Abstract
Grasshoppers (Orthoptera: Acridoidea) are one of the most dangerous agricultural pests. Environmentally benign microbial pesticides are increasingly desirable for controlling grasshopper outbreaks in fragile ecosystems. However, little is known about natural pathogens infecting this pest. Here we profile the rich viral communities in forty-five grasshopper species and report 302 viruses, including 231 novel species. Most of the identified viruses are related to other insect viruses, and small RNA sequencing indicates that some are targeted by host antiviral RNA interference (RNAi) pathway. Our analysis of relationships between host phylogeny and virus diversity suggests that the composition of viromes is closely allied with host evolution. Overall, this study is a first extensive exploration of viruses in grasshoppers and provides a valuable comparative dataset of both academic and applied interest.
Keywords: grasshoppers, virus, virome, diversity, phylosymbiosis
1. Introduction
Grasshoppers (Orthoptera: Acridoidea) are one of the most devastating threats to agriculture throughout human history. They often form patches in the grassland when the density is high, and some can form swarms and migrate long distance, resulting in major economic, social, and environmental impacts on an international scale (Zhang et al. 2019). Even in the 21st century, grasshoppers still cause massive damages that endanger food security and threaten millions of people (Pflüger and Bräunig 2021). Recent events of desert locusts (Schistocerca gregaria) swarms in Arabian Peninsula, East Africa, India, and Pakistan since late 2019 were the worst upsurges seen in last 70 years for some countries (FAO 2020). There are more than 500 documented species of Acrididae that can cause damage to pastures and crops, and about fifty species are considered major pests (Zhang et al. 2019). Most grasshopper controls rely on chemical pesticides, and this has raised many issues about human health, environment, non-target organisms, and biodiversity (Zhang et al. 2019). In recent years, there has been an increased use of alternative biological control methods. A promising control method is using entomopathogenic viruses, which are environmentally benign and species-specific and can spread horizontally and transmit vertically. However, to date, only a few viruses have been isolated and characterized in grasshoppers: entomopoxviruses in Melanoplus sanguinipes and in Oedaleus senegalensis (Henry, Nelson, and Jutila 1969; Afonson et al. 1999) and a picornavirus in Schistocerca americana (Henry and Oma 1973). Melanoplus sanguinipes entomopoxvirus has been investigated for its potential use as biological control agents against orthopteran insects as it infects many grasshopper species (Jaeger and Langridge 1984; Streett, Oma, and Henry 1990). However, the slow-occurring mortality has limited its broad use as microbial insecticides (Erlandson 2008). Thus, there is still a demand for discovering new viral pathogens that could be harnessed to control grasshoppers.
Recent metagenomic studies of a variety of insect species have revealed that they harbor an enormous diversity of RNA viruses (Shi et al. 2016a; Kafer et al. 2019; Wu et al. 2020). Sequencing of viromes of insects has revised the evolutionary history of existing virus families such as Partitiviridae (Webster et al. 2015; Shi et al. 2016a), Flaviviridae (Shi et al. 2016b) and luteo/sobemo-like viruses (Tokarz et al. 2014) and led to discovery of new lineages of viruses (Li et al. 2015; Obbard et al. 2020). Characterizing viromes with known hosts not only provides a better perspective on the taxonomy and evolution of viruses (Tokarz et al. 2014; Webster et al. 2015), but also sheds light on host-association and host-switching of viruses (Li et al. 2015; Shi et al. 2016a). Increasing evidence has shown that the host lineage poses a great influence on the composition of virome, and viruses tend to jump between phylogenetically related host species (Longdon et al. 2014, 2015; Huang et al. 2021).
Orthopteran insects are underrepresented groups in virome studies, with reported viruses belonging to Flaviviridae, Virgaviridae, Narnaviridae, and Partitiviridae in pan-arthropod virome studies (Shi et al. 2016a; Wu et al. 2020). Characterizing viruses of grasshoppers and understanding host-switching of these viruses would be of potential importance for biocontrol decisions in the future. Here, we use a metagenomic approach to characterize viromes associated with forty-five species of grasshoppers including many major agricultural pest species, with the emphasis on better characterizing the diversity and abundance of viruses and understanding their eco-evolutionary relationship with their hosts.
2. Materials and methods
2.1. Sample collection and virus isolation
Grasshoppers were collected with sweep-net from ten locations in Inner Mongolia and Qinghai, China between 2018 and 2021, with 187–3,796 individuals of species. Locusta migratoria were purchased from a grasshopper breeding center in Hebei, China. Species were identified using morphological characteristics and mitochondrial cytochrome c oxidase subunit I gene (COI) sequences.
Crude virus purification was performed for Chorthippus albonemus, Chorthippus brunneus huabeiensis, Dasyhippus barbipes, Bryodema luctuosum luctuosum, and Oedaleus decorus asiaticus collected in Inner Mongolia, C. albonemus collected in Qinghai, and L. migratoria. Briefly, pools of 20–40 grasshoppers of the same species were homogenized in Ringer’s solution, and debris was removed by low-speed centrifugation at 500 × g for 10 min. The supernatant was layered on top of a discontinuous sucrose gradient (30 per cent, 40 per cent, 50 per cent, and 60 per cent w/v) and centrifuged at 64,000 × g for 3 h in A27-8 × 50 ml rotor (Beckman Coulter). The visible virus bands were collected, mixed, and centrifuged for 2 h at 64,000 × g in A27-8 × 50 ml rotors (Beckman Coulter) to sediment virus particles. Viral particles were then suspended in 500 µl of DNase/RNase-Free water (Solarbio).
