar sequence for serovar Typhimurium (42) using the DNASTAR software LaserGene. PCR was performed as described previously (2), and serovar Typhimurium DT104 S3530/96 DNA was used as a template.
TABLE 3.
Primer sequences for PCR
| Primer name | Directiona | 5′-to-3′ DNA sequence of primer | Position of primer (bp)b |
|---|---|---|---|
| marA 387 | F | ATGACGATGTCCAGACGCAACACT | 1433 to 1456 |
| marA 387 | R | CTAGTAGTTGCCATGGTTCAGCGG | 1799 to 1822 |
| marRAB 1.3 | F | ATTCCGCTGGGTCGCTTGA | 1018 to 1036 |
| marRAB 1.3 | R | AGCGCCGCGGTGTTACTC | 1996 to 2313 |
| mar gfp seqc | F | TCATCAACGACCAGGGCAAGAC | 1317 to 1338 |
| mar gfp seq | R | ATCCGCAGCCGTAAAATGAC | 1610 to 1629 |
F, forward; R, reverse.
bp, positions as related to positions in published sequence of serovar Typhimurium (42).
seq, sequencing primer to check insertion of gfp into marA.
DNA hybridization and Southern blotting.
Colony dot blot DNA hybridization studies were performed to test if marA sequence homology was present in the 44 Salmonella serotypes. The serotypes tested were Agama, Agona, Anatum, Arizonae, Binza, Bovis morbificans, Bradenburg, Bredeney, Cubana, Derby, Dublin, Ealing, Enteritidis, Give, Gold Coast, Hadar, Harford, Heidelberg, Idikan, Indiana, Infantis, Kedougou, Kentucky, Lexington, London, Manhattan, Mbandaka, Montevideo, Muenster, Newington, Newport, Ohio, Oranienberg, Orion, Panama, Poona, Pullorum, Senftenberg, Taksony, Tennessee, Thompson, Typhimurium, Virchow, and Wangata. At least two isolates of each serotype were tested. Other strains were also tested as negative controls.
Colony dot blotting was performed using the marA 387-kb probe (from this study) and Southern blotting was performed using the marRAB 1.3-kb probe (from this study). Additionally, Southern blotting was performed using linearized suicide plasmid pLR4 without marRAB or cam. The probes were prepared by PCR. Probe amplicons were labeled using a Rediprime random primer labeling kit (Amersham Life Sciences) according to the manufacturer's instructions for dot blotting or were labeled using a nonradioactive labeling kit (Gene-Images AlkPhos Direct; Amersham Life Sciences) according to the manufacturer's instructions for Southern blotting. Colony dot blot and Southern blot analyses were performed as described previously (26). For Southern blots, DNA was fixed to Hybond N+ membranes (Amersham Life Sciences) by incubation at 80°C for 2 h. For nonradioactive probing, posthybridization washes were performed according to the manufacturer's instructions. For radioactive probing, three washes were performed at 62°C. Wash 1 comprised 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS), and washes 2 and 3 comprised 1× SSC and 0.1% SDS.
Construction of serovar Typhimurium DT104 marA::gfp::cam mutants.
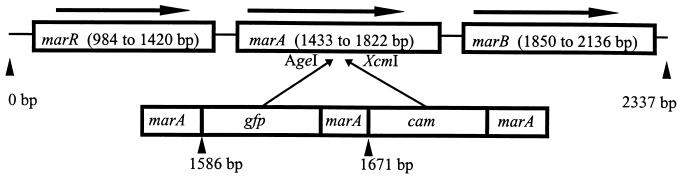
The serovar Typhimurium DT104 marA::gfp::cam reporter mutants (LR1 to LR5) were produced from strain S3992/96 by allelic exchange (Fig. 1). Briefly, the marRAB region amplified with the marRAB 1.3-kb primers (Table 3) was cloned according to the manufacturer's instructions into pCR-script plasmid (Stratagene) to give plasmid pLR1 (Table 2). A 1.1-kb chloramphenicol resistance cassette (Stratagene) was cloned into the XcmI site in the distal part of marA to give plasmid pLR2 (Table 2). The promoterless green fluorescent protein (gfp) gene (14) was obtained by restriction endonuclease digestion of its plasmid vector using PstI and XbaI. The promoterless gfp gene was cloned blunt ended into the AgeI site in the proximal region of marA to give plasmid pLR3 (Table 2). The marA::gfp::cam construct was amplified by marRAB 1.3-kb primers (Table 3) and ligated into the zeocin-resistant pERFORM-Z suicide plasmid (2) to give plasmid pLR4 (Table 2). Plasmid pLR4 was transformed into electrocompetent permissive host E. coli S17(λpir) cells by electroporation. Conjugation with E. coli S17pLR4 as donor and serovar Typhimurium DT104 S3992/96 (Table 1) as recipient was performed by filter mating, with selection made for chloramphenicol. Five putative recombinant clones designated LR1 to LR5 that were zeocin sensitive and chloramphenicol resistant were retained for further analysis.
