Abstract
Toll-like receptors (TLRs) are key sensors that recognize the pathogen-associated molecular patterns (PAMPs) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to activate innate immune response to clear the invading virus. However, dysregulated immune responses may elicit the overproduction of proinflammatory cytokines and chemokines, resulting in the enhancement of immune-mediated pathology. Therefore, a proper understanding of the interaction between SARS-CoV-2 and TLR-induced immune responses is very important for the development of effective preventive and therapeutic strategies. In this review, we discuss the recognition of SARS-CoV-2 components by TLRs and the downstream signaling pathways that are activated, as well as the dual role of TLRs in regulating antiviral effects and excessive inflammatory responses in patients with coronavirus disease 2019 (COVID-19). In addition, this article describes recent progress in the development of TLR immunomodulators including the agonists and antagonists, as vaccine adjuvants or agents used to treat hyperinflammatory responses during SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, innate immune response, Toll-like receptor, immunomodulator
Introduction
Toll-like receptors (TLRs) are members of the pattern recognition receptor (PRR) family, and they play pivotal roles in the activation of innate immune responses and the regulation of cytokine expression (Hedayat et al., 2011; Birra et al., 2020; Debnath et al., 2020). TLRs perform their functions by recognizing distinguishing molecules of invading pathogens, called pathogen-associated molecular patterns (PAMPs), and then initiate innate immune responses via several distinct signaling pathways, thereby limiting infection and promoting adaptive immune responses (Kawai and Akira, 2011). Generally, myeloid differentiation primary response 88 (MyD88) and Toll-IL-1R (TIR)-domain containing adaptor-inducing interferon-β (TRIF) are the two main pathways by which TLRs transduce signals after activation (Kim et al., 2018).
The innate immune system acts as the first line of defense against invading pathogens, including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is the causative viral pathogen of the coronavirus disease 2019 (COVID-19), and the COVID-19 pandemic began in March 2020 (Tabary et al., 2020). The characteristics of COVID-19 are extremely variable, and the disease ranges from an asymptomatic form lasting for a few days or mild to severe forms of interstitial pneumonia which requires ventilation therapy and can lead to death (Khan et al., 2020). TLRs recognize SARS-CoV-2-derived molecules and activate innate immune responses. Many studies have shown that SARS-CoV-2 activates the innate immune system via TLRs, and mediates the upregulation of TLR expression, which contribute to the elimination of the virus (Aboudounya and Heads, 2021; Bortolotti et al., 2021; Sariol and Perlman, 2021). However, TLR activation may act as a double-edged sword, and dysregulated TLR responses may lead to persistent inflammation and tissue destruction (Yokota et al., 2010; Ebermeyer et al., 2021). For instance, the severity of COVID-19 is associated with cytokine storms in patients which could be produced by overactivation of TLR pathways (Tang et al., 2020; Manik and Singh, 2021). Therefore, a better understanding of the relationship between TLRs and SARS-CoV-2 is critical for understanding the immunopathogenesis involved in COVID-19 and for the preventive and therapeutic application of TLR immunomodulators to combat the disease. In this review, we provide an overview of our recent understanding of the roles of TLRs in SARS-CoV-2-induced immune responses, and we discuss the potential prophylactic and/or therapeutic value of TLR agonists or antagonists in COVID-19 patients.
Structure and Classification of Toll-Like Receptors
All TLRs are type I transmembrane proteins that consist of a leucine-rich repeat (LRR) module, a transmembrane region, and a TIR domain. The LRR motif includes 19–25 tandem sequences containing leucine and is responsible for the recognition of PAMPs, while the TIR endodomain functions to initiate intracellular signaling events via downstream adaptors. According to the composition of their subunits, TLRs operate as heterodimers or homodimers. The classification and expression of TLRs differs between species. Thirteen TLRs have been discovered in mice. However, TLR11 has no function, and the TLR12 and TLR13 genes are absent in the human genome. Generally, the TLR family can be divided into two subgroups based on their localization: cell surface TLRs and intracellular TLRs (Kawasaki and Kawai, 2014). Cell surface TLRs reside on the cell membrane and mainly contain TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10, while intracellular TLRs include TLR3, TLR7, TLR8, TLR9, TLR11, TLR12, and TLR13 (Table 1). Intracellular TLRs localize to intracellular compartments, such as the endosomes, endoplasmic reticulum, and lysosomes.
TABLE 1.
Human and murine TLR classification and their natural ligands.
| Classification | TLR name | Gene location | Ligands | References |
| Cell surface TLRs | TLR1 | 5C3.1 (mouse)/4p14 (human) | Triacylated lipopeptides, lipoarabinomannan | Yu and Feng, 2018; Su et al., 2019 |
| TLR2 | 3E3 (mouse)/4q31.3 (human) | Zymosan, peptidoglycan, lipoteichoic acid, endogenous HSP, HMGB1, gp96 | Patidar et al., 2018; Yu and Feng, 2018 | |
| TLR4 | 4C1 (mouse)/9q33.1 (human) | LPS, endogenous HSP, HMGB1, β-defensin 2 | Vaure and Liu, 2014; Patidar et al., 2018 | |
| TLR5 | 1H5 (mouse)/1q41 (human) | Flagellin | Yang and Yan, 2017 | |
| TLR6 | 5C3.1 (mouse)/4p14 (human) | Diacylated lipopeptides, zymosan, lipoteichoic acid | Yu and Feng, 2018 | |
| TLR10 | 4p14 (human) | Unknown | Kawasaki and Kawai, 2014 | |
| Intracellular TLRs | TLR3 | 8B1.1 (mouse)/4q35.1 (human) | dsRNA | Turton et al., 2020 |
| TLR7 | XF5 (mouse)/Xp22.2 (human) | ssRNA | Tanji et al., 2013 | |
| TLR8 | XF5 (mouse)/Xp22.2 (human) | ssRNA | Tanji et al., 2013 | |
| TLR9 | 9F1 (mouse)/3p21.2 (human) | Unmethylated CpG DNA, 5′-xCx DNA | Ohto et al., 2018 | |
| TLR11 | 11C1 (mouse) | Profilin of Toxoplasma gondii | Lester and Li, 2014 | |
| TLR12 | 4D2.2 (mouse) | Profilin of Toxoplasma gondii | Lester and Li, 2014 | |
| TLR13 | XD (mouse) | Bacterial 23S rRNA | Oldenburg et al., 2012 |
Cell surface TLRs mainly recognize microbial membrane components such as lipoproteins, lipids, and proteins. TLR2 forms heterodimers with either TLR1 or TLR6. It recognizes a wide variety of PAMPs including lipoproteins, lipopeptides, peptidoglycans, and zymosan (Yu and Feng, 2018). TLR4 is mainly expressed on cells of the immune system, including macrophages, monocytes, and dendritic cells (DCs). TLR4 recognizes bacterial lipopolysaccharide (LPS) and its activation leads to the synthesis of proinflammatory cytokines and chemokines (Vaure and Liu, 2014). TLR5 is known to specifically sense and recognize bacterial flagellins (Yang and Yan, 2017). The ligands of TLR10 remain unclear but it is believed that human TLR10 collaborates with TLR2 to recognize ligands from Listeria and senses influenza A virus during infection; however, TLR10 has no function in mice due to an insertion of a stop codon (Kawasaki and Kawai, 2014).
