Abstract
Non-alcoholic fatty liver disease (NAFLD), an emerging global health problem affecting 25–30% of the total population, refers to excessive lipid accumulation in the liver accompanied by insulin resistance (IR) without significant alcohol intake. The increasing prevalence of NAFLD will lead to an increasing number of cirrhosis patients, as well as hepatocellular carcinoma (HCC) requiring liver transplantation, while the current treatments for NAFLD and its advanced diseases are suboptimal. Accordingly, it is necessary to find signaling pathways and targets related to the pathogenesis of NAFLD for the development of novel drugs. A large number of studies and reviews have described the critical roles of bile acids (BAs) and their receptors in the pathogenesis of NAFLD. The gut microbiota (GM), whose composition varies between healthy and NAFLD patients, promotes the transformation of more than 50 secondary bile acids and is involved in the pathophysiology of NAFLD through the GM-BAs axis. Correspondingly, BAs inhibit the overgrowth of GM and maintain a healthy gut through their antibacterial effects. Here we review the biosynthesis, enterohepatic circulation, and major receptors of BAs, as well as the relationship of GM, BAs, and the pathogenesis of NAFLD in different disease progression. This article also reviews several therapeutic approaches for the management and prevention of NAFLD targeting the GM-BAs axis.
Keywords: non-alcoholic fatty liver disease (NAFLD), bile acids, gut microbiota, gut microbiota-bile acids axis, disease progression
Introduction
Non-alcoholic fatty liver disease (NAFLD), an emerging global health problem affecting 25–30% of the total population, refers to excessive lipid accumulation in the liver accompanied by insulin resistance (IR) without significant alcohol intake (Caussy et al., 2019; Hrncir et al., 2021). NAFLD, characterized by steatosis, necroinflammatory changes, and varying degrees of liver fibrosis, is the most common cause of chronic liver disease in developed countries (Gottlieb and Canbay, 2019; Ji et al., 2020). Closely related to atherosclerosis, metabolic syndrome, obesity, diabetes, coronary heart disease, and other metabolic diseases, NAFLD’s disease spectrum includes simple steatosis, namely non-alcoholic fatty liver, irreversible non-alcoholic steatohepatitis (NASH), more serious morbid state, like liver fibrosis and cirrhosis, and hepatocellular carcinoma (HCC) (Bhatia et al., 2012; Koliaki et al., 2015; Younossi et al., 2016a; Hrncir et al., 2021). It is estimated that more than 60% of NAFLD patients undergoing liver biopsy have NASH, and about 40% of NASH patients show noticeable symptoms of fibrosis (Younossi et al., 2016b). Approximately 22% of patients with bridging fibrosis (F3) go on to develop cirrhosis, and those with advanced fibrosis are more likely to develop HCC (Sanyal et al., 2019). The increasing prevalence of the disease will be accompanied by an increasing number of patients with cirrhosis and HCC requiring liver transplantation (Friedman et al., 2018). Therefore, discovering an effective NAFLD pharmacotherapy will be a challenge in the field of hepatology.
Gut microbiota (GM), considered as a special organ involved in the pathophysiology of NAFLD, is being intensively studied using various high-throughput techniques (Rinella, 2015). GM in NAFLD patients is dominated by Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria at the phylum level (Loomba et al., 2017). Furthermore, Eubacterium rectale, Bacteroides vulgatus, and Escherichia coli were the most abundant at the species level (Loomba et al., 2017). GM can be regarded as an “endocrine organ” that regulates host physiological functions (Ridlon and Bajaj, 2015). The role of GM in the progression of NAFLD is critical for exploring its pathophysiology, identifying therapeutic targets and pathways, and subsequent appropriate therapy (Aron-Wisnewsky et al., 2020b).
Primary bile acids (BAs) are synthesized from cholesterol firstly in the liver and account for about half of the organic constituent of bile. Once primary BAs enter the gastrointestinal tract, over 50 secondary BAs are formed (Gottlieb and Canbay, 2019; Winston and Theriot, 2020). In other words, the chemical diversity of the BA pool relies on the mutual efforts of the primary BAs-producing host and the secondary BAs-producing GM. BAs in turn have antibacterial effects against certain bacteria (Winston and Theriot, 2020). Numerous studies have shown that BAs act as signaling molecules to regulate glucose, lipid and energy metabolism and the inflammatory response by mediating the intensive connection between the liver and the gut (Mouzaki et al., 2016; Jia et al., 2018; Yamada et al., 2018; Jia and Jeon, 2019; Sun et al., 2019; Sydor et al., 2020; Shao et al., 2021; Simbrunner et al., 2021; Wu et al., 2021). The composition of BAs in NAFLD patients differs from that of healthy controls, and BAs can prevent NAFLD progression by activating BAs receptors (Gottlieb and Canbay, 2019). By regulating or detecting of biomarkers on the “GM-BAs” axis, it can be helpful for the prevention, treatment and diagnosis of some diseases, including NAFLD, diarrhea-irritable bowel syndrome, polycystic ovary, etc.
In this review, we review the bio-synthesis, transport, and major receptors of BAs, the interactions between BAs and the GM, and elucidate the function of BAs as signaling molecules in the gut-liver axis, focusing on their roles in the pathogenesis of each stage of the NAFLD progression. Several management and preventive approaches for NAFLD targeting the GM-BAs axis have been proposed (Jia et al., 2018).
Bile Acids Act as Signal Molecules Between the Gut and Liver
The enterohepatic circulation of BAs affects diverse metabolic and immune functions (Willis et al., 2020). BAs are synthesized in the liver, while GM is responsible for the formation of different BAs biomolecular structures via bacterial biotransformations, including deconjugation, dehydroxylation, oxidation, desulfation, dehydrogenation, and epimerization (Kang et al., 2019; Poland and Flynn, 2021; Wu et al., 2021).
Fates and Functions of Bile Acids
The Enterohepatic Circulation of Bile Acids
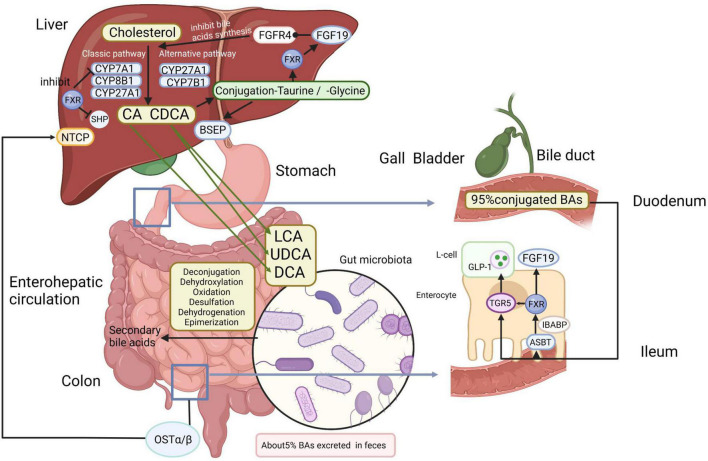
The liver is located at the intersection point of the portal blood flow from the intestinal circulation to peripheral organs (Tilg et al., 2016). The rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1) in the liver initiates the classical synthesis pathway that converts cholesterol to 7α-hydroxycholesterol. 7α-hydroxycholesterol is then transformed to 7α-hydroxy-4-cholesten-3-one (C4), an intermediate for the de novo synthesis of primary BAs, cholic acid (CA) and chenodeoxycholic acid (CDCA) (Chiang and Ferrell, 2018, 2020). Sterol 12α-hydroxylase (CYP8B1) hydroxylates C4 and produces CA to regulate the CA/CDCA ratio, as loss of CYP8B1 results in CDCA production (Chiang and Ferrell, 2020). The alternative synthesis pathway is initiated in the mitochondrial membrane by sterol 27-hydroxylase (CYP27A1) and oxysterol 7α-hydroxylase (CYP7B1). CYP27A1, expressed extra-hepatically in a variety of tissues, catalyzes the production of CDCA or muricholic acids (in mice only), while CYP7B1 is responsible for hydroxylation of 27-hydroxycholesterol and 25-hydroxycholesterol (Ridlon and Bajaj, 2015; Chiang and Ferrell, 2018; Ji et al., 2020). Feedback inhibition of the classic bile acid synthetic enzymes CYP7A1, CYP8B1, and CYP27A1 by BAs is a well-established mechanism that reduces primary BA synthesis in response to an expanded bile acid pool or cholestasis with obviously elevated bile acid concentrations (Copple and Li, 2016). Most primary BAs are conjugated to taurine or glycine so as to enhance solubility and reduce toxicity (Chiang and Ferrell, 2019). Then conjugated CA/CDCA are excreted into the bile via the bile salt export pump (BSEP) across the canalicular (apical) membrane and stored in the cholecyst (Wahlström et al., 2016). Cholecystokinin secreted by the duodenum after meals, prompts gallbladder contraction, and releases BAs into the gut (Li and Chiang, 2014). After reaching the terminal ileum, approximately 95% of BAs are assimilated from the intestine by the apical sodium-dependent bile acid transporter (ASBT), a protein located in the brush border of the enterocyte (Wahlström et al., 2016; Jia et al., 2018). These BAs are associated to the ileal bile acid-binding protein (IBABP), which regulates the transportation of BAs from the intestinal epithelium to the basolateral membrane, where the heteromeric organic solute transporter (OST) promotes the transport of BAs to the portal vein (Jia et al., 2018). In other words, BAs are recirculated to the liver through the portal vein and are mainly taken up by the sodium taurocholate co-transporting polypeptide (NTCP), which is responsible for the absorption of most of the conjugated BAs (Jia et al., 2018). The mechanism through which BAs can return to the liver again through the portal vein is called “enterohepatic circulation” (Figure 1), and exerts noteworthy physiological functions in feedback inhibition of BA synthesis and regulation of systemic lipid homeostasis (Chiang, 2009; Marra and Svegliati-Baroni, 2018). Small amounts of BAs (<5%) escape the uptake mediated by protein transporters in the ileum before entering the colon (Poland and Flynn, 2021). A fraction of primary BAs is deconjugated and dehydroxylated by anaerobic GM in the distal small intestine and colon, resulting in the production of secondary BAs, lithocholic acid (LCA) and deoxycholic acid (DCA), which can be passively uptaken from the large intestine or excreted in the feces (Kwong et al., 2015; Arab et al., 2017).
