Abstract
Nociceptors are specialized sensory neurons that detect damaging or potentially damaging stimuli and are found in the dorsal root ganglia (DRG) and trigeminal ganglia. These neurons are critical for the generation of neuronal signals that ultimately create the perception of pain. Nociceptors are also primary targets for treating acute and chronic pain. Single-cell transcriptomics on mouse nociceptors has transformed our understanding of pain mechanisms. We sought to generate equivalent information for human nociceptors with the goal of identifying transcriptomic signatures of nociceptors, identifying species differences and potential drug targets. We used spatial transcriptomics to molecularly characterize transcriptomes of single DRG neurons from eight organ donors. We identified 12 clusters of human sensory neurons, 5 of which are C nociceptors, as well as 1 C low-threshold mechanoreceptors (LTMRs), 1 Aβ nociceptor, 2 Aδ, 2 Aβ, and 1 proprioceptor subtypes. By focusing on expression profiles for ion channels, G protein–coupled receptors (GPCRs), and other pharmacological targets, we provided a rich map of potential drug targets in the human DRG with direct comparison to mouse sensory neuron transcriptomes. We also compared human DRG neuronal subtypes to nonhuman primates showing conserved patterns of gene expression among many cell types but divergence among specific nociceptor subsets. Last, we identified sex differences in human DRG subpopulation transcriptomes, including a marked increase in calcitonin-related polypeptide alpha (CALCA) expression in female pruritogen receptor–enriched nociceptors. This comprehensive spatial characterization of human nociceptors might open the door to development of better treatments for acute and chronic pain disorders.
INTRODUCTION
Pain is a major medical problem that has been treated for millennia with drugs whose origins can be traced to natural products (1). Although some new mechanism-based therapeutics have recently been approved for treatment of pain, these were developed on the basis of biochemical observations in clinical studies, such as the calcitonin gene-related peptide (CGRP) link to migraine headache (2). There has been an unsatisfying failure to translate preclinical work on peripheral pain mechanisms, which has largely been done in rodents, into effective pain therapeutics (3, 4). A potential explanation for this failure to translate is that important species differences in nociceptor molecular phenotypes exist between mice and humans, an idea partially supported by bulk RNA sequencing (RNA-seq) experiments (5, 6) and other lines of evidence (7, 8). Nociceptors are the first neurons in the pain pathway and express a broad variety of receptors that allow them to respond to stimuli arising from the environment, from local cells native to tissues, and from infiltrating immune cells that may be involved in inflammation or other processes (9–12). These neurons increase their excitability in both acute and chronic pain states, and changes in their excitability phenotype, such as the generation of spontaneous activity, are directly linked to chronic pain states like neuropathic pain (13). Therefore, nociceptors are excellent target cells for acute and chronic pain drugs. In the work described here, we have created a high-resolution map of human sensory neurons in the dorsal root ganglia (DRG), including nociceptors, with the goal of accelerating discovery and/or validation of high-quality drug targets that can be manipulated to improve pain treatment.
Single-cell sequencing of DRG neurons has delineated the molecular architecture of somatosensory neuron subtypes in the mouse (14–16), elucidated their developmental transcriptional paths (17), and characterized how these neurons change phenotype in response to injury (18, 19). However, it is not clear how this information can be applied to humans due to the lack of comprehensive transcriptomic map of human sensory neurons. Most contemporary single-cell profiling studies use nuclear RNA-seq because this technology is scalable, fully commercialized, and widely available (20). However, human DRG neurons are among the largest in the body (20 to 100 μm in diameter) (21) and also have large nuclei, creating challenges for many sequencing platforms. Sensory neurons are also postmitotic cells with large cytoplasmic volumes that contain a high concentration of extranuclear RNA. Sequencing technologies that combine spatial resolution with the ability to accurately sample cytoplasmic RNA may reveal a clearer picture of the full neuronal transcriptome (22), which is important when looking for drug targets that may have low expression. To overcome these technical challenges and fill this gap in knowledge with respect to human sensory neuron transcriptomes, we have conducted spatial sequencing experiments (10x Genomics Visium technology, which uses 55-μm barcoded spots) on human, lumbar DRG obtained from organ donors. We identified one proprioceptor, two Aβ low-threshold mechanoreceptors (LTMRs), one Aβ nociceptor, one Aδ-LTMR, one Aδ high-threshold mechanoreceptor (HTMR), one C-LTMR, and five C-nociceptor subtypes. We have compared our findings to both mouse (16) and nonhuman primate datasets (23), finding not only many similarities but also important differences, many of which have important implications for pain target identification. Because sex differences in pain mechanisms are increasingly recognized (24, 25), we performed our studies with an equal number of male and female samples. We anticipate that our data will advance our understanding of molecular pain mechanisms in humans and create a new path forward for pain and itch therapeutic development (4).
RESULTS
Spatial transcriptomics generates near single-neuron resolution
We generated whole-cell transcriptomes for single neurons, using the 10x Genomics Visium Spatial Gene Expression platform (26, 27). This technology uses 55-μm barcoded spots printed on the capture area of Visium slides. Human DRGs, collected within 4 hours of cross-clamp from neurologically dead organ donors (four female and four male, details on organ donors are provided in table S1), were sectioned into the capture areas of the Visium slides, stained, and imaged (Fig. 1A). After tissue permeabilization, mRNA from each section was bound to barcoded primers and subsequently processed for library preparation and RNA-seq. We obtained, on average, ~52 M reads and detected an average total of ~24,000 genes per section, for a total of ~830 M reads from 16 tissue sections (fig. S1A). Because each section was stained and imaged, the barcoded mRNAs and respective genes’ location can be visualized within each DRG section using Loupe Browser (10x Genomics). In addition, barcoded spots can be selected on the basis of their position on the tissue (fig. S1B). To generate near single-neuron resolution, we selected all barcodes that overlapped a single neuron in all sections and processed them for downstream analysis. From two tissue sections from each donor (total 16 sections), we identified 4356 barcodes that overlap a single neuron (neuronal barcodes, also contain some signal from other surrounding cells) and 12,118 barcodes that directly surround neurons (surrounding barcodes). The remaining 20,725 barcoded spots were classified as other barcodes. Barcodes that overlapped multiple neurons were excluded. We optimized tissue permeabilization to enhance neuronal RNA elution onto the slides to develop neuronally enriched libraries (fig. S2). We detected a higher number of RNA molecules and a higher number of unique genes in the neuronal barcodes (fig. S1C). In addition, neuronal barcodes had a distinct gene expression profile from surrounding and other barcodes (fig. S1D).
Fig. 1. Identification of neuronal subtypes in human DRG using spatial transcriptomics.
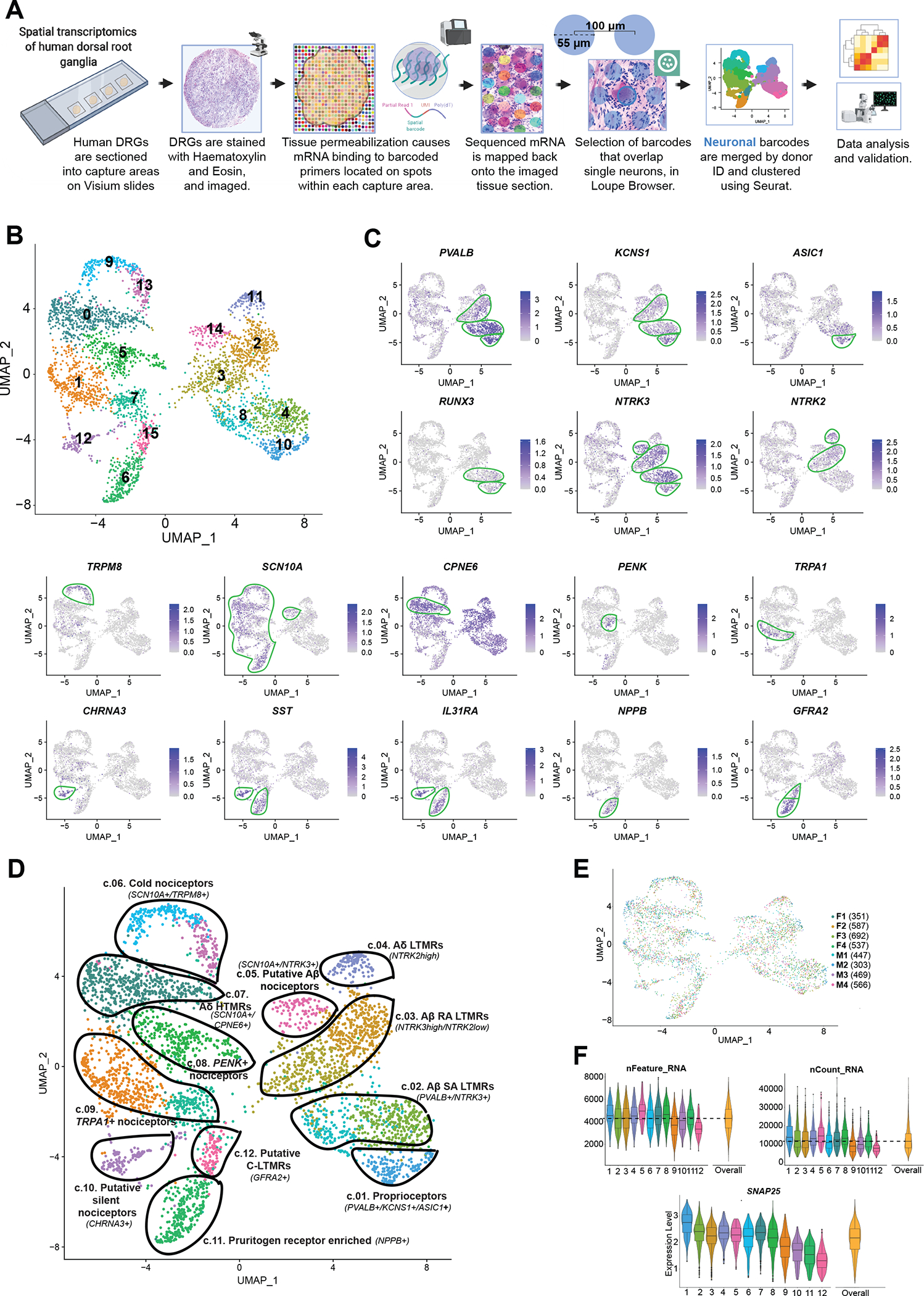
(A) Overview of the workflow and analysis. Neuronal barcodes (barcoded spots that overlap single neurons) were manually selected in Loupe Browser and clustered using Seurat package in R. (B) UMAP plot showing the 16 clusters generated by Seurat’s workflow. (C) UMAP plots of the expression of gene markers that were used to label neuronal clusters. (D) UMAP plot showing the 12 labeled human DRG neuronal clusters that were curated from the original 16 clusters, which are still shown with color coding matching (B). (E) UMAP plot shows the contribution of each donor for cluster formation. The number of barcodes per donor used for clustering is in parenthesis. (F) Violin plots show consistent distributions of the number of detected genes (nFeature_RNA), the counts of unique RNA molecules (nCount_RNA), and the average expression for the neuronal marker SNAP25 across clusters. The numbers on the x axis correspond to cluster numbers.
Neuronal barcodes with both a low number of reads and a low count for the neuronal marker SNAP25 (16, 23, 28) were removed, as described in Materials and Methods. A total of 3952 neuronal barcodes were grouped by donor ID and clustered using Seurat’s anchor integration workflow, followed by graph-based clustering (see Materials and Methods for detailed information and fig. S3) (29). Initially, Seurat generated 16 clusters (Fig. 1B). We highlight several known neuronal markers from the literature that were enriched in these clusters to characterize these subsets of human DRG neurons based on their specific gene enrichment (Fig. 1C and fig. S4). We ultimately selected eight clusters for merging. Each of these was neighboring clusters with highly overlapping gene expression where two clusters were merged into one. This led to 12 final clusters of human DRG neurons (Fig. 1D), which are described in detail below. For data quality purposes, we verified that each individual donor contributed neurons to each cluster and that no individual donor was responsible for any particular cluster (Fig. 1E). The number of genes and unique RNA molecules detected per cluster as well as the average expression distribution of the neuronal marker SNAP25 across clusters is shown in Fig. 1F.
Defining the transcriptomes of human sensory neuron subtypes
DRG neurons are derived from neural crest cells and are responsible for transmitting all somatosensation (touch, proprioception, nociception, and temperature) from the body to the spinal cord and brainstem (30). These neurons have been grouped into two main classes based on the diameter of the cell body and the conduction velocity—A- and C-fibers. Myelinated Aβ-fiber neurons are mostly large-diameter cells that innervate the skin through terminal organs that are responsible for detection of nonnoxious stimuli, in particular, light touch (31, 32). Proprioceptors innervate muscle and other structures and are responsible for communicating signals about the location of our limbs in space. Unmyelinated, small-diameter C-fiber neurons are critical for the detection of most noxious stimuli. Aδ neurons are not only lightly myelinated and have larger diameter than C-fibers but also respond to stimuli in the noxious range. These classes of sensory neurons differentially express specific neurotrophic receptors during development and into adulthood (30).