Intracellular TLRs can recognize nucleic acids derived from foreign pathogens and self-derived nucleic acids in the context of some autoimmune diseases (Schlee and Hartmann, 2016). TLR3 is widely distributed in various epithelial cells, fibroblasts, nerve cells and immune cells. TLR3 mainly recognizes viral double-stranded RNA (dsRNA), self-RNAs derived from damaged cells and the replication intermediates generated during the life cycle of single-stranded RNA (ssRNA) viruses and DNA viruses (Turton et al., 2020). TLR7 is localized to endosomes and is mainly expressed by B cells, monocytes and plasmacytoid DCs (pDCs), while TLR8 expression is closely associated with conventional DCs (cDCs). TLR7/8 can recognize ssRNA and activate MyD88-dependent pathways, which lead to the subsequent production of type I interferons (IFNs) and inflammatory cytokines (Kawai and Akira, 2011; Jung and Lee, 2021). TLR9 recognizes unmethylated CpG-DNA motifs which are frequently presented in viral and bacterial DNA, and activates the innate immune response to eliminate these pathogens. In addition, TLR9 also binds to the DNA sequences that contain cytosine in the second position from the 5′ end (5′-xCx DNA), which cooperatively promotes dimerization and activation of TLR9 in the presence of CpG DNA (Ohto et al., 2018). TLR11 is located to endolysosomes, and TLR12 is predominantly expressed in myeloid cells. These TLRs function in recognizing profilin from the parasites (Lester and Li, 2014). TLR13, the orphan receptor in mice, was found to recognize a conserved 23S ribosomal RNA (rRNA) sequence in bacteria (Wang et al., 2016).
Toll-Like Receptor Recognition of Severe Acute Respiratory Syndrome Coronavirus 2 Components
Structure of Severe Acute Respiratory Syndrome Coronavirus 2
Viral proteins and nucleic acids can serve as PAMPs, which are sensed by TLRs to induce the production of antiviral and inflammatory cytokines. Thus, it is quite necessary to understand the nucleic acid composition and protein structure of SARS-CoV-2. Similar to other coronaviruses, SARS-CoV-2 is a positive-sense single-stranded RNA (+ssRNA) virus and its genome is approximately 29.9 kb in size; this genome mainly encodes four major structural proteins, including the spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins. The S protein consists of the receptor binding subunit S1 and the membrane fusing subunit S2. S1 includes a N-terminal domain (NTD), receptor-binding domain (RBD), subdomain 1 (SD1) and subdomain 2 (SD2). S2 includes a fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), and transmembrane (TM). The rest of the genome includes two major open reading frames (ORFs)-ORF1a and ORF1b, which can be translated to pp1a and pp1b polypeptides, and then cleaved to form 16 non-structural proteins (Zhang et al., 2021).
Toll-Like Receptors Recognize Viral Proteins and RNA of Severe Acute Respiratory Syndrome Coronavirus 2
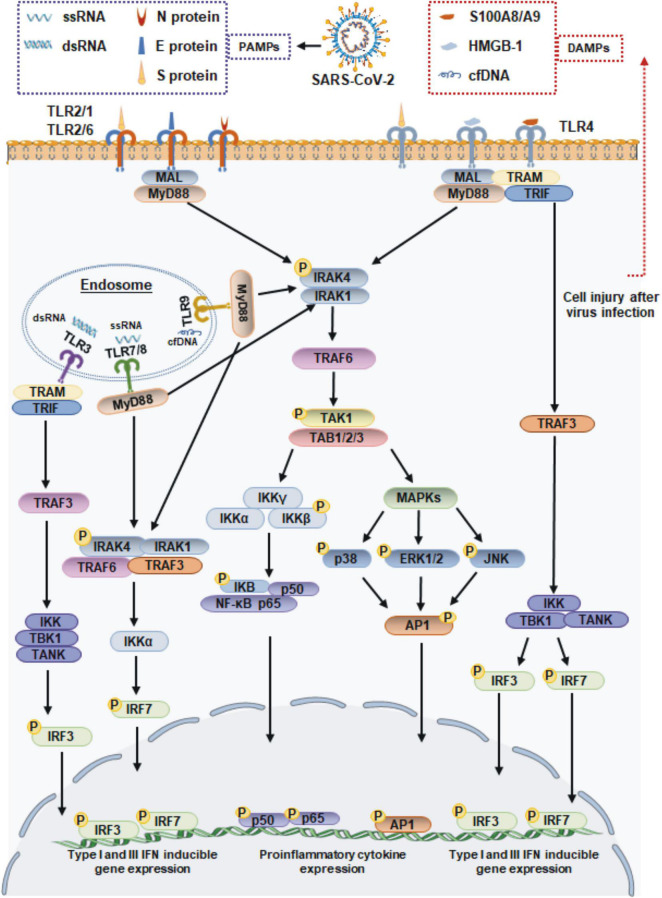
When an infection occurs, viruses enter the body and adhere to cell surfaces. Cell surface TLRs are most likely to be involved in recognizing molecular patterns from SARS-CoV-2 to induce immune responses. The S protein is a major structural protein and is essential for the interaction of SARS-CoV-2 with host cell receptors. In addition to its direct binding to angiotensin-converting enzyme II (ACE2), the S protein is a potent viral PAMP that is sensed by TLR2 in macrophages, monocytes, and lung epithelial cells. Then TLR2 forms heterodimers with TLR1 or TLR6, promoting formation of a complex containing MyD88 with IRAK kinase family members, activating nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling, and ultimately leading to the production of inflammatory cytokines and chemokines (Khan et al., 2021). SARS-CoV-2 infection also increases the expression of TLR2 and other molecules, such as melanoma differentiation-associated gene 5 (MDA5), ACE2 and interferon regulatory factor 3 (IRF3) (Mohanty et al., 2021; Yang et al., 2022), and the expression of TLR2 is positively associated with the severity of COVID-19 (Zheng et al., 2021). In addition to the S protein, the immunogenic properties of other structural proteins have also been investigated. Recent reports have found that TLR2 binds to the SARS-CoV-2 E and N proteins, but only the SARS-CoV-2 N protein induces TLR2 activation, which was not observed with other coronavirus-derived N proteins (Qian et al., 2021; Zheng et al., 2021). TLR4 is well known to recognize bacterial LPS. Actually, TLR4 was found to exhibit a very strong ability to bind to the S protein of SARS-CoV-2 by molecular docking studies (Choudhury and Mukherjee, 2020), and a surface plasmon resonance (SPR) assay confirmed that trimeric SARS-CoV-2 S protein, which is presented on the surface of viral particles, directly binds to TLR4 with a high affinity of ∼300 nM (Zhao et al., 2021). These findings suggest that TLR4 also plays a potential role in the recognition of SARS-CoV-2. This hypothesis has been supported by other evidence demonstrating that the S1 subunit of S protein elicits inflammatory responses in vitro and signals through TLR4. The direct exposure of microglia, macrophages, or TLR4 signaling transgenic HEK293 cells to S1 could activate the MyD88-dependent pathway, resulting in the upregulated expression of proinflammatory cytokines (Shirato and Kizaki, 2021; Frank et al., 2022). Moreover, the responses induced by S1 could be blocked by the TLR4-specific inhibitor Resatorvid or TLR4 siRNA (Shirato and Kizaki, 2021; Zhao et al., 2021). Taken together, these findings suggested that the TLR2 and TLR4 can sense SARS-CoV-2 structural proteins, and they may function independently to induce inflammatory processes (Figure 1).
FIGURE 1.