FIG. 1.
Construction of marA::gfp::cam. Base pair positions refer to positions in the published sequence of serovar Typhimurium (42). The gfp gene was inserted into the AgeI site of marA at bp 1586, and the cam gene was inserted into the XcmI site of marA at bp 1671.
Complementation of strains.
Plasmid pLR1 (Table 2) was electroporated into S3992/96 and LR1 (Table 1), with selection made for ampicillin resistance to give LR6 and LR7, respectively (Table 1).
Induction of marA over time.
The fluorescence and cell optical density of LR1 and its isogenic parent strain were determined at various time points up to 24 h when grown at 30 or 37°C with test substances (see “Antibiotics, chemicals, bacterial growth, and media” above). After washing and resuspension in half the original volume of phosphate-buffered saline were carried out, fluorescence (200-μl volumes in a 96-well plate) was measured using a Millipore CytoFluor 2350 set to an excitation wavelength of 485 nm and emission wavelength of 530 nm. As a negative control, the isogenic parent strain was grown under identical conditions. All experiments were performed in triplicate, and the relative fluorescence was calculated as fluorescence divided by the optical density at 630 nm. A paired t test was used to determine if differences in fluorescence between LR1 and its isogenic parent were statistically significant.
MIC determinations.
MICs of antibiotics were determined by agar doubling dilution using the National Committee for Clinical Laboratory Standards method (34), except that Iso-Sensitest agar was used instead of Mueller-Hinton agar. Inoculated plates were incubated at 37°C overnight before the MIC was recorded. All antibiotic MICs were determined using multiple clones, and MIC experiments were repeated.
For experiments to determine induction of marA in the reporter mutant LR1, growth in broth was determined. As such, for antibiotics and disinfectants used in induction experiments, MICs in broth were determined by a method similar to the doubling dilution method used in agar.
Cyclohexane resistance.
Cyclohexane resistance was determined by the method of Asako et. al. (7). Briefly, bacteria grown overnight at 30°C were inoculated (using a multipoint inoculator) onto duplicate solid agar media in glass petri dishes. Each plate was flooded to a depth of 3 mm (6 ml) with hexane or cyclohexane, sealed with Nescofilm, and incubated overnight at 30°C. Strains that grew in the presence of cyclohexane were deemed cyclohexane resistant.
Preparation and separation of OMPs.
OMP extracts of strains were prepared by a method similar to that of Inokuchi et al. (23). The outer membranes were recovered in 50 μl of 10 mM sodium phosphate buffer (pH 7), and the protein levels were determined and adjusted to 2,000 μg/ml in the final sample buffer. Samples (20 μl) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) at 300 V for 7 h in a 12.5% (wt/vol) gel without urea (36). Marker standards used were carbonic anhydrase (29 kDa), glyceraldehyde-3-phosphate dehydrogenase (36 kDa), and egg albumin (45 kDa). After being run, gels were stained overnight in 0.1% Coomassie blue (Sigma). Strains with plasmid pLR1 (LR6 and LR7) were grown with 50 μg of ampicillin/ml at all stages prior to OMP extraction.
N′-terminal sequencing of proteins.
The identity of OmpC, OmpF, and OmpA bands from SDS-PAGE gels (see Fig. 3) were determined previously (unpublished observations). To identify the unknown protein (see Fig. 3) from SDS-PAGE, the band was transferred by Western blotting (28) onto a polyvinylidene fluoride membrane (Sequi-blot; Bio-Rad). N′-terminal sequencing of the first few amino acids of the unknown protein was carried out with an Applied Biosystems 477A sequencer.