Within the A-fiber group, we identified six subtypes in the human DRG. The first cluster was classified as proprioceptors (cluster 1) based on the expression of parvalbumin (PVALB), neurotrophic receptor tyrosine kinase 3 (NTRK3), and acid-sensing ion channel subunit 1 (ASIC1) and reduction in neurotrophic receptor tyrosine kinase 2 (NTRK2) (33). This cluster was also enriched for potassium voltage-gated channel modifier subfamily S member 1 (KCNS1) and a displayed enriched expression of Runt-related transcription factor (RUNX) family transcription factor 3 (RUNX3), which plays an evolutionarily conserved role in vertebrates in suppressing NTRK2 in A-fiber proprioceptors (Fig. 2A) (34). Aβ slowly adapting (SA) LTMRs (cluster 2) innervate hairy and glabrous skin and terminate on Merkel cells (31, 35). These neurons were enriched in NTRK3 and PVALB and showed lower expression for NTRK2, a pattern of expression consistent with Aβ SA LTMRs in the mouse (36). These neurons were also enriched in receptor activity modifying protein 1 (RAMP1) expression, a receptor component for the CGRP receptor. The end organs of Aβ rapidly adapting (RA) LTMRs are Meissner and Pacinian corpuscles in glabrous skin and lanceolate endings in hairy skin (31). The Aβ RA LTMR subgroup (cluster 3) was likewise identified by expression of NTRK3 and a low expression for NTRK2 (36, 37). Aδ-LTMRs are also known as D-hair afferents and terminate as longitudinal lanceolate endings in hair follicles (31). Aδ-LTMRs (cluster 4) were characterized by their high expression of NTRK2 (36, 37). Mice lacking this subset of Ntrk2-positive neurons are less sensitive to touch and nonresponsive to mechanical stimulation after injury (38). This suggests that Aδ-fibers may be involved in the development of mechanical allodynia. Aδ-fibers have previously been characterized in human skin nerves as similar to “down-hair” Aδ neurons in other species (39). One group of A-fiber neurons expressed both NTRK3 and sodium voltage-gated channel alpha subunit 10 (SCN10A), a voltage-gated sodium channel (VGNaC) that is enriched in nociceptive neurons (40). Therefore, we identified this cluster as putative Aβ nociceptors (cluster 5). Aβ-fibers that respond to noxious stimuli have been reported in other species (41) including monkeys (42). A recent study has demonstrated that humans also have Aβ-fiber nociceptors with nociceptive properties (43). The final cluster of A-fibers (cluster 7) had high expression of NTRK1, Copine 6 (CPNE6), and SCN10A, which is consistent with HTMRs in the mouse and macaque (14, 23). This cluster also expressed calcitonin- related polypeptide alpha (CALCA) and lysophosphatidic acid receptor 3 (LPAR3).
Fig. 2. Enriched gene expression in human DRG neuronal clusters and spatial visualization of neuronal subtypes.
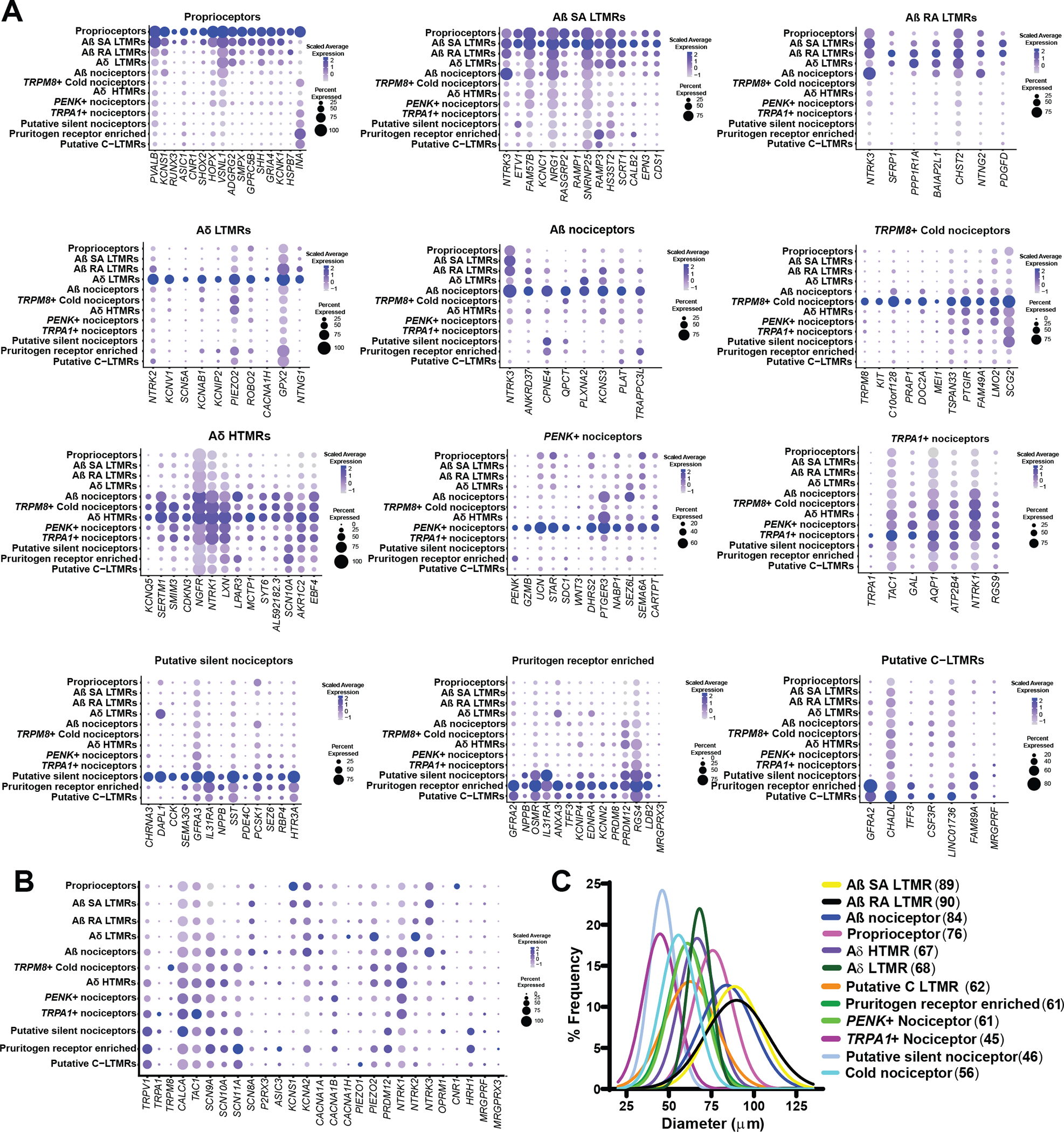
(A) Dot plots showing the top genes for each neuronal subpopulation and how they are expressed across all clusters. The size of the dot represents the percentage of barcodes within a cluster, and the color corresponds to the average expression (scaled data) across all barcodes within a cluster for each gene shown. (B) Dot plot showing the expression of known pain genes and markers across clusters. (C) Neuronal clusters were mapped back to DRG sections to visualize neurons within the DRG. Diameters of neurons with visible nuclei were measured to ascertain and plot cell sizes for each cluster with mean diameter (in micrometers) shown in parentheses. Gaussian curve fits are shown for visualization purposes.
We also identified five subtypes of C-fiber nociceptors and a putative C-LTMR cluster (Fig. 2A). Transient receptor potential cation channel subfamily M member 8 (TRPM8), a known menthol and cold-sensitive channel, labeled the cold nociceptors (cluster 6) (44). This cluster expressed SCN10A but little transient receptor potential cation channel subfamily V member 1 (TRPV1), a unique feature compared to other human nociceptor clusters. Proenkephalin (PENK), an endogenous opioid and precursor to several enkephalins (45), was enriched in another C-nociceptor cluster (cluster 8). This cluster also uniquely expressed the peptide transmitter gene urocortin (UCN) and was enriched for the prostaglandin E2 (PGE2) receptor PTGER3, encoding the prostaglandin E receptor 3 (EP3) that is distinct among PGE2 receptors in producing analgesia upon agonist binding (46). Another cluster of C-fibers was distinguished by transient receptor potential cation channel subfamily A member 1 TRPA1 expression (cluster 9). This subpopulation also showed very high expression for tachykinin precursor 1 (TAC1) (which encodes substance P) and CALCA (47), although these neuropeptides were broadly expressed by all nociceptor clusters (Fig. 2, A and B). This difference in neuropeptide expression is an important distinction between human and rodent sensory neurons, likely indicating that peptidergic and nonpeptidergic subsets of sensory neurons do not exist in humans (7, 8, 48). This notion is further supported by our recent histological work showing that the peptidergic marker CGRP (gene: CALCA) and the nonpeptidergic marker P2X purinoceptor 3 (P2X3R; gene: P2RX3) are highly coexpressed at the mRNA and protein levels in human DRG neurons (8, 48). These neurons also coexpress the nociceptor marker, sodium channel Nav1.8 (gene: SCN10A) (8). Afferent input of CGRP and P2X3R into the dorsal horn is found throughout all of lamina I and II, supporting their high coexpression in human nociceptors (48).
The specific expression of cholinergic receptor nicotinic alpha 3 subunit (CHRNA3) identified a cluster of putative “silent” nociceptors (cluster 10) (Fig. 2A) (49). Silent nociceptors correspond to a subset of C-fibers that innervate joints, viscera, and skin and are often referred to as mechanoinsensitive C-fibers. They are unresponsive to noxious mechanical stimuli under normal conditions but are sensitized and become mechanically sensitive after inflammatory stimulation and likely play key roles in certain pain disorders (49–52). The silent nociceptor cluster expressed a large array of ion channels including the serotonin receptor 5-hydroxytryptamine receptor 3A (HTR3A); purinergic receptors P2X 3, 4, 6, and 7 (P2RX3, P2RX4, P2RX6, and P2RX7); proton receptor acid sensing ion channel subunit 3 (ASIC3); and glutamate receptors such as glutamate ionotropic receptors kainate type subunits 2 to 5 (GRIK2, GRIK3, GRIK4, and GRIK5), glutamate ionotropic receptor delta type subunit 1 (GRID1), glutamate ionotropic receptor N-methyl-D-aspartate (NMDA) type subunit 1 (GRIN1), and glutamate ionotropic receptors α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid type subunit 3 and 4 (GRIA3 and GRIA4) (data file S1), which may underlie the sensitivity of this subset of neurons to inflammatory mediators. These neurons also expressed the H1 histamine receptor gene, histamine receptor H1 (HRH1), which is known to sensitize these neurons to mechanical stimulation and is also a likely pathway for histamine-induced itch in humans (53, 54). Therefore, this subset of C-fibers likely also participates in the generation of itch signals from the periphery. A separate pruritogen receptor–enriched cluster (cluster 11) was classified on the basis of the expression of natriuretic peptide B (NPPB), GDNF family receptor alpha 2 (GFRA2), and interleukin-31 receptor A (IL31RA) (55), although these latter two genes were also found in other populations. Our data also show that sodium voltage-gated channel alpha subunit 11 (SCN11A) has a very high expression in this subpopulation. Nav1.9 (SCN11A) gain of function mutations can lead to congenital insensitivity to pain or partial loss of pain sensation. Studies in mice have reported that the mutation causes a pruritic phenotype (56, 57). Humans with Nav1.9 mutations report a severe pruritis (56, 58). Mechanisms associated with the enrichment of SCN11A in itch nociceptors may explain this phenotype. A final C-fiber cluster was enriched in GFRA2, a characteristic marker of C-LTMRs in mice (14) and was classified as putative C-LTMRs (cluster 12). This cluster had high similarity in terms of gene expression with the pruritogen receptor–enriched population but had lower expression of NPPB, a marker for itch nociceptors in mice (fig. S5, A and B). A distribution of genes associated with pain across human DRG neuronal subtype clusters is shown in Fig. 2B. Ranked gene expression by gene for all 12 A- and C-fiber clusters is given in data file S1.
Spatial visualization of neuronal subtypes
Lumbar DRG neuronal subtypes did not show any clear spatial organization in any analyzed tissue sections. However, we did use visualization of barcode position in DRG sections to measure neuron diameter associated with each of the 12 clusters (see Materials and Methods). This independent measure validated that Aβ clusters correspond to the largest-diameter neurons in the DRG, whereas C nociceptor clusters were the smallest (Fig. 2C and fig. S6). A∂ clusters were intermediate in size between Aβ- and C-fiber neurons, in line with cell size distributions in all other species where this has been assessed (8, 59–61).