Toll-like receptor-mediated antiviral and inflammatory responses during SARS-CoV-2 infection. TLR1/2/6 and 4 localize to cell membrane, and TLR3, TLR7/8, and 9 localize to endosome surface. The viral proteins of SARS-CoV-2 signal through TLR2 and TLR4 to activate the adaptor MyD88, which subsequently signals via NF-κB and MAPK to promote the expression of proinflammatory cytokines. TLR4 also recruits the adapter protein TRIF, which activates the TRAF3, resulting the IRF3 activation to lead the production of type I and III IFN. The ssRNA or dsRNA replication intermediates of SARS-CoV-2 are recognized by TLR3 and TLR7/8, respectively. The TLR7/8 recruit MyD88, and TLR3 via TRIF molecule. The proinflammatory cytokines induced by DAMPs accumulating during SARS-CoV-2 infection are driven by the transcription factor NF-κB. MAL, MyD88 adapter-like; TRAF, tumor necrosis factor receptor-associated factor; TRIF, TIR-domain-containing adapter-inducing interferon-β; TRAM, TRIF-related adaptor molecule; IRAK1/4, interleukin-1 receptor associated kinase 1/4; TAK1, transforming growth factor β-activated kinase 1; TAB, TAK1-binding proteins; IKK, IkB kinase; TBK1, TANK-binding kinase 1; ERK1/2, extracellular signal-regulated kinases 1/2; JNK, c-Jun N-terminal kinase; AP-1, activating protein-1.
When the SARS-CoV-2 binds to a host cell receptor such as ACE2, it fuses to the host cell membrane and releases its contents into the cell. Viral ssRNA and its replication intermediate dsRNA, are likely to be initially detected by host cells via TLR7/8 and TLR3 in endosomes. Previous studies showed that SARS-CoV-2 infection of Calu-3/MRC-5 multicellular spheroids induced the activation of both the TLR3 and TLR7/8 RNA sensor pathways (Bortolotti et al., 2021). Subsequently, TLR7/8 signaling engages the IRF-7 and MyD88-NF-κB signaling pathways, leading to the increased production of type I IFNs and proinflammatory cytokines (Jung and Lee, 2021). In particular, TLR3 signaling relies on the TRIF adaptor protein but not the MyD88 adaptor protein. TRIF interacts with TRAF6 and TRAF3, leading to the activation of IRF3, which results in the release of IFN-α and IFN-β (Kawasaki and Kawai, 2014; Figure 1).
Toll-Like Receptor-Mediated Antiviral and Inflammatory Responses Against Severe Acute Respiratory Syndrome Coronavirus 2: A Double-Edged Sword
It is well-known that TLR signaling plays a key role in host defense against many pathogens including SARS-CoV-2. However, TLR activation can act as a double-edged sword, and it may activate immune-mediated pathogenesis instead of inducing an immune response that defends against pathogens.
Toll-Like Receptor Signaling as an Antiviral Mechanism During Severe Acute Respiratory Syndrome Coronavirus 2 Infection
The IFN signaling cascade induced by TLR activation is crucial for controlling SARS-CoV-2 infection. As mentioned above, during SARS-CoV-2 infection, the expression of TLRs, such as TLR3 and TLR7, is elevated (Jung and Lee, 2021), and activated TLR3, TLR4 and TLR7/8 can interact with the signaling adaptor TRIF, inducing the phosphorylation and translocation of IRF3 and IRF7 and the transcription of type I and III IFN genes. Mice lacking the TLR3 gene are prone to SARS-CoV-2 infection and have increased mortality rates (Totura et al., 2015). Additionally, TLR copy numbers also play a role in the production of IFN; for example, the TLR7 and TLR8 genes are located on the X chromosome. Therefore, TLR7/8 copy numbers are higher in women than in men and their biallelic expression leads to the stronger activation of TLR7/8 and more production of type I IFN. These phenomena are correlated with higher resistance to SARS-CoV-2 infection and better prognosis in women (Viveiros et al., 2021). Moreover, loss-of-function variants of TLR7 have recently been reported to underlie a strong predisposition to severe COVID-19 in a small number of males (Fallerini et al., 2021). Both type I and III IFNs can induce an antiviral transcriptional program (Park and Iwasaki, 2020), thus, dysregulation of host IFN responses has been shown to be associated with severe disease progression in COVID-19 patients. For example, pDCs use TLR7 to sense ssRNA fragments from viruses that are rich in guanine and uracil (GU rich), and TLR7 seems to mainly act via the MyD88-TRAF3/6 pathway, inducing the production of type I IFN, IFN-γ, and IFN-λ3 and the upregulation of CD86 expression (Bortolotti et al., 2021; Salvi et al., 2021). However, the pDCs response of COVID-19-infected patients is functionally impaired (Choi and Shin, 2021).
In addition, peripheral blood immune cells from severe COVID-19 patients exhibit diminished type I and III IFN responses but enhanced proinflammatory IL-6 and TNF-α responses (Blanco-Melo et al., 2020; Hadjadj et al., 2020). Analysis of the serum from critical COVID-19 patients showed the levels of proinflammatory cytokines and chemokines are strongly elevated while the levels of type I and III IFNs are undetectable (Park and Iwasaki, 2020). Notably, SARS-CoV-2 also possesses some strategies to escape the innate immune response due to a wide range of viral proteins, such as the M, NSP6, NSP13, NSP15, ORF3b, ORF6, ORF8, and ORF9b proteins, that affect the IFN-mediated antiviral responses (Jiang et al., 2020; Konno et al., 2020; Lei et al., 2020; Han et al., 2021; Sui L. et al., 2021). Therefore, future investigations are still required to obtain a clear understanding of the inhibition of TLR signaling by SARS-CoV-2.
Toll-Like Receptor Signaling as a Part of the Inflammatory Response During Severe Acute Respiratory Syndrome Coronavirus 2 Infection
The activation of TLRs plays a dual role in the progression of COVID-19. It is thought that the production of proinflammatory cytokines is induced by the activation of different TLRs, such as TLR2, TLR4, and TLR7/8, which greatly contributes to the pathogenesis of COVID-19 and its severity. Immunopathological processes that cause death in COVID-19 patients occur due to the interaction of TLRs with virus particles (Patra et al., 2021). TLR2 and TLR4 sense viral proteins, and the expression levels of molecules related to the TLR2- and TLR4-inflammatory signaling molecules are upregulated in in COVID-19 patients, which suggest the involvement of TLR2 and TLR4 signaling in the induction of pathological inflammation during COVID-19 (Sohn et al., 2020). Moreover, several studies have shown that an overwhelming TLR7 response may promote the development of severe COVID-19 (Fallerini et al., 2021; Kayesh et al., 2021), highlighting the clinical importance of the TLR-mediated immune response during SARS-CoV-2 infection. Moreover, other studies on SARS-CoV-2 have revealed the pathological role of TLR4 and TLR7/8 in excessive inflammatory response in COVID-19 patients as it leads to the formation of neutrophil extracellular traps (NETs) and the activation of inflammasomes, leading to acute lung injury (Khadke et al., 2020; Sohn et al., 2020; Veras et al., 2020).
Additionally, obese and overweight individuals exhibit significantly increased TLR expression, which impacts the severity of COVID-19. It is hypothesized that desensitization of TLR signaling may occur due to chronic stimulation in obese and elderly people. TLR/MyD88 signaling, which is enhanced in obese individuals, may contribute to the excessive inflammatory response observed during severe infection with SARS-CoV-2 (Cuevas et al., 2021). Similarly, significantly elevated expression of IL-6 and TNF-α was observed to be associated with TLR expression in obese individuals but not in controls (Kayesh et al., 2021). Moreover, delayed IFN responses fail to control the virus and can cause inflammation and tissue damage. Delayed but considerable type I IFN responses in SARS-CoV-2 infected BALB/c mice trigger the accumulation of monocytes and macrophages as well as the production of proinflammatory cytokines, resulting in lethal pneumonia, vascular leakage, and insufficient T-cell responses (Channappanavar et al., 2016).
