Abstract
Macroautophagy/autophagy is an evolutionally conserved catabolic process in which cytosolic contents, such as aggregated proteins, dysfunctional organelle, or invading pathogens, are sequestered by the double‐membrane structure termed autophagosome and delivered to lysosome for degradation. Over the past two decades, autophagy has been extensively studied, from the molecular mechanisms, biological functions, implications in various human diseases, to development of autophagy‐related therapeutics. This review will focus on the latest development of autophagy research, covering molecular mechanisms in control of autophagosome biogenesis and autophagosome–lysosome fusion, and the upstream regulatory pathways including the AMPK and MTORC1 pathways. We will also provide a systematic discussion on the implication of autophagy in various human diseases, including cancer, neurodegenerative disorders (Alzheimer disease, Parkinson disease, Huntington's disease, and Amyotrophic lateral sclerosis), metabolic diseases (obesity and diabetes), viral infection especially SARS‐Cov‐2 and COVID‐19, cardiovascular diseases (cardiac ischemia/reperfusion and cardiomyopathy), and aging. Finally, we will also summarize the development of pharmacological agents that have therapeutic potential for clinical applications via targeting the autophagy pathway. It is believed that decades of hard work on autophagy research is eventually to bring real and tangible benefits for improvement of human health and control of human diseases.
Keywords: autophagy, cancer, cardiovascular diseases, metabolic diseases, neurodegenerative diseases, SARS‐CoV‐2
Macroautophagy/autophagy is an evolutionally conserved catabolic process induced under various stress conditions to maintain intracellular homeostasis. Dysregulation of autophagy is closely associated with various human diseases. This review will focus on the molecular mechanisms in control of autophagy, the implication of autophagy in various human diseases, and the development of pharmacological agents that target autophagy.
1. INTRODUCTION
Autophagy refers to a process in which the intracellular components such as abnormal proteins, damaged organelles, foreign pathogens, and other cellular components are degraded via lysosome. This catabolic process is evolutionarily conserved from yeast to mammalian cells. The history of autophagy research spans over 7 decades and marked with two Nobel prizes. 1 , 2 The modern concept of autophagy was coined in 1960s by Christian de Duve after he studied the function of lysosome, the key digestive organelle that he discovered in 1950s, which earned him the Nobel Prize in Physiology or Medicine in 1974. However, in the following 4 decades after the discovery of lysosome, the autophagy research remained a relatively small field, mainly due to limited research tools and lack of understanding of its molecular mechanisms. In 1990s, the ground‐breaking work by Yoshinori Ohsumi identified a series of autophagy‐related genes (Atgs, originally termed as Apgs) in control of autophagy using yeast as the model organism for studies. 3 The discovery of Atgs and subsequent work on the molecular mechanisms elucidating the function of the proteins encoded by those Atgs earned Ohsumi the Nobel Prize in Physiology or Medicine in 2016. At present, a total of more than 40 Atgs have been identified and the autophagy field is still expanding, from the molecular mechanisms to biological functions and implications in health and disease. 4 , 5 , 6
In mammalian cells, autophagy has been traditionally classified into the following three main types, macroautophagy, microautophagy, and chaperone‐mediated autophagy (CMA). Among them, macroautophagy is featured by the formation of a unique double‐membrane organelle, the autophagosome. 7 , 8 In contrast, both microautophagy and CMA bypass autophagosome formation and the cargos are directly delivered to lysosome. 9 , 10 At present, the majority of the autophagy research is on macroautophagy, or referred as autophagy hereafter in this review. On the other hand, depending on the nature of the cargos, autophagy can be categorized into general/nonselective and selective autophagy. For nonselective autophagy, the cellular cargos are engulfed into the autophagosomes randomly, a process usually induced by general stress conditions such as nutrient starvation. 11 In contrast, selective autophagy refers to selective degradation of specific cargos, and so far, there are many types of selective autophagy being studied, such as mitophagy (selective degradation of mitochondria), endoplasmic reticulum (ER)‐phagy (selective degradation of ER), aggrephagy (selective degradation of protein aggregates), and xenophagy (selective degradation of invaded pathogens), just to name a few. 12 , 13 , 14 , 15 At present, it has been well studied that autophagy have important functions in various biological processes, such as cell survival and cell death, inflammation and immunity, development and differentiation, metabolic homeostasis, and so on. As such, autophagy is known to be closely implicated in the pathogenesis of human diseases. 16 , 17 In this review, we will mainly focus on nonselective macroautophagy to provide a systematic discussion on the latest development on the molecular mechanisms, the implication of autophagy in important human diseases including cancer, neurodegeneration, metabolic diseases, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, cardiovascular diseases, and aging. Moreover, we will also discuss the therapeutic potential of targeting autophagy in human diseases. Finally, we will highlight the challenges the autophagy research field is facing and the directions of future study.
2. MOLECULAR MECHANISMS IN CONTROL OF AUTOPHAGY
2.1. ATGs and autophagosome biogenesis
The biogenesis of autophagosome is coordinated by the core ATG (autophagy related) proteins (Table 1). These ATG proteins form different complexes to drive the initiation, nucleation, expansion, and closure of autophagosome (Figure 1). Briefly, these complexes include: (i) the ULK1 complex, consisting of ULK1 (unc‐51 like autophagy activating kinase 1), RB1CC1/FIP200 (RB1 inducible coiled‐coil 1), ATG13 ang ATG101; (ii) the class III lipid kinase complex I (PI3KC3–C1), formed by PIK3C3/VPS34 (phosphatidylinositol 3‐kinase catalytic subunit type 3), PIK3R4/VPS15/p150 (phosphoinositide‐3‐kinase regulatory subunit 4), BECN1/Beclin 1, ATG14/ATG14L and NRBF2 (nuclear receptor binding factor 2); (iii) the WIPI (WD‐repeat protein interacting with phosphoinositides) family; (iv) the two ubiquitin‐like conjugation systems that facilitate conjugation of mATG8 to membrane‐resident phosphatidylethanolamine (PE); (v) the ATG9 vesicles. 45 , 46 , 47
TABLE 1.
Autophagy core machinery and their functions
| Autophagy core machinery | Protein name | Functions | References |
|---|---|---|---|
| The ULK1 complex | ULK1/ATG1 | Serine/threonine kinase; phosphorylates different components of the autophagy machinery to initiate autophagy. | 18 , 19 |
| ATG13 | Interacts with ULK1; bridges ULK1 to RB1CC1. | 19 , 20 , 21 | |
| ATG101 | Heterodimerizes with ATG13 via their HORMA domain to stabilize the binding of ATG13 to ULK1; enhances ULK1 activity. | 20 , 22 | |
| RB1CC1/FIP200 | Scaffold protein; possibly serves as scaffold for ULK1 and ATG13. | 21 , 23 | |
| The class III lipid kinase complex I (PI3KC3–C1) | PIK3C3/VPS34 | PtdIns3 kinase; generates PtdIns3P. | 24 |
| PIK3R4/VPS15 | Scaffold and protein kinase; binds to PIK3C3 to form the catalytic arm of PI3KC3–C1. | 25 , 26 | |
| BECN1 | Forms the regulatory arm of PI3KC3–C1 via binding to ATG14. | 24 , 27 | |
| ATG14 | Directs PI3KC3–C1 to PAS. | 28 , 29 | |
| The WIPI family and WIPI–ATG2A complex | WIPI1 | PtdIns3P binding protein. | 30 |
| WIPI2 | PtdIns3P binding protein; recruits ATG12–ATG5‐ATG16L1 complex to conjugate LC3/GABARAP to PE. | 30 , 31 | |
| WIPI3 | PtdIns3P binding protein; associates with TSC complex to regulate mTOR activity; complexes with ATG2A to regulate phagophore expansion. | 32 , 33 | |
| WIPI4 | PtdIns3P binding protein; complexes with ATG2A to regulate phagophore expansion. | 32 , 33 , 34 | |
| ATG2A | Complexes with WIPI3/WIPI4 to regulate phagophore expansion. | 33 , 34 , 35 | |
| The two ubiquitin‐like conjugation systems | ATG4 | Cysteine protease to cleave pro‐ATG8s; deconjugation of ATG8s from PE (phosphatidylethanolamine). | 36 , 37 |
| ATG7 | E1‐like enzyme for both the ATG8s conjugation system and ATG12 conjugation system. | 4 , 38 | |
| ATG3 | E2‐like enzyme for the ATG8s conjugation system. | 4 , 38 | |
| ATG10 | E2‐like enzyme that conjugates ATG12 to ATG5. | 38 , 39 | |
| ATG5 | Conjugated by ATG12. | 38 , 39 | |
| ATG12 | The ubiquitin‐like molecule in the ATG12 conjugation system. | 38 , 39 | |
| ATG16L1 | Interacts with the ATG12–ATG5 conjugate to form the ATG12–ATG5‐ATG16L1 complex, which acts as the E3‐like enzyme to conjugate mATG8 to PE. | 40 | |
| mATG8 | The ubiquitin‐like molecule in the ATG8s conjugation system. | 41 , 42 | |
| ATG9 vesicles | ATG9 | Contributes membrane source to the forming phagophore. | 43 , 44 , 45 |
FIGURE 1.
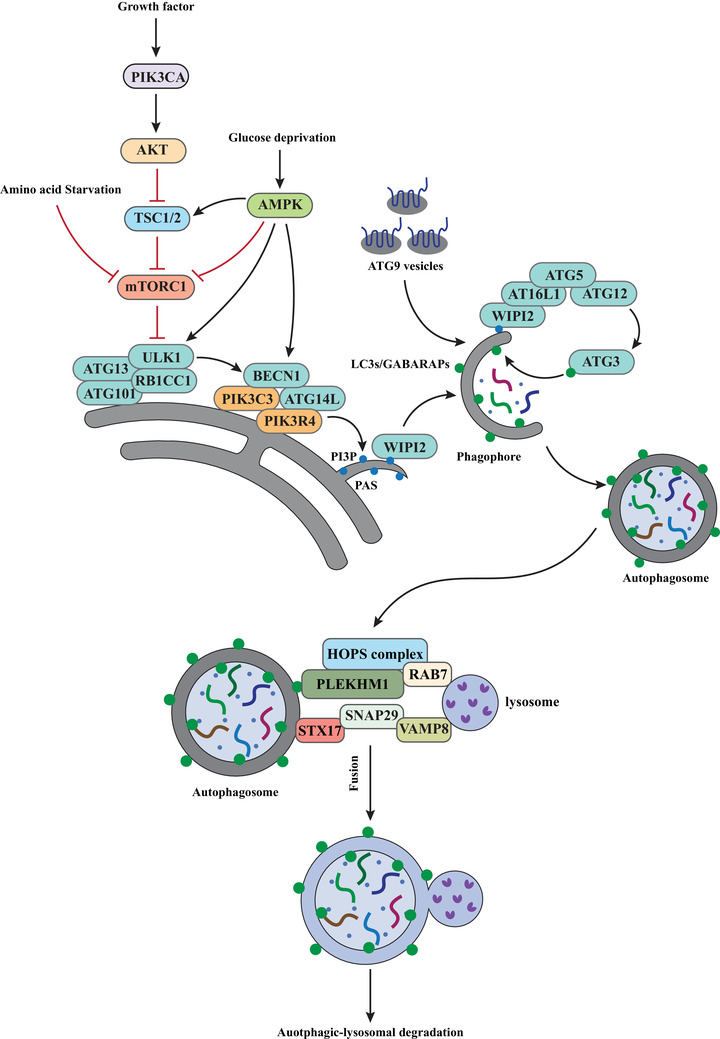
Molecular mechanisms in control of autophagy. Autophagy could be induced by various stress conditions such as amino starvation, glucose depletion, and others. The common target of these signaling pathways is the ULK1 complex. Under normal condition, MTORC1 inhibits ULK1 via phosphorylation. Under stress condition, MTORC1 is suppressed, leading to activation and recruitment of the ULK1 complex to PAS. ULK1 further activates PI3KC3–C1, resulting in generation of PtdIns3P (PI3P). WIPI2 then binds to PtdIns3P and further recruits ATG12–ATG5–ATG16L1 to the phagophore to mediate the lipidation of mATG8, which is essential for elongation and closure of the phagophore membrane. Additionally, the ATG9 vesicles are believe to supply membrane source, which also contributes to elongation of the phagophore membrane. Autophagosome–lysosome fusion is mediated by specific SNARE proteins, the HOPS tethering complex, small GTPases such as RAB7 and their effector PLEKHM1, and other factors detailed in the main text. The molecular mechanisms of these factors in autophagosome–lysosome fusion are discussed in the main text
2.1.1. The ULK1 complex
Autophagy could be triggered under various stressed conditions such as amino acid starvation, glucose depletion, hypoxia, oxidative stress, and others. Upon autophagy induction, ULK1 is activated via autophosphorylation within its kinase domain at Thr180. 48 , 49 , 50 ULK1 is a serine/threonine protein kinase that contains a protein kinase domain at its N‐terminus, while the C‐terminus is responsible for interaction with the C‐terminus of ATG13. 19 ATG101 is fully composed of a HORMA (Hop1, Rev7, Mad2) domain and it heterodimerizes with ATG13 via the HORMA domain at its N terminus, which prevents the proteasomal degradation of ATG13 and strengthens the interaction between ATG13 and ULK1. 20 , 22 On the other hand, ATG13 also bridges ULK1 to the scaffolding subunit RB1CC1. 21 , 23 The assembly of the ULK1 complex is constitutive and not affected much by nutrient deprivation. 21 , 51 There is evidence indicating that the association of ULK1 with ATG13 and RB1CC1 also contributes to its autophosphorylation and protein stability. 51 Once activated, ULK1 in turn phosphorylates ATG13 (S318/S203), RB1CC1 (S943/S986/S1323), and ATG101 (S11/S203). 52 , 53 Further studies are necessary to understand the exact functions of such phosphorylation by ULK1 in autophagy.
Upon activation, the ULK1 complex is recruited to the phagophore assembly site (PAS). So far, how ULK1 complex is recruited remains largely undefined. Both ULK1 and ATG13 contains LC3‐interacting region (LIR), or the ATG8‐interacting motif, which binds to mATG8 (mammalian ATG8, including LC3A/MAP1LC3A (microtubule associated protein 1 light chain 3 alpha), LC3B, LC3C, GABARAP, GABARAPL1 (GABA type A receptor‐associated protein like 1) and GABARAPL2). 54 , 55 While mutation of the LIR motif in ATG13 shows marginal effects on autophagy initiation, 56 mutation of the LIR motif in ULK1 causes a significant defect in autophagy initiation, 54 indicating that the interaction of ULK1 with mATG8 is important for autophagy induction. While lipidation of mATG8 is normally considered as downstream of ULK1 complex during autophagy, it is believed that GABARAP may act upstream to recruit ULK1 complex to initiate autophagy.
Apart from GABARAP, the small GTPase RAB1A (homolog of Ypt1 in yeast) has also been reported to recruit ULK1 complex to PAS in yeast and mammalian cells. 57 , 58 As an effector of RAB1A, C9orf72 (C9orf72–SMCR8 complex subunit) interacts with the ULK1 complex and mediates the translocation of ULK1 complex to PAS via RAB1A. 57 Moreover, the ER contact proteins VAPA (VAMP‐associated protein A) and VAPB also play important roles in ULK1 complex recruitment via direct interaction with ULK1 and RB1CC1. 59 It appears that multiple mechanisms are involved in ULK1 recruitment and it would be important to understand how these mechanisms are coordinated with each other to precisely regulate autophagy initiation.
2.1.2. The class III lipid kinase complex I (PI3KC3–C1)
PI3KC3–C1 consists of the lipid kinase PIK3C3, the scaffold and potential protein kinase PIK3R4, the regulatory subunit BECN1, and ATG14, and is responsible for production of PtdIns3P/PI3P, a key lipid metabolite essential for autophagy. 24 The complex adopts a V shape model, with PIK3C3 and PIK3R4 forming the right arm of the V with catalytic functions, and BECN1 and ATG14 forming the left arm of the V with regulatory functions. 25 PIK3C3 contains a C2 domain, a helical domain, and a kinase domain. The C2 domain is critical for the binding with PIK3R4 as it has a helical insertion that directly contacts the WD40 domain of PIK3R4. 26 This WD40 domain of PIK3R4 is also a part of the left arm that serves as a docking site for BECN1 and ATG14. 60 The central coiled‐coil domain of BECN1 regulates its binding to the coiled‐coil regions of ATG14. 27 NRBF2 has been discovered as the fifth subunit and its interaction with BECN1 and ATG14 is triggered upon autophagy induction such as amino acid starvation, resulting in enhanced kinase activity and dimerization of PI3KC3–C1. 61 , 62 , 63 Intriguingly, an inhibitory role of NRBF2 in autophagy is also reported. 64
Similar to ULK1, the subunits of PI3KC3–C1 (including PIK3C3, BECN1, and ATG14) also contain LIR motif with preferential bindings to GABARAP and GABARAPL1 and such bindings help scaffolding PI3KC3–C1 on membranes to promote efficient autophagosome formation. 65 Upon autophagy induction, these subunits are phosphorylated by ULK1 including PIK3C3 (S249), BECN1 (S15/30/96/279/337), and ATG14 (S29). 52 , 66 , 67 Phosphorylation of PIK3C3 at S249 enhances its bindings to GABARAP, GABARAPL1, and LC3C. 65 Phosphorylation of Beclin‐1 (S15) and ATG14 (S29) by ULK1 has been shown to enhance the activity of PI3KC3–C1. 66 , 67 A more recent study shows that ULK1 also phosphorylates PIK3R4 (S813/S861/S865/S879/S1039/S1289) and mutations of these sites impair the activity of PI3KC3–C1 and autophagy. 68 Importantly, mutation of the ATG14 LIR motif inhibits the phosphorylation of ATG14 at S29 and BECN1 at S96, thereby inhibiting the activity of PI3KC3–C1. 65 It appears that the binding of ATG14 to GABARAP and GABARAPL1 is essential for PI3KC3–C1 recruitment and activation following autophagy induction, which is supported by earlier studies which show that ATG14 targets PI3KC3–C1 to PAS. 28 , 29 Recruitment and activation of PI3KC3–C1 results in phosphorylation of phosphatidylinositol to produce PtdIns3P essential for autophagosome nucleation and recruitment of downstream effectors. 69 , 70
2.1.3. The WIPI family and WIPI–ATG2A complex
Following the activation of PI3KC3–C1 and production of PtdIns3P, ZFYVE1/DFCP1 (zinc finger FYVE‐type containing 1), and the WIPI/PROPPIN (β‐propellers that bind polyphosphoinositides) family act as PtdIns3P effectors to recruit downstream autophagy machinery. 69 , 71 Although DFCP1 puncta has been widely used as a marker for omegasome, its functions in autophagy remains largely undefined. On the other hand, WIPI proteins bind to downstream effector proteins via their seven bladed β‐propellers and bind to PtdIns3P via their Phe‐Arg‐Arg‐Gly (FRRG) motifs. 72 , 73 There are four members of the WIPI family including WIPI1, WIPI2, WIPI3/WDR45B, and WIPI4/WDR45. 74 Among these WIPI proteins, the role of WIPI2 in autophagy is relatively well established. Six isoforms of WIPI2 have been reported thus far. 30 , 75 Among them, WIPI2B and WIPI2D have been shown to be essential for recruitment of the ATG12–ATG5–ATG16L1 complex to mediate the conjugation of LC3B to PE to promote phagophore expansion (and possibly sealing). 31 , 75 Consistently, downregulation or dysfunction of WIPI2 has been linked to defective autophagy. 76 , 77 Recently, it has been shown that WIPI2 positively regulates the proteasomal degradation of outer mitochondrial proteins and mitophagy via recruiting VCP (valosin‐containing protein) to damaged mitochondria. 78 Thus, such observations advance our understanding on WIPI2 functions and reveals the critical role of WIPI2 in the ubiquitin‐proteasome and autophagy–lysosome systems to maintain cellular homeostasis.
As for other WIPIs, WIPI1 was reported to function upstream of LC3B lipidation, while its exact mechanism remains unknown. 79 WIPI3 and WIPI4 were reported to function upstream of autophagy via regulating STK11/LKB1 (serine/threonine kinase 11)–AMPK (AMP‐activated protein kinase)–TSC2 (TSC complex subunit 2) axis. 32 , 80 Recently, it has been reported that WIPI3, similar to WIPI2, is also able to interact with ATG16L1 to promote LC3B lipidation. 75 Accumulating evidence from recent studies show that both WIPI3 and WIPI4 can interact with ATG2A via the WIR (WIPI‐interacting‐region) motif. 33 , 81 , 82 , 83 Further studies show that the WIPI3/WIPI4–ATG2A complex serve to establish the contact sites between ER and phagophore, enabling the transportation of lipids to promote phagophore expansion. 33 , 34 , 35 Moreover, WIPI3/WIPI4–ATG2A complex is also involved in autophagosome–lysosome fusion, 84 adding to the complexity of the WIPIs functions.
Up to date, how WIPI proteins are recruited to omegasome, the phagophore nucleating from a subdomain of the ER, is less understood. RAB11A (RAB11A, member RAS oncogene family) has been reported to recruit WIPI2 to PtdIns3P‐enriched membrane structures to mediate LC3B lipidation. 85 TRAPPC11 (trafficking protein particle complex subunit 11) has been shown to recruit WIPI4–ATG2A complex to promote phagophore expansion. 86 Whether WIPI3–ATG2A complex is recruited via a similar mechanism remains to be further tested.
2.1.4. The two ubiquitin‐like conjugation systems
The two ubiquitin‐like conjugation systems refer to the mATG8 and ATG12 systems in which mATG8 is conjugated to PE, while ATG12 is conjugated to ATG5 in a ubiquitination‐like manner. 38 , 39 mATG8 is firstly cleaved by ATG4 and thereby exposes a glycine residue. 87 , 88 Among the four isoforms of ATG4 reported, ATG4B is able to cleave all members of the mATG8, while ATG4A is more specific for GABARAPs. 36 , 37 Cleaved mATG8 is subsequently conjugated to PE with the action of ATG7 (E1), ATG3 (E2) and the ATG12–ATG5–ATG16L1 complex (E3). 4 In the ATG12 system, the covalent conjugation of ATG12 to ATG5 relies on ATG7 (E1) and ATG10 (E2), and the resultant ATG12–ATG5 conjugates further interact with ATG16L1 to form the ATG12–ATG5–ATG16L1 complex that acts as a E3‐like enzyme to conjugate mATG8 to PE. 40 mATG8 lipidation is essential for phagophore expansion and possibly closure. 89 , 90
While the recruitment of ATG16L1 has been reported to be mediated by WIPI2, it has been recently showed that ATG16L1 contains two membrane‐binding regions and can support mATG8 lipidation even in the absence of WIPI2. 40 In addition, ULK1‐mediated phosphorylation of ATG16L1 at S278 is important for this recruitment. However, whether this phosphorylation is important for ATG16L1 interaction with WIPI2 needs to be further investigated.
2.1.5. ATG9 vesicles
ATG9 is the only transmembrane protein of the autophagy core machineries and is predicted to contain six transmembrane helices. 43 There are two members of ATG9 in mammalian system including ATG9A and ATG9B, which are the main connection between the Golgi complex and autophagosome biogenesis. 91 ATG9 resides in the Golgi complex and follows the traffic via the endosomal system including early, sorting, and late endosomes. 44 , 45 As a result, the ATG9 vesicles are believed to be derived from the Golgi complex and the Golgi–endosomal system. 44 , 45 The ATG9 vesicles are mobilized to PAS by TRAPPIII (trafficking protein particle III) complex. 92 Phosphorylation of ATG9 at S761 is mediated differentially by ULK1 and AMPK under basal and stress conditions and is required for ATG9 localization at autophagosomal structures. 93
At present, the exact functions of the ATG9 vesicles have been one of the leading questions in the autophagy field and remain mysterious. The ATG9 vesicles are believed to contribute to autophagosome expansion via supplying the membrane source. 44 , 94 Such a notion is supported by the observations in yeast that ATG9 vesicles function as seeds for phagophore growth. 95 Further studies are needed to further understand the precise functions of ATG9 in autophagosome biogenesis.
2.2. Autophagosome–lysosome fusion
The final step of autophagy is the fusion between the double‐membrane autophagosome and lysosome, which leads to degradation of the autophagic cargo by the lysosomal hydrolases (Figure 1). 96 This process is regulated by a series of factors including: (i) phosphoinositides, such as PtdIns3P; (ii) small GTPases, such as RAB7; (iii) mATG8 proteins; (iv) tethering factors, such as the HOPS (homotypic fusion and vacuole protein sorting) tethering complex; (v) SNARE (SNAP (Soluble NSF Attachment Protein) receptor) complexes; (vi) motor adaptors, such as PLEKHM1 (pleckstrin homology and RUN domain containing M1). 97 Due to space restraint, we will focus the discussion on the tethering complex and the SNARE complex.
2.2.1. The HOPS complex
The HOPS complex is the core tethering complex for autophagosome–lysosome fusion. It consists of six subunits, including VPS11, VPS16, VPS18, VPS33A, VPS39, and VPS41. 98 All these subunits are able to interact with STX17 (syntaxin‐17), one of the key SNARE proteins present at the completed autophagosome. 99 On the other hand, RABs are small GTPases to mediate membrane fusion via binding to tethering factors. 100 Among them, RAB7 is the best studied for autophagosome–lysosome fusion. RAB7 is localized to late endosomes and lysosomes. 101 , 102 Importantly, RAB7 binds to two subunits of HOPS, VPS39, and VPS41. 103 , 104 Formation of the RAB7–HOPS complex is critical for membrane tethering to support SNAREs‐mediated autophagosome–lysosome fusion. 105
The molecular mechanisms underlying the recruitment of HOPS complex to autophagosome remain largely unknown. UVRAG (UV radiation resistance associated) is a regulatory subunit of PI3KC3‐C2 (the class III lipid kinase complex II) which is involved in autophagosome–lysosome fusion. 25 This complex has been shown to interact with and recruit the HOPS complex to autophagosome to promote autophagosome–lysosome fusion. 106 Moreover, a study from Dikic's laboratory reveals a RAB7 effector protein PLEKHM1 as a key positive regulator for the HOPS complex recruitment via simultaneous binding to LC3 and RAB7. 104 The exact function of the HOPS tethering complex in the process of autophagosome–lysosome fusion remains to be further investigated, especially in model organisms such as yeast and drosophila.
2.2.2. SNAREs
SNAREs are the main players in control of membrane‐mediated transport via vesicle fusion 107 and have been shown to be the core machinery for autophagosome–lysosome fusion. 108 Structurally, SNAREs can be classified into Q‐SNAREs (those having a Gln/Q residue) and R‐SNAREs (those having an Arg/R residue). 109 The Q‐SNAREs can be furthered classified into Q‐a, Q‐b, and Q‐c SNAREs based on the amino acid sequence within the SNARE domain. 110 , 111 The R‐SNARE and Q‐SNARE protein on separate membranes form the trans‐SNARE complex for membrane fusion. 108 For instance, the Qa‐SNARE protein STX17 localizes to the autophagosome, the R‐SNARE protein VAMP8 (vesicle‐associated membrane protein 8) localizes to lysosome/late endosome and by recruiting the Qbc‐SNARE protein SNAP29 (synaptosome associated protein 29), these SNAREs form the trans‐SNARE complex to mediate autophagosome–lysosome fusion. 108 Other trans‐SNARE complexes, including STX17–SNAP29–VAMP7 and YKT6 (YKT6 v‐SNARE homolog)–SNAP29–STX17, are also reported to regulate autophagosome–lysosome fusion. 97
In addition to HOPS and SNAREs, one key ATG, ATG14, has also been shown to play a key role in regulating autophagosome–lysosome fusion: ATG14 binds to STX17, which contributes to stabilization of STX17–SNAP29 complex and promotes membrane tethering and enhancing membrane fusion. 112 Given that phosphorylation of STX17 regulates its functions 113 and that ATG14 is a key component of the ULK1 complex, 50 it would be interesting to test whether ULK1 plays a role in control of STX17 phosphorylation and functions. O‐linked beta‐N‐acetylglucosamine (O‐GlcNAc) modification of SNAP29 has been reported to undermine its association with STX17, which inhibits the formation of the trans‐SNARE complex. 114 Of note, O‐GlcNAcylation of SNAP29 is controlled by nutrient status and is reduced by starvation. 114 More studies are needed to better understand how precisely SNAREs are recruited and how their functions are regulated in the course of autophagosome–lysosome fusion.
2.3. Upstream regulators of autophagy
MTORC1 (mammalian target of rapamycin complex 1) and AMPK are the two key regulatory factors that work upstream in control of autophagy. In the following sections, we will focus on these two key protein kinases and their roles in regulation of autophagy.