FIGURE 1.
Bile acids (Bas) metabolism and enterohepatic circulation. In the liver, primary BAs (CA and CDCA) are synthesized by enzymatic catalysis from cholesterol. CYP7A1 initiates the classical synthesis pathway and the ratio of CA/CDCA is regulated by CYP8B1. CYP27A1 and CYP7B1 are mainly responsible for the alternative synthesis pathway. Then conjugated CA/CDCA with taurine or glycine are excreted across the canalicular (apical) membrane into the bile by the BSEP and stored in the gallbladder before being released to the duodenum, where primary BAs are transformed into secondary BAs by gut microbiota via deconjugation, dehydroxylation, oxidation, desulfation, dehydrogenation, and epimerization. Most BAs are reabsorbed in the ileum by the ASBT and then connected to the IBABP. After that, BAs are transported to the portal vein via OSTα/β and are taken up mainly by NTCP. In enteroendocrine cells, FXR enables TGR5 to induce the secretion of GLP-1 in intestinal L-cells. The combination of BAs to FXR not only inhibits the FXR-SHP pathway, accelerating lipid synthesis, but also induces FGF19 in the liver and ilea, and FGF19 binds to the FGFR4 on the hepatocytes, inhibiting BA synthesis. Approximately 5% of BAs are lost in fecal excretion. BAs, bile acids; CA, cholic acid; CDCA, chenodeoxycholic acid; CYP7A1, cholesterol 7α-hydroxylase; CYP8B1, sterol 12α-hydroxylase; CYP27A1, sterol 27-hydroxylase; CYP7B1, oxysterol 7α-hydroxylase; BSEP, bile salt export pump; ASBT, apical sodium-dependent bile acid transporter; IBABP, ileal bile acid-binding protein; OST, organic solute transporter; NTCP, sodium taurocholate co-transporting polypeptide; FXR, farnesoid X receptor; TGR5, Takeda G protein-coupled receptor 5; GLP-1, glucagon-like peptide-1; SHP, small heterodimer partner; FGF19, fibroblast growth factor 15/19; FGFR4, fibroblast growth factor receptor 4. Created with BioRender.com.
Signaling Receptors of Bile Acids
Bile acids were under study over the past several decades and were well-recognized to function as endogenous signaling molecules to regulate metabolic processes, such as liver function and cholesterol metabolism by diverse receptors (de Aguiar Vallim et al., 2013; Arab et al., 2017). Specific receptors mediate the regulation of BAs, including the members of the nuclear receptor superfamily and G protein-coupled receptor superfamily, such as farnesoid X receptor (FXR), vitamin D receptor (VDR), constitutive androstane receptor (CAR), pregnane X receptor (PXR) Takeda G protein-coupled receptor 5 (TGR5) as well as sphingosine-1-phosphate receptor 2 (S1PR2) (Arab et al., 2017; Chiang and Ferrell, 2018). The receptors are expressed not only in enterohepatic tissues but also outside of the liver-gut system to mediate distinct functions throughout the body (Copple and Li, 2016; Chávez-Talavera et al., 2017). Much current knowledge about BAs is concerned with FXR and TGR5 (Arab et al., 2017).
Farnesoid X receptor is activated by different BAs (CDCA > DCA > LCA≫CA) and in turn firmly regulates the synthesis and transport of BAs via negative feedback inhibition (de Aguiar Vallim et al., 2013; Wahlström et al., 2016). The binding of BAs to ileal FXR induces the expression of fibroblast growth factor 15/19 (FGF15/19, FGF15 in mice and FGF19 in humans), which binds to the FGF receptor 4 (FGFR4) on the hepatocytes and inhibits the synthesis of BAs (Jia et al., 2018). It is consistent with the fact that the lack of either FGF15/19 or FGFR4 exhibits the increase of hepatic CYP7A1 mRNA and a larger BA pool size (Copple and Li, 2016). Thus, FXR sustains the homeostasis of BAs and avoids the overaccumulation of toxic BAs concentrations in liver cells (Stanimirov et al., 2015). In terms of transport of BAs, ileal FXR activated by unconjugated BAs reduces ASBT expression and causes IBABP and OSTα/β expression (Jia et al., 2018). Therefore, the deficiency of FXR activity induces the disorder of metabolic and host BA regulation (Guzior and Quinn, 2021). As for the other members of the nuclear receptor superfamily, both PXR and VDR are involved in the metabolism and detoxification of BAs and xenobiotics (Chiang, 2009). CAR contributes to the BA homeostasis with PXR and VDR synergistically (Jia et al., 2018).
The expression of TGR5 is different over diverse anatomical compartments, including the liver, gallbladder as well as small and large intestine (Keitel and Häussinger, 2018; Simbrunner et al., 2021). TGR5 shows a higher affinity for secondary BAs than primary BAs and induces disparate intracellular signaling pathways (Sorrentino et al., 2020; Willis et al., 2020). TGR5 is highly expressed in immune cells as well, including macrophages (hepatic Kupffer cells), where BAs induce Toll-like receptor (TLR) signaling (Willis et al., 2020). FXR and TGR5 are reported to colocalize in enteroendocrine cells, and FXR enables TGR5 to induce glucagon-like peptide-1 (GLP-1) secretion, thus stimulating pancreatic β-cells to secrete insulin and connecting BAs to glucose metabolism (Chiang and Ferrell, 2019). In the ileum and colon, another member of the G protein-coupled receptor superfamily, S1PR2, is expressed and activated by conjugated primary BAs (GCA, TCA, GCDCA, and TCDCA) to regulate inflammation, cancer development, and some liver diseases (Guo et al., 2016a; Fiorucci et al., 2021). S1PR2 triggers proliferation via ERK1/2 and AKT signaling pathways, which are crucial in the regulation of glucose and lipid metabolism in the liver (Yu et al., 2018). Therefore, regulated expression of BA signaling receptors is essential for hepatic homeostasis.
Interactions Between Gut Microbiota and Bile Acids
Bacteriostasis of Bile Acids
Bile salts are effective antibacterial agents in the intestine that inhibit the overgrowth of bacteria and protect a healthy intestinal tract (Betrapally et al., 2016; Sannasiddappa et al., 2017). Apparent leakage of intracellular potassium from Staphylococcus aureus cells was detected after being exposed to bile salts (Sannasiddappa et al., 2017). Furthermore, Spirochaetes, Gonococcus, and Meningococcus were reported susceptible to BAs (Stacey and Webb, 1947). Both conjugated and unconjugated BAs were demonstrated to inhibit Escherichia coli, Klebsiella spp., and Enterococcus spp., while unconjugated BAs were found to have stronger antibacterial capability than conjugated BAs when analyzed with S. aureus, several Lactobacillus species and Bifidobacterium species (Sung et al., 1993; Kurdi et al., 2006). Unconjugated primary BAs can disturb membranes and trigger intracellular damage (Guzior and Quinn, 2021). It is because the pKa of unconjugated BAs is between 5 and 6.5, however, the conjugation of glycine and taurine lowers pKa and ionizes the conjugates at a neutral pH, leading to the difficulty to cross cell membranes (Sannasiddappa et al., 2017; Guzior and Quinn, 2021; Poland and Flynn, 2021). In addition, antibacterial secondary BAs limit the growth of BAs-intolerant bacteria via breaking the cell membrane integrity of bacteria (Kanmani et al., 2020). Moreover, BAs were proved to be entero-protective, possibly via their detergent properties and via FXR activation, which prevents bacterial over proliferation in the distal small intestine (Betrapally et al., 2016). Thus, the size and composition of the BA pool are regarded as vital factors in regulating the structure of GM (Dahiya et al., 2017).
Gut Microbiota and Its Effect on Bile Acids
The adult colon is estimated to contain 200 grams of bacteria. Most bacteria colonizing the gut are obligate anaerobes, while facultatively anaerobic bacteria, archaea and yeast are less abundant (Ridlon et al., 2014). The composition and function of microbiota are affected by diverse host and environmental factors including circadian rhythm, lifestyle, and geographic location (Kolodziejczyk et al., 2019). Seminal studies led to the discovery of the role of GM in regulating the synthesis and metabolism of BAs by adjusting their biomolecular structure.
Glyco-conjugated and tauro-conjugated CA and CDCA are deconjugated in the intestine because anaerobic microbiota can transform conjugated BAs to the unconjugated BAs by bile salt hydrolases (BSHs), and these free BAs are able to trigger BA signaling receptors (Chiang, 2009; Wu et al., 2021). Many bacteria can hydrolyze bile salts (Horác̆ková et al., 2018). For example, choloylglycine hydrolase, a BSH hydrolyzing the amide bond between glycine/taurine and the steroid nucleus of BAs, exits in bacterial genera Lactobacillus, Bifidobacterium, Clostridium, and Bacteroides (Horác̆ková et al., 2018).
The activity of 7α-dehydroxylase in GM removes the 7α-hydroxy group to produce DCA and LCA. The gene encoding 7α-dehydroxylase is present in Clostridium scindens (Chiang, 2009; Jia and Jeon, 2019; Guzior and Quinn, 2021). BSH-rich bacteria from the genera Enterobacter, Clostridium, and Enterococcus are able to form more secondary BAs (Jia et al., 2018). In humans, approximately 2% of CDCA is converted to its 7β epimer, ursodeoxycholic acid (UDCA), a highly soluble and non-toxic secondary BA formed via bacterial 7β-hydroxysteroid dehydrogenase (7β-HSDH) (Chiang and Ferrell, 2019). The activities of BSH and 7α/β-HSDH are abundant in the genera Clostridium, Bifidobacterium, Enterococcus, Lactobacillus, Listeria, and Bacteroides of the phylum Firmicutes and Bacteroidetes. Thus, GM above regulates the synthesis of secondary BAs (Chiang and Ferrell, 2019). Furthermore, HSDHs are present in various GM which can oxidize or reduce the hydroxy groups at the 3-, 7-, and 12- carbons of BAs (Ridlon et al., 2016). The differential composition of BAs in germ-free mice and conventional mice demonstrated that GM not only balances the metabolism of secondary BAs, but also hinders BA synthesis by alleviating the inhibition of FXR in the ileum (Sayin et al., 2013). A recent prospective trial identified that a high-fat diet could cause slight increases in the total BAs, especially DCA in plasma and taurodeoxycholate in liver tissues, along with the elevated abundance of Blautia, Coprococcus, Intestinimonas, Lactococcus, Roseburia, and Ruminococcus (Lin et al., 2019).