FIG. 3.
OMPs of parent and LR1 complemented with plasmid pLR1. Lane 1 and 6, standards (sizes as shown); 2, parent; 3, LR1; 4, parent complemented with plasmid pLR1; 5, LR1 complemented with plasmid pLR1.
Comparisons of protein sequences.
Comparisons of N′-terminal protein sequences were made using the Jalview multiple alignment editor (http://www.ebi.ac.uk/∼michele/jalview/). Comparisons were to OMPs of specific interest, such as OmpC, OmpF, OmpA, and TolC.
The N′-terminal sequences were also compared using BLAST search programs (BLASTp and BLASTn) (3), which are available online at the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The unknown N′-terminal protein sequence was compared to sequences within the E. coli database only.
RESULTS
The marA region is present in Salmonella.
It is widely assumed that the marRAB region is conserved among Salmonella serotypes. Colony dot blot DNA hybridization studies confirmed the presence of sequences that shared homology with serovar Typhimurium marA for all 44 Salmonella serotypes tested (data not shown). Two E. coli strains hybridized with this probe, but other non-Salmonella strains tested did not hybridize (results not shown). Also, Southern hybridizations were performed to test if marRAB in Salmonella was present as a single copy and in a conserved region of the Salmonella genome. Chromosomal DNA from Salmonella serotypes Arizonae (n = 1), Dublin (n = 3), Enteritidis (n = 3), Hadar (n = 1), Infantis (n = 1), Kedougou (n = 1), Newport (n = 1), Typhimurium (n = 5), and Virchow (n = 1) was digested to completion with HindIII and probed. All serotypes hybridized a single fragment of approximately 3.5 kb except for serotype Arizonae strain S170/97, for which a single fragment of approximately 4.5 kb hybridized (results not shown).
Construction of marA::gfp::cam mutants.
In order to study the regulation of the marRAB region and the phenotype of a mar null mutant, marA::gfp::cam reporter mutants LR1 to LR5 were constructed by allelic exchange in serovar Typhimurium DT104 S3992/96. The correct orientation of the gfp gene in the marRAB::gfp::cam construct was confirmed by sequencing (results not shown), using custom-made primers (Table 3). To confirm successful allelic exchange in colonies that were chloramphenicol resistant but zeocin sensitive, Southern blotting was performed (results not shown). Chromosomal DNA digested to completion with HindIII and PvuII was hybridized with the marRAB 1.3-kb probe and the marRAB::gfp::cam probe. HindIII did not cut within gfp, cam, or the marRAB 1.3-kb region, and a single, larger DNA fragment was seen for the recombinants (5.2 kb) than for that of the parent (3.5 kb). PvuII did not cut in the marRAB 1.3-kb region but cut in both gfp and cam, and so a single product was seen for the parent and three products were seen for the recombinants. The combined lengths of gfp and cam were approximately 1.8 kb, so the predicted increases in the sizes of the hybridizing region were observed (results not shown). Plasmid pLR4 without the marA::gfp::cam construct did not hybridize Southern blots of putative reporter mutants that were chloramphenicol resistant but zeocin sensitive (results not shown), thus confirming the absence of suicide plasmid DNA in the reporter mutants.
marA was induced by salicylate but not by antibiotics.
Tests were done to assess what compounds or conditions induced marA. Salicylate (2.5 to 20 mM) increased fluorescence of the marA::gfp reporter mutant LR1 two- to fourfold (P < 0.05 to P < 0.01) (Table 4 and Fig. 2), whereas other substances, including nine antibiotics or simulated environmental stresses such as disinfectants (see Materials and Methods), did not. For these, the mean fluorescence of LR1 ranged from 0.6 to 1.7 times that of the parent strain, and these differences were not significant at P < 0.05 (results not shown).
TABLE 4.
Induction of fluorescence in reporter mutant LR1
| Salicylate concn (mM) | Growth condition (°C) | Relative increasea in fluorescence (SEM) | P valueb |
|---|---|---|---|
| 2.5 | 37 | 1.56 (0.16) | 0.031 |
| 5 | 30 | 2.30 (0.16) | 0.004 |
| 5 | 37 | 2.40 (0.16) | 0.006 |
| 10 | 30 | 4.00 (0.47) | 0.001 |
| 10 | 37 | 2.50 (0.12) | 0.005 |
| 20 | 30 | 4.00 (0.16) | 0.006 |
| 20 | 37 | 4.55 (0.41) | 0.008 |
Mean increase in relative fluorescence of LR1 compared to parent strain under identical growth conditions. SEM, standard error of the mean.
