Abstract
We report the first case of a primary renal undifferentiated sarcoma harboring an SS18::POU5F1 gene fusion. The patient was a 38 year‐old male diagnosed with a 5 cm renal tumor which invaded the adrenal gland and extended into the renal vein. Microscopically, the neoplasm had a predominantly undifferentiated round cell morphology, with areas of rhabdoid and spindle cell growth. Similar to the previously reported cases with this fusion, by immunohistochemistry the neoplasm expressed S100 protein and epithelial markers (diffuse EMA, focal cytokeratin), suggesting the possibility of a myoepithelial phenotype. This report documents another example of a fusion‐positive undifferentiated soft tissue sarcoma occurring as a primary renal neoplasm, adding to the already broad list of such entities. It highlights the crucial role of molecular analysis in establishing a specific diagnosis given the overlapping morphology and immunophenotypes such entities may exhibit.
Keywords: renal, sarcoma, SS18::POU5F1
1. INTRODUCTION
The differential diagnosis of undifferentiated small round cell tumors of the kidney in children and young adults is broad. In patients under the age of 10, considerations revolve around the most common pediatric renal neoplasm, Wilms tumor (nephroblastoma), as well as entities such as clear cell sarcoma of the kidney, rhabdoid tumor, cellular congenital mesoblastic nephroma, intrarenal neuroblastoma, and malignant lymphoma. 1 , 2 , 3 , 4 , 5 In older children and young adults, more common entities in the differential include Ewing sarcoma family of tumors and synovial sarcoma. 6 , 7 , 8 In some cases, molecular pathology is absolutely essential for the diagnosis, as the immunohistochemical profiles of entities may overlap. For example, both Ewing sarcoma and poorly differentiated synovial sarcoma label diffusely for CD99, while CIC::DUX4 sarcomas of the kidney label for WT1 which could lead to their misdiagnosis as Wilms tumor. 9
We report a primary undifferentiated malignant neoplasm of the kidney which demonstrated an SS18::POU5F1 gene fusion that has recently been described in only four previously reported cases, all outside of the kidney. 10 , 11 , 12 The morphology and immunohistochemical profile in the setting of a mass centered on the kidney lead to a broad differential diagnosis.
2. METHODS
2.1. IRB approval
This study was approved by the Institutional Review Boards at our institutions.
2.1.1. Immunohistochemistry
Immunohistochemistry for HMB45, cathepsin k, cytokeratins AE1/3 and Cam5.2, epithelial membrane antigen (EMA), PAX8, S100 protein, melan A, desmin, smooth muscle actin, TFE1, OCT3/4, and TFE3 were performed as previously described. 13 , 14
2.1.2. DNA and RNA sequencing
DNA and RNA sequencing were performed by the Genomics and Molecular Pathology Core at UT Southwestern Medical Center. 15 Briefly, tumor hematoxylin and eosin slides were examined and marked by two pathologists (P.A. and D.N.P.) for subsequent macro‐dissection, nucleic acid isolation, and molecular testing. Areas enriched with tumor were then scraped from adjacent 5 μm thick formalin‐fixed paraffin embedded (FFPE) sections. Extraction and purification were performed using Qiagen Allprep kits (Qiagen, Germantown, MD). Libraries were prepared using KAPA Hyperplus kits (Roche Sequencing and Life Science Kapa Biosystems, Wilmington, MA) with genomic regions of interest captured by custom probes covering all exons of 1516 cancer‐related genes. The libraries were sequenced using Sequencing by Synthesis (SBS) paired‐end cluster generation on the Illumina NextSeq 550 platform (Illumina Inc., San Diego, CA. DNA Sequence reads were aligned to reference genome GRCh38. Single nucleotide variants (SNVs), insertions and deletions were called using Strelka2, MuTect2, Freebayes and Platypus. Copy number alterations were called using CNVKit. Fusions were called using the STAR‐Fusion algorithm with the RNASeq reads (https://github.com/bcantarel/school). All fusions and variant calls were classified according to the Association for Molecular Pathology/American Society for Clinical Oncology/College of American Pathologists guidelines 16 and manually inspected using Integrated Genomics Viewer version 2.3.4 (IGV; Broad Institute, MIT Harvard, Cambridge, MA).