2.3.1. MTORC1
MTOR is a serine/threonine protein kinase belonging to PI3K‐related protein kinases family that responds to growth factor and nutrients. 115 In mammalian cells, MTOR forms two distinctive complexes known as MTORC1 and MTORC2, with MTOR as the catalytic subunit. 116 MTORC1 is assembled by MTOR, MLST8 (MTOR‐associated protein, LST8 homolog), RAPTOR (regulatory associated protein of MTOR complex 1), DEPTOR (DEP domain‐containing MTOR‐interacting protein), PRAS40 (proline‐rich Akt substrate 40), and FKBP12/FKBP1A (FKBP prolyl isomerase 1A), while MTORC2 is assembled by MTOR, MLST8, DEPTOR, RICTOR (RPTOR independent companion of MTOR complex 2), MAPKAP1 (MAPK associated protein 1), and PRR5/5L (proline rich 5/5L). 46 , 116
The role of MTORC1 in autophagy has been extensively studied over the past decades and it has been revealed as a crucial negative regulator for autophagy. MTORC1 can be activated via various stimuli. For example, in response to amino acid, MTORC1 is activated by the GTP‐bound RHEB (ras homolog enriched in brain) and lysosomal translocation via a protein complex called regulator and V‐ATPases. 115 In response to growth factor such as insulin, PIK3CA–AKT pathway is activated, which then inhibits TSC1/2 (tuberous sclerosis 1/2) and release RHEB (Ras homolog enriched in brain), a small GTPase, from this complex. Subsequently, the active GTP‐bound RHEB mediates the activation of MTORC1. 117 MTORC1 suppresses autophagy mainly through the following mechanisms: (i) phosphorylation of the autophagosome core machineries including ULK1 (S637/S757), ATG13 (S258), ATG14 (S3/S383/S440/T233), NRBF2 (S113/S120), and WIPI2 (S395) to inhibit autophagosome biogenesis 118 ; (ii) phosphorylation of TFEB (transcription factor EB) (S122/S142/S211) to transcriptionally suppress autophagy. 119 As a result, various MTOR inhibitors (such as rapamycin) have been reported to effectively induce autophagy. 120
2.3.2. AMPK
AMPK is a serine/threonine kinase and exists as an obligate heterotrimer consist of a catalytic subunit (α) and two regulatory subunits (β and γ). 121 AMPK is an important cellular energy sensor that can be activated by phosphorylation at T172 mediated by STK11/LKB1 (serine/threonine kinase 11) upon glucose deprivation. 122 AMPK positively regulates autophagy via multiple mechanisms. First, AMPK inhibits MTORC1 directly via phosphorylating RAPTOR (S722). 123 Second, AMPK activating TSC2 via phosphorylation (T1227/S1345), thereby inhibiting MTORC1 indirectly. 124 Third, AMPK promotes autophagy by activating the ULK1 complex or PI3KC3‐C1 via phosphorylating ULK1 (S317/S777), 125 BECN1 (S91/S94/T388), 126 , 127 and ATG13 (S224). 128
Interestingly, AMPK‐mediated phosphorylation of other targets could also inhibit autophagy. For instance, phosphorylation of PIK3C3 (T163/S165) by AMPK suppresses the production of PtdIns3P, which could be reversed by overexpression of ATG14. 127 Moreover, it has been reported that WIPI4–ATG2A complex interacts with AMPK under fed condition, while upon glucose deprivation, WIPI4–ATG2A complex disassociates from AMPK and is recruited to nascent autophagosome. 32 While the biological implication of such interaction has not been studied, findings from this study raised the possibility that AMPK may inhibit the WIPI4–ATG2 complex function via phosphorylation under fed condition.
3. IMPLICATION OF AUTOPHAGY IN HEALTH AND DISEASES
As one of the most intricate processes in cells, the roles of autophagy has been very fascinating and is not limited to a basic starving response, but also extended to various “controversial pair” roles like “antiapoptotic versus proapoptotic,” 129 “antibacterial versus probacterial,” 130 , 131 and “antitumorigenic versus protumorigenic.” 132 , 133 As a result, dysregulation of autophagy is closely associated with various human diseases. 16 In this section, we will discuss the recent insights into the role of autophagy in the most pervasive human diseases, including cancer, neurodegenerative diseases, metabolic diseases, viral infections, cardiovascular diseases, and aging. Understanding the implication of autophagy in these diseases is crucial for moving autophagy research from bench to bed for clinical applications in diagnosis and therapy.
3.1. Autophagy in cancer
Autophagy has long been considered as a double‐edged sword in cancer. At the early initiation stage, autophagy acts as tumor suppressor via removal of potentially harmful cytosolic contents and damaged organelles, thereby avoiding cell injury such as DNA mutation. At the stage of progression, autophagy acts as a survival mechanism to sustain tumor viability under stressful microenvironment, which also contributes to therapeutic resistance. Therefore, in‐depth understanding of the roles of autophagy during various stages of carcinogenesis and in tumor therapeutic responses will provide important therapeutic strategies to eliminate cancer cells, reverse drug resistance and prevent recurrence.
3.1.1. Autophagy in cancer initiation and progression
The discovery of a monoallelic deficiency of the key autophagy gene BECN1 in breast, ovarian, and prostate malignancies led to the hypothesis that autophagy is a tumor suppressive mechanism. 134 , 135 Such hypothesis is supported by studies in cell‐based assays and animal models which showed enhanced tumorigenesis via genetical suppression of autophagy. 136 , 137 Consistently, pharmacological interventions to block autophagic flux also results in the appearance of early tumor lesions in various preclinical tumor models. 138 The anticancer effects have been linked to the roles of autophagy in various cellular processes, including maintaining genomic integrity, suppressing oxidative stress, and inhibiting NRF2/NFE2L2 (nuclear factor (erythroid‐derived 2)‐like 2) overactivation. 16 For example, Holdgaard et al. 139 identified selective autophagy as an essential centrosome‐regulating process in mitosis. Moreover, increasing evidence supports the notion that the oncosuppressive functions of autophagy are partly due to its ability to attenuate the inflammatory response and counteract the establishment of an inflammatory milieu. 140 , 141 , 142 , 143
Different from the above studies, there are reports showing that autophagy promotes initiation, progression or adaptive responses in tumor cells, indicating the protumor function of autophagy. For instance, DeVorkin et al. 144 found that tumor development was greatly slowed in mice deficient in Atg5, Atg14, or Atg16L1. Yamamoto et al. 145 discovered that inhibiting autophagy enhances antitumor T cell responses and therefore suppresses tumor growth. Vera‐Ramirez et al. 146 reported that autophagy is a crucial strategy for the survival of disseminated dormant breast cancer (BC) cells, while inhibition of autophagy in quiescent BC cells causes apoptosis. These conflicting functions may reflect a fact that the exact role of autophagy in cancer depends on an array of factors, including the stage of the tumor, the type of the tumor and genetic context of the host such as the mutated status of p53. 147
3.1.2. Autophagy in tumor immunosurveillance
The roles of autophagy in preventing malignant transformation have been linked to its functions in regulation of the microenvironment and activation of immunosurveillance (Figure 2).
FIGURE 2.
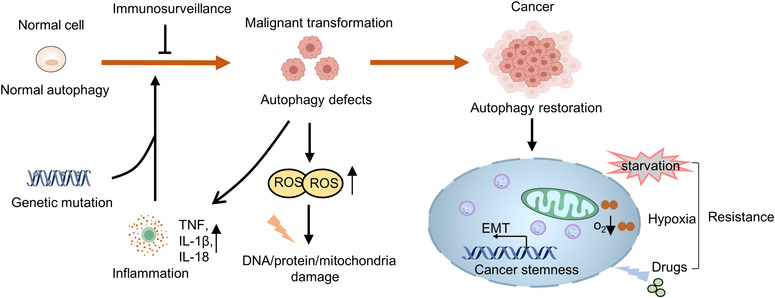
The implication of autophagy in carcinogenesis. Autophagy may act as a powerful barrier to prevent transformation of normal cells into tumor cells. Upon autophagy deficiency, misfold proteins or dysfunctional organelles accumulates, which may cause genomic defects that accompanies transformation. On the other hand, autophagy acts as a mechanism for transformed cells to get adapted to various cellular stress responses, promote metastasis, and maintain tumor stemness
As autophagy is essential for the removal of cytotoxic contents to maintain the intracellular homeostasis, impaired autophagy may result in accumulation of cytotoxic factors and the subsequent formation of premalignant microenvironment. 148 One good example to support such notion is the generation of reactive oxygen species (ROS) due to impaired autophagy. 149 ROS causes cell injury via multiple mechanisms such as modification and dysfunctions of proteins, DNA or lipids, and increased risk of DNA mutation, all of which increase the risk of malignant transformation. 150 Additionally, ROS is also involved in production of proinflammatory cytokines such as tumor necrosis factor. 140 As autophagy is a main pathway for inflammasomes degradation, impairment of autophagy further exacerbates inflammation via promoting maturation and secretion of proinflammatory factors such as IL‐Iβ and IL‐18. 151 Moreover, excessive ROS also impairs mitochondria and promotes the release of mitochondrial DNA, which also induces the expression of inflammation‐related genes via activating the cGAS (cyclic GMP–AMP synthase)–STING (stimulator of interferon response cGAMP interactor 1) signaling pathway. 152 , 153 Chronic inflammation has been found as a key factor that contributes to malignant transformation. These findings together suggest the importance of autophagy in fine‐tuning inflammation and prevents the formation of pre‐malignant microenvironment.
Elimination of malignant cells by immune cells is another key part of immunosurveillance. Autophagy indeed plays multiple roles in regulating immune cell functions. First, autophagy is actually involved in renewal, differentiation, and homeostasis of immune cells, including dendritic cells (DCs) and T cells. 154 , 155 Second, autophagy also modulates MHC class I and class II antigen presentation to promote cancer cell death. 156 Third, autophagy is also essential for long‐term survival of CD8+ cytotoxic T lymphocytes upon acquisition of a memory phenotype. 157 Consistently, defective autophagy is associated with reduced immune response. 158 , 159 The roles of autophagy in immunosurveillance may thus contribute to prevention of malignant transformation.
3.1.3. Autophagy in cancer stem cells
Cancer stem cells (CSCs) refer to a group of cancer cells that possess characteristics of stem cells with an ability to form a new cancer. CSCs are usually featured with high potency in proliferation, resistance to therapeutics, and are responsible for metastasis and relapse of cancer. 160 Determining the molecular mechanisms underlying CSC survival is therefore critical for overcoming the current challenges in anticancer therapies. Although the concept of CSCs is still in dispute, over the recent decade, the functional implication of autophagy has been suggested in CSCs from a wide variety of tissues, including pancreas, liver, 161 breast, 162 ovarian, 163 and brain. 164 , 165 In most cases, it is believed that autophagy defects impair the self‐renewal capacity of CSCs. 166 Furthermore, autophagic flux has been revealed to be significantly higher in CSCs than in bulk tumor cells. 162 , 167 Consistently, increased autophagic flux has been linked to CSCs‐mediated tumorigenesis, such as the development of leukemia 168 and BC. 162 Furthermore, autophagy can regulate the stemness of tumor cells, 169 and suppression of autophagy with chloroquine (CQ) reduces the CSCs populations and thereby sensitizes cancer cells to chemotherapy. 170 Intriguingly, another study shows that autophagy suppresses the CSCs properties of glioma cells. 171 As autophagy and tumor stemness maintenance are closely related to metastasis and chemoresistance, targeting autophagy and CSCs may be the key direction to prevent tumor resistance and recurrence.
3.2. Autophagy in neurodegenerative diseases
Autophagy is essential for maintaining the homeostatic demands of neurons, both at the level of the central and peripheral nervous systems. 172 , 173 Based on the observations that neurodegenerative disorders occur in autophagy‐defective mice, it has been hypothesized that autophagy defect is an important etiological factor for neurodegenerative diseases in humans. Pathologically, most neurodegenerative diseases are associated with accumulation of protein aggregates, including mutant α‐synuclein in Parkinson disease (PD), Aβ and C‐terminal fragments of the amyloid precursor protein (APP) in Alzheimer disease (AD), pathogenic mutant huntingtin (mHtt) in Huntington disease (HD), and mutant SOD1 (superoxide dismutase 1) as well as TDP‐43/TARDBP (TAR DNA binding protein) in amyotrophic lateral sclerosis (ALS). 172 , 174 These protein aggregates are toxic drivers of neurological lesions and are supposed to be degraded by the autophagy–lysosome pathway. 172 Consistently, gene mutations of autophagic receptors (e.g., SQSTM1 (sequestosome 1), OPTN (optineurin), NBR1 (neighbor of BRCA1 gene 1)) are closely associated with neurodegenerative diseases. 175 , 176 , 177 As a result, modulating autophagy is believed to be a promising strategy for treating neurodegenerative diseases. For example, autophagy activation of the aggregating receptor SQSTM1 promotes the clearance of mHtt, insoluble tau, and Aβ42. 178 , 179 Conversely, inhibition of autophagy with 3‐methyladenine (3‐MA) results in accumulation of mHtt aggregates. 180 , 181 Here, we review the recent advances in understanding the pathogenesis of neurodegenerative diseases associated with defective autophagy and discuss the therapeutic interventions for these diseases via targeting autophagy.
3.2.1. Alzheimer disease
AD, the most common neurodegenerative disorder, is characterized by the progression from episodic memory problems to severe cognitive decline. 182 , 183 So far, no effective therapy is available to block or slow down AD progression, and the exact mechanisms underlying the disease remain mysterious. 184 Intracellular microtubule associated protein tau (MAPT)/tau tangles and extracellular beta amyloid peptide [Ab] plaques in the brains of patients with AD are the early pathological features that gradually lead to neuronal cell death and cognitive decline. 17 In fact, the connections between autophagy and AD originated from the observation of accumulated immature autophagic vacuoles in AD brains. 185 Multilayered brain proteomic analysis revealed that the autophagy cargo receptor SQSTM1 accumulates in AD, suggesting impaired autophagic flux. 186 Moreover, functional autophagy is required for removal of soluble and aggregated variants of MAPT/tau. 187 , 188 Consistently, genetic variations of CTSD (cathepsin D), a lysosomal peptidase involved in clearance of aggregated proteins such as tau, is associated with an increased risk of AD. 189 Clearly, these studies indicate the important role of autophagy in AD via targeting MAPT/tau for lysosomal degradation. Interestingly, accumulating MAPT/tau tangles can in turn perturb the retrograde axonal transport of autophagosomes by interfering with the dynein–DCTN (dynactin) complex, ultimately triggering the deleterious accumulation of MAPT/tau‐containing autophagic vesicles. 190
Aβ originates from the processing of its APP and its accumulation is also a hallmark of AD. 191 Autophagy is known to play an important role in Aβ quality control. First, it has been found that induction of ATG5‐dependent autophagy enhances the degradation of APP. 192 Second, autophagy also modulates Aβ clearance. Enhancing autophagy by chemical reagents or genetic engineering techniques significantly reduces the deposition of intracellular Aβ and extracellular amyloid in mice brain and improves cognitive ability. 193 , 194 , 195 Moreover, activation of autophagy reduces the burden of Aβ plaque in rodents. 172 , 196 , 197 Third, it has been revealed that autophagy is not only required for intracellular degradation of Aβ via lysosome, but also for Aβ secretion. 198 , 199 These results thus suggest that autophagy plays multiple roles in intracellular Aβ clearance.
In the course of AD pathogenesis, accumulated Aβ leads to ROS production which causes mitochondrial damage, and mitophagy is the essential mechanism to eliminate damaged mitochondria. 14 , 183 , 200 In fact, mitochondrial dysfunction is also defined as one of the early pathological features of AD. This is supported by the evidence from AD postmortem brain tissues that mitochondrial dysfunction is one of the initial pathological events in AD. 201 Similarly, mitophagy is impaired in induced pluripotent stem cell‐derived human AD neurons, animal AD models, and the hippocampus of AD patients. 183 , 202 As a result, mitophagy activation alleviates both Aβ and tau pathologies and improves the cognitive functions of Caenorhabditis elegans and mouse AD models. 203 Although further investigation is required to elucidate the relationship between mitophagy and AD pathogenesis, mitophagy‐mediated clearance of dysfunctional mitochondria appears to be a promising therapeutic target for AD intervention.
3.2.2. Parkinson disease
PD is the second most common neurodegenerative disease. 204 PD is pathologically defined by the loss of dopaminergic neurons in the substantia nigra (SN) and the prevalence of proteinaceous Lewy bodies, characterized by pathological α‐synuclein inclusions in the dopaminergic neurons of the SN. 17 , 205 It is well known that α‐synuclein carrying pathogenic mutations are degraded by the autophagy‐lysosome system. 206 Knockout of Atg7 induces an age‐dependent increase in the formation of Sqstm1 α‐synuclein inclusion bodies in dopaminergic neurons of aged mice, leading to motor dysfunction. 207 Conversely, α‐synuclein inclusions impair the autophagic pathway. 208 , 209 , 210 , 211 These studies collectively suggest the close correlation between defective autophagy and α‐synuclein aggregates accumulation (Figure 3).
FIGURE 3.
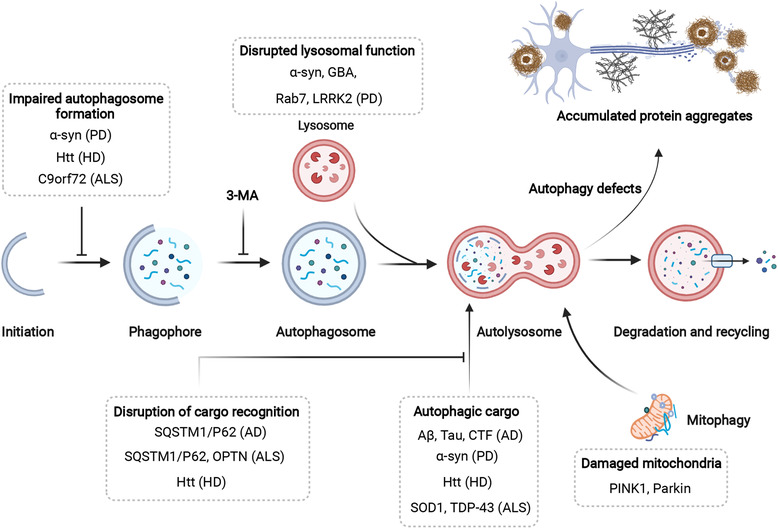
The mechanisms and therapeutic targets of autophagy in neurodegenerative diseases. An increasing number of genes associated with neurodegenerative diseases, especially AD, PD, HD, and ALS, act at different steps throughout the autophagic process. Their proposed sites of action, as well as associated neurodegenerative diseases are indicated. The protein aggregates, supposed to be degraded by the autophagy–lysosome pathway, accumulate and contribute to neurodegenerative diseases when autophagy is defective. AD, Alzheimer's disease; PD, Parkinson's disease; HD, Huntington's disease; ALS, Amyotrophic lateral sclerosis
Glucocerebrosidase (GBA) is a lysosomal enzyme for degrading glucosylceramide, and mutations in GBA are one of the most common genetic risk factors for PD. 212 PD‐associated GBA mutations (N370S and L444P) reduced its protein levels and enzymatic activity and impaired its trafficking from the ER to the lysosomes, leading to ER stress, accumulation of lysosomal lipids, and ultimately autophagy–lysosome dysfunction. 213 , 214 It has been demonstrated that the GBA interacts with α‐synuclein to promote α‐synuclein degradation and prevent α‐synuclein aggregation, while lysosomal membrane‐bound α‐synuclein is able to inhibit lysosomal functions and further exacerbates PD. 215 , 216
Besides, mutations in LRRK2/PARK8 (leucine‐rich repeat kinase 2) are genetically linked to autosomal‐dominant PD and have also been extensively studied for its involvement in aberrant autophagy. More than 40 pathogenic LRRK2 mutations have been reported in patients with PD. 217 Loss of LRRK2 function leads to striking defects in the endolysosomal and autophagy pathways. 218 Interestingly, many pathogenic mutations in LRRK2 are gain‐of‐function mutations, such as G2019S and R1441C, which increase its kinase activity but impair autophagic degradation, similar to LRRK2 deficiency. 219 , 220 , 221 Further studies are needed to understand the exact roles of LRRK2 in autophagy and its implication in PD pathogenesis.
Mutations in LRRK2, α‐synuclein, and GBA contribute to increased or impaired autophagy, but they also contribute to mitochondrial dysfunction. 204 It had been shown that patient cells with G2019S mutation in LRRK2 showed an increase in the number of mitochondrial fragments and a decrease in mitochondrial membrane potential. 222 , 223 Ryan et al. 224 demonstrated that neurons expressing mutant α‐synuclein also exhibit fragmented mitochondria, as well as exposed cardiolipin on outer mitochondrial membrane, which initiates mitophagy. Patients with GBA1 deficiency and mouse models of GBA1 knockout exhibit mitochondrial dysfunction. 225 , 226 , 227
Given the close association of mitochondrial dysfunction with PD, defective mitophagy (selective autophagy of damaged mitochondria) has emerged as one of the main causes contributing to PD progression. 228 Mutations in the genes encoding for PINK1 (PTEN (phosphatase and tensin homolog)‐induced putative kinase‐1) and Parkin have been found in some familial forms of PD. 229 The field of PINK1–Parkin‐dependent mitophagy has significantly expanded over this decade. In this pathway, PINK1 is stabilized and accumulates on the mitochondria upon mitochondria depolarization. Subsequently, ubiquitin phosphorylated by PINK1 at Ser65 serves as docking sites for Parkin binding, which partly activate Parkin E3 ligase activity. 230 , 231 , 232 Subsequent phosphorylation of Parkin by PINK1 within its ubiquitin‐like domain fully activates its E3 ligase activity. 233 , 234 In this way, PINK1 and Parkin form a feedforward loop to amplify the mitophagy signal to mediate clearance of damaged mitochondria. 235 , 236 Impairment of PINK1–Parkin signaling pathway leads to accumulation of damaged mitochondria, which causes oxidative stress and toxic burdens that could lead to neuronal cell death. 228 A recent study from Youle's group 237 reports a strong inflammatory phenotype in both the PINK1−/− and Parkin−/− mice due to the release of mtDNA and the subsequent activation of the cGAS–STING pathway. While the exact roles of PINK1 and Parkin in the pathogenesis of PD is still not well understood given the fact that mice lacking either PINK1 or parkin do not develop substantial PD‐relevant phenotypes, this study implies that PINK1–Parkin‐dependent mitophagy mitigates PD via fine‐tuning innate immunity response.
3.2.3. Huntington disease
HD is an incurable, autosomal‐dominant progressive neurodegenerative disease, which is mainly manifested by cognitive dysfunction, behavioral disturbances, and severe motor dysfunction. 238 HD is caused by the expansion of the CAG repeat within a single gene huntingtin (HTT), which encodes a large protein with an extended polyglutamine (polyQ) tail. 239 The abnormal expansion of a polyQ repeat in exon 1 produces mHtt proteins, leading to cytotoxicity in the striatum and cortex and inducing progressive motor deficits accompanied by the accumulation of autophagosomes. 240 , 241 There is substantial evidence that autophagy is dysfunctional in HD. For example, it has been found that expression of the truncated N‐terminal huntingtin fragment (HTT552) activates the autophagy/lysosomal degradation pathway via increasing LC3‐II, BECN1, and CTSB/CTSL expression. 242 Importantly, significant differences have emerged between the functions of wild‐type and mutant HTT in regulating the autophagy process. 243 Compared with wild‐type HTT, mutant HTT has multiple roles in autophagy inhibition. 244 For instance, Martinez‐Vicente et al. 245 found that mutant HTT negatively affects autophagosomal cargo recognition through dysregulated recruitment of SQSTM1. Rui et al. 246 demonstrated that mutant HTT disrupts the ability of HTT to bind and activate ULK1.
In fact, mHtt aggregation status and HD progression can be modulated by autophagy. 179 , 247 Through a mouse model, it has been reported that heterozygous loss of the autophagy adaptor protein Alfy/Wdfy3 (WD repeat and FYVE domain containing 3) impedes the clearance of mHtt and promotes HD progression. 248 In an unbiased in vivo genome‐wide screening, Wertz et al. 249 found that many ATGs appear to prevent mHtt toxicity and HD progression. Taken together, identifying the precise regulatory mechanisms of autophagy in HD will be essential for the development of rational therapeutic interventions via targeting autophagy.
3.2.4. Amyotrophic lateral sclerosis
ALS is a rare and devastating neurodegenerative disease, characterized by the selective death of motor neurons controlling the voluntary muscles. 250 , 251 Pathologically, ALS is featured by cytoplasmic ubiquitin‐positive inclusion formation in the brain and spinal cord and the aberrant amassing of misfolded proteins including SOD1 and TDP‐43. 17 Besides, inherited genetic mutations in TDP‐43, sarcoma (FUS), and UBQLN2 (ubiquilin 2) also cause ALS. 252 , 253 Numerous studies suggest that autophagy is closely implicated in the pathogenesis of ALS. For example, autophagy induction mitigates ALS via TDP‐43 clearance. 254 Aggregation‐prone SOD1 and TDP‐43 fail to be disposed of upon mutation of SQSTM1. 175 , 255 Moreover, SQSTM1 was also found to colocalize with TDP‐43 inclusions and its deficiency exacerbated ALS symptoms caused by the insoluble protein aggregates. 256 , 257 UBQLN2, a genetic risk factor for ALS, is also closely associated with autophagy. It has been reported that UBQLN2 forms a complex with LC3 and facilitates autophagosome–lysosome fusion. 258 Besides, Chen et al. 259 demonstrated that selectively expressing mutant UBQLN2P497H in motor neurons compromises autophagy–lysosome fusion and is sufficient to trigger ALS progression in rats. Conversely, interventions to activate autophagy, such as depletion of the transcription factor XBP1 (X‐box binding protein 1) and pharmacological modulation of HSPB8 (heat shock protein family B (small) member 8) expression in the nervous system, counteract ALS symptomatology by promoting autophagic clearance of SOD1. 260 , 261 These studies clearly suggest the importance of autophagy in the clearance of misfolded proteins that contribute to ALS progression.
In addition, C9orf72 (hexonucleotide repeat amplification of the intron of Chromosome 9 ORF 72) is the most common genetic cause for ALS. 262 Compared with healthy individuals, ALS patients typically carry more than 100 hexanucleotide repeats. 263 Although the cellular function of C9orf72 remains unclear, increasing evidence has suggested that C9orf72 affects ALS pathogenesis by modulating autophagy via various mechanisms. It has been reported that C9orf72 functionally interacts with multiple members of the Rab small GTPases family to positively regulate autophagy, while loss of C9orf72 impairs autophagosome biogenesis in neuronal cells. 264 C9orf72 is also reported to be associated with the autophagy core machinery such as the RB1CC1–ULK1 complex to promote autophagosome biogenesis. 265 As a result, deficiency of C9orf72 also leads to autophagic degradation blockade, leading to accumulation of cytotoxic contents and ultimately neuronal cell death. 266 , 267 These results suggest an important role of C9orf72 in control of autophagy and its implications in ALS pathogenesis.
3.3. Autophagy in metabolic diseases
Overnutrition and reduced energy expenditure, mirrored by aberrant activation of the trophic axis (e.g., insulin signaling), contribute to the development of metabolic diseases such as obesity, insulin resistance, and type 2 diabetes (T2D). Mechanistically, excess fat accumulation causes insulin resistance and elevated serum free fatty acid levels, leading to systemic lipotoxicity and β‐cell dysfunction. 268 In fact, autophagy responds to minimal oscillations in intracellular and extracellular metabolism, thereby maintaining a tightly regulated balance between the anabolic and catabolic pathways. 269 For instance, the essential molecular players of cellular energy status, such as MTORC1 and AMPK, are involved in nutrient deprivation‐induced autophagy. Autophagy performs inherent metabolic tasks in major organs such as adipose tissue, liver, and the exocrine pancreas and participates in maintaining energy balance in the body. 270 As a result, dysregulated autophagic flux contributes to the pathogenesis and progression of metabolic diseases. In this section, due to space limitation, we will focus on the implications of autophagy in obesity and T2D.
3.3.1. Obesity
As a metabolic disorder, obesity is clinically manifested by an excessively sustained positive energy balance. An overly simplistic view holds that the pathogenesis of obesity underlies the dramatic accumulation of autophagic substrates such as lipid droplets, protein aggregates, and damaged mitochondria, and that a defective autophagic process accelerates obesity development. 271 In fact, this view is easily confuted because multiple intracellular and extracellular factors are closely related to the pathogenesis and progression of obesity.
At present, there is evidence indicating that autophagy is repressed under obesogenic conditions. For instance, in mice with prolonged feeding of a high‐fat diet, the expression levels of Atg5 and Atg7 in the liver are significantly downregulated, indicating that autophagy is impaired. 272 Consistent with this finding, an obesity‐induced increase in cytoplasmic calcium concentration in hepatocytes impedes fusion between autophagosomes and lysosomes. 273 Conversely, restoring the expression levels of Atg7 and Tfeb in liver prevents weight gain and metabolic syndrome in diet‐induced and genetically obese mouse models. 272 , 274 Furthermore, autophagy can also contribute to appetite. It has been revealed that hypothalamic inhibition of autophagy increased energy intake and reduced energy expenditure. 275 However, genetic obliteration of Atg7 in hypothalamus contributes to a lean phenotype. 276 Intriguingly, autophagy is involved in the conversion of white adipose tissue into brown adipose tissue. Upon adipose‐specific deletion of Atg7, the white adipose tissue decreases, metabolism is enhanced, and these mutant mice obtain a certain resistance to obesity caused by high‐fat diet. 277 Besides, autophagy can also influence the obesogenic phenotype through inflammatory reactions. 278 , 279 An in‐depth investigation regarding the specific role of autophagy in the pathogenesis of obesity is necessary in order to fully exploit its therapeutic potential in prevention and treatment of obesity.
3.3.2. Type 2 diabetes
T2D clinically manifests with the appearance of insulin resistance and relative insulin deficiency (Figure 4A). Oxidative stress, inflammation, ER stress, and mitochondrial dysfunction in pancreatic β‐cells, liver, skeletal muscle, and adipose tissue are closely related to the pathogenesis of T2D. 268 In addition, aging is also a recognized risk factor for T2D. Notably, autophagy appears to have etiological significance (Figure 4B). On the one hand, autophagy‐deficient and insulin‐responsive tissues fail to effectively alleviate oxidative stress and ER stress caused by persistent stimulation of insulin. 271 , 280 On the other hand, the homeostatic functions of pancreatic β‐cells require the participation of autophagy. 281 , 282 It has been demonstrated that genetic ablation of Atg7 in β‐cells results in islet degeneration and impaired glucose tolerance, accompanied by reduced insulin secretion, suggesting that autophagy is necessary to maintain the structure, mass, and function of pancreatic β‐cells. 281 , 282 Besides, Yamamoto et al. 280 revealed that autophagy hyperactivation enhances insulin signaling, but the insulin storage and secretion as well as glucose tolerance could be reduced by selective sequestration and degradation of insulin granule vesicles by autophagy in β‐cells. Unlike most cell types, short‐term starvation in pancreatic β‐cells triggers nascent granule degradation and Golgi membrane‐associated degradation mechanisms and it inhibits autophagy via activation of MTOR, thereby inhibiting insulin secretion and preventing hypoglycemia under physiological starvation conditions. 283 , 284
FIGURE 4.