2.2. RNA sequencing and reads assembly
Total viral RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Viral RNA quality was examined using NanoDrop 2000 (ThermoFisher Scientific, Waltham, MA) and Agilent 2100 bioanalyzer (ThermoFisher Scientific, CA, USA). Residual ribosomal RNA was depleted using Ribo-Zero kits (Epicentre, Madison, WI). Libraries were constructed using a TruSeq total RNA library preparation kit (Illumina), and paired-end (250–300 bp) sequencing was performed on the Hiseq-PE150 platform (Illumina, Sandiego, CA). Additionally, RNA-seq data of thirty-nine grasshoppers collected worldwide were retrieved from the National Center for Biotechnology Information (NCBI) database (Supplementary Table S1). Sequencing reads were quality trimmed using Trimmomatic (Bolger, Lohse, and Usadel 2014) and de novo assembled using the Trinity version 2.8.6 with minimum contig length set at 200 nt (Grabherr et al. 2011).
2.3. Discovery of viral sequences
The assembled contigs were compared to reference viral protein database (taxid: 10239) downloaded from NCBI using Diamond BLASTx version 2.0.11 (Huson and Buchfink 2015), with e-value cut-off of 1 × 10–5. To eliminate sequences that can be mapped to both virus and host, the putative viral contigs were then compared to the non-redundant protein database of NCBI using Diamond BLASTx version 2.0.11 (Huson and Buchfink 2015). Contigs with credible, significant BLAST hits (e-value < 1 × 10−5) to only viral proteins were kept for further analysis. To detect highly divergent viruses, open reading frames (ORFs) were predicted using the open-source NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). Predicted amino acid (aa) sequences with less than 200 aa length were removed from the following analysis. To reduce redundancy, aa sequences were grouped based on sequence identity using the CD-HIT package version 4.6.5 (Fu et al. 2012). Predicted ORFs without any BLASTx hits were searched for homologous proteins in the protein families database and the RNA-dependent RNA polymerase (RdRp) database of RNA viruses using HMMER version 3.3 (Eddy 2011). Viruses would be considered as novel species based on International Committee on Taxonomy of Viruses (ICTV) species demarcation criteria (https://talk.ictvonline.org/ictv-reports), e.g. for Dicistroviridae and Iflaviridae, if the aa sequence of capsid protein has less than 90 per cent identity compared to known isolates and strains, it is considered as a new species (detailed in Supplementary Table S1).
The structure of complete or near-complete viral genomes was annotated after comparing them against the genome of the closest virus relative using BLASTp. To estimate virus abundance in each library, Salmon version 1.4.0 (Patro et al. 2017) was used to calculate the number of transcripts per million (TPM) of each contig, which was normalized by sequencing depth (total number of reads) and sequence length.
2.4. Phylogenetic analysis
The RdRp or polyproteins of viruses discovered in this study were aligned with sequences of known viruses from the same families using MAFFT version 7.158 with the E-INS-i algorithm (Katoh and Standley 2013). Maximum likelihood (ML) phylogenetic trees were constructed using IQ-TREE version 1.6.12 with 1,000 bootstraps (Nguyen et al. 2015) and the best aa substitution models were determined using Raxml version 2.0 (Kozlov et al. 2019). COI sequences of grasshoppers were aligned using MAFFT version 7.158 with the E-INS-i algorithm (Katoh and Standley 2013). ML trees of the grasshoppers were constructed using the same method described above.
2.5. Phylosymbiosis analysis
To test if there is an association between host phylogeny and their viral composition, we performed phylosymbiosis analysis on five grasshopper species collected from the same location and their viromes. Host phylogeny was constructed using mitochondrial COI gene sequences. The host phylogeny was consistent with phylogeny constructed based on genome data in Song et al. (2020). The viral dendrogram was generated from Bray–Curtis beta diversity of the viral metagenomes. In brief, the R packages Vegan version 2.5-6 (Oksanen et al. 2020) was used to calculate Bray–Curtis distances using the virus abundance in each library (TPM). A clustergram of host viromes was constructed employing unweighted pair-group method with arithmetic means (UPGMA) (Leigh et al. 2018). The topological similarity and significance between the host phylogeny and the virome clustergram was determined by calculating a congruence index described in De Vienne et al. (2007). Spodoptera frugiperda was set as the outgroup for the analysis (Xu et al. 2020).
2.6. Investigation of viral prevalence in natural grasshopper populations
To survey the prevalence of viruses discovered in this study, we examined the presence of fifty-three virus species in three wild grasshopper populations sampled in nine locations in Qinghai Province, China, with 255–565 individuals tested for each population. Total virus RNA was prepared as described above. Reverse transcription was carried out using the PrimeScript RT reagent kit (Takara) using random hexamers, and polymerase chain reaction (PCR) was performed with PrimeSTAR Max DNA Polymerase (Takara). Primers were designed based on the assembled viral contigs. We tried to design degenerate primers based on conserved positions for similar viruses and specific primers for less conserved virus sequences (Supplementary Table S2).
We also checked the prevalence of forty-two viruses in D. barbipes and B. luctuosum luctuosum populations collected in 2019 and 2021, respectively. Specific primers were designed for each of the identified viruses in D. barbipes and B. luctuosum luctuosum (Supplementary Table S3).
2.7. Small RNA Sequencing and data analysis
Locusta migratoria were collected and used for small RNA sequencing. Total RNA was extracted and small RNA sequencing was performed using the high-throughput Illumina Nova6000 sequencing. Two samples were sequenced with 1.194 Gb and 1.292 Gb small RNA (sRNA) data generated. The same RNA samples were also used for metagenomic sequencing. The sRNA sequences obtained were put through quality control and then mapped to L. migratoria genome (Wang et al. 2014) to remove host sequences. Filtered reads were then mapped to various viral contigs using Bowtie 2 version 2.4.4 (Langmead and Salzberg 2012). The distribution of sRNAs was analyzed using R package viRome.