2.2. Fluorescence in situ hybridization
Fluorescence in Situ Hybridization (FISH) on interphase nuclei from paraffin‐embedded 4‐micron sections was performed using the Abbott Molecular Vysis LSI SS18 Break Apart FISH Probe Kit, covering and flanking the SS18 gene (https://www.molecularcatalog.abbott/int/en/Vysis‐LSI‐SS18‐Dual‐Color‐Break‐Apart‐Probe‐Kit). The first 650 kb probe labeled in SpectrumOrange is telomeric to the SS18 gene. The second probe labeled in SpectrumGreen is 1044 kb is centromeric to the SS18 gene. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a split‐apart signal. Nuclei with incomplete set of signals were omitted from the score. 17
3. RESULTS
3.1. Gross appearance and morphology
The patient was a 38 year old male who presented with a renal mass and underwent radical nephrectomy. On gross examination, he was found to have a poorly delineated, tan 5 cm renal neoplasm which grossly invaded through the renal capsule into the adrenal gland and extended through the renal sinus to the renal vein. Microscopically, the neoplasm demonstrated sheet‐like growth, engulfing native glomeruli and tubules similar to the aggressive intrarenal growth pattern of lymphoma or small cell carcinoma. There was extensive peritheliomatous necrosis. Microscopically, the neoplasm involved the renal sinus fat and blood vessels, extended into the renal vein to involve the renal vein margin, and invaded perirenal fat and the adrenal gland (Figure 1). The neoplastic cells were predominantly of an undifferentiated small round cell phenotype, associated with high mitotic activity (>5 mitoses per high power field). In some areas, the neoplasm demonstrated more epithelioid morphology, with eosinophilic cytoplasm and eccentric round nuclei with open chromatin and prominent nucleoli, creating a rhabdoid appearance. In other areas, the neoplastic cells showed a nested architecture (such as in their invasion of the adrenal), while in focal areas (approximately 5%) the neoplastic cells were spindled with eosinophilic cytoplasmic tails, raising the possibility of rhabdomyoblastic differentiation. By immunohistochemistry, the neoplastic cells were diffusely immunoreactive for PAX8 and EMA. S100 protein and TLE1 showed multifocal patchy immunoreactivity. There was focal staining for pan‐cytokeratin AE1/3 (Figure 1). Markers that were negative included WT1, desmin, smooth muscle actin, GATA3, cytokeratin 7, cytokeratin 20, OCT3/4, CD99, and NKX2.2. INI1 and BRG1 expressions were retained.
FIGURE 1.

The neoplasm is a highly permeative undifferentiated cancer which extends between native glomeruli and tubules, similar to the pattern of intrarenal lymphoma (A). In the direct extension into the adrenal gland (right), the neoplastic cells are somewhat more cohesive and demonstrate more rhabdoid cytologic features, such as vesicular chromatin, prominent nucleoli, and eccentric cytoplasm (B). At higher power, the neoplasm mainly has an undifferentiated small round blue cell appearance, with minimal cytoplasmic differentiation. The neoplasm demonstrates peritheliomatous growth around capillaries with necrosis highlighted at the upper left (C). Mitotic figures are readily identifiable. Focal areas of this neoplasm demonstrate striking rhabdoid cytologic features, including vesicular chromatin, prominent nucleoli, and hyaline cytoplasmic inclusions (D). Other foci demonstrate transitions to spindle growth with eosinophilic cytoplasmic extensions which raised the possibility of rhabdomyoblastic differentiation (E). The neoplastic cells demonstrate diffuse strong immunoactivity for PAX8 (F) and for EMA (G), along with patchy immunoreactivity for S100 protein (H).
3.2. Molecular studies
RNA‐seq revealed a fusion transcript composed of exon 10 of SS18 and exon 5 of POU5F1 (Figure 2). The SS18 gene rearrangement was confirmed by break apart FISH (Figure 3).
FIGURE 2.
