Summary
The regulatory influence of ubiquitin is vast, encompassing all cellular processes, by virtue of its central roles in protein degradation, membrane trafficking, and cell signaling. But how does ubiquitin, a 76 amino acid peptide, carry out such diverse, complex functions in eukaryotic cells? Part of the answer is rooted in the high degree of complexity associated with ubiquitin polymers, which can be “read” and processed differently depending on topology and cellular context. However, recent evidence indicates that post-translational modifications on ubiquitin itself enhance the complexity of the ubiquitin code. Here, we review recent discoveries related to the regulation of the ubiquitin code by phosphorylation. We summarize what is currently known about phosphorylation of ubiquitin at Ser65, Ser57 and Thr12, and we discuss the potential for phospho-regulation of ubiquitin at other sites. We also discuss accumulating evidence that ubiquitin-like modifiers, such as SUMO, are likewise regulated by phosphorylation. A complete understanding of these regulatory codes and their complex lexicon will require dissection of mechanisms that govern phosphorylation of ubiquitin and ubiquitin-like proteins, particularly in the context of cellular stress and disease.
Keywords: ubiquitin, phosphorylation, signalling, stress responses, proteostasis, mitophagy, protein degradation
Introduction
Post-translational modification of proteins by conjugation to ubiquitin (Ub) is critical for cellular adaptations that involve remodeling the proteome in response to a changing environment or exposure to stress. Decades of intense research have been dedicated to interrogating ubiquitin, and yet we still do not fully understand how this 76 amino acid peptide exhibits such broad utility as a regulatory modifier. Part of the answer is rooted in the high degree of complexity associated with Ub polymers, which can be “read” and processed differently depending on topology and cellular context. This is referred to as the ubiquitin code. One layer of this code is based on the formation of linkage-specific polymers by the Ub conjugation enzymes (E1-E2-E3 cascades) [1–3] combined with regulated disassembly of Ub polymers by deubiquitylases (DUBs) [4]. However, recent evidence indicates the code is further enhanced by the formation of mixed and branched polymers [5, 6] and by post-translational modifications (PTMs) on Ub itself [7, 8]. These recent findings reveal that the ubiquitin code is more complex than previously appreciated. A complete understanding of its lexicon will require a deeper interrogation of how complex topologies and ubiquitin PTMs contribute to modified substrates’ regulatory fate.
One emerging paradigm for lexicon expansion of the ubiquitin code involves phosphorylation of ubiquitin itself (Figure 1). The first report describing the detection of phosphorylated ubiquitin was published in 2003 [9] and subsequently numerous studies have reported proteomic evidence of Ub phosphorylation at multiple sites. However, understanding the biological functions of ubiquitin phosphorylation has been slow, likely due to the technical challenges associated with detecting phosphorylated ubiquitin species. However, enthusiasm for the biology of ubiquitin phosphorylation was bolstered in 2014 when a series of studies reported that Ser65-phosphorylated ubiquitin regulates PINK1-Parkin-mediated mitophagy [10–13], providing the first evidence of functional regulation of ubiquitin by phosphorylation. Since then, additional roles for phosphorylated ubiquitin have been reported (Table 1), and there is emerging evidence that phosphorylation of ubiquitin-like modifiers (UBLs) also confers functional regulation. In this review, we summarize recent findings related to the regulation of Ub and UBLs by phosphorylation, discuss critical knowledge gaps that remain, and propose experimental directions and strategies for catalyzing discoveries that will enhance our understanding of ubiquitin phosphorylation and the regulatory lexicon of the ubiquitin code.
Figure 1.

Phosphorylation has the potential to regulate many aspects of ubiquitin function. The blue box highlights several important questions that remain to be answered in this new arena of regulatory biology.
Table 1.
Summary of phosphorylation sites on ubiquitin. Phosphorylation of human ubiquitin is represented in the top rows of the table (shaded green) while yeast ubiquitin is represented in the bottom rows of the table (shaded orange). This table represents phosphorylation events that are the best characterized (to date) and have regulatory significance with regards to ubiquitin function. Other phosphorylation sites on ubiquitin have been detected, but the significance of other events remains to be elucidated.
| Site | Biological function | Kinases | References | |
|---|---|---|---|---|
| Ubiquitin (human) | Ser65 | Regulation of mitophagy | PINK1 | [11, 12, 74] |
| Ser57 | Unknown | MARK1–4, SIK1–2 PKA, PKC, PKG |
[36] | |
| Thr12 | DNA damage response | unknown | [56] | |
| Ubiquitin (yeast) | Ser65 | unknown | unknown | |
| Ser57 | stress responses ubiquitin homeostasis | Vhs1, Sks1 Gin4, Kcc4 |
[35, 36] | |
| Thr12 | unknown | unknown | ||
Ser65 Phosphorylation of Ubiquitin
Ser65 phosphorylation (pS65) is the most extensively studied phosphorylation event on Ub based on its role in mitochondrial homeostasis regulation [10, 14, 15]. When mitochondria become damaged in healthy cells, mitochondrial proteins in the damaged region are conjugated to pS65-Ub, which targets destruction by autophagosomal degradation (mitophagy) in a tightly regulated signaling cascade. AQUA-based quantification data revealed that pS65-Ub is <0.1% of the total Ub in unperturbed human cells but the level increases to 2% in cells treated with mitochondria-damaging agents. Global mitochondrial depolarization enriches pS65 ubiquitylation of proteins at the outer mitochondrial membrane (OMM), reaching roughly 20% of the total mitochondrial Ub [13]. In yeast, pS65-Ub comprises <0.5% of total Ub at a steady-state, approximately as abundant as pSer57 [16]. Outside its role in maintaining mitochondrial health, the biology of pS65-Ub in a cellular context is poorly understood. Although Ser65 is non-essential as demonstrated by alanine substitution [17], it is part of the β5-bulge adjacent to Lys63 [18] (Figure 2), a site for K63-linked Ub polymerization that modulates DNA repair [19], cell-cycle control [20], transcriptional regulation [21], endocytic trafficking [22], and inflammatory signaling [23].
Figure 2.