Furthermore, damage to host cell caused by SARS-CoV-2 infection can also lead to the release of endogenous danger-associated molecular patterns (DAMPs). These endogenous self-antigens, such as high-mobility group box 1 (HMGB1) and heat shock proteins (HSPs), activate MAPK and NF-κB signaling, which triggers an inflammatory response. In COVID-19 patients, HMGB1 can be released from dying cells and functions as a pro-inflammatory inducer to bind with TLR4, leading to the production of IL-1β, IL-6, and TNF-α (Yang et al., 2015; Cicco et al., 2020). Another DAMP shown to regulate inflammation during SARS-CoV-2 infection is the S100A8/A9 complex, which is released from host neutrophils and has been proposed to be a potential biomarker in COVID-19 patients. Mechanistically, the S100A8/A9 complex is also an endogenous ligand of TLR4 on DCs and mediates host proinflammatory responses (Mellett and Khader, 2022). TLR9 is not directly involved in recognition of SARS-CoV-2. However, cell-free DNA (cfDNA) production that is triggered by tissue injury during virus infection, cfDNA can subsequently act as a DAMP and exacerbates inflammation via TLR9. Recent studies have reported that elevated levels of cfDNA in COVID-19 patients are strongly correlated with COVID-19 disease severity (Andargie et al., 2021; Cavalier et al., 2021). Moreover, the increased cfDNA levels in COVID-19 patients generates excessive mitochondrial ROS (mtROS) production in a concentration-dependent manner (Andargie et al., 2021). Taken together, these findings suggest that host molecules that are released during SARS-CoV-2 infection may function independently as DAMPs, resulting in a more severe inflammatory response (Figure 1).
Toll-Like Receptor Agonists as Adjuvants for Coronavirus Disease 2019 Vaccines
More efficient and safe vaccines are still a critical need for combating COVID-19. In vaccine development, adjuvants are required to increase antigen recognition and enhance the magnitude and durability of the elicited immune responses. TLR agonists are considered important molecules for triggering and enhancing rapid and long-term innate immune responses, and they have been extensively studied for use as vaccine adjuvants against cancer and microbial infections. Compared to the first-generation adjuvants, such as aluminum adjuvant, TLR agonist adjuvants could guide DC maturation to elicit a stronger T-cell response. Various TLR agonists, including Pam3CSK4, poly(I:C), monophosphoryl lipid A (MPLA), resiquimod (R848), and CpG oligonucleotide (ODN), are currently under investigation for use as vaccine adjuvants to prevent SARS-CoV-2 infection. The TLR agonists currently under development for COVID-19 vaccines are listed in Table 2.
TABLE 2.
Toll-like receptor agonists as vaccine adjuvants in COVID-19 vaccine formulation.
| TLR agonists | Platform | Adjuvant | Antigen | Formulation | Immunological response |
Route | Animal model or clinical trial | References | ||
| Nab | sIgA | T-cell response | ||||||||
| TLR 1/2 | Peptide vaccine | XS15 | T-cell epitopes from viral protein | Montanide ISA 51 VG | Weak | NA | CD4+ T and IFN-γ response | SC | Phase II trial | Rammensee et al., 2021; Heitmann et al., 2022 |
| TLR 1/2 and TLR3 | Subunit vaccine | L-pampo | RBD and S1 antigens | NA | Strong | NA | IFN-γ response | IM | BALB/c and ferret | Jeong et al., 2021 |
| TLR3 | Adenovirus-based vaccine | dsRNA | SARS-CoV-2 S and N gene | Adenovirus | Strong | Moderate | NA | Oral | Hamsters and phase I trial | Tiboni et al., 2021 |
| TLR3 | Subunit vaccine | PIKA | Trimeric S antigen | NA | Strong | NA | Balanced Th1/Th2 and IFN-γ response | IM | Rabbits, mice, and non-human primates | Liu Y. et al., 2021 |
| TLR3 and TLR9 | Subunit vaccine | CpG ODN + poly I:C + IL-15 | S1 protein | PLGA or DOTAP | Weak | Strong | CD4+ T response | IM, IN | Rhesus macaques | Sui Y. et al., 2021 |
| TLR4 | Subunit vaccine | MPLA + PUUC | S1 protein | Polymer nanoparticles | Strong | Strong | Memory T-cell response | IN, IM | BALB/c | Atalis et al., 2022 |
| TLR-4 | Subunit vaccine | MPLA + GM-CSF | SARS-CoV-2 N/S1/S2 proteins | MSRs | Strong | NA | CD4+ and CD8+ T response | SC | BALB/c | Langellotto et al., 2021 |
| TLR7/8 | Subunit vaccine | Alhydroxiquim-II | Trimeric spike antigen | NA | Strong | NA | CD4+ T | IM | C57BL/6, rabbits, horses | Counoupas et al., 2022 |
| TLR7/8 | Subunit vaccine | R848 | S1 protein | Nanoparticle decorated erythrocytes | Strong | NA | CD4+ T response | IV | C57BL/6 | Wang et al., 2021 |
| TLR7/8 | Inactivated vaccine | Chemisorbed Algel | Inactivated antigen | NA | Strong | NA | CD4+ T and Th1-biased responses | IM | Mice, rats, and rabbits | Ganneru et al., 2021 |
| TLR-7 or TLR-9 | Subunit vaccine | AS37-Alum or CpG 1018-Alum | RBD antigen | Self-assembling protein nanoparticle | Strong | NA | CD4+ T response | IM | Rhesus macaques and phase II trial | Arunachalam et al., 2021; Richmond et al., 2021 |
| TLR9 | mRNA vaccine | CpG SD-101 | RBD mRNA | CART | Strong | NA | CD4+ and CD8+ T | IV, IM | BALB/c | Haabeth et al., 2021 |
| TLR-9 | Subunit vaccine | CpG 7909-Alum | Trimeric S antigen | NA | Strong | NA | CD4+ T response | IM | BALB/c and monkeys | Liu H. et al., 2021 |
NA, not available; Nab, naturalization antibody; SC, subcutaneous; IM, intramuscular; IV, intravenous; IN, intranasal; PUUC, RIG-I agonist; PLGA, poly(lactic-co-glycolic acid); DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; CART, charge-altering releasable transporters.
Toll-Like Receptor 1/2 Agonists
The TLR1/TLR2 ligand Pam3CSK4 is a synthetic triacylated lipopeptide and can activate the proinflammatory transcription factor NF-κB. A new water-soluble synthetic Pam3CSK4-derivative, named XS15, is being used in combination with peptides from the SARS-CoV-2 spike protein to develop a vaccine format that induces CD4+ T-cell responses against peptides predicted to bind to HLA-DR (Rammensee et al., 2021). In a phase I open-label trial, a peptide-based vaccine CoVac-1, which is composed of T-cell epitopes derived from various SARS-CoV-2 proteins, was combined with XS15 and emulsified in Montanide ISA 51 VG. This vaccine induced profound SARS-CoV-2-specific T-cell responses targeting multiple vaccine peptides in all the study participants. Moreover, the interferon (IFN)-γ T-cell responses induced by CoVac-1 persisted in the follow-up analyses and surpassed those detected after vaccination with approved vaccines (Heitmann et al., 2022). In addition, in a study of a SARS-CoV-2 subunit vaccine, a combination of TLR1/2 and TLR3 agonists (L-pampo) was found to be a potent adjuvant for eliciting a neutralization antibody response and an antigen-specific cellular immune response against SARS-CoV-2, resulting in a substantially decreased viral load in a ferret model (Jeong et al., 2021).
