Abstract
Approximately 2.3 million people are suffering from human immunodeficiency virus (HIV)/hepatitis C virus (HCV) co-infection worldwide. Faster disease progression and increased mortality rates during the HIV/HCV co-infection have become global health concerns. Effective therapeutics against co-infection and complete infection eradication has become a mandatory requirement. The study of small non-coding RNAs in cellular processes and viral infection has so far been beneficial in various terms. Currently, microRNAs are an influential candidate for disease diagnosis and treatment. Dysregulation in miRNA expression can lead to unfavorable outcomes; hence, this exact inevitable nature has made various studies a focal point. A considerable improvement in comprehending HIV and HCV mono-infection pathogenesis is seen using miRNAs. The prominent reason behind HIV/HCV co-infection is seen to be their standard route of transmission, while some pieces of evidence also suspect viral interplay between having a role in increased viral infection. This review highlights the involvement of microRNAs in HIV/HCV co-infection, along with their contribution in HIV mono- and HCV mono-infection. We also discuss miRNAs that carry the potentiality of becoming a biomarker for viral infection and early disease progression.
Keywords: co-infection, HCV, HIV, infectious disease, microRNA
Introduction
In 1983, a French virologist Luc Montagnier and his team first reported about a virus, which later became identified as human immunodeficiency virus type 1 or HIV-1 in the field of medical virology. 1 Just after which another two groups of researchers collected enough evidence, from isolated samples, that claimed HIV to be the causative reason behind the acquired immunodeficiency syndrome, acronym as AIDS.2,3 Characteristics like consisting of two positive strands of RNA encoding for numerous proteins and viral microRNAs (vmiRNAs) make HIV stand out among the other viruses. Researchers have, so far, discovered five HIV-1 microRNAs and 15 proteins, which facilitate various viral processes via direct or indirect mechanisms. 4 The envelope protein of HIV-1, gp120, carries out efficient binding with the CD4 receptor present on several cells. This leads to a conformational change that in turn allows the viral protein to attach itself with the present co-receptors further. Usually, chemokine receptors like CCR5 and CXCR4 act as co-receptors. A reverse transcriptional process on the HIV-1 genome, followed by its entry, results in double-stranded DNA formation. This DNA is then imported into the nucleus and integrated within the host cell genome via multiple intertwined mechanisms. As the name suggests, HIV infection leads to slow and irreparable exhaustion of immune CD4+ T cells, hence opening a door for any kind of opportunistic disease to enter and infect the human body. Initially, in the process of finding a promising treatment for HIV, scientists faced a lot of challenges, but currently, combined antiretroviral therapy (cART) is giving out positive outcomes to an extent. However, complete elimination of HIV-1 is something yet to achieve.
On the contrary, HCV is also a blood-borne virus, like HIV, belonging to the family Flaviviridae and genus hepacivirus. HCV is one of the significant causes of liver disease across the globe. WHO states that nearly 37 million and 70 million people worldwide annually gets affected by HIV and HCV, respectively. It consists of a single-stranded and positive-sense RNA containing 5′ and 3′ UTR. The absence of a 5′ cap and polyA tail makes the genome of HCV significantly different, generally seen to follow the lytic cycle for proliferation. 5 The genome, when processed, synthesizes a 3010 amino acid long polyprotein. When further cleaved, it results in three structural (core, E1, and E2) and seven non-structural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins. 5 Core protein helps in the formations of the capsid, which also contains glycoproteins E1 and E2. The internalization of HCV is mediated by receptors present in host cells. A dedicated structure named membranous web is utilized for cytoplasmic HCV replication. 6 Being a hepatotropic virus, HCV causes significant damage to the liver by altering its basic cellular processes. Many researchers have claimed miRNAs (microRNAs) to have an essential role in viral–host interaction.5,7 Although the first- and second-generation protease and polymerase inhibitors Boceprevir, Telaprevir, and Sofosbuvir were approved by FDA as the direct-acting antiviral agents, no vaccine to date is present against HCV infection. The reason is its nature of being immensely diverse. It is found to develop more than 30% divergence, at the amino acid level, between 7 main HCV genotypes. 8 Reports say nearly 20% of patients infected with HCV can successfully clear out the virus, but the majority of patients end up with severe complications, over the years, like cirrhosis, fibrosis, or hepatocellular carcinoma (HCC), leading them to liver failure and death. 9
Both HIV and HCV are infectious and can spread through blood, hence sharing a common transmission route. Usage of contaminated needles, syringes, or drug injection equipment is always seen to be the primary reason behind getting an infection of HCV or HIV. This leads to a higher chance of getting infected by HIV and HCV co-infection. Currently, available data show that about 36.7 million people are HIV infected globally, among which 2.3 million reported having HCV infection, past or present. Interestingly, 1.36 million co-infection patients were ones who were treated with intravenous drugs. 10 The natural history of both the viruses significantly alters during the co-infection. While the HCV viral RNA count, liver fibrosis, and other clinical features of hepatic dysfunction develop more rapidly in HIV-infected patients, the disease progression toward AIDS also observed much faster in HIV/HCV co-infection than in HIV alone. 11