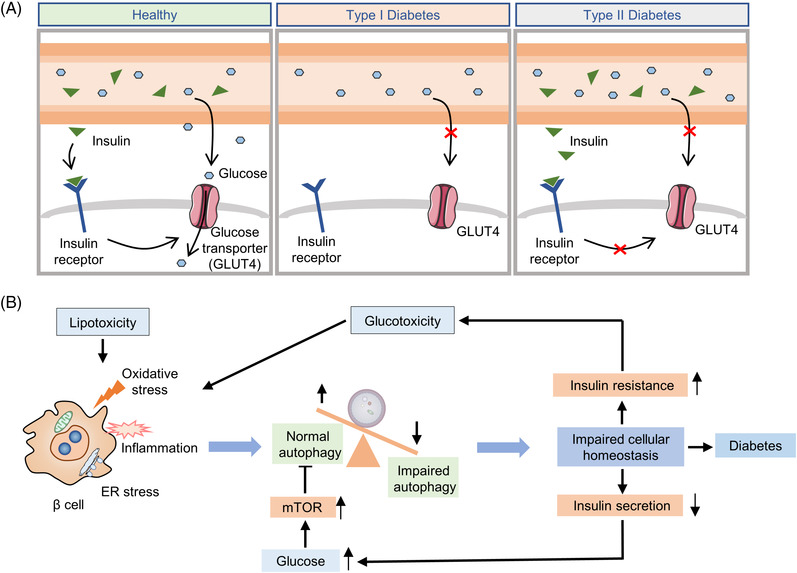
Pathogenesis of diabetes and the implication of autophagy in diabetes. (A) The schematic diagram of type I diabetes and type II diabetes; (B) the effects of insulin resistance and insulin secretion on β‐cell autophagy. Various stress and inflammatory responses to lipotoxicity can induce polyubiquitination of proteins and cytoplasmic aggregation of damaged organelles in β‐cells. Impaired autophagy leads to cellular dysfunction, affects insulin secretion, results in insulin resistance, and promotes diabetes progression
Given the established role of autophagy in maintaining β‐cells homeostasis under stressful conditions, intervention in autophagy early in the development of diabetes may help patients retain β‐cells mass and prevent disease progression. 285 Recently, Cheng et al. found that a simulated fasting diet promoted β‐cells regeneration in a mouse model of T2D. This effect can be mimicked by simultaneous inhibition of the autophagy negative regulators MTORC1 and PKA (protein kinase A), suggesting that autophagy may play a role in islet reprogramming. 286 Indeed, mimicking glycolipid toxicity in cultured β cells can inhibit autophagy by reducing lysosome‐related gene expression, impairing autophagosome–lysosomal fusion, or reducing lysosomal acidification. 287 , 288 , 289 Furthermore, activated autophagy under stress conditions and blockade of autophagic flux leads to the accumulation of defective lysosomes, causing β‐cells death. 290 Metformin, rosiglitazone and GLP‐1 mimetics are highly effective drugs for treatment of diabetes and have been shown to effectively induce autophagy in β cells. 291 As a key inhibitor of autophagy, a large number of studies have been conducted to explore whether MTORC1 can be inhibited as a strategy for diabetes treatment. 292 , 293 , 294 Overall, caution and thoughtful design should be taken into consideration for diabetes therapies via targeting autophagy. Further studies are needed to elucidate the mechanisms of autophagy in cell survival and apoptosis to develop therapeutics that can selectively promote β‐cells survival.
3.4. Autophagy in viral infection
One key aspect of the biological functions of autophagy is its regulatory role in pathogen infection, inflammation, and immunity. So far, the relationship between autophagy and infection has been extensively studied and there are rather complicated relationships between these two important processes. 295 , 296 First, autophagy can target invaded pathogens such as bacteria and viruses for clearance. Autophagy thus serves as a potent anti‐infection and anti‐inflammatory mechanism. Second, certain strains of microbes can make use of the autophagy machinery or process to survive, replicate and resist to the immunity. Autophagy thus acts as a proinfection and proinflammatory mechanism. Third, reciprocally invaded microbes have diverse impacts on the autophagy process in the host cells, either promotes or inhibits autophagy in a context‐dependent manner. Fourth, autophagy can directly or indirectly regulate both innate and adaptive immunity and the associated inflammatory processes. And finally, targeting autophagy (either promoting or inhibiting) has been an attractive approach in treatment of various forms of infections and related infectious diseases. 297 , 298 , 299
At present, all of the above points have been extensively discussed in literature. In this review, we will mainly focus on the implication of autophagy in coronavirus disease of 2019 (COVID‐19) caused by SARS‐CoV‐2. Since the outbreak at the end of 2019, SARS‐CoV‐2 has quickly spread to the whole globe, causing a major pandemic with severe loss to both life and economy. Based on WHO (https://covid19.who.int/), up to date (April 20, 2022), there are more than 500 million confirmed cases, with more than 6.2 million deaths worldwide. Despite the enormous efforts in control of this pandemic, including vaccination and lockdown of major cities around the world, there is no clear sign of control, especially with the emerging variants with increased transmissibility and pathogenicity. 300 , 301
Biologically, SARS‐CoV‐2 is a type of RNA viruses with a positive‐sense single‐stranded RNA genome. Its genome encodes 11 genes with 14 open reading frames (ORFs) that produce 16 nonstructural proteins, four structural proteins, and nine accessory proteins. 302 As shown in Figure 5, the replication process of SARS‐CoV‐2 has also been well established, consisting of the following six steps 303 , 304 : (i) binding: the virus first binds to the host cell surface via the interaction between the viral S protein and host receptor angiotensin‐converting enzyme‐2 (ACE2); (ii) endocytosis: the virus enters into the host cell's endocytic pathway (endosomes and lysosomes) via fusion of viral membrane with the host cell membrane; (iii) transcription and translation of viral proteins from the viral RNAs; (iv) viral replication within the replicative membranous compartment; (v) nucleocapsid packaging: virus assembly and package at the ER and/or the Golgi complex; and finally (vi) budding and egress: release of new virions via exocytosis.
FIGURE 5.
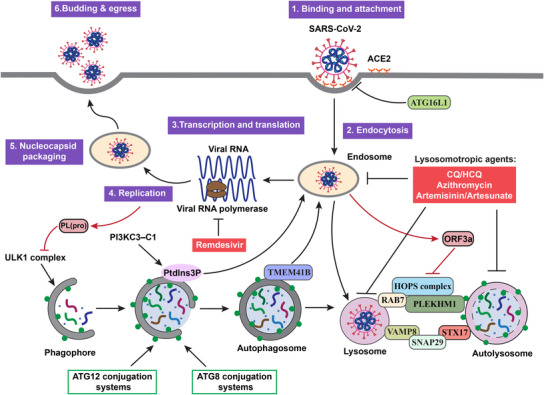
Targeting the autophagy‐lysosome pathway as novel therapeutic strategies for COVID‐19. The replication cycle of SARS‐CoV‐2 consists of six consecutive steps, as well as the cross‐talks of this cycle with the autophagy–lysosome pathway are presented. The red boxes indicate that the lysosomotropic agents targeting the autophagy–lysosome pathway are under development as therapeutics against COVID‐19
One important research topic in the study of SARS‐CoV‐2 and COVID‐19 is the role of the autophagy and lysosome system in the pathogenicity of this deadly disease. Up to date, there is accumulating evidence demonstrating the reciprocal nature of interaction between autophagy and SARS‐CoV‐2 and COVID‐19. 305 , 306 , 307 , 308 Here, we will provide an update on the effect of autophagy on SARS‐CoV‐2 and COVID‐19 and the reciprocal effects of SARS‐CoV‐2 on autophagy. Understanding such intricate relationship is important for development of novel therapeutic approaches of COVID‐19 by targeting the autophagy pathway.
3.4.1. The effects of autophagy on SARS‐CoV‐2 and COVID‐19
Similar to the effects of autophagy on many other types of viruses, autophagy also has dual effects against SARS‐CoV‐2: proviral and antiviral effects. At present, there are multiple lines of evidence suggesting that the autophagy machinery or the autophagy process can assist SARS‐CoV‐2 in the cycle of viral replication as highlighted in Figure 5.
First, ACE2 is the receptor at the host cell's plasma membrane that is able to directly bind to the receptor‐binding domain of the spike protein of SARS‐CoV‐2. 309 , 310 Two recent studies independently identified the presence of LIR motifs in the tails of ACE2, 311 , 312 indicating the possible involvement of LC3 in the initial step of viral infection, although the exact significance of this LIR motifs in the viral infection remains to be further determined.
Second, mechanistically, there are novel molecular links connecting autophagy with SARS‐CoV‐2 replication. For instance, by using genome‐wide CRISPR‐Cas9 screening, TMEM41B (transmembrane protein 41B), an ER‐localized transmembrane protein, has been identified as a novel autophagy regulator required for autophagosome formation. 313 , 314 , 315 Interestingly, two recent studies demonstrated that TMEM41B is required for SARS‐CoV‐2 infection, based on the observations that deletion of TMEM41B markedly reduced infectivity, which could be fully restored with the reconstitution of TMEM41B. 316 , 317 With increasing research on SARS‐CoV‐2 and autophagy, we expect to see more such common regulators to be discovered.
Third, although the canonical autophagy proteins such as BECN1, ATG5, and ATG7 are not required for viral infection of SARS‐CoV‐2 in the host cells, 316 the class III PI3K (phosphoinositide 3‐kinase) or PIK3C3, a key regulator in autophagy, has been shown to play an important role in viral replication of SARS‐CoV‐2: (i) PI3K is activated in cells infected with SARS‐CoV‐2 to produce PtdIns3P 318 ; (ii) inhibition of PI3K kinase activity led to suppression of SARS‐CoV‐2 replication in human airway epithelial cells. 319 Intriguingly, other autophagy inhibitors such as ULK1 inhibitor SBI0206965 and lysosome inhibitor hydroxyl chloroquine (HCQ) failed to exert similar inhibitory effects on SARS‐CoV‐2 replication, suggesting the possibility that it is PIK3C3, but not autophagy per se that may participate the replication process of SARS‐CoV‐2. 320 Obviously more work is needed to establish the therapeutic value of targeting the autophagy machinery in treating COVID‐19.
At present, relatively little is known about the possible antiviral effect of autophagy against SARS‐CoV‐2. ATG16L1 is one of the key ATGs in control of autophagosome biogenesis by forming a protein complex with ATG12–ATG5 in mediating LC3 lipidation. 321 In response to SARS‐CoV‐2 infection, its receptor ACE2 is able to recruit ATG16L1 to form exosome‐like vesicles called defensosomes that are capable of blocking viral entry. 322 Moreover, it is known that an ATG16L1 allele (ATG16L1‐T300A) was associated with impaired autophagy/xenophagy and reduced ability in clearing pathogens. 323 Interestingly, people bearing the ATG16L1 T300 allele expressed higher level of ACE2 compared with ATG16L1 A300, suggesting that ATG16L1 T300 is associated with a greater probability of SARS‐CoV‐2 infection, 324 although the mechanisms linking ATG16L1 and ACE2 remains unknown.
3.4.2. The reciprocal effects of SARS‐CoV‐2 on autophagy
What we have discussed above is the effect of autophagy on SARS‐CoV‐2. Interestingly, there is accumulating evidence suggesting that SARS‐CoV‐2 has reciprocal effect on autophagy, either activate or suppress via a variety of mechanisms. For instance, papain‐like protease PL(pro), a viral protease from SARS‐CoV‐2, is able to disrupt formation of ULK1–ATG13 complex and eventually suppress starvation‐induced autophagy at the early stage of autophagosome biogenesis 325 (Figure 5).
However, majority of the evidence on the inhibitory effect of SARS‐CoV‐2 on autophagy is actually on the late stage, especially on fusion of autophagosome with lysosome, a process known to be controlled by a group of protein complexes including SNARE (STX17–SNAP29–VAMP8), HOPS (VPS39, VPS11), and ATGs (ATG14). 96 , 97 Multiple studies have pointed to ORF3a, an accessory viral protein from SARS‐CoV‐2 that is capable of suppressing the autophagic flux by disrupting autophagosome–lysosome fusion. 326 , 327 , 328 , 329 , 330 Mechanistically, ORF3a is able to interact with VPS39, an important component of the HOPS complex in control of autophagosome–lysosome fusion. 331 , 332 On the other hand, ORF3a is also capable of inhibiting autophagy and promoting lysosomal exocytosis and extracellular egress of SARS‐CoV‐2. 333
Moreover, another viral protein from SARS‐CoV‐2, NSP6, has also been found to impair the autophagic flux at the late stage by targeting ATP6AP1 (ATPase H+ transporting accessory protein 1), a key component of the lysosomal V‐ATPase in control of lysosomal acidity. 334 Similarly, SARS‐CoV‐2 and other CoVs such as MHV are enriched in the endocytic organelles including endosomes and lysosomes, leading to lysosome deacidification and suppression of autophagy at the late stage. 335
In addition to the inhibitory effects on autophagy by SARS‐CoV‐2, there are reports demonstrating the opposite effects on autophagy. For instance, infection with SARS‐CoV‐2 enhanced LAMP2a expression and upregulated autophagic flux. 336 The autophagy‐promoting activity of SARS‐CoV‐2 has also been demonstrated in several animal models in vivo, including human ACE2 transgenic mice. 337 Such results thus provide experimental basis for using autophagy inhibitors including CQ for treatment of COVID‐19, a topic to be discussed in the section below.
3.4.3. Targeting autophagy as a potential therapeutic strategy for treatment of COVID‐19
Since the outbreak of COVID‐19 more than 2 years ago, the efforts in developing effective therapeutic drugs for clinical application never stop, although with only limited success, in comparison with the much more successful vaccination program. The initiation excitement with Remdesivir did not last that long. Remdesivir was approved by United States Food and Drug Administration (US FDA) while WHO provide the opposite advice on its clinical application in treatment of COVID‐19, mainly due to its inconsistent and controversial clinical outcomes. 338 , 339 , 340 , 341 In December 2021, another oral medication PAXLOVID (nirmatrelvir + ritonavir, inhibitors of viral proteases) has received US FDA's emergency use authorization for COVID‐19.
Among many types of antiviral drugs tested against COVID‐19 so far, the lysosomotropic agents (CQ/HCQ) have received much attention. 342 Pharmacologically, both CQ and HCQ are with lysosomotropic property, capable of enriching inside the endolysosomes and chemically neutralizing the pH, leading to suppression of the degradative function of the endolysosomal system. Based on this unique pharmacological activity, CQ/HCQ has a long history of treating malaria and also has been widely used as autophagy inhibitors by suppressing lysosomal function. 343 , 344 In fact, CQ/HCQ are the only autophagy inhibitors in clinical trials in autophagy‐related diseases such as cancer. 345 Based on the existing knowledge of the close implication of the autophagy‐lysosome system in the replication cycle of SARS‐CoV‐2, 306 CQ/HCQ quickly caught people's attention as potential therapeutic agents against COVID‐19. Unfortunately, despite the strong evidence from preclinical studies (cell culture and animal models), the outcomes of many clinical trials using CQ/HCQ are largely disappointing, accompanied by significant side effects. 346 , 347 , 348 Thus, US FDA revoked its authorization for the emergency use of CQ/HCQ in COVID‐19 patients (https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19‐update‐fda‐revokes‐emergency‐use‐authorization‐chloroquine‐and).
In addition to CQ/HCQ, several other lysosomotropic agents have been investigated. One example is azithromycin, a commonly used antibiotic that has similar lysosomotropic property as CQ/HCQ and this pharmacology effect is related to its antibacterial and antiviral function. 349 , 350 Recent work has indicated that azithromycin is able to block the entry of SARS‐CoV‐2 via disruption of the fusion process between viral and vacuolar membranes, a process related to its lysosomotropic property. 351 Moreover, numerous clinical trials have been conducted, using either azithromycin alone or in combination with CQ/HCQ in treatment of COVID‐19. 352 , 353 , 354 , 355 , 356 Despite the therapeutic benefits of using azithromycin alone or in combination with CQ/HCQ in treatment of COVID‐19, more work is needed to move azithromycin from bench to bed.
Artemisinin and its related compounds from the extracts of the medicinal plant, Artemisia annua L., have been widely used as an antimalarial drug and the main researcher of this wonder drug, Youyou Tu, won the Nobel Prize in Physiology or Medicine in 2015. Interestingly, it has been reported that artesunate, a derivative of artemisinin, has lysosomotropic property and is able to activate lysosomal function. 357 At present, there is evidence demonstrating the antiviral activity of artemisinin and some of its derivatives such as artesunate, artemether, and dihydroartemisinin against SARS‐CoV‐2, from in vitro cell‐based assays, to animal models and clinical trials, either alone or in combination with other therapeutic agents. 358 , 359 , 360 Understanding the potential therapeutic of artemisinin and its derivatives in COVID‐19 is important, especially for control of this pandemic in less‐developed countries and regions such as Africa where malaria and COVID‐19 constitute two major public health challenges. 361 , 362
3.5. Autophagy in cardiovascular diseases
Cardiovascular disease is the leading cause of death in the developed world. 363 Increasing number of studies have linked autophagy to cardiovascular health and disease (Figure 6). Generally, basal autophagy plays a vital role in maintaining the intracellular homeostasis of the major cardiovascular cell types, including cardiomyocytes, cardiac fibroblasts, endothelial cells, vascular smooth muscle cells, and macrophages via clearance of superfluous or damaged cellular components. 364 Moreover, autophagy is also crucial for cardiac progenitor cell differentiation. 365 , 366 It has been further shown that depletion of essential ATGs impairs cardiac morphogenesis. 367 These findings collectively reflect the beneficial effects of autophagy on cardiovascular health and thus dysregulation of autophagy is believed to play a role in the pathogenesis of various cardiovascular disorders. 368 Here, we mainly focus on the roles of autophagy in cardiac ischemia/reperfusion injury, cardiac hypertrophy, and dilated cardiomyopathies.
FIGURE 6.
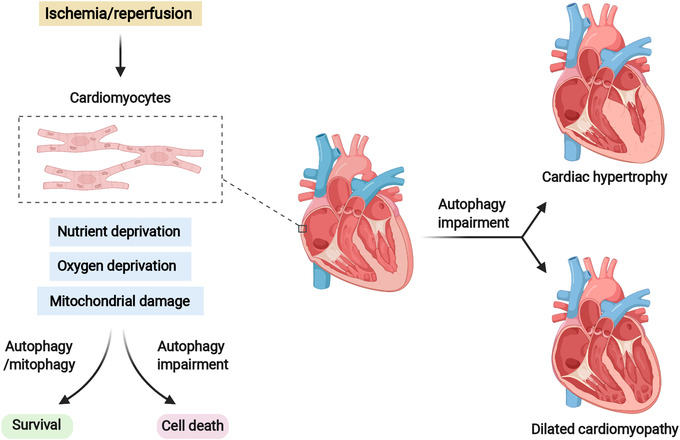
The protective effects of autophagy on cardiovascular diseases. Autophagy is essential for maintaining the intracellular homeostasis of cardiomyocytes under both basal and stress conditions. In response to cardiac ischemia/reperfusion, autophagy acts as a survival mechanism for replenishment of metabolic substrates and removal of damaged organelle such as damaged mitochondria. Impairment of autophagy results in cardiomyocyte cell death and exacerbates ischemia/reperfusion injury. Dysregulation of autophagy is also related to other cardiovascular diseases such as cardiac hypertrophy and dilated cardiomyopathy
3.5.1. Autophagy in ischemia/reperfusion injury
Ischemic heart disease is one of the leading causes for sudden cardiac death worldwide. During ischemia, the supplies of nutrients and oxygen are limited, which is accompanied by mitochondrial damage. 369 In response to ischemia, AMPK is activated, leading to suppression of MTORC1 and hence activation of autophagy. 370 Functionally, cardiomyocyte autophagy acts as a cytoprotective mechanism for replenishment of metabolic substrates and removal of damaged organelle such as mitochondria. 371 Accordingly, enhancing autophagy via activating AMPK has been shown to protect against ischemic injury, 372 , 373 , 374 while suppression of autophagy promotes cardiomyocyte cell death. 375 As damaged mitochondria are responsible for releasing ROS, selective removal of damaged mitochondria by mitophagy has been proposed as a key mechanism to protect against ischemia injury. 376 , 377 Consistently, loss of Parkin or Pink1, the two key players in mitophagy, increases the heart's vulnerability to ischemic injury in mouse model. 378 , 379
During reperfusion, the level of nutrients and oxygen are restored to normal level. Intriguingly, it has been reported that autophagy is still activated during reperfusion via an AMPK–MTORC1‐independent mechanism. 370 However, whether autophagy is beneficial or detrimental under such condition is a subject of debate, with no consensus reached so far. On one hand, it has been shown that reperfusion injury is blunted by activation of autophagy flux, 380 , 381 , 382 suggestive of the protective effect of autophagy. Of note, the cytoprotective effects of autophagy on cardiomyocyte involves the suppression of ROS production. 380 Consistently, activation of mitophagy also attenuates ROS production and cardiomyocyte cell death in response to reperfusion. 383 Impaired autophagy contributes to cardiomyocyte cell death and exacerbates reperfusion injury. 384 , 385 , 386 On the other hand, there is evidence suggesting that autophagic cell death is involved in reperfusion injury 387 and pharmacologic inhibition of excessive autophagy protects against myocardial reperfusion injury. 375 , 388 Thus, further studies are necessary to define whether increased autophagy in response to reperfusion is an epiphenomenon or a causative factor in cardiomyocyte cell death.
3.5.2. Autophagy in cardiac hypertrophy
Cardiac hypertrophy refers to the abnormal enlargement or thickening of the heart muscle, which is defined as an adaptive response and accompanied by other cardiovascular diseases such as hypertension, ischemic disease, and heart failure (HF). 389 The first study on cardiac hypertrophy and autophagy dated 1983, in which reduced autophagic vacuoles were observed in ventricular hypertrophy derived from supravalvular aortic constriction. 390 Cardiac‐specific deficiency of Atg5 reveals the importance of autophagy in maintaining cardiomyocyte size and global cardiac structure and function. 391 Moreover, in adult mice, temporally controlled cardiac‐specific deficiency of Atg5 led to various cardiomyopathies including cardiac hypertrophy, left ventricular dilatation, and contractile dysfunction. 391 Mutations of LAMP2 were also reported in association with severe cardiac hypertrophy. 392 Deficiency of Foxo32 (also known as MAFbx or Atrogin‐1), a muscle‐specific gene required for muscle atrophy, impairs autophagic flux, and results in heart hypertrophy in mice. 393
As the key mechanism to remove dysfunctional mitochondria to ensure cardiomyocyte energetic and metabolic needs, mitophagy also plays a pivotal role in preventing cardiac hypertrophy. 394 Mice with Parkin or Pink1 depletion exhibit enhanced cardiac hypertrophy and contractile dysfunction in response to pressure overload. 395 It has also been reported that mice deficient in Pink1 develop early left ventricular dysfunction and pathological cardiac hypertrophy. 396 Intriguingly, BNIP3, a key player involved in Parkin‐independent mitophagy, appears to accelerate the progression of cardiac hypertrophy. 397 Given that BNIP3 is also involved in other cellular processes such as inflammation, apoptosis, and necrosis, 398 BNIP3‐mediated mitophagy could be an epiphenomenon associated with cardiac hypertrophy progression. Further study is necessary to determine whether BNIP3‐mediated mitophagy is a compensatory mechanism to sustain the heart function in the course of cardiac hypertrophy.
Misfolded proteins and intracellular aggregates have been shown to contribute to cardiac hypertrophy. 399 One good example is the small heat shock protein alphaB‐crystallin (CryAB) whose missense mutation (CryABR120G) causes aberrant Desmin and CryAB aggregation and results in cardiac hypertrophy. 400 At cellular level, it has been shown that CryABR120G triggers ATG7‐depedent autophagy, leading to clearance of CryABR120G aggregates in cardiomyocytes. 401 , 402 Similarly, Becn1 haploinsufficiency results in reduction of autophagy and promotes HF progression in αMHC‐CryABR120G mice. 401
The above‐mentioned findings collectively connect impaired autophagy to cardiac hypertrophy. Activation of autophagy at the whole‐body levels by nutritional (such as caloric restriction [CR] or spermidine that operates as a CR mimetic [CRM]) or pharmacological (such as rapamycin) interventions shows beneficial effects on aged‐dependent cardiac hypertrophy reversal. 403
3.5.3. Autophagy in dilated cardiomyopathy
Dilated cardiomyopathy (DCM) refers to a syndrome manifested by cardiac enlargement and poor systolic function of the left ventricular, with an ejection fraction <45%, which is a major cause for HF. 404 The evidence of autophagy in heart diseases was first reported in tissue samples from DCM patients. 405 However, the exact role of autophagy in DCM in this study was not studied. A more recent study shows that the autophagic vacuoles are identified in the cardiomyocytes with myofilament changes in the left ventricular. 406 Importantly, the number of autophagic vacuoles in cardiomyocytes is associated with a better HF prognosis in patients with DCM, 406 suggesting the importance of autophagy in prevention of HF resulted from DCM. Conversely, mice with cardiac‐specific deficiency of Atg5 develop left ventricular dilatation and contractile dysfunction. 391 A mutation in the LAMP2 gene (c.928G>A mutation) previously found in patients with hypertrophic cardiomyopathy is shown to be associated with severe DCM. 407 Moreover, CryABR120G has also been reported in DCM. In mice model, expression of CryABR120G results in aberrant mitochondrial‐sarcomere architecture and defective mitochondrial function, leading to apoptosis of cardiomyocyte and eventually DCM and HF. 408 Given the importance of autophagy in clearance of protein aggregates, it is believed that autophagy possesses beneficial effects in prevention of DCM.
3.6. Autophagy in aging
Aging is a biological process characterized by time‐dependent cellular and functional decline and a main risk factor associated with the progress of many disorders such as cancer, cardiovascular diseases, and neurodegenerative diseases, leading to reduced life quality of the organisms. 409 Diverse stress response pathways, including autophagy, promote growth and delay aging by limiting tissue damage, promoting tissue repair and alleviating environmental pressures. 410 , 411 Despite the complex relationship between autophagy and aging, a growing body of research supports the critical role of autophagy in promoting longevity and delaying aging (Figure 7). As a conserved degradation pathway, autophagy promotes cell survival by removing aged molecules and mobilizing energy substances. 412 Impaired autophagy causes diverse cellular dysfunctions that predispose individuals to age‐related diseases and exacerbate the aging process, whereas enhanced autophagy prolongs lifespan and improves health span. 413 Therefore, an in‐depth understanding of the cellular biological functions of autophagy in aging is critical for developing autophagy‐targeted therapies for aging and aging‐related diseases.
FIGURE 7.
Systemic antiaging effects of autophagy. Autophagy may increase organismal fitness by eliminating protein aggregates, damaged organelles, apoptotic corpses. In addition, autophagy may contribute to the clearance of intracellular pathogens and enhancing the function of antigen‐presenting cells to attenuate age‐related dysfunctions and extend lifespan
One important observation in autophagy and aging study is that aging is accompanied by reduced lysosomal protease activity and reduced expression of several ATGs in rodent and Drosophila. 414 , 415 , 416 Similarly, in C. elegans, the autophagy activity generally declines with aging, whereas long‐lived mutants of Daf‐2 and Glp‐1 regulate autophagy in different spatiotemporally specific ways to prolong lifespan. 417 In drosophila, loss‐of‐function mutations of Atg7 or Atg8 promote the accumulation of ubiquitin‐positive aggregates in neurons and increase sensitivity to stress, thereby shortening the drosophila lifespan. 415 , 418 In mice, disruption of the BECN1–BCL2 complex is an effective mechanism for enhancing autophagy, preventing premature aging, promoting longevity, and improving health. 419 In humans, the expression levels of ATG5, ATG7, and BECN1 are downregulated with aging. 420 These studies collectively suggest anti‐aging as a primordial function of autophagy.
Among the various signaling networks regulating autophagy, the roles of MTOR and AMPK in regulation of aging and lifespan attract particular attention. 413 Upregulation of TOR signaling and insulin/IGF‐1 (IIS) pathway has been widely reported during aging, and inhibition of TOR signaling is sufficient to promote longevity in an autophagy‐dependent manner. 421 Similarly, inhibition of AMPK activity also occurs during aging, and its activation prolongs the lifespan of C. elegans by elevating autophagy. 422 Given the critical roles of MTOR and AMPK in control of nutrient metabolism, CR without malnutrition is an effective intervention to delay aging and prevent aging‐related diseases in most species and autophagy is likely to be one of the underlying mechanisms, whereas autophagy inhibition diminishes the life‐extending effects of CR. 423 , 424 , 425 CR‐induced autophagy may involve multiple mechanisms, mainly via inhibiting MTOR signaling. Consistently, CR‐like lifespan extension can be achieved with the MTOR inhibitor rapamycin. 426 In addition, rapamycin may extend lifespan by inhibiting inflammation and reducing protein imbalances that lead to aging. 427 , 428 These results suggest that MTOR inhibitors can be used to treat age‐related diseases, but their side effects, such as immunosuppression and insulin resistance, limit their widespread application as an anti‐aging therapy. 427 This has driven the development of MTOR inhibitors with superior pharmacodynamics and agents that mimic the effects of CR, the so‐called CRMs, such as metformin, spermidine, and resveratrol. 413 Applying precision medicine for drug adaptation and artificial intelligence‐based dosing strategies to promote longevity by enhancing autophagy is thus promising.
4. MODULATION OF AUTOPHAGY AS THERAPEUTIC STRATEGY IN HUMAN DISEASES
As we discussed in the previous section, approaches modulating autophagy are demanded by its important role in human health and disease. Based on the intention of different clinical applications, both autophagy activators and inhibitors have been developed and tested in the past several decades (Figure 8). Here, we reviewed and grouped the known autophagy modulators in this section.