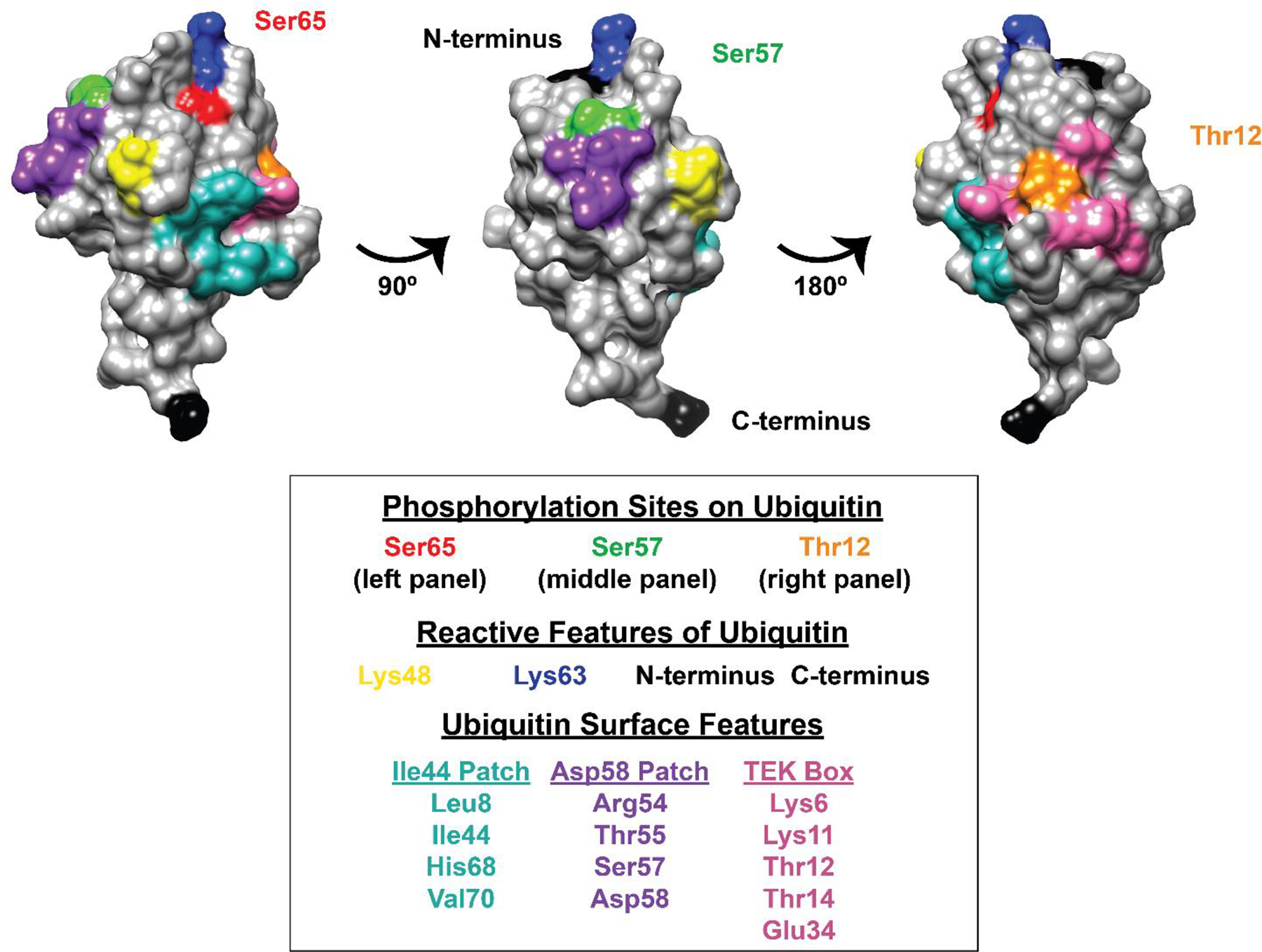
Analysis of phosphorylation sites relative to other functional surfaces on ubiquitin. The structure of ubiquitin (PDB ID: 1UBQ) was analyzed using UCSF Chimera (ver. 1.15), positioning the molecule around a vertical N-to-C terminal axis. (N- and C-termini are colored black.) Sites of phosphorylation are colored as follows: Ser65 = red, Ser57 = green, Thr12 = orange. Functional surface features of ubiquitin are colored as follows: K48 = yellow, K63 = dark blue, Ile44 hydrophobic patch residues = teal, Asp58 patch residues = purple, TEK box residues = pink. The molecule is rotated to provide views focused on Ser65 (left), Ser57 (middle) and Thr12 (right).
PINK1 (PTEN induced putative kinase 1) is the only kinase known to phosphorylate the Ser65 residue of free and conjugated Ub, which has been demonstrated in vitro and in vivo [10, 11, 24]. In the absence of mitochondrial damage, PINK1 engages with the mitochondrial import machinery. It is subsequently cleaved and degraded, but a damage-induced loss of mitochondrial import results in accumulation of PINK1 on the mitochondrial outer membrane where it can phosphorylate ubiquitin and activate Parkin-mediated mitophagy. Crystal structure (PDB 6EQI) analysis of PINK1 in complex with Ub (a T66V/L67N variant) revealed that Ub forms a bipartite interaction with the N lobe and the activation segment in the C lobe of the PINK kinase domain [25]. Tyr198 of the N lobe forms a hydrogen bond with Gly47 of Ub and hydrophobic interactions with Ile44 and Val70. The activation segment (residues 360–385) of the C lobe contacts the Ser65-containing loop of Ub, orienting Ser65 to a phospho-accepting position [25]. These interactions underlie the complex coordination between mitochondrial import and activation of mitophagy. Recently, the phosphatases PTEN-L (phosphatase and tensin homolog-long isoform) and PPEF2 (protein phosphatase with EF-hand domain 2) were found to antagonize the PINK1-dependent mitophagy by dephosphorylating pS65-Ub on the OMM [26–29]. Although pS65-Ub is routinely detected in yeast, there are no apparent structural homologs of PINK1 or PTEN-L/PPEF2 in yeast, and their functional analogs are yet to be identified.
S65 phosphorylation of Ub significantly impacts its structural and biophysical properties. Spectra generated by high-resolution 2D-NMR analysis reveal that pS65-Ub has two non-identical Ub conformations in dynamic equilibrium [30]. The major conformation comprises 70% of the population and resembles the unmodified Ub with notable resonance perturbations at the vicinity of Ser65. Crystallographic analysis of this significant species reveals that the phosphoryl group of pS65 forms a hydrogen bond with the backbone amide of Gln62, an interaction conserved in the unmodified Ub that links the hydroxyl group of Ser65 and Gln62. This major conformation of pS65-Ub exhibits a similar overall secondary structure compared to unmodified Ub, and the principal UBD recognition site, the hydrophobic patch including Ile44, is not grossly altered. In contrast, the remaining 30% of the population exhibits a significantly changed conformation compared to the unmodified Ub. This minor conformation exhibits a two-amino acid β5-strand slippage downstream of the polypeptide chain, causing the C-terminal tail’s contraction into the Ub core by two amino acids. The slippage stabilizes Leu73 and Arg74 residues and alters the interaction of the β5 with β1 and β3 at the core. A new set of contacts are formed between Ile44-Val70, Arg42-Arg72, Leu69-Phe4 and Leu71-Lys6, resulting in extension of the solvent-exposed loop that contains the pSer65, which could provide an opportunity for new surface interactions [30].
Given the drastic conformational perturbations associated with this minor conformation, it is not surprising that pS65-Ub exhibits some biochemical loss of function concerning specific conjugating enzymes. The Ser65-containing surface of Ub does not directly interact with E1 (activating) or E2 (conjugating) enzymes; and in vitro biochemical analysis did not reveal any loss of function with respect to thioester bond formation between pS65-Ub and E1 or E2 enzymes (13 of the 19 human E2 enzymes tested) [30, 31]. However, the discharge from E2 and Ub polymer assembly are altered in a subset of E2 enzymes. For example, the autoubiquitylation of human E2 enzymes UBE2R1(cdc34), UBE2E1(UbcH6) and UBE2T is significantly reduced with pS65-Ub. In addition, pS65-Ub impairs the production of unanchored K11-linked polyubiquitin by UBE2D1 [30]. When human E2 UBE2N was paired with the E2-fold proteins UBE2V1(UEVIA) or UBE2V2(Mms2), the assembly of pS65-Ub into free K63-linked chains was impaired [30]. Similarly, S65E phosphomimetic Ub impaired polymerization by yeast E2 enzymes Ubc1 and the Ubc13/Mms2 complex [16], with the notable caveat that phosphomimetic Ub is not a perfect proxy for pS65-Ub. Structural analysis of the yeast Ubc13/Mms2 complex shows that the phosphoryl group of pSer65 would clash with Glu18 of Mms2, thereby altering the orientation of Ub Lys63 at the active site of Mms2 [30]. In addition to its effects on E2 discharge, pS65-Ub can also impact specific E3 ligases. Ser65 phosphorylation inhibits conjugation reactions using the E2 enzymes UBE2D1 and UBE2D3 and the E3 ligase TRAF6, while the same E2 enzymes paired with the E3 ligase cIAP1 revealed no such defect compared to unmodified ubiquitin. In addition, Ser65 phosphorylation inhibits the formation of M1-linked (linear) polyUb chains by the E3 HOIP [30]. All of these biochemical analyses indicate that pS65-Ub can modify specific conjugation reactions in cells. Still, the regulatory impact remains unclear given the low abundance of unanchored pS65-Ub monomers relative to the pool of free ubiquitin. Future studies will need to address whether E3 ligases use pS65-Ub in conjugation reactions in cells.
