Abstract
Diallyl trisulfide (DATS) is an attractive anti-cancer phytochemical with in vitro and in vivo growth inhibitory effects against different solid tumors including breast cancer. We have shown previously that an immortalized mammary epithelial cell line (MCF-10A) is resistant to growth inhibition by DATS. In this study, we performed RNA-seq analysis using a breast cancer cell line (SK-BR-3) and MCF-10A cells to gain insights into cancer selective effects of DATS. The Gene Ontology analysis revealed upregulation of genes associated with actin cytoskeleton but downregulation of mitochondria-related genes in the SK-BR-3 human breast cancer cell line but not in the non-oncogenic MCF-10A cell line upon treatment with DATS. Quantitative real-time reverse transcription polymerase chain reaction confirmed DATS-mediated upregulation of several actin cytoskeleton-related genes in the SK-BR-3 cell line. The DATS treatment dose-dependently disrupted actin cytoskeleton in the SK-BR-3 cell line, whereas the MCF-10A cell line was more resistant to this effect. The DATS treatment caused a marked increase in phosphorylation of dynamin-1-like (DRP1) protein in the SK-BR-3 cell line. However, the DATS-mediated apoptosis was not affected by genetic deletion of DRP1 protein. The Reactome pathway analysis showed downregulation of genes associated with citric acid cycle in the SK-BR-3 cell line but not in the MCF-10A cells. However, expression of aconitase 2 or dihydrolipoamide S-succinyltransferase was not affected by DATS treatment. In conclusion, this study reveals that actin cytoskeleton is a novel target of DATS in the SK-BR-3 cell line, which may explain its inhibitory effect on breast cancer cell migration.
Keywords: Breast cancer, Diallyl trisulfide, Chemoprevention
INTRODUCTION
Cancer chemoprevention using natural agents or synthetic agents is attractive for reducing morbidity and mortality in patients with breast cancer, which is a leading cause of cancer-related deaths in American women [1,2]. Clinical use of selective estrogen receptor modulators and aromatase inhibitors for chemoprevention of luminal-type breast cancer attests to the merit of this approach [3,4]. Because of their favorable safety profile, bioactive phytochemicals from edible plants are appealing for chemoprevention of breast cancer and other malignancies [5]. Phytochemicals with anti-cancer activity have been identified from many edible plants including soy (isoflavones such as genistein), tomato (lycopene), broccoli (sulforaphane and other isothiocyanates), and Allium vegetables such as garlic (organosulfur compounds including diallyl trisulfide [DATS]) [5-7]. Some of these phytochemicals or edible plants have been tested clinically; these include lycopene, sulforaphane, and DATS [5,8,9]. For example, in an interventional study with garlic component DATS (200 mg synthetic allitridum in combination with 100 μg selenium) given every other day for one month of each year during November 1989 to December 1991, the relative cancer risk was 0.67 (95% confidence interval 0.43-1.03) [9]. Epidemiological data is generally supportive of the preventive effect of garlic on cancer risk [10]. The epidemiological studies were instrumental for identification of bioactive phytochemicals from garlic and other Allium family vegetables [10].
The bioactive phytochemicals these include garlic are generated upon cutting or chewing this vegetable. These include water-soluble (e.g., S-allyl cysteine) and lipid-soluble (e.g., DATS) organosulfur compounds. The lipid-soluble compounds are relatively more effective in inhibiting proliferation/survival of breast cancer cells when compared to water-soluble organosulfur compounds. Among various lipid-soluble organosulfur compounds, DATS is relatively more effective than diallyl disulfide or diallyl monosulfide in inhibiting survival of cancer cells [11]. In breast cancer cells, the DATS treatment causes G2/M phase and mitotic arrest, apoptotic cell death, and inhibition of cell migration [12-15]. The DATS-induced apoptosis in breast cancer cells is triggered by generation of reactive oxygen species (ROS) [13,14]. The mechanism underlying DATS-mediated inhibition of cell cycle progression or cell migration in breast cancer is not fully understood, but checkpoint kinase 1 and cell division cycle 25C were implicated in G2/M phase mitotic arrest in prostate cancer cells [16,17]. It is highly likely that similar mechanisms are operative in DATS-mediated cell cycle arrest in breast cancer cells. The inhibitory effect of DATS treatment on self-renewal of breast cancer stem-like cells has also been demonstrated [18,19]. A role for Wnt/β-catenin and FoxQ1 transcription factor has been shown in suppression of breast cancer stem-like cells following DATS treatment [18,19]. In addition to FoxQ1, DATS treatment was shown to inhibit expression or activity of other oncogenic transcription factors, including activator protein 1, estrogen receptor-α, signal transducer and activator of transcription 3, and hypoxia-inducible factor-1α [20-23]. The DATS treatment potentiated chemotherapeutic efficacy of doxorubicin in an experimentally induced mammary cancer model by decreasing Notch 1 [24]. The DATS administration has shown in vivo efficacy in rodent models of breast cancer [13,18,24].
We have shown previously that an immortalized non-oncogenic human mammary epithelial cell line (MCF-10A) is resistant to growth inhibition and apoptosis induction by DATS treatment, but these effects were observed in human breast cancer cells belonging to the luminal-type (MCF-7), triple-negative (MDA-MB-231), and human epidermal growth factor receptor 2+(HER2+) (BRI-JM04) subtypes following exposure to this phytochemical [14]. The mechanisms underlying breast cancer-selective effects of DATS is not known. In this study, we have tried to address this gap in knowledge by determining the effect of DATS treatment on gene expression changes using MCF-10A and SK-BR-3 human breast cancer cells. The SK-BR-3 cells are well-characterized representative of HER2+ subtype, and hence expected to be sensitive to growth inhibition and apoptosis induction following DATS treatment similar to the Her2+ murine BRI-JM04 cells.
MATERIALS AND METHODS
Reagents
The DATS (purity 99.2%) was purchased from LKT Laboratories (St. Paul, MN, USA), dissolved in dimethyl sulfoxide (DMSO; 28 mM stock), and stored at –80°C. Cell culture media was from MediaTech (Manassas, VA, USA). FBS was obtained from Atlanta Biologicals (Norcross, GA, USA). Anti-aconitase 2 (ACO2) antibody and anti-dihydrolipoamide S-succinyltransferase (DLST) antibody were from Cell Signaling Technology (Danvers, MA, USA). Anti-β-actin antibody and propidium iodide (PI) were from Sigma Aldrich (St. Louis, MO, USA). Anti-dynamin-1-like (DRP1), anti-phospho (S616)-DRPl, and anti-mitofusin 2 (MFN2) antibodies were from Cell Signaling Technology; anti-fission, mitochondrial 1 (FIS1) antibody was from Proteintech (Rosemont, IL, USA); and anti-optic atrophy protein 1 (OPA1) antibody was from Abcam (Waltham, MA, USA). Fluorescein isothiocyanate Annexin V Apoptosis Detection Kit was from BD Biosciences (San Jose, CA, USA).