In addition to the known transformations of BAs by GM (deconjugation, dehydroxylation, oxidation, desulfation, dehydrogenation, and epimerization), anabolic reactions suggest that the microbiome conjugates CA by unique amino acids, thereby creating newly identified BAs (phenylalanocholic acid, tyrosocholic acid, and leucocholic acid) (Quinn et al., 2020). Although the novel finding indeed deepens our understanding of BAs diversity in our gut, the specific implications for the host and the microbial dynamics are not fully understood (Guzior and Quinn, 2021).
Gut Microbiota in the Pathogenesis of Non-alcoholic Fatty Liver Disease
The Pathogenesis of Non-alcoholic Fatty Liver Disease
Non-alcoholic fatty liver disease is defined as steatosis, which influences >5% of hepatic cells and is accompanied by IR with a lack of excessive alcohol intake (Li et al., 2019). Steatosis refers to either large droplets or a mixture of large and small droplets (Bedossa, 2017). Nine to twenty percent of early-stage NASH patients may develop cirrhosis within 5–10 years, and part of them continue to develop HCC (F3/F4-staged fibrosis patients have about 20% incidence of HCC within 5 years) (Bessone et al., 2019). NAFLD is related to many features of the metabolic syndrome, including IR, obesity, hyperlipidemia, and hypertension, and increases the possibility of cardiovascular disease and type 2 diabetes mellitus, but the pathogenesis of NAFLD is complex and not fully understood (Hrncir et al., 2021). It is currently believed that NAFLD is caused by complex interactions of genetic susceptibility, IR, environmental factors, and GM disorders, in addition to the well-known “two-hit” theory or the “parallel multiple-hit theory” (Kolodziejczyk et al., 2019; Zhou and Fan, 2019; Hrncir et al., 2021). In the two-hit theory, the first “hit” includes aberrant triglyceride accumulation and glucose metabolism in hepatocytes, and the second “hit” refers to the proinflammatory cytokines induced by bacterial endotoxin and oxidative stress relevant to reactive oxygen species (Li et al., 2019). According to the “parallel multiple hit theory,” IR is the “first hit” that results in elevated hepatocellular free fatty acids, a major pathogenic factor that makes the liver more vulnerable to further hits, including mitochondrial dysfunction caused by oxidative stress, endoplasmic reticulum stress, TLR4-dependent inflammatory cytokine release, and iron overload (Bessone et al., 2019).
The Roles of Gut Microbiota in Non-alcoholic Fatty Liver Disease
Recent studies suggest that the compositional and functional diversity of GM may play a role in the progression of NAFLD, as GM diversity in NAFLD patients remains lower than that in healthy subjects (Tilg et al., 2016; Shen et al., 2017; Hrncir et al., 2021). The GM in patients of NAFLD and advanced diseases detected in different studies are listed in Table 1, categorized by phylum, family, and genus. Increased levels of intestinal permeability and inflammation were observed in NAFLD patients with GM dysbiosis. Higher abundances of inflammation-associated Streptococcus and Lactobacillus, Anaerobacter, and Escherichia, and decreased abundances of Ruminococcaceae and Faecalibacterium prausnitzii were observed in NAFLD patients (Jiang et al., 2015; Duarte et al., 2018). Abnormal GM composition caused by factors such as lifestyle, diet, medication, and environment can alter the immune system and the integrity of intestinal mucosa directly and regulate the levels of inflammatory cytokines and fatty acid oxidation, which contributes to liver diseases (Leung et al., 2016; Zheng et al., 2018; Hrncir et al., 2021).
TABLE 1.
Differential gut microbiota between patients with different progression of non-alcoholic fatty liver disease.
| Patients | Composition | Sex (female/male) | Age (years) | Sequencing method | Phylum | Family | Genus | References |
| NAFLD vs. healthy controls (HC) | 39 participants including 13 NAFLD patients and 26 HC | NAFLD: 7/6 HC: 12/14 |
NAFLD: 13.6 ± 3.0 HC: 13.3 ± 2.7 |
16S rRNA | Epsilonproteobacteria, Gammaproteobacteria↑ | / | Prevotella↑ | Michail et al., 2015 |
| NAFLD vs. HC | 72 participants including 18 NAFLD patients and 54 HC | NAFLD: 12/6 HC: 39/15 | NAFLD: 54.0 ± 14.9 HC: 45.9 ± 19.9 |
16S rRNA | / | Rikenellaceae, Mogibacterium, Peptostreptococcaceae↓ | Streptococcus, Bacillus, Lactococcus↑ and Catenibacterium, Pseudomonas↓ | Caussy et al., 2019 |
| NAFLD vs. HC | 85 participants including 53 NAFLD patients and 32 HC | NAFLD: 27/26 HC: 27/5 |
NAFLD: 48 (22–72) HC: 41 (26–52) |
16S rRNA | Lentisphaerae↓ | Ruminococcaceae↓ | Alternatively, Escherichia, Anaerobacter, Lactobacillus, Streptococcus, Clostridium XI? and Alistipes, Prevotella, Oscillibacter, Odoribacter, Flavonifractor↓ | Jiang et al., 2015 |
| NAFLD vs. HC | 60 participants including 30 NAFLD patients and 30 HC | NAFLD: 17/13 HC: 17/13 |
NAFLD: 49 (34–57) HC: 51 (47–56) |
Multitag pyrosequencing | Lachnospiraceae↑ and Ruminococcaceae ↓ | / | Lactobacillus, Dorea, Robinsoniella, Roseburia↑ and Oscillibacter ↓ | Raman et al., 2013 |
| NAFLD vs. HC | 126 participants including 43 NAFLD patients and 83 HC | NAFLD: 7/36 HC: 13/70 |
NAFLD: 47 (34.5–61.0) HC: 40.5 (33.0–52.0) |
16S rRNA | Bacteroidetes↑ and Firmicutes↓ | / | Coprococcu, Pseudobutyrivibrio, Moryella, Roseburia, Anaerosporobacter, Anaerotruncus, Ruminococcus, Lactobacillus↓ | Wang et al., 2016 |
| NAFLD vs. HC | 81 participants including 27 NAFLD patients and 54 HC | NAFLD: 6/21 HC: 31/23 |
NAFLD: 12.04 ± 2.78 HC: 10.24 ± 2.51 |
16S rRNA | Actinobacteria↑ and Bacteroidetes↓ | Rikenellaceae↓ | Bradyrhizobium, Anaerococcus, Peptoniphilus, Propionibacterium acnes, Dorea, Ruminococcus↑ and Oscillospira↓ | Del Chierico et al., 2017 |
| NAFLD vs. HC | 47 participants including 25 NAFLD patients and 22 HC | NAFLD: 6/19 HC: 5/17 |
NAFLD: 45.5 ± 10.1 HC: 50.5 ± 9.5 |
16S rDNA | Proteobacteria, Fusobacteria↑ and Bacteroidetes↓ | Lachnospiraceae, Enterobacteriaceae, Erysipelotrichaceae, Streptococcaceae↑ and Prevotellaceae, Ruminococcaceae↓ | Escherichia Shigella, Lachnospiraceae Incertae Sedis, Blautia↑ and Prevotella↓ | Shen et al., 2017 |
| NASH vs. HC | 38 participants including 16 NASH patients and 22 HC | NASH: 7/9 HC: 13/9 |
NASH: 51 ± 9 HC: 44 ± 10 |
16S rRNA | / | / | Parabacteroides, Allisonella↑ and Faecalibacterium, Anaerosporobacter↓ | Wong et al., 2013 |
| NASH vs. HC | 39 participants including 22 NASH patients and 17 HC | NASH: 12/10 HC: 7/10 |
NASH: 47 (29–68) HC: 36 (23–58) |
qPCR | Percentage of Bacteroidetes↓ | / | / | Mouzaki et al., 2013 |
| NASH vs. HC | 80 participants including 26 NASH patients and 54 HC | NASH: 15/11 HC: 31/23 |
NASH: 12.27 ± 2.47 HC: 10.24 ± 2.51 |
16S rRNA | / | / | Dorea, Ruminococcus, Blautia↑ and Oscillospira↓ | Del Chierico et al., 2017 |
| NASH vs. HC | 28 participants including 6 NASH patients and 22 HC | NASH: not listed HC: 5/17 |
NASH: not listed HC: 50.5 ± 9.5 |
16S rDNA | / | Lachnospiraceae↑ | Blautia↑ | Shen et al., 2017 |
| NASH vs. HC | 38 participants including 22 NASH patients and 16 HC | NASH: 10/13 HC: 6/10 |
NASH: 13.6 ± 3.5 HC: 14.4 ± 1.8 |
16S rRNA | Bacteroidetes, Proteobacteria↑ and Firmicutes↓ | Prevotellaceae, Alcaligenaceae, Enterobacteriaceae↑ and Bifidobacteriaceae, Rikenellaceae, Ruminococcaceae, Lachnospiraceae↓ | Prevotella, Peptoniphilus↑ and Bifidobacterium, Oscillospira, Roseburia, Alistipes, Blautia, Coprococcus, Ruminococcus↓ | Zhu et al., 2013 |
| NASH vs. HC | 30 participants including 10 NASH and 20 HC | NASH: 4/6 HC: 7/13 |
NASH: 61 (52–70) HC: 55 (47–64) |
16S rRNA | / | / |
Bacteroides↑ and Prevotella↓ |
Boursier et al., 2016 |
| Fibrosis vs. HC | 47 participants including 27 fibrosis patients and 20 HC | Fibrosis: 12/15 HC: 7/13 |
Fibrosis: 62 (56–67) HC: 55 (47–64) |
16S rRNA | / | / | Ruminococcus, Bacteroides↑ and Prevotella↓ | Boursier et al., 2016 |
| Fibrosis vs. NAFLD | 86 participants including 14 fibrosis patients and 72 NAFLD patients | Fibrosis: 12/2 NAFLD: 36/36 |
Fibrosis: 63.4 ± 3 NAFLD: 49.3 ± 12.6 |