Toll-Like Receptor 4 Agonists
The TLR4 ligand LPS can regulate inflammation and effector T-cell differentiation. MPLA is a modified form of LPS that exhibits strong immune stimulatory activity but avoids most inflammatory toxicity. It has been approved for utilization as an adjuvant in vaccines against human papilloma virus and hepatitis B virus. In a previous vaccine study, MPLA-adjuvanted truncated spike protein fused with Fc of human IgG (S377-588-Fc) induced a significantly higher titer of specific IgG antibodies against Middle East respiratory syndrome coronavirus than did the alum-adjuvanted protein. In particular, MPLA-adjuvanted S377-588-Fc protein elicited a stronger Th2 (IgG1)-biased response (Zhang et al., 2016). Recently, a biomaterial COVID-19 vaccine based on mesoporous silica rods (MSRs) and loaded with MPLA, granulocyte-macrophage colony-stimulating factor (GM-CSF), and SARS-CoV-2 viral protein antigens was shown to slowly release their cargo and form subcutaneous scaffolds that recruited and activated antigen-presenting cells (APCs) at the local site to generate adaptive immune responses (Langellotto et al., 2021). The SARS-CoV-2 is still mutating; however, the MPLA-adjuvanted antigens like S-trimer/MPLA, RBD/MPLA, and S1/MPLA remain to induce a strong humoral and cellular immune responses against spike variants, including alpha, beta, gamma, delta, and omicron (Wang et al., 2022). Another TLR4 agonist, inulin acetate (InAc), which is a plant-based polymer, has been reported to induce high IgG1, IgG2a, and sIgA titers against antigens in serum after intranasal immunization using antigen-loaded InAc nanoparticles (InAc-NPs); this approach resulted in a strong memory response indicative of both humoral and cellular immune activation, and may be useful in the development of a COVID-19 vaccine (Bakkari et al., 2021).
Toll-Like Receptor 7/8 Agonists
Toll-Like Receptor 7/8 activation may be investigated as an additional strategy for anti-SARS-CoV-2 vaccines address the current challenge of viral escape. TLR7 stimulation may help viral clearance through Th1 antiviral responses as well as exert beneficial broncho-vasodilatory activity (Khalifa and Ghoneim, 2021). As a synthetic and selective ligand for TLR7, imiquimod has been approved for human-papillomavirus-induced genital and perianal warts. An imiquimod analog, resiquimod (R848), was proven to be a dual TLR7 and TLR8 synthetic agonist that elicits prominent IFN-α/β and IL-6 responses and robust cytotoxic T-cell (CTL) and B-cell proliferation; thus, it is particularly suited for antiviral immune responses. Indeed, the cytokine profiles induced by R848, are almost identical to the profiles induced by the licensed mRNA vaccines against COVID-19 (Rossmann et al., 2021). In a SARS-CoV-2 virus-mimetic nanoparticle vaccine, the SARS-CoV-2 spike protein S1 subunit and R848 were attached to erythrocytes and injected into mice, resulting in greater maturation and activation of APCs, production of specific IgG antibodies, and systemic antiviral T-cell responses than the nanoparticles alone (Wang et al., 2021). However, R848 adjuvanticity should stress more on vaccine formulation. A recent study showed that R848 conjugated to multilamellar liposomes rather than forming a linear structure, resulting in stronger immunostimulatory activity (Liang et al., 2020). Moreover, an R848-encapsulating poly lactic-co-glycolic acid (PLGA) nanoparticle can reduce the excessive level of inflammatory cytokines induced by free R848 (Chen et al., 2021), which could be beneficial for providing an appropriate immune response and long-term safety in vaccine development.
Toll-Like Receptor 3/9 Agonists
Optimal protection against coronavirus probably involves neutralizing antibodies and CD8+ T cells. Among the TLR agonists, the TLR3 and TLR9 ligands, poly(I:C) and CpG ODN significantly augment the CD8+ T-cell responses to a greater extent than other adjuvants. Thus, CpG ODN and poly(I:C) have been utilized as adjuvants in influenza vaccines. Studies have demonstrated that CpG ODN or PIKA [a stabilized derivative of poly(I:C)] can stimulate enhanced IgG production in animals have been used in the context of immunization with an inactivated SARS-CoV-2 vaccine (Gai et al., 2008; Gupta and Gupta, 2020). Moreover, compared with the alum adjuvanted SARS-CoV-2 S1 vaccine, intranasal boosting with nanoparticles, formulating with CpG 1018 and poly(I:C), elicits higher dimeric IgA production, IFN-α production, and T-cell activation in rhesus macaques (Sui Y. et al., 2021). In another study, a recombinant S trimeric protein adjuvanted with PIKA was reported to induce high titers of SARS-CoV-2 neutralizing antibodies and to protect non-human primates from virus challenge (Liu Y. et al., 2021). Another CpG ODN 7909-adjuvanted SARS-CoV-2 vaccine, 202-COV, is a S-protein subunit vaccine formulated with aluminum hydroxide; that elicits robust neutralizing antibody responses and substantial CD4+ T-cell responses in both mice and non-human primates; this vaccine is currently being investigated in phase II studies of COVID-19 (Liu H. et al., 2021). CpG ODN is capable of inducing both cellular and humoral immune responses, and it preferentially induces Th1-biased responses. Mice that were immunized with mRNA encoding the spike protein of SARS-CoV-2, co-formulated with CpG ODN, developed therapeutically relevant levels of RBD-specific neutralizing antibodies in both circulation and lung bronchial fluids. In addition, vaccination elicited strong and long-lasting RBD-specific Th1 T-cell responses including CD4+ and CD8+ T-cell memory responses (Haabeth et al., 2021).
Toll-Like Receptor Signaling Inhibitors Protect Against Hyperinflammatory Response in Severe Acute Respiratory Syndrome Coronavirus 2 Infection
Uncontrolled TLR-mediated inflammation has been suggested to contribute to immunopathological consequences in COVID-19 patients. It is quite obvious that targeted manipulation of TLR signaling decreases excessive inflammatory responses. TLR4 is one of the major contributors to SARS-CoV-2 infectivity and pathogenesis, and its antagonists are capable of inhibiting the harmful effects of TLR4 signaling. For example, Lipid X is a Lipid A biosynthetic precursor that can competitively bind to TLR4 to block cytokine production via the downstream signaling pathways. Therefore, many TLR4 modulators, both natural and synthetic, can be investigated in the context of COVID-19 treatment. TAK242 is a potent and selective TLR4 antagonist that inhibits the release of inflammatory cytokines from human THP-1 cells after exposure to SARS-CoV-2. Therefore, it is postulated that TAK242 may improve patient outcomes by dampening the inflammatory response and preventing systemic infection in patients with COVID-19 (Kate Gadanec et al., 2021).
The intracellular RNA sensors TLR3 and TLR7/8 are thought to be involved in the hyperinflammatory responses induced by SARS-CoV-2 infection. A TLR7/8 antagonist, Enpatoran (M5049), is a potent dual TLR7/8 inhibitor that is expected to cease the hyperinflammatory milieu in symptomatic patients with COVID-19. Furthermore, Merck has already initiated a phase II randomized, controlled clinical study evaluating the efficacy and safety of M5049 in the COVID-19 patient population (Khalifa and Ghoneim, 2021; Port et al., 2021). Similarly, famotidine, a specific histamine H2 receptor antagonist, can inhibit TLR3 expression in SARS-CoV-2 infected cells and reduce TLR3-dependent NF-κB and IRF3 signaling, subsequently controlling antiviral and inflammatory responses (Mukherjee et al., 2021) and reducing the risk of intubation and death in hospitalized patients with COVID-19 (Freedberg et al., 2020). Another TLR signaling inhibitor, the PPARα agonist oleoylethanolamide (OEA), was reported to attenuate TLR3-induced hyperthermia and reduce the expression of hyperthermia-related genes including IL-1β, iNOS, COX2, and m-PGES in the hypothalamus (Flannery et al., 2021).