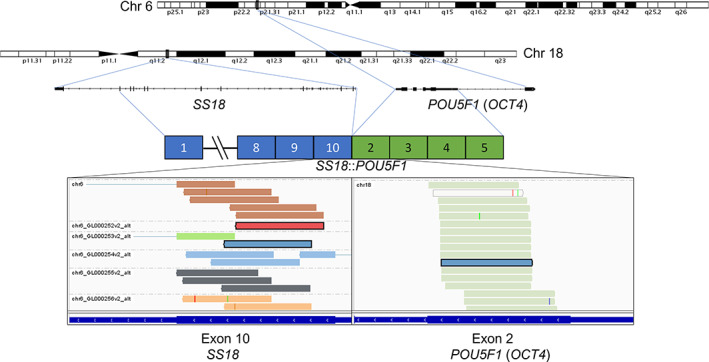
SS18::POU5F1 gene fusion. Schematic representation of interchromosomal fusion SS18::POU5F1. Gene loci of SS18 and POU5F1, also known as OCT4, are shown within chromosomes 18 and 6, respectively. RNA‐seq revealed fusion transcripts consisting of exon 10 of SS18 (chr 18q11.2; ENST00000415083.7) and exon 5 of POU5F1 (chr 6p21.33; ENST00000259915.13). Displayed are the discordant reads grouped by mate chromosome (indicated in the top left corner of the IGV screenshots).
FIGURE 3.
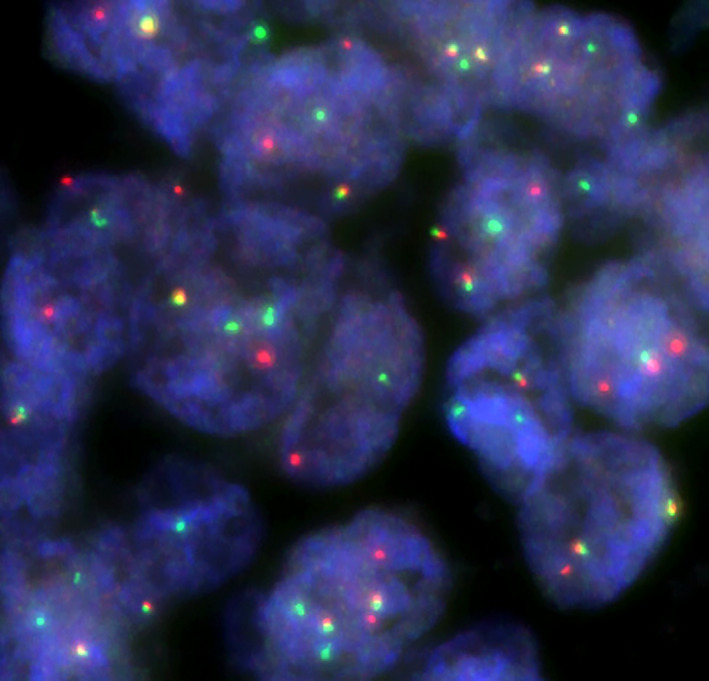
SS18 Break Apart FISH. The cells demonstrate splitting of the 5′ SS18 probe (red) and the 3′ SS18 probe (green), yielding a complex abnormal signal pattern.
4. DISCUSSION
Only four prior cases of undifferentiated malignant neoplasms with the SS18::POU5F1 gene fusion have been reported 10 , 11 , 12 (Table 1). While we have no follow up on our case, two of three previously reported cases have developed systemic metastases and died of disease. Including our case, in all except one, the fusion transcript involved exon 10 of SS18 fused to exon 2 of POU5F1. Liu et al reported an individual case with SS18 exon 9 fused to POU5F1 exon 2. 12 These neoplasms have all been described previously as having small round cell to epithelioid morphology. By immunohistochemistry, all five cases demonstrated at least focal labeling for EMA and S100 protein, which would suggest a myoepithelial line of differentiation (Table 1). Focal cytokeratin labeling has also been seen. Stains for WT1, muscle markers, and melanocytic markers have consistently been negative, and markers of undifferentiated carcinoma (INI1, BRG1) have been intact in all the cases tested.