Whole-genome shotgun sequencing |
Proteobacteria↑ and Firmicutes↓ |
/ | / | Loomba et al., 2017 |
| Cirrhosis vs. HC | 80 participants including 26 cirrhotic patients and 54 HC | Cirrhosis: 20/6 HC: 39/15 |
Cirrhosis: 65.1 ± 9.8 HC: 45.9 ± 19.9 |
16S rRNA | / | Enterobacteriaceae↑ and Rikenellaceae, Mogibacterium, Peptostreptococcaceae↓ | Streptococcus, Megasphaera, Gallibacterium↑ and Catenibacterium, Pseudomonas↓ | Caussy et al., 2019 |
| Cirrhosis vs. HC | 181 participants including 98 cirrhotic patients and 83 HC | Cirrhosis: 33/65 HC: 35/48 |
Cirrhosis: 50 ± 11 HC: 42 ± 9 |
/ |
Proteobacteria, Fusobacteria↑ and Bacteroidetes↓ |
/ | Veillonella, Streptococcus, Clostridium, Prevotella↑ and Bacteroides, Eubacterium, Alistipes↓ | Qin et al., 2014 |
| Cirrhosis vs. HC | 61 participants including 47 cirrhotic patients and 14 HC | Early cirrhosis: 6/17 Advanced cirrhosis: 4/20 HC: 4/10 |
Early cirrhosis: 55 ± 2 Advanced cirrhosis: 54 ± 5 HC: 52 ± 5 |
Multi-tagged pyrosequencing | / | Enterobacteriaceae, Veillonellaceae↑ and Lachonospiraceae, Ruminococcaceae↓ | Blautia↓ | Kakiyama et al., 2013 |
| Cirrhosis vs. HC | 60 participants including 36 cirrhotic patients and 24 HC | Cirrhosis: 11/25 HC: 10/14 |
Cirrhosis: 49 ± 11 HC: 46 ± 8 |
16S rRNA | Proteobacteria, Fusobacteria↑ and Bacteroidetes↓ | Enterobacteriaceae, Veillonellaceae, Streptococcaceae↑ and Lachnospiraceae↓ | / | Chen et al., 2011 |
| Cirrhosis vs. HC | 244 participants including 219 cirrhotic patients and 25 HC | Compensated Outpatients: 92/29 Decompensated Outpatients: 40/14 Inpatients: 31/13 HC: 17/8 |
Compensated Outpatients: 57.5 ± 6.1 Decompensated Outpatients: 56.8 ± 6.8 Inpatients: 55.9 ± 6.7 HC: 55.7 ± 8.5 |
Multi-tagged pyrosequencing | / | Staphylococcae, Enterococceae, Enterobacteriaceae↑ and Clostridiales XIV, Lachnospiraceae, Ruminococcaceae, Rikenellaceae↓ | / | Bajaj et al., 2014 |
| Cirrhosis vs. HC | 58 participants including 30 cirrhotic patients and 28 HC | Cirrhosis: 7/23 HC: 6/22 |
Cirrhosis: 49 ± 8 HC: 52 ± 9 |
16S rRNA | / | / | Veillonella, Megasphaera, Dialister, Atopobium, Prevotella↑ and Neisseria, Haemophilus, SR1 genera incertae sedis↓ | Chen et al., 2016 |
| Cirrhosis vs. HC | 40 participants including 20 cirrhotic patients and 20 HC | Cirrhosis: 8/12 HC: 11/9 |
Cirrhosis: 60.62 ± 10.46 HC: 60 ± 14 |
16S rRNA |
Proteobacteria, Bacteroidetes, Cyanobacteria↑ and Verrucomicrobia, Tenericutes, Euryarchaeota↓ |
Enterobacteriaceae, Lactobacillaceae, Pasteurellaceae, Rikenellaceae, Prevotellaceae, Bacteroidaceae, Porphyromonadaceae, Barnesillaceae, Streptococcaceae, Enterococcaceae, Veillonellaceae↑ and Verrucomicrobiaceae, Methanobacteriaceae↓ | Lactobacillus, Haemophilus, Klebsiella, Prevotella, Parabacteroides, Phascolarctobacterium, Veillonella, Enterococcus, Pseudomonas, Streptococcus, Bacteroides, Atopobium, Dialister, Ruminococcus, Christensenella↑ and Akkermansia, Methanobrevibacter↓ | Ponziani et al., 2019 |
| HCC vs. Cirrhosis | 41 participants including 21 HCC patients and 20 cirrhotic patients | HCC: 3/18 Cirrhosis: 8/12 |
HCC: 66.38 ± 6.67 Cirrhosis: 60.62 ± 10.46 |
16S rRNA | Bacteroidetes↑ | Bacteroidaceae, Streptococcaceae, Enterococcaceae, Gemellaceae↑ and Verrucomicrobiaceae, Bifidobacteriaceae↓ | Phascolarctobcterium, Enterococcus, Streptococcus, Gemella, Bilophila↑ and Akkermansia, Bifidobacterium, Dialister, Collinsella, Adlercreutzia↓ | Ponziani et al., 2019 |
The role of GM dysbiosis in NAFLD and its advanced disease progression is shown in Figure 2. GM regulates glucose and lipid metabolism through intestinal metabolites, including BAs and short-chain fatty acids (SCFAs), which are altered and involved in the pathogenesis of NAFLD (Chen and Vitetta, 2020; Hu et al., 2020). Normal GM produces 50–100 mmol/l per day of SCFAs, including acetate, propionate, and butyrate, which are produced by fermentation from fibers, balancing lipid metabolism, and stimulating incretin hormone production (Leung et al., 2016; Schoeler and Caesar, 2019). Altered synthesis of multiple SCFAs due to carbohydrate consumption and gut dysbiosis may contribute to NAFLD through multiple mechanisms (Leung et al., 2016). Butyrate is a relevant energy source for colonocytes, and plays an important role in anti-inflammation, so a reduction in the butyrate-producing microbiota in the gut may further cause hepatic steatosis (Park et al., 2016). Besides, a positive correlation was found between the DNA of Lactobacillus gasseri and Lactobacillus taiwanensis and lipid droplets in the liver (Mokhtari et al., 2017). In conclusion, GM and its metabolites influence the hepatic metabolism of carbohydrates and lipids, as well as the balance of inflammation, thereby affecting NAFLD and its progression (Kolodziejczyk et al., 2019).
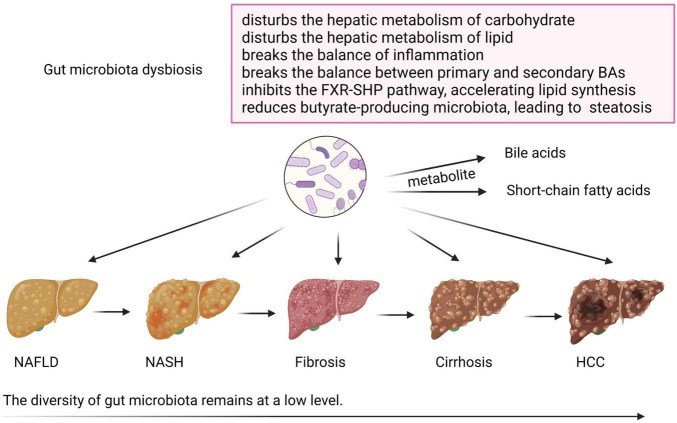
FIGURE 2.
The role of gut microbiota dysbiosis in non-alcoholic fatty liver disease (NAFLD) and its advanced disease progression. During the progression of NAFLD, the diversity of gut microbiota remains at a low level. Gut microbiota dysbiosis disturbs the hepatic metabolism of carbohydrates and lipids and breaks the balance of inflammation. Disturbed gut microbiota also changes the normal metabolism of bile acids (BAs) and short-chain fatty acids, and may cause NAFLD via breaking the balance between primary and secondary BAs, inhibiting the FXR-SHP pathway, which accelerates lipid synthesis, and reducing butyrate-producing microbiota, which causes steatosis. Created with BioRender.com.
Treatment of Non-alcoholic Fatty Liver Disease via Gut Microbiota
It was reported in laboratory research and clinical applications that the disturbed GM structure can be improved by administration of antibiotics, supplement probiotics and prebiotics, or operation of fecal microbial transplantation (FMT) (Hu et al., 2020). For example, the administration of prebiotics can improve body weight, lipid metabolism, IR, and chronic inflammation by modulating GM (Sun et al., 2018). Akkermansia muciniphila, a major propionate-producing bacterium, is recommended as a probiotic and an essential gut symbiont to maintain the metabolism homeostasis and alleviate obesity, diabetes, and liver diseases (Mokhtari et al., 2017; Wang et al., 2019). FMT, a GM-altering approach, has been reported to reduce high-fat diet-induced steatosis, increase insulin sensitivity in patients with metabolic syndrome and improve the cognitive performance in cirrhotic patients (Zhou et al., 2017; Moreno-Gonzalez and Beraza, 2021). Thus, studying the role of GM in NAFLD and the effect of specific intestinal flora in NAFLD patients will further help to diagnose and stratify patients and find therapeutic targets (Kolodziejczyk et al., 2019).
Gut Microbiota-Bile Acids Axis in the Progression of Non-alcoholic Fatty Liver Disease
Gut Microbiota-Bile Acids Axis in Non-alcoholic Fatty Liver Disease
Composition of Bile Acids Altered by Gut Microbiota Leads to Liver Diseases
The composition of the BA pool shows great plasticity because of the modification by GM, thus affecting digestion, nuclear receptor binding, bile fluid toxicity, and solubility, which displays considerable differences between normal individuals and patients with liver diseases (Simbrunner et al., 2021). Due to their cytolytic effects, BAs may be highly toxic when accumulating at high concentrations in the liver (Wu et al., 2021). The hydrophobicity of BAs, which is influenced by the type of conjugation, and the number, orientation, and position of hydroxyl groups, is an important determinant of the toxicity of BAs, and a hydrophobic BA pool is more efficient for intestinal cholesterol absorption (Perez and Briz, 2009; Chávez-Talavera et al., 2017; Molinaro et al., 2018). In addition, excessive secondary BAs were demonstrated to induce reactive oxygen and defective function of mitochondria, leading to DNA apoptosis of cells in the liver (Wu et al., 2021). Therefore, analyzing how GM-modified BAs modulate the host is conducive to understanding the pathogenesis of liver diseases.