P value for comparison of relative fluorescence of LR1 and parent.
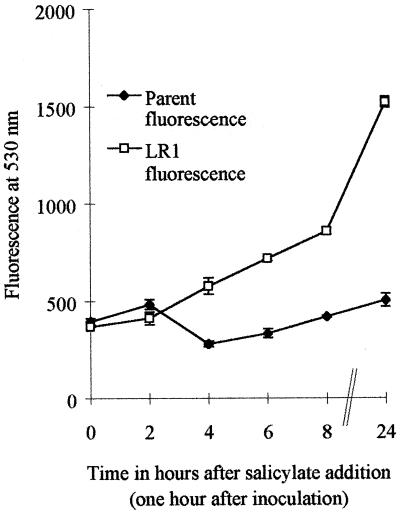
FIG. 2.
Induction of marA in reporter mutant LR1 grown with 10 mM salicylate.
With regard to the antibiotics tested, LR1 was more sensitive than the parent by 1 dilution to nalidixic acid and ciprofloxacin when grown without salicylate (Table 5). However, growth with salicylate induced decreased sensitivity to most antibiotics tested, more so for the parent than for LR1 (Table 5). Here was clear evidence that salicylate, the inducer of marA, was associated with the Mar-like phenotype.
TABLE 5.
MICs for LR1 and its parent before and after passage with salicylate or tetracycline and with and without plasmid expressing marA
| Strain | Growtha | Result typeb | MIC (μg/ml)c
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TET | CHL | AMP | AMC | CFP | STR | NAL | CIP | ERY | SUL | |||
| Parent | LB broth | MIC | 1.25 | 5 | 1.25 | 1.25 | 0.63 | 20 | 2.5 | 0.02 | 80 | 160 |
| 2.5 mM Sal | MIC | 2.5 | 10 | 2.5 | NT | 1.25 | 20 | 5 | 0.04 | NT | NT | |
| 10 mM Sal | MIC | 5 | 10 | 2.5 | NT | 1.25 | 20 | 20 | 0.16 | NT | NT | |
| Ex TET 50 | MIC | 20 | NT | 10 | 5 | 5 | 10 | 20 | 0.63 | 320 | 640 | |
| LR6 | AMP 50 μg/ml | MIC | 5 | NT | NT | NT | NT | NT | 10 | NT | NT | NT |
| LR1 | LB broth | MIC min | 1.25 | NT | 1.25 | 1.25 | 0.31 | 10 | 2.5 | 0.02 | 80 | 160 |
| MIC max | 1.25 | NT | 1.25 | 1.25 | 0.63 | 20 | 2.5 | 0.02 | 160 | 320 | ||
| 2.5 mM Sal | MIC | 1.25 | 80d | 1.25 | NT | 0.31 | 10 | 2.5 | 0.02 | NT | NT | |
| 10 mM Sal | MIC | 2.5 | 80d | 1.25 | NT | 0.63 | 20 | 5 | 0.04 | NT | NT | |
| Ex TET 50 | MIC min | 10 | NT | 1.25 | 1.25 | 0.63 | 5 | 10 | 0.08 | 320 | 160 | |
| MIC max | 10 | NT | 5 | 1.25 | 0.63 | 5 | 20 | 0.16 | 320 | 320 | ||
| LR7 | AMP 50 μg/ml | MIC | 5 | NT | NT | NT | NT | NT | 10 | NT | NT | NT |
Strains were normally grown in LB broth prior to MIC determinations. LB broths were supplemented as shown with 2.5 and 10 mM salicylate (Sal) or with 50 μg of ampicillin/ml (to maintain plasmid pLR1). Ex Tet 50, strains grown for eight passages with increasing concentrations of tetracycline (TET) up to 50 μg/ml.
MIC min, lowest recorded MIC; MIC max, highest recorded MIC.