MicroRNAs: the game-changer in viral pathogenesis
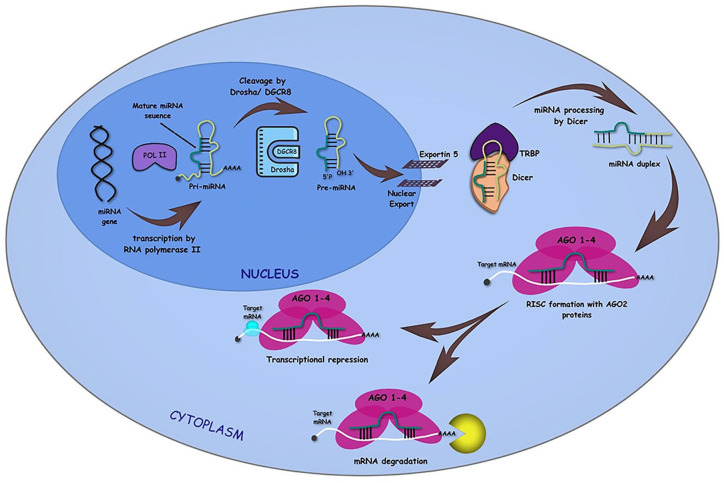
Till now, about 3000 miRNAs have been identified in mammalian genome. 12 microRNAs are 18–22 nucleotide long, non-coding RNAs. They play a vital role in different cellular activities like proliferation, apoptosis, and differentiation by controlling gene expression. They are highly conserved in human genomes. When required, RNA polymerase II transcripts form primary miRNAs (pri-miRNAs). This Pri-miRNA consists of 5′-end cap and poly-A tail, further refined by ribonuclease Drosha to form precursor miRNA (pre-miRNA). This hairpin structured Pre-miRNA is then transported to the cytoplasm with the help of Exportin-5. There it is cleaved by Dicer to form mature miRNA (Figure 1). 13 Although miRNAs form not more than 1% of the human genome but are capable of targeting approximately 60% of mRNA present in the genome. 13 miRNA recognizes the target mRNA via forming an RNA-inducing silencing complex (RISC). RISC consists of mature miRNA associated with Argonaute/EIF2 C (AGO) and GW182 family proteins. 14 miRNA transcription is monitored by many different cellular factors. Transcription factors like p53, MYC, ZEB1/2, and myoblast determination proteins 1 also play a role in regulating miRNA transcription. Along with which, epigenetic factors like DNA methylation and histone modification are also a part of miRNA regulation. 5 miRNA regulates gene expression either by mRNA degradation or by translational repression. It induces de-adenylation, which leads to mRNA breakage. It also hinders protein synthesis via translation repression at the site of cap recognition or by initiating before-time removal of ribosomes.13,14 It targets mRNA via complementary base-pairs between 5′end of miRNA and 3′ UTR of mRNA. Hence, the second to seventh nucleotide section at 5′end of miRNA is called ‘miRNA Seed’. This is a crucial region for target recognition. Every miRNA is seen to have many different targets, ones that share almost identical sequence at their 5′end belong to one miRNA seed family. Therefore, different miRNAs from the same family share common mRNA as targets. 14 Similarly, even though miRNAs of the miR-199 family are located on different genes, they share the same seed region. As an outcome, they all share a common target, and their upregulation in HCV infection is seen to promote fibrosis. 15 Thus, one mRNA can be monitored by many different miRNAs; this connective nature of miRNA hence demonstrates the idea of cells’ vast regulatory network. Virus–host interaction also involves specific steps altering cellular processes. One of which is to hijack and modulate cellular miRNA expression. They conduct this either by impairing miRNA pathways via interference with cellular proteins or producing their own miRNAs to promote viral mRNA expression.16,17 This ability of viral RNA to efficiently hijack and modify miRNA expression favors viral infectivity and protects against immune responses.5,7,18 They share an intertwined complex relationship. For instance, human Rhinovirus (RV) is responsible for causing respiratory infections. Bioinformatic analysis has proven miR-128 and miR-155 to have a role against RV infection. 19 They are reported to contribute to the innate immune response in various ways and inhibit RV infection. At the same time, upregulation of let-7c in A549 cells promotes replication of type A influenza virus by suppressing M1 protein. 19 Many studies have been done on possible involvement of miRNAs in SARS-CoV infection. 20 Upregulation of three miRNAs, miR-17, miR-574-5p, and miR-214, in infected BASCs showed antiviral activities. On the contrary, downregulation of miR-223 and miR-98 by SARS-CoV proteins was also observed. 21 Finally, the question is, can the viral genome also encode for viral miRNAs? Researchers have found that many viruses like herpesviruses, Heliothis virescens ascovirus (HvAV), and simian virus 40 (SV40) or the virus families like herpesvirus, polyomavirus, papillomavirus, and retroviruses encode their own miRNAs. Nearly 530 viral miRNAs or vmiRNAs have been identified so far. However, HCV miRNAs are currently unestablished.22,23 In this context, it would be noteworthy that HIV-1 has been shown to encode at least six viral miRNAs to date, but the controversies still persist over the expression of these viral miRNAs. 24
Figure 1.
The Biogenesis of microRNAs by Canonical Pathway: Maturation of miRNA sequence by transcription of primary miRNAs or Pri-miRNAs from the miRNA gene by RNA polymerase II followed by the cleavage of Pri-miRNAs to precursor or Pre-miRNAs through Drosha/DGCR8. The Pre-miRNAs are then exported from nucleus to cytoplasm with the help of Exportin 5. In the cytoplasm, Dicer cleavage of the hairpin structure takes place, where one of the strands of the miRNA duplex is loaded onto the RISC via the Argonaut or Ago proteins. Finally, the RISC-loaded miRNAs regulate the repression of gene expression by base complementarity inducing transcriptional repression or mRNA degradation.