Receptors of Bile Acids Affect Non-alcoholic Fatty Liver Disease
Bile acids have been revealed as signaling molecules, most likely to induce NAFLD through FXR and TGR5 (Shao et al., 2021). A recent study of methionine and choline-deficient diet-induced mouse model of NASH showed that hepatic FXR signaling was inhibited and CYP7A1 mRNA was upregulated. In contrast, a study concluded that BAs deconjugated by GM induced FXR signaling in the gut, decreased CYP7A1 expression, and inhibited the FXR-small heterodimer partner (SHP) pathway, resulting in accelerated lipid synthesis, and even liver disease (Park et al., 2016). FXR is highly expressed in tissues involved in the enterohepatic circulation of BAs, with the highest expression in the ileum, so dysbiosis of GM may break the balance between primary and secondary BAs, resulting in the interruption of FXR signaling and leading to far-reaching metabolic consequences (Schaap et al., 2014; Hrncir et al., 2021). Elevated ratio of secondary to primary BAs (DCA/CDCA) is known to dysregulate lipid and glucose metabolism by FXR through various target genes (Abdelmalek, 2021; Hrncir et al., 2021).
In the biliary tract, TGR5 expression is critical to prevent BAs overload (Holter et al., 2020). It not only regulates glucose homeostasis and body weight by regulating the expression of GLP-1, but also contributes to anti-inflammatory effects (Wang et al., 2019; Rao et al., 2020). Secondary BAs bind to TGR5 and inhibit NLRP3 inflammasome activation and inflammatory cytokines production, including interleukin-1β (IL-1β) and IL-18 via the TGR5-cAMP-PKA axis, thereby improving insulin and glucose tolerance (Guo et al., 2016b; Perino et al., 2021).
Gut Microbiota-Bile Acids Axis in Non-alcoholic Steatohepatitis
The diagnosis of NASH is different from NAFLD, requiring the presence of steatosis, hepatocellular ballooning, and lobular inflammation with a certain degree of fibrosis on liver biopsy (Friedman et al., 2018). Liver inflammation causes a positive-feedback mechanism that affects BA synthesis (Ridlon et al., 2013). The levels of total BAs, CA, and CDCA in fecal and the levels of GCA, GCDCA, and TCA in plasma were significantly higher in NASH patients compared with those of healthy individuals (Kalhan et al., 2011; Mouzaki et al., 2016). According to a recent study, alterations in the expression of BAs metabolizing enzymes and transporters resulted in an increase in total BAs in plasma, thereby protecting from the hepatotoxicity during the progression to NASH, but such alterations may contribute to the development of liver fibrosis (Suga et al., 2019). The activated FXR helps converting cholesterol into BAs, storing glucose, reducing fat accumulation and inflammation, and preventing hepatic fibrosis. Impaired FXR activation contributes to a lower level of FGF15/19. Drugs targeting the FXR-FGF15/19 axis are efficacious in the treatment of NASH (Abdelmalek, 2021; Hrncir et al., 2021). Obeticholic acid (6α-ethyl-chenodeoxycholic acid) is a semi-synthetic analogue of CDCA approved by the FDA to treat primary biliary cholangitis (Pellicciari et al., 2002). It can bind to the FXR with high affinity to regulate BA synthesis and transport, modulate hepatic lipid, carbohydrate metabolism and immune function, and improve the histological features of NASH effectively (Tølbøl et al., 2018; Hrncir et al., 2021). However, several dose-dependent side effects such as pruritus, increased cholesterol and LDL along with lower HDL in plasma were observed in NASH patients treated with an FXR agonist (Fiorucci et al., 2020). INT-767, a dual FXR and TGR5 agonist, potently increases intracellular Ca2+ level, cAMP concentration, and GLP-1 secretion and regulates gene expression involved in the BA synthesis, thus it exerts potent therapeutic benefits against NASH (Pathak et al., 2017; Wang et al., 2018).
The impairment of the intestinal epithelial barrier is an early event in NASH pathogenesis (Mouries et al., 2019). Proinflammatory cytokines, including TNF-α, IL-6, and IFN-γ were increased in the gut mucosa biopsies (Jiang et al., 2015). Proinflammatory bacteria are consistently associated with gut permeability, leading to the transportation of bacterial components to the liver, accelerating simple steatosis and the progression to NASH (Schnabl and Brenner, 2014; Jiang et al., 2015). At the phylum level, Bacteroides was increased in abundance in NASH patients, compared to healthy controls (Kolodziejczyk et al., 2019). Members of the genus Escherichia have been reported to be elevated in NASH patients, increasing blood ethanol concentrations (Zhu et al., 2013). Later studies found that endogenous alcohol produced by Escherichia coli increases gut permeability (Jiang et al., 2015).
Gut Microbiota-Bile Acids Axis in Liver Fibrosis
Fibrosis is the consequence of a long-time process of hepatocyte damage and is divided into four stages, F0 to F4 (Shen et al., 2017; Marra and Svegliati-Baroni, 2018). Fibrosis is induced by resident cells in the liver, including hepatocytes, Kupffer cells, and hepatic stellate cells (HSCs) (Kanmani et al., 2020). An earlier experiment demonstrated that a higher ratio of secondary to primary BAs decreased and higher conjugated cholate increased the likelihood of significant fibrosis (F ≥ 2) (Puri et al., 2018). Contrary to the results of this experiment, recent studies have showed that the abundance of total BAs in the plasma of fibrotic individuals is higher, which is positively correlated with the stage of exacerbation of fibrosis due to higher levels of primary BAs rather than secondary BAs (Nimer et al., 2021). Taurodeoxycholate and glycodeoxycholate, which are conjugated 12a-hydroxylated (12a-OH) BAs, were found to increase in mice with liver fibrosis due to their activation of HSCs (Xie et al., 2021).
Compositions of GM in fibrosis patients were determined in a prospective study via whole-genome shotgun sequencing of DNA extracted from stool samples and revealed that the abundance of Firmicutes was higher in moderate NAFLD while Proteobacteria was higher in advanced fibrosis (Loomba et al., 2017). Another study determined the composition of GM in stool samples by 16S ribosomal RNA gene sequencing, demonstrating that the abundance of Bacteroides and Ruminococcus was increased in fibrosis patients, whereas Prevotella abundance was decreased (Boursier et al., 2016). In several studies, researchers found that specific intestinal bacteria are correlated with fibrosis severity and primary BAs, such as Ruminococcus bromii, F. prausnitzii, Roseburia intestinalis, and Megamonas (Lee et al., 2020). For example, F. prausnitzii, one of the butyrate-producing bacteria, has the ability of immunomodulation and is highly sensitive to a slight increase in bile salts concentrations (Duarte et al., 2018; Bromke and Krzystek-Korpacka, 2021). Moreover, UDCA treatment may increase the abundance of F. prausnitzii and Megamonas (Liu et al., 2020). Sterilization of the gut led to less bacteria-derived LPS in plasma, which might ameliorate liver fibrosis in mice, thus improving intestinal disorder with non-absorbable antibiotics or strengthening the intestinal barrier may ameliorate liver fibrosis (Mazagova et al., 2015; Kanmani et al., 2020).
Gut Microbiota-Bile Acids Axis in Liver Cirrhosis
Decreased conversion of primary to secondary BAs in feces is more common in patients with cirrhosis, which is associated with the abundance of certain GMs (Kakiyama et al., 2013). In a recent review on BAs, TCA, GCA, TCDCA, and GCDCA were described as BA biomarkers in liver cirrhosis (Yang et al., 2019). Some species in Ruminococcaceae and Blautia are known to produce 7α-dehydroxylase, leading to higher DCA levels in the stool of patients with cirrhosis (Lin et al., 2019). Changes in the ratio of Bacteroides/Firmicutes and elevation of Gram-negative bacteria are pathogenic (Kakiyama et al., 2013; Shao et al., 2021). Compared with healthy controls, patients with cirrhosis had fewer Bacteroidetes and Firmicutes but higher abundances of Proteobacteria and Fusobacteria (Chen et al., 2011; Qin et al., 2014). Moreover, studies have shown that pathogenic Enterobacteriaceae as well as strains from oral microbiota, such as Veillonella and Streptococcus were enriched in patients with cirrhosis, consistent with the spread of bacteria from the mouth to the gut (Chen et al., 2011; Qin et al., 2014; Yu and Schwabe, 2017). The GM composition of patients with cirrhosis showed relatively lower levels of Lachnospiraceae, Ruminococcaceae, Clostridialies XIV, and F. prausnitzii and a higher abundance of Bacteroidaceae (Bajaj et al., 2014; Aron-Wisnewsky et al., 2020a; Moreno-Gonzalez and Beraza, 2021). The microbial signature of cirrhosis showed high abundance of Megasphaera, Dialister, Atopobium, Prevotella, and Gallibacterium (Chen et al., 2016; Caussy et al., 2019).
Gut Microbiota-Bile Acids Axis in Hepatocellular Carcinoma
Hepatocellular carcinoma is one of the top three causes of cancer mortality worldwide (Schwabe and Greten, 2020). TLR4 activation is involved in hepatocarcinogenesis through HSCs, macrophages, and hepatocytes, resulting in a chronic inflammatory state. TLR4 also promotes fibrosis and induces the expression of epiregulin in HSCs, a potent HCC-promoting hepatocyte mitogen (Schwabe and Greten, 2020). TLR4 activation and GM-induced proliferation are required for HCC promotion, and gut sterilization targeting advanced stages of hepatocarcinogenesis may ameliorate HCC symptoms (Dapito et al., 2012). Compared with NASH-cirrhotic patients, NASH-HCC patients have a distinct GM composition, accompanied by reduced diversity but elevated abundances of anti-inflammatory-associated bacteria, Bifidobacterium and Blautia, and higher levels of Enterococcus, Ruminococcus, Bacteroides, Phascolarctobacterium, and Oscillospira (Moreno-Gonzalez and Beraza, 2021). In a recent study, Lactobacilli was found to be associated with BAs levels in serum samples and liver injury in NASH-HCC patients (Sydor et al., 2020).