3. Results
3.1. Abundant and divergent viruses identified in grasshoppers
Ten ribosome-depleted total RNAs extracted from twenty to forty individuals of six species were non-stranded sequenced, generating 8.69 Gb to 15.59 Gb sequence data for each sample. Different percentages of reads could be mapped to viruses: 21.77 per cent for O. decorus asiaticus, 23.46 per cent for C. albonemus in Qinghai, China, 43.82 per cent for D. barbipes, 57.46 per cent for C. albonemus in Inner Mongolia, 86.40 per cent for C. brunneus huabeiensis, 94.05 per cent for B. luctuosum luctuosum, and 3.29 per cent for L. migratoria. Additional selected publicly available transcriptomic data of thirty-nine species of grasshoppers collected across the world were also included in the analysis (Supplementary Table S1). These datasets were generated from crushed whole grasshoppers or specific tissues, and total bases obtained vary between 1.5 Gb and 17.8 Gb.
Through a BLASTX search with assembled sequences, a total of 588 candidate viral contigs were identified. Using identifiable RdRp sequences or other relatively conserved genes, we were able to assign 296 putative viruses to forty-four virus families while six viruses were identified as unclassified viruses (Fig. 1A, Supplementary Table S1). The number of viruses identified in each host species varied a lot, with C. albonemus collected from Qinghai containing as many as fifty-one viruses. Significantly fewer viruses were identified in publicly available data that used poly(A)-mRNA-enriched sequencing, with smaller sequencing depth or used lab-reared insects (Fig. 1A).
Figure 1.
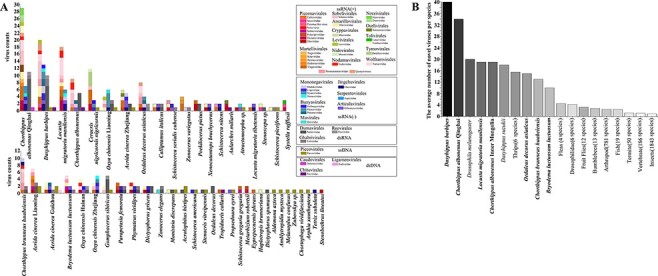
Viruses associated with grasshoppers. A. Virome compositions of grasshopper species. B. The average number of novel viruses identified in different hosts by metagenomic sequencing. The species marked in bold are samples sequenced in this study.
In total, 134 positive single-stranded RNA (+ssRNA) viruses, 74 negative single-stranded RNA (−ssRNA) viruses, 58 double-stranded RNA (dsRNA) viruses, 8 double-stranded DNA (dsDNA) viruses, and 23 single-stranded DNA (ssDNA) viruses were identified in our study (Fig. 1A, Supplementary Table S1). RNA viruses were the dominant type, comprising up to 88 per cent of the identified viruses. Picorna-like viruses including viruses mainly from Iflaviridae, Dicistroviridae, Solinviviridae, and Polycipiviridae were present in twenty grasshopper hosts, accounting for 26 per cent of +ssRNA viruses (Fig. 1A). Mononega-like viruses and bunya-like viruses were the most common −ssRNA viruses identified, representing up to 30 per cent of all −ssRNA viruses identified and present in more than 36 per cent grasshopper hosts in this study. Partiti-like viruses were found in 42 per cent grasshopper hosts and were the most common dsRNA viruses. Twelve contigs that can only be mapped to entomopoxviruses (dsDNA viruses) but not the hosts were found in six grasshoppers. Three of these contigs were annotated as spheroidin gene, which is often used for phylogenetic analysis of entomopoxviruses (Thézé et al. 2013). Oedaleus asiaticus entomopoxvirus and two potential new entomopoxviruses were present in our samples based on spheroidin sequences. Contigs of twenty-three parvo-like ssDNA viruses including sequences of Viral polypeptide 4 (VP4) and nonstructural protein 1 (NS1) were identified and presented in eighteen grasshopper hosts (Fig. 1A).
The majority of newly identified viruses were highly divergent from previously reported viral sequences: 68 per cent viruses shared less than 50 per cent aa identity with their most closely related RdRp sequence (Supplementary Table S1). Based on ICTV species demarcation criteria, 231 viruses can be considered as novel species (details in Supplementary Table S1). This is a large number of novel viruses identified when compared to similar studies of other organisms (Fig. 1B) and has substantially enriched the number of recorded orthopteran viruses (Li et al. 2015; Webster et al. 2015, 2016; Medd et al. 2018; Shi et al. 2018; Harvey et al. 2019; Kafer et al. 2019; Pascall et al. 2019; Lay et al. 2020; Chiapello et al. 2021; Geoghegan et al. 2021). Novel viruses found in this study are named after their host species, related virus family like, followed by a number (e.g. Chorthippus albonemus chu-like virus 1). If one virus infects more than one host species, the genus or family names of multiple hosts were used (e.g. Gomphocerinae chu-like virus). Complete or near-complete genome sequences were obtained for seventy-one novel viral species belonging to twenty-two families and tentative genome structures are shown in Fig. 2. Viruses from the same family tend to share similar genome structures with exceptions of Virgaviridae, Flaviviridae, and Totiviridae (Fig. 2). Virgaviridae showed a great flexibility in genome size and arrangement. Flaviviridae contains both typical segmented genome and a substantially larger unsegmented genome (Paraskevopoulou et al. 2021). Toti-like virus in grasshoppers could either encode a capsid protein or a novel proline–alanine-rich protein, as described in a previous study (Spear et al. 2010).
Figure 2.