Cell lines
The SK-BR-3 human breast cancer and MCF-10A human mammary epithelial cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained as suggested by the supplier. The SK-BR-3 cell line was last authenticated by us in March, 2017. Immortalized mouse embryonic fibroblasts (MEFs) from DRP1 knockout mice (DRP1–/–) and wild-type mice (DRP1+/+) were a generous gift from Dr. Katsuyoshi Mihara (Kyushu University, Fukuoka, Japan). The MEFs were cultured in Dulbecco’s modified essential medium supplemented with 10% FBS and antibiotics. The MEFs were not authenticated by us.
RNA-seq analysis
The RNA-seq analysis was done by Novogene (Sacramento, CA, USA). The results were obtained by the Gene Ontology (GO) and the Reactome pathway analyses as described by us previously [15,25]. The data has been submitted to Gene Expression Omnibus (GSE200872).
Quantitative real-time reverse transcription (qRT)-PCR
The cells were plated at a density of 5 × 105 cells/dish in 6-cm dishes, allowed to attach overnight, and then treated with DMSO (control) or indicated doses of DATS for specified time points. Total RNA isolation, reverse transcription, and qRT-PCR were performed as described previously [25]. Primers for calcium/calmodulin-dependent serine protein kinase (CASK), eukaryotic translation elongation factor 1 alpha 1 (EEF1A1), guanylate binding protein 2 (GBP2), intersectin 1 (ITSN1), and utrophin (UTRN) were obtained from GeneCopoeia (Rockville, MD, USA). Primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as follows. GAPDH-forward: 5ʹ-GGACCTGACCTGCCGTCTAGAA-3ʹ; GAPDH-reverse: 5’-GGTGTCGCTGTTGAAGTCAGAG-3’. The PCR conditions were as follows: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute and 72°C for 30 seconds. Gene expression levels were calculated using the method of Livak and Schmittgen [26].
Confocal microscopy for visualization of the filamentous actin (F-actin) cytoskeleton
The MCF-10A or SK-BR-3 cells were plated on coverslip in 24-well plate at a density of 5 × 104 cells/well, allowed to attach, and then treated with DMSO (control) or indicated doses of DATS for 24 hours. After treatment, the cells were washed with PBS and fixed with 4% paraformaldehyde for 10 minutes and then permeabilized with 0.5% Triton X-100 containing PBS for 5 minutes. The cells were incubated with Rhodamine Phalloidin (Cytoskeleton, Inc., Denver, CO, USA) for 30 minutes at 37°C in the dark after washing with PBS. The cells were counterstained with 4’,6-diamidino-2-phenylindole and mounted and then imaged at ×40 oil objective magnification using Nikon A1 confocal microscope (Nikon, Tokyo, Japan).
Western blotting
The MCF-10A or SK-BR-3 cells were plated in 6-cm dish at a density of 5 × 105 cells/dish and allowed to attach. The cells were treated with DMSO (control) or indicated doses of DATS for specified time points. Preparation of whole cell lysates and immunoblotting procedures were done as described by us previously [27].
Determination of apoptosis
Apoptotic cell death was quantitated by flow cytometry following staining with Annexin V and PI essentially as described by us previously [28]. Briefly, the MEFs were plated in 6-well plates at a density of 2 × 105 cells/well and incubated overnight to allow attachment. The MEFs were then treated with DMSO (control) or 20 or 40 μM DATS for 24 hours. The MEFs were collected by trypsinization, suspended in Annexin V/PI solution, incubated for 30 minutes at room temperature with gentle shaking in the dark, and analyzed using a CytoFLEXflow cytometer (Beckman Coulter Life Sciences, Brea, CA, USA).
Statistical analysis
GraphPad Prism (version 8.0.0; GraphPad Software, San Diego, CA, USA) was used to perform statistical analyses. Statistical significance of difference for two group comparison was determined by unpaired Student’s t-test. For dose response comparisons, one-way analysis of variance (ANOVA) followed by Dunnett’s test was used. One-way ANOVA with Bonferroni’s test was used for multiple group comparisons. A P-value of < 0.05 was considered statistically significant.
RESULTS
RNA-seq analysis
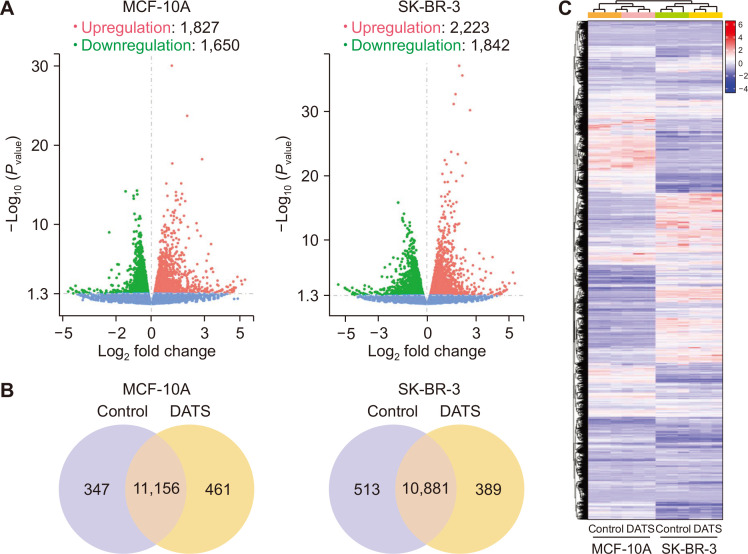
Volcano plots in Figure 1A show distribution of differentially expressed genes between control and DATS-treated MCF-10A or SK-BR-3 cells. The DATS treatment resulted in upregulation of 1,827 genes and downregulation of 1,650 genes in MCF-10A cells. Upregulation of 2,223 genes but downregulation of 1,842 genes was observed in SK-BR-3 cells following DATS treatment. The Venn diagrams for control and DATS-treated MCF-10A or SK-BR-3 cells show unique and overlapping gene expression changes (Fig. 1B). The heatmaps of three biological replicates of control and DATS-treated MCF-10A and SK-BR-3 cells are shown in Figure 1C demonstrating data consistency and DATS-mediated gene expression changes.
