Abstract
Background
One of the main challenges in personalized medicine is to establish and apply a large number of variants from genomic databases into clinical diagnostics and further facilitate genome-driven drug repurposing. By utilizing biological chronic hepatitis B infection (CHB) risk genes, our study proposed a systematic approach to use genomic variants to drive drug repurposing for CHB.
Method
The genomic variants were retrieved from the Genome-Wide Association Study (GWAS) and Phenome-Wide Association Study (PheWAS) databases. Then, the biological CHB risk genes crucial for CHB progression were prioritized based on the scoring system devised with five strict functional annotation criteria. A score of ≥ 2 were categorized as the biological CHB risk genes and further shed light on drug target genes for CHB treatments. Overlapping druggable targets were identified using two drug databases (DrugBank and Drug-Gene Interaction Database (DGIdb)).
Results
A total of 44 biological CHB risk genes were screened based on the scoring system from five functional annotation criteria. Interestingly, we found 6 druggable targets that overlapped with 18 drugs with status of undergoing clinical trials for CHB, and 9 druggable targets that overlapped with 20 drugs undergoing preclinical investigations for CHB. Eight druggable targets were identified, overlapping with 25 drugs that can potentially be repurposed for CHB. Notably, CD40 and HLA-DPB1 were identified as promising targets for CHB drug repurposing based on the target scores.
Conclusion
Through the integration of genomic variants and a bioinformatic approach, our findings suggested the plausibility of CHB genomic variant-driven drug repurposing for CHB.
Keywords: Bioinformatics, Chronic hepatitis B, Drug repurposing, Genomic variants
Highlights
-
•
The feasibility of utilizing genomic variants can provide novel biological insight and facilitate genomic-driven drug repurposing for chronic hepatitis B.
-
•
The use of genomic information can be translated into clinical implementation through genomic-based therapies and guidance for drug discovery.
-
•
Our findings suggested the plausibility of chronic hepatitis B genomic-driven drug repurposing for chronic hepatitis B.
1. Introduction
Chronic Hepatitis B (CHB) infection has been the most prevalent viral infectious disease in the world. Notably, the hepatitis B virus is approximately five times more infectious than the hepatitis C virus, and far more stable than the hepatitis C virus [1]. Although the vaccination program against human hepatitis B virus (HBV) has been widely implemented, HBV infection remains a serious problem in public health burden across the world. As reported by the World Health Organization (WHO) in 2018, around 257 million new cases as HBV carriers and roughly 887,000 deaths due to HBV infection [2]. HBV infection that is not monitored and not treated properly can lead to the progression of acute and chronic hepatitis, as well as the development of liver cirrhosis and hepatocellular carcinoma (HCC) [3].
Currently, two groups of antiviral drugs are therapies that are available for CHB. These two classes of drugs (alpha interferon (IFN-α) and nucleos(t)ide analogues) have been approved by the Food and Drug Administration (FDA) to be used for CHB therapies. Unfortunately, the drug resistance of currently available CHB drugs, resulting in poor treatment effects that cannot be ignored. Besides, current therapies challenges are not totally eradicated, with the viral covalently closed circular DNA (cccDNA) as a source of CHB antigens [4]. Despite substantial research, there are presently no specific medications can halt CHB progression.
Developing new drugs requires an estimated 15 years of time and more than $1 billion in funds to bring the new drug to market [5]. Moreover, the U.S FDA ultimately approves fewer than 5% of new molecules entering phase I clinical trials [6]. Most drugs failed in Phase II clinical trials, and at least 50% of these were due to a lack of efficacy, while 25% were due to toxicity [7]. Because of the lack of new clinical drugs, an emerging new approach to identify new drug targets is drug repurposing, also known as drug repositioning. This concept is an alternative approach for finding novel indications for existing drugs or drugs in clinical trials [8]. The favourable outlook for drug repositioning is based on drug makers' efforts to meet demand, particularly for illnesses where the choice of drug is limited or otherwise costly to treat [9]. Drug repositioning is critical in drug development since it reduces overall development costs. Furthermore, drug repositioning reduces development risk because the repositioned pharmaceuticals' safety has previously been shown in humans and other preclinical animals. Aside from that, because the safety evaluation and formulation development methods are already in place, this strategy has a substantially shorter development timetable [8]. Repurposing sildenafil citrate for erectile dysfunction is the most successful example of drug repurposing [10]. When Pfizer repurposed Sildenafil for the treatment of erectile dysfunction under the name Viagra, it became the market leader in that category with a market share of 47% in 2012, with global sales of $2.05 billion [11]. Therefore, in such circumstances, drug repurposing is an alternative and promising strategy to accelerate the discovery of a new drug for CHB.