miRNAs in HIV lifecycle
HIV replication
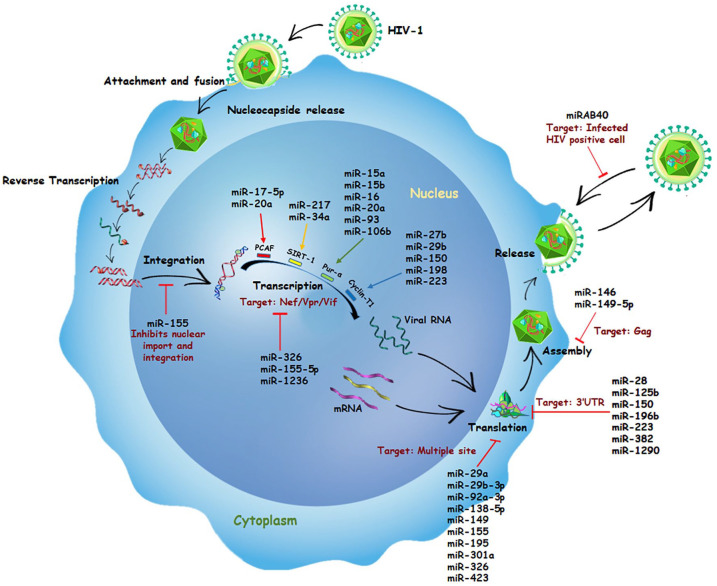
For decades, HIV has successfully kept scientists busy with their research on finding its every possible cure. One of the properties that make HIV stand out is its ability to encode its own microRNAs. One such viral miR, hiv1-miR-H1, is seen to promote HIV replication in macrophages successfully. 25 It is found near NF-κB sites in the LTR region. It enhances the infection and cell damage by targeting apoptosis antagonizing transcription factor (AATF), leading to apoptosis. It also targets a few cellular proteins like c-Myc and Dicer. 25 Likewise, the viral genome also encodes for transacting responsive hairpin (TAR) consisting of a terminal repeat region. After asymmetrical slicing by Dicer, the miRNAs produced from these regions are claimed to be distinctively crucial because of their varied role in the HIV life cycle. 26 They participate in various processes like dimerization, replication, transcription, and translation. In 2008, researchers first established the location of HIV-1 TAR to be present at 5′ end in a stem-loop structure. 26 The same group later established another unique characteristic of TAR miRs, where they use AGO proteins to manipulate cellular mRNA expression. 27 In addition to other studies, bioinformatics showed the higher possibility of TAR having complementary sites within genes of apoptosis-like Caspase 8, Aiolos, and Nucleophosmin (NPM)/B23. In conclusion, HIV-1 TAR is completely capable of causing an imbalance between apoptosis and cell viability in T cells by influencing gene expression. 27 Some studies also claim its presence in monocytes-derived macrophages, responsible for monitoring viral replication rate via regulation of negative regulatory factor. 28 Alongside, many cellular miRNAs also play a vital role in either enhancing or inhibiting viral replication (Figure 2). 25 One of them is miR-32, whose role is to cause decrement in the expression of tumor necrosis factor-receptor-associated factor 3 (TRAF3). Hence when upregulated by HIV TAT protein, it leads to overexpression of interferon regulatory factors 3 and 7, opening the door for HIV-1 to affect innate immune response. 25 Similarly, two cellular miRNAs, miR-217 and miR-34a, activated by TAT, tend to repress Sirtuin 1, which is known to cleave HIV-1 LTR, hence favoring viral transactivation. 25 Some cellular miRNAs often favor viral infection by targeting host cellular proteins and mRNA expression. One such example can be seen when miR-132 interacts with methyl-CpG-binding protein 2 (MeCP2), a transcriptional repressor, and enhances viral proliferation. 25
Figure 2.
Role of miRNAs in HIV replication: The figure describes the mode of action of various cellular and viral miRNAs that regulate the HIV replication. While the cellular miRNAs, miR-28, miR-29, miR-92a, miR-125, miR-138, miR-146, miR-149, miR-150, miR-155, miR-195, miR-196b, miR-223, miR-301, miR-326, miR-382, miR-423, miR-1236, and miR-1290, and the viral miRNA, miRAB40 block the HIV replication, the cellular miRNAs, miR-15, miR-16, miR-17, miR-20a, miR-27b, miR-34a, miR-93, miR-106b, miR-198, and miR-217 are reported to be promoting the viral replication. Both the viral and the cellular miRNAs regulate the host and the viral gene expression or HIV-positive cells to modify the replication process either as a host response trying to fight the infection or to block or establish, or both, the infection within the host at different stages of virus lifecycle.
HIV latency
The period in HIV infection, about 6 weeks later, with no signs and symptoms is also called asymptomatic HIV infection. Here, the virus is still active and replicating at a meager rate, yet fully capable of progressing the infection to its 3rd stage, that is, AIDS. These latent proviruses manage to escape immune responses, and even current ART treatments fail to target them. Like any other process, this also employs various viral and cellular miRNAs. Numerous studies investigated miRNA differences between elite controllers and chronic HIV samples in order to target essential miRNAs for diagnostic or therapeutic uses. One such study found concentrated amounts of miR-125 and miR-150 present in cells undergoing latent infection, possibly because of their notable role as HIV protein suppressors. 29 At the same time, another study showed contrary results where they found these same miRNAs to be downregulated in HIV elite controllers compared with chronically infected samples. 30 A similar study done by Reynoso R. revealed increased expression of miR-146a-5p, miR-33a-5p, and miR-29b-3p in those HIV-positive patients who controlled viral replication in the absence of ART. 31 A lot of miRNAs like miR-28, miR-223, miR-382, miR-125b, and miR-150 exhibit their role in maintaining latency period, while other studies have also added another two miRNAs to that list, miR-196b and miR-1290.29,32 These miRNAs simply bind to 3′ UTR of HIV mRNAs and contribute to HIV latency.29,32 Furthermore, a recent study showed the contribution of host miRNAs in dysregulation of the p53 signaling pathway during HIV infection that drives latent infection. 33 Along with cellular miRNAs, HIV encoded viral miR-TAR, miR-N367, and miR-H1 have also established their roles in HIV latency, and on the contrary, miR-H3 has proved its role in activation of the latent reservoirs. 34 miR-N367 is located within Nef gene region in the HIV genome. 35 Nef protein is responsible for the easy production and release of virions from the infected cell. 25 Hence, its degradation led by hiv1-miR-N367 helps in viral latency. 25 The presence of such transcriptionally silent virus in resting CD4 + cells increases the chances of reoccurrences. Hence, other than ART treatments, it has become essential to target factors like miRNAs that are responsible for viral latency. This new therapeutic approach might help us find a definite cure for HIV.