Figure 1. RNA-seq revealed transcriptome alteration by DATS treatment in the MCF-10A human mammary epithelial cell line and the SK-BR-3 human breast cancer cell line.
(A) Volcano scatter plots from MCF-10A and SK-BR-3 cell lines displaying genes upregulated or downregulated by 20 µM DATS treatment for 16 hours. (B) Venn diagram showing common and unique genes affected by DATS in each cell line. (C) Heatmap diagram showing differentially expressed genes by DATS treatment in MCF-10A and SK-BR-3 cell lines. DATS, diallyl trisulfide.
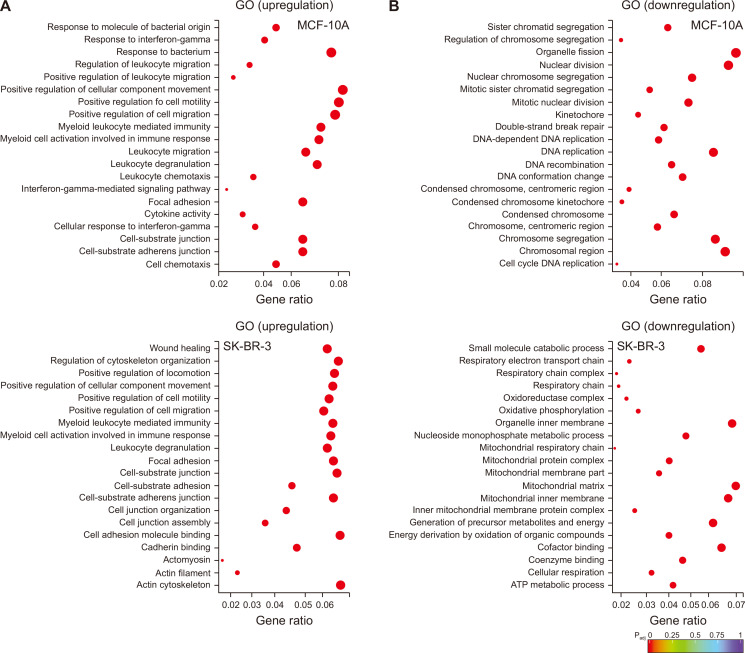
The RNA-seq data were analyzed by GO term analysis tool, which aims to identify biological processes, cellular locations, and molecular functions. The genes upregulated by DATS treatment in MCF-10A cells were associated with positive regulation of cell motility and migration, myeloid leukocyte mediated immunity, myeloid cell activation, leukocyte degranulation, migration, and chemotaxis, focal adhesion, cell-substrate adherens junction, and cell chemotaxis (Fig. 2A). Many of these terms were also similarly affected by DATS treatment in SK-BR-3 cells, including positive regulation of cell motility and migration, myeloid cell activation, leukocyte degranulation, focal adhesion, etc. (Fig. 2A). On the other hand, upregulation of genes associated with regulation of cytoskeleton organization and actin cytoskeleton was observed only in DATS-treated SK-BR-3 cells (Fig. 2A).
Figure 2. The GO term enrichment analysis for differentially expressed genes in the DATS-treated MCF-10A human mammary epithelial cell line and the SK-BR-3 human breast cancer cell line.
The GO term enrichment analysis of (A) upregulated or (B) downregulated genes by DATS treatment in MCF-10A (upper panel) and SK-BR-3 (lower panel) cell lines. DATS, diallyl trisulfide; GO, Gene Ontology.
The DATS-treatment resulted in downregulation of genes associated with sister chromatid segregation, organelle fission, nuclear division, nuclear chromosome segregation, DNA replication, centromeric region, etc. in MCF-10A cells (Fig. 2B). The GO terms downregulated by DATS treatment were quite distinct in the SK-BR-3 cell line when compared to MCF-10A cells and included small molecule catabolic process, organelle inner membrane, mitochondrial protein complex, mitochondrial matrix, mitochondrial inner membrane, energy derivation by oxidation, cofactor binding, coenzyme binding, cellular respiration, ATP metabolic process, etc. (Fig. 2B). These results indicate that DATS uniquely targets some pathways in SK-BR-3 cells.
DATS-induced disruption of F-actin cytoskeleton in SK-BR-3 cells
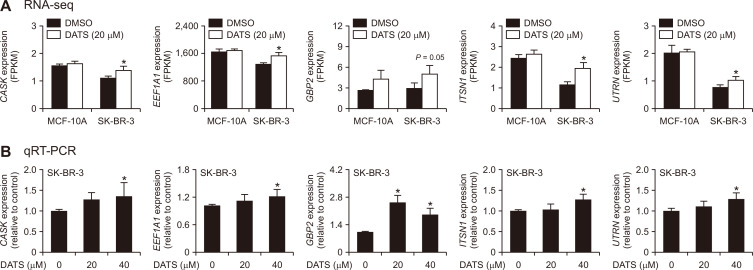
Table 1 lists DATS-mediated changes in genes associated with actin cytoskeleton in SK-BR-3 cells. We examined DATS-mediated changes in expression of several cytoskeleton-related genes from the RNA-seq data (Fig. 3A). The RNA-seq data showed significant upregulation of CASK, EEF1A1, GBP2, ITSN1, and UTRN expression upon DATS treatment in the SK-BR-3 cell line (Fig. 3A). None of these genes were significantly upregulated by DATS treatment in MCF-10A cells (data not shown). The qRT-PCR confirmed the RNA-seq data as every gene was significantly upregulated by DATS treatment in SK-BR-3 cells (Fig. 3B). However, additional work is necessary to determine their role in DATS-induced disruption of actin cytoskeleton.
Table 1.