Genomic information has a good prospect to facilitate drug repurposing by integrating the genomic variants and bioinformatic approach. Genome-Wide Association Study (GWAS) and Phenome-Wide Association Study (PheWAS) databases are two common databases providing genomic variants for various diseases. Several genetic loci have been identified as hereditary risks for sporadic CHB using GWAS [12] and PheWAS [13]. GWAS and PheWAS data have provided essential biological insights into the molecular underpinnings of CHB. However, clinical translation of genomic findings from GWAS and PheWAS has remained limited. These two databases were promising to be used for genomic-driven drug repurposing, including CHB therapies. In this study, we used GWAS and PheWAS to find CHB-associated single nucleotide polymorphisms (SNPs) and identified biological CHB risk genes using a functional annotation-based scoring system. The candidates of drug repurposing were then demonstrated by overlapping the drug target genes and the drug candidates for CHB using a drug database.
2. Materials and methods
2.1. Study design for genomic variants retrieved from GWAS and PheWAS catalogues
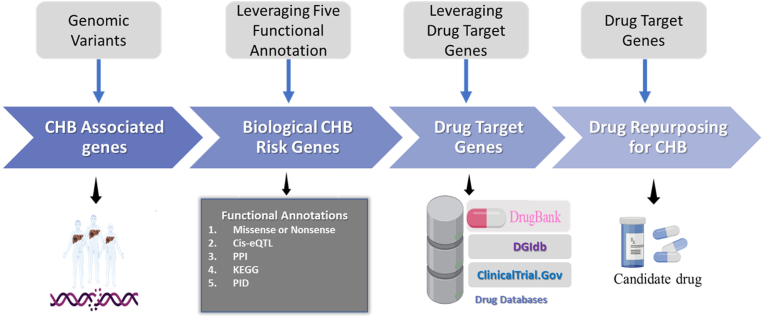
This study utilizes the genomic variants as the source of SNPs. SNPs is one area of the functional genomic variants that are well studied. Our study utilized the use of CHB genomic variants from GWAS (https://www.ebi.ac.uk/gwas) [14] and the PheWAS database (https://phewascatalog.org/) [15]. These two databases provided genomic variants of risk genes for several diseases, including for SNPs associated with CHB progression. The accession date of the two databases is February 25, 2022. The detailed information related to the process of genomic variants acquired from the databases was depicted in Fig. 1.
Fig. 1.
The use of genomic variants to drive drug repurposing for chronic hepatitis B (CHB).
The SNPs which met the criteria were prioritized for further analysis. Herein, we prioritize the SNPs associated with the CHB based on the threshold GWAS p-value <5 × 10−8 and PheWAS algorithm p-value < 0.01. GWAS and PheWAS are two popular genomic databases that provide information on common genomic variants and particular diseases. Through this database also we can understand more detail how the genomes contribute to the disease or the biology of disease. Further, we utilized the HaploReg version 4.1 to expand the SNPs profiled and encoded the associated genes (CHB-associated genes). We set the criterion of r2 > 0.8 in the Asian population to expand the variants profiled [16]. In order to prioritize these genes, we used five functional annotation criteria. Genes were classified as “biological CHB risk genes” if they met two out of three criteria (score ≥ 2). Our next step was to map biological CHB risk genes according to the DrugBank and Drug-Gene Interaction Database (DGIdb) version 4.0 to identify potential drugs to be repurposed for CHB [17,18]. To further identify the clinical status of drugs, we used ClinicalTrials.gov (https://clinicaltrials.gov/) accessed on January 30, 2022. Furthermore, we used the PubMed database to check whether the candidate drugs that we identified have been proven in preclinical investigations.