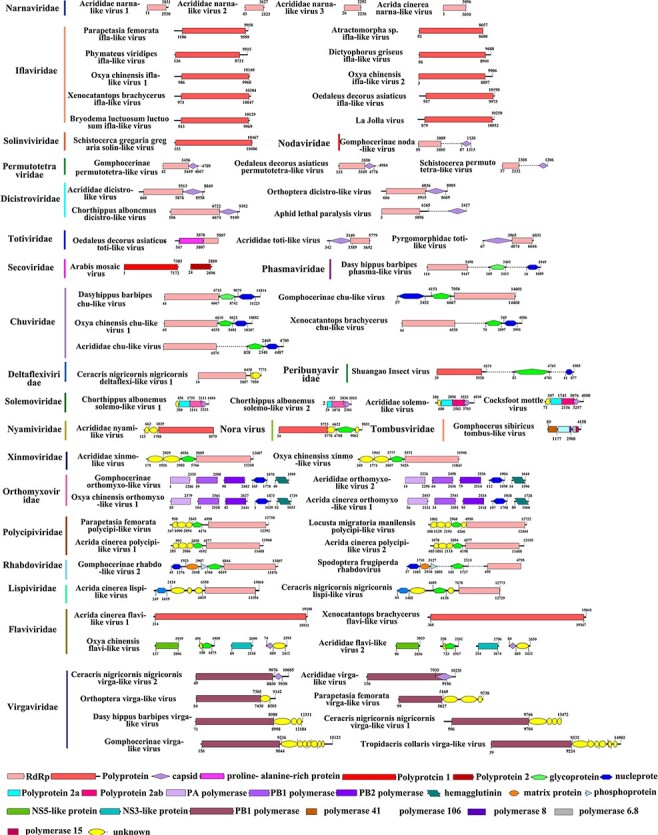
Annotations of complete or near-complete viral genomes. Viral proteins are colored according to their putative functions. For unsegmented viruses, different functional proteins that lack overlapping sequence regions are connected by dotted lines.
3.2. Phylogenetic analysis reveals previously unknown arthropod-associated virus groups
Contigs of RdRps along with known viruses and uncurated viral sequences from the NCBI Transcriptome Shotgun Assemblies database were grouped, and phylogenetic trees were generated following the optimization of alignments. Phylogenetic trees were not constructed for families where only one virus was identified. Seventeen trees were generated for twenty-seven virus families (Figs 3, 4, detailed trees are shown in Figs S1–14). Most of the identified viruses were relatives of known or suspected insect viruses and they belonged to Iflaviridae, Chuviridae, Polycipiviridae, Solinviviridae, Permutotetraviridae, Nodaviridae, Lispiviridae, Rhabdoviridae, Phenuiviridae, Phasmaviridae, Reoviridae, and Poxviridae. Some were related to viral sequences from plants and vertebrates. We tried to test the co-occurrence of these viruses and their potential non-insect hosts (Wallace 2021) and found that the Acrididae solemo-like virus was consistently co-occurring with Triticum aestivum in eleven of our samples (correlation coefficient = 0.738, P < 0.001), suggesting that it likely infects T. aestivum rather than grasshoppers. Many other potential plant or vertebrate virus sequences were abundant in our samples, which indicates that grasshoppers can harbor high copies of plant and vertebrate viruses, which they may acquire through feeding on virus-contaminated food.
Figure 3.
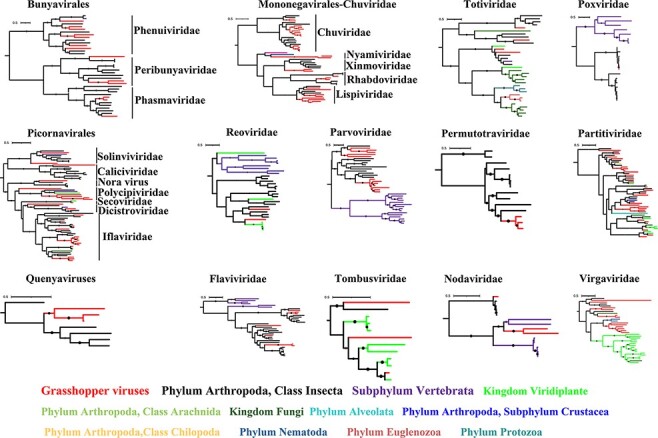
ML phylogenies of viruses in grasshoppers. The viruses described in this study are marked in red and novel viruses are marked with red pentagrams. The larger the solid dot on the branch, the larger the bootstraps.
Figure 4.
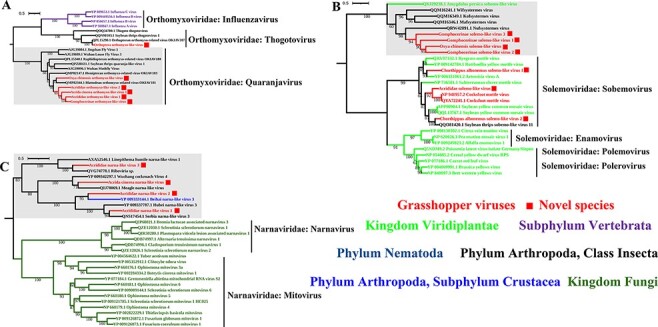
ML phylogenies of Orthomyxoviridae, Solemoviridae, and Narnaviridae. (A) Phylogenetic tree of Orthomyxoviridae constructed using PB1 polymerase sequences. (B) Phylogenetic tree of Solemoviridae constructed using replicase sequences. (C) Phylogenetic tree of Narnaviridae constructed using RdRp sequences. The viruses described in this study are marked in red and novel viruses have red solid square at the back of their names.
Interestingly, we found that certain viral families that were considered to only infect plant, fungus, or vertebrate hosts had formed a separate clade containing viruses discovered in arthropods. For example, five novel orthomyxo-like viruses grouped together and formed a separate clade with viruses found in flies, thrips, and other blattodean and hemipteran insects (Fig. 4A); four novel solemo-like viruses grouped together with viruses found in termites (Fig. 4B); and four novel narna-like viruses found in this study formed a distinct clade together with viruses discovered in Linepithema ants, cockroaches, and other arthropods (Fig. 4C). These arthropod-associated virus clades may expand the host range and fill evolutionary gaps of these virus families that were previously thought to be plant/fungus/vertebrate specific.