A list of actin cytoskeleton-associated genes significantly (P ≤ 0.05) upregulated by DATS treatment in the SK-BR-3 cell line
| Gene name | MCF-10A | SK-BR-3 | Gene description | |||
|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | |||
| ACKR2 | 1.24 | 0.4100 | 1.48 | 0.0135 | Atypical chemokine receptor 2 | |
| ACTB | 1.26 | 0.2035 | 1.25 | 0.0306 | Actin beta | |
| ACTBL2 | 2.49 | 0.1142 | 3.11 | 0.0062 | Actin, beta like 2 | |
| ACTR10 | 1.16 | 0.1731 | 1.26 | 0.0002 | Actin related protein 10 homolog | |
| AFAP1L1 | 1.13 | 0.1995 | 2.98 | 0.0033 | Actin filament associated protein 1 like 1 | |
| ANLN | –1.45 | 0.0188 | 1.50 | 0.0040 | Anillin actin binding protein | |
| ARHGAP21 | 1.06 | 0.4300 | 1.50 | 0.0313 | Rho GTPase activating protein 21 | |
| ARPC2 | 1.20 | 0.2129 | 1.43 | 0.0047 | Actin related protein 2/3 complex subunit 2 | |
| ASAP1 | 1.08 | 0.3273 | 1.44 | 0.0261 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | |
| BCAR1 | 1.37 | 0.0851 | 1.33 | 0.0364 | BCAR1, Cas family scaffold protein | |
| BIN1 | –1.24 | 0.0375 | 1.57 | 0.0074 | Bridging integrator 1 | |
| BIN2 | Not detected | 4.54 | 0.0042 | Bridging integrator 2 | ||
| CAP2 | 1.05 | 0.3896 | 1.30 | 0.0029 | Cyclase associated actin cytoskeleton regulatory protein 2 | |
| CAPZA2 | 1.15 | 0.1962 | 1.26 | 0.0010 | Capping actin protein of muscle Z-line subunit alpha 2 | |
| CASK | 1.06 | 0.1780 | 1.29 | 0.0281 | Calcium/calmodulin dependent serine protein kinase | |
| CD2AP | 1.05 | 0.4486 | 1.34 | 0.0332 | CD2 associated protein | |
| CDC42BPA | 1.12 | 0.0983 | 1.32 | 0.0136 | CDC42 binding protein kinase alpha | |
| CFL2 | 1.12 | 0.3922 | 1.72 | 0.0019 | Cofilin 2 | |
| CORO1C | 1.10 | 0.2186 | 1.44 | 0.0099 | Coronin 1C | |
| COTL1 | 1.22 | 0.0791 | 1.65 | 0.0102 | Coactosin like F-actin binding protein 1 | |
| CRK | 1.11 | 0.0882 | 1.24 | 0.0046 | CRK proto-oncogene, adaptor protein | |
| CRYAB | 1.74 | 0.2224 | 3.39 | 0.0476 | Crystallin alpha B | |
| CTNNA1 | 1.17 | 0.0796 | 1.26 | 0.0251 | Catenin alpha 1 | |
| CTTN | 1.17 | 0.1458 | 1.22 | 0.0478 | Cortactin | |
| DBN1 | 1.30 | 0.0556 | 1.68 | 0.0127 | Drebrin 1 | |
| DCTN6 | 1.17 | 0.2016 | 1.37 | 0.0053 | Dynactin subunit 6 | |
| EEF1A1 | 1.02 | 0.4486 | 1.18 | 0.0101 | Eukaryotic translation elongation factor 1 alpha 1 | |
| EPB41L2 | –1.03 | 0.7086 | 10.17 | 0.0362 | Erythrocyte membrane protein band 4.1 like 2 | |
| FER | –1.08 | 0.0055 | 1.55 | 0.0080 | FER tyrosine kinase | |
| FLNA | 1.04 | 0.6271 | 1.39 | 0.0427 | Filamin A | |
| GAS2L3 | –1.30 | 0.2667 | 1.84 | 0.0060 | Growth arrest specific 2 like 3 | |
| GAS7 | 1.01 | 0.9813 | 4.36 | 0.0023 | Growth arrest specific 7 | |
| GBP2 | 1.63 | 0.0703 | 1.79 | 0.0502 | Guanylate binding protein 2 | |
| GMFB | 1.00 | 0.9908 | 1.35 | 0.0180 | Glia maturation factor beta | |
| ITSN1 | 1.08 | 0.2819 | 1.71 | 0.0070 | Intersectin 1 | |
| KLHL2 | –1.03 | 0.6052 | 1.88 | 0.0196 | Kelch like family member 2 | |
| LIMA1 | 1.38 | 0.0708 | 1.67 | 0.0167 | LIM domain and actin binding 1 | |
| LIMCH1 | 1.27 | 0.0641 | 1.30 | 0.0277 | LIM and calponin homology domains 1 | |
| MICAL1 | 1.43 | 0.0692 | 1.60 | 0.0105 | Microtubule associated monooxygenase, calponin and LIM domain containing 1 | |
| MYL12A | 1.38 | 0.2107 | 1.58 | 0.0002 | Myosin light chain 12A | |
| MYL12B | 1.32 | 0.2112 | 1.50 | 0.0052 | Myosin light chain 12B | |
| MYO10 | 1.13 | 0.1758 | 1.63 | 0.0263 | Myosin X | |
| MYO7B | 1.84 | 0.0956 | 2.33 | 0.0106 | Myosin VIIB | |
| MYOM1 | –1.17 | 0.7071 | 4.96 | 0.0243 | Myomesin 1 | |
| NCKAP1 | –1.02 | 0.4588 | 1.46 | 0.0005 | NCK associated protein 1 | |
| NDC1 | –1.44 | 0.0180 | 1.27 | 0.0013 | NDC1 transmembrane nucleoporin | |
| PAWR | –1.02 | 0.6527 | 1.33 | 0.0533 | Pro-apoptotic WT1 regulator | |
| PDCD6IP | 1.12 | 0.1263 | 1.32 | 0.0080 | Programmed cell death 6 interacting protein | |
| PEAK1 | –1.03 | 0.8576 | 1.52 | 0.0068 | Pseudopodium enriched atypical kinase 1 | |
| PLS1 | 1.05 | 0.4272 | 1.62 | 0.0407 | Plastin 1 | |
| PLS3 | 1.13 | 0.1160 | 1.56 | 0.0057 | Plastin 3 | |
| RDX | –1.12 | 0.0244 | 1.38 | 0.0146 | Radixin | |
| SEPT7 | 1.04 | 0.5335 | 1.33 | 0.0197 | Septin 7 | |
| SH2B2 | 1.37 | 0.1005 | 1.54 | 0.0090 | SH2B adaptor protein 2 | |
| SPECC1 | 1.04 | 0.5224 | 1.32 | 0.0092 | Sperm antigen with calponin homology and coiled-coil domains 1 | |
| SPTA1 | Not detected | 4.03 | 0.0117 | Spectrin alpha, erythrocytic 1 | ||
| TLN2 | 1.00 | 0.9898 | 1.95 | 0.0002 | Talin 2 | |
| TMOD1 | 1.16 | 0.7239 | 4.16 | 0.0002 | Tropomodulin 1 | |
| TPM1 | 1.28 | 0.4564 | 1.69 | 0.0016 | Tropomyosin 1 | |
| TPM3 | 1.14 | 0.3401 | 1.23 | 0.0051 | Tropomyosin 3 | |
| TPM4 | 1.29 | 0.1956 | 1.37 | 0.0163 | Tropomyosin 4 | |
| TWF1 | 1.04 | 0.5886 | 1.27 | 0.0408 | Twinfilin actin binding protein 1 | |
| UTRN | 1.03 | 0.7276 | 1.35 | 0.0499 | Utrophin | |
| WDR1 | 1.29 | 0.2219 | 1.37 | 0.0064 | WD repeat domain 1 | |
| YES1 | –1.01 | 0.7511 | 1.41 | 0.0069 | YES proto-oncogene 1, Src family tyrosine kinase | |
Figure 3. Effect of DATS treatment on actin cytoskeleton-associated genes.