2.2. Biological CHB risk genes
The biological CHB risk genes were derived from functional annotation categories that met two or more of the criteria (i.e., had a score of ≥ 2). The current study used functional annotations and scoring criteria adopted from Okada et al. (2015) which were predicted candidate drugs for rheumatoid arthritis [19]. The following five criteria were used to evaluate each gene in the CHB risk loci. The first annotation is missense mutation will be considered in this step. We first mapped the variants onto the corresponding genes with missense/non-sense mutations as one of the non-synonymous changes in the single base substitution of a different amino acid in the resulting protein. The SNPs encoded the genes annotated as missense mutations which were retrieved from HaploReg version 4.1 [20]. For the second annotation criterion, we considered cis-expression quantitative trait locus (cis-eQTL) to prioritize the CHB associated genes. We utilized this annotation with the knowledge that functional rules of variants affect protein expression. The variants may cause changes in gene expression in the direction of the tissues involved (i.e., our analyses focused on the whole blood and heart). If the identified variants cause an upregulation of gene X, leading to an increased risk of a disease, then an inhibitor of its protein product may be considered a repositioning candidate. The CHB associated genes with significant effects on whole blood and heart tissues. For this step, we leveraged the HaploReg version 4.1 integrated with the GTEx portal database to links between the CHB genomic variants and cis-eQTL [21]. We hypothesized that CHB genetic variants prioritization using five functional annotation criteria will enable us to translate the risk genes to meaningful insights on CHB pathogenesis.
A third functional annotation criterion was based on protein-protein interactions (PPIs): genes had been prioritized by biological process gene. If the genes involved in the biological protein networks are related in CHB pathogenesis, then it is hypothesized to be important to inhibit the protein. An FDR of q <0.05 was considered significant [22]. For the fourth functional annotation criterion, we used molecular pathway criteria. In this criterion, the genes were prioritized based on the data in the Kyoto Encyclopaedia of Genes and Genomes (KEGG). Gene scores are determined by the number of matched criteria for each gene. The five annotation as the last annotation criterion is the primary immuno-deficiency (PID) diseases, which are innate immune diseases reported to be associated with CHB. Notably, genes overlapping with the PID can play a causal role in CHB pathogenesis [23,24].
2.3. Drug target genes overlapped using drug databases
To identify candidate drugs for CHB, we overlapped the biological CHB risk genes to DrugBank and DGIdb (data released on February 27, 2022). DrugBank (www.drugbank.ca) and DGIdb ( www.dgidb.org) are two databases that not only provide bioinformatics and chemoinformatics information about drugs but also offer a large collection of druggable targets from publications, databases, and other sources online [17,18].
Several criteria were used to investigate the candidates for CHB, including drugs with pharmacological activity, human effectiveness, approved annotations, clinical trials, or experimental drugs. Additionally, each drug was checked through ClinicalTrials.gov (https://clinicaltrials.gov/; accessed on February 29, 2022) to see if it was currently being investigated for CHB or other diseases.
2.4. Statistical analysis
We performed all analytical workflows in this study using the R Studio 4.0.3 program (RStudio, 250 Northern Ave, Boston, MA 02210). Missense and cis-expression quantitative trait loci (cis-eQTLs) were evaluated in R using the haploR library. WebGestalt R package was used to perform the over-representation analysis (ORA), which included PPI network and molecular pathway analysis. The enrichment analysis for PID was conducted using a hypergeometric test; the criterion for significance was a p-value of 0.05. Visualization of drug under clinical and preclinical investigation was utilized program RAWGraphs visualization [25]. While for the candidate of drug repurposing was visualized by using R language. Chord diagram was built using R with the circlize package.
3. Results
3.1. Identification of SNPs associated with CHB
A total of 207 CHB-associated SNPs were extracted from GWAS and PheWAS catalogues (Table S1). To obtain more candidate variants encoded by the gene, we subsequently expanded the catalogue using HaploReg version 4.1. Therefore, we identified 5.757 CHB-associated SNPs. Among these, we encoded the SNPs for a total of 220 genes. For this step, we categorized the CHB-associated genes. Further analyses were prioritized using these genes to identify biological CHB risk genes through five functional annotations criteria.