Ser65 phosphorylation of ubiquitin is also reported to affect the recognition and activity of DUBs. One study measured the effect of Ser65 phosphorylation on DUB activities in vitro (31 human DUBs were tested) and found that the majority were strongly repressed in all di-Ub linkage types tested [31]. Similar observations were recorded using representative members from different DUB families and tested with phospho-tetra-Ub or high-MW phospho-polyUb as substrates [30]. Importantly, recent studies have reported that Ser65 phosphorylation inhibits the activity of USP30 [32, 33] – a mitochondria-localized DUB that antagonizes Parkin-mediated mitophagy. Surprisingly, Ser65 phosphorylation mildly enhanced some DUB activities, including OTULIN activity towards linear Ub polymers and USP4 and USP5 activity toward both Lys11 and Lys48 Ub polymers [31]. Ser65 phosphorylation also strongly enhanced the activity of USP16 on Lys33-linked polymers and OTUB1 and USP16 on Lys48linked polymers [31]. Taken together, these in vitro data indicate Ser65 phosphorylation of Ub has the potential to either repress or activate DUB activities, depending on the specific DUB and the linkage type of the polymer.
While several studies have reported biochemical data indicating how Ser65 phosphorylation of Ub alters conjugation and deconjugation reactions in vitro, less is known about its ability to modify recognition by UBDs and thus regulate the functional outcome of ubiquitin modification. One study used quantitative proteomics to compare interaction profiles of wildtype and S65E mono-ubiquitin in yeast [16]. This study found that S65E phosphomimetic Ub exhibits reduced interaction with specific UBD-containing proteins, such as the CUE-containing Cue5, UBA-containing Rup1, GAT-containing Tom1, and UIM-containing Vps27 [16]. In contrast, the UBA-containing Dsk2, Rad23 and Ede1 exhibit greater interaction with S65E Ub compared to WT Ub [16]. This analysis highlights the potential for Ser65 phosphorylation to modify UBD interactions and thus the effector outcomes of ubiquitylation.
Given all the biochemical data reported for how pS65-Ub affects conjugation, deconjugation, and ubiquitin-binding in vitro, there is strong potential for this modification to regulate various ubiquitin-dependent processes in a biological context. A recent study performed proteomic analysis of immunoprecipitated pS65-Ub and identified nuclear proteins involved in DNA repair, including DHX9, PRKDC XRCC6 and ɣH2A.X [29], suggesting that pS65-Ub may be associated with sites of DNA damage. Other pS65-Ub-associated proteins identified in this study include CSN1, MYCB2, PRS6/7 and PSA6, which are involved in the proteasome or autophagy pathways [29]. These findings emphasize the potential for Ser65 phosphorylation to regulate other ubiquitin-dependent processes. Still, additional functional and mechanistic analyses will be required to elucidate additional regulatory roles for this modification of ubiquitin. Some of the critical remaining questions that will fuel future research on pS65-Ub include:
What pathways (besides mitophagy) are regulated by pS65-Ub?
What kinases generate pS65-Ub in eukaryotes that lack PINK1 (e.g., yeast)?
How does Ser65 phosphorylation change how the ubiquitin code is “read” by UBDs?
Ser57 phosphorylation of ubiquitin
pS57-Ub was the first phospho-Ub species discovered in a pioneering proteomic analysis of the ubiquitin-modified proteome of yeast [9]. Since then, other studies have reported pS57-Ub in yeast [34–36] as well as in different human cells and tissues – including breast cancer tissues [37], melanoma cells [38], and lung adenocarcinoma cells [39, 40]. However, alanine scanning mutagenesis of ubiquitin had revealed that Ser57 is not required for the essential functions of ubiquitin in yeast [17]. Indeed, a more recent deep mutational scanning analysis of ubiquitin revealed that the Ser57 position is fairly unique in that mutations at this position do not confer sensitivity across a panel of chemical stressors [41]. More directed studies have uncovered phenotypes associated with both S57A (phosphorylation resistant) and S57D (phosphomimetic) Ub variants. We found that S57D-Ub was associated with several stress-resistance phenotypes, conferring resistance to heat stress and toxic amino acid analogs such as canavanine and thialysine [35, 36]. Interestingly, S57D-Ub also conferred hypersensitivity to hydroxyurea, while expression of S57A-Ub conferred hypersensitivity to heat and oxidative stress [36]. Together, these findings underscore the importance of the Ser57 position during conditions of stress. Interestingly, we also found that S57D-Ub exhibits a decreased half-life compared to WT ubiquitin. This finding is consistent with observations of reduced ubiquitin half-life in ppz phosphatase mutants, which exhibit elevated levels of pS57-Ub [35]. Following up on this observation, we found that S57D-Ub inhibits the activity of Doa4 [35] – a DUB that recycles ubiquitin before intraluminal vesicle formation in the ESCRT pathway. Based on these findings, we proposed that Ser57 phosphorylation regulates ubiquitin recycling efficiency in the endosomal sorting process.
In the absence of defined Ser57 ubiquitin kinases, some studies have analyzed pS57-Ub biochemical properties in vitro using purified recombinant pS57-Ub generated by biosynthetic incorporation of phosphoserine during translation in E. coli. One such study found that pS57-Ub is capable of hyper-activating Parkin in vitro, which is surprising given that pS65-Ub is a well-established Parkin activator in cells. This study raises the intriguing possibility that pS57-Ub could drive Parkin-mediated mitophagy independently of PINK1 and pS65-Ub [42], although this hypothesis is yet to be tested in a cellular context. Another study looked more broadly at how different phosphorylated Ub species affect conjugation cascades and DUB activities. This study found that Ser57 phosphorylation did not impact E2 charging nor the formation of linkage-specific polymers in conjugation assays using various E3 ubiquitin ligases [31]. However, when incorporated into dimers of different linkage types, pS57-Ub inhibited the activity of several DUBs – suggesting that Ser57 phosphorylation may regulate deconjugation, albeit in a way that is highly specific to the DUB and the linkage type of the polymer. (By comparison, pS65-Ub was a much more potent inhibitor of DUB activities in vitro [31].) Additional studies will be required to test if the biochemical effects of Ser57 phosphorylation reported in vitro correspond to the regulation of ubiquitin polymers in cells.
These biochemical studies make robust predictions about the regulatory consequences phosphorylation should have regarding ubiquitin conjugation and deconjugation, but how Ser57 phosphorylation may regulate interactions with UBDs remains unknown. NMR analysis of S57D-Ub did not reveal significant structural perturbations associated with this phosphomimetic substitution [35], suggesting that phosphorylation is unlikely to disrupt the structure of ubiquitin grossly. Most UBDs that have been characterized structurally interact with Ub on its hydrophobic patch (Leu8, Ile44, and Val70) [43]. However, relative to the hydrophobic patch, Ser57 is located on the opposite side of Ub (Figure 2), so phosphorylation at this position is unlikely to have a broad impact on Ub-UBD interactions. Interestingly, a few UBDs interact with the Asp58 patch on Ub, which includes Ser57 [44, 45]. For example, an A20-type zinc finger (A20-ZnF) domain from Rabex-5, a Rab5 GEF that regulates membrane fusion at recycling endosomes, interacts with the Asp58 patch of Ub [46, 47]. In these structures, Ser57 of Ub was observed to form polar interactions with Asn28 of Rabex-5. Likewise, a different A20-ZnF domain from ZNF216, a protein that regulates NF-κB signaling, similarly binds to Ub at the Asp58 patch [45]. In both examples, the Asp58 patch engages in multiple polar interactions with the A20-ZnF domain. Importantly, UBDs that bind the Asp58 patch do not compete with interactions at the Ile44 hydrophobic patch, allowing for individual ubiquitin moieties to function as scaffolds in multi-protein complexes [48]. Together, these structural studies reveal the strong potential for Ser57 phosphorylation to regulate interactions occurring at the Asp58 patch, which could have implications for how ubiquitin functions as a scaffold in the regulation of processes including endocytic trafficking and cell signaling.