Changes in BAs may cause metabolic disorders, hepatic lesions, resistance to apoptosis, and high proliferation, which contribute to tumorigenesis (Wu et al., 2021). A preclinical rodent model has shown that the toxic effects of secondary BAs transformed by GM led to the development of HCC (Moreno-Gonzalez and Beraza, 2021). For example, in HSCs, the enterohepatic circulation of DCA, synergizing with the TLR2 agonist, lipoteichoic acid, a component of Gram-positive gut microbes, induces the senescence-associated secretory phenotype, which suppresses anti-tumor immunity and promotes tumor-promoting factors, such as cyclooxygenase-2 (COX-2), followed by COX-2-mediated prostaglandin E2 (PGE2) inhibition of antitumor immunity through PGE2 receptor subtype 4 (EP4), resulting in HCC in mice (Yoshimoto et al., 2013; Loo et al., 2017). Excessive COX-2 expression and PGE2 production were also detected in HSCs of HCC patients without cirrhosis and NASH (Loo et al., 2017). It was demonstrated in a NASH-HCC model that the secondary BAs accumulation affected by GM also induces carcinogenesis through mTOR signaling activation in hepatocytes (Yamada et al., 2018). Furthermore, the expression of CXCL16 may affect the abundance of CDCA in non-tumor liver tissues of HCC patients, while it is negatively correlated with the secondary BA, glycolithocholic acid (Ma et al., 2018). Mice depleted of Gram-positive bacteria with vancomycin showed accumulation of hepatic natural killer T (NKT) cells and reduction in HCC. However, administration of secondary BAs or colonization of bile acid–metabolizing bacteria reversed NKT cell accumulation and tumor growth (Ma et al., 2018). In the same research, Clostridium scindens colonization was found to mediate BA conversion and reduce hepatic NKT cells by suppressing CXCL16 expression in hepatic sinusoidal endothelial cells to affect the growth of HCC (Ma et al., 2018). Rifaximin, a non-absorbable antibiotic, may target the gut–liver axis and moderately reduce HCC development (Dapito et al., 2012; Yu and Schwabe, 2017).
Conclusion
Growing evidence suggests that BAs and GM play critical roles in the pathogenesis of NAFLD. BAs are endogenous molecules and therapeutic targets in liver diseases, effectively maintain cholesterol and lipid homeostasis in the gut, regulate metabolic signaling through multiple receptors, and act as the intermedium of complex molecular crosstalk between humans and their gut microbiota (Suga et al., 2019). GM intervenes in the pathogenesis of NAFLD by transporting its substances or metabolites (Ji et al., 2019). In the gut, interactions between GM and BAs contribute to the development of disease. Further studies of the GM-BAs axis are necessary to explore the pathogenic mechanism of metabolic diseases, such as NAFLD. Currently, most studies on the GM-BAs axis are based on mouse models, yet there is a vast chasm between humans and mice in the immune system, BAs metabolism, GM, and etc. Thus studies taken on humans are important for a better understanding of the role of GM-BAs axis in the progression of NAFLD (Shao et al., 2021). The gut-liver axis has attracted much attention in the studies of metabolic diseases, and the prevention and treatment of NAFLD targeting the gut-liver axis may be imperative in the future (Ji et al., 2020). The complex roles of BAs and the host microbiome in NAFLD are just beginning to be understood. More molecular mechanisms and signaling pathways for the prevention, diagnosis and treatment of NAFLD need to be validated and applied clinically.
Author Contributions
YN and ML edited the manuscript. YX, QW, XYG, and YL searched the references. TXZ, CX, and TZ prepared the figures and table. X-JG and MZ were responsible for the supervision of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
- NAFLD
non-alcoholic fatty liver disease
- IR
insulin resistance
- NASH
non-alcoholic steatohepatitis
- HCC
hepatocellular carcinoma
- BAs
bile acids
- GM
gut microbiota
- CYP7A1
cholesterol 7α-hydroxylase
- C4
7α-hydroxy-4-cholesten-3-one
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- CYP8B1
sterol 12α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- CYP7B1
oxysterol 7α-hydroxylase
- BSEP
bile salt export pump
- ASBT
apical sodium-dependent bile acid transporter
- IBABP
ileal bile acid-binding protein
- OST
organic solute transporter
- NTCP
sodium taurocholate co-transporting polypeptide
- LCA
lithocholic acid
- DCA
deoxycholic acid
- FXR
farnesoid X receptor
- VDR
vitamin D receptor
- CAR
constitutive androstane receptor
- PXR
pregnane X receptor
- TGR5
Takeda G protein-coupled receptor 5
- S1PR2
sphingosine-1-phosphate receptor 2
- SHP
small heterodimer partner
- FGF15/19
fibroblast growth factor 15/19
- FGFR4
fibroblast growth factor receptor 4
- TLR
toll-like receptor
- GLP-1
glucagon-like peptide-1
- BSHs
bile salt hydrolases
- UDCA
ursodeoxycholic acid
- 7β-HSDH
7β-hydroxysteroid dehydrogenase
- SCFAs
short-chain fatty acid
- FMT
fecal microbial transplantation
- HSCs
hepatic stellate cells
- COX-2
cyclooxygenase-2
- PGE2
prostaglandin E2
- EP4
PGE2 receptor subtype
- NKT
natural killer T.
Funding
This work was supported by Innovation Project for Undergraduates of Shanghai University of Traditional Chinese Medicine (2020SHUTCM134) and also sponsored by the General Program of National Natural Science Foundation of China (82074083).
References
- Abdelmalek M. F. (2021). Nonalcoholic fatty liver disease: another leap forward. Nat. Rev. Gastroenterol. Hepatol. 18 85–86. 10.1038/s41575-020-00406-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab J. P., Karpen S. J., Dawson P. A., Arrese M., Trauner M. (2017). Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology 65 350–362. 10.1002/hep.28709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Warmbrunn M. V., Nieuwdorp M., Clément K. (2020b). Nonalcoholic Fatty Liver Disease: modulating Gut Microbiota to Improve Severity? Gastroenterology 158 1881–1898. 10.1053/j.gastro.2020.01.049 [DOI] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Vigliotti C., Witjes J., Le P., Holleboom A. G., Verheij J., et al. (2020a). Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17 279–297. 10.1038/s41575-020-0269-9 [DOI] [PubMed] [Google Scholar]
- Bajaj J. S., Heuman D. M., Hylemon P. B., Sanyal A. J., White M. B., Monteith P., et al. (2014). Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 60 940–947. 10.1016/j.jhep.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedossa P. (2017). Pathology of non-alcoholic fatty liver disease. Liver Int. 37(Suppl. 1), 85–89. 10.1111/liv.13301 [DOI] [PubMed] [Google Scholar]
- Bessone F., Razori M. V., Roma M. G. (2019). Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell Mol. Life Sci. 76 99–128. 10.1007/s00018-018-2947-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrapally N. S., Gillevet P. M., Bajaj J. S. (2016). Changes in the Intestinal Microbiome and Alcoholic and Nonalcoholic Liver Diseases: causes or Effects? Gastroenterology 150 1745.e–1755.e. 10.1053/j.gastro.2016.02.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia L. S., Curzen N. P., Calder P. C., Byrne C. D. (2012). Non-alcoholic fatty liver disease: a new and important cardiovascular risk factor? Eur. Heart J. 33 1190–1200. 10.1093/eurheartj/ehr453 [DOI] [PubMed] [Google Scholar]
- Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F., et al. (2016). The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63 764–775. 10.1002/hep.28356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromke M. A., Krzystek-Korpacka M. (2021). Bile Acid Signaling in Inflammatory Bowel Disease. Int. J. Mol. Sci. 22:22169096. 10.3390/ijms22169096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C., Tripathi A., Humphrey G., Bassirian S., Singh S., Faulkner C., et al. (2019). A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 10:1406. 10.1038/s41467-019-09455-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Talavera O., Tailleux A., Lefebvre P., Staels B. (2017). Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 152 1679.e–1694.e. 10.1053/j.gastro.2017.01.055 [DOI] [PubMed] [Google Scholar]
- Chen J., Vitetta L. (2020). Gut Microbiota Metabolites in NAFLD Pathogenesis and Therapeutic Implications. Int. J. Mol. Sci. 21:21155214. 10.3390/ijms21155214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ji F., Guo J., Shi D., Fang D., Li L. (2016). Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep. 6:34055. 10.1038/srep34055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang F., Lu H., Wang B., Chen Y., Lei D., et al. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 54 562–572. 10.1002/hep.24423 [DOI] [PubMed] [Google Scholar]
- Chiang J. Y. (2009). Bile acids: regulation of synthesis. J. Lipid Res. 50 1955–1966. 10.1194/jlr.R900010-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. L., Ferrell J. M. (2018). Bile Acid Metabolism in Liver Pathobiology. Gene Expr. 18 71–87. 10.3727/105221618x15156018385515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. L., Ferrell J. M. (2019). Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu Rev. Nutr. 39 175–200. 10.1146/annurev-nutr-082018-124344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. L., Ferrell J. M. (2020). Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 318 G554–G573. 10.1152/ajpgi.00223.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple B. L., Li T. (2016). Pharmacology of bile acid receptors: evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol. Res. 104 9–21. 10.1016/j.phrs.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya D. K., Renuka, Puniya M., Shandilya U. K., Dhewa T., Kumar N., et al. (2017). Gut Microbiota Modulation and Its Relationship with Obesity Using Prebiotic Fibers and Probiotics: a Review. Front. Microbiol. 8:563. 10.3389/fmicb.2017.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapito D. H., Mencin A., Gwak G. Y., Pradere J. P., Jang M. K., Mederacke I., et al. (2012). Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 21 504–516. 10.1016/j.ccr.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim T. Q., Tarling E. J., Edwards P. A. (2013). Pleiotropic roles of bile acids in metabolism. Cell Metab. 17 657–669. 10.1016/j.cmet.2013.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F., Nobili V., Vernocchi P., Russo A., De Stefanis C., Gnani D., et al. (2017). Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65 451–464. 10.1002/hep.28572 [DOI] [PubMed] [Google Scholar]
- Duarte S. M. B., Stefano J. T., Miele L., Ponziani F. R., Souza-Basqueira M., Okada L., et al. (2018). Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: a prospective pilot study. Nutr. Metab. Cardiovasc. Dis. 28 369–384. 10.1016/j.numecd.2017.10.014 [DOI] [PubMed] [Google Scholar]