FIGURE 8.
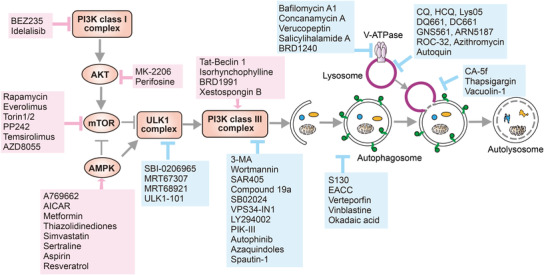
Autophagy activators and inhibitors. Mechanistic targets of autophagy activators and inhibitors. The major targets and related compounds of autophagy activation (arrows and boxes in red) and inhibition (arrows and boxes in blue) have been demonstrated
4.1. Activation of autophagy
Activation of autophagy has been mainly achieved via activation of AMPK or/and suppression of PIK3CA/AKT/MTOR signaling (Table 2).
TABLE 2.
Known autophagy pharmacological activators and their potential application in disease
| Class | Agent | Mechanism of action | Biological effects and application in disease | References |
|---|---|---|---|---|
| AMPK activator | A769662 | Allosterically activating AMPK | Type‐II diabetes and associated metabolic disorders (preclinical) | 429 , 430 |
| AICAR | Allosterically activating AMPK via its metabolite ZMP, which is an AMP mimetic | Type‐II diabetes (clinical trials); intestinal barrier function and epithelial differentiation (preclinical) | 431 , 432 | |
| Metformin | Inhibiting RCC1 (respiratory chain complex‐1) to raise the cellular ATP/AMP ratio; upregulating of SIRT1 expression | First‐line antidiabetic drug (approved); potential application of in cancer treatment (clinical trials), including osteosarcoma, prostate cancer, and so on | 433 , 434 , 435 | |
| Thiazolidinediones | Inhibiting RCC1, to raise the cellular ATP/AMP ratio | Potential application of in cancer treatment (preclinical); diabetes, obesity (clinical trials) | 436 , 437 | |
| Simvastatin | Raising cellular ATP/AMP ratio via inhibiting cholesterol biosynthesis | Antihypercholesterolaemia (approved); cardioprotective effects (approved); potential application of in cancer treatment (clinical trials) | 438 , 439 , 440 | |
| Sertraline | Antagonizing the mitochondrial VDAC1 (voltage‐dependent anion channel 1), resulting in reduced cellular ATP level | Antidepressant drug (approved) | 441 | |
| Resveratrol | Enhancing CamKKβ‐AMPK; SIRT1 signaling; upregulating cytoplasmic (de)acetylation reactions | Antioxidant (over‐the‐counter (OTC) supplements); antiaging effects, including anti‐inflammatory, cancer prevention, antidiabetic, cardioprotective, and neuroprotective properties (clinical trials) | 442 , 443 , 444 , 445 , 446 , 447 , 448 | |
| MTORC1 inhibitor | Rapamycin and rapalogs (Sirolimus) | Destabilization of MTOR–RAPTOR complex | Life‐span extension (preclinical studies); to prevent transplant rejection and to treat a rare pulmonary disease (approved); neuroprotective properties (preclinical); anticancer, including breast cancer; advanced pancreatic; non‐small‐cell lung cancer (clinical trials) | 193 , 449 , 450 , 451 |
| Everolimus (RAD‐001) | Destabilization of MTOR–RAPTOR complex | Anticancer effects (approved for breast cancer and renal cell carcinoma treatment), kidney transplant failure and rejection | 452 , 453 , 454 , 455 | |
| Temsirolimus | Destabilization of MTOR–RAPTOR complex | Potential treatment for treatments of LMNA cardiomyopathy, Huntington disease (preclinical); anticancer effects (approved for advanced renal cell carcinoma treatment) | 456 , 457 , 458 , 459 | |
| Torin1/2 | An ATP‐competitive inhibitor for MTOR | Experiment tools | 460 , 461 , 462 | |
| PP242 | An ATP‐competitive inhibitor for MTOR | Reduce tumor size in xenograft models (preclinical) | 460 , 463 , 464 , 465 | |
| AZD8055 | An ATP‐competitive inhibitor for MTOR | Advanced solid malignancies (clinical trials) | 188 , 466 , 467 , 468 , 469 | |
| EN6 | Targeting the ATP6V1A subunit of the lysosomal V‐ATPase, which stimulates MTORC1 via the Rag GTPase | Clears TDP‐43 aggregates, a causative agent in frontotemporal dementia (preclinical) | 470 | |
| AcCoA inhibitor | Hydroxycitrate acid | Competitive inhibiting the ATP citrate lyase (ACLY) to block the AcCoA synthesis, which activates MTOR vis EP300‐induced acetylation | Potential anticancer effects; immunosurveillance (OTC supplements) | 471 |
| Triethylenetetramine | A Cu(II)‐selective chelator, which depleting AcCoA via activating of SAT1; | Wilson's disease (approved); corrects diet‐induced metabolic syndrome; prevention or treatment of obesity (preclinical); cancer prevention (clinical trials) | 472 | |
| PI3K and MTOR inhibitor | BEZ235 | Binding to the ATP‐binding clefts of PI3K class I and MTOR kinase | Promote survival of drug‐resistant glioma; T‐cell acute lymphoblastic leukemia; hepatocellular carcinoma (clinical trials) | 473 , 474 , 475 |
| PI3K inhibitor | Idelalisib (CAL‐101) | Small‐molecule inhibitor of the delta isoform (p110δ) of PI3K | Chronic lymphocytic leukemia (approved) | 476 , 477 |
| AKT inhibitor | MK‐2206 | Allosterically inhibiting AKT in a pH‐domain‐dependent inhibitor | Anticancer effect, including colorectal cancer, melanoma, and advanced liver cancer (clinical trials), inhibiting COVID‐19 propagation in vitro (preclinical) | 478 , 479 , 480 |
| Perifosine | Allosterically inhibiting Akt in a pH‐domain‐dependent inhibitor | Chronic lymphocytic leukemia, small lymphocytic lymphoma (clinical trials) | 481 | |
| Multiple targets (AMPK or MTOR related) | Aspirin (and salicylate) | Activating AMPK; inhibiting EP300, COX (cyclooxygenase)‐1, COX‐2 and MTOR; | Anti‐inflammatory, antimicrobial, antipyretic, and analgesic drug (approved); cardiovascular diseases (clinical trials); anticancer (clinical trials) | 482 , 483 , 484 |
| Spermidine | Inhibiting acetyltransferases (such as EP300) leading to increased cytoplasmic deacetylation reactions; inhibiting MTOR; activating AMPK | Antioxidant; antiaging effects, including anti‐inflammatory, cancer prevention (clinical trials), antidiabetic, cardioprotective (clinical trials), and neuroprotective properties | 442 , 471 , 485 , 486 , 487 , 488 , 489 , 490 , 491 | |
| EGCG | CaMKKβ–AMPK signaling; MTOR, HATs, and KDACs | Potential therapeutic reagent to prevent cardiovascular complication, metabolic syndromes and potential anticancer effect (clinical trials) | 492 , 493 | |
| Curcumin | AMPK‐MTOR; TFEB‐lysosome pathway | Potential anticancer effect (clinical trials) | 494 , 495 , 496 | |
| Trehalose | Inhibition of glucose transporter SLC2A (also known as GLUT) to induce AMPK‐dependent autophagy; low‐grade lysosomal stress‐mediated TFEB activation | Neurodegenerative diseases, cardioprotective effects, atherosclerosis, hepatic steatosis; bipolar disorders, dry eye syndrome and vascular ageing, Alzheimer disease, nonalcoholic fatty live, infertility (clinical trials) | 497 , 498 , 499 , 500 , 501 , 502 , 503 , 504 , 505 | |
| Cucurbitacin | Mitochondrial ROS; ERK–MTOR–STAT3 | Potential anticancer, anti‐inflammatory, antibacterial and antidepressant effects (preclinical) | 506 , 507 , 508 | |
| Quercetin | Modulation of AKT‐MTOR signaling and hypoxia‐induced factor 1α (HIF‐1α) signaling, mitochondria ROS, SIRT1 | Antitumor, antioxidant, anti‐inflammatory, antidiabetic, hepatoprotective, and cardioprotective; COVID‐19 treatment (clinical trials) | 509 , 510 | |
| ROS enhancer | Antimycobacterial antibiotics (isoniazid or pyrazinamide) | Activation of cellular and mitochondrial ROS | Tuberculosis infection (approved) | 511 |
| BECN1 activator | Tat‐BECN1 | A cell‐permeable peptide derived from BECN1; interacting with a negative regulator of autophagy, GAPR‐1 | Antivirual, anticancer, cardioprotective, cerebrovascular, and cardiovascular protective, neuroprotective properties (preclinical) | 512 , 513 , 514 , 515 , 516 , 517 , 518 , 519 , 520 , 521 , 522 , 523 |
| Isorhynchophylline | Dependent on BECN1 function | Neuroprotective properties against neurodegenerative diseases including Parkinson disease (preclinical) | 524 | |
| BRD1991 | Bcl‐2–BECN1 PPI (protein–protein interaction) inhibitors | Experimental agents | 525 | |
| Xestospongin B | The IP(3)R antagonist inhibit IP(3)R interaction with BECN1 | Experimental agents; potential anticancer effects (preclinical) | 526 , 527 , 528 , 529 | |
| Inositol monophosphatase (IMPase) inhibitor | Carbamazepine | Reduction in Ins(1,4,5)P3 and inositol levels; a voltage‐dependent sodium channels blocker | Reduces hepatic fibrosis; fatty liver conditions; neuroprotection (preclinical); treatment of seizures and bipolar disorders (approved) | 530 , 531 , 532 , 533 |
| Lithium | Depletion of free inositol, reduction of IP3 levels, and increase in levels of Beclin1; inhibitor of GSK3β. | Bipolar disorder (approved); neurodegenerative disorders, including huntingtin or Parkinson's disease (clinical trials) | 534 , 535 , 536 , 537 , 538 | |
| TFEB/TFE3 activator | 3,4‐Dimethoxychalcone (3,4‐DC) | Stimulating the translocation of TFEB and TFE3 into nuclei; | Cardioprotective effects, and improved the efficacy of anticancer chemotherapy (preclinical) | 539 |
| Thiostrepton | Activation of TFEB and TFE3 | Stimulate anticancer immune responses of immunogenic cell death (ICD)‐inducing chemotherapeutics (clinical trials) | 540 | |
| HDACI inhibitors | Trichostatin A | FOXO1‐dependent pathways | Anti‐cancer property (clinical trials) | 541 , 542 , 543 |
| Suberoylanilide hydroxamic acid | FOXO1‐dependent pathways | Cutaneous T‐cell lymphoma treatment (approved); epilepsy, Cushing's disease, breast cancer with a combination with pembrolizumab, tamoxifen (clinical trials) | 541 , 544 | |
| p38 kinases inhibitor | SB202190 | p38alpha/beta kinases inhibitor; TFEB/TFE3‐dependent autophagy and lysosomal biogenesis | Treatment of colorectal cancer (preclinical) | 545 , 546 |
| Ca2+ channel antagonist | Fluspirilene | Ca2+ channel antagonist | Antipsychotic drugs (approved) | 547 , 548 |
| Verapamil | L‐type Ca2+ channel antagonist | Nonalcoholic fatty liver, extends lifespan (preclinical); ischemic stroke; type 1 diabetes (clinical trials) | 273 , 549 , 550 , 551 | |
| Felodipine | Ca2+ channel antagonist | Hypertension, cardiovascular diseases (clinical trials); neurodegenerative disorders | 552 | |
| Loperamide | An opioid receptor agonist that also inhibits voltage‐gated P/Q‐type Ca2+ channels; ER stress | Anticancer property (clinical trials) | 551 , 553 , 554 | |
| Amiodarone | Ca2+ channel antagonist | Antiarrhythmic agent (approved); hepatitis B virus‐associated hepatocellular carcinoma (preclinical) | 555 , 556 | |
| Cholesterol depleting agent | MBCD | Enhancing lysosomal V‐ATPase assembly and promoting VAMP3‐mediated autophagosome formation | Experiment agents | 557 , 558 |
| Mitophagy inducer | Urolithin A | Reducing mitochondria membrane potential | Antiaging effects via clearance of dysfunctional mitochondria (preclinical) | 559 |
| Selective autophagy | Autophagy‐targeting chimera (AUTAC) | Hijack autophagy to selectively degrade destines substrate via a standalone tag, S‐Guanylation | Experimental tools | 560 |
| Unclear mechanism | SMER‐28 | Unclear (independent of MTORC1) | Clearance of aggregation‐prone proteins, including amyloid‐β (preclinical) | 561 |
| BRD5631 | Unclear (independent of MTORC1) | Bacterial clearance; suppresses NPC1‐induced cell death (preclinical) | 562 | |
| 10‐NCP | Unclear (independent of MTORC1) | Against toxicity in a Huntington disease model (preclinical) | 563 | |
| Alpha‐lipoic acid | Upregulation of autophagy‐related genes | Antiaging effects (as a nutritional supplement), including anti‐inflammatory, cancer prevention, antidiabetic, cardioprotective, and neuroprotective properties, atherosclerosis, hypertension and Alzheimer's disease; nonalcoholic fatty liver disease (clinical trials) | 564 |
4.1.1. Activation of AMPK signaling
AMPK, the major cellular energy sensor, responses to the changes of cellular ATP/AMP ratio. Activation of AMPK provides a powerful target to trigger cellular autophagy. 565 AMPK can be allosterically activated by small molecules through blockage of AMPK T172 dephosphorylation (e.g., A769662). 429 , 430 Another direct approach to stimulate AMPK activity is the utilization of AMP mimetics. 566 For example, 5‐aminoimidazole‐4‐carboxamide ribonucleoside, an adenosine monophosphate, which can be metabolized to an AMP mimetic, is able to cause allosteric activation of AMPK. 431 , 432 Indirect activation of AMPK can be achieved via downregulation of the cellular ATP/AMP ratios, which usually resulted from agents blocking ATP production, such as thiazolidinediones, simvastatin and sertraline. 436 , 437 , 438 , 439 , 440 , 441 Both direct and indirect activators of AMPK have been shown to induce autophagy potently and perform therapeutic importance to treat diabetes and related metabolic disorders. 565
In addition, lifespan extension, or the so‐called anti‐aging function, is one of the most attracting features of AMPK activators. Both AMPK and autophagy are robustly stimulated by CR and excises, which are known strategies to promote health and prolong lifespan in a wide range of animal models with substantial evidences. 567 The concept of CRMs has been proposed to describe compounds that mimic some of the anti‐aging effects of CR. The key features of CRMs embrace activation of autophagy and global cellular proteins deacetylation. 568 As a potent approach to activate autophagy, the anti‐aging effects of a number of AMPK activators have been tested and grouped into CRMs, including metformin, resveratrol, aspirin, and spermidine. 434 , 442 , 447 , 482 , 483 , 487 These AMPK‐related CRMs enhance protein deacetylation via induction of an NAD(+)‐dependent deacetylase SIRT1 (sirtuin 1) or inhibition of acetyltransferase EP300 (E1A binding protein p300), which provides additional health benefits in both autophagy‐dependent and ‐independent manners. 434 , 442 , 447 , 482 , 483 , 487
Here, we described a good example to show the function of AMPK activation and the subsequent induction of autophagy: metformin, the first‐line antidiabetic drug, functions as an indirect AMPK activator by suppressing RCC1 (respiratory chain complex‐1) to decrease the cellular ATP/AMP ratio. 433 , 434 , 435 Metformin has also been found to induce autophagy via both AMPK‐dependent and SIRT1‐dependent pathways. Autophagy and SIRT1 have been found to be downregulated in a T2D mouse model. 433 Therefore, in addition to the energy sensing pathways orchestrated by AMPK, metformin also restores autophagy and SIRT1, which improve diabetic symptoms and contributes to lifespan extension. 433 , 434 , 435 AMPK activation, autophagy induction, and protein deacetylation can be considered as the “trinity” mechanisms efficiently counteracting aging‐associated features, such as cardiovascular, neurodegenerative, malignant, and metabolic diseases. There are still amounts of open questions remained, such as the causal relationship amongst this “trinity,” the criteria to select the appropriate targeting population for different CRMs, and the possible strategies to achieve stronger effects when combining CRMs and other aging‐related signaling regulators.
4.1.2. Inhibition of PIK3CA/AKT/MTOR signaling
PIK3CA/AKT/MTOR signaling is the most‐described inhibitory signaling of autophagy. 569 Small molecules suppressing the element in this pathway, including AKT, MTOR, and PIK3CA complex, can be used as strong inducers of autophagy. There are mainly two direct strategies to target MTOR activity. First, reagents working as rapamycin, the most famous MTOR inhibitor, allosterically block MTOR kinase activity through destabilizing the MTOR–RAPTOR complex. 193 , 449 , 450 , 451 Second, small molecules, such as Torin1/2, PP242, and AZD8055, target the ATP binding site on MTOR, hence suppressing its kinase activity in an ATP‐competitive manner. 460 , 461 , 462 , 463 , 464 , 465 In addition to MTOR inhibition, the kinase activities of PI3K or AKT can be allosterically inhibited through small molecules targeting the delta isoform (p110δ) of PI3K or the PH‐domain of AKT, respectively. 476 , 477 , 478 , 479 , 480
Suppression of PIK3CA/AKT/MTOR signaling has been applied to the treatments of different types of cancers. Idelalisib (CAL‐101) is the first approved PI3K inhibitor applied in chronic lymphocytic leukemia. 476 , 477 Everolimus and temsirolimus, two MTOR inhibitors, have been approved for treatments for chronic myeloid leukemia and prostate cancer patients. 452 , 453 , 454 , 455 The anticancer efficacies of AZD8055, rapamycin, and rapamycin‐related analogs have been evaluated in a number of Phase I and II clinical trials. 188 , 193 , 449 , 450 , 451 , 466 , 467 , 468 , 469 However, special cautions are needed to interpret the role of autophagy in these anticancer therapies based on suppression of PIK3CA/AKT/MTOR signaling. Autophagy can be used as a mechanism for cancer cells survival under unfavorable microenvironment and enhancement of tumor immune evasion, which largely increases the possibility of drug resistance. 145 , 570 Therefore, the synergistic efficacy of the suppression of PIK3CA/AKT/MTOR signaling with autophagy inhibitors has been extensively tested from tumor cell biology to clinical trials, which shows great antitumor potential and needs to be further validated in the future. 571 , 572 , 573 , 574
4.1.3. Other autophagy activators
Autophagy induction is also druggable through viable targets independent of MTOR signaling, such as BECN1 promotion, HDAC inhibition, TFEB activation, and Ca2+ channel antagonist. For instance, 3,4‐DC and thiostrepton stimulate autophagy via activation of TFEB/TFE3, the master transcription factor for lysosome‐related genes and ATGs. 539 , 540 These agents have been found to provide additional benefits for the antitumor effects of immunogenic cell death (ICD) inducers, which may indicate the role of autophagy in antitumor immunity. 539 , 540
Several Ca2+ channel blockers, including fluspirilene, verapamil, loperamide, and amiodarone, have been found to induce autophagy potently. 273 , 547 , 548 , 549 , 550 , 551 , 552 , 553 , 554 , 555 , 556 Although the exact mechanisms are not clearly defined, there are reports indicating that the Ca2+ channel antagonists‐induced autophagosome formation relates to calpain‐mediated cleavage of ATG5. 273 , 552
Inositol monophosphatase (IMPase) inhibitors, including the approved mood‐stabilizing agents, have been reported to perform neuroprotective activity via induction of autophagy. The IMPase inhibitors induce autophagy in a way dependent on decreasing free inositol or myo‐inositol‐1,4,5‐trisphosphate (IP3) levels. 526 , 527 , 528 , 529 The molecular mechanism underlying the inositol signaling‐regulated autophagy is MTOR independent but is still not fully understood. Studies using an IP3 receptor blockage (Xestospongin B) have demonstrated that IP3 receptor modulates autophagy via its direct interaction with BECN1, which may hint the obligation of BECN1 in IMPase inhibitor‐mediated autophagy. 526 , 527 , 528 , 529 Interestingly, other BECN1 activators, such as Tat‐BECN1 and Isorhynchophylline, have also been identified to play a neuroprotective role in preclinical studies, which indicates the great potential of BECN1‐depedent autophagy in treatment of neurodegenerative diseases.
4.2. Suppression of autophagy
Unlike autophagy activating strategies, which mainly focusing on kinases activity modulation (such as AMPK and MTOR), the strategies inhibiting autophagy usually target the relatively late stages of autophagy, including inhibition of autophagosome formation (both initiation and maturation processes) and lysosomal degradation (Table 3).
TABLE 3.
Known autophagy pharmacological inhibitors and their potential application in disease
| Class | Agent | Mechanism of action | Biological effects and application in disease | References |
|---|---|---|---|---|
| ULK inhibitor | SBI‐0206965 | Pyrimidine analogues inhibiting ULK1 activity in an ATP‐competitive manner; inhibiting AMPK activity in a type IIb inhibitor manner | Potential anticancer effects; diabetic therapy development; traumatic, and neurodegenerative disorders (preclinical) | 52 , 280 , 575 , 576 , 577 , 578 , 579 |
| MRT67307 and MRT68921 | Small molecule inhibitors (also known as TBK1 inhibitor) | Potential anticancer effects (preclinical) | 579 , 580 | |
| ULK1‐101 | A small molecule inhibitor | Sensitizes KRAS mutant lung cancer cells to nutrient stress (preclinical) | 581 | |
| PI3K class III complex inhibitor | 3‐MA | A dual inhibitor of PI3K (class I and III) | Experimental tool | 582 , 583 , 584 , 585 , 586 |
| Wortmannin | The sterol‐like fungal metabolite inhibiting PI3K (class I and III) reversibly | Experimental tool | 582 , 587 , 588 , 589 | |
| SAR405 | Inhibiting the ATP‐binding cleft of PIK3 class III (with higher selectivity when compared with wortmannin and 3‐MA) | Enhancing the antitumor efficacy of cisplatin or mTOR inhibitors; improves anti‐PD‐1/PD‐L1 immunotherapy; promotes host cell death during latency reversal of HIV (preclinical) | 590 , 591 , 592 , 593 , 594 , 595 | |
| SB02024 | Selectively inhibiting PIK3C3/VPS34 | Improves the sensitivity of breast cancer cells to Sunitinib; improves anti‐PD‐1/PD‐L1 immunotherapy (preclinical) | 595 , 596 | |
| VPS34‐IN1 | Selective cell permeable PIK3C3/VPS34 inhibitor in nanomolar level | Anticancer effects including acute myeloid leukemia and CNS tumor; SARS‐CoV‐2 inhibitors; (preclinical) | 319 , 597 , 598 | |
| LY294002 | A reversible inhibitor of PI3K | Promoting antitumor effect of different chemotherapy drugs (preclinical) | 587 , 599 , 600 | |
| PIK‐III | Inhibiting PIK3C3/VPS34 activity via binding to a unique hydrophobic pocket (with higher selectivity when compared with wortmannin and 3‐MA) | Treatment for CML patients; SARS‐CoV‐2 inhibition (preclinical) | 319 , 594 , 601 | |
| Autophinib | Inhibiting PIK3C3/VPS34 in an ATP‐competitive manner | Potential anticancer effects in cell level (preclinical) | 602 , 603 , 604 , 605 | |
| Azaquindoles | Inhibiting PIK3C3/VPS34 in an ATP‐competitive manner | Experimental agents | 606 | |
| Spautin‐1 | Targeting the BECN1 subunit of PIK3C3/VPS34 complexes via Inhibition of USP10 and USP13 | Anticancer effects (preclinical) | 607 , 608 , 609 | |
| Compound 19a | Inhibiting the Beclin 1‐ATG14L PPI, which is required for PI3KC3‐C1 formation | Experimental agents | 610 | |
| ATG4B inhibitor | NSC185058 | ATG4B antagonist | Enhancing the antitumor effects of radiotherapy or temozolomide (TMZ) in glioma treatment (preclinical) | 611 , 612 , 613 |
| FMK‐9a | Suppressing ATG4B through formation of a covalent bond with Cys74 | Experimental agents | 614 , 615 | |
| PP2A inhibitor | Okadaic acid | Enhancing phosphorylated ATG4B (Ser316) via inhibition of PP2A; Inducing cytokeratin cytoskeleton disruption | Experimental agents | 616 , 617 , 618 |
| ATG7 inhibitor | Pyrazolopyrimidine sulfamate compounds | Targeting ATG7 and covalently binding to ATG3 | Experimental agents | 619 |
| STX17 inhibitor | EACC | Suppressing the translocation of STX17 on autophagosomes | Experimental agents | 620 , 621 , 622 |
| LC3 inhibitor | S130 | Blocking the recycling of LC3‐I via inhibiting the delipidation of LC3‐II | Potential anticancer effects (preclinical) | 623 |
| GRAMD1A inhibitor | Autogramins | Disturbing the cholesterol distribution via competing with cholesterol binding to the GRAMD1A, which is required for autophagosome initiation | Experimental agents | 624 |
| Cytoskeletal protein inhibitor | CA‐5f | Inhibiting autophagy–lysosome fusion via inhibiting cytoskeletal proteins and membrane traffic proteins | A novel late‐stage autophagy inhibitor with potential clinical application for NSCLC therapy (preclinical) | 625 |
| Vinblastine | Destabilizing microtubule function to suppress autophagosome maturation | Treatment on AML and recurrent solid tumor with sirolimus (clinical trials) | 626 , 627 , 628 | |
| Small GTPase modulating agent | Thapsigargin | Inhibiting recruitment of RAB7‐mediated autophagosome–lysosome fusion | Thasigargin based prodrugs mipsagargin is in Phase II clinical testing for solid tumor (clinical trials) | 629 , 630 |
| Vacuolin‐1 | Inhibiting RAB5A‐mediated autophagosome–lysosome fusion; PIKfyve ihibitor | Potential anti‐COVID‐19 effects; inhibition of migration and metastasis of cancer cells (preclinical) | 631 , 632 , 633 , 634 , 635 | |
| Lysosomotropic agent | CQ and HCQ | Blocking lysosomal acidification | Antimalarial agent (approved); anticancer effects, alone or combined with other chemotherapy drugs (clinical trials); antiviral effects including COVID‐19 (clinical trials) | 306 , 319 , 558 , 594 , 636 , 637 , 638 , 639 |
| Lys05 | A dimeric form of CQ | Treatment for CML patients (preclinical) | 594 , 640 | |
| DQ661 | A dimeric quinacrine | Potential anti‐cancer effects (preclinical) | 641 | |
| DC661 | A dimeric CQ | Potential anticancer effects (preclinical) | 642 | |
| GNS561 | Inhibiting PPT1 function, which results in lysosomal Zn2+ accumulation and blockage of cathepsin enzyme activity | Liver cancers treatment (clinical trials); COVID‐19 treatment (clinical trials) | 643 , 644 , 645 | |
| ARN5187 | A lysosomotropic REV–ERBβ ligand | Potential anticancer effects | 646 | |
| ROC‐325 | A compound with structural motifs of both HCQ and lucanthone | Superior preclinical anticancer activity; augments the antileukemic activity of azacitidine | 647 , 648 | |
| Azithromycin | Blocking lysosomal acidification | With poorly understood anti‐inflammatory properties; potential antitumor effects | 649 , 650 , 651 | |
| Autoquin | Blocking lysosomal acidification | Experimental agent | 652 | |
| V‐ATPase inhibitors | Bafilomycin A1 | Inhibiting V‐ATPase by blocking the dissociated V(1)‐ATPase; suppression Ca‐P60A/SERCA‐dependent autophagosome–lysosome fusion | Experimental agent | 558 , 637 , 653 |
| Concanamycin A | Highly potent V‐ATPase inhibitors via binding to V(0) subunit c | Antiviral effects, including HIV, influenza (preclinical) | 654 , 655 , 656 | |
| Verucopeptin | Interacting with ATP6V1G to block V‐ATPase activity | Potential therapeutics against multidrug‐resistant cancers (preclinical) | 657 | |
| Salicylihalamide A | Blocking the ATPase activity by inhibiting the V(0) domain | Experimental agents | 658 | |
| BRD1240 | Suppresses V‐ATPase function | Experimental agents | 659 | |
| Lysosome function inhibitor | Lucanthone (Miracil D) | Inducing lysosomal membrane permeabilization (unclear molecular mechanism) | Antischistosomal drug (approved); potential anticancer effect (clinical trials for brain tumor and brain metastases) | 660 , 661 |
| PIKfyve inhibitor | Apilimod | Disrupts lysosomal homeostasis and intracellular trafficking; Inducing secretory autophagy | Treatment of non‐Hodgkin lymphoma (clinical trials); treatment of COVID‐19 (clinical trials) | 662 , 663 , 664 |
| TRPML1 inhibitor | ML‐SI1 | Small molecule inhibits Mucolipin TRP channel 1 (TRPML1) as the key lysosomal Ca2+ channel | Experimental agent | 665 |
| Na(+),K(+)‐ATPase inhibitor | Cardiac glycosides | Antagonists of Na(+),K(+)‐ATPase, inhibit autotic cell death | Extensively used in the past for treatment of heart failure and arrhythmia treatment inhibited autosis death, which might occur in liver and heart of patients with severe anorexia nervosa clinical potential of CGs in anticancer and other therapies | 523 , 666 , 667 , 668 |
| Mitophagy inhibitor | Mdivi‐1 | A pharmacological inhibitor of DRP1, inhibit mitochondria fission | Chronic obstructive pulmonary disease (COPD); pressure overload‐induced heart failure; type 2 diabetes | 669 , 670 , 671 |
| Unclear mechanism | Oxautin‐1 | Inhibiting both autophagosome biogenesis and autophagosome maturation | Experiment agents | 672 |
| Clomipramine | Blocking autophagolysosomal fluxes | Treatment of psychiatric disorders (approved); Potential anticancer effects (preclinical) | 626 , 673 | |
| Verteporfin | Disturbing the shape of double membrane autophagosome structure | A benzoporphyrin derivative used in photodynamic therapy (approved) | 674 , 675 |
4.2.1. Inhibition of autophagy initiation and autophagosome formation
Small molecules and natural products targeting ULK1 complex and PI3KC3–C1 are two major approaches to suppress autophagy initiation, which results in the blockage of autophagosome biogenesis. These reagents have been reported to have great potentials in anticancer, antidiabetic therapy, and treatment of neurodegenerative disorders. However, special attention should be taken since these kinase inhibitors are commonly with a broad inhibition of kinase activity, which may bring undesired impacts or even results opposing to its original intention.