Validation of spatial transcriptome-defined subtypes with RNAscope
Our spatial transcriptomic approach provides detailed insight into the types of neurons present in the human DRG, but there are limitations, such as the lack of pure single neuronal transcriptomes for any given barcode. We have previously demonstrated that RNAscope in situ hybridization technology offers highly sensitive detection of neuronal mRNAs in human DRG (8). As a validation tool, we conducted RNAscope experiments on human DRG tissue sections for several mRNAs that showed high abundance in specific neuronal clusters: PR/SET domain 12 (PRDM12), NPPB, somatostatin (SST), NTRK1–3, PVALB, LPAR3, PENK, TRPM8, GFRA2, MAS-related GPR family member X1 (MRGPRX1), and tyrosine hydroxylase (TH). We assessed their coexpression with nociceptor-enriched genes SCN10A, TRPV1, and CALCA or with other cluster-specific markers (Fig. 3A). The nociceptor population (SCN10A+, TRPV1+, or CALCA+) comprised ~60 to 70% of all human sensory neurons and were small in diameter (average = 54 μm) (Fig. 3, B and C). PRDM12, a gene that is essential for human pain perception (62), was expressed in ~74% of DRG neurons that also coexpressed CALCA (Fig. 3D). CALCA mRNA was detected in all neuronal clusters and surrounding/other barcodes in the Visium data, likely because CALCA mRNA localizes to axons (63) explaining its widespread detection. Smaller subdivisions of nociceptors such as the putative silent and pruritogen receptor–enriched nociceptor populations (NPPB+ or SST+) amounted to ~30% of the population that also coexpressed SCN10A (Fig. 3, E and F). NTRK1, which is most abundant in the nociceptor clusters, was found in 68% of the neuronal population and colocalized with SCN10A (Fig. 3G). NTRK2, which was enriched in the Aδ LTMR cluster, a cluster that is depleted of SCN10A, was detected in medium-sized neurons (Fig. 3C) and showed little coexpression with SCN10A (Fig. 3H). The proprioceptor, Aβ-LTMR and Aβ nociceptor marker, NTRK3, was found in larger-sized neurons and showed slightly higher coexpression with SCN10A than NTRK2, most likely due to its presence in the SCN10A+ Aβ nociceptor cluster (Fig. 3I). In the Visium dataset, LPAR3 was enriched in the Aδ HTMR and Aβ nociceptor clusters but was also lowly expressed in other nociceptor clusters, all of which express TRPV1. Similarly, LPAR3 was expressed in 80% of all sensory neurons, most of which were TRPV1-positive (fig. S7, A to C). PVALB, which was highly enriched in the proprioceptor and Aβ SA LTMR clusters, was found in ~45% of sensory neurons, half of which were TRPV1-negative (fig. S7, A to C). The cold nociceptor cluster marker, TRPM8, was found in ~50% of sensory neurons, whereas PENK, which was enriched in a different cluster (PENK nociceptors), was found in ~35% of sensory neurons (fig. S7, B to E). Similar to Visium, these two genes did show some overlap (21.2%) using RNAscope but were also detected in separate populations (fig. S7, B to E).
Fig. 3. RNAscope in situ hybridization and functional validation on human DRG.
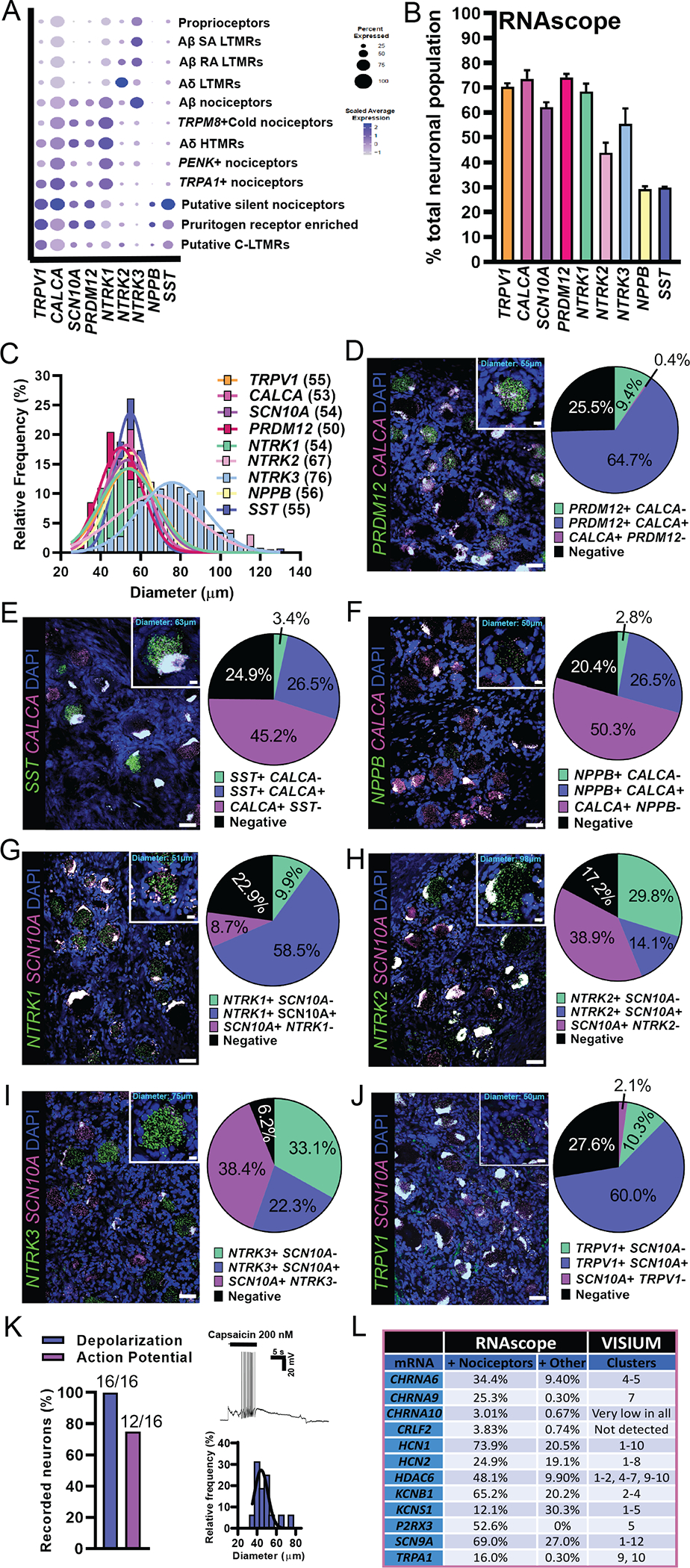
(A) Visualization of Visium gene expression for markers that were used for RNAscope analysis. (B) Percentage of neurons expressing each target compared to the total neuronal population. (C) Size distribution of all target-positive neurons. Gaussian mean in micrometer diameter in parentheses. (D to J) Merged image for the target of interest (green) with a nociceptor marker (magenta) and DAPI (blue) is shown. Scale bars, 50 μm. Inset for each panel shows a blowup of a single neuron. Scale bars, 10 μm. Population distribution of each neuronal marker is shown in the pie chart. (K) The TRPV1 agonist, capsaicin (200 nM), was applied to small-diameter human DRG neurons in vitro causing depolarization (100%) and action potential firing (75%). (L) RNAscope data are summarized from (8) and compared to findings from Visium sequencing. The neuronal cluster for each target is listed. Clusters: 1, proprioceptors; 2, Aβ SA LTMR; 3, Aβ RA LTMR; 4, Aδ LTMR; 5, Aβ nociceptors; 6, TRPM8+ cold nociceptors; 7, Aδ HTMR; 8, PENK+ nociceptors; 9, TRPA1+ nociceptors; 10, putative silent nociceptors; 11, pruritogen receptor enriched; 12, putative C-LTMRs.
GFRA2, which was expressed in the putative C-LTMR cluster, was found in ~33% of small-sized human sensory neurons and highly coexpressed MRGPRX1 (fig. S8, A to E). MRGPRX1 was only detected in a few barcodes, all of which were in the C-LTMR cluster; however, we detected more MRGPRX1+ neurons using RNAscope, many of which were positive for GFRA2 (fig. S8, A to E). TH, the mouse marker for C-LTMRs (14), showed little to no expression in human DRG using Visium and RNAscope (fig. S8, A to D). For further assessment of this cluster, we assessed GFRA2 expression in combination with NPPB as we classified this cluster as a potential C-LTMR population due to its expression of GFRA2 and depletion of NPPB (fig. S5). We observed neurons that were copositive for GFRA2 and NPPB (~27.3%) and a smaller population of GFRA2-expressing neurons that were negative for NPPB (7.4%). This GFRA2-positive, NPPB-negative population likely represents the C-LTMR cluster 12 (fig. S8, F to H).
We have previously reported that TRPV1 mRNA is more widely expressed in human nociceptors than in mouse (8), and TRPV1 was detected in all nociceptor clusters with Visium spatial sequencing, with the exception of cold nociceptors where it was slightly expressed. Using SCN10A as a nociceptor marker, we again observed that TRPV1 was found in most nociceptors (Fig. 3J). We next determined whether these neurons were functionally responsive to the TRPV1 ligand, capsaicin. Application of capsaicin depolarized all small-sized, dissociated human DRG neurons and caused action potential firing in 75% (Fig. 3K). We concluded that RNAscope, spatial sequencing, and functional analysis support broad expression of TRPV1 in human nociceptors. As a final validation, previously published RNAscope findings substantiated the proposed neuronal subclusters from Visium sequencing (Fig. 3L) (8). For example, we previously proposed KCNS1 as a marker of human Aβ neurons due to its expression in large-sized neurons that were negative for CALCA and P2RX3 (8). KCNS1 was also enriched in Aβ clusters using the spatial transcriptomic approach.
Sex differences in human sensory neurons
Molecular differences between male and female sensory neurons have been reported in defined population cell sequencing experiments in rodents (64) and inferred from bulk RNA-seq on human DRGs (5), but our knowledge of sex differences in neuronal gene expression in the human DRG is limited. On the basis of our results, it is apparent that males and females have the same DRG neuronal subtypes, because neuronal barcodes from both sexes were clearly represented in all clusters (Fig. 4A). We then looked for sex differences within the overall population of neuronal barcodes and within each specific cluster. With the spatial sequencing approach, neuronal barcodes include mRNA from surrounding cells. To overcome detection of generic sex differences contributed by other cell types, we performed statistical tests on surrounding barcodes (overall surrounding barcodes and specific to each neuronal cluster). Genes were considered to be differentially expressed (DE) if fold change (FC) ≥ 1.33 and adjusted P < 0.05. We considered genes to be DE specifically in neurons if they were not DE in the respective surrounding barcodes (fig. S9). Similar to findings in the mouse where sex differences in the neuronal population were small (64), we identified only 44 genes with sex differential expression in the neuronal barcodes pooled together by sex (Fig. 4B and data file S2). However, this approach pools together expression data for transcriptomically diverse neurons, creating variation that is a product mostly of different cellular phenotypes. To overcome this issue, we looked at potential sex differences in gene expression within each neuronal subtype. Here, we found more neuronally enriched DE genes (Fig. 4C, data files S3 and S14 for neuronal barcodes, and data files S15 to S27 for surrounding barcodes). The pruritogen receptor–enriched population had the highest number of DE genes (96), suggesting potential molecular differences in mechanisms of pruritis between men and women. We performed gene set enrichment analysis for DE genes in all neuronal subpopulations using the gene ontology (GO) enrichment analysis resource PANTHER (65). We did not identify any GO terms for DE genes in proprioceptors, A∂ LTMRs, cold nociceptors, A∂ HTMRs, PENK+ nociceptors, TRPA1+ nociceptors, putative silent nociceptors, or C-LTMRs. We identified GO terms for pruritogen receptor–enriched subtype, 84 for Aβ SA LTMRs and 6 for Aβ RA LTMRs (data file S28). We were particularly interested in potential sex differences in human nociceptors and, thus, focused on the pruritogen receptor–enriched cluster. A main finding in that cluster was the higher expression of CALCA, which encodes the CGRP protein, found in female pruritogen receptor–enriched neurons (Fig. 4D). This finding was validated in RNAscope experiments examining CALCA expression in NPPB-positive neurons from male and female organ donors (Fig. 4E and fig. S10).
Fig. 4. Sex differences in gene expression within human DRG neuronal populations.