3.3. Phylosymbiosis detected between grasshoppers and their viromes
Although environmental factors are considered to play an essential role, host genetics and evolutionary history may also affect the composition of the host’s virome. If hosts influence a sufficient amount of the composition of virome, then hosts with greater genetic divergence may exhibit more distinguishable viral composition (Brooks et al. 2016; Leigh et al. 2018). In this study, host phylogeny showed a significant congruence with the branching pattern of the viral dendrogram (Icong = 1.31, P < 0.05) (Fig. 5). This result suggests a phylosymbiotic relationship between grasshopper host and viral beta diversity, meaning that evolutionary changes in the host are associated with ecological changes in the virome (Brooks et al. 2016).
Figure 5.

Phylosymbiosis between five grasshopper species and their viromes. The host phylogeny was constructed based on the COI gene, and the UPGMA hierarchical cluster relationships of the viromes were based on Bray–Curtis beta diversity distances. Spodoptera frugiperda and its virome data were used as outgroups for the analysis (Xu et al. 2020).
3.4. Natural prevalence of grasshopper viruses
To get an idea of how prevalent grasshopper viruses are in nature, we applied PCR tests on various natural populations of grasshoppers collected in different years. Dasyhippus barbipes and B. luctuosum luctuosum are dominant species in south Inner Mongolia from May to June. To test if the viral composition of these species varies in different years, we collected D. barbipes and B. luctuosum luctuosum in 2019 and 2021 separately. Through PCR tests, 67 per cent of the viruses identified in 2019 were found in B. luctuosum luctuosum collected in 2021, while 22 per cent of viruses identified in 2019 were detected in D. barbipes grasshoppers in 2021 (Supplementary Table S3). We also tested three grasshopper species collected from Qinghai for viruses identified in the same species collected from Inner Mongolia. Only three viruses out of fifty-three viruses tested could be detected in Qinghai populations. The natural infection rates of these three viruses are provided in Table 1. Viruses present in populations collected in both provinces tend to infect multiple host species. For example, Acrididae narna-like virus 1 was present in eight host species, Acrididae narna-like virus 3 was present in four host species, and Acrididae permutotetra-like virus was present in six host species (Supplementary Table S1). This result indicates that these viruses are common in grasshoppers and may be transmitted horizontally across different species.
Table 1.
The average natural infection rates of viruses in the wild.
| Host\Virus | Acrididae narna-like virus 1 | Acrididae permutotetra-like virus | Gomphocerinae permutotetra-like virus |
|---|---|---|---|
| Chorthippus dubius | 63% | 46% | 19% |
| Chorthippus albonemus | 43% | 52% | 17% |
| Chorthippus fallax | 0.00 | 28% | 0.00 |
3.5. Antiviral RNAi against various viruses in Locusta migratoria
Different from intensively studied RNAi response in Drosophila melanogaster, previous study did not find typical small interfering RNA (siRNA) 21 nt peak or piwi interacting RNA (piRNA) pattern in the distribution of virus-derived siRNAs (vsiRNAs) in L. migratoria (Lewis et al. 2018). To explore siRNA-based antiviral immunity in grasshoppers, we carried out small RNA sequencing on the same L. migratoria samples which we used for virus RNA-seq. Among twenty-five viruses that were found in L. migratoria, sRNAs were successfully mapped to contigs of seven viruses after filtering out host genome sequences. sRNAs mapped to Acrididae narna-like virus 2 and Acrididae xinmo-like virus showed an obvious enrichment in 22 nt (Fig. 6). Five more viruses including three +ssRNA viruses, one −ssRNA virus, and one dsRNA virus did not show obvious 21 nt or 22 nt peak (Fig. 6). We did not find virus-derived piRNA bearing the signature of ping-pong amplification. Notably, no sRNA was found to be mapped on many viruses, such as Oedipodinae noda-like virus, Acrididae solemo-like virus, and Drosophila A virus, even they were highly abundant in the host. These results suggest that antiviral RNAi pathways are actively involved in response to some viruses, and the distribution of sRNA may vary for different viruses.
Figure 6.
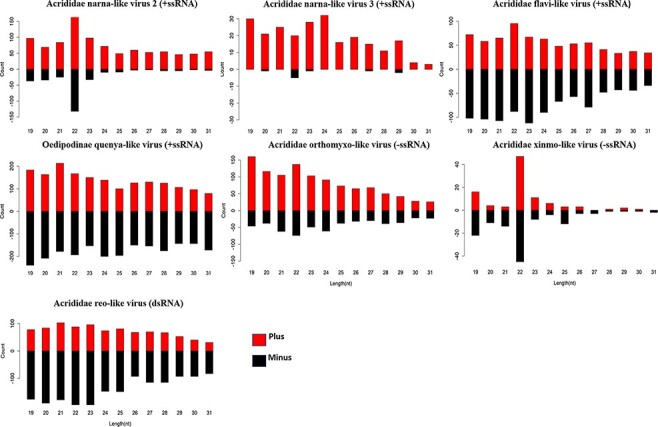
Profile of vsiRNAs. vsiRNAs derived from seven viruses identified in grasshoppers.
4. Discussion
In this study, we present the first virome survey of notorious insect pests and demonstrate that they harbor a diverse range of viruses. Overall, 271 RNA viruses and thirty-one DNA viruses were identified in forty-five species of grasshoppers. These viruses are quite divergent from previously known species, and 231 of them can be considered as novel species. Although the potential of these viruses be used as biological control agents is currently unclear, there are some good candidates. For example, entomopoxviruses and densoviruses have been registered as biocontrol agents (Abd-Alla, Meki, and Demirbas-Uzel 2020), and we identify six entomopoxviruses and twenty novel densoviruses infecting six and fifteen host species, respectively. Further isolation and pathogenicity assays are required to evaluate the potential use of these viruses as microbial control agents.