A critical limitation to the investigation of pS57-Ub has been a general lack of understanding of the enzymes and physiological conditions that trigger its production in cells. Recently, we reported the identification of the first kinases capable of phosphorylating ubiquitin at the Ser57 position. Specifically, we screened for yeast kinases capable of phosphorylating ubiquitin in an E. coli co-expression system. This effort initially uncovered a single yeast kinase – the Snf1-related kinase Vhs1 – as a Ser57 ubiquitin kinase. However, subsequent rounds of screening uncovered additional yeast Snf1-related kinases capable of phosphorylating ubiquitin at the Ser57 position – including Sks1 (the Vhs1 paralog), Gin4, and Kcc4 [36]. Importantly, both Sks1 and Vhs1 were capable of generating pS57-Ub when expressed in yeast. Furthermore, phenotypes associated with Vhs1 or Sks1 overexpression were suppressed in the presence of S57A-Ub, while phenotypes observed in Δsks1Δvhs1 mutant cells were suppressed in the presence of S57D-Ub. These genetic interactions between Vhs1, Sks1 and the Ser57 position of ubiquitin indicate that the function of Sks1 and Vhs1 in yeast is related to pS57-Ub. Importantly, we also reported that human MARK and SIK kinases, members of the human family of Snf1-related kinases and homologous to Vhs1 and Sks1, exhibit Ser57 ubiquitin kinase activity in vitro. Another recent study performed large-scale in vitro screening to identify human ubiquitin kinases. This study found that monoubiquitin is generally recalcitrant to phosphorylation across a panel of human kinases [49], suggesting it may have evolved to be a poor kinase substrate by itself. However, several AGC kinases – including members of the PKA, PKC, PKG and RSK families – were capable of phosphorylating the Ser57 position of ubiquitin when GST was fused to both ubiquitin and kinase, providing a dimerization platform [49]. These results indicate that some AGC family kinases can phosphorylate ubiquitin when brought into proximity by binding to protein-ubiquitin conjugates. Interestingly, the substitution of amino acids flanking Ser57 (T55R and D58L) to create a PKA consensus recognition motif resulted in a ubiquitin variant that was a strong PKA substrate, independent of GST dimerization [49]. Thus, the suboptimal recognition motif is overcome only by recruitment via protein-protein interactions. Combined, these recent studies reveal a small number of Ser57 ubiquitin kinases – including MARK kinases which operate on ubiquitin polymers directly and AGC kinases that operate on ubiquitin when recruited to ubiquitin by other protein-protein interactions – but much additional work will be required to fully understand how these kinases function in a biological context.
PINK1-mediated production of pS65-Ub is tightly regulated and highly context-dependent, and there are indications that cellular production of pS57-Ub is likewise tightly regulated and context-dependent. First, deletion of many Ser57 ubiquitin kinases in yeast results in various stress-related phenotypes [36], suggesting they operate in the cellular response to different types of stress. For example, while both Sks1 and Vhs1 are required for the yeast response to oxidative stress, Vhs1 regulates sensitivity to canavanine and thialysine (toxic analogs of arginine and lysine, respectively). In contrast, Sks1 regulates sensitivity to hydroxyurea [36], illustrating that these kinases may have partially overlapping but also distinct cellular functions. Second, the in vitro activities of these kinases exhibit nuanced differences in terms of specificity. For example, both Vhs1 and Sks1 showed activity toward mono-Ub but more robust activity toward polymers. Specifically, Vhs1 exhibited the greatest activity toward linear (M1-linked) and K29-linked ubiquitin polymers, while Sks1 exhibited the greatest activity toward linear (M1-linked) and K63-linked polymers [36]. Furthermore, MARK kinases exhibited activity toward linear ubiquitin tetramers, but not monomers, in vitro. Linkage specificity analysis of this activity revealed that MARK2 could phosphorylate linear, K11-, K63-, or K29-linked ubiquitin tetramers [36]. These linkage-type specificities may underlie unique regulatory functions and are consistent with the emerging picture that ubiquitin phosphorylation is tightly regulated and occurs in highly context-dependent manners in cells. Finally, the localization of these kinases is not well-characterized, and we hypothesize that stress-dependent subcellular localization may play an essential role in regulating localized production of pS57-Ub in cells. Ultimately, a combination of genetic analysis, biochemical characterization, and functional studies in cells will be required to define the specific contexts in which these kinases operate.
Currently, phosphatases that operate on pS57-Ub are still not defined. Our initial interest in pS57-Ub derived analysis of a phosphatase mutant – the ppz mutant of yeast, which lacks both Ppz1 and Ppz2. Yeast ppz mutants exhibit defects in the maintenance of ubiquitin levels (i.e., ubiquitin homeostasis) associated with increased levels of pS57-Ub [35]. While it is tempting to speculate that these function as Ser57 ubiquitin phosphatases in cells, we have been unable to express and purify recombinant Ppz complexes in vitro and thus unable to test if these Ppz phosphatases operate directly on phospho-ubiquitin. Therefore, further analysis will be required to define which phosphatases operate on pS57-Ub biochemically and in cells.
In recent years, progress has been made towards understanding the functional significance of pS57-Ub, but many important questions remain:
What specific pathways are targeted by Ser57 ubiquitin kinases in cells?
How do different kinases operate on different ubiquitin forms – including monomeric, unanchored polymeric, and protein-conjugated ubiquitin?
What subcellular and stress-dependent contexts are associated with pS57-Ub production?
What phosphatases operate on pS57-Ub?
How does Ser57 phosphorylation change how the ubiquitin code is “read” by UBDs?
pT12-Ub and other phosphorylated ubiquitin species
Compared to pS65-Ub and pS57-Ub, significantly less is known about other phosphorylated ubiquitin species. Many additional phosphorylation sites on ubiquitin have been detected in proteomic studies, including Thr7, Thr12, Thr14, Ser20, Thr22, Tyr59 and Ser66 (summarized in [50] and [51]). In vitro studies revealed that pS20-Ub converts UBE3C from a dual-specificity E3 ubiquitin ligase to a K48-specific ligase [31]. pS20-Ub was also reported to inhibit the activity of some DUBs, including cleavage of linear (M1-linked) di-Ub by CYLD, cleavage of K33-linked di-Ub by USP6 and USP25, and cleavage of K63-linked di-Ub by TRABID [31]. Additionally, pS20-Ub broadly inhibited cleavage of K48-linked di-Ub across a panel of DUBs [31]. This biochemical analysis underscores the regulatory potential of ubiquitin phosphorylation at Ser20, although the biological significance of pS20-Ub has not been determined.
Ubiquitin phosphorylated at Thr12 (pT12-Ub) was first detected in proteomic experiments comparing tissue samples from hepatocellular carcinoma and healthy patients [52]. Subsequent studies identified pT12-Ub in several different biological contexts, including: (i) proteomic analysis of nuclear proteins from Epstein-Barr virus-transformed B-lymphocyte cells [53], (ii) phosphoproteomic analysis of HeLa and K562 cell lines [54], and (iii) phosphoproteomic analysis of mitotically arrested HeLa cells [55]. Structurally, Thr12, along with Lys6, Lys11, Thr14, and Glu34 all contribute to a surface feature known as the TEK box [3], which is proposed to play a role in orienting ubiquitin for the formation of K11 linkages. For example, the TEK box is critical for forming K11-linked ubiquitin polymers by APC [57]. Thus, phosphorylation at Thr12 has the potential to regulate the formation of K11 linkages by APC and, by extension, the cell cycle. Another potential function of pT12-Ub involves the activity of HOIL-1, one of the subunits of the LUBAC complex. HOIL-1 was recently reported to catalyze the formation of oxyester bonds between ubiquitin and Ser or Thr residues on substrates [58]. Interestingly, HOIL-1 generated ubiquitin dimers in vitro that were conjugated at the Thr12 position [58], raising the interesting possibility that pThr12-Ub could antagonize ester-linked polymerization of Ub by atypical E3 ligases. By extension, phosphorylation at other positions could likewise regulate ester-linked polymerization of ubiquitin at other Ser and Thr residues. The development of new tools and reagents to detect pT12-Ub will be needed to determine if this type of regulation occurs in cells.