The early identified ULK inhibitors, MRT67307 and MRT68921, have been screened from a closely related series of analogues generated during the original TBK1 (TANK‐binding kinase 1) screen, hence the side effects of TBK1 inhibition are unavoidable for these drugs. 580 Another early identified ULK inhibitor SBI‐0206965 also demonstrates a potent AMPK inhibitory effect. 576 , 577 The recently reported ULK‐101 is considered as the most promising ULK inhibitor with a significantly higher potency and selectivity when comparing to the previous inhibitors, while further investigation in vivo is required to proceed with preclinical testing. 581
3‐MA, a pan‐PI3K inhibitor widely used as a useful experiment tool to block early stage of autophagy, has been found to block the autophagy‐promoting PI3K complex (class III) transiently, while inhibits the autophagy‐suppressing PI3K complex (class I) persistently. In this scenario, 3‐MA plays a time‐dependently dual role in autophagy modulation. The similar problem of poor selectivity is also unavoidable when using another PI3K inhibitor, wortmannin. 582 Therefore, to avoid the side effects caused by PIK3CA inhibition, small molecules with higher specificity for PIK3C3 are screened and selected, such as SAR405, VPS34‐IN1, PIK‐III, and SB02024. 319 , 590 , 591 , 592 , 593 , 594 , 595 , 596 , 597 , 598 , 601 These reagents have been found to inhibit autophagy and sensitize cancer response to immunotherapy (anti‐PD‐1/PD‐L1) or chemotherapy (sunitinib and cisplatin) in preclinical studies. 595 , 596 However, PIK3C3 targeting strategy is still not perfect for specific autophagy inhibition, since PIK3C3 is the key component for PI3KC3–C1 for autophagy initiation and PI3KC3–C2 for endosomal trafficking. 676 Suppression of PIK3C3 also alters endosomal pathways in addition to autophagy inhibition. Recently, compound 19a has been screened and studied as a small molecule, which only disturbs the interaction between BECN1 and ATG14 without interference with PI3KC3–C2. 610 This study demonstrates the viability of targeting protein–protein interaction (PPI) to control cellular autophagy process.
In addition to these kinases targeting approaches, diverse novel targets specified for autophagosome formation have been developed, including inhibition of ATG4B, ATG7, and alteration of autophagosome lipid composition by inhibiting lipid transport proteins. 611 , 612 , 613 , 614 , 615 , 619 , 624 Although further validation of the potency and selectivity are still needed, these tools provide great opportunities to observe the detailed modulating processes on autophagosome formation depending on ATG4B, ATG7, or lipid transportation.
4.2.2. Inhibition of autophagosome maturation and lysosome fusion
The enclosing process of double membrane autophagosomes and their fusion with lysosomes can be targeted for autophagy inhibition. Agents blocking cytoskeleton and membrane transport proteins, such as CA‐5f and vinblastine, destabilize cellular organelles (including autophagosomes and lysosomes) movements, resulting in autophagy inhibition via the disturbance of autophagosome–lysosome fusion. 625 , 626 , 627 , 628 However, these agents are usually with unwanted side effects and unavoidable cytotoxicity because of the broad impacts caused by cytoskeleton inhibition. An alternative approach to block autophagosome formation is to modulate the activity of small GTPases required for the autophagosomes and lysosomes membrane fusion, but also with the problem of strong side effects. For example, thapsigargin, blocking agent for RAB7 recruitment, is used to be considered as an inhibitor for autophagosome fusion. However, it also induces ER stress and inhibits SERCA pump. 629 , 630 Vacuolin‐1, which activates RAB5A GTPase to block autophagosome–lysosome fusion, also causes strong side effects via suppressing protein sorting and endocytic pathways simultaneously. 631 , 632 , 633 , 634 , 635
More specific strategies targeting autophagosome maturation are proposed and studied recently. EACC blocks the recruitment of STX17 on autophagosome membranes, which provide an interesting tool to understand the specific function of STX17 on autophagosome formation. 620 , 621 , 622 In addition, S130 has been reported to decrease LC3‐II delipidation and hence suppresses the recycling of LC3, which has been validated via in vivo model and shows inhibitory effects on colon cancer cells. 623 Vertepofin, a US FDA‐approved drug that sensitizes pancreas cancer patients to gemcitabine treatment, inhibits autophagosome maturation via disturbance of double membrane structure, possibly via its interaction with LIR motifs. 674 , 675
4.2.3. Suppression of lysosome activity
Autophagy is known as a one‐way ticket to lysosome degradation. Suppression of lysosome activity significantly blocks autophagic degradation. Here, we described the main lysosomal targeting strategies.
Lysosomotropic drugs are the weak bases compounds incorporated and accumulated into cellular acidic organelles (lysosomes and late endosomes), leading to increased acidic pH value, destabilization of lysosomal membranes, and subsequent blockade of lysosomal functions. 677 CQ and HCQ, as the most‐studied lysosomotropic agents, are widely used as traditional antimalaria drugs, and draw more and more attentions because of its possible therapeutic anti‐COVID‐19 efficacy. 306 The rationale to combine autophagy inhibitors with chemo‐, radio‐, or immune‐antitumor therapy has been proposed based on the importance of autophagy in tumor resistance. As the only autophagy inhibitory drugs approved by US FDA, CQ spurred multiple preclinical studies and clinical trials to investigate their anticancer efficacies alone or in combination with other anticancer treatments. However, although CQ/HCQ have been proved to be safely tolerated in a relatively high dosage and preliminarily effective in a subset of patients (such as patients with melanoma, colon cancer, and kidney cancer), 678 there are still a number of open questions left: (1) the suitable in vivo autophagic marker to confirm whether autophagy is fully blocked by CQ/HCQ; (2) the criteria to select the sensitive population of CQ; (3) side effects including cardiomyopathy. Derivatives modified based on the structure of CQ and other antimalaria drugs have been systematically explored to optimize the potency on autophagy inhibition. Further profiling is needed for this new generation of lysosomotropic agents, including Lys05, DQ661, DC661, lucanthone, and ROC‐325. 594 , 640 , 641 , 642 , 647 , 648 , 660 , 661
V‐ATPase inhibitors prevent lysosome acidification via blockage of lysosomal proton pump activity directly. Multiple nature products, including bafilomycin A1, concanamycin A, and salicylihalamide A, are screened and identified as V‐ATPase inhibitors to suppress autophagic degradation with high potency. 558 , 637 , 653 , 654 , 657 , 658 The lysosomal located V‐ATPase are also required for nutrient sensing and MTORC1 activation. These features may provide additional advantages of V‐ATPase inhibitors in cancer treatment. For instance, a novel identified V‐ATPase inhibitor, verucopeptin, leads to the disassociation of MTOR from lysosome membrane, hence suppressing the MTORC1 activity in addition to autophagy degradation. Verucopeptin has been shown to effectively prevent multidrug resistance and cancer progression in vivo. 657
In addition to the lysosomotropic and V‐ATPase inhibiting strategies, there are multiple mechanistic targets to antagonize lysosome proteolysis, such as the key lysosomal Ca2+ channel TRPML1 (mucolipin TRP channel 1) and lysosomal membrane permeabilization. 649 , 650 , 651 , 665
Several lysosome function inhibitors, including CQ and bafilomycin A1, routinely serve as useful tools to investigate autophagic flux and study the function of autophagy in different physiological or pathological situations. However, it should be cautious when using the lysosomal inhibitors in autophagy studies. These reagents also disturb other lysosome involving cellular processes in addition to autophagic degradation, including endocytosis, phagocytosis, and nutrient sensing. Multiple approaches and carefully designed controls are needed to determinate autophagic flux, especially when the lysosome‐related processes are also involved.
5. SUMMARY AND PERSPECTIVES
The modern era of autophagy research started more than 70 years ago when Christian de Duve discovered lysosome in 1950s and coined the term autophagy in 1960s, while the real breakthrough came only when the ATGs were identified in yeast by Yoshinori Ohsumi in 1990s. In the past near 3 decades, we have observed tremendous progresses in the autophagy field, from the molecular mechanisms, biological functions, and implications in health and diseases. The autophagy research covers almost all the model organisms, from plant, yeast, C. elegans, all the way to mouse and human. In particular, the availability of various transgenic animal models in which ATGs and related genes are manipulated have provided valuable resources for deeper understanding of autophagy and its association with human diseases. More importantly, the autophagy research has moved from basic mechanistic research to translational studies, with numerous autophagy inhibitors or activators being in preclinical and clinical experiments, and we have attempted to summarize all the above points to showcase the importance of autophagy.
However, despite the remarkable achievements, there are equally important challenges and bottlenecks in autophagy research. 6 First, in comparison with macroautophagy, which is autophagosome dependent, relatively little is known about microautophagy in which various cargos are delivered to lysosome for degradation independent of the canonical autophagy machinery and autophagosome formation. 10 For example, how many distinct types of microautophagy exist in eukaryotes? What are the molecular mechanisms unique to microautophagy? Are there any specific proteins that mediate the invagination of autophagic cargos into the lysosome? What factors that trigger microautophagy instead of macroautophagy? How do microautophagy and macroautophagy coordinate with each other to maintain the cellular homeostasis? Answering these questions may help to expand the scope of autophagy research and provide new insights into the biological functions and importance of microautophagy in diseases.
Second, there is increasing appreciation to the importance of selective autophagy. At present, a variety of selective autophagy have been identified, from mitophagy (mitochondria), ER‐phagy (ER), aggrephagy (protein aggregates), lipophagy (lipid droplets), lysophagy (lysosome), glycophagy (glycogen), ferritinophagy (ferritin), just to name a few. 679 These selective forms of autophagy are often featured in specific type of cells/tissues, induced by specific stimuli and mediated by specific adaptors, thus with particularly relevance to certain type of diseases. One good example is mitophagy and its close implication in neurodegenerative disorders such as PD. 236 , 680 Obviously, much more work is needed to understand these forms of selective autophagy, from molecular mechanisms to functional implications in health and disease.
Third, only limited number of transgenic animal models are available that mimic development of autophagy‐related disease via either deletion or overexpression of ATGs. Such animal models are indeed important for understanding the exact role of autophagy in disease, as well as for development of autophagy‐targeted therapeutics. In addition to mouse models, establishment of other model organisms such as zebrafish, drosophila and C. elegans will also be useful.
Fourth, the autophagy research field still faces one technical challenge: lack of proper tools for measuring autophagic flux in vivo, especially in human specimen. Such tools are actually critical for translating the knowledge of autophagy from bench to bed, as we need to know whether autophagy activators or inhibitors should be applied to patients with either impaired or activated autophagy under specific disease conditions.
Last, despite the tremendous efforts in developing autophagy‐specific therapeutic agents, most of them are still at the earlier stage of development, mainly preclinical. The only autophagy inhibitor in clinical trials is the repurposed antimalaria drug CQ/HCQ based on their lysosomotropic property and inhibitory effect on lysosomal function. 344 , 681 It is expected that more such autophagy‐targeted agents, both for macroautophagy and microautophagy, nonselective and selective autophagy, will be entering the clinical stage and bring real benefits to patients.
CONFLICTS OF INTERESTS
Canhua Huang is an editorial board member of MedComm. The paper was handled by another Editor and has undergone a rigorous peer‐review process. Author Canhua Huang was not involved in the journal's review of/or decisions related to this manuscript. The other authors have no conflicts of interest to declare.
ETHICS STATEMENT
Not applicable.
AUTHOR CONTRIBUTIONS
H. M. S., C. W., and W. H. conceived the structure of the manuscript. G. L., Y. W., Y. S., and H. M. S. drafted initial manuscript, with contribution from C. W., W. H., C. H. H. H. M. S., G. L., Y. S., Y. W., Z. Z., C. W., W. H., and C. H. H. revised the manuscript. G. L., Y. W., Y. S., and Z. Z. prepared the figures. All authors read and approved the final manuscript. G. L., Y. W., and Y. S. contributed equally to this study.
ACKNOWLEDGMENTS
We apologized for failing to cite many excellent works due to space limits. This work was supported the following research grants: UM‐MYRG2020‐00022‐FHS and Macau Science and Technology Development Fund (FDCT0078/2020/A2 and 0031/2021/A1) to H. M. S. Figures 3, 4, 6, and 7 were created by BioRender (BioRender.com).
Lu G, Wang Y, Shi Y, et al. Autophagy in health and disease: From molecular mechanisms to therapeutic target. MedComm. 2022;3:e150. 10.1002/mco2.150
Contributor Information
Weifeng He, Email: whe761211@hotmail.com.
Chuang Wang, Email: wangchuang@nbu.edu.cn.
Han‐Ming Shen, Email: hmshen@um.edu.mo.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7(9):847‐849. [DOI] [PubMed] [Google Scholar]
- 2. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsukada M, Ohsumi Y. Isolation and characterization of autophagy‐defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1‐2):169‐174. [DOI] [PubMed] [Google Scholar]
- 4. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458‐467. [DOI] [PubMed] [Google Scholar]
- 5. Maruyama T, Alam JM, Fukuda T, et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat Struct Mol Biol. 2021;28(7):583‐593. [DOI] [PubMed] [Google Scholar]
- 6. Mizushima N, White E, Rubinsztein DC. Breakthroughs and bottlenecks in autophagy research. Trends Mol Med. 2021;27(9):835‐838. [DOI] [PubMed] [Google Scholar]
- 7. Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20(5):521‐527. [DOI] [PubMed] [Google Scholar]
- 8. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaushik S, Cuervo AM. The coming of age of chaperone‐mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schuck S. Microautophagy ‐ distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci. 2020;133(17) [DOI] [PubMed] [Google Scholar]
- 11. Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36(13):1811‐1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin M, Liu X, Klionsky DJ. SnapShot: Selective Autophagy. Cell. 2013;152(1‐2):368‐368 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farré JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17(9):537‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20(3):233‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zellner S, Schifferer M, Behrends C. Systematically defining selective autophagy receptor‐specific cargo using autophagosome content profiling. Mol Cell. 2021; 81(6):1337‐1354.e8 [DOI] [PubMed] [Google Scholar]
- 16. Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383(16):1564‐1576. [DOI] [PubMed] [Google Scholar]
- 17. Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO j. 2021;40(19):e108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132‐139. [DOI] [PubMed] [Google Scholar]
- 19. Noda NN, Fujioka Y. Atg1 family kinases in autophagy initiation. Cell Mol Life Sci. 2015;72(16):3083‐3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi S, Kim DJ, Stjepanovic G, Hurley JH. Structure of the human Atg13‐Atg101 HORMA heterodimer: an interaction hub within the ULK1 Complex. Structure. 2015;23(10):1848‐1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hosokawa N, Hara T, Kaizuka T, et al. Nutrient‐dependent mTORC1 association with the ULK1‐Atg13‐FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981‐1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5(5):649‐662. [DOI] [PubMed] [Google Scholar]
- 23. Hara T, Takamura A, Kishi C, et al. FIP200, a ULK‐interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Funderburk SF, Wang QJ, Yue Z. The Beclin 1‐VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20(6):355‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohashi Y, Tremel S, Williams RL. VPS34 complexes from a structural perspective. J Lipid Res. 2019;60(2):229‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rostislavleva K, Soler N, Ohashi Y, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350(6257):aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X, He L, Che KH, et al. Imperfect interface of Beclin1 coiled‐coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA. 2011;108(19):7769‐7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3‐kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360‐5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome‐anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506‐522. [DOI] [PubMed] [Google Scholar]
- 31. Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12‐5‐16L1. Mol Cell. 2014;55(2):238‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bakula D, Muller AJ, Zuleger T, et al. WIPI3 and WIPI4 beta‐propellers are scaffolds for LKB1‐AMPK‐TSC signalling circuits in the control of autophagy. Nat Commun. 2017;8:15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graef M. Membrane tethering by the autophagy ATG2A‐WIPI4 complex. Proc Natl Acad Sci USA. 2018;115(42):10540‐10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chowdhury S, Otomo C, Leitner A, et al. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A‐WIPI4 complex. Proc Natl Acad Sci USA. 2018;115(42):E9792‐E9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osawa T, Ishii Y, Noda NN. Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells. 2020;25(1):65‐70. [DOI] [PubMed] [Google Scholar]
- 36. Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286(9):7327‐7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Woo J, Park E, Dinesh‐Kumar SP. Differential processing of Arabidopsis ubiquitin‐like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA. 2014;111(2):863‐868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1‐10. [DOI] [PubMed] [Google Scholar]
- 39. Ohsumi Y, Mizushima N. Two ubiquitin‐like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15(2):231‐236. [DOI] [PubMed] [Google Scholar]
- 40. Lystad AH, Carlsson SR, de la Ballina LR, et al. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy‐related processes. Nat Cell Biol. 2019;21(3):372‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nieto‐Torres JL, Leidal AM, Debnath J, Hansen M. Beyond Autophagy: The Expanding Roles of ATG8 Proteins. Trends Biochem Sci. 2021;46(8):673‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432(1):80‐103. [DOI] [PubMed] [Google Scholar]
- 43. Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matoba K, Kotani T, Tsutsumi A, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020;27(12):1185‐1193. [DOI] [PubMed] [Google Scholar]
- 45. Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14(2):207‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu G, Wang L, Zhou J, Liu W, Shen HM. A destiny for degradation: interplay between cullin‐RING E3 ligases and autophagy. Trends Cell Biol. 2021;31(6):432‐444. [DOI] [PubMed] [Google Scholar]
- 47. Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349‐364. [DOI] [PubMed] [Google Scholar]
- 48. Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440(2):283‐291. [DOI] [PubMed] [Google Scholar]
- 49. Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185(3):871‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy. 2013;9(2):124‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297‐12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Egan DF, Chun MG, Vamos M, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59(2):285‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grasso D, Renna FJ, Vaccaro MI. Initial steps in mammalian autophagosome biogenesis. Front Cell Dev Biol. 2018;6:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kraft C, Kijanska M, Kalie E, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin‐like protein Atg8 regulates autophagy. EMBO J. 2012;31(18):3691‐3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alemu EA, Lamark T, Torgersen KM, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3‐interacting region (LIR) motifs. J Biol Chem. 2012;287(47):39275‐39290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wallot‐Hieke N, Verma N, Schlutermann D, et al. Systematic analysis of ATG13 domain requirements for autophagy induction. Autophagy. 2018;14(5):743‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Webster CP, Smith EF, Bauer CS, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35(15):1656‐1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J, Menon S, Yamasaki A, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci USA. 2013;110(24):9800‐9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao YG, Liu N, Miao G, Chen Y, Zhao H, Zhang H. The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr Biol. 2018;28(8):1234‐1245 e4. [DOI] [PubMed] [Google Scholar]
- 60. Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu J, He L, Behrends C, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L‐linked phosphatidylinositol‐3 kinase III activity. Nat Commun. 2014;5:3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J. 2014;461(2):315‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Young LN, Cho K, Lawrence R, Zoncu R, Hurley JH. Dynamics and architecture of the NRBF2‐containing phosphatidylinositol 3‐kinase complex I of autophagy. Proc Natl Acad Sci USA. 2016;113(29):8224‐8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhong Y, Morris DH, Jin L, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein‐containing Beclin 1‐Vps34 complex architecture and reducing intracellular phosphatidylinositol‐3 phosphate levels. J Biol Chem. 2014;289(38):26021‐26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Birgisdottir AB, Mouilleron S, Bhujabal Z, et al. Members of the autophagy class III phosphatidylinositol 3‐kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15(8):1333‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin‐1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741‐750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wold MS, Lim J, Lachance V, Deng Z, Yue Z. ULK1‐mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington's disease models. Mol Neurodegener. 2016;11(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mercer TJ, Ohashi Y, Boeing S, et al. Phosphoproteomic identification of ULK substrates reveals VPS15‐dependent ULK/VPS34 interplay in the regulation of autophagy. EMBO J. 2021;40(14):e105985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Proikas‐Cezanne T, Takacs Z, Donnes P, Kohlbacher O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci. 2015;128(2):207‐217. [DOI] [PubMed] [Google Scholar]
- 70. Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chang C, Jensen LE, Hurley JH. Autophagosome biogenesis comes out of the black box. Nat Cell Biol. 2021;23(5):450‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Krick R, Busse RA, Scacioc A, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta‐propeller protein family. Proc Natl Acad Sci USA. 2012;109(30):E2042‐E2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two‐site recognition of phosphatidylinositol 3‐phosphate by PROPPINs in autophagy. Mol Cell. 2012;47(3):339‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Proikas‐Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI‐1alpha (WIPI49), a member of the novel 7‐bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation‐induced autophagy. Oncogene. 2004;23(58):9314‐9325. [DOI] [PubMed] [Google Scholar]
- 75. Fracchiolla D, Chang C, Hurley JH, Martens S. A PI3K‐WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol. 2020;219(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu G, Yi J, Gubas A, et al. Suppression of autophagy during mitosis via CUL4‐RING ubiquitin ligases‐mediated WIPI2 polyubiquitination and proteasomal degradation. Autophagy. 2019;15(11):1917‐1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stavoe AK, Gopal PP, Gubas A, Tooze SA, Holzbaur EL. Expression of WIPI2B counteracts age‐related decline in autophagosome biogenesis in neurons. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lu G, Tan HWS, Schmauck‐Medina T, et al. WIPI2 positively regulates mitophagy by promoting mitochondrial recruitment of VCP. Autophagy. 2022:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764‐776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bakula D, Mueller AJ, Proikas‐Cezanne T. WIPI beta‐propellers function as scaffolds for STK11/LKB1‐AMPK and AMPK‐related kinase signaling in autophagy. Autophagy. 2018;14(6):1082‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ren J, Liang R, Wang W, Zhang D, Yu L, Feng W. Multi‐site‐mediated entwining of the linear WIR‐motif around WIPI beta‐propellers for autophagy. Nat Commun. 2020;11(1):2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bueno‐Arribas M, Blanca I, Cruz‐Cuevas C, Escalante R, Navas MA, Vincent O. A conserved ATG2 binding site in WIPI4 and yeast Hsv2 is disrupted by mutations causing beta‐propeller protein‐associated neurodegeneration. Hum Mol Genet. 2021;31(1):111‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Osawa T, Noda NN. Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 2019;28(6):1005‐1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ji C, Zhao H, Chen D, Zhang H, Zhao YG. beta‐propeller proteins WDR45 and WDR45B regulate autophagosome maturation into autolysosomes in neural cells. Curr Biol. 2021;31(8):1666‐1677 e6. [DOI] [PubMed] [Google Scholar]
- 85. Puri C, Vicinanza M, Ashkenazi A, et al. The RAB11A‐positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P‐RAB11A. Dev Cell. 2018;45(1):114‐131 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stanga D, Zhao Q, Milev MP, Saint‐Dic D, Jimenez‐Mallebrera C, Sacher M. TRAPPC11 functions in autophagy by recruiting ATG2B‐WIPI4/WDR45 to preautophagosomal membranes. Traffic. 2019;20(5):325‐345. [DOI] [PubMed] [Google Scholar]
- 87. Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane‐binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin‐like key regulators of autophagy. Essays Biochem. 2013;55:51‐64. [DOI] [PubMed] [Google Scholar]
- 89. Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE‐16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20(4):444‐454. [DOI] [PubMed] [Google Scholar]
- 90. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728‐741. [DOI] [PubMed] [Google Scholar]
- 91. Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010;584(7):1319‐1326. [DOI] [PubMed] [Google Scholar]
- 92. Lamb CA, Nuhlen S, Judith D, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35(3):281‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Weerasekara VK, Panek DJ, Broadbent DG, et al. Metabolic‐stress‐induced rearrangement of the 14‐3‐3zeta interactome promotes autophagy via a ULK1‐ and AMPK‐regulated 14‐3‐3zeta interaction with phosphorylated Atg9. Mol Cell Biol. 2014;34(24):4379‐4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Maeda S, Yamamoto H, Kinch LN, et al. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol. 2020;27(12):1194‐1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sawa‐Makarska J, Baumann V, Coudevylle N, et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 2020;369(6508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39(2):61‐71. [DOI] [PubMed] [Google Scholar]
- 97. Lorincz P, Juhasz G. Autophagosome‐lysosome fusion. J Mol Biol. 2020;432(8):2462‐2482. [DOI] [PubMed] [Google Scholar]
- 98. van der Kant R, Jonker CT, Wijdeven RH, et al. Characterization of the mammalian CORVET and HOPS complexes and their modular restructuring for endosome specificity. J Biol Chem. 2015;290(51):30280‐30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome‐lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25(8):1327‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Muller MP, Goody RS. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases. 2018;9(1‐2):5‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo‐lysosomal traffic. Cell Signal. 2011;23(3):516‐521. [DOI] [PubMed] [Google Scholar]
- 102. Guerra F, Bucci C. Multiple roles of the small GTPase Rab7. Cells. 2016;5(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C‐Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE‐dependent docking and fusion. J Cell Biol. 2000;151(3):551‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McEwan DG, Popovic D, Gubas A, et al. PLEKHM1 regulates autophagosome‐lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57(1):39‐54. [DOI] [PubMed] [Google Scholar]
- 105. Lurick A, Gao J, Kuhlee A, et al. Multivalent Rab interactions determine tether‐mediated membrane fusion. Mol Biol Cell. 2017;28(2):322‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liang C, Lee JS, Inn KS, et al. Beclin1‐binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10(7):776‐787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631‐643. [DOI] [PubMed] [Google Scholar]
- 108. Itakura E, Kishi‐Itakura C, Mizushima N. The hairpin‐type tail‐anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256‐1269. [DOI] [PubMed] [Google Scholar]
- 109. Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q‐ and R‐SNAREs. Proc Natl Acad Sci USA. 1998;95(26):15781‐15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323(5913):474‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18(9):3463‐3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kumar S, Gu Y, Abudu YP, et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019;49(1):130‐144 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Guo B, Liang Q, Li L, et al. O‐GlcNAc‐modification of SNAP‐29 regulates autophagosome maturation. Nat Cell Biol. 2014;16(12):1215‐1226. [DOI] [PubMed] [Google Scholar]
- 115. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21(4):183‐203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Condon KJ, Sabatini DM. Nutrient regulation of mTORC1 at a glance. J Cell Sci. 2019;132(21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dossou AS, Basu A. The emerging roles of mTORC1 in macromanaging autophagy. Cancers (Basel). 2019;11(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37(11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125(1):25‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hardie DG, Carling D, Gamblin SJ. AMP‐activated protein kinase: also regulated by ADP? Trends Biochem Sci. 2011;36(9):470‐477. [DOI] [PubMed] [Google Scholar]
- 122. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577‐590. [DOI] [PubMed] [Google Scholar]
- 125. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang D, Wang W, Sun X, et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy. 2016;12(9):1447‐1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1‐2):290‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Puente C, Hendrickson RC, Jiang X. Nutrient‐regulated phosphorylation of ATG13 inhibits starvation‐induced autophagy. J Biol Chem. 2016;291(11):6026‐6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mariño G, Niso‐Santano M, Baehrecke EH, Kroemer G. Self‐consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15(2):81‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mostowy S. Autophagy and bacterial clearance: a not so clear picture. Cell Microbiol. 2013;15(3):395‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Czyzyk‐Krzeska MF, Meller J, Plas DR. Not all autophagy is equal. Autophagy. 2012;8(7):1155‐1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651‐662. [DOI] [PubMed] [Google Scholar]
- 135. Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672‐676. [DOI] [PubMed] [Google Scholar]
- 136. Poillet‐Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33(11‐12):610‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809‐1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Galluzzi L, Pietrocola F, Bravo‐San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. Embo j. 2015;34(7):856‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Holdgaard SG, Cianfanelli V, Pupo E, et al. Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat Commun. 2019;10(1):4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Monkkonen T, Debnath J. Inflammatory signaling cascades and autophagy in cancer. Autophagy. 2018;14(2):190‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cannizzo ES, Clement CC, Morozova K, et al. Age‐related oxidative stress compromises endosomal proteostasis. Cell Rep. 2012;2(1):136‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013‐1022. [DOI] [PubMed] [Google Scholar]