miRNAs directly target HIV-1 genome
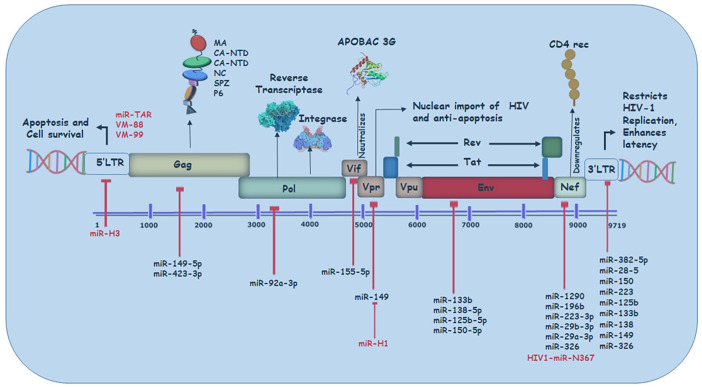
As previously discussed, miRNAs regulate viral processes either by interfering with cellular mechanisms or by directly targeting the viral genome (Figure 3).16–18 Likewise other viral proteins, HIV Vpr protein is crucial in promoting viral infection via different mechanisms. 36 However, miR-149 is seen to target and repress Vpr protein expression. Interestingly, miR-149 is found to be, in turn, targeted by HIV-miR-H1 in order to facilitate viral infection. 25 Similarly, another HIV structural polyprotein Gag is targeted by host miR-423 and miR-146a. 37 Here, miR-146a is observed to inhibit the process of Gag-multimerization successfully. This way, it hinders virion production and budding. 37 Many studies have discovered various miRNAs that carry complementary sites with Nef gene region in the HIV genome. miRNAs like miR-29a and miR-29b-3p are found to have a common target, that is, Nef.25,31 In comparison, miR-423 and miR-301a bind with Vif region and interfere with the viral process, 25 though exact mechanisms of these microRNAs and the effect of their interactions are not fully discovered. Furthermore, Zhang et al. have reported the existence of a novel HIV-1-encoded viral miRNA called miR-H3 located in the region of the HIV-1 RNA genome that encodes for reverse transcriptase enzyme. The miR-H3 targets the TATA box in HIV-1 5′ LTR region to upregulate the promoter activity. It represents another HIV-1-encoded element, in addition to TAR, that activates viral transcription via cis regulation targeting viral genome. 38
Figure 3.
miRNAs targeting HIV genome: Various miRNAs have been reported to interact directly with the HIV genome; however, they impede viral replication upon binding. The binding of those cellular (black) and viral (red) miRNAs within the 5′ or 3′ LTR or the HIV ORF has been depicted along with their respective viral gene targets.
miRNAs in HIV antiviral activities
Given the fact that miRNAs are an intrigued part of cellular mechanisms, hence, viruses tend to hijack and use them for their benefit. 16 However, some miRNAs are found to exhibit antiviral properties by interfering with several pathways inhibiting protein expression or by targeting HIV dependency factors (HDFs).37,39 Multiple studies demonstrated how the dysregulation of miR-7-3p, miRNAs-30, miRNA-125b, miRNA-150, miR-186, miR-210, and miR-222 has a negative impact on viral infection.40–43 These miRNAs were found to target genes of Dicer, NTNG1, EFNB2, CXCL12, or HRB (HIV-1 Rev-binding protein), and HIV-EP2 (HIV-1 enhancer-binding protein 2).40,42 The downregulation of these genes eventually leads to reduce viral expression.40–42 Few other microRNAs exhibiting antiviral activities are listed here (Table 1).25–27,29,31,35,38,44–51
Table 1.
miRNAs exhibiting anti-HIV-1 activity.
| microRNA | Mechanism | Outcome |
|---|---|---|
| Cellular microRNAs | ||
| miR-155 44 | Targets various HDFs: ADAM metallopeptidase domain 10 (ADAM10) Nucleoporin Mr 153,000 (Nup153) Transportin 3 (TNPO3) Lens epithelium derived growth factor (LEDGF/p75) |
Repression of all these HDFs hinders HIV-1 import and integration |
| miR-326 25 | Dicer–dependent | Restricts viral replication |
| miR-126 45 | Regulates expression of TLR9 and TLR7 Regulates pDC homeostasis and survival |
Helps in type 1 IFN-mediated innate immune response |
| miR- 20a 46 miR-17-5p 46 | P300/CBP-associated factor (PCAF) | Hinders Tat-mediated LTR activation and inhibits viral replication |
| miR-29 25,31 | Targets HIV Nef gene | Interfere with viral replication |
| miR-149 25 | Targets HIV Vpr protein | Inhibiting viral infection |
| miR-1236 47 | Targets Vpr-binding protein | Inhibits Vpr-mediated cell cycle arrest and viral replication |
| miR-198 48 miR-27b 48 | Targets a Tat cofactor, Cyclin-T1 | Inhibits HIV-1 via repression of cyclin-T1 |
| miR-191-5p 49 | Targets and inhibits expression of CCR1 and NUP50 | Inhibits viral replication via target repression. |
| miR-28 29,50 miR-125b 29,50 miR-150 29,50 miR-223 29,50 miR-382-5p 29,50 | Target the 3′ UTR of HIV-1 mRNA | Decreases CD4 + T-cell activation |
| vmiRNAs | ||
| vmiR88 51 vmiR99 51 | Mediates TNF-α release | Facilitates immune activation |
| miR-H1 25 | Targets AATF, c-Myc and Dicer | Promotes HIV replication |
| miR-TAR 26,27 | Targets AGO proteins, Caspase 8, Aiolos, and NPM/B23 | Manipulate cellular mRNA expression and imbalance between apoptosis and cell viability |
| miR-H3 38 | Targets TATA box of HIV-1 5′ LTR region | Activates viral transcription |
| miR-N367 25,35 | Degradation of HIV Nef protein | Maintenance of viral latency |
pDC, plasmacytoid dendritic cells.