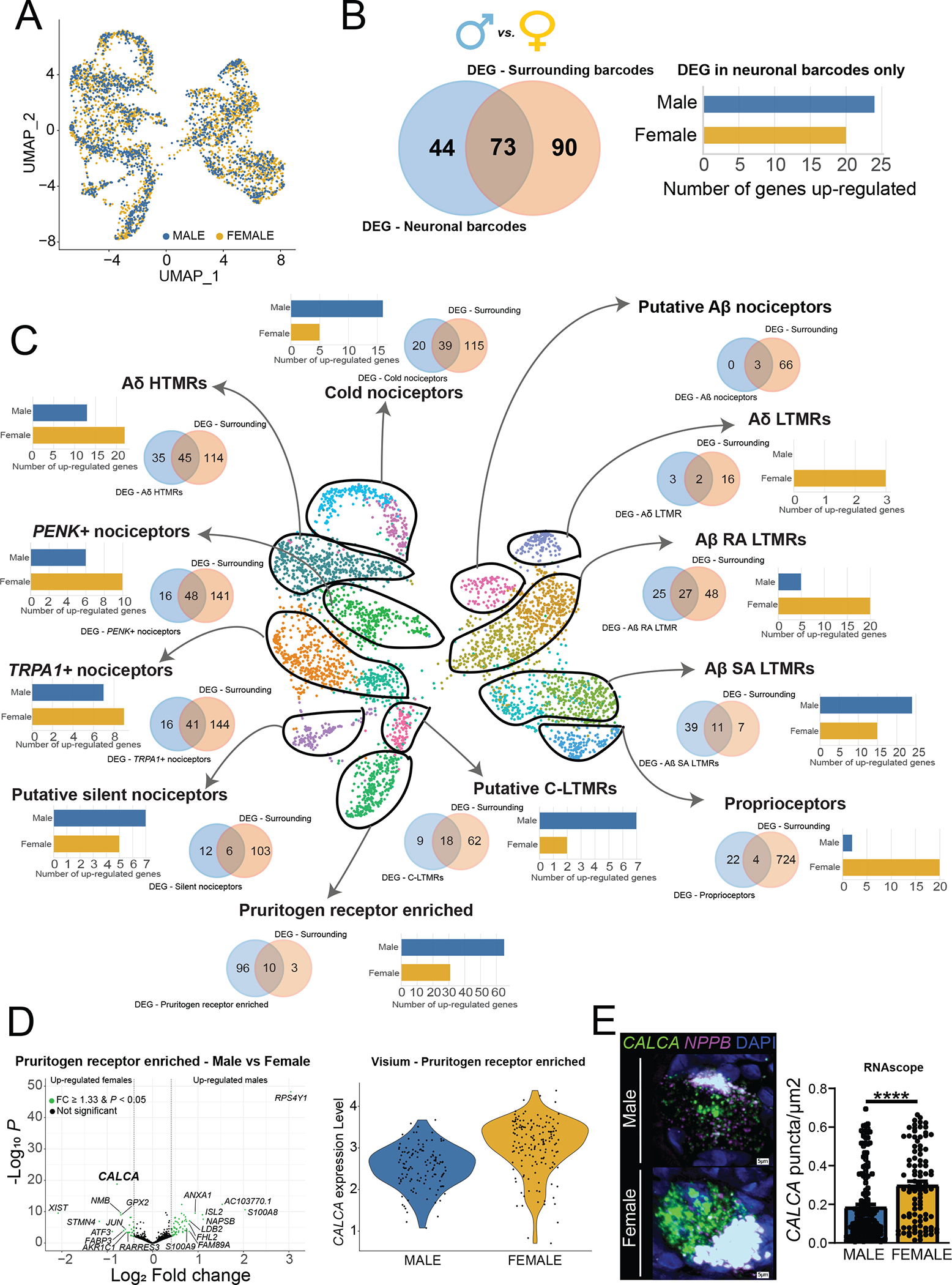
(A) UMAP showing male and female barcodes in all clusters. (B) Venn diagram showing the overlap between the number of DE genes in the overall neuronal population (blue, left) and the overall surrounding population of barcodes (beige, right). Bar plot shows the number of up-regulated genes per sex after removing genes that were also DE in surrounding barcodes. (C) Venn diagrams show the overlap between the number of DE genes in each neuronal subtype (blue, left) and the respective surrounding population (beige, right). Bar plots show the number of up-regulated genes per sex in each cluster after removing genes that were also DE in the respective surrounding barcodes. (D) Volcano plot shows DE genes in the pruritogen receptor–enriched population after removing DE genes in surrounding barcodes (we highlighted the top 10 genes in each sex ranked by log2 fold change). Violin plot shows CALCA expression in individual barcodes in males and females within the pruritogen receptor–enriched population. (E) RNAscope for CALCA mRNA colocalized with NPPB, a marker of pruritogen receptor–enriched nociceptors, with quantification of differences in expression between male and female neurons for amount of CALCA expression in the dot plot. Representative image scale bars, 5 μm. DEG, differentially expressed gene. Genes were considered to be DE if FC ≥ 1.33 and adjusted P < 0.05. ****P < 0.0001.
Similarities and differences between human and mouse DRG neurons with a focus on pharmacological targets
Next, we examined expression of individual genes within gene families, such as ion channels, G protein–coupled receptors (GPCRs), and tyrosine receptor kinases, that are involved in transduction of nociceptive signals by nociceptors and are considered important pharmacological targets for existing or potential drugs. We made comparisons between our spatial transcriptomic dataset from human DRG and mouse single-neuron data from DRG that is publicly available at mousebrain.org (16). Most preclinical studies are conducted in rodents (in particular, mice) so the comparative expression maps that follow can be used to directly assess similarities and differences in sensory neuron gene expression profiles between mice and humans.
VGNaCs are the foundation of the ability of neurons to carry action potentials, and sensory neurons express a unique subset of these genes (66). We observed that VGNaC genes have very similar expression patterns in human and mouse (Fig. 5A), demonstrating that the expression of α subunits that encode the pore-forming unit of the channel is conserved. An exception among this family was the sodium voltage-gated channel beta subunit 4 (SCN4B) gene, which encodes the β4 subunit of the VGNaC. This β subunit is critical for resurgent currents that are key contributors to excitability (67, 68). In mouse, Scn4b was found mostly in A-fiber neurons, consistent with previous studies (67, 68), yet in human, SCN4B mRNA was distributed among all sensory neuron types. Because β4 subunits regulate resurgent currents through Nav1.8 channels (69), and these two genes are more highly coexpressed in human nociceptors, this could potentially contribute to enhanced resurgent Nav1.8 currents in those cells, a hypothesis that could be tested in future experiments.
Fig. 5. Expression of VGNaC channel and GPCR genes in human and mouse datasets.
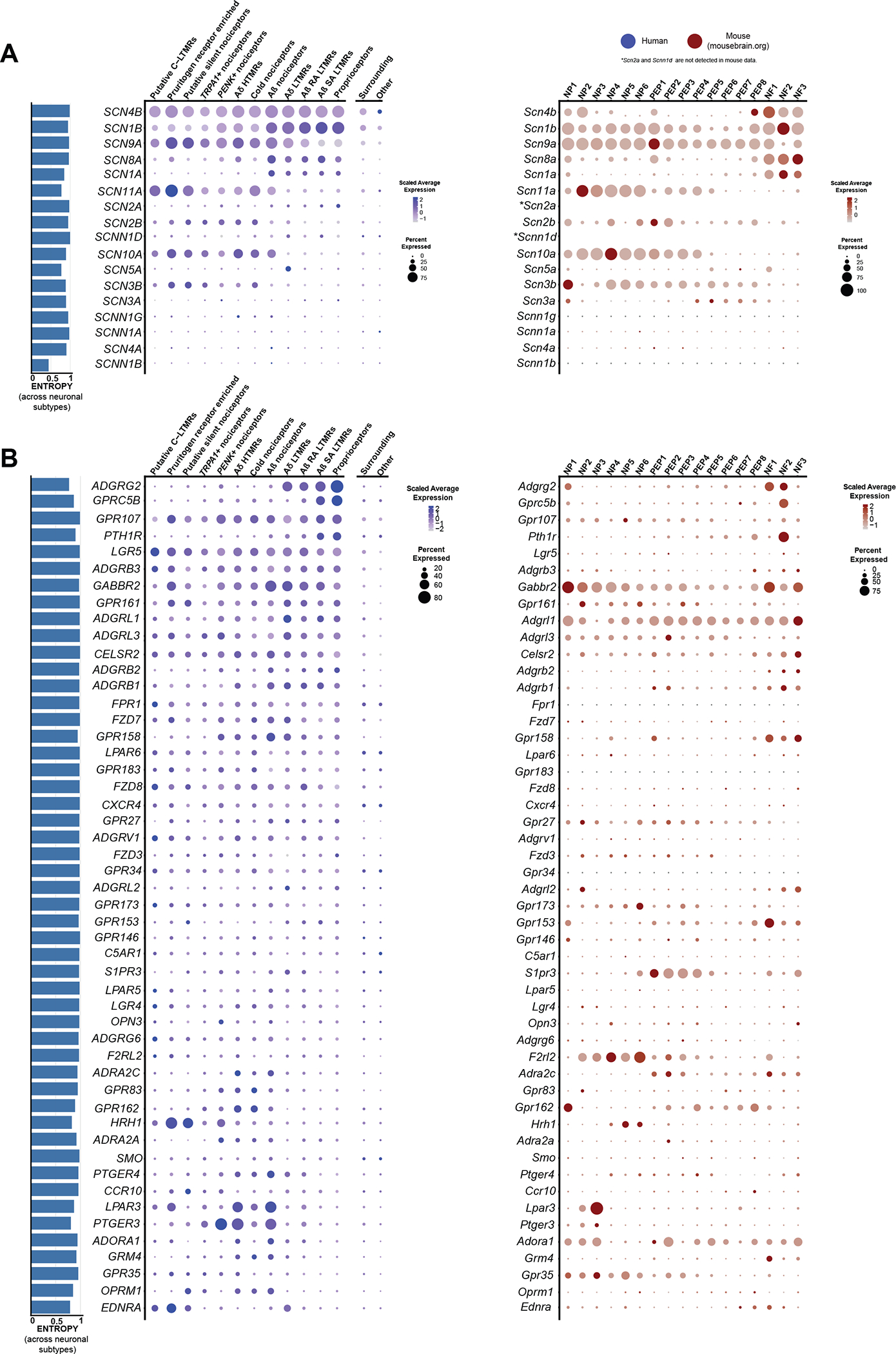
(A) Dot plots showing the expression of VGNaC channel genes in human spatial transcriptomic (in blue) and mouse single-cell experiments (in red). (B) Dot plots showing the expression of GPCR genes in human spatial transcriptomic (in blue) and mouse single-cell experiments (in red). The size of the dot represents the percentage of barcodes within a cluster, and the color corresponds to the average expression (scaled data) across all barcodes within a cluster for each gene shown. Normalized entropy was used as a measure of “specificity of neuronal subtype”, where a score of 0 means a gene is specific to one neuronal subtype and 1 means that a gene has uniform distribution across neuronal subtypes.
GPCRs are the largest family of receptors in the mammalian genome and have diverse roles in nociceptors ranging from inflammation detection to cell adhesion. These receptors are also important targets for therapeutic development. We compared the expression and distribution of the top 50 most highly expressed GPCRs in human DRG to their homologs in mouse. Whereas some GPCRs showed consistent patterns of expression, many were divergent suggesting important differences in expression across species for this family of receptors. Two notable differences were the PTGER3 and LPAR3 genes (Fig. 5B). PTGER3 was enriched in the PENK+ nociceptor population in humans and was also expressed by several other nociceptor subtypes, whereas it was restricted to a subset of nonpeptidergic neurons in mice. Given the potential for this prostaglandin receptor as an antinociceptive target, this could be important for therapeutic purposes with EP3 agonists (46). LPAR3 is a receptor for lysophosphatidic acid and has been associated with neuropathic pain (70). This GPCR was broadly expressed in nociceptor subtypes in humans but was again restricted to nonpeptidergic nociceptors in mice. Receptors of the metabotropic glutamate receptor family (GRM) also showed divergent expression across species (Fig. 5B), consistent with the previous observation that group I GRM family genes are not detected in human DRG (5). Some GPCRs did show strong conservation of expression, for instance, gamma-aminobutyric acid (GABA) type B receptor subunit 2 (GABBR2), encoding a subunit of the GABAB receptor complex, which is likely the most highly expressed Gαi-coupled receptor in sensory neurons in both humans and mice.