TET, tetracycline; CHL, chloramphenicol; AMP, ampicillin; AMC, Co-amoxiclav (amoxicillin:clavulanic acid, 2:1); CFP, cefoperazone; STR, streptomycin; NAL, nalidixic acid; CIP, ciprofloxacin, ERY, erythromycin; SUL, sulfadiazine. NT, not tested. Experiments were performed with multiple clones and/or repeated with identical or comparable results.
Resistant to chloramphenicol due to insertion of cam in LR1.
LR1 could be trained to Mar-like resistance.
Passage of E. coli with tetracycline is reported to select for mar constitutive mutants that have increased multiple antibiotic resistance (19, 20). We hypothesized that LR1 would not yield such derivatives, due to insertional inactivation of marA, if the marRAB was the only operon involved in low-level antibiotic resistance in Salmonella. To test this, LR1 and its parent were passaged eight times with tetracycline, and both showed increased resistance up to 16 times higher to tetracycline, ampicillin, nalidixic acid, ciprofloxacin, and erythromycin, as compared to the strains before passage with tetracycline (Table 5). However, LR1 showed a lower increase in resistance than its parent against these antibiotics (Table 5). The parent strain also showed some increase in resistance to cefoperazone. Little or no increase in resistance after antibiotic passage was observed against the antibiotics streptomycin and sulfadiazine for either LR1 or its parent. Complementation of the parent and LR1 with pLR1 gave the Mar phenotype with respect to low-level antibiotic resistance (Table 5). This level of resistance was not as high as that seen for strains passaged eight times with tetracycline. This further suggests that non-mar mechanisms were involved in the acquisition of resistance after multiple passage with tetracycline.
Cyclohexane resistance is induced by salicylate.
Organic solvent tolerance in E. coli has been shown to be linked to mutations in marR, to overexpression of marA, soxS, and rob, to upregulation of acrAB, and to increased resistance to numerous antibiotics (6, 7, 47). We hypothesized that LR1 would remain cyclohexane sensitive even under inducing conditions, due to insertional inactivation of marA if the marRAB was involved in cyclohexane resistance in Salmonella. Both LR1 and its parent were sensitive to cyclohexane when grown on LB agar medium (results not shown). However, the parent strain but not LR1 became cyclohexane resistant when grown on LB agar medium supplemented with salicylate. Complementation of the parent and LR1 with pLR1 led to cyclohexane resistance in both strains (data not shown).
Both LR1 and its parent became resistant to cyclohexane after multiple (eight) passage with tetracycline (results not shown), which suggested that cyclohexane resistance in Salmonella was not controlled by marRAB exclusively.
OMP profiles were not altered by the marA::gfp::cam mutation.
Induction of mar in E. coli has been reported to lead to down regulation of OmpF (10, 12). To investigate this in Salmonella, OMP profiling was performed for the parent and LR1 when grown in normal media or media supplemented with 5 mM salicylate and when complemented with the plasmid pLR1 (media supplemented with 50 μg of ampicillin/ml to maintain the plasmid). The OMP profiles of the parent and LR1 when grown in normal media were similar (Fig. 3, lanes 2 and 3), and previous work has confirmed the major bands shown to be OmpC, OmpF, and OmpA (unpublished observations). Growth of the parent and LR1 with salicylate did cause down regulation of OmpF (results not shown). These data showed that the mutation of marA was not correlated with altered OmpF levels.
Complementation of the parent and LR1 strains with plasmid pLR1 did not affect the levels of OmpC, OmpF, and OmpA (Fig. 3, lanes 4 and 5), but an unknown protein of ca. 50 kDa was up regulated in the parent. N′-terminal sequencing of this novel protein yielded the sequence APCDNTIFA. When this sequence was compared with Salmonella OmpF, OmpC, OmpA, and TolC proteins using an alignment program (http://www.ebi.ac.uk/∼michele/jalview/), it showed the most similarity to OmpA. The sequence APCDNTIFA was identical to six of the nine amino acids of the OmpA sequence (amino acids 22 to 30). The OmpA sequence to which the N′-terminal sequence showed some homology immediately followed a classic type 1 cleavage sequence (AQA).
The APCDNTIFA sequence showed similarity to 9 E. coli proteins using the BLASTP search programs and to 19 proteins using the TBLASTN search programs (3; http://www.ncbi.nlm.nih.gov). However, the results of these searches were generally inconclusive, although it was interesting that some homology was shown to the E. coli multiple drug resistance proteins EmrY and EmrK (44).