In addition, there are some other miRNAs, which show antiviral properties by sharing a common target that is Purine-rich element-binding protein alpha. This protein plays a vital role in Tat-mediated LTR activation and hence in viral replication. It has been observed to be targeted by nearly six miRNAs, that is, miR-15a, miR-15b, miR-16, miR-20a, miR-93, and miR-106b. 52
HCV infection and role of miRNAs
HCV entry
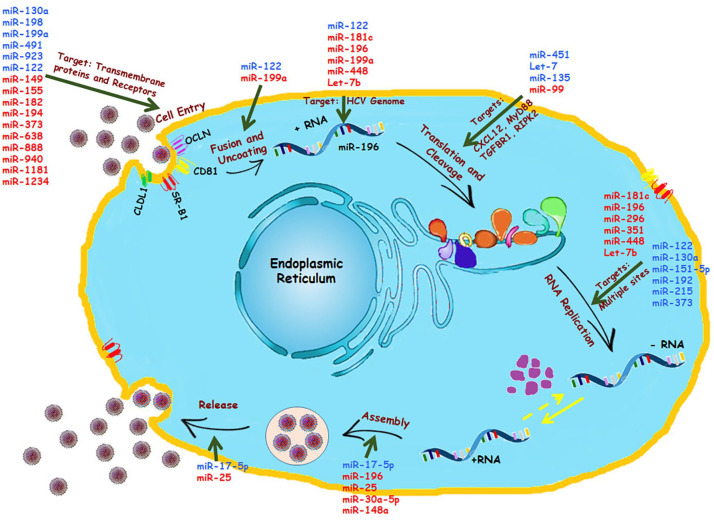
HCV tropism is generally found in human and chimpanzee liver cells, allowing the researchers to be suspicious about various intracellular factors favoring HCV entry and infectivity. Few studies had shown the importance of SCARB1, CD81, claudin-1 (CLDN1), and occludin (OCLN) in facilitating viral entry. 53 This was again proven when scientists were successful in creating a mouse model for HCV infection. 54 These factors allowed the HCV to enter, yet no sustained chronic infection was observed later. On the contrary, when scientists allowed miR-122 to be abnormally over-expressed in mouse embryonic fibroblasts, it showed increased subgenomic HCV replication (Figure 4). 55 In contrast, another study unveiled an antiviral property of miR-122 due to its binding efficiency with OCLN. Over-expression of mimic miR-122 resulted in downregulation of OCLN protein by ~80%. Hence, a notable decrease in expression of OCLN in chronic HCV patients can be blamed upon inhibiting the property of miR-122. 56 Along with miR-122, miR-200c is also observed to efficiently target 3′ UTR of OCLN and repress its activity. 57 However, different miRNAs exhibit different effects on HCV entry receptors like CLDN1. The role of miR-155 and miR-182 showed conflicting results in terms of interaction with claudin-1 (Figure 4). miR-182 is claimed to inhibit CLDN-1 mRNA expression in the infected cell line, hampering HCV infection. 58 Nevertheless, a positive correlation between miR-155 expression and CLDN1 is seen in diseases like colorectal cancer. 59 The role of their interaction in HCV infection requires further research. A study on 28 HCV liver biopsies and bioinformatic analysis revealed the inhibitory effect of miR-194 on the CD81 receptor. Successful treatment of miR-194 mimics in different HCV-infected cell lines resulted in decreased expression of CD81 protein (Figure 4). 60 CD81 is an essential tool during the process of HCV viral entry and infection; hence, its inhibition by miR-194 might open up new therapeutic approaches in HCV treatment. In addition, epigallocatechin gallate (EGCG), one of the components found in green tea extract, is known for its antiviral effects against HCV. 61 Along with its other activities, EGCG hinders HCV infection by repressing CD81 expression with the help of miR-194.60,61 Similarly, EGCG is also found to promote expressions of other miRNAs like miR-548a. A recent publication stated that miR-548a consists of 2 binding sites on CD81, providing it a tool to inhibit protein expression. Hence, ECGC and miR-548a can be linked with the host cellular defense mechanism. 62 HCV infection also enhances miR-130a expression, which in turn inhibits endogenous interferon-induced transmembrane protein 1 (IFITM1) expression in a hepatoma cell line (Figure 4). 63 However, on the other side, enhanced expression of the same miR-130a was observed as a result of IFN treatment and claimed for restoring the act of IFN α/β. 64 Future research is needed to study the contradictory nature of this miRNA.
Figure 4.
Cellular miRNAs controlling HCV lifecycle: Several miRNAs have been identified for controlling HCV replication and pathogenesis, as illustrated. These miRNAs are known for their targets in the various phases of viral lifecycles: Cell entry, Fusion and Uncoating of viral RNA, Translation and Cleavage of the viral polyprotein, RNA replication, Assembly of the viral genome, and Release of progeny viruses. The upregulated miRNAs are shown in blue, and the downregulated miRNAs are shown in red.
HCV replication
Involvement of various cellular miRNAs in viral replication has always been a field of interest for researchers (Figure 4). Utilizing Bioinformatics to study and identify different miRNAs and their role in HCV infection has also been proven beneficial. With the help of this resource, scientists identified miR-199a-3p to have complimentary seed sequences with domain II of HCV internal ribosomal entry site, which is conserved among all HCV genotypes. Hence, its expression can possibly suppress HCV replication. 65 Similarly, binding sites on NS5B and 5′ UTR of HCV genome for let7-b were also established. This allowed let7-b to act against HCV replication and restrict its accumulation. 66 Ectopic expression of miR-196 is linked with decreased viral load with two possible explanations (1) by targeting Bach1 (2) by targeting NS5a region of HCV genome. 67 Interestingly, Bach1 is believed to be responsible for HMOX-1 downregulation by managing its enhancer availability. 67 Heme oxygenase 1 is a cytoprotective protein with antioxidant properties. It is seen to be underexpressed in patients with chronic HCV infection. However, it has an important role against the infection as its overexpression leads to a subsequent decrease in viral replication.5,67 Similarly, another miRNA induced by IFN, miR-448, targets core sequences of the viral genome and hence holds a negative correlation with viral replication in the cell. 67 In addition, miR-181c levels are observed to be inversely proportional to the proliferation rate of HCV due to its compatibility with E1 and NS5a regions. It inhibits the infection by altering CCAAT/ enhancer-binding protein β (C/EBPβ). 68
miR-122: protagonist or antagonist?