3.2. Identification of biological CHB risk genes through functional annotation criteria
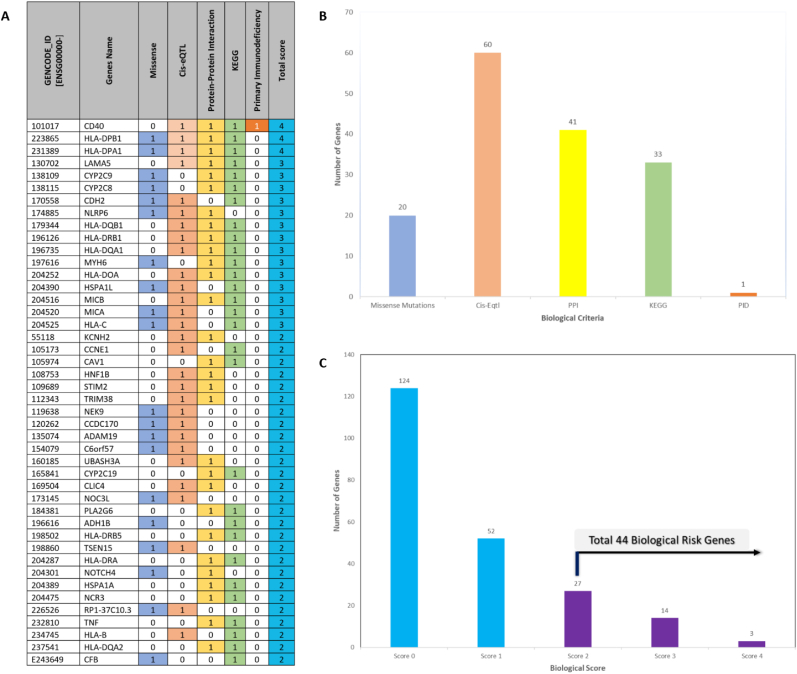
To determine the biological CHB risk genes, we used five functional annotation criteria. Using scores ranging from 0 to 5 for each gene, each gene was scored according to the number of criteria is satisfied. Genes with CHB risk missense variants (n = 20); genes with cis-eQTL effects (n = 60); enriched genes prioritized by PPI (n = 41) and genes with a molecular pathway (n = 31); PID (n = 1). Our biological score threshold of two was set so as to find a much higher number of biological CHB risk genes. In order to prove that the pipeline works, the gene scores were evaluated.
We found that top five genes had a score above 3 and were prioritized as the most biological CHB risk genes, including CD40, HLA-DPB1, HLA-DPA1, LAMA5, and CYP2C9 (Fig. 2A). A distribution of CHB associated genes for each of the five criteria of functional annotations is shown in Fig. 2B. While the distribution of score in each of functional annotation was depicted in Fig. 2C the genes with cis-eQTL are much higher than other functional annotations. It was indicated that the CHB risk genes are mostly higher expressed in whole blood and heart tissues. According to the scoring system, we highlighted that the score 0 was dominant compared to scores more than 2 (Table S2). It is indicated that the average of CHB associated genes does not match with functional annotations. Herein we identified 44 genes with a score ≥ 2 and thereafter categorized as biological CHB risk genes.
Fig. 2.
Chronic hepatitis B (CHB) genomic-drug repurposing process. (A) Five criteria of functional annotation-derived CHB biological risk genes. (B) Bar chart showing the number of genes and scores for each criterion. (C) Bar chart showing the scores of five functional annotations and number of biological CHB risk genes.