- Fiorucci S., Biagioli M., Sepe V., Zampella A., Distrutti E. (2020). Bile acid modulators for the treatment of nonalcoholic steatohepatitis (NASH). Expert Opin. Investig. Drugs 29 623–632. 10.1080/13543784.2020.1763302 [DOI] [PubMed] [Google Scholar]
- Fiorucci S., Carino A., Baldoni M., Santucci L., Costanzi E., Graziosi L., et al. (2021). Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 66 674–693. 10.1007/s10620-020-06715-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. L., Neuschwander-Tetri B. A., Rinella M., Sanyal A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24 908–922. 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb A., Canbay A. (2019). Why Bile Acids Are So Important in Non-Alcoholic Fatty Liver Disease (NAFLD) Progression. Cells 8:8111358. 10.3390/cells8111358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Chen W. D., Wang Y. D. (2016a). TGR5, Not Only a Metabolic Regulator. Front. Physiol. 7:646. 10.3389/fphys.2016.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., et al. (2016b). Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 45 802–816. 10.1016/j.immuni.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Guzior D. V., Quinn R. A. (2021). Review: microbial transformations of human bile acids. Microbiome 9:140. 10.1186/s40168-021-01101-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holter M. M., Chirikjian M. K., Govani V. N., Cummings B. P. (2020). TGR5 Signaling in Hepatic Metabolic Health. Nutrients 12:12092598. 10.3390/nu12092598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horác̆ková S., Plocková M., Demnerová K. (2018). Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol. Adv. 36 682–690. 10.1016/j.biotechadv.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Hrncir T., Hrncirova L., Kverka M., Hromadka R., Machova V., Trckova E., et al. (2021). Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 9:9050957. 10.3390/microorganisms9050957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Ye C., Leung E. L., Zhu L., Hu H., Zhang Z., et al. (2020). Bletilla striata oligosaccharides improve metabolic syndrome through modulation of gut microbiota and intestinal metabolites in high fat diet-fed mice. Pharmacol. Res. 159:104942. 10.1016/j.phrs.2020.104942 [DOI] [PubMed] [Google Scholar]
- Ji Y., Yin Y., Li Z., Zhang W. (2019). Gut Microbiota-Derived Components and Metabolites in the Progression of Non-Alcoholic Fatty Liver Disease (NAFLD). Nutrients 11:11081712. 10.3390/nu11081712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Yin Y., Sun L., Zhang W. (2020). The Molecular and Mechanistic Insights Based on Gut-Liver Axis: nutritional Target for Non-Alcoholic Fatty Liver Disease (NAFLD) Improvement. Int. J. Mol. Sci. 21:21093066. 10.3390/ijms21093066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Jeon C. O. (2019). Promotion and induction of liver cancer by gut microbiome-mediated modulation of bile acids. PLoS Pathog. 15:e1007954. 10.1371/journal.ppat.1007954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Xie G., Jia W. (2018). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15 111–128. 10.1038/nrgastro.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wu N., Wang X., Chi Y., Zhang Y., Qiu X., et al. (2015). Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 5:8096. 10.1038/srep08096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiyama G., Pandak W. M., Gillevet P. M., Hylemon P. B., Heuman D. M., Daita K., et al. (2013). Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58 949–955. 10.1016/j.jhep.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan S. C., Guo L., Edmison J., Dasarathy S., McCullough A. J., Hanson R. W., et al. (2011). Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 60 404–413. 10.1016/j.metabol.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. D., Myers C. J., Harris S. C., Kakiyama G., Lee I. K., Yun B. S., et al. (2019). Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 26 27.e–34.e. 10.1016/j.chembiol.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanmani P., Suganya K., Kim H. (2020). The Gut Microbiota: how Does It Influence the Development and Progression of Liver Diseases. Biomedicines 8:8110501. 10.3390/biomedicines8110501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keitel V., Häussinger D. (2018). Role of TGR5 (GPBAR1) in Liver Disease. Semin. Liver Dis. 38 333–339. 10.1055/s-0038-1669940 [DOI] [PubMed] [Google Scholar]
- Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., et al. (2015). Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 21 739–746. 10.1016/j.cmet.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Zheng D., Shibolet O., Elinav E. (2019). The role of the microbiome in NAFLD and NASH. EMBO Mol. Med. 11:201809302. 10.15252/emmm.201809302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi P., Kawanishi K., Mizutani K., Yokota A. (2006). Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 188 1979–1986. 10.1128/jb.188.5.1979-1986.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong E., Li Y., Hylemon P. B., Zhou H. (2015). Bile acids and sphingosine-1-phosphate receptor 2 in hepatic lipid metabolism. Acta Pharm. Sin. B. 5 151–157. 10.1016/j.apsb.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., You H. J., Bajaj J. S., Joo S. K., Yu J., Park S., et al. (2020). Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 11:4982. 10.1038/s41467-020-18754-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C., Rivera L., Furness J. B., Angus P. W. (2016). The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 13 412–425. 10.1038/nrgastro.2016.85 [DOI] [PubMed] [Google Scholar]
- Li T., Chiang J. Y. (2014). Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66 948–983. 10.1124/pr.113.008201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yang H., Zhao Q., Wang X., Zhang J., Zhao X. (2019). Polyphenol-Rich Loquat Fruit Extract Prevents Fructose-Induced Nonalcoholic Fatty Liver Disease by Modulating Glycometabolism, Lipometabolism, Oxidative Stress, Inflammation, Intestinal Barrier, and Gut Microbiota in Mice. J. Agric. Food Chem. 67 7726–7737. 10.1021/acs.jafc.9b02523 [DOI] [PubMed] [Google Scholar]
- Lin H., An Y., Tang H., Wang Y. (2019). Alterations of Bile Acids and Gut Microbiota in Obesity Induced by High Fat Diet in Rat Model. J. Agric. Food Chem. 67 3624–3632. 10.1021/acs.jafc.9b00249 [DOI] [PubMed] [Google Scholar]
- Liu T., Song X., Khan S., Li Y., Guo Z., Li C., et al. (2020). The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: an old story, yet mesmerizing. Int. J. Cancer. 146 1780–1790. 10.1002/ijc.32563 [DOI] [PubMed] [Google Scholar]
- Loo T. M., Kamachi F., Watanabe Y., Yoshimoto S., Kanda H., Arai Y., et al. (2017). Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE(2)-Mediated Suppression of Antitumor Immunity. Cancer Discov. 7 522–538. 10.1158/2159-8290.Cd-16-0932 [DOI] [PubMed] [Google Scholar]
- Loomba R., Seguritan V., Li W., Long T., Klitgord N., Bhatt A., et al. (2017). Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 25 1054.e–1062.e. 10.1016/j.cmet.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:5931. 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra F., Svegliati-Baroni G. (2018). Lipotoxicity and the gut-liver axis in NASH pathogenesis. J. Hepatol. 68 280–295. 10.1016/j.jhep.2017.11.014 [DOI] [PubMed] [Google Scholar]
- Mazagova M., Wang L., Anfora A. T., Wissmueller M., Lesley S. A., Miyamoto Y., et al. (2015). Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. Faseb. J. 29 1043–1055. 10.1096/fj.14-259515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michail S., Lin M., Frey M. R., Fanter R., Paliy O., Hilbush B., et al. (2015). Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 91 1–9. 10.1093/femsec/fiu002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtari Z., Gibson D. L., Hekmatdoost A. (2017). Nonalcoholic Fatty Liver Disease, the Gut Microbiome, and Diet. Adv. Nutr. 8 240–252. 10.3945/an.116.013151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro A., Wahlström A., Marschall H. U. (2018). Role of Bile Acids in Metabolic Control. Trends Endocrinol. Metab. 29 31–41. 10.1016/j.tem.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Moreno-Gonzalez M., Beraza N. (2021). The Role of the Microbiome in Liver Cancer. Cancers 13:113102330. 10.3390/cancers13102330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouries J., Brescia P., Silvestri A., Spadoni I., Sorribas M., Wiest R., et al. (2019). Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 71 1216–1228. 10.1016/j.jhep.2019.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M., Comelli E. M., Arendt B. M., Bonengel J., Fung S. K., Fischer S. E., et al. (2013). Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58 120–127. 10.1002/hep.26319 [DOI] [PubMed] [Google Scholar]
- Mouzaki M., Wang A. Y., Bandsma R., Comelli E. M., Arendt B. M., Zhang L., et al. (2016). Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One 11:e0151829. 10.1371/journal.pone.0151829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimer N., Choucair I., Wang Z., Nemet I., Li L., Gukasyan J., et al. (2021). Bile acids profile, histopathological indices and genetic variants for non-alcoholic fatty liver disease progression. Metabolism 116:154457. 10.1016/j.metabol.2020.154457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. Y., Kim S. J., Ko E. K., Ahn S. H., Seo H., Sung M. K. (2016). Gut microbiota-associated bile acid deconjugation accelerates hepatic steatosis in ob/ob mice. J. Appl. Microbiol. 121 800–810. 10.1111/jam.13158 [DOI] [PubMed] [Google Scholar]
- Pathak P., Liu H., Boehme S., Xie C., Krausz K. W., Gonzalez F., et al. (2017). Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J. Biol. Chem. 292 11055–11069. 10.1074/jbc.M117.784322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicciari R., Fiorucci S., Camaioni E., Clerici C., Costantino G., Maloney P. R., et al. (2002). 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45 3569–3572. 10.1021/jm025529g [DOI] [PubMed] [Google Scholar]
- Perez M. J., Briz O. (2009). Bile-acid-induced cell injury and protection. World J. Gastroenterol. 15 1677–1689. 10.3748/wjg.15.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino A., Demagny H., Velazquez-Villegas L., Schoonjans K. (2021). Molecular Physiology of Bile Acid Signaling in Health, Disease, and Aging. Physiol. Rev. 101 683–731. 10.1152/physrev.00049.2019 [DOI] [PubMed] [Google Scholar]
- Poland J. C., Flynn C. R. (2021). Bile Acids, Their Receptors, and the Gut Microbiota. Physiology 36 235–245. 10.1152/physiol.00028.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponziani F. R., Bhoori S., Castelli C., Putignani L., Rivoltini L., Del Chierico F., et al. (2019). Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 69 107–120. 10.1002/hep.30036 [DOI] [PubMed] [Google Scholar]