microRNAs are seen to participate in viral infections via direct and indirect interactions. Some have complementary sequences to those of viral genome, giving it the advantage of either enhancing or inhibiting the expression of viral RNA via direct interaction, while some miRNAs have an indirect effect on viral RNA by controlling host factors necessary for viral infection. HCV is seen to have its most effect on hepatic cells where miR-122 is dominantly expressed. It nearly occupies 70% of the miRNA population, making its involvement in HCV infection quite obvious. The other way around, it can also be one of the responsible intracellular factors behind the prominent nature of HCV in liver cells. Usually, decrement of miR-122 in liver cells results in alterations within cholesterol biosynthesis. It is also seen to have an inverse effect on fatty acid oxidation. 69 The relationship between miR-122 and HCV was first studied in 2005. Studies revealed two binding sites for miR-122 on 5′ UTR of HCV genome. Unlikely binding of 3′ miRNA nucleotides, along with the 5′ region, helps in stabilizing the viral genome inside the host cell. This nucleotide annealing resulted in 3′ overhangs covering the 5′ end of viral RNA and protecting it from degradation. 70 This mechanism then helps miR-122 to promote viral replication and translation in the host cell. Another mechanism miR-122 might use to protect the viral genome is targeting exonuclease Xrn1. 71 miRNAs form RISC and AGO proteins to carry out translational repression. Now, as miR-122 is involved in HCV RNA enhancement, it made researchers question the role of AGO2 proteins in this process. AGO2 proteins turned out to be one of the requirements, as an apparent reduction in HCV levels was seen after exhaustion of AGO2 from infected cells. 72 Later, it was also observed that the cells lacking miR-122 also showed successful replication of HCV, thus questioning the role of miR-122 in HCV replication. Hence to answer this question, researchers took non-hepatic cells (lacks miR-122), infecting them with HCV along with miR-122 being exogenously expressed. They ended up concluding that miR-122 might not be entirely essential but does play a supporting role in viral infection as a considerable increment in HCV replication was found in cells.71,73 Another evidence supporting this theory was obtained when exogenous expression of miR-122 was also conducted in a non-permissive cell line, and the results turned out to be no different. It showed successful infectious virion production and replication.71,74 Some studies also suggest miR-122 has a triphasic relationship with viral RNA levels of HCV infection. Researchers observed an increment in miR-122 levels during acute HCV infection (first four weeks). Later, an inverse correlation was observed between viral RNA and miR-122 in the following 10–14 weeks and eventually rise in miR-122 levels during HCV clearance and alanine leucine transaminase (ALT) normalization. 75 Not only this but also miR-122 is seen to cause an increment in oxidative stress indirectly; hence, in another study, expression levels of heme oxygenase-1 gene, Bach1 and miR-122 were compared in liver biopsy samples of patients suffering from CHC. The results revealed the expression of HMOX 1 to be directly proportional to miR-122 due to antioxidant response against excess ROS production. 76 The indulgence of miR-122 with HCV translation and infection was clearly visible through all such different experiments. Hence, many researches could correlate miR-122 levels with disease progression in patients. Serum level of miR-122 and miR-21 is claimed to be possible biomarker to detect the presence of CHC infection due to their correlation with increased necro inflammatory response.71,77 However, the specificity of miR-122 for HCV-mediated liver disease (like fibrosis) is controversial.
Can dysregulation in miRNAs alter crucial pathways for HCV infection?
Some miRNAs particularly target host cellular pathways to enhance HCV replication and infection. One such pathway that plays a role in cell proliferation, survival, and growth is PI3k/Akt pathway, targeted by miR-491, miR-320c, and miR-483-3p. Dysregulation of these miRNAs helps elude the immune system and HCV enhancement. 78 IFN system and innate immune response go hand in hand, and miRNAs are also seen to be an essential part of this circle. One of the mechanisms cell uses to fight viral infection involves type-1 interferons, and HCV is seen to successfully defeat this mechanism by using several miRNAs (Table 2).5,67 For instance, miR-122 inhibits IFN signaling pathway to favor HCV replication. Hence, using antisense oligonucleotides of miR-122 to carry out miRNA silencing induced methylation at SOCS3 gene promoter, resulting in a decrease of SOCS3 levels. This decrement then intensified IFN-induced ISRE activity in the host. 79 In addition to miR-122, upregulation of some other miRNAs like miR-758 in infected cells is also responsible for reduced IFN signaling. They carry out this by eliminating ongoing activities of toll-like receptors 3 and 7, 80 while some miRNAs favor immune responses induced by IFN like miR-221 and miR-30 cluster either by targeting SOCS1/3 proteins and genes, respectively.81,82 A study revealed that nearly 30 miRNAs were differentially expressed in response to IFN α, β and γ treatment given to HCV-infected cells. Among which, eight are almost complementary to the seed sequence of the HCV genome.5,67,68 Further studies showed that some miRNAs like miR-196, miR-296, miR-351, miR-431, and miR-448 exhibited antiviral effects by inhibiting viral replication (Table 2).5,15,63,65,67,83–89
Table 2.
miRNAs involved in HCV infection, liver disease, and HCC.