Recently, pT12-Ub has garnered attention for its role in regulating the DNA damage response [56]. DNA damage triggers ubiquitin modification of histone H2A at Lys13 and Lys15, via the RNF168 ubiquitin ligase. Ubiquitin modification of H2A at Lys15 is recognized by 53BP1, a factor that promotes non-homologous end-joining. Interestingly, DNA damage-induced production of pT12-Ub was reported to block 53BP1 recruitment and promote homologous recombination. Other DNA repair factors, such as RAD51, were still able to bind in the presence of pT12-Ub. Structural analysis revealed that phosphorylation at Thr12 disrupts the Ub-53BP1 interface [56].To date, specific kinases and phosphatases that operate on Thr12 have not been reported.
Phosphorylation of Ubiquitin-like Modifiers
Ubiquitin-like proteins (UBLs) play essential roles in cellular function [59]. Humans encode 9 UBL families: ATG8, ATG12, FAT10, FUBI, ISG15, NEDD8, SUMO, UFM1, URM1 [60] (Figure 3A). Sequence alignment analysis reveals limited conservation at sites corresponding to Ser57 and Ser65 ubiquitin (Figure 3B–C). For example, sequence analysis of yeast UBLs revealed that the site corresponding to Ser57 in yeast was often either a potential phosphorylation site (Ser, Thr) or a phosphomimetic residue (Glu) (Figure 3B). This trend was not observed in human UBLs (Figure 3C), although it is noteworthy that the human UBLs contained an Ala residue at the site corresponding to Ser57. Interestingly, yeast and human SUMO homologs all contain phosphomimetic Asp at the position corresponding to Ser65 in ubiquitin. Overall, this analysis suggests that the potential for phospho-regulation of UBLs at sites corresponding to Ser57 and Ser65 of Ub is limited.
Figure 3.
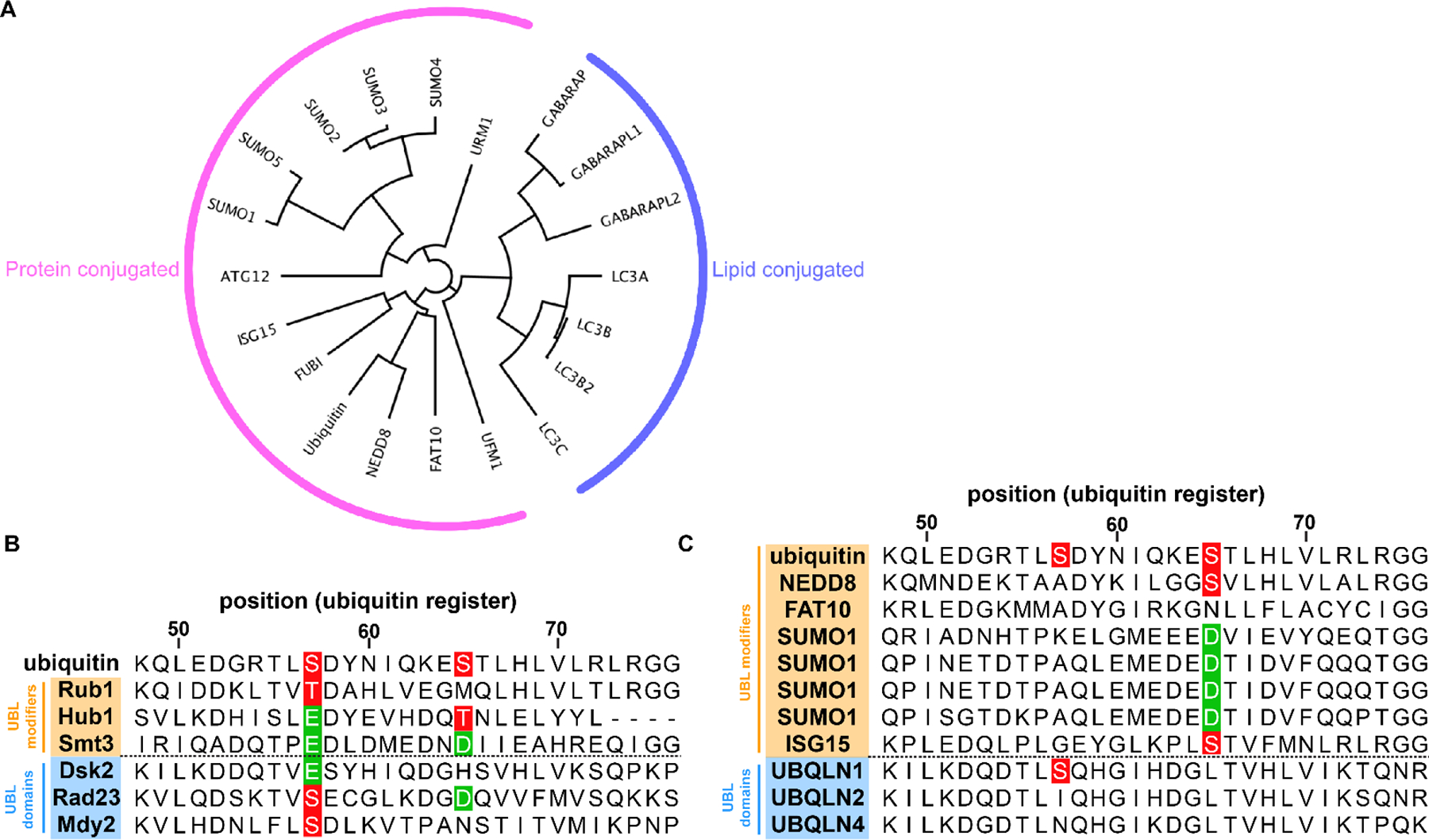
Analyzing the potential for phosphoregulation of UBLs. (A) Phylogenetic relationship of ubiquitin and human UBLs. Branch lengths are proportional to phylogenetic distances. (B and C) Yeast (B) or human (C) amino acid sequences for ubiquitin (top row), ubiquitin-like (UBL) modifiers (shaded orange) or UBL domains in larger proteins (shaded blue) were aligned using Clustal Omega. The alignment was visualized using Jalview, a portion of which (corresponding to the indicated register for ubiquitin) is shown. Highlighted are the positions corresponding to Ser57 and Ser65 in ubiquitin. Positions shaded red indicate a residue that can be phosphorylated (Ser or Thr) while positions shaded green are phosphomimetic (Glu or Asp). For several yeast UBLs (Rub1, Rad23 and Mdy2) the position corresponding to Ser57 is a potential site of phosphorylation, while for others (Hub1, Smt3 and Dsk2) this residue is a phosphomimic (glutamate).
For most human UBLs phosphorylated species have been detected in proteomic studies (Table 2). ATG8 phosphorylation sites were partially characterized, and several kinases for ATG8 family members are known. PKA phosphorylates Ser12 on LC3A, and the phosphomimetic S12D variants reduced LC3A recruitment to autophagosomes, while S12A substitutions increased recruitment to autophagosomes [61]. LC3B and LC3B2 are phosphorylated on T6, T29 and T50. PKC phosphorylates T6 and T29, but the phosphorylation of these sites does not affect autophagy. In contrast, STK3, STK4 [62], NEK9 and PKCζ [63] phosphorylate T50 to induce autophagy [62]. TBK1, a kinase known for its significant role in autophagy [64–66] and innate immune signaling [67, 68], can phosphorylate LC3C and GABARAPL2 (LC3C at S93 and S96; GABARAPL2 at S87 and S88) [69]. LC3C and GABARAPL2 phosphorylation ensure the steady formation of autophagosomes. Phosphorylation of LC3C and GABARAPL2 blocks their interaction with ATG4 and subsequently prevents the removal of LC3C and GABARAPL2 from premature autophagosomes [69]. Impaired autophagy has been linked to several diseases, including neurodegeneration, inflammatory disorders and cancer [70]. Therefore, uncontrolled phosphorylation of ATG8 family members might be crucial to understand the molecular pathways resulting in these pathological phenotypes.