- 143. Moscat J, Karin M, Diaz‐Meco MT. p62 in cancer: Signaling adaptor beyond autophagy. Cell. 2016;167(3):606‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. DeVorkin L, Pavey N, Carleton G, et al. Autophagy regulation of metabolism is required for CD8(+) T cell anti‐tumor immunity. Cell Rep. 2019;27(2):502‐513.e5. [DOI] [PubMed] [Google Scholar]
- 145. Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC‐I. Nature. 2020;581(7806):100‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Vera‐Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018;9(1):1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rosenfeldt MT, O'Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296‐300. [DOI] [PubMed] [Google Scholar]
- 148. Zhong Z, Sanchez‐Lopez E, Karin M. Autophagy, inflammation, and immunity: a Troika governing cancer and its treatment. Cell. 2016;166(2):288‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Li L, Tan J, Miao Y, Lei P, Zhang Q. ROS and autophagy: Interactions and molecular regulatory mechanisms. Cell Mol Neurobiol. 2015;35(5):615‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cordani M, Donadelli M, Strippoli R, Bazhin AV, Sanchez‐Alvarez M. Interplay between ROS and autophagy in cancer and aging: from molecular mechanisms to novel therapeutic approaches. Oxid Med Cell Longev. 2019;2019:8794612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yamazaki T, Bravo‐San Pedro JM, Galluzzi L, Kroemer G, Pietrocola F. Autophagy in the cancer‐immunity dialogue. Adv Drug Deliv Rev. 2021;169:40‐50. [DOI] [PubMed] [Google Scholar]
- 152. Zhao Y, Liu B, Xu L, et al. ROS‐induced mtDNA release: the emerging messenger for communication between neurons and innate immune cells during neurodegenerative disorder progression. Antioxidants (Basel). 2021;10(12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Quan Y, Xin Y, Tian G, Zhou J, Liu X. Mitochondrial ROS‐Modulated mtDNA: a potential target for cardiac aging. Oxid Med Cell Longev. 2020;2020:9423593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19(3):170‐183. [DOI] [PubMed] [Google Scholar]
- 155. Xia H, Green DR, Zou W. Autophagy in tumour immunity and therapy. Nat Rev Cancer. 2021;21(5):281‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Crotzer VL, Blum JS. Autophagy and its role in MHC‐mediated antigen presentation. J Immunol. 2009;182(6):3335‐3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Puleston DJ, Zhang H, Powell TJ, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Seto S, Tsujimura K, Horii T, Koide Y. Autophagy adaptor protein p62/SQSTM1 and autophagy‐related gene Atg5 mediate autophagosome formation in response to Mycobacterium tuberculosis infection in dendritic cells. PLoS One. 2013;8(12):e86017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32(2):227‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675‐699. [DOI] [PubMed] [Google Scholar]
- 161. Song YJ, Zhang SS, Guo XL, et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient‐deprived tumor microenvironment. Cancer Lett. 2013;339(1):70‐81. [DOI] [PubMed] [Google Scholar]
- 162. Gong C, Bauvy C, Tonelli G, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem‐like/progenitor cells. Oncogene. 2013;32(18):2261‐2272, 2272e.1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Peng Q, Qin J, Zhang Y, et al. Autophagy maintains the stemness of ovarian cancer stem cells by FOXA2. J Exp Clin Cancer Res. 2017;36(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhang D, Zhao Q, Sun H, et al. Defective autophagy leads to the suppression of stem‐like features of CD271(+) osteosarcoma cells. J Biomed Sci. 2016;23(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Buccarelli M, Marconi M, Pacioni S, et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018;9(8):841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F. Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ. 2019;26(4):690‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Cojoc M, Mäbert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015;31:16‐27. [DOI] [PubMed] [Google Scholar]
- 168. Mortensen M, Watson AS, Simon AK. Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. Autophagy. 2011;7(9):1069‐1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Li L, Wang Y, Jiao L, et al. Protective autophagy decreases osimertinib cytotoxicity through regulation of stem cell‐like properties in lung cancer. Cancer Lett. 2019;452:191‐202. [DOI] [PubMed] [Google Scholar]
- 170. Choi DS, Blanco E, Kim YS, et al. Chloroquine eliminates cancer stem cells through deregulation of Jak2 and DNMT1. Stem Cells (Dayton, Ohio). 2014;32(9):2309‐2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lei Y, Zhang D, Yu J, Dong H, Zhang J, Yang S. Targeting autophagy in cancer stem cells as an anticancer therapy. Cancer Lett. 2017;393:33‐39. [DOI] [PubMed] [Google Scholar]
- 172. Menzies FM, Fleming A, Caricasole A, et al. Autophagy and neurodegeneration: Pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015‐1034. [DOI] [PubMed] [Google Scholar]
- 173. Mallucci GR, Klenerman D, Rubinsztein DC. Developing therapies for neurodegenerative disorders: Insights from protein aggregation and cellular stress responses. Annu Rev Cell Dev Biol. 2020;36:165‐189. [DOI] [PubMed] [Google Scholar]
- 174. Deng Z, Purtell K, Lachance V, Wold MS, Chen S, Yue Z. Autophagy receptors and neurodegenerative diseases. Trends Cell Biol. 2017;27(7):491‐504. [DOI] [PubMed] [Google Scholar]
- 175. Deng Z, Lim J, Wang Q, et al. ALS‐FTLD‐linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti‐oxidative stress pathway. Autophagy. 2020;16(5):917‐931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. McCauley ME, Baloh RH. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 2019;137(5):715‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Odagiri S, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in alpha‐synucleinopathy. Acta Neuropathol. 2012;124(2):173‐186. [DOI] [PubMed] [Google Scholar]
- 178. Caccamo A, Ferreira E, Branca C, Oddo S. p62 improves AD‐like pathology by increasing autophagy. Mol Psychiatry. 2017;22(6):865‐873. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 179. Fu Y, Wu P, Pan Y, et al. A toxic mutant huntingtin species is resistant to selective autophagy. Nat Chem Biol. 2017;13(11):1152‐1154. [DOI] [PubMed] [Google Scholar]
- 180. Ravikumar B, Duden R, Rubinsztein DC. Aggregate‐prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11(9):1107‐1117. [DOI] [PubMed] [Google Scholar]
- 181. Tsvetkov AS, Arrasate M, Barmada S, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Natu Chem Biol. 2013;9(9):586‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Citron M. Alzheimer's disease: strategies for disease modification. Nat Rev Drug Disco. 2010;9(5):387‐398. [DOI] [PubMed] [Google Scholar]
- 183. Kerr JS, Adriaanse BA, Greig NH, et al. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40(3):151‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. 2022 Alzheimer's disease facts and figures. Alzheimer's Dement. 2022;18(4):700‐789. [DOI] [PubMed] [Google Scholar]
- 185. Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno‐electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113‐122. [DOI] [PubMed] [Google Scholar]
- 186. Bai B, Wang X, Li Y, et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer's disease progression. Neuron. 2020;105(6):975‐991.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Berger Z, Ravikumar B, Menzies FM, et al. Rapamycin alleviates toxicity of different aggregate‐prone proteins. Hum Mol Genet. 2006;15(3):433‐442. [DOI] [PubMed] [Google Scholar]
- 188. Silva MC, Nandi GA, Tentarelli S, et al. Prolonged tau clearance and stress vulnerability rescue by pharmacological activation of autophagy in tauopathy neurons. Nat Commun. 2020;11(1):3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Papassotiropoulos A, Bagli M, Kurz A, et al. A genetic variation of cathepsin D is a major risk factor for Alzheimer's disease. Ann Neurol. 2000;47(3):399‐403. [PubMed] [Google Scholar]
- 190. Butzlaff M, Hannan SB, Karsten P, et al. Impaired retrograde transport by the Dynein/Dynactin complex contributes to Tau‐induced toxicity. Hum Mol Genet. 2015;24(13):3623‐3637. [DOI] [PubMed] [Google Scholar]
- 191. van Harten AC, Visser PJ, Pijnenburg YA, et al. Cerebrospinal fluid Aβ42 is the best predictor of clinical progression in patients with subjective complaints. Alzheimer's Dementia. 2013;9(5):481‐487. [DOI] [PubMed] [Google Scholar]
- 192. Cavieres VA, González A, Muñoz VC, et al. Tetrahydrohyperforin inhibits the proteolytic processing of amyloid precursor protein and enhances its degradation by Atg5‐dependent autophagy. PLoS One. 2015;10(8):e0136313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Caccamo A, Majumder S, Richardson A, Strong R, Oddo S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid‐beta, and Tau: effects on cognitive impairments. J Biol Chem. 2010;285(17):13107‐13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Heckmann BL, Teubner BJW, Tummers B, et al. LC3‐associated endocytosis facilitates β‐amyloid clearance and mitigates neurodegeneration in Murine Alzheimer's disease. Cell. 2019;178(3):536‐551.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Huang Y, Happonen KE, Burrola PG, et al. Microglia use TAM receptors to detect and engulf amyloid β plaques. Nat Immunol. 2021;22(5):586‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Boland B, Kumar A, Lee S, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosc. 2008;28(27):6926‐6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Meng T, Lin S, Zhuang H, et al. Recent progress in the role of autophagy in neurological diseases. Cell Stress. 2019;3(5):141‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Nilsson P, Loganathan K, Sekiguchi M, et al. Aβ secretion and plaque formation depend on autophagy. Cell Rep. 2013;5(1):61‐69. [DOI] [PubMed] [Google Scholar]
- 199. Nilsson P, Sekiguchi M, Akagi T, et al. Autophagy‐related protein 7 deficiency in amyloid β (Aβ) precursor protein transgenic mice decreases Aβ in the multivesicular bodies and induces Aβ accumulation in the Golgi. Am J Pathol. 2015;185(2):305‐313. [DOI] [PubMed] [Google Scholar]
- 200. Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362(4):329‐344. [DOI] [PubMed] [Google Scholar]
- 201. Kobro‐Flatmoen A, Lagartos‐Donate MJ, Aman Y, Edison P, Witter MP, Fang EF. Re‐emphasizing early Alzheimer's disease pathology starting in select entorhinal neurons, with a special focus on mitophagy. Ageing Res Rev. 2021;67:101307. [DOI] [PubMed] [Google Scholar]
- 202. Pradeepkiran JA, Reddy PH. Defective mitophagy in Alzheimer's disease. Ageing Res Rev. 2020;64:101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Fang EF, Hou Y, Palikaras K, et al. Mitophagy inhibits amyloid‐β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. 2019;22(3):401‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Lizama BN, Chu CT. Neuronal autophagy and mitophagy in Parkinson's disease. Mol Aspects Med. 2021;82:100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Singleton AB, Farrer M, Johnson J, et al. alpha‐Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. [DOI] [PubMed] [Google Scholar]
- 206. Bellomo G, Paciotti S, Gatticchi L, Parnetti L. The vicious cycle between alpha‐synuclein aggregation and autophagic‐lysosomal dysfunction. Mov Disord. 2020;35(1):34‐44. [DOI] [PubMed] [Google Scholar]
- 207. Sato S, Uchihara T, Fukuda T, et al. Loss of autophagy in dopaminergic neurons causes Lewy pathology and motor dysfunction in aged mice. Sci Rep. 2018;8(1):2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Yan JQ, Yuan YH, Chu SF, Li GH, Chen NH. E46K Mutant α‐Synuclein is degraded by both proteasome and macroautophagy pathway. Molecules (Basel, Switzerland). 2018;23(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha‐Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278(27):25009‐25013. [DOI] [PubMed] [Google Scholar]
- 210. Hoffmann AC, Minakaki G, Menges S, et al. Extracellular aggregated alpha synuclein primarily triggers lysosomal dysfunction in neural cells prevented by trehalose. Sci Rep. 2019;9(1):544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Moors TE, Paciotti S, Ingrassia A, et al. Characterization of brain lysosomal activities in GBA‐related and Sporadic Parkinson's disease and dementia with lewy bodies. Mol Neurobiol. 2019;56(2):1344‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Do J, McKinney C, Sharma P, Sidransky E. Glucocerebrosidase and its relevance to Parkinson disease. Mol Neurodegener. 2019;14(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. García‐Sanz P, Orgaz L, Bueno‐Gil G, et al. N370S‐GBA1 mutation causes lysosomal cholesterol accumulation in Parkinson's disease. Mov Disord. 2017;32(10):1409‐1422. [DOI] [PubMed] [Google Scholar]
- 214. Velayati A, Yu WH, Sidransky E. The role of glucocerebrosidase mutations in Parkinson disease and Lewy body disorders. Curr Neurol Neuroscie Rep. 2010;10(3):190‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Yap TL, Velayati A, Sidransky E, Lee JC. Membrane‐bound α‐synuclein interacts with glucocerebrosidase and inhibits enzyme activity. Mol Genet Metab. 2013;108(1):56‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Mazzulli JR, Xu YH, Sun Y, et al. Gaucher disease glucocerebrosidase and α‐synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146(1):37‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Esteves AR, Cardoso SM. LRRK2 at the crossroad between autophagy and microtubule trafficking: Insights into Parkinson's disease. Neuroscientist. 2017;23(1):16‐26. [DOI] [PubMed] [Google Scholar]
- 218. Dodson MW, Leung LK, Lone M, Lizzio MA, Guo M. Novel ethyl methanesulfonate (EMS)‐induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo. Dis Models Mech. 2014;7(12):1351‐1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Kett LR, Dauer WT. Leucine‐rich repeat kinase 2 for beginners: six key questions. Cold Spring Harb Perspect Med. 2012;2(3):a009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Ramonet D, Daher JP, Lin BM, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6(4):e18568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Sánchez‐Danés A, Richaud‐Patin Y, Carballo‐Carbajal I, et al. Disease‐specific phenotypes in dopamine neurons from human iPS‐based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4(5):380‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Walter J, Bolognin S, Antony PMA, et al. Neural stem cells of Parkinson's disease patients exhibit aberrant mitochondrial morphology and functionality. Stem Cell Rep. 2019;12(5):878‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75(22):2017‐2020. [DOI] [PubMed] [Google Scholar]
- 224. Ryan T, Bamm VV, Stykel MG, et al. Cardiolipin exposure on the outer mitochondrial membrane modulates α‐synuclein. Nat Commun. 2018;9(1):817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. de la Mata M, Cotán D, Oropesa‐Ávila M, et al. Pharmacological chaperones and coenzyme Q10 treatment improves mutant β‐glucocerebrosidase activity and mitochondrial function in neuronopathic forms of gaucher disease. Sci Rep. 2015;5:10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Li H, Ham A, Ma TC, et al. Mitochondrial dysfunction and mitophagy defect triggered by heterozygous GBA mutations. Autophagy. 2019;15(1):113‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Osellame LD, Rahim AA, Hargreaves IP, et al. Mitochondria and quality control defects in a mouse model of Gaucher disease–links to Parkinson's disease. Cell Metabo. 2013;17(6):941‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Vives‐Bauza C, Przedborski S. Mitophagy: the latest problem for Parkinson's disease. Trends Mol Med. 2011;17(3):158‐165. [DOI] [PubMed] [Google Scholar]
- 229. Malpartida AB, Williamson M, Narendra DP, Wade‐Martins R, Ryan BJ. Mitochondrial dysfunction and mitophagy in Parkinson's disease: From mechanism to therapy. Trends Biochem Sci. 2021;46(4):329‐343. [DOI] [PubMed] [Google Scholar]
- 230. Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143‐153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162‐166. [DOI] [PubMed] [Google Scholar]
- 232. Kazlauskaite A, Kondapalli C, Gourlay R, et al. Parkin is activated by PINK1‐dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460(1):127‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Shiba‐Fukushima K, Imai Y, Yoshida S, et al. PINK1‐mediated phosphorylation of the Parkin ubiquitin‐like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Kondapalli C, Kazlauskaite A, Zhang N, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2(5):120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Ordureau A, Sarraf SA, Duda DM, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56(3):360‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Wang L, Qi H, Tang Y, Shen HM. Post‐translational modifications of key machinery in the control of mitophagy. Trends Biochem Sci. 2020;45(1):58‐75. [DOI] [PubMed] [Google Scholar]
- 237. Sliter DA, Martinez J, Hao L, et al. Parkin and PINK1 mitigate STING‐induced inflammation. Nature. 2018;561(7722):258‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Tabrizi SJ, Ghosh R, Leavitt BR. Huntingtin lowering strategies for disease modification in Huntington's disease. Neuron. 2019;102(4):899. [DOI] [PubMed] [Google Scholar]
- 239. Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83‐98. [DOI] [PubMed] [Google Scholar]
- 240. Heng MY, Duong DK, Albin RL, et al. Early autophagic response in a novel knock‐in model of Huntington disease. Hum Mol Genet. 2010;19(19):3702‐3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Erie C, Sacino M, Houle L, Lu ML, Wei J. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington's disease. Eur J Neurosci. 2015;42(3):1941‐1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Wu JC, Qi L, Wang Y, et al. The regulation of N‐terminal Huntingtin (Htt552) accumulation by Beclin1. Acta Pharmacol Sin. 2012;33(6):743‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Ashkenazi A, Bento CF, Ricketts T, et al. Polyglutamine tracts regulate beclin 1‐dependent autophagy. Nature. 2017;545(7652):108‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Ochaba J, Lukacsovich T, Csikos G, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci USA. 2014;111(47):16889‐16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Martinez‐Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13(5):567‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Rui YN, Xu Z, Patel B, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17(3):262‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Korac J, Schaeffer V, Kovacevic I, et al. Ubiquitin‐independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126(Pt 2):580‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Fox LM, Kim K, Johnson CW, et al. Huntington's disease pathogenesis is modified in vivo by Alfy/Wdfy3 and selective macroautophagy. Neuron. 2020;105(5):813‐821.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Wertz MH, Mitchem MR, Pineda SS, et al. Genome‐wide in vivo CNS screening identifies genes that modify CNS neuronal survival and mHTT toxicity. Neuron. 2020;106(1):76‐89.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Bhatt JM, Gordon PH. Current clinical trials in amyotrophic lateral sclerosis. Expert Opin Investig Drugs. 2007;16(8):1197‐1207. [DOI] [PubMed] [Google Scholar]
- 251. Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: an update. Mol Neurodegener. 2013;8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Williams KL, Warraich ST, Yang S, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(10):2527.e3‐10. [DOI] [PubMed] [Google Scholar]
- 253. Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X‐linked juvenile and adult‐onset ALS and ALS/dementia. Nature. 2011;477(7363):211‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Barmada SJ, Serio A, Arjun A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10(8):677‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Brady OA, Meng P, Zheng Y, Mao Y, Hu F. Regulation of TDP‐43 aggregation by phosphorylation and p62/SQSTM1. J Neurochem. 2011;116(2):248‐259. [DOI] [PubMed] [Google Scholar]
- 256. Hadano S, Mitsui S, Pan L, et al. Functional links between SQSTM1 and ALS2 in the pathogenesis of ALS: cumulative impact on the protection against mutant SOD1‐mediated motor dysfunction in mice. Hum Mol Genet. 2016;25(15):3321‐3340. [DOI] [PubMed] [Google Scholar]
- 257. Lattante S, de Calbiac H, Le Ber I, Brice A, Ciura S, Kabashi E. Sqstm1 knock‐down causes a locomotor phenotype ameliorated by rapamycin in a zebrafish model of ALS/FTLD. Hum Mol Genet. 2015;24(6):1682‐1690. [DOI] [PubMed] [Google Scholar]
- 258. Rothenberg C, Srinivasan D, Mah L, et al. Ubiquilin functions in autophagy and is degraded by chaperone‐mediated autophagy. Hum Mol Genet. 2010;19(16):3219‐3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Chen T, Huang B, Shi X, Gao L, Huang C. Mutant UBQLN2(P497H) in motor neurons leads to ALS‐like phenotypes and defective autophagy in rats. Acta Neuropathol Commun. 2018;6(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Hetz C, Thielen P, Matus S, et al. XBP‐1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23(19):2294‐2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Crippa V, Sau D, Rusmini P, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum Mol Genet. 2010;19(17):3440‐3456. [DOI] [PubMed] [Google Scholar]
- 262. Almeida S, Gao FB. Lost & found: C9ORF72 and the autophagy pathway in ALS/FTD. EMBO J. 2016;35(12):1251‐1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Dols‐Icardo O, García‐Redondo A, Rojas‐García R, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Hum Mol Genet. 2014;23(3):749‐754. [DOI] [PubMed] [Google Scholar]
- 264. Tang BL. C9orf72's interaction with Rab GTPases‐modulation of membrane traffic and autophagy. Front Cell Neurosc. 2016;10:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Sullivan PM, Zhou X, Robins AM, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy‐lysosome pathway. Acta Neuropathol Commun. 2016;4(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Boivin M, Pfister V, Gaucherot A, et al. Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders. Embo J. 2020;39(4):e100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Sellier C, Campanari ML, Julie Corbier C, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin‐2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35(12):1276‐1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Kitada M, Koya D. Autophagy in metabolic disease and ageing. Nat Rev Endocrinol. 2021;17(11):647‐661. [DOI] [PubMed] [Google Scholar]
- 269. Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344‐1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Lim YM, Lim H, Hur KY, et al. Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun. 2014;5:4934. [DOI] [PubMed] [Google Scholar]
- 271. Pietrocola F, Bravo‐San Pedro JM. Targeting autophagy to counteract Obesity‐associated oxidative stress. Antioxidants (Basel). 2021;10(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Park HW, Park H, Semple IA, et al. Pharmacological correction of obesity‐induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation‐induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF‐kappaB pathway. J Biol Chem. 2011;286(37):32324‐32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Kaushik S, Rodriguez‐Navarro JA, Arias E, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14(2):173‐183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose‐specific deletion of autophagy‐related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106(47):19860‐19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Matsuzawa‐Ishimoto Y, Hwang S, Cadwell K. Autophagy and inflammation. Annu Rev Immunol. 2018;36:73‐101. [DOI] [PubMed] [Google Scholar]
- 279. Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19(8):477‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Yamamoto S, Kuramoto K, Wang N, et al. Autophagy differentially regulates insulin production and insulin sensitivity. Cell Rep. 2018;23(11):3286‐3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Ebato C, Uchida T, Arakawa M, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high‐fat diet. Cell Metab. 2008;8(4):325‐332. [DOI] [PubMed] [Google Scholar]
- 282. Jung HS, Chung KW, Won Kim J, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318‐324. [DOI] [PubMed] [Google Scholar]
- 283. Goginashvili A, Zhang Z, Erbs E, et al. Insulin granules. Insulin secretory granules control autophagy in pancreatic β cells. Science. 2015;347(6224):878‐882. [DOI] [PubMed] [Google Scholar]
- 284. Yamaguchi H, Honda S, Torii S, et al. Wipi3 is essential for alternative autophagy and its loss causes neurodegeneration. Nat Commun. 2020;11(1):5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Marasco MR, Linnemann AK. β‐cell autophagy in diabetes pathogenesis. Endocrinology. 2018;159(5):2127‐2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Cheng CW, Villani V, Buono R, et al. Fasting‐mimicking diet promotes Ngn3‐Driven β‐cell regeneration to reverse diabetes. Cell. 2017;168(5):775‐788.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Cnop M, Abdulkarim B, Bottu G, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63(6):1978‐1993. [DOI] [PubMed] [Google Scholar]
- 288. Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in β‐cells. J Biol Chem. 2011;286(49):42534‐42544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Mir SU, George NM, Zahoor L, Harms R, Guinn Z, Sarvetnick NE. Inhibition of autophagic turnover in β‐cells by fatty acids and glucose leads to apoptotic cell death. J Biol Chem. 2015;290(10):6071‐6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Zummo FP, Cullen KS, Honkanen‐Scott M, Shaw JAM, Lovat PE, Arden C. Glucagon‐like Peptide 1 protects pancreatic β‐cells from death by increasing autophagic flux and restoring lysosomal function. Diabetes. 2017;66(5):1272‐1285. [DOI] [PubMed] [Google Scholar]
- 291. Jiang Y, Huang W, Wang J, et al. Metformin plays a dual role in MIN6 pancreatic β cell function through AMPK‐dependent autophagy. Int J Biol Sci. 2014;10(3):268‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Tanemura M, Ohmura Y, Deguchi T, et al. Rapamycin causes upregulation of autophagy and impairs islets function both in vitro and in vivo. Am J Transplant. 2012;12(1):102‐114. [DOI] [PubMed] [Google Scholar]
- 293. Vergès B, Cariou B. mTOR inhibitors and diabetes. Diabetes Res Clin Pract. 2015;110(2):101‐108. [DOI] [PubMed] [Google Scholar]
- 294. Fraenkel M, Ketzinel‐Gilad M, Ariav Y, et al. mTOR inhibition by rapamycin prevents beta‐cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes. 2008;57(4):945‐957. [DOI] [PubMed] [Google Scholar]
- 295. Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14(2):243‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Riebisch AK, Mühlen S, Beer YY, Schmitz I. Autophagy‐A story of bacteria interfering with the host cell degradation machinery. Pathogens. 2021;10(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Pang Y, Wu L, Tang C, Wang H, Wei Y. Autophagy‐inflammation interplay during infection: balancing pathogen clearance and host inflammation. Front Pharmacol. 2022;13:832750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Paik S, Jo EK. An interplay between autophagy and immunometabolism for host defense against mycobacterial infection. Front Immunol. 2020;11:603951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. DeGrace MM, Ghedin E, Frieman MB, et al. Defining the risk of SARS‐CoV‐2 variants on immune protection. Nature. 2022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Burkert FR, Lanser L, Bellmann‐Weiler R, Weiss G. Coronavirus disease 2019: Clinics, treatment, and prevention. Front Microbiol. 2021;12:761887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2020:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS‐CoV‐2: Structure, biology, and Structure‐based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Bello‐Perez M, Sola I, Novoa B, Klionsky DJ, Falco A. Canonical and noncanonical autophagy as potential targets for COVID‐19. Cells. 2020;9(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci. 2020;16(10):1724‐1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Shroff A, Nazarko TY. The molecular interplay between human coronaviruses and autophagy. Cells. 2021;10(8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Pereira G, Leão A, Erustes AG, et al. Pharmacological modulators of autophagy as a potential strategy for the treatment of COVID‐19. Int J Mol Sci. 2021;22(8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS‐CoV‐2. Antiviral Res. 2020;177:104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Scialo F, Daniele A, Amato F, et al. ACE2: The major cell entry receptor for SARS‐CoV‐2. Lung. 2020;198(6):867‐877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Mészáros B, Sámano‐Sánchez H, Alvarado‐Valverde J, et al. Short linear motif candidates in the cell entry system used by SARS‐CoV‐2 and their potential therapeutic implications. Sci Signal. 2021;14(665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Kliche J, Kuss H, Ali M, Ivarsson Y. Cytoplasmic short linear motifs in ACE2 and integrin β(3) link SARS‐CoV‐2 host cell receptors to mediators of endocytosis and autophagy. Sci Signal. 2021;14(665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Morita K, Hama Y, Izume T, et al. Genome‐wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol. 2018;217(11):3817‐3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Moretti F, Bergman P, Dodgson S, et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018;19(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17(4):e2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Hoffmann HH, Schneider WM, Rozen‐Gagnon K, et al. TMEM41B is a pan‐flavivirus host factor. Cell. 2021;184(1):133‐148.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Schneider WM, Luna JM, Hoffmann HH, et al. Genome‐scale identification of SARS‐CoV‐2 and pan‐coronavirus host factor networks. Cell. 2021;184(1):120‐132.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Twu WI, Lee JY, Kim H, et al. Contribution of autophagy machinery factors to HCV and SARS‐CoV‐2 replication organelle formation. Cell Rep. 2021;37(8):110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Williams CG, Jureka AS, Silvas JA, et al. Inhibitors of VPS34 and fatty‐acid metabolism suppress SARS‐CoV‐2 replication. Cell Rep. 2021;36(5):109479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Yuen CK, Wong WM, Mak LF, et al. Suppression of SARS‐CoV‐2 infection in ex‐vivo human lung tissues by targeting class III phosphoinositide 3‐kinase. J Med Virol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092‐2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Ching KL, de Vries M, Gago J, et al. ACE2‐containing defensosomes serve as decoys to inhibit SARS‐CoV‐2 infection. bioRxiv. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. The ATG16L1‐T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2015;64(10):1546‐1552. [DOI] [PubMed] [Google Scholar]
- 324. Rayner JO, Roberts RA, Kim J, et al. AR12 (OSU‐03012) suppresses GRP78 expression and inhibits SARS‐CoV‐2 replication. Biochem Pharmacol. 2020;182:114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Mohamud Y, Xue YC, Liu H, et al. The papain‐like protease of coronaviruses cleaves ULK1 to disrupt host autophagy. Biochem Biophys Res Commun. 2021;540:75‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Kern DM, Sorum B, Mali SS, et al. Cryo‐EM structure of SARS‐CoV‐2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Miao G, Zhao H, Li Y, et al. ORF3a of the COVID‐19 virus SARS‐CoV‐2 blocks HOPS complex‐mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Yang S, Tian M, Johnson AN. SARS‐CoV‐2 protein ORF3a is pathogenic in Drosophila and causes phenotypes associated with COVID‐19 post‐viral syndrome. bioRxiv. 2020; [Google Scholar]
- 329. Zhang Y, Sun H, Pei R, et al. The SARS‐CoV‐2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Stukalov A, Girault V, Grass V, et al. Multilevel proteomics reveals host perturbations by SARS‐CoV‐2 and SARS‐CoV. Nature. 2021;594(7862):246‐252. [DOI] [PubMed] [Google Scholar]