| miRNAs | Viral & Cellular Targets | Mechanism |
|---|---|---|
| miR-196 67 | NS5A; Bach1 | Annealing between host protein and viral RNA |
| miR-448 5 | Core | NR |
| miR-29 67 | Collagen and ECM protein | NR |
| miR-296, miR-351, miR-431 5 | NR | NR |
| miR-130a 63 | IFITM1 | NR |
| miR-373 83 | JAK and IRF9 | Impairs JAK/STAT pathway |
| miR-124 84 | ROCK2 and EZH2 | Cytoskeletal modification and EMT |
| miR-155 85 | Wnt signaling | Inhibits apoptosis and promotes proliferation |
| miR-221/222 86 | Tumor suppressor p27, p57 and PTEN | Fibrosis progression and carcinogenesis |
| miR-199a-3p15,65 | HCV IRES-Domain II | Inhibits HCV replication |
| miR-449a 87 | NOTCH signaling & C-C ligand 2 activity | Altered expression of protein regulator (YKL40), fibrosis development |
| miR-107 87 | C-C ligand 2 activity | Promote inflammation and fibrosis. |
| miR-21 88 | SMAD7 | Fibrosis development (HSCs) via TGF-β signaling |
| miR-200c 89 | FAP1 and Src signaling | Increased expression of collagen and fibroblast factor |
Role of miRNAs in HIV/HCV co-infection
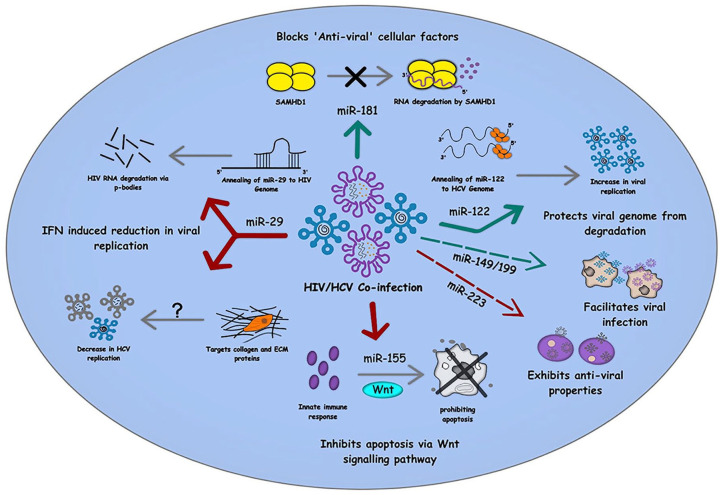
Developing treatments for HIV-1 efficiently promises a healthy life span to patients, while on the contrary, severe liver diseases in HIV/HCV co-infection patients have been proven fatal. 90 Researchers observed that HIV has a significant role in the prognosis of HCV infection. The average time span between HCV-mono-infection and occurrence of cirrhosis is estimated to be 25–30 years usually, while in co-infection, the time span dramatically decreases, and a faster occurrence of cirrhosis or HCC is observed. Clinical data show co-infected patients have increased HCV viral load compared with mono-infected patients. 91 HIV-1 infection is so far claimed to be a reason behind the increment of viral load in co-infected patients.92,93 Although the counter effect of HCV on HIV infection is not significantly seen yet, some studies found HCV to enhance HIV-1 infection in macrophages with the help of different pathways. 92 HCV seems to use TLR2-, JNK-, and MEK1/2-dependent pathways to induce increased HIV viral replication. 94 Along with this, slow recovery of the immune system in co-infected patients is commonly seen compared with mono-infected patients. 95 Alongside, the effect of ART on liver disease progression in co-infected patients is still controversial. Some studies show no significant effect of ART in causing liver damage, with a lesser chance of liver failure, while some claim ART is associated with increased liver transaminase or frank liver decomposition.96,97 As a result, the severity of liver illnesses such as fibrosis and cirrhosis has grown. HIV-1 is significantly different from HCV as it is a retrovirus that majorly targets CD4+ T cells and macrophages. It contains two copies of positive-sense, single-stranded RNA, which encodes nine genes. Unlike HCV infection, HIV RNA undergoes reverse transcription and forms DNA which is then integrated within the host cell genome, 94 while on the other end, HCV RNA directly undergoes replication and transcription without any DNA intermediate. 6 So far, both HIV-1 and HCV infections are seen to have some effect on one another. Along with this, toxicity caused due to various treatments or protein interactions can also be seen as effects that cause increased progression toward liver diseases in co-infection. However, to achieve reliable diagnostic or therapeutic outcomes, the study of underlining mechanisms is necessary. microRNAs are one such non-detachable part of cellular mechanisms. Hence, the role of miRNA in co-infection is an emerging field of research. The scientists have already established several miRNAs and their roles concerning HIV-1 and HCV mono-infection. Although sex-specific biasness has been observed in the miRNA expression of HIV/HCV-co-infected patients, role of some miRNAs is believed to be conserved in the mono-infections and also has their potential contribution in HIV/HCV co-infection 98 (Figure 5).
Figure 5.
Role of cellular miRNAs in HIV/HCV co-infection: The figure summarizes the involvement of various miRNAs in the different cellular pathways commonly involved in HIV/HCV co-infection. While miR-29 and miR-155 involved in IFN-mediated suppression of viral replication, miR-122 and miR-181 inhibit apoptosis and other antiviral cellular factors to maintain a pro-viral environment within the infected cells. The red arrow denotes the inhibitory effect of miRNAs, and the green arrow represents pro-viral effects, whereas the dashed arrows signify the role of miRNAs that are yet to be examined mechanistically in HIV/HCV co-infection.