Toll-like receptor antagonists are not only used as individual drugs but may also be used in combination with immunomodulatory drugs in severe cases of COVID-19 to enhance potential synergistic effects and possibly reduce adverse effects. Currently, most TLR antagonists are being investigated in clinical trials to evaluate their efficacy in reducing detrimental immune effects without causing a drastic change in their basal levels to maintain the immune homeostasis.
Conclusion
Toll-like receptors are important constituents of the innate immune system and can recognize a wide variety of PAMPs from viruses. This review has described the various TLRs that are involved in the immunopathogenesis of SARS-CoV-2 infection and the effects of application of TLR immunomodulators in patients with COVID-19. Of the TLRs that have been identified, TLR7/TLR8 and TLR3 are intracellular receptor that sense viral ssRNA, and dsRNA replication intermediates, respectively. TLR2 and TLR4 reside on the cell surface and are activated by SARS-CoV-2 glycoproteins. TLR signaling elicits antiviral and proinflammatory cytokine production through MyD88-dependent and/or TRIF-dependent pathways. The activation of the innate immune response often contributes to viral clearance and disease resolution. Therefore, TLR agonists could be formulated as adjuvants for use with the S protein or RBD to enhance neutralizing antibody production and T-cell responses against SARS-CoV-2 infection since they elicit timely and optimal TLR responses. However, dysregulated immune signaling may lead to the detrimental production of proinflammatory cytokines and chemokines that cause severe disease. Thus, the use of TLR antagonists might exert a beneficial effect, by attenuating deleterious hyperinflammatory responses in severe COVID-19 patients. However, most of these new therapeutics or combination strategies for anti-SARS-CoV-2 infection are currently being studied in clinical trials of various phases.
Author Contributions
JD, YBW, and HRW contributed to the writing of the manuscript. MY and YS provided the ideas and revised the draft manuscript. ZG, YW, MF, SS, PZ, and HW approved the version to be published. MF provided a professional English language revision. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81902111) and the Department of Finance of Jilin Province (grant number JLSWSRCZX2020-00107).
References
- Aboudounya M. M., Heads R. J. (2021). COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflamm. 2021:8874339. 10.1155/2021/8874339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andargie T. E., Tsuji N., Seifuddin F., Jang M. K., Yuen P. S., Kong H., et al. (2021). Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight 6:e147610. 10.1172/jci.insight.147610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P. S., Walls A. C., Golden N., Atyeo C., Fischinger S., Li C., et al. (2021). Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594 253–258. 10.1038/s41586-021-03530-2 [DOI] [PubMed] [Google Scholar]
- Atalis A., Keenum M. C., Pandey B., Beach A., Pradhan P., Vantucci C., et al. (2022). Nanoparticle-delivered TLR4 and RIG-I agonists enhance immune response to SARS-CoV-2 subunit vaccine. bioRxiv[Preprint]. 10.1101/2022.01.31.478507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkari M. A., Valiveti C. K., Kaushik R. S., Tummala H. (2021). Toll-like Receptor-4 (TLR4) Agonist-Based Intranasal Nanovaccine Delivery System for Inducing Systemic and Mucosal Immunity. Mol. Pharm. 18 2233–2241. 10.1021/acs.molpharmaceut.0c01256 [DOI] [PubMed] [Google Scholar]
- Birra D., Benucci M., Landolfi L., Merchionda A., Loi G., Amato P., et al. (2020). COVID 19: a clue from innate immunity. Immunol. Res. 68 161–168. 10.1007/s12026-020-09137-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B. E., Liu W. C., Uhl S., Hoagland D., Moller R. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181 1036–1045.e9. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti D., Gentili V., Rizzo S., Schiuma G., Beltrami S., Strazzabosco G., et al. (2021). TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 9:1820. 10.3390/microorganisms9091820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier E., Guiot J., Lechner K., Dutsch A., Eccleston M., Herzog M., et al. (2021). Circulating Nucleosomes as Potential Markers to Monitor COVID-19 Disease Progression. Front. Mol. Biosci. 8:600881. 10.3389/fmolb.2021.600881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A. R., Vijay R., Mack M., Zhao J., Meyerholz D. K., et al. (2016). Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 19 181–193. 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Huang Q., Zhao T., Sui L., Wang S., Xiao Z., et al. (2021). Nanotherapies for sepsis by regulating inflammatory signals and reactive oxygen and nitrogen species: new insight for treating COVID-19. Redox Biol. 45:102046. 10.1016/j.redox.2021.102046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Shin E. C. (2021). Roles of Type I and III Interferons in COVID-19. Yonsei Med. J. 62 381–390. 10.3349/ymj.2021.62.5.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. (2020). In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 92 2105–2113. 10.1002/jmv.25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicco S., Cicco G., Racanelli V., Vacca A. (2020). Neutrophil Extracellular Traps (NETs) and Damage-Associated Molecular Patterns (DAMPs): two Potential Targets for COVID-19 Treatment. Mediators Inflamm. 2020:7527953. 10.1155/2020/7527953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counoupas C., Pino P., Stella A. O., Ashley C., Lukeman H., Bhattacharyya N. D., et al. (2022). High-Titer Neutralizing Antibodies against the SARS-CoV-2 Delta Variant Induced by Alhydroxyquim-II-Adjuvanted Trimeric Spike Antigens. Microbiol. Spectr. 10:e0169521. 10.1128/spectrum.01695-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas A. M., Clark J. M., Potter J. J. (2021). Increased TLR/MyD88 signaling in patients with obesity: is there a link to COVID-19 disease severity? Int. J. Obes. 45 1152–1154. 10.1038/s41366-021-00768-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath M., Banerjee M., Berk M. (2020). Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 34 8787–8795. 10.1096/fj.202001115R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebermeyer T., Cognasse F., Berthelot P., Mismetti P., Garraud O., Hamzeh-Cognasse H. (2021). Platelet Innate Immune Receptors and TLRs: a Double-Edged Sword. Int. J. Mol. Sci. 22:7894. 10.3390/ijms22157894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., et al. (2021). Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. eLife 10:e67569. 10.7554/eLife.67569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery L. E., Kerr D. M., Hughes E. M., Kelly C., Costello J., Thornton A. M., et al. (2021). N-acylethanolamine regulation of TLR3-induced hyperthermia and neuroinflammatory gene expression: a role for PPARalpha. J. Neuroimmunol. 358:577654. 10.1016/j.jneuroim.2021.577654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M. G., Nguyen K. H., Ball J. B., Hopkins S., Kelley T., Baratta M. V., et al. (2022). SARS-CoV-2 spike S1 subunit induces neuroinflammatory, microglial and behavioral sickness responses: evidence of PAMP-like properties. Brain Behav. Immun. 100 267–277. 10.1016/j.bbi.2021.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg D. E., Conigliaro J., Wang T. C., Tracey K. J., Callahan M. V., Abrams J. A., et al. (2020). Famotidine Use Is Associated With Improved Clinical Outcomes in Hospitalized COVID-19 Patients: a Propensity Score Matched Retrospective Cohort Study. Gastroenterology 159 1129–1131.e3. 10.1053/j.gastro.2020.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai W., Zou W., Lei L., Luo J., Tu H., Zhang Y., et al. (2008). Effects of different immunization protocols and adjuvant on antibody responses to inactivated SARS-CoV vaccine. Viral Immunol. 21 27–37. 10.1089/vim.2007.0079 [DOI] [PubMed] [Google Scholar]