TABLE 1.
SS18::POU5F1 fusion cases in the literature
| Case | Age/sex | Clinical | Outcome | Morphology | IHC+ | IHC‐ | Exons fused |
|---|---|---|---|---|---|---|---|
| Antonescu 2020 Case #1 10 | 18/M | 4 cm pancreatic mass with liver metastases | DOD 31 months (systemic metastases) | Small round cell/epithelioid | EMA (diffuse); AE1/3, Cam5.2, S100, TLE1, CD99 (all focal), synaptophysin (rare). Ki67 90% | P40, p63, S100, SOX10, HMB45, desmin, WT‐1, GATA3, chromogranin, hCG, INSM‐1, hPL, trypsin, chymotrypsin, PR, OCT4, CD30, SALL4. | SS18 exon 10‐POU5F1 exon 2 |
| Antonescu 2020 Case #2 10 | 29/F | 2.5 cm subcutaneous groin mass thought to be cyst | NED 10 months with adjuvant chemotherapy | Small round cell/epithelioid | S100 (focal), EMA (weak focal) | Pan Cytokeratin, CK7, CK20, CK8/18, SOX10, HMB45, melan A, WT1, CD117, CD99, TLE1, STAT6, BCOR, desmin, myogenin, synaptophysin, TTF1, PLAP, HCG, ER, CD3, CD20, CD30, CD31, CD34, CD43, MUM1, MPO, TdT. INI1, BRG1, BRM1 retained. | SS18 exon 10‐POU5F1 exon 2 |
| Shenoy 2020 11 | 11/F | 5 cm back mass with femoral lymph node metastases | Not known | Small round cell/epithelioid | AE1/3, EMA, S100 | CD99, desmin, myogenin, WT‐1, ERG, CD45, SOX10, HMB45, p63, GFAP, calponin, TLE1. INI1 retained. | SS18 exon 10‐POU5F1 exon 2 |
| Liu 2021 12 | 18/F | 5 cm groin | DOD 8 months (systemic metastases) | Small round cell/epithelioid | Pan Keratin, EMA, CD31, BCl2, vimentin, CD99, S100 (focal) | CD34, HMB45, MyoD1, myogenin, ERG, Factor 8. | SS18 exon 9‐POU5F1 exon 2 |
| Current case | 38/M | 5 cm renal mass with renal vein and adrenal metastasis (pT4) | Not known | Small round cell/epithelioid/ rhabdoid/spindle | EMA, PAX8; S100, TLE1 (patchy) AE1/3 (focal) | GATA3, Ck7, CK20, WT1, desmin, actin, CD99, NKX2.2, Oct3/4. INI1, BRG1 intact. | SS18 exon 10‐POU5F1 exon 2 |
The predominantly undifferentiated morphology, along with focal areas of rhabdoid and spindle cell growth, raised multiple diagnostic pitfalls in the context of renal neoplasia. First, the diffuse PAX8 and EMA labeling, along with the focal AE1/3 labeling, could be considered sufficient for the diagnosis of an undifferentiated renal cell carcinoma, given WT‐1, GATA3, and cytokeratin 7 negativity which would argue against nephroblastoma or urothelial origin. It should be noted that PAX8 immunoreactivity is, in the setting of small round cell tumors, not specific for renal cell carcinoma, as it may be seen in sarcomas such as clear cell sarcoma of the kidney or rarely in primary renal synovial sarcoma. 18 Undifferentiated renal cell carcinomas often show rhabdoid morphology and demonstrate loss of INI1 or BRG1 (SMARCB4 and SMARCA4); however, both proteins were intact in our neoplasm. 19 The undifferentiated appearance, along with the focal areas of eosinophilic cytoplasm that suggested rhabdomyoblastic differentiation, raised the possibility of blastemal‐predominant Wilms tumor with rhabdomyoblastic differentiation. However, the absence of WT1 immunoreactivity or immunohistochemical evidence of skeletal muscle differentiation (desmin or myogenin) helped exclude that possibility. Renal Ewing sarcoma is likely more common than blastemal Wilms tumor in our patient's age group; however, the absence of CD99 or NKX2.2 immunoreactivity excluded this possibility. Poorly differentiated synovial sarcoma also was a significant concern, given the focal spindle cell growth and rhabdoid features, both of which are well described in that entity. In fact, the rearrangement of SS18 in the current neoplasm could further mislead if only break‐apart FISH for SS18 were performed on this case, as could the patchy TLE1 immunohistochemical labeling. It should be noted that the strong diffuse EMA staining and lack of CD99 immunoreactivity are unusual for a diagnosis of poorly differentiated synovial sarcoma.