3.3. An integrated approach for repurposing CHB drugs
We identified potential drugs for CHB by mapping biological CHB risk genes to drug databases such as DrugBank and DGIdb. A total of 44 biological CHB risk genes were screened based on the scoring system from five functional annotations. Interestingly, we found that 6 druggable targets which overlapped with 18 drugs with status of undergoing clinical trial for CHB (Fig. 3). In addition, 9 druggable targets overlapped with 20 drugs which were undergoing preclinical investigation for CHB (Fig. 4). Besides, we ultimately identified 8 druggable targets which overlapped with 25 drugs promising to be repurposed for CHB (Fig. 5). Taken together, these results provided the biological plausibility of utilizing old drugs (drug repurposing/drug repositioning) to be repurposed as indications to CHB pharmacotherapy.
Fig. 3.
Identification of 6 drug targets and 18 drugs under clinical trial for CHB.
Fig. 4.
Identification of 9 druggable targets that overlapped with 20 drugs undergoing preclinical investigation for CHB.
Fig. 5.
Identification of 25 drugs promising to be repurposed for CHB which overlapped with 8 drug target genes.
4. Discussion
Recent advances in the field of genomic research have significantly improved our understanding of the pathophysiology of a variety of disorders (biology of disease), including CHB disease. The purpose of genetic research was not only to find disease risk genes but also to understand disease biology and apply it to clinical practice [4]. Despite the extensive catalogues of genetic risk loci for a variety of human phenotypes, little is known about how to integrate genetic research findings into biological risk genes [4,26]. Our study attempted to integrate the genetics of CHB with diverse biological resources to aid CHB drug repurposing. We utilized functional and bioinformatics annotations of CHB risk variants and integrated current CHB genetic findings with the complete catalogue of approved drugs for CHB and other diseases. Herein, we used GWAS and the PheWAS catalogues to comprehensively determine CHB susceptibility genes.
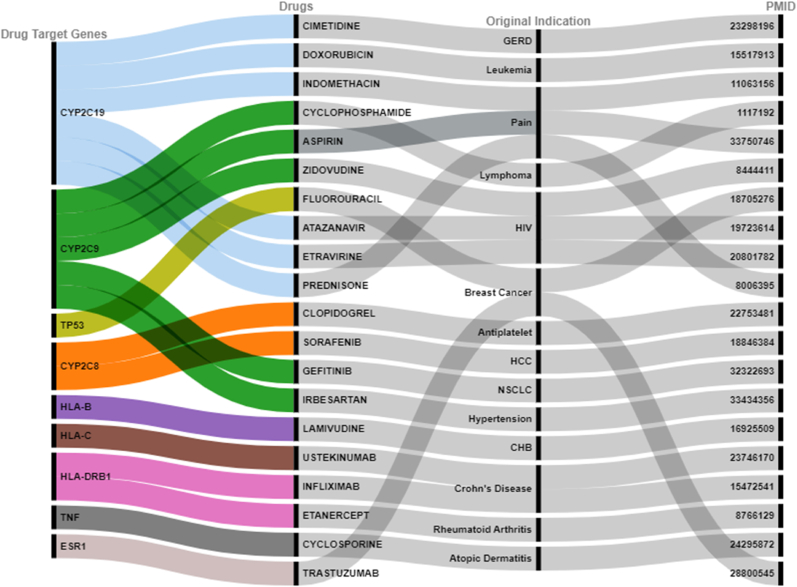
We employed 5 biological criteria to prioritize the genomic variants and identify biological CHB risk genes for identification of druggable targets for CHB. Among 220 CHB associated genes, 44 genes were determined as the biological CHB risk genes. We interpreted the findings into two categories. First, our result suggested the possibility with status of undergoing a clinical trial for CHB. According to our findings, in total 6 druggable targets (NOS3, KCNH2, HNF1B, HLA-B, EGFR, CYP2C19) were identified to be overlapped with 18 drugs. Interestingly, some of the drugs that we identified are antiviral e.g., HIV [lamivudine, efavirenz, etravirine, atazanavir, abacavir, zidovudine, nevirapine], Hepatitis C [ribavirin], and some of the drugs are antibacterial e.g., antituberculosis [rifampin] (Fig. 3).