Viruses from Iflaviridae, Lispiviridae, Virgaviridae, Permutotetraviridae, Totiviridae, Narnaviridae, Solemoviridae, Chuviridae, Orthomyxoviridae, Partitiviridae, Flaviviridae, and Parvoviridae were abundantly present in many grasshopper species. Multispecies infection was seen more often in viruses from families mentioned above, suggesting that viruses from these families have the potential to infect a broad host range and cross-species transmission. We also found arthropod-infecting virus clades in vertebrate-specific virus family Orthomyxoviridae, plant-specific virus family Solemoviridae, and fungus-specific virus family Narnaviridae. This highlights the role of insects in connecting the evolution of viruses between organisms of different kingdoms. Furthermore, viruses that are more related to plant and vertebrate viruses were found with high abundance, for example, Acrididae solemo-like virus and Locusta migratoria manilensis noda-like virus. This provides a possibility that viruses may transmit from insects back to plant and animal hosts by contact or feeding (Walters 1951; Nunamaker et al. 2003; Drolet et al. 2009). Thus, grasshoppers not only harbor abundant viruses that are close relatives to viruses infecting vertebrate, plant, and fungus, but also have the potential to facilitate the transmission of plant and vertebrate viruses.
Whether host phylogeny plays an essential role in shaping the virome composition remains an intriguing question. Based on the five grasshopper species that were analyzed, significant topology congruence was found between host phylogeny and virus Bray–Curtis beta dendrogram. This suggests that in natural environment, phylosymbiotic relationship may exist between these grasshoppers and their viromes. Phylosymbiosis was proposed to describe the eco-evolutionary pattern, whereby the ecological relatedness of host-associated microbial communities closely aligns with host evolution (Brooks et al. 2016). Under the phylosymbiosis hypothesis, host-associated microbial communities form as a result of interactions with host, instead of being stochastically assembled through environmental acquisition. Therefore, in common environment, there will be congruence between the host phylogeny and microbial community dendrogram (Brooks et al. 2016). Studies found that there are survival and performance reductions in animals containing a heterospecific microbiome, indicating the biological importance of this congruence. Previous studies have found phylosymbiosis between hosts and microbial communities (mostly bacteria and fungi) in a diverse range of systems under controlled regimes (Brucker and Bordenstein 2013; Brooks et al. 2016) and in natural environments (Easson and Thacker 2014; Sanders et al. 2014). In plant and animal systems, significant phylosymbiosis was also found between hosts and virus communities (Roossinck 2005; Minot et al. 2011). Our study presents a new case of phylosymbiosis between insect hosts and their viromes in nature, and it provides a potential system to study underlying genetic and biochemical mechanisms.
RNA interference pathway has been shown as a major antiviral strategy used by many insects. By analysis of metagenomics and sRNA data, we could show that the antiviral RNAi may play an essential role in defense against viruses in L. migratoria. The virus-derived interfering RNA (viRNA) profile of L. migratoria shows a 22 nt peak for RNA viruses (Wei et al. 2009). Similar viRNA distributions were observed in other insects, such as in sandflies (Belda et al. 2019), whiteflies (Huang et al. 2021), thrips (Chiapello et al. 2021), and bumblebees (Pascall et al. 2019). It is possible that the 21 nt peak we see in Brachycera species such as flies and mosquitoes is unusual for insects (Obbard 2018; Zhang et al. 2022). Further studies are required to investigate whether different sRNA patterns derived from viruses exist in insects and if there is an alternative cleavage pattern of RNAi proteins.
Like all other metagenomic studies, our work has several limitations. For instance, our virus identification is purely based on sequence homology search. With high divergence, only the most conserved sequences are recognizable at the protein level. Indeed, for many novel viruses found in this study, especially those with segmented genomes, we had difficulty in identifying other proteins besides the RdRp. Moreover, we had difficulty in determining if these identified viruses are grasshopper-infecting. Additional sRNA sequencing would be useful in solving this issue in the future. Nevertheless, this study is a significant addition to our understanding of the abundance and diversity of insect-associated viruses and their evolutionary interactions with insect hosts, providing a rich resource for developing biological control agents for controlling grasshopper pests.
Supplementary Material
Acknowledgements
We thank Li Aomei, Wang Xiaoyu, Wang Huiping, Nie Weilei, Ume Hani, Yang Kejia, Tian Jingjing, and Feng Mingyue for their assistance in collecting samples, and Frank Jiggins and Darren Obbard for their helpful comments on an earlier manuscript version, and thank the two anonymous reviewers for their suggestions and comments.
Contributor Information
Yao Xu, Department of Entomology, China Agricultural University, No. 2 Yuanmingyuan West Road, Haidian District, Beijing 100193, China.
Jingyi Jiang, Department of Entomology, China Agricultural University, No. 2 Yuanmingyuan West Road, Haidian District, Beijing 100193, China.
Xiaoju Lin, Department of Entomology, China Agricultural University, No. 2 Yuanmingyuan West Road, Haidian District, Beijing 100193, China.
Wangpeng Shi, Department of Entomology, China Agricultural University, No. 2 Yuanmingyuan West Road, Haidian District, Beijing 100193, China.
Chuan Cao, Department of Entomology, China Agricultural University, No. 2 Yuanmingyuan West Road, Haidian District, Beijing 100193, China.
Data availability
The raw reads of RNA-seq and sRNA data generated in this study were deposited in NCBI Sequence Read Archive (SRA) with accession numbers SRR17030292–SRR17030301, SRR18059444, and SRR18054818. Sequences of all identified novel viruses from this study and sequence alignments used for constructing phylogeny trees have been deposited in GitHub https://github.com/yaoxu2019/viruses-associated-with-grasshoppers.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
This work was supported by The 2115 Talent Development Program of China Agricultural University.
Conflict of interest:
None declared.