(A) Expression of CASK, EEF1A1, GBP2, ITSN1, and UTRN in MCF-10A and SK-BR-3 cells after 16-hour treatment with DMSO (control) or 20 µM DATS. Results are shown as mean ± SD (n = 3) and statistical analysis was done by unpaired Student’s t-test. (B) qRT-PCR analysis of the genes shown in panel (A) in the SK-BR-3 cell line treated for 16 hours with DMSO (control) or the indicated doses of DATS. Combined results from two independent experiments are shown as mean ± SD (n = 6). Statistical analysis was done by one-way ANOVA followed by Dunnett’s test. qRT-PCR, quantitative real-time reverse transcription-PCR; SD, standard deviation; DMSO, dimethyl sulfoxide, DATS, diallyl trisulfide. *P < 0.05,
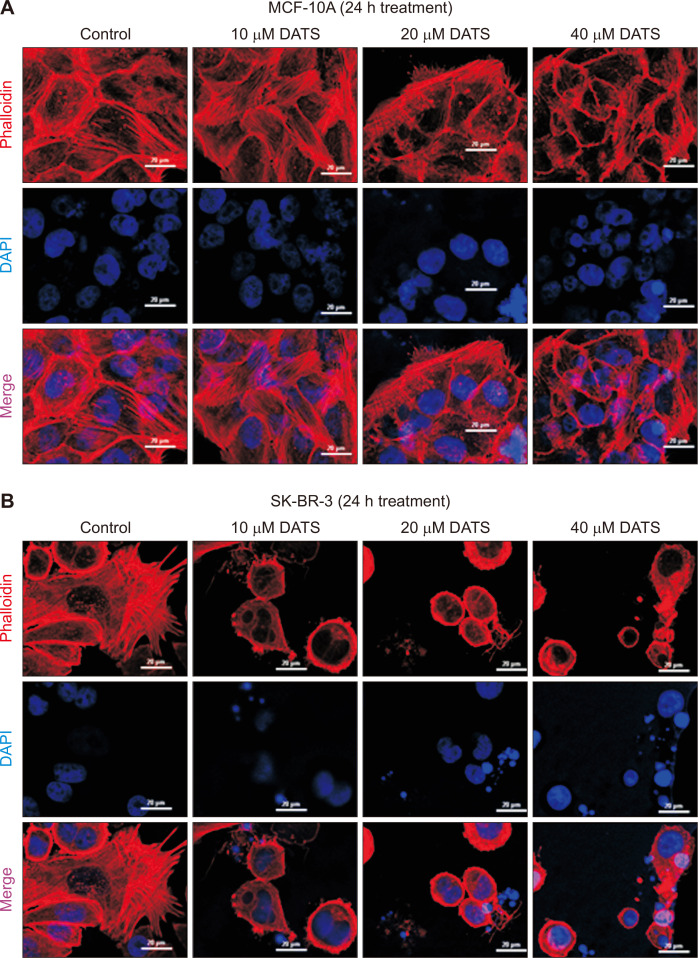
Next, we performed confocal microscopy to visualize F-actin cytoskeleton in MCF-10A (Fig. 4A) and SK-BR-3 cells (Fig. 4B) stained with phalloidin. The F-actin cytoskeleton was barely disturbed by DATS treatment in the MCF-10A cell line especially at higher concentrations (Fig. 4A). On the other hand, the DATS-treated SK-BR-3 cells exhibited severely impaired F-actin cytoskeleton.
Figure 4. DATS-caused disruption of actin cytoskeleton in the SK-BR-3 cell line.
Representative confocal microscopic images showing phalloidin-labeled actin filaments in MCF-10A (A) and SK-BR-3 (B) cell lines after 24 hours of treatment with DMSO (control) or indicated doses of DATS. Scale bar, 20 µm. ×40 oil objective magnification. DATS, diallyl trisulfide; DMSO, dimethyl sulfoxide; DAPI, 4’,6-diamidino-2-phenylindole.
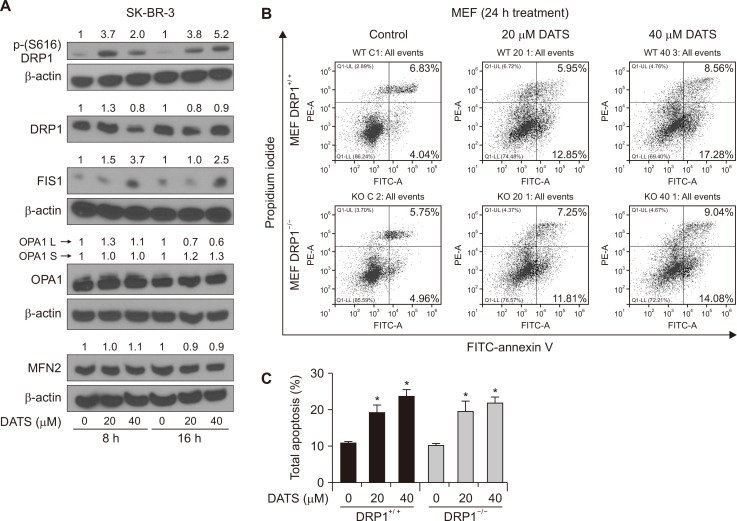
The effect of DATS treatment on proteins related to mitochondrial fission and fusion
Because DATS treatment downregulated mitochondria-related genes in SK-BR-3 cells but not in MCF-10A, we determined expression of proteins related to mitochondrial fission (DRP1 and FIS1) and fusion (OPA1 and MFN2). The level of DRP1, OPA1, and MFN2 was not affected by DATS treatment (Fig. 5A). On the other hand, DATS treatment caused an increase in phosphorylation of DRP1 and the protein level of FIS1 (Fig. 5A). The PINK1 kinases causes S616 phosphorylation of DRP1 to regulate mitophagy and mitochondrial dynamics [29]. We used MEFs from wild-type and DRP1 knockout mice to investigate its role in DATS-mediated apoptosis. Figure 5B shows flow histograms for control and DATS-treated MEFs. The DATS treatment increased apoptosis in both DRP1+/+ and DRP1–/– MEFs nearly equally (Fig. 5C). These results indicate that DRP1 protein is dispensable for DATS-induced apoptosis.