As a result of the wide‐spread application of RNA sequencing in clinical practice, an increasing number of undifferentiated neoplasms have been molecularly characterized, which has exposed gray zones in defining tumor entities based on morphology or genomic signatures. One such example is the neoplasms at the crossroads of malignant myoepithelial tumors and Ewing sarcoma. 20 By current criteria, our case and those previously reported with the SS18‐POU5F1 gene fusion could be considered in the spectrum of high grade myoepithelial carcinomas with a round cell/undifferentiated phenotype given the co‐expression of S100 and epithelial markers (the currently used gold standard for myoepithelial differentiation), and involvement of POU5F1‐related fusions (POU5F1 is known to be fused to EWS in myoepithelial carcinomas 17 ). Along these lines, the original study by Antonescu et al. demonstrated clustering of the sarcomas with the SS18::POU5F1 gene fusion with three myoepithelial tumors with EWSR1::POU5F1 or FUS::POU5F1 fusions, but not with two synovial sarcomas with SS18 rearrangements. However, the SS18::POU5F1 sarcomas did not cluster with other myoepithelial neoplasms with non‐POU5F1 fusions, such as EWSR1::ZNF44 or FUS::KLF17. Hence, whether the SS18‐POU5F1 neoplasms represent primitive sarcomas or are more closely related to high grade myoepithelial carcinomas remains unclear, especially given the known imprecision of morphologic criteria for the latter. Further studies are need to address this issue.
AUTHOR CONTRIBUTIONS
Pedram Argani, Doreen Palsgrove, Jeffrey A. Gagan, Andres Matoso, and Cristina R. Antonescu conceived the study, Pedram Argani and John M. Gross performed data analysis, Pedram Argani wrote the manuscript. Doreen Palsgrove, Jeffrey SoRelle, and Jeffrey Gagan performed and interpreted RNA sequencing. Cristina R. Antonescu and Yanming Zhang performed and interpreted fluorescence in situ hybridization.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We thank Norman Barker MA, MS, for expert photographic assistance. Supported in part by: P50 CA217694 (Cristina R. Antonescu), P50 CA 140146‐01 (Cristina R. Antonescu), P30 CA 008748 (Cristina R. Antonescu); Cycle for Survival (Cristina R. Antonescu), Kristin Ann Carr Foundation (Cristina R. Antonescu), Dahan Translocation Carcinoma Fund and Joey's Wings (Pedram Argani).