The characterization of expression of interleukins (ILs) and their receptors in neuronal subpopulations can reveal how their ligands may interact with different populations of sensory neurons in different species. IL31RA, for instance, was more broadly expressed in human DRG neurons than in mouse where the gene was restricted to itch nociceptors (fig. S11), as shown previously using in situ hybridization (71). Anti-inflammatory IL receptors, IL-4 receptor (IL4R), IL-10 receptor subunit alpha (IL10RA), and IL-13 receptor subunit alpha 1 (IL13RA1) showed broader expression across human sensory neuron subtypes than in mice, where Il4r was not detected. Other genes such as IL-6 cytokine family signal transducer (IL6ST) showed conserved expression in humans and mice.
We examined expression of other gene families across human and mouse neuronal subtypes including: ASICs (fig. S12), anoctamins (fig. S13), aquaporins (fig. S14), calcium channels (fig. S15), chloride channels (fig. S16), cholinergic receptors (fig. S17), ionotropic GABA receptors (fig. S18), gap-junction/connexins (fig. S19), ionotropic glutamate receptors (fig. S20), glycine receptors (fig. S21), neuropeptide genes (fig. S22), potassium channels (fig. S23), ionotropic purinergic receptors (fig. S24), transient receptor potential channels (fig. S25), and transcription factors involved in neuronal differentiation (fig. S26). We also looked at the expression of genes that encode for proteins that are part of the understudied druggable genome (figs. S27 to S29) (72). We detected 56 understudied GPCRs (out of 117) (fig. S27), 49 understudied ion channels (out of 62) (fig. S28), and 133 kinases (out of 150) (fig. S29) in human DRG neuronal subtypes. Last, we created an expression map of the genes with lowest normalized entropy representing genes with the greatest variance of expression across neuronal subtypes (fig. S30).
Comparison of human and nonhuman primate sensory neuron subtypes
Next, we took advantage of a recently published single-cell dataset from nonhuman primate DRG to compare neuronal subtypes between human and macaque. Distinct orthology was identified between human and macaque subpopulations, but several orthologs had a many-to-one mapping (for example, between A-LTMR in rhesus and Aδ LTMR and Aβ RA LTMR in humans), suggesting that some of the macaque subpopulations could be further subdivided into distinct populations. The subpopulation orthology and corresponding gene expression clusters are shown in Fig. 6 (A to C). Comparison of human and mouse neuronal subpopulation transcriptomes shows many important changes in neuronal DRG expression (8, 48, 73), which are consistent with macaque and mouse differences (23). On the basis of analysis of the most lineage-restricted human DRG genes in Fig. 6, expression enrichment was found to be broadly conserved in humans and macaque, but regulatory divergences in some important sensory genes were also observed.
Fig. 6. Orthologous neuronal populations between human and macaque.
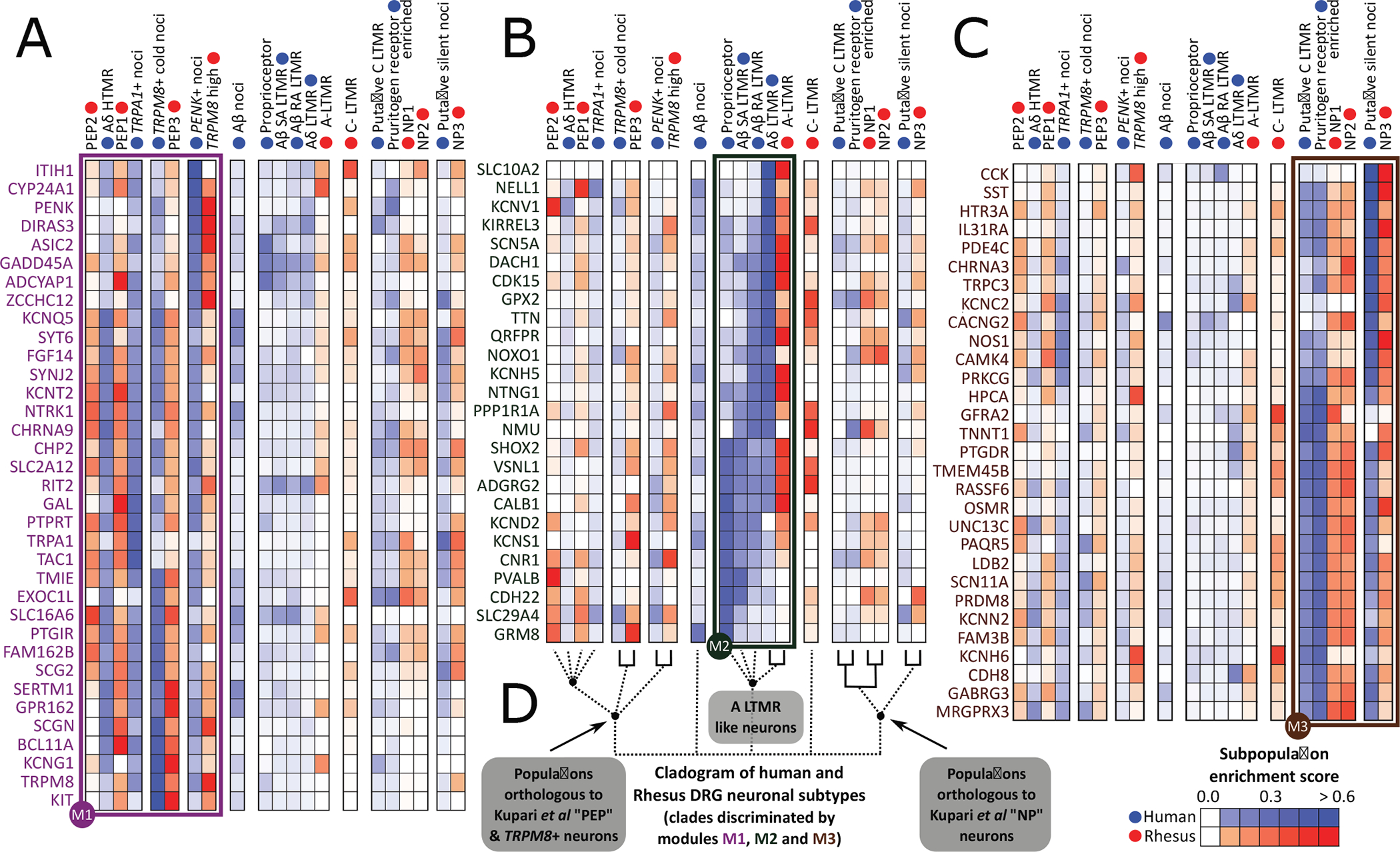
(A to C) Gene modules showing expression patterns of lineage-restricted human DRG neuronal genes that have high dynamic range of expression in macaque DRG neuronal populations (from Smart-seq2). (D) Orthology among neuronal populations based on the hDRG lineage-restricted genes, with strong orthologies indicated with solid lines.
PVALB gene expression in humans was enriched in proprioceptors and Aβ SA LTMRs but not in the corresponding macaque LTMR populations (Fig. 6B). Instead, it was enriched in macaque peptidergic PEP2 population, which is transcriptionally closer to the human Aδ HTMRs and TRPA1+ nociceptors where PVALB is de-enriched. This species difference is particularly important because Pvalb is a marker of A-fiber LTMR neurons in rodents (74). The neuronal calcium sensor HPCA (hippocalcin) was enriched in human pruritogen receptor–enriched, silent nociceptors and putative C-LTMRs, and their macaque orthologs (nonpeptidergic subpopulations; Fig. 6C). However, in macaques, it was additionally enriched in TRPM8+ and PEP1 subpopulations but de-enriched in the human orthologs of these populations. Last, at the population level, macaque C-LTMRs showed enrichment for some human pruritogen receptor–enriched neuron and putative C-LTMR genes [GFRA2, potassium voltage-gated channel subfamily H member 6 (KCNH6), and transmembrane protein 45B (TMEM45B)], as well as enrichment for some human A-LTMR genes [neuromedin U (NMU), glutathione peroxidase 2 (GPX2), and kirre-like nephrin family adhesion molecule 3 (KIRREL3)] (Fig. 6, B and C). Human putative C-LTMRs, on the other hand, were de-enriched for all of the analyzed human A-LTMR genes. This suggests that macaque C-LTMRs are transcriptionally divergent from all identified human subpopulations, including the human A-LTMRs and putative C-LTMR populations. Markers for Aβ nociceptors are not enriched in specific macaque populations and, hence, are not shown (along with some additional markers for human putative C-LTMRs and human pruritogen receptor enriched neurons) in Fig. 6. The complete data for all 111 analyzed gene markers can be found in data file S30.
DISCUSSION
Our work demonstrates that spatial transcriptomics can be used to generate near single-neuron resolution to define molecular profiles of neuronal subtypes in the human DRG. Our findings demonstrate not only many similarities but also substantial differences between mice, where most single-nociceptor transcriptome work has been done (15, 16, 18), and humans. Some of these differences may be explained by technical issues related to sequencing methods; however, our demonstration of more consistent similarities between macaque and human, where different sequencing techniques were also applied (23), makes this possibility less likely.
An important outcome of our experiments is the ability to now directly assess target expression across species with single-neuron resolution. We lay out these expression profiles for most pharmacologically relevant targets in mouse and human DRGs. This expression map can allow investigators to initiate DRG-focused target identification efforts with human neuronal transcriptome insight and then make data-driven choices about model species and testing paradigms that best fit the chosen development pipeline.
An area where evolutionary divergence between mouse and human sensory neurons is most substantial is in neuropeptide, TRPV1 and NTRK1 expression. This is in line with previous in situ hybridization work (8, 75). In mice and rats, these genes are developmentally regulated with expression in all nociceptors in early development and then silencing in specific populations after postnatal target innervation (17, 76, 77). In contrast to rodents, most human nociceptors share expression of these genes suggesting a blending of many of the markers of peptidergic and nonpeptidergic nociceptors that are found in other species, in particular, the mouse. This indicates that the peptidergic and nonpeptidergic nomenclature is unlikely to have utility for describing human nociceptors (7, 8, 48, 75). Our findings suggest that development programs that silence TPRV1 and NTRK1 expression in subgroups of nociceptive sensory neurons are not engaged in humans.
We found important differences in receptor and neuropeptide expression in the pruritogen receptor–enriched population with more widespread expression of many markers that have been identified in mouse, particularly into the silent nociceptor and putative C-LTMR subtypes. This is consistent with previous studies showing species differences in expression of pruritogen receptor genes such as the IL-31 receptor (71). It is also consistent with a recent study comparing macaque and human DRG expression of pruritogen receptors [MAS-related GPR family member D (MRGRPD) and MRGPRX1 that demonstrated coexpression with TRPV1] (78). This contrasts with mouse experiments where Mrgprd is expressed by a subset of neurons that are devoid of Trpv1 (79). Klein and colleagues (78) also demonstrated that histamine creates a greater area of flare and wheal in human volunteers than MRGPRD or MRGPRX1 agonists. This finding is explained by the expansion of neuronal populations that express HRH1, the H1 histamine receptor, in human DRG. We do not find expression for most known markers of C-LTMRs in human DRG such as TH using both Visium and RNAscope. This also includes a lack of markers that were recently identified in a nuclear sequencing study from macaques (23), making this population challenging to identify in our study. The exception was GFRA2+ expression, which marks C-LTMRs across species (16, 23), enabling putative identification of C-LTMRs in human DRG. This subpopulation was validated with RNAscope (GFRA2+/NPPB−).
Our spatial transcriptomic characterization of human DRG neuronal subtypes should facilitate discoveries in the pain and sensory neuroscience field. One advance is the identification of sets of markers that can be used to molecularly phenotype subtypes of sensory neurons that can be sampled through skin biopsies and other methods from neuropathy patients. Although there are clear indications of pathology in sensory neurons indicated from clinical skin biopsy studies, these are almost always grouped into small and large fiber neuropathies, but further distinctions are not made. Our work enables greater mechanistic insight from routine clinical tests. The finding that neuronal transcriptomes in the DRG are stable unless frank axonal injury has occurred (18) suggests that our dataset can be used for this purpose almost immediately. Our dataset can also be used to mine for pharmacological targets that can be used to specifically manipulate the excitability of different subsets of nociceptors. This offers the possibility for the development of pain targets that are identified based entirely on human transcriptomic data. Our dataset contains both male and female samples. We highlight sex differences (for example, greater CGRP expression in the pruritogen receptor–enriched population in females) that may be important considerations for therapeutic development. Last, this dataset can be a foundation to more thoroughly vet targets that have been found in studies of peripheral nerves in animal pain models. Our findings might make it possible for conservation of gene expression in human nociceptors to be a first step in derisking pain targets for future drug development (4).