- 331. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Miao G, Zhao H, Li Y, et al. ORF3a of the COVID‐19 virus SARS‐CoV‐2 blocks HOPS complex‐mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56(4):427‐442.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Chen D, Zheng Q, Sun L, et al. ORF3a of SARS‐CoV‐2 promotes lysosomal exocytosis‐mediated viral egress. Dev Cell. 2021;56(23):3250‐3263.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Sun X, Liu Y, Huang Z, et al. SARS‐CoV‐2 non‐structural protein 6 triggers NLRP3‐dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Ghosh S, Dellibovi‐Ragheb TA, Kerviel A, et al. β‐coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183(6):1520‐1535.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Verma R, Saha S, Kumar S, Mani S, Maiti TK, Surjit M. RNA‐protein interaction analysis of SARS‐CoV‐2 5' and 3' untranslated regions reveals a role of Lysosome‐associated membrane protein‐2a during viral infection. mSystems. 2021;6(4):e0064321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Shang C, Zhuang X, Zhang H, et al. Inhibition of autophagy suppresses SARS‐CoV‐2 replication and ameliorates pneumonia in hACE2 transgenic mice and xenografted human lung tissues. J Virol. 2021;95(24):e0153721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 ‐ final report. N Engl J Med. 2020;383(19):1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct‐acting antiviral that inhibits RNA‐dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785‐6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Agarwal A, Rochwerg B, Lamontagne F, et al. A living WHO guideline on drugs for covid‐19. BMJ. 2020;370:m3379. [DOI] [PubMed] [Google Scholar]
- 341. Moreno S, Alcázar B, Dueñas C, González Del Castillo J, Olalla J, Antela A. Use of antivirals in SARS‐CoV‐2 infection. Critical review of the role of remdesivir. Drug Des Dev Ther. 2022;16:827‐841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Chen Y, Li MX, Lu GD, Shen HM, Zhou J. Hydroxychloroquine/chloroquine as therapeutics for COVID‐19: truth under the mystery. Int J Biol Sci. 2021;17(6):1538‐1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double‐edged sword of autophagy. Cancer Res. 2013;73(1):3‐7. [DOI] [PubMed] [Google Scholar]
- 345. Rosenfeld MR, Ye X, Supko JG, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359‐1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Younis NK, Zareef RO, Al Hassan SN, Bitar F, Eid AH, Arabi M. Hydroxychloroquine in COVID‐19 patients: pros and cons. Front Pharmacol. 2020;11:597985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Tang D, Li J, Zhang R, Kang R, Klionsky DJ. Chloroquine in fighting COVID‐19: good, bad, or both? Autophagy. 2020;16(12):2273‐2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Abella BS, Jolkovsky EL, Biney BT, et al. Efficacy and safety of hydroxychloroquine vs placebo for pre‐exposure SARS‐CoV‐2 prophylaxis among health care workers: a randomized clinical trial. JAMA Intern Med. 2020; 181(2):195‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Tyteca D, Van Der Smissen P, Mettlen M, et al. Azithromycin, a lysosomotropic antibiotic, has distinct effects on fluid‐phase and receptor‐mediated endocytosis, but does not impair phagocytosis in J774 macrophages. Exp Cell Res. 2002;281(1):86‐100. [DOI] [PubMed] [Google Scholar]
- 350. Tian AL, Wu Q, Liu P, et al. Lysosomotropic agents including azithromycin, chloroquine and hydroxychloroquine activate the integrated stress response. Cell Death Dis. 2021;12(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Du X, Zuo X, Meng F, et al. Direct inhibitory effect on viral entry of influenza A and SARS‐CoV‐2 viruses by azithromycin. Cell Prolif. 2021;54(1):e12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 353. Gyselinck I, Janssens W, Verhamme P, Vos R. Rationale for azithromycin in COVID‐19: an overview of existing evidence. BMJ Open Respir Res. 2021;8(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Ghazy RM, Almaghraby A, Shaaban R, et al. A systematic review and meta‐analysis on chloroquine and hydroxychloroquine as monotherapy or combined with azithromycin in COVID‐19 treatment. Sci Rep. 2020;10(1):22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Fiolet T, Guihur A, Rebeaud ME, Mulot M, Peiffer‐Smadja N, Mahamat‐Saleh Y. Effect of hydroxychloroquine with or without azithromycin on the mortality of coronavirus disease 2019 (COVID‐19) patients: a systematic review and meta‐analysis. Clin Microbiol Infect. 2021;27(1):19‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Echeverría‐Esnal D, Martin‐Ontiyuelo C, Navarrete‐Rouco ME, et al. Azithromycin in the treatment of COVID‐19: a review. Expert Rev Anti Infect Ther. 2020:1‐17. [DOI] [PubMed] [Google Scholar]
- 357. Yang ND, Tan SH, Ng S, et al. Artesunate induces cell death in human cancer cells via enhancing lysosomal function and lysosomal degradation of ferritin. J Biol Chem. 2014;289(48):33425‐33441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Uckun FM, Saund S, Windlass H, Trieu V. Repurposing Anti‐malaria phytomedicine artemisinin as a COVID‐19 drug. Front Pharmacol. 2021;12:649532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Krishna S, Augustin Y, Wang J, et al. Repurposing antimalarials to tackle the COVID‐19 pandemic. Trends Parasitol. 2021;37(1):8‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Cao R, Hu H, Li Y, et al. Anti‐SARS‐CoV‐2 Potential of artemisinins in vitro. ACS Infect Dis. 2020;6(9):2524‐2531. [DOI] [PubMed] [Google Scholar]
- 361. Wang J, Xu C, Wong YK, et al. Preparedness is essential for malaria‐endemic regions during the COVID‐19 pandemic. Lancet. 2020;395(10230):1094‐1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Ataba E, Dorkenoo AM, Nguepou CT, et al. Potential emergence of plasmodium resistance to artemisinin induced by the use of artemisia annua for malaria and COVID‐19 prevention in Sub‐African region. Acta Parasitol. 2022;67(1):55‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Go AS, Mozaffarian D, Roger VL, et al. Heart disease and stroke statistics–2014 update: a report from the American Heart Association. Circulation. 2014;129(3):e28‐e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Lavandero S, Chiong M, Rothermel BA, Hill JA. Autophagy in cardiovascular biology. J Clin Invest. 2015;125(1):55‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Zhang J, Liu J, Huang Y, et al. FRS2alpha‐mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110(4):e29‐e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Lampert MA, Orogo AM, Najor RH, et al. BNIP3L/NIX and FUNDC1‐mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy. 2019;15(7):1182‐1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Lee E, Koo Y, Ng A, et al. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 2014;10(4):572‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Schiattarella GG, Hill JA. Therapeutic targeting of autophagy in cardiovascular disease. J Mol Cell Cardiol. 2016;95:86‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Lesnefsky EJ, Chen Q, Tandler B, Hoppel CL. Mitochondrial dysfunction and myocardial Ischemia‐reperfusion: Implications for novel therapies. Annu Rev Pharmacol Toxicol. 2017;57:535‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP‐activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100(6):914‐922. [DOI] [PubMed] [Google Scholar]
- 371. Ma S, Wang Y, Chen Y, Cao F. The role of the autophagy in myocardial ischemia/reperfusion injury. Biochim Biophys Acta. 2015;1852(2):271‐276. [DOI] [PubMed] [Google Scholar]
- 372. Hamacher‐Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281(40):29776‐29787. [DOI] [PubMed] [Google Scholar]
- 373. Lu Q, Liu J, Li X, et al. Empagliflozin attenuates ischemia and reperfusion injury through LKB1/AMPK signaling pathway. Mol Cell Endocrinol. 2020;501:110642. [DOI] [PubMed] [Google Scholar]
- 374. Zhang Y, Wang Y, Xu J, et al. Melatonin attenuates myocardial ischemia‐reperfusion injury via improving mitochondrial fusion/mitophagy and activating the AMPK‐OPA1 signaling pathways. J Pineal Res. 2019;66(2):e12542. [DOI] [PubMed] [Google Scholar]
- 375. Valentim L, Laurence KM, Townsend PA, et al. Urocortin inhibits Beclin1‐mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40(6):846‐852. [DOI] [PubMed] [Google Scholar]
- 376. Yang M, Linn BS, Zhang Y, Ren J. Mitophagy and mitochondrial integrity in cardiac ischemia‐reperfusion injury. Biochim Biophys Acta Mol Basis Dis. 2019;1865(9):2293‐2302. [DOI] [PubMed] [Google Scholar]
- 377. Wu M, Lu G, Lao YZ, et al. Garciesculenxanthone B induces PINK1‐Parkin‐mediated mitophagy and prevents ischemia‐reperfusion brain injury in mice. Acta Pharmacol Sin. 2021;42(2):199‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Siddall HK, Yellon DM, Ong SB, et al. Loss of PINK1 increases the heart's vulnerability to ischemia‐reperfusion injury. PLoS One. 2013;8(4):e62400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Dorn GW, 2nd . Parkin‐dependent mitophagy in the heart. J Mol Cell Cardiol. 2016;95:42‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Xie M, Cho GW, Kong Y, et al. Activation of Autophagic flux blunts cardiac Ischemia/Reperfusion injury. Circ Res. 2021;129(3):435‐450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Yang J, He J, Ismail M, et al. HDAC inhibition induces autophagy and mitochondrial biogenesis to maintain mitochondrial homeostasis during cardiac ischemia/reperfusion injury. J Mol Cell Cardiol. 2019;130:36‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Zhang Y, Liu D, Hu H, Zhang P, Xie R, Cui W. HIF‐1alpha/BNIP3 signaling pathway‐induced‐autophagy plays protective role during myocardial ischemia‐reperfusion injury. Biomed Pharmacother. 2019;120:109464. [DOI] [PubMed] [Google Scholar]
- 383. Wu H, Ye M, Liu D, et al. UCP2 protect the heart from myocardial ischemia/reperfusion injury via induction of mitochondrial autophagy. J Cell Biochem. 2019;120(9):15455‐15466. [DOI] [PubMed] [Google Scholar]
- 384. Ma X, Liu H, Foyil SR, et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125(25):3170‐3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Kanamori H, Takemura G, Goto K, et al. The role of autophagy emerging in postinfarction cardiac remodelling. Cardiovasc Res. 2011;91(2):330‐339. [DOI] [PubMed] [Google Scholar]
- 386. Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia‐reperfusion injury. Autophagy. 2012;8(9):1394‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Sadoshima J. The role of autophagy during ischemia/reperfusion. Autophagy. 2008;4(4):402‐403. [DOI] [PubMed] [Google Scholar]
- 388. Wu S, Chang G, Gao L, et al. Trimetazidine protects against myocardial ischemia/reperfusion injury by inhibiting excessive autophagy. J Mol Med (Berl). 2018;96(8):791‐806. [DOI] [PubMed] [Google Scholar]
- 389. Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109(13):1580‐1589. [DOI] [PubMed] [Google Scholar]
- 390. Dammrich J, Pfeifer U. Cardiac hypertrophy in rats after supravalvular aortic constriction. II. Inhibition of cellular autophagy in hypertrophying cardiomyocytes. Virchows Arch B Cell Pathol Incl Mol Pathol. 1983;43(3):287‐307. [DOI] [PubMed] [Google Scholar]
- 391. Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619‐624. [DOI] [PubMed] [Google Scholar]
- 392. Maron BJ, Roberts WC, Arad M, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Zaglia T, Milan G, Ruhs A, et al. Atrogin‐1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J Clin Invest. 2014;124(6):2410‐2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Vasquez‐Trincado C, Garcia‐Carvajal I, Pennanen C, et al. Mitochondrial dynamics, mitophagy and cardiovascular disease. J Physiol. 2016;594(3):509‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Han K, Hassanzadeh S, Singh K, et al. Parkin regulation of CHOP modulates susceptibility to cardiac endoplasmic reticulum stress. Sci Rep. 2017;7(1):2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN‐inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci USA. 2011;108(23):9572‐9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Du Q, Zhu B, Zhai Q, Yu B. Sirt3 attenuates doxorubicin‐induced cardiac hypertrophy and mitochondrial dysfunction via suppression of Bnip3. Am J Transl Res. 2017;9(7):3360‐3373. [PMC free article] [PubMed] [Google Scholar]
- 398. Gao A, Jiang J, Xie F, Chen L. Bnip3 in mitophagy: Novel insights and potential therapeutic target for diseases of secondary mitochondrial dysfunction. Clin Chim Acta. 2020;506:72‐83. [DOI] [PubMed] [Google Scholar]
- 399. McLendon PM, Robbins J. Proteotoxicity and cardiac dysfunction. Circ Res. 2015;116(11):1863‐1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400. Wang X, Osinska H, Klevitsky R, et al. Expression of R120G‐alphaB‐crystallin causes aberrant desmin and alphaB‐crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89(1):84‐91. [DOI] [PubMed] [Google Scholar]
- 401. Tannous P, Zhu H, Johnstone JL, et al. Autophagy is an adaptive response in desmin‐related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105(28):9745‐9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109(2):151‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Bravo‐San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120(11):1812‐1824. [DOI] [PubMed] [Google Scholar]
- 404. McNally EM, Mestroni L. Dilated cardiomyopathy: Genetic determinants and mechanisms. Circ Res. 2017;121(7):731‐748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65(11):965‐968. [DOI] [PubMed] [Google Scholar]
- 406. Saito T, Asai K, Sato S, et al. Autophagic vacuoles in cardiomyocytes of dilated cardiomyopathy with initially decompensated heart failure predict improved prognosis. Autophagy. 2016;12(3):579‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Gourzi P, Pantou MP, Gkouziouta A, et al. A new phenotype of severe dilated cardiomyopathy associated with a mutation in the LAMP2 gene previously known to cause hypertrophic cardiomyopathy in the context of Danon disease. Eur J Med Genet. 2019;62(1):77‐80. [DOI] [PubMed] [Google Scholar]
- 408. Maloyan A, Sanbe A, Osinska H, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha‐B‐crystallin desmin‐related cardiomyopathy. Circulation. 2005;112(22):3451‐3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Aman Y, Schmauck‐Medina T, Hansen M, et al. Autophagy in healthy aging and disease. Nat Aging. 2021;1(8):634‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504‐512. [DOI] [PubMed] [Google Scholar]
- 411. López‐Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461‐472. [DOI] [PubMed] [Google Scholar]
- 413. Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age‐related disease. Nat Cell Biol. 2018;20(12):1338‐1348. [DOI] [PubMed] [Google Scholar]
- 414. Sarkis GJ, Ashcom JD, Hawdon JM, Jacobson LA. Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech Ageing Dev. 1988;45(3):191‐201. [DOI] [PubMed] [Google Scholar]
- 415. Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176‐184. [DOI] [PubMed] [Google Scholar]
- 416. Demontis F, Perrimon N. FOXO/4E‐BP signaling in Drosophila muscles regulates organism‐wide proteostasis during aging. Cell. 2010;143(5):813‐825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Juhász G, Erdi B, Sass M, Neufeld TP. Atg7‐dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21(23):3061‐3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Fernández Á F, Sebti S, Wei Y, et al. Disruption of the beclin 1‐BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558(7708):136‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Lipinski MM, Zheng B, Lu T, et al. Genome‐wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107(32):14164‐14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genetics. 2008;4(2):e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Kaushik S, Tasset I, Arias E, et al. Autophagy and the hallmarks of aging. Ageing Res Rev. 2021;72:101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423. Mattison JA, Colman RJ, Beasley TM, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424. Kapahi P, Kaeberlein M, Hansen M. Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res Rev. 2017;39:3‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Weindruch R, Walford RL. Dietary restriction in mice beginning at 1 year of age: effect on life‐span and spontaneous cancer incidence. Science. 1982;215(4538):1415‐1418. [DOI] [PubMed] [Google Scholar]
- 426. Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age‐related disease. Nature. 2013;493(7432):338‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Steffen KK, Dillin A. A ribosomal perspective on proteostasis and aging. Cell Metab. 2016;23(6):1004‐1012. [DOI] [PubMed] [Google Scholar]
- 429. Cool B, Zinker B, Chiou W, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3(6):403‐416. [DOI] [PubMed] [Google Scholar]
- 430. Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D. Defining the mechanism of activation of AMP‐activated protein kinase by the small molecule A‐769662, a member of the thienopyridone family. J Biol Chem. 2007;282(45):32539‐32548. [DOI] [PubMed] [Google Scholar]
- 431. Paquette M, El‐Houjeiri L, CZ L, et al. AMPK‐dependent phosphorylation is required for transcriptional activation of TFEB and TFE3. Autophagy. 2021;17(12):3957‐3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Sun X, Yang Q, Rogers CJ, Du M, Zhu MJ. AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell Death Differ. 2017;24(5):819‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433. Song YM, Lee YH, Kim JW, et al. Metformin alleviates hepatosteatosis by restoring SIRT1‐mediated autophagy induction via an AMP‐activated protein kinase‐independent pathway. Autophagy. 2015;11(1):46‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434. Martin‐Montalvo A, Mercken EM, Mitchell SJ, et al. Metformin improves healthspan and lifespan in mice. Nat Commun. 2013;4:2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Howell JJ, Hellberg K, Turner M, et al. Metformin inhibits hepatic mTORC1 signaling via Dose‐dependent mechanisms involving AMPK and the TSC complex. Cell Metab. 2017;25(2):463‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436. Yan J, Yang H, Wang G, et al. Autophagy augmented by troglitazone is independent of EGFR transactivation and correlated with AMP‐activated protein kinase signaling. Autophagy. 2010;6(1):67‐73. [DOI] [PubMed] [Google Scholar]
- 437. Fediuc S, Pimenta AS, Gaidhu MP, Ceddia RB. Activation of AMP‐activated protein kinase, inhibition of pyruvate dehydrogenase activity, and redistribution of substrate partitioning mediate the acute insulin‐sensitizing effects of troglitazone in skeletal muscle cells. J Cell Physiol. 2008;215(2):392‐400. [DOI] [PubMed] [Google Scholar]
- 438. Hsieh CC, Li CY, Hsu CH, et al. Mitochondrial protection by simvastatin against angiotensin II‐mediated heart failure. Br J Pharmacol. 2019;176(19):3791‐3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Zhao H, Ji Z, Tang D, Yan C, Zhao W, Gao C. Role of autophagy in early brain injury after subarachnoid hemorrhage in rats. Mol Biol Rep. 2013;40(2):819‐827. [DOI] [PubMed] [Google Scholar]
- 440. Bruiners N, Dutta NK, Guerrini V, et al. The anti‐tubercular activity of simvastatin is mediated by cholesterol‐driven autophagy via the AMPK‐mTORC1‐TFEB axis. J Lipid Res. 2020;61(12):1617‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Hwang HY, Shim JS, Kim D, Kwon HJ. Antidepressant drug sertraline modulates AMPK‐MTOR signaling‐mediated autophagy via targeting mitochondrial VDAC1 protein. Autophagy. 2021;17(10):2783‐2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Morselli E, Marino G, Bennetzen MV, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. Morselli E, Maiuri MC, Markaki M, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin‐1‐dependent induction of autophagy. Cell Death Dis. 2010;1(1):e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high‐calorie diet. Nature. 2006;444(7117):337‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191‐196. [DOI] [PubMed] [Google Scholar]
- 446. Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686‐689. [DOI] [PubMed] [Google Scholar]
- 447. Park SJ, Ahmad F, Philp A, et al. Resveratrol ameliorates aging‐related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148(3):421‐433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Shaito A, Posadino AM, Younes N, et al. Potential adverse effects of resveratrol: A literature review. Int J Mol Sci. 2020;21(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Bjedov I, Toivonen JM, Kerr F, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450. Robida‐Stubbs S, Glover‐Cutter K, Lamming DW, et al. TOR signaling and rapamycin influence longevity by regulating SKN‐1/Nrf and DAF‐16/FoxO. Cell Metab. 2012;15(5):713‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451. Viscomi MT, D'Amelio M, Cavallucci V, et al. Stimulation of autophagy by rapamycin protects neurons from remote degeneration after acute focal brain damage. Autophagy. 2012;8(2):222‐235. [DOI] [PubMed] [Google Scholar]
- 452. Haritunians T, Mori A, O'Kelly J, Luong QT, Giles FJ, Koeffler HP. Antiproliferative activity of RAD001 (everolimus) as a single agent and combined with other agents in mantle cell lymphoma. Leukemia. 2007;21(2):333‐339. [DOI] [PubMed] [Google Scholar]
- 453. Ranek MJ, Kokkonen‐Simon KM, Chen A, et al. PKG1‐modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature. 2019;566(7743):264‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454. Wolpin BM, Hezel AF, Abrams T, et al. Oral mTOR inhibitor everolimus in patients with gemcitabine‐refractory metastatic pancreatic cancer. J Clin Oncol. 2009;27(2):193‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455. Mártinez JM, Pulido LB, Bellido CB, et al. Rescue immunosuppression with mammalian target of rapamycin inhibitor drugs in liver transplantation. Transplant Proc. 2010;42(2):641‐643. [DOI] [PubMed] [Google Scholar]
- 456. Choi JC, Muchir A, Wu W, et al. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med. 2012;4(144):144ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Patel PH, Senico PL, Curiel RE, Motzer RJ. Phase I study combining treatment with temsirolimus and sunitinib malate in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2009;7(1):24‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458. Dreyling M, Jurczak W, Jerkeman M, et al. Ibrutinib versus temsirolimus in patients with relapsed or refractory mantle‐cell lymphoma: an international, randomised, open‐label, phase 3 study. Lancet. 2016;387(10020):770‐778. [DOI] [PubMed] [Google Scholar]
- 459. Mascarenhas L, Chi YY, Hingorani P, et al. Randomized phase II trial of bevacizumab or temsirolimus in combination with chemotherapy for first relapse rhabdomyosarcoma: A report from the children's oncology group. J Clin Oncol. 2019;37(31):2866‐2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. Zhou J, Tan SH, Nicolas V, et al. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome‐lysosome fusion. Cell Res. 2013;23(4):508‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Liu Q, Xu C, Kirubakaran S, et al. Characterization of Torin2, an ATP‐competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 2013;73(8):2574‐2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Hösel M, Huber A, Bohlen S, et al. Autophagy determines efficiency of liver‐directed gene therapy with adeno‐associated viral vectors. Hepatology. 2017;66(1):252‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463. Seo SU, Woo SM, Lee HS, Kim SH, Min KJ, Kwon TK. mTORC1/2 inhibitor and curcumin induce apoptosis through lysosomal membrane permeabilization‐mediated autophagy. Oncogene. 2018;37(38):5205‐5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464. Nam HY, Han MW, Chang HW, et al. Radioresistant cancer cells can be conditioned to enter senescence by mTOR inhibition. Cancer Res. 2013;73(14):4267‐4277. [DOI] [PubMed] [Google Scholar]
- 465. Li P, He J, Yang Z, et al. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2020;16(7):1186‐1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Huang S, Yang ZJ, Yu C, Sinicrope FA. Inhibition of mTOR kinase by AZD8055 can antagonize chemotherapy‐induced cell death through autophagy induction and down‐regulation of p62/sequestosome 1. J Biol Chem. 2011;286(46):40002‐40012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Chresta CM, Davies BR, Hickson I, et al. AZD8055 is a potent, selective, and orally bioavailable ATP‐competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70(1):288‐298. [DOI] [PubMed] [Google Scholar]
- 468. Willems L, Chapuis N, Puissant A, et al. The dual mTORC1 and mTORC2 inhibitor AZD8055 has anti‐tumor activity in acute myeloid leukemia. Leukemia. 2012;26(6):1195‐1202. [DOI] [PubMed] [Google Scholar]
- 469. Zhou M, Xu W, Wang J, et al. Boosting mTOR‐dependent autophagy via upstream TLR4‐MyD88‐MAPK signalling and downstream NF‐κB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine. 2018;35:345‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Chung CY, Shin HR, Berdan CA, et al. Covalent targeting of the vacuolar H(+)‐ATPase activates autophagy via mTORC1 inhibition. Nat Chem Biol. 2019;15(8):776‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471. Pietrocola F, Pol J, Vacchelli E, et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell. 2016;30(1):147‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Castoldi F, Hyvönen MT, Durand S, et al. Chemical activation of SAT1 corrects diet‐induced metabolic syndrome. Cell Death Differ. 2020;27(10):2904‐2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473. Fan QW, Cheng C, Hackett C, et al. Akt and autophagy cooperate to promote survival of drug‐resistant glioma. Sci Signal. 2010;3(147):ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474. Chiarini F, Grimaldi C, Ricci F, et al. Activity of the novel dual phosphatidylinositol 3‐kinase/mammalian target of rapamycin inhibitor NVP‐BEZ235 against T‐cell acute lymphoblastic leukemia. Cancer Res. 2010;70(20):8097‐8107. [DOI] [PubMed] [Google Scholar]
- 475. Thomas HE, Mercer CA, Carnevalli LS, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med. 2012;4(139):139ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476. Ikeda H, Hideshima T, Fulciniti M, et al. PI3K/p110{delta} is a novel therapeutic target in multiple myeloma. Blood. 2010;116(9):1460‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. Furman RR, Sharman JP, Coutre SE, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370(11):997‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. Cheng Y, Ren X, Zhang Y, et al. eEF‐2 kinase dictates cross‐talk between autophagy and apoptosis induced by Akt Inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK‐2206. Cancer Res. 2011;71(7):2654‐2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479. Gassen NC, Papies J, Bajaj T, et al. SARS‐CoV‐2‐mediated dysregulation of metabolism and autophagy uncovers host‐targeting antivirals. Nat Commun. 2021;12(1):3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Xing Y, Lin NU, Maurer MA, et al. Phase II trial of AKT inhibitor MK‐2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019;21(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481. Fu L, Kim YA, Wang X, et al. Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of major components in the mTOR axis and induces autophagy. Cancer Res. 2009;69(23):8967‐8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482. Hawley SA, Fullerton MD, Ross FA, et al. The ancient drug salicylate directly activates AMP‐activated protein kinase. Science. 2012;336(6083):918‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483. Pietrocola F, Castoldi F, Markaki M, et al. Aspirin recapitulates features of caloric restriction. Cell Rep. 2018;22(9):2395‐2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484. Castoldi F, Humeau J, Martins I, et al. Autophagy‐mediated metabolic effects of aspirin. Cell Death Discov. 2020;6(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485. Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11(11):1305‐1314. [DOI] [PubMed] [Google Scholar]
- 486. Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359(6374) [DOI] [PubMed] [Google Scholar]
- 487. Eisenberg T, Abdellatif M, Schroeder S, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22(12):1428‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488. Pietrocola F, Lachkar S, Enot DP, et al. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 2015;22(3):509‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Gupta VK, Scheunemann L, Eisenberg T, et al. Restoring polyamines protects from age‐induced memory impairment in an autophagy‐dependent manner. Nat Neurosci. 2013;16(10):1453‐1460. [DOI] [PubMed] [Google Scholar]
- 490. Chrisam M, Pirozzi M, Castagnaro S, et al. Reactivation of autophagy by spermidine ameliorates the myopathic defects of collagen VI‐null mice. Autophagy. 2015;11(12):2142‐2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491. Michiels CF, Kurdi A, Timmermans JP, De Meyer GRY, Martinet W. Spermidine reduces lipid accumulation and necrotic core formation in atherosclerotic plaques via induction of autophagy. Atherosclerosis. 2016;251:319‐327. [DOI] [PubMed] [Google Scholar]
- 492. Zhang Y, Yang ND, Zhou F, et al. (‐)‐Epigallocatechin‐3‐gallate induces non‐apoptotic cell death in human cancer cells via ROS‐mediated lysosomal membrane permeabilization. PLoS One. 2012;7(10):e46749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493. Kim HS, Montana V, Jang HJ, Parpura V, Kim JA. Epigallocatechin gallate (EGCG) stimulates autophagy in vascular endothelial cells: a potential role for reducing lipid accumulation. J Biol Chem. 2013;288(31):22693‐22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494. Kong D, Zhang Z, Chen L, et al. Curcumin blunts epithelial‐mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020;36:101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495. Zhang J, Wang J, Xu J, et al. Curcumin targets the TFEB‐lysosome pathway for induction of autophagy. Oncotarget. 2016;7(46):75659‐75671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496. Han J, Pan XY, Xu Y, et al. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy. 2012;8(5):812‐825. [DOI] [PubMed] [Google Scholar]
- 497. Rodríguez‐Navarro JA, Rodríguez L, Casarejos MJ, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39(3):423‐438. [DOI] [PubMed] [Google Scholar]
- 498. Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9(9):1308‐1320. [DOI] [PubMed] [Google Scholar]
- 499. Rusmini P, Cortese K, Crippa V, et al. Trehalose induces autophagy via lysosomal‐mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019;15(4):631‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500. Sciarretta S, Yee D, Nagarajan N, et al. Trehalose‐induced activation of autophagy improves cardiac remodeling after myocardial infarction. J Am Coll Cardiol. 2018;71(18):1999‐2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501. Evans TD, Jeong SJ, Zhang X, Sergin I, Razani B. TFEB and trehalose drive the macrophage autophagy‐lysosome system to protect against atherosclerosis. Autophagy. 2018;14(4):724‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Jeong SJ, Stitham J, Evans TD, et al. Trehalose causes low‐grade lysosomal stress to activate TFEB and the autophagy‐lysosome biogenesis response. Autophagy. 2021;17(11):3740‐3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503. Sergin I, Evans TD, Zhang X, et al. Exploiting macrophage autophagy‐lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8:15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504. Berglund R, Guerreiro‐Cacais AO, Adzemovic MZ, et al. Microglial autophagy‐associated phagocytosis is essential for recovery from neuroinflammation. Sci Immunol. 2020;5(52) [DOI] [PubMed] [Google Scholar]
- 505. DeBosch BJ, Heitmeier MR, Mayer AL, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016;9(416):ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506. Zhang T, Li Y, Park KA, et al. Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase‐dependent apoptosis. Autophagy. 2012;8(4):559‐576. [DOI] [PubMed] [Google Scholar]
- 507. Ni Y, Wu S, Wang X, et al. Cucurbitacin I induces pro‐death autophagy in A549 cells via the ERK‐mTOR‐STAT3 signaling pathway. J Cell Biochem. 2018;119(7):6104‐6112. [DOI] [PubMed] [Google Scholar]
- 508. Yuan G, Yan SF, Xue H, Zhang P, Sun JT, Li G. Cucurbitacin I induces protective autophagy in glioblastoma in vitro and in vivo. J Biol Chem. 2014;289(15):10607‐10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509. Wang K, Liu R, Li J, et al. Quercetin induces protective autophagy in gastric cancer cells: involvement of Akt‐mTOR‐ and hypoxia‐induced factor 1α‐mediated signaling. Autophagy. 2011;7(9):966‐978. [DOI] [PubMed] [Google Scholar]
- 510. Han X, Xu T, Fang Q, et al. Quercetin hinders microglial activation to alleviate neurotoxicity via the interplay between NLRP3 inflammasome and mitophagy. Redox Biol. 2021;44:102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511. Kim JJ, Lee HM, Shin DM, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11(5):457‐468. [DOI] [PubMed] [Google Scholar]
- 512. Shoji‐Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy‐inducing peptide. Nature. 2013;494(7436):201‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513. Cinque L, Forrester A, Bartolomeo R, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528(7581):272‐275. [DOI] [PubMed] [Google Scholar]