Argani P, Matoso A, Gross JM, et al. Primary renal sarcoma with SS18::POU5F1 gene fusion. Genes Chromosomes Cancer. 2022;61(9):572‐577. doi: 10.1002/gcc.23053
Funding information Dahan Translocation Carcinoma Fund and Joey's Wings; Kristin Ann Carr Foundation, Grant/Award Numbers: P30 CA 008748, P50 CA 140146‐01, P50 CA217694
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Knezevich SR, Garnett MJ, Pysher TJ, et al. ETV6‐NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58:5046‐5048. [PubMed] [Google Scholar]
- 2. Rubin BP, Chen CJ, Morgan TW, et al. Congenital mesoblastic nephroma t(12;15) is associated with ETV6‐NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998;153:1451‐1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Argani P, Fritsch M, Kadkol SS, et al. Detection of the ETV6‐NTRK3 chimeric RNA of infantile fibrosarcoma/cellular congenital mesoblastic nephroma in paraffin‐embedded tissue: application to challenging pediatric renal stromal tumors. Mod Pathol. 2000;13:29‐36. [DOI] [PubMed] [Google Scholar]
- 4. Fan R. Primary renal neuroblastoma: a clinical pathologic study of 8 cases. Am J Surg Pathol. 2012;36:94‐100. [DOI] [PubMed] [Google Scholar]
- 5. Dhull VS, Mukherjee A, Karunanithi S, Durgapal P, Bal C, Kumar R. Bilateral primary renal lymphoma in a pediatric patient: staging and response evaluation with 18F‐FDG PET/CT. Rev Esp Med Nucl Imagen Mol. 2015;34:49‐52. [DOI] [PubMed] [Google Scholar]
- 6. Quezado M, Benjamin DR, Tsokos M. EWS/FLI‐1 fusion transcripts in three peripheral primitive neuroectodermal tumors of the kidney. Hum Pathol. 1997;28:767‐771. [DOI] [PubMed] [Google Scholar]
- 7. Jimenez RE, Folpe AL, Lapham RL, et al. Primary Ewing's sarcoma/primitive neuroectodermal tumor of the kidney. A clinicopathologic and immunohistochemical analysis of 11 cases. Am J Surg Pathol. 2002;26:320‐327. [DOI] [PubMed] [Google Scholar]
- 8. Argani P, Faria PA, Epstein JI, et al. Primary renal synovial sarcoma. Morphologic and molecular delineation of an entity previously included among embryonal sarcomas of the kidney. Am J Surg Pathol. 2000;24:1087‐1096. [DOI] [PubMed] [Google Scholar]
- 9. Mangray S, Kelly DR, LeGuellec S, et al. Clinicopathologic features of a series of primary renal CIC‐rearranged sarcomas with comprehensive molecular analysis. Am J Surg Pathol. 2018;42:1360‐1369. [DOI] [PubMed] [Google Scholar]
- 10. Antonescu CR, Agaram NP, Sung YS, Zhang L, Dickson BC. Undifferentiated round cell sarcomas with novel SS18‐POU5F1 fusions. Genes Chromosomes Cancer. 2020;59:620‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shenoy A, Newsom K, Gray B, et al. Malignant round cell tumor with SS18‐POU5F1 fusion: is it a myoepithelial neoplasm, a synovial sarcoma or a new entity? Histopathology. 2020;77:681‐684. [DOI] [PubMed] [Google Scholar]
- 12. Liu Z, Yuan H, Han M. Therapeutic response of soft tissue sarcoma with novel SS18‐POU5F1 fusion: a case report. Front Oncol. 2021;11:666946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Argani P, Zhong M, Reuter VE, et al. TFE3‐fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol. 2016;40:723‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR. RBM10‐TFE3 renal cell carcinoma: a potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false‐negative TFE3 FISH. Am J Surg Pathol. 2017;41:655‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Argani P, Reuter VE, Eble JN, et al. Biphasic hyalinizing psammomatous renal cell carcinoma (BHP RCC): a distinctive neoplasm associated with somatic NF2 mutations. Am J Surg Pathol. 2020;44:901‐916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Antonescu CR, Zhang L, Chang NE, et al. EWSR1‐POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty‐six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114‐1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Argani P, Kao Y‐C, Zhang L, et al. Primary renal sarcomas with BCOR‐CCNB3 gene fusion: a report of two cases showing histologic overlap with clear cell sarcoma of kidney, suggesting further link between BCOR‐related sarcomas of the kidney and soft tissues. Am J Surg Pathol. 2017;41:1702‐1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agaimy A, Cheng L, Egevad L, et al. Rhabdoid and undifferentiated phenotype in renal cell carcinoma: analysis of 32 cases indicating a distinctive common pathway of dedifferentiation frequently associated with SWI/SNF complex deficiency. Am J Surg Pathol. 2017;41:253‐262. [DOI] [PubMed] [Google Scholar]
- 20. Suurmeijer AJH, Dickson BC, Swanson D, et al. A morphologic and molecular reappraisal of myoepithelial tumors of soft tissue, bone, and viscera with EWSR1 and FUS gene rearrangements. Genes Chromosomes Cancer. 2020;59:348‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