Figure 5. DATS treatment increased phosphorylation of DRP1 in the SK-BR-3 human breast cancer cell line.
(A) Western blotting for total DRP1, phospho-(S616)DRP1, OPA1, FIS1, MFN2, and β-Actin in SK-BR-3 cells after 8- or 16 hours of treatment with DMSO (control) or the indicated doses of DATS. Numbers on top of the immunoreactive bands represent changes in protein levels relative to corresponding DMSO-treated control. Results were comparable in replicate experiments. (B) Representative flow histograms depicting apoptotic fraction in DRP1+/+ and DRP1–/– MEFs after 24 hours of treatment with DMSO (control) or specified doses of DATS. (C) Percentage of total apoptotic fraction (early + late apoptotic cells) (mean ± SD; n = 3). Significant (*P < 0.05) compared with respective DMSO-treated control by one-way ANOVA followed by Bonferroni’s multiple comparisons. Experiments were repeated with comparable results. DRP1, dynamin-1-like; OPA1, optic atrophy protein 1; F1S1, fission, mitochondrial 1; FITC, fluorescein isothiocyanate; MFN2, mitofusin 2; MEFs, mouse embryonic fibroblasts; DATS, diallyl trisulfide; DMSO, dimethyl sulfoxide.
The effect of DATS treatment on expression of citric acid cycle proteins
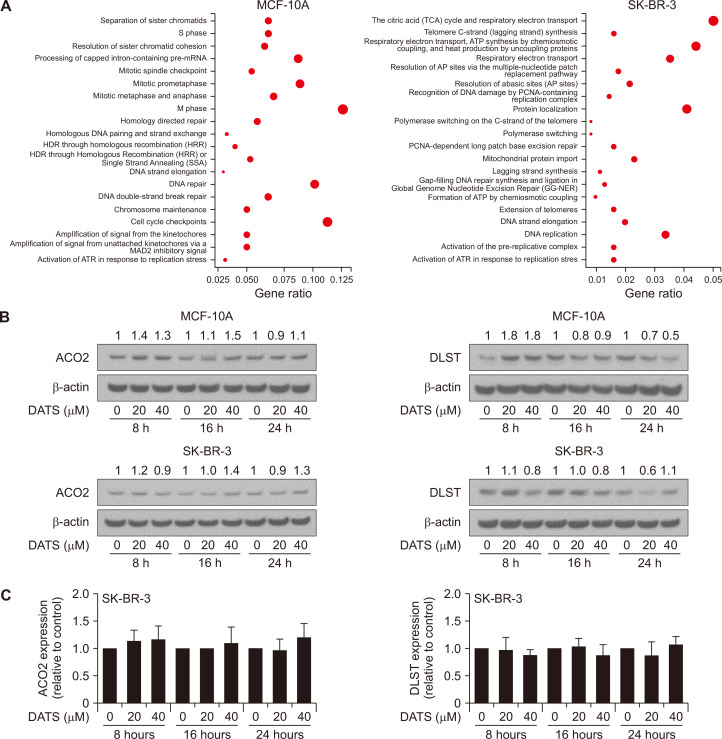
The Reactome pathway analysis of the RNA-seq data showed SK-BR-3-selective downregulation of citric acid cycle-associated genes after DATS treatment (Fig. 6A). However, expression of ACO2 or DLST proteins was not affected meaningfully by DATS treatment in either MCF-10A or SK-BR-3 cells (Fig. 6B and 6C). The citric acid cycle is mediated by other proteins including isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, succinyl-CoA synthetase, succinate dehydrogenase, fumrase, malate dehydrogenase, and citrate synthetase. Further work is necessary to determine the effect of DATS on expression of these enzyme proteins.
Figure 6. Effect of DATS on expression of tricarboxylic acid cycle-associated proteins.
(A) Reactome pathway enrichment analysis of downregulated genes by DATS treatment in MCF-10A and SK-BR-3 cell lines. (B) Immunoblot analysis for ACO2 and DLST in MCF-10A and SK-BR-3 cells treated with DMSO (control) or indicated doses of DATS for specified time points. The numbers above bands represent fold change in expression relative to corresponding DMSO-treated controls. (C) Quantification of ACO2 and DLST expression in SK-BR-3 cells. Results combined from three independent experiments are shown as mean ± SD (n = 3) and statistical analysis was done by one-way ANOVA followed by Dunnett’s test. DATS, diallyl trisulfide; DMSO, dimethyl sulfoxide, DLST, dihydrolipoamide S-succinyltransferase; ACO2, aconitase 2; HDR, Homology directed repair; ATR, ataxia telangiectasia and Rad3 related; AP, abasic sites; PCNA, proliferating cell nuclear antigen.
DISCUSSION
The RNA-seq analysis reveals common and unique pathways affected by DATS treatment in MCF-10A and SK-BR-3 cells. Determination of the relevance of DATS-induced modulation of genes involved in each pathway to the anti-cancer effect of this phytochemical is beyond the scope of this publication. However, some inferences can be drawn based on published literature. For example, DATS-mediated cell cycle arrest, apoptosis induction, and inhibition of cell migration are mediated by generation of ROS [13,14,17]. The DATS-induced apoptosis in breast cancer cells is significantly attenuated by overexpression of copper, zinc-superoxide dismutase and manganese-superoxide [14]. However, the precise mechanism by which ROS are generated after DATS treatment is not known. The mitochondrial electron transport chain is one of the sources of ROS generation. Inhibition of complex I and complex III leads to ROS generation. Notably, the GO analysis of the RNA-seq data revealed downregulation of genes associated with respiratory electron transport chain in DATS-treated SK-BR-3 cells but not in MCF-10A cells (Fig. 2B). In this context, the RNA-seq data showed statistically significant decrease in expression of complex I subunit genes NDUFA10, NDUFA11, NDUFB1, NDUFS3, and NDUFV3 in only DATS-treated SK-BR-3 cells (data not shown). It is possible that DATS treatment inhibits complex I leading to ROS generation. However, further work is necessary to explore this possibility.