- Ganneru B., Jogdand H., Daram V. K., Das D., Molugu N. R., Prasad S. D., et al. (2021). Th1 skewed immune response of whole virion inactivated SARS CoV 2 vaccine and its safety evaluation. iScience 24:102298. 10.1016/j.isci.2021.102298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta T., Gupta S. K. (2020). Potential adjuvants for the development of a SARS-CoV-2 vaccine based on experimental results from similar coronaviruses. Int. Immunopharmacol. 86:106717. 10.1016/j.intimp.2020.106717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haabeth O. A. W., Lohmeyer J. J. K., Sallets A., Blake T. R., Sagiv-Barfi I., Czerwinski D. K., et al. (2021). An mRNA SARS-CoV-2 Vaccine Employing Charge-Altering Releasable Transporters with a TLR-9 Agonist Induces Neutralizing Antibodies and T Cell Memory. ACS Cent. Sci. 7 1191–1204. 10.1021/acscentsci.1c00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. (2020). Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369 718–724. 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zhuang M. W., Deng J., Zheng Y., Zhang J., Nan M. L., et al. (2021). SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med. Virol. 93 5376–5389. 10.1002/jmv.27050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayat M., Netea M. G., Rezaei N. (2011). Targeting of Toll-like receptors: a decade of progress in combating infectious diseases. Lancet Infect. Dis. 11 702–712. 10.1016/s1473-3099(11)70099-8 [DOI] [PubMed] [Google Scholar]
- Heitmann J. S., Bilich T., Tandler C., Nelde A., Maringer Y., Marconato M., et al. (2022). A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 601 617–622. 10.1038/s41586-021-04232-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S. K., Heo Y. K., Jeong J. H., Ham S. J., Yum J. S., Ahn B. C., et al. (2021). COVID-19 Subunit Vaccine with a Combination of TLR1/2 and TLR3 Agonists Induces Robust and Protective Immunity. Vaccines 9:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. W., Zhang H. N., Meng Q. F., Xie J., Li Y., Chen H., et al. (2020). SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 17 998–1000. 10.1038/s41423-020-0514-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H. E., Lee H. K. (2021). Current Understanding of the Innate Control of Toll-like Receptors in Response to SARS-CoV-2 Infection. Viruses 13:2132. 10.3390/v13112132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kate Gadanec L., Qaradakhi T., McSweeney K., Ashiana Ali B., Zulli A., Apostolopoulos V. (2021). Dual targeting of Toll-like receptor 4 and angiotensin-converting enzyme 2: a proposed approach to SARS-CoV-2 treatment. Future Microbiol. 16 205–209. 10.2217/fmb-2021-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34 637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Kawai T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5:461. 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayesh M. E. H., Kohara M., Tsukiyama-Kohara K. (2021). An Overview of Recent Insights into the Response of TLR to SARS-CoV-2 Infection and the Potential of TLR Agonists as SARS-CoV-2 Vaccine Adjuvants. Viruses 13:2302. 10.3390/v13112302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadke S., Ahmed N., Ahmed N., Ratts R., Raju S., Gallogly M., et al. (2020). Harnessing the immune system to overcome cytokine storm and reduce viral load in COVID-19: a review of the phases of illness and therapeutic agents. Virol. J. 17:154. 10.1186/s12985-020-01415-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa A. E., Ghoneim A. I. (2021). Potential value of pharmacological agents acting on toll-like receptor (TLR) 7 and/or TLR8 in COVID-19. Curr. Res. Pharmacol. Drug Discov. 2:100068. 10.1016/j.crphar.2021.100068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Shafiei M. S., Longoria C., Schoggins J., Savani R. C., Zaki H. (2021). SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. bioRxiv [Preprint]. 10.1101/2021.03.16.435700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Siddique R., Shereen M. A., Ali A., Liu J., Bai Q., et al. (2020). Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: biology and Therapeutic Options. J. Clin. Microbiol. 58:e00187–20. 10.1128/jcm.00187-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. Y., Shim H. J., Kim S. Y., Heo S., Youn H. S. (2018). Differential regulation of MyD88- and TRIF-dependent signaling pathways of Toll-like receptors by cardamonin. Int. Immunopharmacol. 64 1–9. 10.1016/j.intimp.2018.08.018 [DOI] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., et al. (2020). SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 32:108185. 10.1016/j.celrep.2020.108185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langellotto F., Dellacherie M. O., Yeager C., Ijaz H., Yu J., Cheng C. A., et al. (2021). A Modular Biomaterial Scaffold-Based Vaccine Elicits Durable Adaptive Immunity to Subunit SARS-CoV-2 Antigens. Adv. Healthc. Mater. 10:e210137. 10.1002/adhm.202101370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. (2020). Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11:3810. 10.1038/s41467-020-17665-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester S. N., Li K. (2014). Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 426 1246–1264. 10.1016/j.jmb.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Zhu H., Wang X., Jing B., Li Z., Xia X., et al. (2020). Adjuvants for Coronavirus Vaccines. Front. Immunol. 11:589833. 10.3389/fimmu.2020.589833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhou C., An J., Song Y., Yu P., Li J., et al. (2021). Development of recombinant COVID-19 vaccine based on CHO-produced, prefusion spike trimer and alum/CpG adjuvants. Vaccine 39 7001–7011. 10.1016/j.vaccine.2021.10.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Dai L., Feng X., Gao R., Zhang N., Wang B., et al. (2021). Fast and long-lasting immune response to S-trimer COVID-19 vaccine adjuvanted by PIKA. Mol. Biomed. 2:29. 10.1186/s43556-021-00054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manik M., Singh R. K. (2021). Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 94 869–877. 10.1002/jmv.27405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellett L., Khader S. A. (2022). S100A8/A9 in COVID-19 pathogenesis: impact on clinical outcomes. Cytokine Growth Factor Rev. 63 90–97. 10.1016/j.cytogfr.2021.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty M. C., Varose S. Y., Sawant U. P., Fernandes M. M. (2021). Expression of innate immune response genes in upper airway samples of SARS-CoV-2 infected patients: a preliminary study. Indian J. Med. Res. 153 677–683. 10.4103/ijmr.IJMR_131_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R., Bhattacharya A., Bojkova D., Mehdipour A. R., Shin D., Khan K. S., et al. (2021). Famotidine inhibits toll-like receptor 3-mediated inflammatory signaling in SARS-CoV-2 infection. J. Biol. Chem. 297:100925. 10.1016/j.jbc.2021.100925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U., Ishida H., Shibata T., Sato R., Miyake K., Shimizu T. (2018). Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation. Immunity 48 649–658.e4. 10.1016/j.immuni.2018.03.013 [DOI] [PubMed] [Google Scholar]
- Oldenburg M., Krüger A., Ferstl R., Kaufmann A., Nees G., Sigmund A., et al. (2012). TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337 1111–1115. 10.1126/science.1220363 [DOI] [PubMed] [Google Scholar]