It was considered possible that changes to the bacterial surface induced by mutation, salicylate induction, or overexpression of the novel 50-kDa protein alter other observable phenotypes. No alterations to cell surface hydrophobicity or acid tolerance between LR1 and its isogenic parent were observed under the conditions tested (results not shown).
DISCUSSION
All serotypes of Salmonella tested possessed marA, and Southern blot analysis showed one copy of a marRAB-like sequence in a conserved region of the chromosome. Salmonella marA is reported to share 86% sequence homology with the same region of E. coli (42), and this would explain the hybridization of the serovar Typhimurium marA probe with E. coli under the stringency conditions used.
Southern blot analysis and sequencing confirmed the insertion of gfp and cam into marA in the reporter mutant LR1. Induction of marA in the reporter mutant showed that background fluorescence increased in a linear fashion with increased growth over time. However, when LR1 and the isogenic parent strain were grown with salicylate, the reporter mutant clearly showed a greater increase in fluorescence, which was most noticeable 24 h after inoculation. Salicylate was both an inhibitor of growth and an inducer of marA, and interpretation of induction was dependent upon a comparison of relative fluorescence of the parent and LR1 under identical conditions. Only salicylate enhanced the transcription of marA significantly. Previous studies with E. coli showed that chloramphenicol and tetracycline, but not ampicillin, nalidixic acid, or norfloxacin, caused an increase in marA hybridizable mRNA (22). None of these antibiotics caused a significant increase in marA transcription in this study; however, Northern blotting may be needed to detect low levels. Alternatively, the inducers of marA in Salmonella differ from those for E. coli. For E. coli, salicylate has been shown to bind to MarR, inhibiting the formation of the MarR-marO complex (27). It is assumed that salicylate induces mar in Salmonella in a similar way.
Despite numerous papers on the function of mar, the natural inducer of the mar locus is still not known (1). It has been postulated that mar originated from primal chromosomal genes involved in responding to the hostile environment of antibiotic-producing soil microbes (24). This would explain partially the reported ability of antibiotics such as tetracycline and chloramphenicol to induce mar in E. coli (22). Other workers (29) have postulated that naturally occurring plant breakdown products such as naphthoquinones are the true substrates for intrinsic resistance systems such as mar, emrRAB, sox, and acrAB. While salicylate is the most potent inducer of mar in E. coli (10), naphthoquinones such as plumbagin and menadione are effective inducers at lower concentrations (10, 39).
The increased induction of marA transcription when LR1 was grown with 10 mM rather than 2.5 mM salicylate correlated with slight increases in the MICs of MAR-type antibiotics (i.e., antibiotics to which expression of mar confers some resistance, such as β-lactams, chloramphenicol, quinolones, and tetracyclines) when LR1 was grown with 10 mM rather than 2.5 mM salicylate. The lack of (or reduced) increase in MICs for LR1 when grown with salicylate compared to those for its parent implicates the involvement of mar in low-level antibiotic resistance in Salmonella. This was further endorsed by complementation studies with plasmid pLR1, which constitutively expresses marA, as demonstrated by constitutive expression of gfp from pLR3. In LR1 and its parent, pLR1 increased tetracycline and nalidixic acid MICs fourfold. For E. coli, salicylate has been shown to induce antibiotic resistance by a mar-dependent and a mar-independent pathway (10), and our results suggest that salicylate functions in Salmonella in a similar way. The mar-independent pathway was evident in that growth with salicylate caused increased antibiotic resistance for both LR1 and its parent. The mar-dependent pathway was suggested in that the plasmid expressing marA caused an 8-fold increase in resistance to nalidixic acid, whereas growth with 10 mM salicylate caused a 16-fold increase in resistance to nalidixic acid.
The effect of growing strains with salicylate on increased fluoroquinolone resistance in Staphylococcus aureus has been reported (21), and fears have been expressed that salicylate used therapeutically could give rise to sufficient levels in vivo (5 mM) to induce antibiotic resistance (8, 21). This could apply to Salmonella also. Fluoroquinolone antibiotics are the agents of choice for treating bacteremic enteric fevers due to Salmonella (15, 41, 46). For ciprofloxacin, MICs following growth with 2.5 and 10 mM salicylate increased by about 2 to 4 times and up to 8 times (MIC maximum, 0.31 μg/ml), respectively. Salicylate and acetylsalicylate (aspirin) are chemically similar, and growth of E. coli with both salicylate and acetylsalicylate has been shown to cause increased resistance to ampicillin, tetracycline, and nalidixic acid (38). Whether aspirin treatment of fever accompanying systemic Salmonella infection could make the difference between antibiotics curing and failing to cure a life-threatening infection is a matter of conjecture.