Secondly, we classified the candidate drugs based on the drugs undergoing preclinical investigation for CHB (Fig. 4). Interestingly, 9 druggable targets overlapped with 20 drugs. And the 20 promising drugs were supported by preclinical investigation. Several drugs were not only supported by preclinical investigation but also it was supported by clinical trials, such as Sorafenib as the HCC treatment and Lamivudine as the drug for CHB we identified here. We also identified several drugs for cancer, such as trastuzumab and fluorouracil. Besides, we ultimately identified 8 druggable targets which overlapped with 25 drugs promising to be repurposed for CHB (Fig. 5). Taken together, these results provided the biological plausibility of utilizing old drugs (drug repurposing/drug repositioning) to be repurposed for CHB treatment.
Noteworthy, among the eight promising targets that overlapped with 25 drugs to be repurposed for CHB, CD40 and HLA-DPB1 were identified as highly promising targets for CHB treatment since they achieved a high systemic score on functional annotations. CD40 has an essential role in the pathogenesis of CHB. Regulating the expression of CD40 may assist in predicting clinical outcomes and lead to the development of new therapies for CHB [27]. The induction of efficient virus-specific CD8+ T-cell responses, which are necessary for viral clearance, requires the activation of CD40 signaling [28,29]. In a transgenic mouse model, the CHB antigen-specific CD8+ T cell depletion might be recovered by CD40-mediated mDC activation. As a result, CD40 signaling is important in antiviral immune responses [30]. This study found five drugs overlapped with CD40, including streptozocin, hydroquinone, fenofibrate, imatinib, and atorvastatin. Meanwhile, HLA-DPB1 is linked to two drugs: aspirin and clozapine. HLA-DPB1 significantly impacts the likelihood of CHB infection, and CHB infection is linked to a reduction in HLA-DPB1 expression. The expression of HLA-DPB1 is essential in the control of CHB [31]. The development of therapies that increase the expression of HLA-DPB1 may prove helpful in the treatment of CHB. The greater expression of HLA-DPB1 could facilitate the clearance of CHB [32].
It is important to note that this study has limitations that should be considered. This retrospective study showed that the identified biological CHB risk genes were associated with clinical variables; therefore, further functional studies in vitro and in vivo (in primary basic/pre-clinical research or clinical studies) and validations are needed to determine the clinical value of our findings especially to alleviate the problems of drug resistance in the CHB medications. However, the strength of our current study lies in providing new insights in pharmacogenomic-guided pharmacotherapy based on the biological CHB risk profiles of CHB patients, thereby bridging the areas of genomic medicine and traditional personalized trials for CHB treatment.
5. Conclusion
Our findings reveal an integrated framework for the identification of druggable targets for CHB using functional genomic variants and bioinformatics. Our approaches aim to narrow down the list of candidate drugs before conducting preclinical investigation and clinical trial studies, thus providing new opportunities for CHB treatment, providing a strong basis for drug discovery efforts, and identifying gene targets and drug candidates for CHB. In summary, our pipeline found CD40 and HLA-DPB1 as promising targets for drug repositioning to treat CHB. To establish the biological mechanisms of CD40 and HLA-DPB1 in CHB, additional research using animal models and clinical trials is needed. Taken together, our findings showcased the use of genomic information that can be translated into clinical implementation through genomic-based therapies and biomarkers for drug discovery.
Author contributions
L.M.I., W.A and D.A.P conceived and designed the study. L.M.I and W.A performed the computational analysis. L.M.I wrote the manuscript. D.A.P., H.D., R.M., I.N.F., I.N.S., Y.V.A.P and R.C revised the manuscript. L.M.I supervised and coordinated this study. All authors have read and approved of the manuscript and have made significant contributions to this study.
Declaration of competing interest
The authors disclose no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
Acknowledgement
This project was supported by grant from Lembaga Penelitian dan Pengabdian Masyarakat (LPPM) Universitas Ahmad Dahlan (UAD), Yogyakarta, Indonesia: (NO:-099/SP3/LPPM-UAD/VII/2022).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101307.