References
- Abd-Alla A. M. M., Meki I. K., and Demirbas-Uzel G. (2020) ‘Insect Viruses as Biocontrol Agents: Challenges and Opportunities’. In: El-Wakeil, N., Saleh, M., and Abu-hashim, M. (eds) Cottage Industry of Biocontrol Agents and Their Applications, pp. 277–95. Springer: Cham. [Google Scholar]
- Afonson C. L. et al. (1999) ‘The Genome of Melanoplus sanguinipes Entomopoxvirus’, Journal of Virology, 73: 533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda E. et al. (2019) ‘De Novo Profiling of RNA Viruses in AnophelesMalaria Vector Mosquitoes from Forest Ecological Zones in Senegal and Cambodia’, BMC Genomics, 20: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B. (2014) ‘Trimmomatic: A Flexible Trimmer for Illumina Sequence Data’, Genome Analysis, 15: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A. W. et al. (2016) ‘Phylosymbiosis: Relationships and Functional Effects of Microbial Communities across Host Evolutionary History’, PLoS Biology, 14: e2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker R. M., and Bordenstein S. R. (2013) ‘The Hologenomic Basis of Speciation: Gut Bacteria Cause Hybrid Lethality in the Genus Nasonia’, Science, 341: 667–9. [DOI] [PubMed] [Google Scholar]
- Chiapello M. et al. (2021) ‘Complexity and Local Specificity of the Virome Associated with Tospovirus-transmitting Thrips Species’, Journal of Virology, 95: e0059721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vienne D. M. et al. (2007) ‘A Congruence Index for Testing Topological Similarity between Trees’, Bioinformatics, 23: 3119–24. [DOI] [PubMed] [Google Scholar]
- Drolet B. S. et al. (2009) ‘Infection of Melanoplus sanguinipes Grasshoppers following Ingestion of Rangeland Plant Species Harboring Vesicular Stomatitis Virus’, Applied and Environmental Microbiology, 75: 3029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easson C. G., and Thacker R. W. (2014) ‘Phylogenetic Signal in the Community Structure of Host-specific Microbiomes of Tropical Marine Sponges’, Frontiers in Microbiology, 5: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2011) ‘Accelerated Profile HMM Searches’, PLoS Computational Biology, 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson M. (2008) ‘Insect Pest Control by Viruses’. In: Erlandson, M. (ed.) Encyclopedia of Virology, pp. 125–33, Agriculture & Agri-Food Canada: Saskatoon, SK, Canada. [Google Scholar]
- Food and Agriculture Organization of the United Nations . (2020), FAO Appeals for Urgent Support to Fight Worsening Desert Locust Upsurge in the Horn of Africa, <https://www.fao.org/news/story/en/item/1259082/icode/> accessed 30 Jan 2020.
- Fu L. et al. (2012) ‘CD-HIT: Accelerated for Clustering the Next-generation Sequencing Data’, Bioinformatics Oxford, 28: 3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L. et al. (2021) ‘Virome Composition in Marine Fish Revealed by Meta-transcriptomics’, Virus Evolution, 7: veab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. (2011) ‘Trinity: Reconstructing A Full-length Transcriptome without A Genome from RNA-Seq Data’, Nature Biotechnology, 29: 644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey E. et al. (2019) ‘Identification of Diverse Arthropod Associated Viruses in Native Australian Fleas’, Virology, 535: 189–99. [DOI] [PubMed] [Google Scholar]
- Henry J. E., Nelson B. P., and Jutila J. W. (1969) ‘Pathology and Development of the Grasshopper Inclusion Body Virus in Melanoplus sanguinipes’, Journal of Virology, 3: 605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J. E., and Oma E. A. (1973) ‘Ultrastructure of the Replication of the Grasshopper Crystalline-array Virus in Schistocerca americana Compared with Other Picornaviruses’, Journal of Invertebrate Pathology, 21: 273–81. [DOI] [PubMed] [Google Scholar]
- Huang H. J. et al. (2021) ‘Diversity and Infectivity of the RNA Virome among Different Cryptic Species of an Agriculturally Important Insect Vector: Whitefly Bemisia tabaci’, Npj Biofilms and Microbiomes, 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., and Buchfink B. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Jaeger B., and Langridge W. (1984) ‘Infection of Locusta migratoria with Entomopoxviruses from Arphia conspersa and Melanoplus sanguinipes Grasshoppers’, Journal of Invertebrate Pathology, 43: 374–82. [Google Scholar]
- Kafer S. et al. (2019) ‘Re-assessing the Diversity of Negative Strand RNA Viruses in Insects’, PLoS Pathogens, 15: e1008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., and Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A. M. et al. (2019) ‘RAxML-NG: A Fast, Scalable and User-friendly Tool for Maximum Likelihood Phylogenetic Inference’, Bioinformatics, 35: 4453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L. (2012) ‘Fast Gapped-read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay C. et al. (2020) ‘Unmapped RNA Virus Diversity in Termites and Their Symbionts’, Viruses, 12: 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh B. A. et al. (2018) ‘Finer-scale Phylosymbiosis: Insights from Insect Viromes’, mSystems, 3: e00131–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. H. et al. (2018) ‘Pan-arthropod Analysis Reveals Somatic piRNAs as an Ancestral Defence against Transposable Elements’, Nature Ecology & Evolution, 2: 174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. X. et al. (2015) ‘Unprecedented Genomic Diversity of RNA Viruses in Arthropods Reveals the Ancestry of Negative-sense RNA Viruses’, eLife, 4: e05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B. et al. (2015) ‘The Evolution, Diversity, and Host Associations of Rhabdoviruses’, Virus Evolution, 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B. et al. (2014) ‘The Evolution and Genetics of Virus Host Shifts’, PLoS Pathogens, 10: e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medd N. C. et al. (2018) ‘The Virome of Drosophila suzukii, an Invasive Pest of Soft Fruit’, Virus Evolution, 4: vey009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S. et al. (2011) ‘The Human Gut Virome: Inter-individual Variation and Dynamic Response to Diet’, Genome Research, 21: 1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunamaker R. A. et al. (2003) ‘Grasshoppers (Orthoptera: Acrididae) Could Serve as Reservoirs and Vectors of Vesicular Stomatitis Virus’, Journal of Medical Entomology, 40: 957–63. [DOI] [PubMed] [Google Scholar]
- Obbard D. J. (2018) ‘Expansion of the Metazoan Virosphere: Progress, Pitfalls, and Prospects’, Current Opinion in Virology, 31: 17–23. [DOI] [PubMed] [Google Scholar]
- ——— et al. (2020) ‘A New Lineage of Segmented RNA Viruses Infecting Animals’, Virus Evolution, 6: vez061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J. et al. (2020), Vegan: Community Ecology Package <https://CRAN.R-project.org/package=vegan> accessed 17 Apr 2022.