Most conspicuous effect of DATS treatment observed in this study was disruption of the F-actin cytoskeleton. A complete collapse of F-actin cytoskeleton was observed in DATS-treated SK-BR-3 cells (Fig. 4B). On the other hand, F-actin cytoskeleton was maintained in DATS-treated MCF-10A cells (Fig. 4A). Actin cytoskeleton plays a key role in motility of cancer cells [30]. The three isotypes (α, β, and γ) of actin polymerize to form actin filaments that are thin and flexible fibers [30]. The actin filaments are organized into bundles or three-dimensional networks in cancer cells leading to formation of lamellipodia, filopodia, and invadopodia that facilitate cancer cell migration [30]. Cell migration is inhibited by DATS treatment in breast cancer cells [14]. It is possible that disruption of the F-actin cytoskeleton is responsible for DATS-mediated inhibition of breast cancer cell migration.
Even though increased aerobic glycolysis (Warburg effect) is the widely studied metabolic abnormality in other solid tumors, evidence exists for deregulation of citric acid cycle in breast cancer [31]. For example, patients with estrogen receptor+, progesterone receptor+, and HER2+ breast cancers harbor mutations in isocitrate dehydrogenase 1 that is accompanied by increased circulating levels of 2-hydroxyglutarate [32]. The expression of succinate dehydrogenase was shown to depend on the breast cancer subtype and the metastatic phenotype [33]. The expression of pyruvate carboxylase, which converts pyruvate to oxaloacetate, was higher in breast cancer and was found to be essential for growth and invasion of MDA-MB-231 cells [34]. The DATS-mediated inhibition of citric acid cycle may partly be responsible for its inhibitory effect on growth and invasion of breast cancer cells.
The DRP1-mediated mitochondrial fission is responsible for apoptosis induction by a heptamethine cyanine dye [35]. The present study reveals that DATS-mediated apoptosis is not affected by DRP1 protein at least in the SK-BR-3 cell line. We also found an increase in the protein level of FIS1 in SK-BR-3 cell line after DATS treatment. An exact role of FIS1 in apoptosis regulation is unclear [36]. Further work is necessary to determine potential involvement of FIS1 in DATS-induced apoptosis.
In conclusion, the present study identifies common and unique pathways affected by DATS treatment in MCF-10A and SK-BR-3 cells. Further work is required to determine the significance of gene expression changes in anti-cancer mechanisms of DATS, but selective disruption of F-actin cytoskeleton may partly explain the inhibitory effect of DATS treatment on breast cancer cell migration. We also propose that DATS treatment may inhibit complex I in SK-BR-3 cells to generate ROS that is responsible for apoptosis induction [13,14].
Footnotes
FUNDING
This study was supported by the National Cancer Institute at the National Institutes of Health grant (No. R01 CA219180) (to SVS). This study used the UPMC Hillman Cancer Center Flow Cytometry Facility supported by the National Cancer Institute at the National Institutes of Health grant (No. P30 CA047904).
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Cazzaniga M, Bonanni B. Breast cancer chemoprevention: old and new approaches. J Biomed Biotechnol. 2012;2012:985620. doi: 10.1155/2012/985620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel project P-1 study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 4.Goss PE, Ingle JN, Ales-Martinez JE, Cheung AM, Chlebowski RT, Wactawski-Wende J, et al. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med. 2011;364:2381–91. doi: 10.1056/NEJMoa1103507. Erratum in: N Engl J Med 2011;365:1361. [DOI] [PubMed] [Google Scholar]
- 5.Kotecha R, Takami A, Espinoza JL. Dietary phytochemicals and cancer chemoprevention: a review of the clinical evidence. Oncotarget. 2016;7:52517–29. doi: 10.18632/oncotarget.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fimognari C, Lenzi M, Hrelia P. Chemoprevention of cancer by isothiocyanates and anthocyanins: mechanisms of action and structure-activity relationship. Curr Med Chem. 2008;15:440–7. doi: 10.2174/092986708783503168. [DOI] [PubMed] [Google Scholar]
- 7.Shukla Y, Kalra N. Cancer chemoprevention with garlic and its constituents. Cancer Lett. 2007;247:167–81. doi: 10.1016/j.canlet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Alumkal JJ, Slottke R, Schwartzman J, Cherala G, Munar M, Graff JN, et al. A phase II study of sulforaphane-rich broccoli sprout extracts in men with recurrent prostate cancer. Invest New Drugs. 2015;33:480–9. doi: 10.1007/s10637-014-0189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Li HQ, Wang Y, Xu HX, Fan WT, Wang ML, et al. An intervention study to prevent gastric cancer by micro-selenium and large dose of allitridum. Chin Med J (Engl) 2004;117:1155–60. [PubMed] [Google Scholar]
- 10.Fleischauer AT, Arab L. Garlic and cancer: a critical review of the epidemiologic literature. J Nutr. 2001;131:1032S–40S. doi: 10.1093/jn/131.3.1032S. [DOI] [PubMed] [Google Scholar]
- 11.Xiao D, Choi S, Johnson DE, Vogel VG, Johnson CS, Trump DL, et al. Diallyl trisulfide-induced apoptosis in human prostate cancer cells involves c-Jun N-terminal kinase and extracellular-signal regulated kinase-mediated phosphorylation of Bcl-2. Oncogene. 2004;23:5594–606. doi: 10.1038/sj.onc.1207747. [DOI] [PubMed] [Google Scholar]
- 12.Malki A, El-Saadani M, Sultan AS. Garlic constituent diallyl trisulfide induced apoptosis in MCF7 human breast cancer cells. Cancer Biol Ther. 2009;8:2175–85. doi: 10.4161/cbt.8.22.9882. [DOI] [PubMed] [Google Scholar]