The study has some limitations
The most important limitation is that the spatial transcriptomic approach only approximates single neuron transcriptomes in the human DRG. We did not use a single-cell sequencing approach but instead used 10x Genomics Visium spatial transcriptomics. The neuronal barcodes used for downstream analyses were selected on the basis of their spatial location (that is, overlapping a single neuron). Combining our findings with nuclear sequencing will improve resolution. Human DRG neurons are too large for many of the standard current single-cell technologies, but single-nucleus sequencing can give single-cell resolution and has recently been used on human DRG (80). As more such studies emerge, they can be combined with spatial transcriptomics, which also provides profiling of cytoplasmic RNAs, to complement one another and provide a comprehensive picture of human DRG neuronal transcriptomes. In relation to this point, while this paper was in revision, a new single-nucleus sequencing study of the human DRG was published (80). This study identifies neuronal subtypes in human DRG that are similar to ours and also identifies species differences in comparison to mice that parallel our findings. One area of divergence is in the identification of C-LTMRs. It is likely that human psychophysical and pharmacological studies will be required to fully delineate the molecular phenotype of these neurons. Because we have sampled from eight organ donors, our data cannot account for possible difference in gene expression across the human population or at earlier stages of life. Future studies can use our foundation to address these important questions. Last, as we have mentioned throughout the manuscript, species comparisons rely on different sequencing techniques and different postmortem intervals, so this should be considered in interpreting the data reported here.
MATERIALS AND METHODS
Study design
The main goal of this study was to molecularly characterize human DRG sensory neurons. For this purpose, human lumbar DRG tissues were collected from organ donors within 4 hours of cross-clamp and from neurologic determination of death donors. Donor information is provided in table S1. Samples were randomly selected on the basis of sex and age from our internal tissue bank. All human tissue procurement procedures were approved by the Institutional Review Boards at the University of Texas at Dallas. To achieve near single-neuron resolution, we used the 10x Visium Spatial Transcriptomics technology. The DRG tissues were sectioned onto Visium slides, stained, and imaged. We performed tissue permeabilization, which caused the mRNAs to bind to the barcoded oligo primers, and the remainder of the protocol was followed according to the manufacturer’s instructions. Samples were sequenced in a NextSeq500. After selecting barcodes that overlapped single neurons, the data were processed using Seurat’s integration workflow, followed by graph-based clustering. Neuronal barcodes with low counts (<2000) and low expression of neuronal marker SNAP25 were not included. The main markers for each cluster were validated using RNAscope. We used DRGs collected from four males and four females and looked for sex differences. We also looked for species differences by comparing our data with publicly available datasets of mice and macaques. Sample sizes were determined on the basis of the number of neuronal barcodes necessary for unsupervised clustering workflow. A formal pre hoc power analysis was not possible because no human DRG spatial transcriptomes were available for estimation of within and between group gene expression variabilities between sensory neuronal subtypes. We used at least two donors in RNAscope analyses as previously reported (8), which allowed us to sample over 300 neurons. Further experiment details on donor and neuron counts from each RNAscope experiment can be found in table S2, and biological replicates and/or technical replicates are noted here in Materials and Methods. Experiments were done under blinded conditions except where noted. Details on blinding for image analysis are provided here in Materials and Methods.
Tissue preparation
DRGs used for Visium and RNAScope were frozen on dry ice at the time of extraction and stored in a −80°C freezer. The human DRGs were gradually embedded in optimal cutting temperature (OCT) in a cryomold by adding small volumes of OCT over dry ice to avoid thawing. DRGs used for Visium were cryosectioned onto SuperFrost Plus charged slides at 10 μm. To sample a larger subset of neurons, two sections were used from each donor, and each 10-μm section was 200 μm apart. DRGs used for RNAscope were sectioned at 20 μm. DRGs used for primary neuronal cultures were placed in artificial cerebral spinal fluid over ice at the time of surgical extraction and transported immediately to the University of Texas at Dallas for processing.
Visium spatial gene expression
Visium tissue optimization and spatial gene expression protocols were followed exactly as described by 10x Genomics (https://10xgenomics.com/) using hematoxylin and eosin (H&E) as the counterstain. Optimal permeabilization time was obtained at 12-min incubation with permeabilization enzyme. Imaging was conducted on an Olympus vs120 slide scanner. DRGs from donors 1 to 8 were used. mRNA library preparation and sequencing (Illumina Nextseq 500) were done at the Genome Center in the University of Texas at Dallas Research Core Facilities.
Visium spatial RNA-seq: Mapping raw counts and alignment of barcoded spots with imaged sections
The output data of each sequencing run (Illumina BCL files) were processed using the Space Ranger (v1.1) pipelines provided by 10x Genomics. Samples were demultiplexed into FASTQ files using Space Ranger’s mkfastq pipeline. Space Ranger’s count pipeline was used to align FASTQ files with bright-field microscope images previously acquired, detect barcode/unique molecular identifier (UMI) counting, and map reads to the human reference transcriptome (Gencode v.27 and GRCh38.p10) (81). This pipeline generates, for each sample, feature-barcode matrices that contain raw counts and places barcoded spots in spatial context on the slide image (cloupe files). Gene expression with spatial context can then be visualized by loading cloupe files onto the Loupe Browser (v4.2.0, 10x Genomics). The space ranger output statistics for raw data can be found in data file S25.
RNAscope in situ hybridization
RNAscope in situ hybridization multiplex v1 and v2 were performed as instructed by Advanced Cell Diagnostics (ACD) and as previously described (8). A table of all probes, combinations, and donor tissues used is shown in table S2. All tissues were checked for RNA quality using a positive control probe cocktail (ACD), which contains probes for high-, medium-, and low-expressing mRNAs that are present in all cells (ubiquitin C > peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). A negative control probe against the bacterial DapB gene (ACD) was used to reference nonspecific/background label. The donor number for the DRGs that were used in each experiment is indicated in table S2.
RNAscope imaging and analysis
DRG sections were imaged on an Olympus FV3000 confocal microscope at ×20 or ×40 magnification. The acquisition parameters were set on the basis of guidelines for the FV3000 provided by Olympus. In particular, the gain was kept at the default setting 1, high voltage ≤ 600, offset = 4, and laser power ≤ 20%. Large globular structures and/or signal that autofluoresced at the 488, 550, and 647 wavelengths (appears white in the overlay images) was considered to be background lipofuscin and was not analyzed. Aside from adjusting brightness/contrast, we performed no digital image processing to subtract background. We have previously attempted to optimize automated imaging analysis tools for our purposes, but these tools were designed to work with fresh, low-background rodent tissues, not human samples taken from older organ donors. Hence, we chose to implement a manual approach in our imaging analysis in which we used our own judgment of the negative/positive controls and target images to assess mRNA label.
For the RNAscope experiments, the same analysis procedure was conducted as previously described (8). Two to three ×20 images were acquired of each human DRG section, and three sections were imaged per human donor. The raw image files were brightened and contrasted in Olympus CellSens software (v1.18) and then analyzed manually one neuron at a time for expression of each mRNA. Images were not analyzed in a blinded fashion. Cell diameters were measured using the polyline tool. Total neuron counts were acquired by counting all probe-labeled neurons and all neurons that were clearly outlined by 4′,6-diamidino-2-phenylindole (DAPI) (satellite cell) signal and contained lipofuscin in the overlay image. For each section, the neuronal counts for each target/subpopulation from all images were summed, and the population percentages were calculated for that section. The population percentages from three sections were then averaged to yield the final population value for each donor. Pie charts represent the average of all of three donors. The total number of neurons assessed is indicated in the figure captions and represents the sum of all neurons analyzed between all three donors. Because TRPV1 and SCN10A signal was assessed independently in three different experiments (in combination with NTRK1, NTRK2, and NTRK3), the data from all three experiments were combined. For those cases, if the same donor was used in each experiment, then their population values were averaged. The same procedure was applied to CALCA, which was used multiple times (in combination with PRDM12, SST, and NPPB).
For the RNAscope experiment shown in Fig. 4 (CALCA/NPPB in males versus females), ~10 NPPB-positive neurons from each section were imaged at ×40 magnification, and three sections were imaged per donor (totaling ~30 neurons per donor). The ×40 images were then cropped to show only a single NPPB-positive neuron, and the file names were blinded by a nonaffiliated person. The blinded experimenter brightened and contrasted the images in Olympus CellSens and then drew regions of interest (ROIs) around the soma (not to include the larger mass of lipofuscin) (fig. S9). The area of the CALCA mRNA signal within the ROI was analyzed using the Count and Measure tool, which highlights the mRNA puncta using a thresholded detection. Because RNAscope fluorescence intensity reflects the number of probe pairs bound to each molecule, a manual threshold was applied to each image so that all mRNA signals were highlighted within the ROI. Because each mRNA puncta in the ACD protocol averages 1.5 μm2, the CALCA area measurements were divided by 1.5 to determine the number of puncta and then divided by the area of the ROI to yield CALCA mRNA puncta per square micrometer. Given the low expression and detection of NPPB in the human DRG, we combined all the data from all donors (five males and three females), which is reflected in the graph. Graphs were generated using GraphPad Prism version 8 (GraphPad Software Inc. San Diego, CA, USA). A relative frequency distribution histogram with a fitted Gaussian distribution curve was generated using the diameters of all mRNA-positive neurons detected in all experiments. Images in the figures are pseudo-colored. The raw values for all image analyses can be found in data file S31.
Human DRG cultures and electrophysiology
Human DRG cultures were prepared as described (82) from three donors (donors 2, 6, and 7). All electrophysiology experiments were performed between days in vitro (DIV)5 and DIV7. Experiments were performed using a MultiClamp 700B (Molecular Devices) patch-clamp amplifier and PClamp 9 acquisition software (Molecular Devices) at room temperature. Recordings were sampled at 20 kHz and filtered at 3 kHz (Digidata 1550B, Molecular Devices). Pipettes (outer diameter, 1.5 mm; inner diameter, 1.1 mm; BF150–110-10, Sutter Instruments) were pulled using a PC-100 puller (Narishige) and heat-polished to 2- to 3-megohm resistance using a microforge (MF-83, Narishige). Series resistance was typically 5 megohm and was compensated up to 70%. Data were analyzed using Clampfit 10 (Molecular Devices). All neurons included in the analysis had a resting membrane potential (RMP) more negative than −40 mV. In current-clamp mode, cells were held at RMP for the duration of the experiment. The pipette solution contained the following: 120 mM K-gluconate, 6 mM KCl, 4 mM adenosine 5′-triphosphate–Mg, 0.3 mM guanosine 5′-triphosphate–Na, 0.1 mM EGTA, 10 mM Hepes, and 10 mM phosphocreatine (pH 7.2) (adjusted with N-methyl glucamine), and osmolarity was ~290 mOsm. The external solution contained the following: 135 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM KCl, 10 mM glucose, and 10 mM Hepes (pH 7.4) (adjusted with N-methyl glucamine), and osmolarity was adjusted to ~315 mOsm with sucrose. Cells were dialyzed for 3 to 5 min after break-in with the internal solution before commencing recordings. The cells were continuously perfused with the external solution using a ValveLink 8 perfusion system. Stock solution of capsaicin was diluted to 200 nM in the external solution and applied directly to the patched neuron using the perfusion system. After 10 s of baseline recording of spontaneous activity in current clamp mode, capsaicin was applied for 10 s to test for depolarization of the neuron or action potential generation. The raw values plotted in Fig. 3K can be found in data file S31.
Selection of neuronal barcodes in Loupe Browser
We manually selected all barcoded spots that overlapped neurons in the Loupe Browser (v4.2.0, 10x Genomics) and exported as csv files for each sample. Surrounding barcodes were computationally obtained on the basis of neuronal barcode’s coordinates. To avoid duplicates and keep data consistent, barcodes could only have one classification. For instance, if a surrounding barcode was also overlapping a neuron, it was removed from the surrounding barcodes. By exclusion, barcodes that were not labeled neuronal or were not directly surrounding a barcode were labeled as “other barcodes.” For downstream analysis, we used neuronal barcodes that overlapped only single neurons. We observed that 3.19% of these neuronal barcodes overlapped the same neuron, with 0.66% being assigned to different clusters. After determination of neuronal clusters, all H&E images of each donor section were loaded into the Loupe Browser, and the barcodes for the identified neuronal clusters in that section were remapped for visualization purposes. An image of the overlaid barcodes on the tissue section was saved and then stacked with the high-resolution H&E image in CellSens. Each neuron and barcode was visualized on the Loupe Browser image, and then the same neuron was found by toggling to the high-resolution image. Neuronal diameters for neurons with visible nuclei were then measured using the polyline tool.