Abbreviations
- cccDNA
Covalently closed circular DNA
- CHB
Chronic Hepatitis B infection
- cis-eQTL
cis-expression Quantitative Trait Locus
- DGIdb
Drug-Gene Interaction Database
- FDA
Food and Drug Administration
- GWAS
Genome-Wide Association Study
- HBV
Human Hepatitis B virus
- HCC
Hepatocellular Carcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ORA
Over-Representation Analysis
- PheWAS
Phenome-Wide Association Study
- PID
Primary Immuno-deficiency
- PPIs
Protein-Protein Interactions
- SNP
Single Nucleotide Polymorphism
- WHO
World Health Organization
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- 1.Liu Z., Hou J. Hepatitis B virus (HBV) and hepatitis C virus (HCV) dual infection. Int J Med Sci. 2006;3(2):57–62. doi: 10.7150/ijms.3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health, O . 18 July 2018. Hepatitis B.https://www.who.int/news-room/fact-sheets/detail/hepatitis-b 17 June 2019. Available from: [Google Scholar]
- 3.Jefferies M., et al. Update on global epidemiology of viral hepatitis and preventive strategies. World journal of clinical cases. 2018;6(13):589–599. doi: 10.12998/wjcc.v6.i13.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green E.D., Guyer M.S. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 5.Sirota M., et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S.M., et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 7.Arrowsmith J. Trial watch: phase II failures: 2008-2010. Nat Rev Drug Discov. 2011;10(5):328–329. doi: 10.1038/nrd3439. [DOI] [PubMed] [Google Scholar]
- 8.Pushpakom S., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 9.Chong C.R., Sullivan D.J., Jr. New uses for old drugs. Nature. 2007;448(7154):645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 10.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 11.Phillips D.J. Forbes; 2013. Pfizer's Expiring Viagra Patent Adversely Affects Other Drugmakers Too. [Google Scholar]
- 12.Muhammad Irham L., et al. Evaluation for the genetic association between store-operated calcium influx pathway (STIM1 and ORAI1) and human hepatocellular carcinoma in patients with chronic hepatitis B infection. Biology. 2020;9(11) doi: 10.3390/biology9110388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kässens J.C., Wienbrandt L., Ellinghaus D. BIGwas: single-command quality control and association testing for multi-cohort and biobank-scale GWAS/PheWAS data. GigaScience. 2021;10(6) doi: 10.1093/gigascience/giab047. p. giab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buniello A., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–d1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denny J.C., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart D.S., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–d1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freshour S.L., et al. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–d1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada Y., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Research. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonsdale J., et al. The genotype-tissue expression (GTEx) project. Nature Genetics. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y., et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–w205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Herz W., et al. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zea-Vera A.F. Common variable immunodeficiency and chronic hepatitis B: therapeutic challenge. Clin Res Hepatol Gastroenterol. 2020;44(2):e38–e40. doi: 10.1016/j.clinre.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Mauri M., et al. Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter. Association for Computing Machinery; Cagliari, Italy: 2017. RAWGraphs: a visualisation platform to create open outputs. p. Article 28. [Google Scholar]
- 26.Webb S. Genomics moves into the clinic. Biotechniques. 2012;52(2):73–76. doi: 10.2144/000113806. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C., et al. Association of CD40 -1C/T polymorphism in the 5'-untranslated region with chronic HBV infection. Cell Physiol Biochem. 2015;35(1):83–91. doi: 10.1159/000369677. [DOI] [PubMed] [Google Scholar]
- 28.Bourgeois C., Rocha B., Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297(5589):2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 29.Bachmann M.F., et al. Maintenance of memory CTL responses by T helper cells and CD40-CD40 ligand: antibodies provide the key. Eur J Immunol. 2004;34(2):317–326. doi: 10.1002/eji.200324717. [DOI] [PubMed] [Google Scholar]
- 30.Isogawa M., et al. CD40 activation rescues antiviral CD8⁺ T cells from PD-1-mediated exhaustion. PLoS Pathog. 2013;9(7) doi: 10.1371/journal.ppat.1003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou G., et al. Variation and expression of HLA-DPB1 gene in HBV infection. Immunogenetics. 2021;73(3):253–261. doi: 10.1007/s00251-021-01213-w. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien T.R., et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 2011;12(6):428–433. doi: 10.1038/gene.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.