- 13.Na HK, Kim EH, Choi MA, Park JM, Kim DH, Surh YJ. Diallyl trisulfide induces apoptosis in human breast cancer cells through ROS-mediated activation of JNK and AP-1. Biochem Pharmacol. 2012;84:1241–50. doi: 10.1016/j.bcp.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Chandra-Kuntal K, Lee J, Singh SV. Critical role for reactive oxygen species in apoptosis induction and cell migration inhibition by diallyl trisulfide, a cancer chemopreventive component of garlic. Breast Cancer Res Treat. 2013;138:69–79. doi: 10.1007/s10549-013-2440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahm ER, Kim SH, Mathan SV, Singh RP, Singh SV. Mechanistic targets of diallyl trisulfide in human breast cancer cells identified by RNA-seq analysis. J Cancer Prev. 2021;26:128–36. doi: 10.15430/JCP.2021.26.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman-Antosiewicz A, Singh SV. Checkpoint kinase 1 regulates diallyl trisulfide-induced mitotic arrest in human prostate cancer cells. J Biol Chem. 2005;280:28519–28. doi: 10.1074/jbc.M501443200. [DOI] [PubMed] [Google Scholar]
- 17.Xiao D, Herman-Antosiewicz A, Antosiewicz J, Xiao H, Brisson M, Lazo JS, et al. Diallyl trisulfide-induced G2-M phase cell cycle arrest in human prostate cancer cells is caused by reactive oxygen species-dependent destruction and hyperphosphorylation of Cdc 25 C. Oncogene. 2005;24:6256–68. doi: 10.1038/sj.onc.1208759. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Kaschula CH, Priedigkeit N, Lee AV, Singh SV. Forkhead box Q1 is a novel target of breast cancer stem cell inhibition by diallyl trisulfide. J Biol Chem. 2016;291:13495–508. doi: 10.1074/jbc.M116.715219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Meng Y, Xie C, Zhu J, Wang X, Li Y, et al. Diallyl trisulfide inhibits breast cancer stem cells via suppression of Wnt/β-catenin pathway. J Cell Biochem. 2018;119:4134–41. doi: 10.1002/jcb.26613. [DOI] [PubMed] [Google Scholar]
- 20.Hahm ER, Singh SV. Diallyl trisulfide inhibits estrogen receptor-α activity in human breast cancer cells. Breast Cancer Res Treat. 2014;144:47–57. doi: 10.1007/s10549-014-2841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Zhu P, Wang Y, Wei Z, Tao L, Zhu Z, et al. Antimetastatic therapies of the polysulfide diallyl trisulfide against triple-negative breast cancer (TNBC) via suppressing MMP2/9 by blocking NF-κB and ERK/MAPK signaling pathways. PLoS One. 2015;10:e0123781. doi: 10.1371/journal.pone.0123781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Z, Shan Y, Tao L, Liu Y, Zhu Z, Liu Z, et al. Diallyl trisulfides, a natural histone deacetylase inhibitor, attenuate HIF-1α synthesis, and decreases breast cancer metastasis. Mol Carcinog. 2017;56:2317–31. doi: 10.1002/mc.22686. [DOI] [PubMed] [Google Scholar]
- 23.Kim SH, Hahm ER, Singh KB, Singh SV. Diallyl trisulfide inhibits leptin-induced oncogenic signaling in human breast cancer cells but fails to prevent chemically-induced luminal-type cancer in rats. J Cancer Prev. 2020;25:1–12. doi: 10.15430/JCP.2020.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsherbiny NM, El-Sherbiny M, Zaitone SA. Diallyl trisulfide potentiates chemotherapeutic efficacy of doxorubicin in experimentally induced mammary carcinoma: role of Notch signaling. Pathol Res Pract. 2020;216:153139. doi: 10.1016/j.prp.2020.153139. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, Hahm ER, Singh KB, Shiva S, Stewart-Ornstein J, Singh SV. RNA-seq reveals novel mechanistic targets of withaferin A in prostate cancer cells. Carcinogenesis. 2020;41:778–89. doi: 10.1093/carcin/bgaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Xiao D, Srivastava SK, Lew KL, Zeng Y, Hershberger P, Johnson CS, et al. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis. 2003;24:891–7. doi: 10.1093/carcin/bgg023. [DOI] [PubMed] [Google Scholar]
- 28.Hahm ER, Kim SH, Singh KB, Singh SV. RNA-seq reveals novel cancer-selective and disease subtype-independent mechanistic targets of withaferin A in human breast cancer cells. Mol Carcinog. 2021;60:3–14. doi: 10.1002/mc.23266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han H, Tan J, Wang R, Wan H, He Y, Yan X, et al. PINK1 phosphorylates Drp1S616 to regulate mitophagy-independent mitochondrial dynamics. EMBO Rep. 2020;21:e48686. doi: 10.15252/embr.201948686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer Lett. 2009;284:122–30. doi: 10.1016/j.canlet.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Harrelson JP, Lee MW. Expanding the view of breast cancer metabolism: promising molecular targets and therapeutic opportunities. Pharmacol Ther. 2016;167:60–73. doi: 10.1016/j.pharmthera.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Fathi AT, Sadrzadeh H, Comander AH, Higgins MJ, Bardia A, Perry A, et al. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. Oncologist. 2014;19:602–7. doi: 10.1634/theoncologist.2013-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HM, Jung WH, Koo JS. Site-specific metabolic phenotypes in metastatic breast cancer. J Transl Med. 2014;12:354. doi: 10.1186/s12967-014-0354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phannasil P, Thuwajit C, Warnnissorn M, Wallace JC, MacDonald MJ, Jitrapakdee S. Pyruvate carboxylase is up-regulated in breast cancer and essential to support growth and invasion of MDA-MB-231 cells. PLoS One. 2015;10:e0129848. doi: 10.1371/journal.pone.0129848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Q, Liu W, Zhang Q, Huang J, Hu C, Liu Y, et al. Dynamin-related protein 1-mediated mitochondrial fission contributes to IR-783-induced apoptosis in human breast cancer cells. J Cell Mol Med. 2018;22:4474–85. doi: 10.1111/jcmm.13749. Erratum in: J Cell Mol Med 2022;26:3309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ihenacho UK, Meacham KA, Harwig MC, Widlansky ME, Hill RB. Mitochondrial fission protein 1: emerging roles in organellar form and function in health and disease. Front Endocrinol (Lausanne) 2021;12:660095. doi: 10.3389/fendo.2021.660095.7f3518f105834c6e875afa31df4302b2 [DOI] [PMC free article] [PubMed] [Google Scholar]