Passage of the parent and LR1 with tetracycline gave rise to increased antibiotic resistance for specific antibiotics, as has been observed for E. coli following passage with tetracycline (19). There was little increase in resistance for streptomycin, but resistance was seen primarily against tetracycline, chloramphenicol, ampicillin, cefoperazone, and quinolones, a pattern of resistance also reported for mar mutants of E. coli (7). Therefore, multiple-antibiotic-resistant mutants of Salmonella with a similar phenotype to the classical mar mutants can arise from passage with antibiotics such as tetracycline but independently of marA.
Salicylate induced cyclohexane resistance in the parent strain but not the reporter mutant LR1. Plasmid pLR1, which constitutively expressed marA, made LR1 and its parent cyclohexane resistant in the absence of salicylate. These results clearly implicate mar in inducible cyclohexane resistance. However, results also showed that cyclohexane resistance could occur independently of mar, since the reporter mutant LR1 became cyclohexane resistant after passage with tetracycline.
Salicylate down regulated OmpF through mar-independent mechanisms as have been observed for E. coli (10), and complementation studies, using plasmid pLR1 expressing marA, gave expected results with respect to low-level multiple antibiotic resistance and cyclohexane resistance. However, the OMP profiles of complemented strains were unexpected, because a ca. 50-kDa protein was up regulated. In E. coli, TolC is an OMP that has a molecular mass of 51 kDa (4, 32, 33, 35) and is thought to link the acrAB efflux pump to the outside of the membrane (16). As such, one would expect TolC to be up regulated when mar is up regulated (16, 18, 33, 47). Homology searches were equivocal and this protein requires more detailed analysis.
ACKNOWLEDGMENTS
The work described in this paper was funded by the Ministry of Agriculture, Fisheries, and Food.
We thank Laura Piddock, Clifford Wray, and Rob Davies for advice and Russell Collighan for considerable expert guidance on cloning.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen-Vercoe E, Woodward M J. Adherence of Salmonella enterica serovar Enteritidis to chick gut explant; the role of flagella but not fimbriae. J Med Microbiol. 1999;48:1–10. doi: 10.1099/00222615-48-8-771. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zheng Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aono R, Kobayashi H. Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl Environ Microbiol. 1997;63:3637–3642. doi: 10.1128/aem.63.9.3637-3642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asako H, Kobayashi K, Aono R. Organic solvent tolerance of Escherichia coli is independent of OmpF levels in the membrane. Appl Environ Microbiol. 1999;65:294–296. doi: 10.1128/aem.65.1.294-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asako H, Nakajima H, Kobayshi K, Kobayshi M, Aono R. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl Environ Microbiol. 1997;64:1428–1433. doi: 10.1128/aem.63.4.1428-1433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Axon J M C, Huskisson E C. Use of aspirin in inflammatory diseases. In: Vane J R, Botting R M, editors. Aspirin and other salicylates. London, England: Chapman & Hall; 1992. pp. 295–320. [Google Scholar]
- 9.Bauer J, Deely D, Bramman J, Viola J, Weiner M P. pCRSCRIPTTM sk(+) cloning system: a simple and fast method for PCR cloning. Strategies. 1992;5:62–64. [Google Scholar]
- 10.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S P, McMurray L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen S P, Yan W, Levy S B. A multidrug resistance regulatory chromosome is widespread among enteric bacteria. J Infect Dis. 1993;168:484–488. doi: 10.1093/infdis/168.2.484. [DOI] [PubMed] [Google Scholar]
- 14.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (gfp) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 15.Dutta P, Rasaily R, Saha M R, Mitra U, Bhattacharya S K, Bhattacharya M K, Lahiri M. Ciprofloxacin for treatment of severe typhoid fever in children. Antimicrob Agents Chemother. 1993;37:1197–1199. doi: 10.1128/aac.37.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambino L, Gracheck S J, Miller P F. Overexpression of the marA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1993;175:2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George A M. Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol Lett. 1996;139:1–10. doi: 10.1111/j.1574-6968.1996.tb08172.x. [DOI] [PubMed] [Google Scholar]