- Puri P., Daita K., Joyce A., Mirshahi F., Santhekadur P. K., Cazanave S., et al. (2018). The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 67 534–548. 10.1002/hep.29359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513 59–64. 10.1038/nature13568 [DOI] [PubMed] [Google Scholar]
- Quinn R. A., Melnik A. V., Vrbanac A., Fu T., Patras K. A., Christy M. P., et al. (2020). Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579 123–129. 10.1038/s41586-020-2047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Ahmed I., Gillevet P. M., Probert C. S., Ratcliffe N. M., Smith S., et al. (2013). Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 11 .e861–.e863. 10.1016/j.cgh.2013.02.015 [DOI] [PubMed] [Google Scholar]
- Rao J., Yang C., Yang S., Lu H., Hu Y., Lu L., et al. (2020). Deficiency of TGR5 exacerbates immune-mediated cholestatic hepatic injury by stabilizing the β-catenin destruction complex. Int. Immunol. 32 321–334. 10.1093/intimm/dxaa002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Alves J. M., Hylemon P. B., Bajaj J. S. (2013). Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 4 382–387. 10.4161/gmic.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Bajaj J. S. (2015). The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm. Sin B. 5 99–105. 10.1016/j.apsb.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., Hylemon P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7 22–39. 10.1080/19490976.2015.1127483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Kang D. J., Hylemon P. B., Bajaj J. S. (2014). Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30 332–338. 10.1097/mog.0000000000000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella M. E. (2015). Nonalcoholic fatty liver disease: a systematic review. Jama 313 2263–2273. 10.1001/jama.2015.5370 [DOI] [PubMed] [Google Scholar]
- Sannasiddappa T. H., Lund P. A., Clarke S. R. (2017). In Vitro Antibacterial Activity of Unconjugated and Conjugated Bile Salts on Staphylococcus aureus. Front. Microbiol. 8:1581. 10.3389/fmicb.2017.01581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A. J., Harrison S. A., Ratziu V., Abdelmalek M. F., Diehl A. M., Caldwell S., et al. (2019). The Natural History of Advanced Fibrosis Due to Nonalcoholic Steatohepatitis: Data From the Simtuzumab Trials. Hepatology 70 1913–1927. 10.1002/hep.30664 [DOI] [PubMed] [Google Scholar]
- Sayin S. I., Wahlström A., Felin J., Jäntti S., Marschall H. U., Bamberg K., et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17 225–235. 10.1016/j.cmet.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Schaap F. G., Trauner M., Jansen P. L. (2014). Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11 55–67. 10.1038/nrgastro.2013.151 [DOI] [PubMed] [Google Scholar]
- Schnabl B., Brenner D. A. (2014). Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146 1513–1524. 10.1053/j.gastro.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler M., Caesar R. (2019). Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 20 461–472. 10.1007/s11154-019-09512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe R. F., Greten T. F. (2020). Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J. Hepatol. 72 230–238. 10.1016/j.jhep.2019.08.016 [DOI] [PubMed] [Google Scholar]
- Shao J. W., Ge T. T., Chen S. Z., Wang G., Yang Q., Huang C. H., et al. (2021). Role of bile acids in liver diseases mediated by the gut microbiome. World J. Gastroenterol. 27 3010–3021. 10.3748/wjg.v27.i22.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Zheng R. D., Sun X. Q., Ding W. J., Wang X. Y., Fan J. G. (2017). Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 16 375–381. 10.1016/s1499-3872(17)60019-5 [DOI] [PubMed] [Google Scholar]
- Simbrunner B., Trauner M., Reiberger T. (2021). Therapeutic aspects of bile acid signalling in the gut-liver axis. Aliment Pharmacol. Ther. 2021:16602. 10.1111/apt.16602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G., Perino A., Yildiz E., El Alam G., Bou Sleiman M., Gioiello A., et al. (2020). Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 159 956.e–968.e. 10.1053/j.gastro.2020.05.067 [DOI] [PubMed] [Google Scholar]
- Stacey M., Webb M. (1947). Studies on the antibacterial properties of the bile acids and some compounds derived from cholanic acid. Proc. R. Soc. Med. 134 523–537. 10.1098/rspb.1947.0029 [DOI] [PubMed] [Google Scholar]
- Stanimirov B., Stankov K., Mikov M. (2015). Bile acid signaling through farnesoid X and TGR5 receptors in hepatobiliary and intestinal diseases. Hepatobiliary Pancreat. Dis. Int. 14 18–33. 10.1016/s1499-3872(14)60307-6 [DOI] [PubMed] [Google Scholar]
- Suga T., Yamaguchi H., Ogura J., Shoji S., Maekawa M., Mano N. (2019). Altered bile acid composition and disposition in a mouse model of non-alcoholic steatohepatitis. Toxicol. Appl. Pharmacol. 379:114664. 10.1016/j.taap.2019.114664 [DOI] [PubMed] [Google Scholar]
- Sun L., Ma L., Ma Y., Zhang F., Zhao C., Nie Y. (2018). Insights into the role of gut microbiota in obesity: pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell 9 397–403. 10.1007/s13238-018-0546-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Pang Y., Wang X., Wu Q., Liu H., Liu B., et al. (2019). Ablation of gut microbiota alleviates obesity-induced hepatic steatosis and glucose intolerance by modulating bile acid metabolism in hamsters. Acta Pharm. Sin. B. 9 702–710. 10.1016/j.apsb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J. Y., Shaffer E. A., Costerton J. W. (1993). Antibacterial activity of bile salts against common biliary pathogens. Effects of hydrophobicity of the molecule and in the presence of phospholipids. Dig. Dis. Sci. 38 2104–2112. 10.1007/bf01297092 [DOI] [PubMed] [Google Scholar]
- Sydor S., Best J., Messerschmidt I., Manka P., Vilchez-Vargas R., Brodesser S., et al. (2020). Altered Microbiota Diversity and Bile Acid Signaling in Cirrhotic and Noncirrhotic NASH-HCC. Clin. Transl. Gastroenterol. 11:e00131. 10.14309/ctg.0000000000000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H., Cani P. D., Mayer E. A. (2016). Gut microbiome and liver diseases. Gut 65 2035–2044. 10.1136/gutjnl-2016-312729 [DOI] [PubMed] [Google Scholar]
- Tølbøl K. S., Kristiansen M. N., Hansen H. H., Veidal S. S., Rigbolt K. T., Gillum M. P., et al. (2018). Metabolic and hepatic effects of liraglutide, obeticholic acid and elafibranor in diet-induced obese mouse models of biopsy-confirmed nonalcoholic steatohepatitis. World J. Gastroenterol. 24 179–194. 10.3748/wjg.v24.i2.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A., Sayin S. I., Marschall H. U., Bäckhed F. (2016). Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24 41–50. 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Wang B., Jiang X., Cao M., Ge J., Bao Q., Tang L., et al. (2016). Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci. Rep. 6:32002. 10.1038/srep32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., He Q., Wang G., Xu X., Hao H. (2018). FXR modulators for enterohepatic and metabolic diseases. Exp. Opin. Ther. Pat. 28 765–782. 10.1080/13543776.2018.1527906 [DOI] [PubMed] [Google Scholar]
- Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., et al. (2019). Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 26 222.e–235.e. 10.1016/j.celrep.2018.12.028 [DOI] [PubMed] [Google Scholar]
- Willis K. A., Gomes C. K., Rao P., Micic D., Moran E. R., III, Stephenson E., et al. (2020). TGR5 signaling mitigates parenteral nutrition-associated liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 318 G322–G335. 10.1152/ajpgi.00216.2019 [DOI] [PubMed] [Google Scholar]
- Winston J. A., Theriot C. M. (2020). Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11 158–171. 10.1080/19490976.2019.1674124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V. W., Tse C. H., Lam T. T., Wong G. L., Chim A. M., Chu W. C., et al. (2013). Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis–a longitudinal study. PLoS One 8:e62885. 10.1371/journal.pone.0062885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Feng J., Li J., Yu Q., Ji J., Wu J., et al. (2021). The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 133:111036. 10.1016/j.biopha.2020.111036 [DOI] [PubMed] [Google Scholar]
- Xie G., Jiang R., Wang X., Liu P., Zhao A., Wu Y., et al. (2021). Conjugated secondary 12α-hydroxylated bile acids promote liver fibrogenesis. EBioMed. 66:103290. 10.1016/j.ebiom.2021.103290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Takashina Y., Watanabe M., Nagamine R., Saito Y., Kamada N., et al. (2018). Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget 9 9925–9939. 10.18632/oncotarget.24066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Khan G. J., Wu Z., Wang X., Zhang L., Jiang Z. (2019). Bile acid homeostasis paradigm and its connotation with cholestatic liver diseases. Drug Discov. Today. 24 112–128. 10.1016/j.drudis.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Yoshimoto S., Loo T. M., Atarashi K., Kanda H., Sato S., Oyadomari S., et al. (2013). Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499 97–101. 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- Younossi Z. M., Blissett D., Blissett R., Henry L., Stepanova M., Younossi Y., et al. (2016a). The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64 1577–1586. 10.1002/hep.28785 [DOI] [PubMed] [Google Scholar]
- Younossi Z. M., Koenig A. B., Abdelatif D., Fazel Y., Henry L., Wymer M. (2016b). Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64 73–84. 10.1002/hep.28431 [DOI] [PubMed] [Google Scholar]
- Yu L. X., Schwabe R. F. (2017). The gut microbiome and liver cancer: mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 14 527–539. 10.1038/nrgastro.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Jiang Z., Zhang L. (2018). Bile acid regulation: a novel therapeutic strategy in non-alcoholic fatty liver disease. Pharmacol. Ther. 190 81–90. 10.1016/j.pharmthera.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Zheng J., Yuan X., Cheng G., Jiao S., Feng C., Zhao X., et al. (2018). Chitosan oligosaccharides improve the disturbance in glucose metabolism and reverse the dysbiosis of gut microbiota in diabetic mice. Carbohydr. Polym. 190 77–86. 10.1016/j.carbpol.2018.02.058 [DOI] [PubMed] [Google Scholar]
- Zhou D., Fan J. G. (2019). Microbial metabolites in non-alcoholic fatty liver disease. World J. Gastroenterol. 25 2019–2028. 10.3748/wjg.v25.i17.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Pan Q., Shen F., Cao H. X., Ding W. J., Chen Y. W., et al. (2017). Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci. Rep. 7:1529. 10.1038/s41598-017-01751-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Baker S. S., Gill C., Liu W., Alkhouri R., Baker R. D., et al. (2013). Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57 601–609. 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]