Table 2:
Phosphorylation sites of UBLs according to PhosphoSitePlus [75, 76]. UBLs, except for FUBI and ATG8, are reversibly conjugated to lysine residues on target proteins via their C-terminal glycine motifs. FUBI is classified as a protein conjugating UBL, but no FUBI substrates are known to date [77]. ATG8 family members are not conjugated to proteins but the lipid phosphatidylethanolamine (PE) to induce autophagy [78, 79].
| Family | Proteins | Phosphorylation sites | Substrate |
|---|---|---|---|
| ATG8 | LC3A LC3B LC3B2 LC3C GABARAP GABARAPL1 GABARAPL2 |
S12; S92 T6; T29; T50 T6; T29; T50 S93; S96 Y49; Y61 Y5; Y49; Y61 S10; S39; S87; S88 |
lipid (PE) |
| ATG12 | Atg12 | S41; T65; T70 | protein (lysine) |
| FAT10 | FAT10 | S64; T73; T77; S109; S110; S111; Y148; Y161 | protein (lysine) |
| FUBI | FUBI | --- | unknown |
| ISG15 | ISG15 | S22; S26; S50 | protein (lysine) |
| NEDD8 | NEDD8 | T7; T9; T20; Y45; T55; Y59; S65 | protein (lysine) |
| SUMO | SUMO1 SUMO2 SUMO3 SUMO4 SUMO5 |
S2; S9; S10; S32; T76 T12; S28; T38; S54 S2; T12; S27; T37; S53 S28 --- |
protein (lysine) |
| UFM1 | UFM1 | Y18; S72 | protein (lysine) |
| URM1 | URM1 | --- | protein (lysine) |
Compared to ATG8 family members, much less is known about phosphorylation of protein-conjugated UBLs. Due to the high number of proteomics datasets available, phosphorylation sites are known for most of these UBLs (Table 1). However, the function and regulation of these phosphorylation events remain unknown. The SUMO family is the best-studied UBL family, and the Mann lab and colleagues described the first SUMO phosphorylation site in 2008. They observed conserved phosphorylation of SUMO at Ser2 in humans and lower eukaryotes such as drosophila and yeast [71]. SUMO phosphorylation at Ser2 is of particular interest since this residue is not conserved amongst all SUMO family members. Ser2 is present in SUMO1, SUMO3 and SUMO5 but not in SUMO2 or SUMO4. Ser2 SUMO phosphorylation might play a role in differentiating SUMO2 and SUMO3, which differ by only three amino acids. SUMO5 is highly expressed in testes and peripheral blood leukocytes but is missing in many other human tissues and cell lines [72]. This highly restricted expression might be the reason why SUMO5 phosphorylation has not been described so far.
The regulatory significance of UBL phosphorylation remains understudied, and deciphering its significance will require new research. Most proteomics datasets revealed UBL phosphorylation under steady-state conditions. Thereby, datasets inherently lack information about context-specific UBL phosphorylation dynamics. ISG15 expression, for instance, is induced by interferon stimulation [73], making it a potential blind spot of phospho-proteomic analysis. One of the significant tasks of future research will be to identify the upstream signaling events resulting in UBL phosphorylation. The cell-type and stimuli-specific expression of UBLs likely results in an incomplete picture of the UBL phosphorylation landscape. The majority of UBL phosphorylation sites were identified in laboratory cell lines, which only approximate cellular behavior in vivo. The origin of the cell lines and their signaling context should be considered when studying UBL phosphorylation. In addition to the upstream signaling events, it will be of equal interest to elucidate UBL phosphorylation’s biological function. Phosphomimetic UBL variants might shed some light on their biological function, but these mutations cannot reflect the spatiotemporal changes of UBL phosphorylation. In this context, identifying the UBL-targeting kinases and phosphatases will be the key to understand the biochemical and physiological functions of UBL phosphorylation.
Concluding Remarks and Remaining Questions
Despite decades of intense research on the ubiquitin code, the significance of Ub phosphorylation has only recently come into focus. So why has it taken so long to appreciate this feature of the ubiquitin code? One reason is the low abundance of phosphorylated ubiquitin species in cells. Given the abundance of ubiquitin, its essential regulatory functions in eukaryotic cells, and the potential for phosphorylation to modify these functions, it is tempting to speculate that ubiquitin may have evolved to avoid recognition by kinases to maintain phosphorylated species at low stoichiometry. Consistent with this speculation, kinase activity toward ubiquitin appears to be tightly regulated and highly localized. For example, PINK1 exhibits very low activity and is degraded until mitochondrial damage triggers its accumulation and activation on the mitochondrial outer membrane, leading to localized production of pS65-Ub in the proximity of aberrant import complexes. Such deliberate, context-dependent production of phosphorylated ubiquitin underscores its regulatory potency as well as the challenges associated with dissecting its functions.
One factor limiting progress in this field is a lack of reagents for detection of pUb species. Although many pUb species can be detected by mass spectrometry, commercial antibodies are currently only available for the detection of pS65-Ub. These antibodies have been critical for advancing the understanding of the PINK1-Parkin pathway and the role pS65-Ub plays in mitophagy. Unfortunately, similar antibodies are not commercially available for other pUb species. Although antibodies detecting pS57-Ub have been described and validated they lack the sensitivity for detection in biological samples [35, 36]. Development of sensitive, commercially-available antibodies for recognition of a variety of pUb species will be an important step forward that will catalyze new mechanistic insights at this important frontier.
Despite these limitations, several themes are emerging that could help to guide ongoing and future research in this area. One emerging theme is that ubiquitin phosphorylation appears to be involved in stress responses: pS65-Ub regulates mitophagy in response to mitochondrial damage, pS57-Ub regulates the oxidative stress response, and pT12-Ub regulates the DNA damage response. All of these stress responses were previously known to be Ub-dependent, but the phosphorylation of ubiquitin in response to specific stressors suggests its function is being modified to optimize the cellular response to various insults and environmental challenges. It is tempting to speculate that ubiquitin phosphorylation may provide a mechanism for rapid activation of stress responses that must be tightly regulated and kept “off” in normal circumstances but must also be switched on quickly to restrict cellular damage in conditions of stress. This is definitely the case for pS65-Ub in mitophagy [13], but additional work will be required to determine if ubiquitin phosphorylation at other positions likewise facilitates feedforward activation during the response to cellular stress.
Generally, an improved understanding of how and where pUb species are produced in cells will require identifying kinases and phosphatases that regulate production in response to stress. Substantial progress will likely require the development of new tools and reagents for the detection and quantification of pUb species. Although several studies have revealed biochemical insights into how Ub phosphorylation can influence conjugation and deconjugation reactions in vitro, much work is still needed to pinpoint which reactions are regulated in a physiological context. Furthermore, it remains unclear to what extent phosphorylation regulates Ub recognition. In this review, we have used structural information to speculate how specific phosphorylation events on Ub might interfere with known ubiquitin binding domains. Additionally, some ubiquitin binding domains may specifically recognize pUb species, and the discovery of such domains could help elucidate how phosphorylation regulates Ub biology. These challenges and unknowns provide a fertile frontier ready for exciting discoveries that have potential to transform our understanding of the ubiquitin code and its role in proteome remodeling and the cellular stress response.