- 19.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George A M, Levy S B. Gene in the major co-transduction gap of Escherichia coli K-12 linkage map required for expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:41–48. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafson J E, Candelaria P V, Fisher S A, Goodridge J P, Lichocik T M, McWilliams T M, Price C T D, O'Brien F G, Grubb W B. Growth in the presence of salicylate increases fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:990–992. doi: 10.1128/aac.43.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hächler H, Cohen S P, Levy S B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inokuchi K, Itoh M, Mizushima S. Domains involved in osmoregulation of the ompF gene in Escherichia coli. J Bacteriol. 1985;164:585–590. doi: 10.1128/jb.164.2.585-590.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch A L. Evolution of antibiotic resistance gene function. Microbiol Rev. 1981;45:355–378. doi: 10.1128/mr.45.2.355-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma D, Cook D N, Albeti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch C F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 27.Martin R G, Rosner J L. Binding of purified multiple antibiotic resistant repressor protein (MarR) to mar operator sequence. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 29.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple antibiotic resistance in Escherichia coli. Mol Micobiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller P F, Gambino L F, Sulavik M C, Gracheck S J. Genetic relationship between soxRS and mar loci promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994;38:1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moken M C, McMurray L M, Levy S B. Selection of multiple-antibiotic-resistant (Mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morona R, Reeves P. The tolC locus of Escherichia coli K-12 affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona R, Manning P A, Reeves P. Identification and characterization of the TolC protein, an outer membrane protein from Escherichia coli. J Bacteriol. 1983;153:693–699. doi: 10.1128/jb.153.2.693-699.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial sensitivity testing for bacteria that grow aerobically. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 35.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickard D, Li J, Roberts M, Maskell D, Hone D, Levine M, Dougan G, Chatfield S. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect Immun. 1994;62:3984–3993. doi: 10.1128/iai.62.9.3984-3993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piddock L J V, Griggs D J, Hall M C, Jin Y F. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob Agents Chemother. 1993;37:662–666. doi: 10.1128/aac.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosner J L. Non-inheritable resistance to chloramphenicol and other antibiotics induced by salicylates and other chemotactic repellents in Escherichia coli K-12. Microbiology. 1985;82:8771–8774. doi: 10.1073/pnas.82.24.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seone A S, Levy S B. Characterization of marR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3417. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. A broad host-range mobilisation system for in vivo genetic engineering: transportation mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 41.Smith M D, Duong N M, Hoa N T, Wain J, Ha H D, Diep T S, Day N P, Hien T T, White N J. Comparison of ofloxacin and ceftriaxone for short-course treatment of enteric fever. Antimicrob Agents Chemother. 1994;38:1716–1720. doi: 10.1128/aac.38.8.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulavik M C, Dazer M, Miller P F. The Salmonella typhimurium mar locus: molecular and genetic analysis and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 44.Tanabe H, Yamasaki K, Furue M, Yamamoto K, Katoh A, Yamamoto M, Yoshioka S, Tagami H, Aiba H, Utsumi R. Growth phase dependent transcription of emrYK, a homolog of multidrug efflux emrAB genes of Escherichia coli. J Gen Appl Microbiol. 1997;43:257–263. doi: 10.2323/jgam.43.257. [DOI] [PubMed] [Google Scholar]
- 45.Toro C S, Lobos S R, Calderon I, Rodriguez M, Mora G C. Clinical isolate of a porinless Salmonella typhi resistant to high levels of chloramphenicol. Antimicrob Agents Chemother. 1990;34:1715–1719. doi: 10.1128/aac.34.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F, Gu X J, Zhang M F, Tai T Y. Treatment of typhoid fever with ofloxacin. J Antimicrob Chemother. 1989;23:785–788. doi: 10.1093/jac/23.5.785. [DOI] [PubMed] [Google Scholar]
- 47.White D G, Goldman J D, Demple B, Levy S B. Role of the acrB locus in organic solvent tolerance meditated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidern M, Smith S S, Michael M Z, Graham M W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]