- Paraskevopoulou S. et al. (2021) ‘Viromics of Extant Insect Orders Unveil the Evolution of the Flavi-like Superfamily’, Virus Evolution, 7: veab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascall D. J. et al. (2019) ‘Host Evolutionary History Predicts Virus PrevalenceÚcross Bumblebee Species’, BioRxiv, 498717.doi: 10.1101/498717 [DOI] [Google Scholar]
- Patro R. et al. (2017) ‘Salmon: Provides Fast and Bias-aware Quantification of Transcript Expression’, Nature Methods, 14: 417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflüger H. J., and Bräunig P. (2021) ‘One Hundred Years of Phase Polymorphism Research in Locusts’, Journal of Comparative Physiology A, 207: 321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck M. J. (2005) ‘Symbiosis versus Competition in Pant Virus Evolution’, Nature Reviews. Microbiology, 3: 917–24. [DOI] [PubMed] [Google Scholar]
- Sanders J. G. et al. (2014) ‘Stability and Phylogenetic Correlation in Gut Microbiota: Lessons from Ants and Apes’, Molecular Ecology, 23: 1268–83. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2016a) ‘Redefining the Invertebrate RNA Virosphere’, Nature, 540: 539–43. [DOI] [PubMed] [Google Scholar]
- ——— et al. (2016b) ‘Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses’, Journal of Virology, 90: 659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2018) ‘The Evolutionary History of Vertebrate RNA Viruses’, Nature, 556: 197–202. [DOI] [PubMed] [Google Scholar]
- Song H. et al. (2020) ‘Phylogenomic Analysis Sheds Light on the Evolutionary Pathways Towards Acoustic Communication in Orthoptera’, Nature Communications, 11: 4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear A. et al. (2010) ‘Plant-feeding Insects Harbor Double-stranded RNA Viruses Encoding A Novel Proline-alanine Rich Protein and A Polymerase Distantly Related to that of Fungal Viruses’, Virology, 404: 304–11. [DOI] [PubMed] [Google Scholar]
- Streett D. A., Oma E. A., and Henry J. E. (1990) ‘Cross Infection of Three Grasshopper Species with the Melanoplus sanguinipes Entomopoxvirus’, Journal of Invertebrate Pathology, 56: 419–21. [Google Scholar]
- Thézé J. et al. (2013) ‘New Insights into the Evolution of Entomopoxvirinae from the Complete Genome Sequences of Four Entomopoxviruses Infecting Adoxophyes honmai, Choristoneura biennis, Choristoneura rosaceana, and Mythimna separata’, Journal of Virology, 87: 7992–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R. et al. (2014) ‘Virome Analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis Ticks Reveals Novel Highly Divergent Vertebrate and Invertebrate Viruses’, Journal of Virology, 88: 11480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. A. (2021) ‘The Discovery, Distribution, and Diversity of DNA Viruses Associated with Drosophila melanogaster in Europe’, Virus Evolution, 7: veab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters H. (1951) ‘Grasshopper Transmission of Three Plant Viruses’, Science, 113: 36–7. [DOI] [PubMed] [Google Scholar]
- Wang X. et al. (2014) ‘The Locust Genome Provides Insight into Swarm Formation and Long-distance Flight’, Nature Communications, 5: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. L. et al. (2015) ‘The Discovery, Distribution, and Evolution of Viruses Associated with Drosophila melanogaster’, PloS Biology, 13: e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2016) ‘Twenty-Five New Viruses Associated with the Drosophilidae (Diptera)’, Evolutionary Bioinformatics, 12: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. et al. (2009) ‘Characterization and Comparative Profiling of the Small RNA Transcriptomes in Two Phases of Locust’, Genome Biology, 10: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. (2020) ‘Abundant and Diverse RNA Viruses in Insects Revealed by RNA-Seq Analysis: Ecological and Evolutionary Implications’, mSystems, 5: e00039–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. et al. (2020) ‘The Discovery of Viruses Associated with the Fall Armyworm Spodoptera frugiperda by High-throughput Sequencing’, Journal Plants Protection, 47: 807–14. [Google Scholar]
- Zhang L. et al. (2019) ‘Locust and Grasshopper Management’, Annual Review of Entomology, 64: 15–34. [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. (2022) ‘The Diversity of Viral Community in Invasive Fruit Flies (Bactrocera and Zeugodacus) Revealed by Meta-transcriptomics’, Microbial Ecology, 83: 739–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads of RNA-seq and sRNA data generated in this study were deposited in NCBI Sequence Read Archive (SRA) with accession numbers SRR17030292–SRR17030301, SRR18059444, and SRR18054818. Sequences of all identified novel viruses from this study and sequence alignments used for constructing phylogeny trees have been deposited in GitHub https://github.com/yaoxu2019/viruses-associated-with-grasshoppers.