Acknowledgements
We are grateful to S. Qualls-Histed and T. Graham for critical reading of the manuscript and insightful discussions. We also thank E. MacGurn for help with graphic design. This study was partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 (Project-ID 390873048), TRR237 (Project-ID 369799452), and SFB1403 (Project-ID 414786233). NLH and JAM were supported by NIH grants R01 GM118491 (to JAM) and R21 AG053562 (to JAM).
Abbreviations
- APC
anaphase promoting complex
- AQUA
absolute quantification mass spectrometry
- DUB
deubiquitylase
- ESCRT
endosomal sorting complexes required for transport
- LUBAC
linear ubiquitin assembly complex
- OMM
outer mitochondrial membrane
- MARK
microtubule affinity regulating kinase
- PINK1
PTEN induced putative kinase 1
- PKA
protein kinase A
- PKC
protein kinase C
- PKG
protein kinase G
- pS65-Ub
Ser65-phosphorylated ubiquitin
- pS57-Ub
Ser57-phosphorylated ubiquitin
- pT12-Ub
Thr12-phosphorylated ubiquitin
- PTM
post-translational modification
- SUMO
small ubiquitin-like modifier
- Ub
ubiquitin
- UBD
ubiquitin binding domain
- UBL
ubiquitin-like modifier
- WT
wildtype
Footnotes
Publisher's Disclaimer: This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions. This article may not be enhanced, enriched or otherwise transformed into a derivative work, without express permission from Wiley or by statutory rights under applicable legislation. Copyright notices must not be removed, obscured or modified. The article must be linked to Wiley’s version of record on Wiley Online Library and any embedding, framing or otherwise making available the article or pages thereof by third parties from platforms, services and websites other than Wiley Online Library must be prohibited.
Conflicts of Interest
Eicke Latz is co-founder and consultant of IFM Therapeutics, DiosCure Therapeutics and Odyssey Therapeutics.
For all other authors there are no conflicts of interest to declare.
References
- 1.Oh E, Akopian D & Rape M (2018) Principles of Ubiquitin-Dependent Signaling, Annu Rev Cell Dev Biol. 34, 137–162. [DOI] [PubMed] [Google Scholar]
- 2.Akutsu M, Dikic I & Bremm A (2016) Ubiquitin chain diversity at a glance, J Cell Sci. 129, 875–80. [DOI] [PubMed] [Google Scholar]
- 3.Kulathu Y & Komander D (2012) Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages, Nat Rev Mol Cell Biol. 13, 508–23. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen CP & MacGurn JA (2020) Coupling Conjugation and Deconjugation Activities to Achieve Cellular Ubiquitin Dynamics, Trends Biochem Sci. 45, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer HJ & Rape M (2014) Enhanced protein degradation by branched ubiquitin chains, Cell. 157, 910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haakonsen DL & Rape M (2019) Branching Out: Improved Signaling by Heterotypic Ubiquitin Chains, Trends Cell Biol. 29, 704–716. [DOI] [PubMed] [Google Scholar]
- 7.Herhaus L & Dikic I (2015) Expanding the ubiquitin code through post-translational modification, EMBO Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X & Hunter T (2014) Pink1, the first ubiquitin kinase, EMBO J. 33, 1621–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D & Gygi SP (2003) A proteomics approach to understanding protein ubiquitination, Nat Biotechnol. 21, 921–6. [DOI] [PubMed] [Google Scholar]
- 10.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K & Matsuda N (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin, Nature. 510, 162–6. [DOI] [PubMed] [Google Scholar]
- 11.Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M & Muqit MM (2014) Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65, Biochem J. 460, 127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S & Youle RJ (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity, J Cell Biol. 205, 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, Wells JA, Gygi SP, Schulman BA & Harper JW (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis, Mol Cell. 56, 360–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XL, Feng ST, Wang YT, Yuan YH, Li ZP, Chen NH, Wang ZZ & Zhang Y (2021) Mitophagy, a Form of Selective Autophagy, Plays an Essential Role in Mitochondrial Dynamics of Parkinson’s Disease, Cell Mol Neurobiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark EH, Vázquez de la Torre A, Hoshikawa T & Briston T (2020) Targeting mitophagy in Parkinson’s disease, J Biol Chem, 100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaney DL, Rodríguez-Mias RA & Villén J (2015) Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover, EMBO Rep. 16, 1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sloper-Mould KE, Jemc JC, Pickart CM & Hicke L (2001) Distinct functional surface regions on ubiquitin, J Biol Chem. 276, 30483–9. [DOI] [PubMed] [Google Scholar]
- 18.Mishra P, Prabha CR, Rao CM & Volety S (2011) Q2N and S65D substitutions of ubiquitin unravel functional significance of the invariant residues Gln2 and Ser65, Cell Biochem Biophys. 61, 619–28. [DOI] [PubMed] [Google Scholar]
- 19.Rona G & Pagano M (2019) Mixed ubiquitin chains regulate DNA repair, Genes Dev. 33, 1615–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundar R, Gudey SK, Heldin CH & Landström M (2015) TRAF6 promotes TGFβ-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFβ type I receptor, Cell Cycle. 14, 554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva GM, Finley D & Vogel C (2015) K63 polyubiquitination is a new modulator of the oxidative stress response, Nat Struct Mol Biol. 22, 116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barriere H, Nemes C, Du K & Lukacs GL (2007) Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery, Mol Biol Cell. 18, 3952–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aksentijevich I & Zhou Q (2017) NF-κB Pathway in Autoinflammatory Diseases: Dysregulation of Protein Modifications by Ubiquitin Defines a New Category of Autoinflammatory Diseases, Front Immunol. 8, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Lee S, Peng Y, Bunker E, Giaime E, Shen J, Zhou Z & Liu X (2014) PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions, Curr Biol. 24, 1854–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert AF, Gladkova C, Pardon E, Wagstaff JL, Freund SMV, Steyaert J, Maslen SL & Komander D (2017) Structure of PINK1 in complex with its substrate ubiquitin, Nature. 552, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Wang J, Tang Y & Shen HM (2018) PTEN-L puts a brake on mitophagy, Autophagy. 14, 2023–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Cho YL, Tang Y, Wang J, Park JE, Wu Y, Wang C, Tong Y, Chawla R, Zhang J, Shi Y, Deng S, Lu G, Tan HW, Pawijit P, Lim GG, Chan HY, Fang L, Yu H, Liou YC, Karthik M, Bay BH, Lim KL, Sze SK, Yap CT & Shen HM (2018) PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy, Cell Res. 28, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Lu G & Shen HM (2020) The Long and the Short of PTEN in the Regulation of Mitophagy, Front Cell Dev Biol. 8, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wall CE, Rose CM, Adrian M, Zeng YJ, Kirkpatrick DS & Bingol B (2019) PPEF2 Opposes PINK1-Mediated Mitochondrial Quality Control by Dephosphorylating Ubiquitin, Cell Rep. 29, 3280–3292.e7. [DOI] [PubMed] [Google Scholar]
- 30.Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM & Komander D (2015) Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis, EMBO J. 34, 307–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huguenin-Dezot N, De Cesare V, Peltier J, Knebel A, Kristaryianto YA, Rogerson DT, Kulathu Y, Trost M & Chin JW (2016) Synthesis of Isomeric Phosphoubiquitin Chains Reveals that Phosphorylation Controls Deubiquitinase Activity and Specificity, Cell Rep. 16, 1180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deol KK, Eyles SJ & Strieter ER (2020) Quantitative Middle-Down MS Analysis of Parkin-Mediated Ubiquitin Chain Assembly, J Am Soc Mass Spectrom. 31, 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gersch M, Gladkova C, Schubert AF, Michel MA, Maslen S & Komander D (2017) Mechanism and regulation of the Lys6-selective deubiquitinase USP30, Nat Struct Mol Biol. 24, 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaney DL, Rodríguez-Mias RA & Villén J (2015) Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover, EMBO Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Tumolo JM, Ehlinger AC, Jernigan KK, Qualls-Histed SJ, Hsu PC, McDonald WH, Chazin WJ & MacGurn JA (2017) Ubiquitin turnover and endocytic trafficking in yeast are regulated by Ser57 phosphorylation of ubiquitin, Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hepowit NL, Pereira KN, Tumolo JM, Chazin WJ & MacGurn JA (2020) Identification of ubiquitin Ser57 kinases regulating the oxidative stress response in yeast, Elife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang KL, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyö D, Ellis MJ, Carr SA & CPTAC N (2016) Proteogenomics connects somatic mutations to signalling in breast cancer, Nature. 534, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart SA, Houel S, Lee T, Wang N, Old WM & Ahn NG (2015) A Phosphoproteomic Comparison of B-RAFV600E and MKK1/2 Inhibitors in Melanoma Cells, Mol Cell Proteomics. 14, 1599–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, Chen HY, Nesvizhskii AI, Ishihama Y & Chen YJ (2015) Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics, Nat Commun. 6, 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweppe DK, Rigas JR & Gerber SA (2013) Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors, J Proteomics. 91, 286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavor D, Barlow KA, Asarnow D, Birman Y, Britain D, Chen W, Green EM, Kenner LR, Mensa B, Morinishi LS, Nelson CA, Poss EM, Suresh P, Tian R, Arhar T, Ary BE, Bauer DP, Bergman ID, Brunetti RM, Chio CM, Dai SA, Dickinson MS, Elledge SK, Helsell CVM, Hendel NL, Kang E, Kern N, Khoroshkin MS, Kirkemo LL, Lewis GR, Lou K, Marin WM, Maxwell AM, McTigue PF, Myers-Turnbull D, Nagy TL, Natale AM, Oltion K, Pourmal S, Reder GK, Rettko NJ, Rohweder PJ, Schwarz DMC, Tan SK, Thomas PV, Tibble RW, Town JP, Tsai MK, Ugur FS, Wassarman DR, Wolff AM, Wu TS, Bogdanoff D, Li J, Thorn KS, O’Conchúir S, Swaney DL, Chow ED, Madhani HD, Redding S, Bolon DN, Kortemme T, DeRisi JL, Kampmann M & Fraser JS (2018) Extending chemical perturbations of the ubiquitin fitness landscape in a classroom setting reveals new constraints on sequence tolerance, Biol Open. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.George S, Wang SM, Bi Y, Treidlinger M, Barber KR, Shaw GS & O’Donoghue P (2017) Ubiquitin phosphorylated at Ser57 hyper-activates parkin, Biochim Biophys Acta Gen Subj. 1861, 3038–3046. [DOI] [PubMed] [Google Scholar]
- 43.Dikic I, Wakatsuki S & Walters KJ (2009) Ubiquitin-binding domains - from structures to functions, Nat Rev Mol Cell Biol. 10, 659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raiborg C, Slagsvold T & Stenmark H (2006) A new side to ubiquitin, Trends Biochem Sci. 31, 541–4. [DOI] [PubMed] [Google Scholar]
- 45.Searle MS, Garner TP, Strachan J, Long J, Adlington J, Cavey JR, Shaw B & Layfield R (2012) Structural insights into specificity and diversity in mechanisms of ubiquitin recognition by ubiquitin-binding domains, Biochem Soc Trans. 40, 404–8. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS & Hurley JH (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5, Nat Struct Mol Biol. 13, 264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S & Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin, Cell. 124, 1183–95. [DOI] [PubMed] [Google Scholar]
- 48.Garner TP, Strachan J, Shedden EC, Long JE, Cavey JR, Shaw B, Layfield R & Searle MS (2011) Independent interactions of ubiquitin-binding domains in a ubiquitin-mediated ternary complex, Biochemistry. 50, 9076–87. [DOI] [PubMed] [Google Scholar]
- 49.Kolbe C-C & Latz E (2020) AGC kinase members phosphorylate ubiquitin, bioRxiv, 2020.07.15.204149. [Google Scholar]
- 50.Herhaus L & Dikic I (2015) Expanding the ubiquitin code through post-translational modification, EMBO Rep. 16, 1071–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swatek KN & Komander D (2016) Ubiquitin modifications, Cell Res. 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee HJ, Na K, Kwon MS, Kim H, Kim KS & Paik YK (2009) Quantitative analysis of phosphopeptides in search of the disease biomarker from the hepatocellular carcinoma specimen, Proteomics. 9, 3395–408. [DOI] [PubMed] [Google Scholar]
- 53.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J & Andersen JS (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response, Mol Cell Proteomics. 9, 1314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ & Mohammed S (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome, J Proteome Res. 12, 260–71. [DOI] [PubMed] [Google Scholar]
- 55.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA & Gerber SA (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells, Sci Signal. 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walser F, Mulder MPC, Bragantini B, Burger S, Gubser T, Gatti M, Botuyan MV, Villa A, Altmeyer M, Neri D, Ovaa H, Mer G & Penengo L (2020) Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response, Mol Cell. 80, 423–436.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin L, Williamson A, Banerjee S, Philipp I & Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex, Cell. 133, 653–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelsall IR, Zhang J, Knebel A, Arthur JSC & Cohen P (2019) The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells, Proc Natl Acad Sci U S A. 116, 13293–13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Veen AG & Ploegh HL (2012) Ubiquitin-like proteins, Annu Rev Biochem. 81, 323–57. [DOI] [PubMed] [Google Scholar]
- 60.Hochstrasser M (2009) Origin and function of ubiquitin-like proteins, Nature. 458, 422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cherra SJ, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW & Chu CT (2010) Regulation of the autophagy protein LC3 by phosphorylation, J Cell Biol. 190, 533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A & Hansen M (2015) Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy, Mol Cell. 57, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shrestha BK, Skytte Rasmussen M, Abudu YP, Bruun JA, Larsen KB, Alemu EA, Sjøttem E, Lamark T & Johansen T (2020) NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1, J Biol Chem. 295, 1240–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ & Dikic I (2016) Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria, Proc Natl Acad Sci U S A. 113, 4039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T & Deretic V (2012) TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation, Immunity. 37, 223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N & Randow F (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria, Nat Immunol. 10, 1215–21. [DOI] [PubMed] [Google Scholar]
- 67.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM & Maniatis T (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway, Nat Immunol. 4, 491–6. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka Y & Chen ZJ (2012) STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway, Sci Signal. 5, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herhaus L, Bhaskara RM, Lystad AH, Gestal-Mato U, Covarrubias-Pinto A, Bonn F, Simonsen A, Hummer G & Dikic I (2020) TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease, EMBO Rep. 21, e48317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine B & Kroemer G (2019) Biological Functions of Autophagy Genes: A Disease Perspective, Cell. 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matic I, Macek B, Hilger M, Walther TC & Mann M (2008) Phosphorylation of SUMO-1 occurs in vivo and is conserved through evolution, J Proteome Res. 7, 4050–7. [DOI] [PubMed] [Google Scholar]
- 72.Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML & Yang WM (2016) SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies, Sci Rep. 6, 26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perng YC & Lenschow DJ (2018) ISG15 in antiviral immunity and beyond, Nat Rev Microbiol. 16, 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K & Matsuda N (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin, Nature. [DOI] [PubMed] [Google Scholar]
- 75.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V & Sullivan M (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse, Nucleic Acids Res. 40, D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V & Skrzypek E (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations, Nucleic Acids Res. 43, D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cappadocia L & Lima CD (2018) Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism, Chem Rev. 118, 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T & Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation, Nature. 408, 488–92. [DOI] [PubMed] [Google Scholar]
- 79.Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems, Nat Rev Mol Cell Biol. 2, 211–6. [DOI] [PubMed] [Google Scholar]