Human and mouse neuronal comparison
We compared our Visium DRG neuronal subtypes directly with mouse neuronal subtypes. We used publicly available data from mousebrain.org (16). We examined expression of individual genes within gene families that are involved in transduction of nociceptive signals by nociceptors and are considered important pharmacological targets for existing or potential drugs: VGNaCs, GPCRs, ILs and their receptors, ASICs, anoctamins, aquaporins, calcium channels, chloride channels, cholinergic receptors, ionotropic GABA receptors, gap-junction/connexins, ionotropic glutamate receptors, glycine receptors, neuropeptide genes, potassium channels, ionotropic purinergic receptors, transient receptor potential channels, and transcription factors involved in neuronal differentiation. We also looked at the expression of genes that encode for proteins that are part of the understudied druggable genome (72). We also created an expression map of the genes with the lowest normalized entropy representing genes with the greatest variance of expression across neuronal subtypes. Normalized entropy was calculated for all genes in the human data and is represented in all figures. Normalized entropy was calculated using the scipy.stats.entropy function. As we mentioned throughout the manuscript, different sequencing approaches were used for human and mouse data, so that should be considered when interpreting the data.
Human and macaque transcriptome comparison
On the basis of comparative transcriptomic analysis of mouse and human DRG RNA-seq, we previously found that neuronal subtype-restricted genes were likely to be conserved in expression in the human DRG bulk RNA-seq data (5). Hence, as a starting point for analysis of conservation of lineage-restricted gene expression across subpopulations in human and macaque DRGs, we first identified the top 555 neuronal lineage-restricted coding genes in the hDRG (genes in the lowest 5% of normalized entropy signifying tissue restricted expression; data file S30) of 11,117 medium or high expression genes in the Visium dataset (read count ≥ 3 in one or more cells). Gene expression in the macaque orthologs in Smart-seq2 assay was obtained from literature (23), but many of these genes have low dynamic range (abundance between 0 and 0.1 across subpopulations) likely due to the nature of the Smart-seq assay. A total of 111 lineage-restricted human DRG genes with higher dynamic range in macaque Smart-seq data were used to perform clustering of human and macaque subpopulations (based on expression enrichment scores in subpopulations for these genes), followed by clustering of the genes based on their gene expression patterns to assess conservation of lineage-restricted gene expression patterns in the two species. Of these, 91 genes in three modules are shown in Fig. 6.
Statistical analysis
Visium spatial RNA-seq analysis
Raw count data for the selected neuronal barcodes were obtained from the respective feature-barcode matrices. We used Python (v3.7 with Anaconda distribution), R (v4.0.3), and Seurat (v3.2.2) for data analysis. Before initiating Seurat clustering workflow, data were cleaned by removing barcodes with low counts (<2000). We verified that selected neuronal barcodes that had no expression (count < 1) of the neuronal marker SNAP25 had minimal overlap with neurons, and for that reason, they were also excluded from downstream analysis. The remaining 3952 neuronal barcodes, grouped by donor ID, created a total of eight Seurat objects. The standard Seurat integration workflow was followed (29). This integration workflow can reduce batch effects by identifying pairwise correspondences (named “anchors”) between single barcodes across samples. First, each object was normalized and identified the 2000 most variable features. After identifying anchors, the data were integrated, generating one combined Seurat object. Next, the data were scaled, and the combined Seurat object was further processed following the standard Seurat clustering workflow. Clustering and visualization were performed using Uniform Manifold Approximation and Projection (UMAP) as the dimensionality reduction algorithm. After the first round of clustering, some clusters had these nonneuronal genes as cluster markers: apolipoprotein D (APOD), metallothionein 3 (MT3), myelin protein zero (MPZ), complexin 1 (CPLX1), SPARC like 1 (SPARCL1), interferon alpha inducible protein 27 (IFI27), creatine kinase B (CKB), biliverdin reductase B (BLVRB), secreted phosphoprotein 1 (SPP1), visinin like 1 (VSNL1), C-type lectin domain family 2 member L (CLEC2L), cell cycle exit and neuronal differentiation 1 (CEND1), trans-2,3-enoyl-coa reductase (TECR), heat shock protein, alpha-crystallin–related B6 (HSPB6), small nuclear ribonucleoprotein U11/U12 subunit 25 (SNRNP25), synuclein beta (SNCB), family with sequence similarity 57 member B (FAM57B), adenosine triphosphatase NA+/K+-transporting subunit alpha 3 (ATP1A3), N-acetyltransferase 8 like (NAT8L), matrix Gla protein (MGP), transgelin (TAGLN), dexi homolog (DEXI), fatty acid binding protein 7 (FABP7), TIMP metallopeptidase inhibitor 1 (TIMP1), CD74 molecule (CD74), and vimentin (VIM). These genes were influencing the clustering, and to overcome this, we scored the nonneuronal signal using the “AddModuleScore” function. This function scores the difference between the average expression of each gene set and randomly selected control gene set, across the neuronal barcodes (83). The barcodes were reclustered (resolution = 1), and the nonneuronal signal was regressed out (vars.to.regress = nn_score1). We identified markers for each cluster using the Wilcoxon rank-sum test integrated in Seurat. Some pairs of clusters had a set of neuronal markers that were unique with respect to all the other clusters but were shared between the two of them. Therefore, clusters without clear distinct neuronal markers were merged to generate the final clusters (fig. S3).
Differential expression analysis
To identify neuron-specific sex differences, we conducted differential expression analysis in neuronal barcodes. Because our neuronal barcodes may contain signal originating in the surrounding cells, we performed statistical analysis for the surrounding barcodes. Figure S9A shows the number of neuronal barcodes and surrounding barcodes used for statistical analysis. We combined barcode counts to generate a pseudo-bulk sample for each neuronal cluster, respective surrounding barcodes, and overall neuronal and overall surrounding barcodes. This approach ensures that statistical hypothesis testing is applicable to the tested population of barcodes and not subject to sampling variance within the large number of individual barcodes in each population. In addition, any effect of varying amounts of neuronal mRNA proportion across spots also gets homogenized by such pooling. Genes with less than 10 reads were excluded from each combined sample and removed from downstream analysis. Each dataset was then analyzed using DESeq2 (84), which normalized the raw gene counts (gene counts are divided by barcode-specific normalization factors that are calculated on the basis of the median ratio of gene counts relative to geometric mean per gene) and corrected for batch effect, followed by testing for differential abundance. We performed differential expression analysis using the “DESeq” function (this function performs differential expression analysis based on the negative binominal distribution and Wald statistics). Nominal P values were corrected for multiple testing using the Benjamini-Hochberg (BH) method (85). In addition, we performed shrinkage of the log2 FC (LFC) estimates to generate more accurate LFC. We used the adaptive shrinkage estimator from the “ashr” R package (86) and set the contrast to male versus female as the groups we wanted to compare. Genes were considered to be DE if FC ≥ 1.33 and adjusted P ≤ 0.05. Because mRNA profiles in each spot are admixture of multiple cell types, we considered a gene to be specifically DE in neuronal barcodes if it was not DE in surrounding barcodes. Statistical hypothesis testing results for all tests can be found in data files S2 and 23. For each gene tested, we report baseMean (mean of normalized counts), LFC, lfcSE (standard error of the LFC estimate), P value (Wald test P value), and Padj (BH-adjusted P values). NA represents missing values.
Supplementary Material
Acknowledgments:
We thank the organ donors and their families for enduring gift. We thank E. Vines and the staff at the Southwest Transplant Alliance for coordinating DRG recovery from organ donation surgeries. We also thank members of the Price laboratory, M. Kume, M. Saad Yousuf, A. Balmain, and J. Mwirigi for assistance in DRG recovery. We thank the Genome Center at The University of Texas at Dallas for the services to support our research. We thank M. Chavez and J. Kuate Fotso for creating the website for public access and visualization of our processed data.
Funding:
This work was supported by NIH grants NS111929 to P.M.D. and T.J.P., NS065926 to G.D. and T.J.P., and NS042595 to R.W.G..
Footnotes
Competing interests: D.T.-F. has an equity position in Doloromics. P.R.R. is a cofounder of Doloromics. A.W. is a cofounder of Doloromics. A.C. consults for Recens. P.M.D. consults for Grunenthal as well as Toray, Wex, and OliPass. R.W.G. is a cofounder and part owner of Neruolux. G.D. is a cofounder of Doloromics and Scientific Lead for Schedule 1 Therapeutics and consults for Abbvie. T.J.P. is a cofounder and has an equity position in Doloromics, 4E Therapeutics, and PARMedics and consults for Grunenthal. The authors declare that they have no other competing interest.
Data and materials availability: All data associated with this study are in the paper or the Supplementary Materials. Raw sequencing data are available in the dbGaP repository (phs001158). Public access to processed data is available at sensoryomics.com. All analyses scripts are available at https://github.com/utdal/, and the Loupe Browser files are available at sensoryomics.com.
SUPPLEMENTARY MATERIALS
View/request a protocol for this paper from Bio-protocol.
REFERENCES AND NOTES
- 1.Scholz J, Woolf CJ, Can we conquer pain? Nat. Neurosci. 5 (Suppl), 1062–1067 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Edvinsson L, Haanes KA, Warfvinge K, Krause DN, CGRP as the target of new migraine therapies—Successful translation from bench to clinic. Nat. Rev. Neurol. 14, 338–350 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mogil JS, The translatability of pain across species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renthal W, Chamessian A, Curatolo M, Davidson S, Burton M, Dib-Hajj S, Dougherty PM, Ebert AD, Gereau R. W. t., Ghetti A, Gold MS, Hoben G, Menichella DM, Mercier P, Ray WZ, Salvemini D, Seal RP, Waxman S, Woolf CJ, Stucky CL, Price TJ, Human cells and networks of pain: Transforming pain target identification and therapeutic development. Neuron 109, 1426–1429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ, Comparative transcriptome profiling of the human and mouse dorsal root ganglia: An RNA-seq-based resource for pain and sensory neuroscience research. Pain 159, 1325–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wangzhou A, McIlvried LA, Paige C, Barragan-Iglesias P, Shiers S, Ahmad A, Guzman CA, Dussor G, Ray PR, R. W. t. Gereau, T. J. Price, Pharmacological target-focused transcriptomic analysis of native vs cultured human and mouse dorsal root ganglia. Pain 161, 1497–1517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middleton SJ, Barry AM, Comini M, Li Y, Ray PR, Shiers S, Themistocleous AC, Uhelski ML, Yang W, Dougherty PM, Price TJ, Bennett DL, Studying human nociceptors: From fundamentals to the clinic. Brain 144, 1312–1335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiers S, Klein RM, Price TJ, Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain 161, 2410–2424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wangzhou A, Paige C, Ray PR, Dussor G, Price TJ, Diversity of receptor expression in central and peripheral mouse neurons estimated from single cell RNA sequencing. Neuroscience 463, 86–96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woolf CJ, Ma Q, Nociceptors–Noxious stimulus detectors. Neuron 55, 353–364 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Dubin AE, Patapoutian A, Nociceptors: The sensors of the pain pathway. J. Clin. Invest. 120, 3760–3772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wangzhou A, Paige C, Neerukonda SV, Naik DK, Kume M, David ET, Dussor G, Ray PR, Price TJ, A ligand-receptor interactome platform for discovery of pain mechanisms and therapeutic targets. Sci. Signal. 14, eabe1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, Dussor G, Kim TH, Price TJ, Dougherty PM, Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 142, 1215–1226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P, Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, Ginty DD, Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron 103, 598–616.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S, Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, Ginty DD, The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renthal W, Tochitsky I, Yang L, Cheng YC, Li E, Kawaguchi R, Geschwind DH, Woolf CJ, Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron 108, 128–144.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen MQ, Le Pichon CE, Ryba N, Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. eLife 8, e4979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark R, Grzelak M, Hadfield J, RNA sequencing: The teenage years. Nat. Rev. Genet. 20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Haberberger RV, Barry C, Dominguez N, Matusica D, Human dorsal root ganglia. Front. Cell. Neurosci. 13, 271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz C, Carlen M, Meletis K, Spatial transcriptomics: Molecular maps of the mammalian brain. Annu. Rev. Neurosci. 44, 547–562 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Kupari J, Usoskin D, Parisien M, Lou D, Hu Y, Fatt M, Lonnerberg P, Spangberg M, Eriksson B, Barkas N, Kharchenko PV, Lore K, Khoury S, Diatchenko L, Ernfors P, Single cell transcriptomics of primate sensory neurons identifies cell types associated with chronic pain. Nat. Commun. 12, 1510 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shansky RM, Murphy AZ, Considering sex as a biological variable will require a global shift in science culture. Nat. Neurosci. 24, 457–464 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Rosen S, Ham B, Mogil JS, Sex differences in neuroimmunity and pain. J. Neurosci. Res. 95, 500–508 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J, Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Rao N, Clark S, Habern O, Bridging genomics and tissue pathology: 10x Genomics explores new frontiers with the Visium Spatial Gene Expression solution. Genet. Eng. Biotechnol. News 40, 50–51 (2020). [Google Scholar]