- Park A., Iwasaki A. (2020). Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 27 870–878. 10.1016/j.chom.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patidar A., Selvaraj S., Sarode A., Chauhan P., Chattopadhyay D., Saha B. (2018). DAMP-TLR-cytokine axis dictates the fate of tumor. Cytokine 104 114–123. 10.1016/j.cyto.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Patra R., Chandra Das N., Mukherjee S. (2021). Targeting human TLRs to combat COVID-19: a solution? J. Med. Virol. 93 615–617. 10.1002/jmv.26387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port A., Shaw J. V., Klopp-Schulze L., Bytyqi A., Vetter C., Hussey E., et al. (2021). Phase 1 study in healthy participants of the safety, pharmacokinetics, and pharmacodynamics of enpatoran (M5049), a dual antagonist of toll-like receptors 7 and 8. Pharmacol. Res. Perspect. 9:e00842. 10.1002/prp2.842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Lei T., Patel P. S., Lee C. H., Monaghan-Nichols P., Xin H. B., et al. (2021). Direct Activation of Endothelial Cells by SARS-CoV-2 Nucleocapsid Protein Is Blocked by Simvastatin. J. Virol. 95:e0139621. 10.1128/JVI.01396-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H. G., Gouttefangeas C., Heidu S., Klein R., Preuss B., Walz J. S., et al. (2021). Designing a SARS-CoV-2 T-Cell-Inducing Vaccine for High-Risk Patient Groups. Vaccines 9:428. 10.3390/vaccines9050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond P., Hatchuel L., Dong M., Ma B., Hu B., Smolenov I., et al. (2021). Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet 397 682–694. 10.1016/s0140-6736(21)00241-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann L., Bagola K., Stephen T., Gerards A. L., Walber B., Ullrich A., et al. (2021). Distinct single-component adjuvants steer human DC-mediated T-cell polarization via Toll-like receptor signaling toward a potent antiviral immune response. Proc. Natl. Acad. Sci. U.S..A. 118:e2103651118. 10.1073/pnas.2103651118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi V., Nguyen H. O., Sozio F., Schioppa T., Gaudenzi C., Laffranchi M., et al. (2021). SARS-CoV-2-associated ssRNAs activate inflammation and immunity via TLR7/8. JCI Insight 6:e150542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. (2021). SARS-CoV-2 takes its Toll. Nat. Immunol. 22 801–802. 10.1038/s41590-021-00962-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Hartmann G. (2016). Discriminating self from non-self in nucleic acid sensing. Nat Rev. Immunol. 16 566–580. 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kizaki T. (2021). SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 7:e06187. 10.1016/j.heliyon.2021.e06187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K. M., Lee S. G., Kim H. J., Cheon S., Jeong H., Lee J., et al. (2020). COVID-19 Patients Upregulate Toll-like Receptor 4-mediated Inflammatory Signaling That Mimics Bacterial Sepsis. J. Korean Med. Sci. 35:e343. 10.3346/jkms.2020.35.e343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Wang Y., Wang J., Mifune Y., Morin M. D., Jones B. T., et al. (2019). Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim. J. Med. Chem. 62 2938–2949. 10.1021/acs.jmedchem.8b01583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L., Zhao Y., Wang W., Wu P., Wang Z., Yu Y., et al. (2021). SARS-CoV-2 Membrane Protein Inhibits Type I Interferon Production Through Ubiquitin-Mediated Degradation of TBK1. Front. Immunol. 12:662989. 10.3389/fimmu.2021.662989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y., Li J., Zhang R., Prabhu S. K., Andersen H., Venzon D., et al. (2021). Protection against SARS-CoV-2 infection by a mucosal vaccine in rhesus macaques. JCI Insight 6:e148494. 10.1172/jci.insight.148494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabary M., Khanmohammadi S., Araghi F., Dadkhahfar S., Tavangar S. M. (2020). Pathologic features of COVID-19: a concise review. Pathol. Res. Pract. 216:153097. 10.1016/j.prp.2020.153097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. (2020). Cytokine Storm in COVID-19: the Current Evidence and Treatment Strategies. Front. Immunol. 11:1708. 10.3389/fimmu.2020.01708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji H., Ohto U., Shibata T., Miyake K., Shimizu T. (2013). Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science 339 1426–1429. 10.1126/science.1229159 [DOI] [PubMed] [Google Scholar]
- Tiboni M., Casettari L., Illum L. (2021). Nasal vaccination against SARS-CoV-2: synergistic or alternative to intramuscular vaccines? Int. J. Pharm. 603:120686. 10.1016/j.ijpharm.2021.120686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A. L., Whitmore A., Agnihothram S., Schafer A., Katze M. G., Heise M. T., et al. (2015). Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 6:e00638-15. 10.1128/mBio.00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton H. A., Thompson A. A. R., Farkas L. (2020). RNA Signaling in Pulmonary Arterial Hypertension-A Double-Stranded Sword. Int. J. Mol. Sci. 21:3124. 10.3390/ijms21093124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaure C., Liu Y. (2014). A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5:316. 10.3389/fimmu.2014.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F. P., Pontelli M. C., Silva C. M., Toller-Kawahisa J. E., de Lima M., Nascimento D. C., et al. (2020). SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 217:e20201129. 10.1084/jem.20201129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros A., Rasmuson J., Vu J., Mulvagh S. L., Yip C. Y. Y., Norris C. M., et al. (2021). Sex differences in COVID-19: candidate pathways, genetics of ACE2, and sex hormones. Am. J. Physiol. Heart Circ. Physiol. 320 H296–H304. 10.1152/ajpheart.00755.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chai J., Wang H. (2016). Structure of the mouse Toll-like receptor 13 ectodomain in complex with a conserved sequence from bacterial 23S ribosomal RNA. FEBS J. 283 1631–1635. 10.1111/febs.13628 [DOI] [PubMed] [Google Scholar]
- Wang J., Yin X. G., Wen Y., Lu J., Zhang R. Y., Zhou S. H., et al. (2022). MPLA-Adjuvanted Liposomes Encapsulating S-Trimer or RBD or S1, but Not S-ECD, Elicit Robust Neutralization Against SARS-CoV-2 and Variants of Concern. J. Med. Chem. 65 3563–3574. 10.1021/acs.jmedchem.1c02025 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang X., Yang F., Liu Y., Meng L., Pang Y., et al. (2021). Systemic antiviral immunization by virus-mimicking nanoparticles-decorated erythrocytes. Nano Today 40:101280. 10.1016/j.nantod.2021.101280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. A., Huang Y. L., Chiang B. L. (2022). Innate immune response analysis in COVID-19 and kawasaki disease reveals MIS-C predictors. J. Formos Med. Assoc. 121 623–632. 10.1016/j.jfma.2021.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Ju Z., Ragab A. A., Lundback P., Long W., et al. (2015). MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J. Exp. Med. 212 5–14. 10.1084/jem.20141318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan H. (2017). TLR5: beyond the recognition of flagellin. Cell Mol. Immunol. 14 1017–1019. 10.1038/cmi.2017.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Okabayashi T., Fujii N. (2010). The battle between virus and host: modulation of Toll-like receptor signaling pathways by virus infection. Mediators Inflamm. 2010:184328. 10.1155/2010/184328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Feng Z. (2018). The Role of Toll-Like Receptor Signaling in the Progression of Heart Failure. Mediators Inflamm. 2018:9874109. 10.1155/2018/9874109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Channappanavar R., Ma C., Wang L., Tang J., Garron T., et al. (2016). Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell. Mol. Immunol. 13 180–190. 10.1038/cmi.2015.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xiang R., Huo S., Zhou Y., Jiang S., Wang Q., et al. (2021). Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 6:233. 10.1038/s41392-021-00653-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., et al. (2021). SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 31 818–820. 10.1038/s41422-021-00495-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Karki R., Williams E. P., Yang D., Fitzpatrick E., Vogel P., et al. (2021). TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 22 829–838. 10.1038/s41590-021-00937-x [DOI] [PMC free article] [PubMed] [Google Scholar]