- 514. Sun Y, Yao X, Zhang QJ, et al. Beclin‐1‐dependent autophagy protects the heart during sepsis. Circulation. 2018;138(20):2247‐2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515. Song X, Zhu S, Chen P, et al. AMPK‐Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System X(c)(‐) Activity. Curr Biol. 2018;28(15):2388‐2399.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516. Soria LR, Gurung S, De Sabbata G, et al. Beclin‐1‐mediated activation of autophagy improves proximal and distal urea cycle disorders. EMBO Mol Med. 2021;13(2):e13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. Nah J, Zhai P, Huang CY, et al. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest. 2020;130(6):2978‐2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. Forte M, Bianchi F, Cotugno M, et al. Pharmacological restoration of autophagy reduces hypertension‐related stroke occurrence. Autophagy. 2020;16(8):1468‐1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Vega‐Rubín‐de‐Celis S, Zou Z, Fernández Á F, et al. Increased autophagy blocks HER2‐mediated breast tumorigenesis. Proc Natl Acad Sci USA. 2018;115(16):4176‐4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520. Mathew B, Chennakesavalu M, Sharma M, et al. Autophagy and post‐ischemic conditioning in retinal ischemia. Autophagy. 2021;17(6):1479‐1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. De Risi M, Torromino G, Tufano M, et al. Mechanisms by which autophagy regulates memory capacity in ageing. Aging Cell. 2020;19(9):e13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522. Soria LR, Allegri G, Melck D, et al. Enhancement of hepatic autophagy increases ureagenesis and protects against hyperammonemia. Proc Natl Acad Sci USA. 2018;115(2):391‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523. Liu Y, Shoji‐Kawata S, Sumpter RM, Jr., et al. Autosis is a Na+,K+‐ATPase‐regulated form of cell death triggered by autophagy‐inducing peptides, starvation, and hypoxia‐ischemia. Proc Natl Acad Sci USA. 2013;110(51):20364‐20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Lu JH, Tan JQ, Durairajan SS, et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha‐synuclein in neuronal cells via inducing autophagy. Autophagy. 2012;8(1):98‐108. [DOI] [PubMed] [Google Scholar]
- 525. Chiang WC, Wei Y, Kuo YC, et al. High‐throughput screens to identify autophagy inducers that function by disrupting beclin 1/Bcl‐2 binding. ACS Chem Biol. 2018;13(8):2247‐2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526. Vicencio JM, Ortiz C, Criollo A, et al. The inositol 1,4,5‐trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16(7):1006‐1017. [DOI] [PubMed] [Google Scholar]
- 527. Criollo A, Maiuri MC, Tasdemir E, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14(5):1029‐1039. [DOI] [PubMed] [Google Scholar]
- 528. Decuypere JP, Welkenhuyzen K, Luyten T, et al. Ins(1,4,5)P3 receptor‐mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy. 2011;7(12):1472‐1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529. Cárdenas C, Müller M, McNeal A, et al. Selective vulnerability of cancer cells by inhibition of Ca(2+) transfer from endoplasmic reticulum to mitochondria. Cell Rep. 2016;14(10):2313‐2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530. Lin CW, Zhang H, Li M, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non‐alcoholic fatty liver conditions in mice. J Hepatol. 2013;58(5):993‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Hidvegi T, Ewing M, Hale P, et al. An autophagy‐enhancing drug promotes degradation of mutant alpha1‐antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229‐232. [DOI] [PubMed] [Google Scholar]
- 532. Vasconcelos‐Ferreira A, Carmo‐Silva S, Codêsso JM, et al. The autophagy‐enhancing drug carbamazepine improves neuropathology and motor impairment in mouse models of Machado‐Joseph disease. Neuropathol Appl Neurobiol. 2022;48(1):e12763. [DOI] [PubMed] [Google Scholar]
- 533. Schiebler M, Brown K, Hegyi K, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR‐independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol Med. 2015;7(2):127‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534. Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170(7):1101‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535. Chalecka‐Franaszek E, Chuang DM. Lithium activates the serine/threonine kinase Akt‐1 and suppresses glutamate‐induced inhibition of Akt‐1 activity in neurons. Proc Natl Acad Sci USA. 1999;96(15):8745‐8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536. Shimada K, Motoi Y, Ishiguro K, et al. Long‐term oral lithium treatment attenuates motor disturbance in tauopathy model mice: implications of autophagy promotion. Neurobiol Dis. 2012;46(1):101‐108. [DOI] [PubMed] [Google Scholar]
- 537. Fornai F, Longone P, Cafaro L, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2008;105(6):2052‐2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538. Chang JW, Choi H, Cotman SL, Jung YK. Lithium rescues the impaired autophagy process in CbCln3(Δex7/8/Δex7/8) cerebellar cells and reduces neuronal vulnerability to cell death via IMPase inhibition. J Neurochem. 2011;116(4):659‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539. Chen G, Xie W, Nah J, et al. 3,4‐Dimethoxychalcone induces autophagy through activation of the transcription factors TFE3 and TFEB. EMBO Mol Med. 2019;11(11):e10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540. Wang Y, Xie W, Humeau J, et al. Autophagy induction by thiostrepton improves the efficacy of immunogenic chemotherapy. J Immunother Cancer. 2020;8(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Zhang J, Ng S, Wang J, et al. Histone deacetylase inhibitors induce autophagy through FOXO1‐dependent pathways. Autophagy. 2015;11(4):629‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542. Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy potentiates the anti‐cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6(8):1057‐1065. [DOI] [PubMed] [Google Scholar]
- 543. Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor‐induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA. 2012;109(17):6561‐6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544. Xie M, Kong Y, Tan W, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129(10):1139‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545. Comes F, Matrone A, Lastella P, et al. A novel cell type‐specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 2007;14(4):693‐702. [DOI] [PubMed] [Google Scholar]
- 546. Yang C, Zhu Z, Tong BC, et al. A stress response p38 MAP kinase inhibitor SB202190 promoted TFEB/TFE3‐dependent autophagy and lysosomal biogenesis independent of p38. Redox Biol. 2020;32:101445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547. Xia HG, Zhang L, Chen G, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6(1):61‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548. Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image‐based high‐throughput screen. Proc Natl Acad Sci USA. 2007;104(48):19023‐1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549. Lai JL, Lian YE, Wu JY, Wang YD, Bai YN. Verapamil induces autophagy to improve liver regeneration in non‐alcoholic fatty liver mice. Adipocyte. 2021;10(1):532‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550. Liu W, Lin H, Mao Z, et al. Verapamil extends lifespan in Caenorhabditis elegans by inhibiting calcineurin activity and promoting autophagy. Aging (Albany NY). 2020;12(6):5300‐5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551. Williams A, Sarkar S, Cuddon P, et al. Novel targets for Huntington's disease in an mTOR‐independent autophagy pathway. Nat Chem Biol. 2008;4(5):295‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552. Siddiqi FH, Menzies FM, Lopez A, et al. Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nat Commun. 2019;10(1):1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553. Meyer N, Henkel L, Linder B, et al. Autophagy activation, lipotoxicity and lysosomal membrane permeabilization synergize to promote pimozide‐ and loperamide‐induced glioma cell death. Autophagy. 2021;17(11):3424‐3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554. Zielke S, Kardo S, Zein L, et al. ATF4 links ER stress with reticulophagy in glioblastoma cells. Autophagy. 2021;17(9):2432‐2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555. Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus‐associated hepatocellular carcinoma through degradation of microRNA‐224. Hepatology. 2014;59(2):505‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556. Wu SY, Lan SH, Wu SR, et al. Hepatocellular carcinoma‐related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology. 2018;68(1):141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557. Shi Y, Tan SH, Ng S, et al. Critical role of CAV1/caveolin‐1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy. 2015;11(5):769‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558. Shi Y, Ye Z, Lu G, et al. Cholesterol‐enriched membrane micro‐domaindeficiency induces doxorubicin resistancevia promoting autophagy in breast cancer. Mol Ther Oncolytics. 2021;23:311‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559. Ryu D, Mouchiroud L, Andreux PA, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22(8):879‐888. [DOI] [PubMed] [Google Scholar]
- 560. Takahashi D, Moriyama J, Nakamura T, et al. AUTACs: Cargo‐specific degraders using selective autophagy. Mol Cell. 2019;76(5):797‐810.e10. [DOI] [PubMed] [Google Scholar]
- 561. Sarkar S, Perlstein EO, Imarisio S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3(6):331‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562. Kuo SY, Castoreno AB, Aldrich LN, et al. Small‐molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc Natl Acad Sci USA. 2015;112(31):E4281‐E4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563. Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small‐molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci USA. 2010;107(39):16982‐16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564. Du G, Qiao Y, Zhuo Z, et al. Lipoic acid rejuvenates aged intestinal stem cells by preventing age‐associated endosome reduction. EMBO Rep. 2020;21(8):e49583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565. Carling D. AMPK signalling in health and disease. Curr Opin Cell Biol. 2017;45:31‐37. [DOI] [PubMed] [Google Scholar]
- 566. Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4:3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567. Ruiz R, Pérez‐Villegas EM, Manuel Carrión Á. AMPK function in aging process. Curr Drug Targets. 2016;17(8):932‐941. [DOI] [PubMed] [Google Scholar]
- 568. Madeo F, Carmona‐Gutierrez D, Hofer SJ, Kroemer G. Caloric restriction mimetics against Age‐associated disease: targets, mechanisms, and therapeutic potential. Cell Metab. 2019;29(3):592‐610. [DOI] [PubMed] [Google Scholar]
- 569. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1‐2):11‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570. Rosich L, Colomer D, Roue G. Autophagy controls everolimus (RAD001) activity in mantle cell lymphoma. Autophagy. 2013;9(1):115‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571. Haas NB, Appleman LJ, Stein M, et al. Autophagy inhibition to augment mTOR inhibition: a phase I/II trial of everolimus and hydroxychloroquine in patients with previously treated renal cell carcinoma. Clin Cancer Res. 2019;25(7):2080‐2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572. Rangwala R, Chang YC, Hu J, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391‐1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573. Mehnert JM, Kaveney AD, Malhotra J, et al. A phase I trial of MK‐2206 and hydroxychloroquine in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2019;84(4):899‐907. [DOI] [PubMed] [Google Scholar]
- 574. Rosich L, Xargay‐Torrent S, López‐Guerra M, Campo E, Colomer D, Roué G. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin Cancer Res. 2012;18(19):5278‐5289. [DOI] [PubMed] [Google Scholar]
- 575. Kallergi E, Daskalaki AD, Kolaxi A, et al. Dendritic autophagy degrades postsynaptic proteins and is required for long‐term synaptic depression in mice. Nat Commun. 2022;13(1):680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Ahwazi D, Neopane K, Markby GR, et al. Investigation of the specificity and mechanism of action of the ULK1/AMPK inhibitor SBI‐0206965. Biochem J. 2021;478(15):2977‐2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577. Dite TA, Langendorf CG, Hoque A, et al. AMP‐activated protein kinase selectively inhibited by the type II inhibitor SBI‐0206965. J Biol Chem. 2018;293(23):8874‐8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578. Vahsen BF, Ribas VT, Sundermeyer J, et al. Inhibition of the autophagic protein ULK1 attenuates axonal degeneration in vitro and in vivo, enhances translation, and modulates splicing. Cell Death Differ. 2020;27(10):2810‐2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579. Chaikuad A, Koschade SE, Stolz A, et al. Conservation of structure, function and inhibitor binding in UNC‐51‐like kinase 1 and 2 (ULK1/2). Biochem J. 2019;476(5):875‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580. Petherick KJ, Conway OJ, Mpamhanga C, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)‐dependent autophagy. J Biol Chem. 2015;290(18):11376‐11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581. Martin KR, Celano SL, Solitro AR, et al. A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress. iScience. 2018;8:74‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582. Wu YT, Tan HL, Shui G, et al. Dual role of 3‐methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3‐kinase. J Biol Chem. 2010;285(14):10850‐10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583. Petiot A, Ogier‐Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'‐kinases are involved in signaling pathways that control macroautophagy in HT‐29 cells. J Biol Chem. 2000;275(2):992‐998. [DOI] [PubMed] [Google Scholar]
- 584. Shi Y, Tao M, Ma X, et al. Delayed treatment with an autophagy inhibitor 3‐MA alleviates the progression of hyperuricemic nephropathy. Cell Death Dis. 2020;11(6):467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585. Wu Y, Wang X, Guo H, et al. Synthesis and screening of 3‐MA derivatives for autophagy inhibitors. Autophagy. 2013;9(4):595‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586. Chicote J, Yuste VJ, Boix J, Ribas J. Cell death triggered by the autophagy inhibitory drug 3‐methyladenine in growing conditions proceeds with DNA damage. Front Pharmacol. 2020;11:580343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587. Blommaart EF, Krause U, Schellens JP, Vreeling‐Sindelárová H, Meijer AJ. The phosphatidylinositol 3‐kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243(1‐2):240‐246. [DOI] [PubMed] [Google Scholar]
- 588. Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3‐kinase inhibitor: the role of phosphatidylinositol 3,4,5‐trisphosphate in neutrophil responses. Biochem J. 1993;296(Pt 2):297‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589. Hansen SH, Olsson A, Casanova JE. Wortmannin, an inhibitor of phosphoinositide 3‐kinase, inhibits transcytosis in polarized epithelial cells. J Biol Chem. 1995;270(47):28425‐28432. [DOI] [PubMed] [Google Scholar]
- 590. Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10(12):1013‐1019. [DOI] [PubMed] [Google Scholar]
- 591. Li K, Chen HS, Li D, et al. SAR405, a highly specific VPS34 inhibitor, disrupts auditory fear memory consolidation of mice via facilitation of inhibitory neurotransmission in basolateral amygdala. Biol Psychiatry. 2019;85(3):214‐225. [DOI] [PubMed] [Google Scholar]
- 592. New J, Arnold L, Ananth M, et al. Secretory autophagy in Cancer‐associated fibroblasts promotes head and neck cancer progression and offers a novel therapeutic target. Cancer Res. 2017;77(23):6679‐6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Ishikawa M, Takaseki S, Yoshitomi T, Covey DF, Zorumski CF, Izumi Y. The neurosteroid allopregnanolone protects retinal neurons by effects on autophagy and GABRs/GABA(A) receptors in rat glaucoma models. Autophagy. 2021;17(3):743‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594. Li M, Liu W, Bauch T, et al. Clearance of HIV infection by selective elimination of host cells capable of producing HIV. Nat Commun. 2020;11(1):4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595. Noman MZ, Parpal S, Van Moer K, et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti‐PD‐1/PD‐L1 immunotherapy. Sci Adv. 2020;6(18):eaax7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596. Dyczynski M, Yu Y, Otrocka M, et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018;435:32‐43. [DOI] [PubMed] [Google Scholar]
- 597. Bago R, Malik N, Munson MJ, et al. Characterization of VPS34‐IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3‐phosphate‐binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3‐kinase. Biochem J. 2014;463(3):413‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598. Meunier G, Birsen R, Cazelles C, et al. Antileukemic activity of the VPS34‐IN1 inhibitor in acute myeloid leukemia. Oncogenesis. 2020;9(10):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599. Zhang J, Fan J, Zeng X, et al. Targeting the autophagy promoted antitumor effect of T‐DM1 on HER2‐positive gastric cancer. Cell Death Dis. 2021;12(4):288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600. Kim KY, Park KI, Kim SH, et al. Inhibition of autophagy promotes Salinomycin‐induced apoptosis via reactive oxygen Species‐mediated PI3K/AKT/mTOR and ERK/p38 MAPK‐dependent signaling in human prostate cancer cells. Int J Mol Sci. 2017;18(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601. Dowdle WE, Nyfeler B, Nagel J, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16(11):1069‐1079. [DOI] [PubMed] [Google Scholar]
- 602. Robke L, Laraia L, Carnero Corrales MA, et al. Phenotypic identification of a novel autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew Chem Int Ed Engl. 2017;56(28):8153‐8157. [DOI] [PubMed] [Google Scholar]
- 603. Parekh VV, Pabbisetty SK, Wu L, et al. Autophagy‐related protein Vps34 controls the homeostasis and function of antigen cross‐presenting CD8α(+) dendritic cells. Proc Natl Acad Sci USA. 2017;114(31):E6371‐E6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604. Liu H, Gong X, Yang K. Overexpression of the clock gene Per2 suppresses oral squamous cell carcinoma progression by activating autophagy via the PI3K/AKT/mTOR pathway. J Cancer. 2020;11(12):3655‐3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605. Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy‐mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10(2):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606. Foley DJ, Zinken S, Corkery D, et al. Phenotyping reveals targets of a pseudo‐Natural‐product autophagy inhibitor. Angew Chem Int Ed Engl. 2020;59(30):12470‐12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607. Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608. Kuramoto K, Kim YJ, Hong JH, He C. The autophagy protein Becn1 improves insulin sensitivity by promoting adiponectin secretion via exocyst binding. Cell Rep. 2021;35(8):109184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609. Kargapolova Y, Rehimi R, Kayserili H, et al. Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology. Nat Commun. 2021;12(1):3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610. Pavlinov I, Salkovski M, Aldrich LN. Beclin 1‐ATG14L Protein‐protein interaction inhibitor selectively inhibits autophagy through disruption of VPS34 complex I. J Am Chem Soc. 2020;142(18):8174‐8182. [DOI] [PubMed] [Google Scholar]
- 611. Huang T, Kim CK, Alvarez AA, et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell. 2017;32(6):840‐855.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612. Huang T, Wan X, Alvarez AA, et al. MIR93 (microRNA ‐93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15(6):1100‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613. Akin D, Wang SK, Habibzadegah‐Tari P, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10(11):2021‐2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614. Qiu Z, Kuhn B, Aebi J, et al. Discovery of Fluoromethylketone‐based peptidomimetics as covalent ATG4B (Autophagin‐1) inhibitors. ACS Med Chem Lett. 2016;7(8):802‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615. Chu J, Fu Y, Xu J, et al. ATG4B inhibitor FMK‐9a induces autophagy independent on its enzyme inhibition. Arch Biochem Biophys. 2018;644:29‐36. [DOI] [PubMed] [Google Scholar]
- 616. Pengo N, Agrotis A, Prak K, Jones J, Ketteler R. A reversible phospho‐switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat Commun. 2017;8(1):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617. Holen I, Gordon PB, Seglen PO. Inhibition of hepatocytic autophagy by okadaic acid and other protein phosphatase inhibitors. Eur J Biochem. 1993;215(1):113‐122. [DOI] [PubMed] [Google Scholar]
- 618. Blankson H, Holen I, Seglen PO. Disruption of the cytokeratin cytoskeleton and inhibition of hepatocytic autophagy by okadaic acid. Exp Cell Res. 1995;218(2):522‐530. [DOI] [PubMed] [Google Scholar]
- 619. Huang SC, Adhikari S, Brownell JE, et al. Discovery and optimization of pyrazolopyrimidine sulfamates as ATG7 inhibitors. Bioorg Med Chem. 2020;28(19):115681. [DOI] [PubMed] [Google Scholar]
- 620. Vats S, Manjithaya R. A reversible autophagy inhibitor blocks autophagosome‐lysosome fusion by preventing Stx17 loading onto autophagosomes. Mol Biol Cell. 2019;30(17):2283‐2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621. Simpson CL, Tokito MK, Uppala R, Sarkar MK, Gudjonsson JE, Holzbaur ELF. NIX initiates mitochondrial fragmentation via DRP1 to drive epidermal differentiation. Cell reports. 2021;34(5):108689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622. Ilnytska O, Lai K, Gorshkov K, et al. Enrichment of NPC1‐deficient cells with the lipid LBPA stimulates autophagy, improves lysosomal function, and reduces cholesterol storage. J Biol Chem. 2021;297(1):100813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623. Fu Y, Hong L, Xu J, et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy. 2019;15(2):295‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624. Laraia L, Friese A, Corkery DP, et al. The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat Chem Biol. 2019;15(7):710‐720. [DOI] [PubMed] [Google Scholar]
- 625. Zhang L, Qiang P, Yu J, et al. Identification of compound CA‐5f as a novel late‐stage autophagy inhibitor with potent anti‐tumor effect against non‐small cell lung cancer. Autophagy. 2019;15(3):391‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626. Rossi M, Rotblat B, Ansell K, et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014;5(5):e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627. Adiseshaiah PP, Clogston JD, McLeland CB, et al. Synergistic combination therapy with nanoliposomal C6‐ceramide and vinblastine is associated with autophagy dysfunction in hepatocarcinoma and colorectal cancer models. Cancer Lett. 2013;337(2):254‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628. Musiwaro P, Smith M, Manifava M, Walker SA, Ktistakis NT. Characteristics and requirements of basal autophagy in HEK 293 cells. Autophagy. 2013;9(9):1407‐1417. [DOI] [PubMed] [Google Scholar]
- 629. Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal‐lysosomal fusion mechanism revealed by thapsigargin‐induced autophagy arrest. Research Support, N.I.H., Extramural Research Support, Non‐U.S. Gov't. Mol Cell. 2011;42(6):731‐743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630. Jaskulska A, Janecka AE, Gach‐Janczak K. Thapsigargin‐from traditional medicine to anticancer drug. Int J Mol Sci. 2020;22(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631. Lu Y, Dong S, Hao B, et al. Vacuolin‐1 potently and reversibly inhibits autophagosome‐lysosome fusion by activating RAB5A. Autophagy. 2014;10(11):1895‐1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632. Sano O, Kazetani K, Funata M, Fukuda Y, Matsui J, Iwata H. Vacuolin‐1 inhibits autophagy by impairing lysosomal maturation via PIKfyve inhibition. FEBS Lett. 2016;590(11):1576‐1585. [DOI] [PubMed] [Google Scholar]
- 633. Kang YL, Chou YY, Rothlauf PW, et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS‐CoV‐2. Proc Natl Acad Sci USA. 2020;117(34):20803‐20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634. Ye Z, Wang D, Lu Y, et al. Vacuolin‐1 inhibits endosomal trafficking and metastasis via CapZβ. Oncogene. 2021;40(10):1775‐1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635. Huynh C, Andrews NW. The small chemical vacuolin‐1 alters the morphology of lysosomes without inhibiting Ca2+‐regulated exocytosis. EMBO Rep. 2005;6(9):843‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636. Mauthe M, Orhon I, Rocchi C, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome‐lysosome fusion. Autophagy. 2018;14(8):1435‐1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637. Redmann M, Benavides GA, Berryhill TF, et al. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2017;11:73‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638. Sumpter MD, Tatro LS, Stoecker WV, Rader RK. Evidence for risk of cardiomyopathy with hydroxychloroquine. Lupus. 2012;21(14):1594‐1596. [DOI] [PubMed] [Google Scholar]
- 639. Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16(3):155‐166. [DOI] [PubMed] [Google Scholar]
- 640. McAfee Q, Zhang Z, Samanta A, et al. Autophagy inhibitor Lys05 has single‐agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Research Support, N.I.H., Extramural Research Support, Non‐U.S. Gov't. Proc Natl Acad Sci USA. 2012;109(21):8253‐8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641. Rebecca VW, Nicastri MC, McLaughlin N, et al. A Unified approach to targeting the Lysosome's degradative and growth signaling roles. Cancer Discov. 2017;7(11):1266‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642. Rebecca VW, Nicastri MC, Fennelly C, et al. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 2019;9(2):220‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643. Brun S, Bestion E, Raymond E, et al. GNS561, a clinical‐stage PPT1 inhibitor, is efficient against hepatocellular carcinoma via modulation of lysosomal functions. Autophagy. 2021:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644. Brun S, Pascussi JM, Gifu EP, et al. GNS561, a new autophagy inhibitor active against cancer stem cells in hepatocellular carcinoma and hepatic metastasis from colorectal cancer. J Cancer. 2021;12(18):5432‐5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645. Bestion E, Zandi K, Belouzard S, et al. GNS561 exhibits potent antiviral activity against SARS‐CoV‐2 through autophagy inhibition. Viruses. 2022;14(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646. De Mei C, Ercolani L, Parodi C, et al. Dual inhibition of REV‐ERBβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene. 2015;34(20):2597‐2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647. Carew JS, Espitia CM, Zhao W, et al. Disruption of autophagic degradation with ROC‐325 antagonizes renal cell carcinoma pathogenesis. Clin Cancer Res. 2017;23(11):2869‐2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648. Nawrocki ST, Han Y, Visconte V, et al. The novel autophagy inhibitor ROC‐325 augments the antileukemic activity of azacitidine. Leukemia. 2019;33(12):2971‐2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649. Renna M, Schaffner C, Brown K, et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest. 2011;121(9):3554‐3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650. Toriyama K, Takano N, Kokuba H, et al. Azithromycin enhances the cytotoxicity of DNA‐damaging drugs via lysosomal membrane permeabilization in lung cancer cells. Cancer Sci. 2021;112(8):3324‐3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651. Tsubouchi K, Araya J, Minagawa S, et al. Azithromycin attenuates myofibroblast differentiation and lung fibrosis development through proteasomal degradation of NOX4. Autophagy. 2017;13(8):1420‐1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652. Laraia L, Garivet G, Foley DJ, et al. Image‐based morphological profiling identifies a lysosomotropic, Iron‐sequestering autophagy inhibitor. Angew Chem Int Ed Engl. 2020;59(14):5721‐5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653. Mauvezin C, Nagy P, Juhasz G, Neufeld TP. Autophagosome‐lysosome fusion is independent of V‐ATPase‐mediated acidification. Nat Commun. 2015;6:7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654. Huss M, Ingenhorst G, König S, et al. Concanamycin A, the specific inhibitor of V‐ATPases, binds to the V(o) subunit c. J Biol Chem. 2002;277(43):40544‐40548. [DOI] [PubMed] [Google Scholar]
- 655. Painter MM, Zimmerman GE, Merlino MS, et al. Concanamycin A counteracts HIV‐1 Nef to enhance immune clearance of infected primary cells by cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 2020;117(38):23835‐23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656. Guinea R, Carrasco L. Concanamycin A blocks influenza virus entry into cells under acidic conditions. FEBS Lett. 1994;349(3):327‐330. [DOI] [PubMed] [Google Scholar]
- 657. Wang Y, Zhang L, Wei Y, et al. Pharmacological targeting of vacuolar H(+)‐ATPase via subunit V1G combats multidrug‐resistant cancer. Cell Chem Biol. 2020;27(11):1359‐1370.e8. [DOI] [PubMed] [Google Scholar]
- 658. Xie XS, Padron D, Liao X, Wang J, Roth MG, De Brabander JK. Salicylihalamide A inhibits the V0 sector of the V‐ATPase through a mechanism distinct from bafilomycin A1. J Biol Chem. 2004;279(19):19755‐19763. [DOI] [PubMed] [Google Scholar]
- 659. Aldrich LN, Kuo SY, Castoreno AB, et al. Discovery of a small‐molecule probe for V‐ATPase function. J Am Chem Soc. 2015;137(16):5563‐5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660. Carew JS, Espitia CM, Esquivel JA, 2nd , et al. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D‐mediated apoptosis. J Biol Chem. 2011;286(8):6602‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661. Yoon JY, Woo SM, Seo SU, Song SR, Lee SG, Kwon TK. Lucanthone, autophagy inhibitor, enhances the apoptotic effects of TRAIL through miR‐216a‐5p‐Mediated DR5 upregulation and DUB3‐mediated Mcl‐1 downregulation. Int J Mol Sci. 2021;23(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662. Gayle S, Landrette S, Beeharry N, et al. Identification of apilimod as a first‐in‐class PIKfyve kinase inhibitor for treatment of B‐cell non‐Hodgkin lymphoma. Blood. 2017;129(13):1768‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663. Hessvik NP, Øverbye A, Brech A, et al. PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell Mol Life Sci. 2016;73(24):4717‐4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664. Riva L, Yuan S, Yin X, et al. Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature. 2020;586(7827):113‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665. Samie M, Wang X, Zhang X, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev Cell. 2013;26(5):511‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666. Kheloufi M, Boulanger CM, Codogno P, Rautou PE. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology. 2015;62(2):657‐658. [DOI] [PubMed] [Google Scholar]
- 667. Keller CW, Sina C, Kotur MB, et al. ATG‐dependent phagocytosis in dendritic cells drives myelin‐specific CD4(+) T cell pathogenicity during CNS inflammation. Proc Natl Acad Sci USA. 2017;114(52):E11228‐E11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668. Xie CM, Chan WY, Yu S, Zhao J, Cheng CH. Bufalin induces autophagy‐mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic Biol Med. 2011;51(7):1365‐1375. [DOI] [PubMed] [Google Scholar]
- 669. Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy‐dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014;124(9):3987‐4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670. Zhang X, Yan H, Yuan Y, et al. Cerebral ischemia‐reperfusion‐induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9(9):1321‐1333. [DOI] [PubMed] [Google Scholar]
- 671. Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PLoS One. 2012;7(3):e32388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672. Laraia L, Ohsawa K, Konstantinidis G, et al. Discovery of novel cinchona‐alkaloid‐inspired oxazatwistane autophagy inhibitors. Angew Chem Int Ed Engl. 2017;56(8):2145‐2150. [DOI] [PubMed] [Google Scholar]
- 673. Nguyen HG, Yang JC, Kung HJ, et al. Targeting autophagy overcomes Enzalutamide resistance in castration‐resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33(36):4521‐4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674. Donohue E, Tovey A, Vogl AW, et al. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem. 2011;286(9):7290‐7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675. Donohue E, Thomas A, Maurer N, et al. The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model. J Cancer. 2013;4(7):585‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676. Tremel S, Ohashi Y, Morado DR, et al. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat Commun. 2021;12(1):1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677. Pisonero‐Vaquero S, Medina DL. Lysosomotropic drugs: Pharmacological tools to study lysosomal function. Curr Drug Metab. 2017;18(12):1147‐1158. [DOI] [PubMed] [Google Scholar]
- 678. Chude CI, Amaravadi RK. Targeting autophagy in Cancer: Update on clinical trials and novel inhibitors. Int J Mol Sci. 2017;18(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679. Gubas A, Dikic I. A guide to the regulation of selective autophagy receptors. FEBS J. 2022;289(1):75‐89. [DOI] [PubMed] [Google Scholar]
- 680. Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85(2):257‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681. Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625(1‐3):220‐233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