- 28.Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL, Tran MN, Besich Z, Tippani M, Chew J, Yin Y, Kleinman JE, Hyde TM, Rao N, Hicks SC, Martinowich K, Jaffe AE, Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raible DW, Ungos JM, in Neural Crest Induction and Differentiation (Springer, 2006), pp. 170–180. [Google Scholar]
- 31.Abraira VE, Ginty DD, The sensory neurons of touch. Neuron 79, 618–639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmerman A, Bai L, Ginty DD, The gentle touch receptors of mammalian skin. Science 346, 950–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dionisi C, Rai M, Chazalon M, Schiffmann SN, Pandolfo M, Primary proprioceptive neurons from human induced pluripotent stem cells: A cell model for afferent ataxias. Sci. Rep. 10, 7752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gau P, Curtright A, Condon L, Raible DW, Dhaka A, An ancient neurotrophin receptor code; a single Runx/Cbfβ complex determines somatosensory neuron fate specification in zebrafish. PLOS Genet. 13, e1006884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson RS, Vallbo AB, Tactile sensibility in the human hand: Relative and absolute densities of four types of mechanoreceptive units in glabrous skin. J. Physiol. 286, 283–300 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng C, Furlan A, Zhang M-D, Su J, Lübke M, Lönnerberg P, Abdo H, Sontheimer J, Sundström E, Ernfors P, Termination of cell-type specification gene programs by the miR-183 cluster determines the population sizes of low-threshold mechanosensitive neurons. Development 145, dev165613 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Olson W, Dong P, Fleming M, Luo W, The specification and wiring of mammalian cutaneous low-threshold mechanoreceptors. Wiley Interdiscip. Rev. Dev. Biol. 5, 389–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhandapani R, Arokiaraj CM, Taberner FJ, Pacifico P, Raja S, Nocchi L, Portulano C, Franciosa F, Maffei M, Hussain AF, de Castro Reis F, Reymond L, Perlas E, Garcovich S, Barth S, Johnsson K, Lechner SG, Heppenstall PA, Control of mechanical pain hypersensitivity in mice through ligand-targeted photoablation of TrkB-positive sensory neurons. Nat. Commun. 9, 1640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adriaensen H, Gybels J, Handwerker H, Van Hees J, Response properties of thin myelinated (A-delta) fibers in human skin nerves. J. Neurophysiol. 49, 111–122 (1983). [DOI] [PubMed] [Google Scholar]
- 40.Akopian AN, Sivilotti L, Wood JN, A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379, 257–262 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Djouhri L, Lawson SN, Aβ-fiber nociceptive primary afferent neurons: A review of incidence and properties in relation to other afferent A-fiber neurons in mammals. Brain Res. Rev. 46, 131–145 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Treede R-D, Meyer RA, Campbell JN, Myelinated mechanically insensitive afferents from monkey hairy skin: Heat-response properties. J. Neurophysiol. 80, 1082–1093 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Nagi SS, Marshall AG, Makdani A, Jarocka E, Liljencrantz J, Ridderström M, Shaikh S, O’Neill F, Saade D, Donkervoort S, Reghan Foley A, Minde J, Trulsson M, Cole J, Bönnemann CG, Chesler AT, Catherine Bushnell M, Glone FM, Olausson H, An ultrafast system for signaling mechanical pain in human skin. Sci. Adv. 5, eaaw1297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKemy DD, Neuhausser WM, Julius D, Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature 416, 52–58 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Przewŀocki R, in Hormones, Brain and Behavior (Elsevier, 2002), pp. 691–733. [Google Scholar]
- 46.Natura G, Bar KJ, Eitner A, Boettger MK, Richter F, Hensellek S, Ebersberger A, Leuchtweis J, Maruyama T, Hofmann GO, Halbhuber KJ, Schaible HG, Neuronal prostaglandin E2 receptor subtype EP3 mediates antinociception during inflammation. Proc. Natl. Acad. Sci. U.S.A. 110, 13648–13653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nassini R, Materazzi S, Benemei S, Geppetti P, in Reviews of Physiology, Biochemistry and Pharmacology, vol. 167 (Springer, 2014), pp. 1–43. [DOI] [PubMed] [Google Scholar]
- 48.Shiers SI, Sankaranarayanan I, Jeevakumar V, Cervantes A, Reese JC, Price TJ, Convergence of peptidergic and non-peptidergic protein markers in the human dorsal root ganglion and spinal dorsal horn. J. Comp. Neurol. 529, 2771–2788 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prato V, Taberner FJ, Hockley JRF, Callejo G, Arcourt A, Tazir B, Hammer L, Schad P, Heppenstall PA, Smith ES, Functional and molecular characterization of mechanoinsensitive “silent” nociceptors. Cell Rep. 21, 3102–3115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michaelis M, Häbler HJ, Jäenig W, Silent afferents: A separate class of primary afferents? Clin. Exp. Pharmacol. Physiol. 23, 99–105 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Ochoa JL, Campero M, Serra J, Bostock H, Hyperexcitable polymodal and insensitive nociceptors in painful human neuropathy. Muscle Nerve 32, 459–472 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Serra J, Campero M, Bostock H, Ochoa J, Two types of C nociceptors in human skin and their behavior in areas of capsaicin-induced secondary hyperalgesia. J. Neurophysiol. 91, 2770–2781 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Namer B, Carr R, Johanek LM, Schmelz M, Handwerker HO, Ringkamp M, Separate peripheral pathways for pruritus in man. J. Neurophysiol. 100, 2062–2069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmelz M, Schmidt R, Weidner C, Hilliges M, Torebjork HE, Handwerker HO, Chemical response pattern of different classes of C-nociceptors to pruritogens and algogens. J. Neurophysiol. 89, 2441–2448 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Mishra SK, Hoon MA, The cells and circuitry for itch responses in mice. Science 340, 968–971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salvatierra J, Diaz-Bustamante M, Meixiong J, Tierney E, Dong X, Bosmans F, A disease mutation reveals a role for NaV1.9 in acute itch. J. Clin. Invest. 128, 5434–5447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebbinghaus M, Tuchscherr L, Segond von Banchet G, Liebmann L, Adams V, Gajda M, Hübner CA, Kurth I, Schaible H-G, Gain-of-function mutation in SCN11A causes itch and affects neurogenic inflammation and muscle function in Scn11a+/L799P mice. PLOS ONE 15, e0237101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woods CG, Babiker MOE, Horrocks I, Tolmie J, Kurth I, The phenotype of congenital insensitivity to pain due to the NaV1.9 variant p. L811P. Eur. J. Hum. Genet. 23, 561–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harper AA, Lawson SN, Electrical properties of rat dorsal root ganglion neurones with different peripheral nerve conduction velocities. J. Physiol. 359, 47–63 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper AA, Lawson SN, Conduction velocity is related to morphological cell type in rat dorsal root ganglion neurones. J. Physiol. 359, 31–46 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price TJ, Flores CM, Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J. Pain 8, 263–272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y-C, Auer-Grumbach M, Matsukawa S, Zitzelsberger M, Themistocleous AC, Strom TM, Samara C, Moore AW, Cho LT, Young GT, Weiss C, Schabhuttl M, Stucka R, Schmid AB, Parman Y, Graul-Neumann L, Heinritz W, Passarge E, Watson RM, Hertz JM, Moog U, Baumgartner M, Valente EM, Pereira D, Restrepo CM, Katona I, Dusl M, Stendel C, Wieland T, Stafford F, Reimann F, von Au K, Finke C, Willems PJ, Nahorski MS, Shaikh SS, Carvalho OP, Nicholas AK, Karbani G, McAleer MA, Cilio MR, McHugh JC, Murphy SM, Irvine AD, Jensen UB, Windhager R, Weis J, Bergmann C, Rautenstrauss B, Baets J, De Jonghe P, Reilly MM, Kropatsch R, Kurth I, Chrast R, Michiue T, Bennett DL, Woods CG, Senderek J, Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 47, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toth CC, Willis D, Twiss JL, Walsh S, Martinez JA, Liu WQ, Midha R, Zochodne DW, Locally synthesized calcitonin gene-related Peptide has a critical role in peripheral nerve regeneration. J. Neuropathol. Exp. Neurol. 68, 326–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith-Anttila CJA, Mason EA, Wells CA, Aronow BJ, Osborne PB, Keast JR, Identification of a sacral, visceral sensory transcriptome in embryonic and adult mice. eNeuro 7, ENEURO.0397–ENEU19.2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD, PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bennett DL, Clark AJ, Huang J, Waxman SG, Dib-Hajj SD, The role of voltage-gated sodium channels in pain signaling. Physiol. Rev. 99, 1079–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Barbosa C, Tan ZY, Wang R, Xie W, Strong JA, Patel RR, Vasko MR, Zhang JM, Cummins TR, Navβ4 regulates fast resurgent sodium currents and excitability in sensory neurons. Mol. Pain 11, 10.0.4.162/s12990-015-0063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Theile JW, Cummins TR, Inhibition of Navβ4 peptide-mediated resurgent sodium currents in Nav1.7 channels by carbamazepine, riluzole, and anandamide. Mol. Pharmacol. 80, 724–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao Y, Barbosa C, Pei Z, Xie W, Strong JA, Zhang JM, Cummins TR, Increased resurgent sodium currents in Nav1.8 contribute to nociceptive sensory neuron hyperexcitability associated with peripheral neuropathies. J. Neurosci. 39, 1539–1550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uchida H, Nagai J, Ueda H, Lysophosphatidic acid and its receptors LPA1 and LPA3 mediate paclitaxel-induced neuropathic pain in mice. Mol. Pain 10, 71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cevikbas F, Wang X, Akiyama T, Kempkes C, Savinko T, Antal A, Kukova G, Buhl T, Ikoma A, Buddenkotte J, Soumelis V, Feld M, Alenius H, Dillon SR, Carstens E, Homey B, Basbaum A, Steinhoff M, A sensory neuron-expressed IL-31 receptor mediates T helper cell–dependent itch: Involvement of TRPV1 and TRPA1. J. Allergy Clin. Immunol. 133, 448–460.e7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheils TK, Mathias SL, Kelleher KJ, Siramshetty VB, Nguyen DT, Bologa CG, Jensen LJ, Vidovic D, Koleti A, Schurer SC, Waller A, Yang JJ, Holmes J, Bocci G, Southall N, Dharkar P, Mathe E, Simeonov A, Oprea TI, TCRD and Pharos 2021: Mining the human proteome for disease biology. Nucleic Acids Res. 49, D1334–D1346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moy JK, Hartung JE, Duque MG, Friedman R, Nagarajan V, Loeza-Alcocer E, Koerber HR, Christoph T, Schroder W, Gold MS, Distribution of functional opioid receptors in human dorsal root ganglion neurons. Pain 161, 1636–1649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ichikawa H, Deguchi T, Nakago T, Jacobowitz DM, Sugimoto T, Parvalbumin, calretinin and carbonic anhydrase in the trigeminal and spinal primary neurons of the rat. Brain Res. 655, 241–245 (1994). [DOI] [PubMed] [Google Scholar]
- 75.Rostock C, Schrenk-Siemens K, Pohle J, Siemens J, Human vs. mouse nociceptors—Similarities and differences. Neuroscience 387, 13–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molliver DC, Wright DE, Leitner ML, Parsadanian AS, Doster K, Wen D, Yan Q, Snider WD, IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron 19, 849–861 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Guo A, Simone DA, Stone LS, Fairbanks CA, Wang J, Elde R, Developmental shift of vanilloid receptor 1 (VR1) terminals into deeper regions of the superficial dorsal horn: Correlation with a shift from TrkA to Ret expression by dorsal root ganglion neurons. Eur. J. Neurosci. 14, 293–304 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Klein A, Solinski HJ, Malewicz NM, Ieong HF, Sypek EI, Shimada SG, Hartke TV, Wooten M, Wu G, Dong X, Hoon MA, LaMotte RH, Ringkamp M, Pruriception and neuronal coding in nociceptor subtypes in human and nonhuman primates. eLife 10, e64506 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zylka MJ, Rice FL, Anderson DJ, Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 45, 17–25 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Nguyen MQ, von Buchholtz LJ, Reker AN, Ryba NJ, Davidson S, Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. eLife 10, e71752 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Sala SC, Chrast J, Cunningham F, Domenico TD, Donaldson S, Fiddes IT, Girón CG, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valtcheva MV, Copits BA, Davidson S, Sheahan TD, Pullen MY, McCall JG, Dikranian K, Gereau IV RW, Surgical extraction of human dorsal root ganglia from organ donors and preparation of primary sensory neuron cultures. Nat. Protoc. 11, 1877–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin J-R, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CGK, Kazer SW, Gaillard A, Kolb KE, Villani A-C, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jané-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA, Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benjamini Y, Hochberg Y, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995). [Google Scholar]
- 86.Stephens M, False discovery rates: A new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