miR-122
As mentioned earlier, miR-122 is abundantly found in hepatic cells, which has a significant role in HCV RNA replication. miR-122 having a complimentary seed sequence with HCV gives it an advantage over other miRNAs and thus is suspected of promoting replication by stabilizing HCV genome.70,72,73,99 Hence, upregulation of miR-122 in HCV-infected cells is not new. Its upregulation is also observed in cells infected with HIV-1, though its connection with HIV-1 genome is not yet clear. 100 Thus, an assumption can be placed on the table that HIV-1 infection upregulates miR-122 in various cells like macrophages, which might give HCV an advantage to grow faster in cells other than liver cells during co-infection.100,101
miR-181
Many times, ‘antiviral’ cellular factors are noticed to be often blocked by miRNAs in order to promote viral infection. Likewise, miR-181 is also seen to anneal and inhibit cellular factor SAMHD1, promoting HIV-1 infectivity. It also holds a strong correlation with HCV viral load along with CD4 + T cells. Some studies claim miR-181 to enhance liver fibrosis. As it is found to be upregulated in HIV/HBV co-infection, hence their role in HIV/HCV co-infection can be a part worth exploring. 100
miR-155
As discussed above, its upregulation in HIV, HCV, and HBV can be linked with the defense mechanism of host cells. miR-155 is also reported to inhibit apoptosis via Wnt signaling pathway (Figure 5). This way, eventually, promotes proliferation and HCC.100,102 Its role in the first line defense mechanism justifies its positive correlation with CD4 T cell count and its downregulation in elite controls of HIV and HCV. It was seen to be upregulated in HIV mono-, HCV mono-, and HIV/HCV co-infection while downregulated in ECs. This might give us an opportunity to use miR-155 as a therapeutic approach against co-infection. 100
miR-29
Another miRNA with potential therapeutic advantage is miR-29. It is seen to inhibit HIV-1 infection via different mechanisms: (1) anneals with 3′ UTR of HIV, which is followed by its degradation via p-bodies, 103 and (2) inhibits HIV protein Nef, leading to reduced infectivity.25,31 As earlier discussed, its expression, induced by IFN, has also shown a reduction in HCV replication.5,67
miR-223
Studies have shown miR-223 to be positively correlated with CD4 T-cell count and negatively correlated with HIV load and are responsible for HIV inactivity.100,104 Alongside, it is also downregulated in untreated HCV patients suffering through HCC. 101 Recently, miR-223 was found to be upregulated in HIV-HCV co-infection, but its significant role in co-infection is yet to be discovered. 100
Some miRNAs are seen to be significantly dysregulated in HIV and HCV mono-infection, but their expression and role in co-infection are not entirely studied (Figure 5). A high-throughput sequencing of smRNAs detected dysregulation of four miRNAs viz. hsa-miR-205-5p and hsa-let-7a/b/f-5p, but their role in HIV-HCV co-infection has not been studied yet. 105 Similarly miR-149 is seen to have noticeable upregulation during HCV infection. 106 At the same time, it is also observed to anneal with HIV 3′ UTR and protein Vpr during HIV infection. Similarly, miRNAs of miR-199a family are seen to support liver fibrosis and are found to be downregulated in HCCs. 15 On the contrary, an increment in the expression of miR-199a was noticed upon HIV infection. 107 Though the expression level of miR-146 is found to be higher in HIV-HCV co-infection, the mechanistic role of this miRNA is yet to be analyzed. 100
Based on these data, such miRNAs can possibly have their own significant roles in HIV/HCV co-infection. Hence, allowing us to explore various advantages that they hold.
Biomarkers
All these studies reveal the indispensable role of miRNAs in viral infection and disease progression. Hence, many researchers have been working on the idea of using such miRNAs as diagnostic/therapeutic tools. Altering the expression of miRNAs for disease treatment has so far been a difficult task because of their varied nature in different situations. However, the same nature of being differentially expressed during different disease conditions allows miRNAs to be used as biomarkers. A study covering HIV-HCV co-infected patients serum with significant history of liver injury revealed upregulation of miR-22, miR-34a, and miR-122, 108 whereas another study disclosed miR-99a, miR-100, miR-125b, and miR-192 as the prime culprit for liver fibrosis and disease progression in plasma samples of HIV-HCV co-infected patients. 109 A recent study published in 2021 provided evidences for eight different miRNA that holds the potentiality to become a biomarker for HIV, HCV, or HIV/HCV co-infection. 110 The study included miRNAs like miR-122, miR-155, miR-223, miR-150, miR-199, miR-149, miR-29, and miR-let7.
Similarly, other studies were conducted on differential expression of miRNAs in HCV mono- and HIV/HCV co-infection after liver transplantation. They used liver biopsies of mono- and co-infected patients after 6 months of liver transplantation. These liver samples were affected by HCV but had not yet undergone any severe liver injury like cirrhosis but associated with the processes of viral reinfection, neurologic impairment, and coagulation disorders.111,112 Researchers believed this approach might become helpful in identifying different miRNAs that play a role in the early stage of disease progression.111,112 Results so far claimed miR29b, miR-200c, miR-222, and miR-338-3p can possibly act as biomarkers. Upregulation of miR-200c after transplantation can represent increased chances of reinfection, while downregulation of miR-338-3p signifies a lack of tumor-suppressing activities. 111 In addition, it was stated that the dysregulation of miR-29b and miR-222 might play the important role in impairing the immune response favoring fibrosis in HCV/HIV co-infected patients. 112
Likewise, many different microRNAs are identified as potential biomarkers in liver disease progression representing different pathways/mechanisms. 113 One such is miR-101 which in normal conditions is indulged in prohibiting hepatic stellate cells activation and pro-fibrogenic cytokines release via TGF-β inhibition. 114 Hence, reduced expression of miR-101 can symbolize the faster progression of liver disease in HCV and HIV/HCV co-infected patients.111,114
Conclusion
HIV and HCV are blood-borne infections; hence, this standard route of transmission plays a significant role in increasing cases of co-infection. Virus–host interactions play multiple different mechanisms involving microRNAs. miRNAs being an intrigued part of any cellular process, the study of their dysregulation during infection has always been the cynosure of researchers. Analyzing the role of differentially expressed miRNAs might allow us to comprehend various ongoing complex mechanisms and interactions in HIV/HCV co-infection. This prominent nature can further be used to develop an early-stage non-invasive diagnostic tool for co-infection. However, there are still many controversial questions that remain unanswered. What role does HCV hold in HIV infection? Do HIV-1 microRNAs play any role in HCV infection? What effects do ART or HCV treatment have on disease progression and pathogenesis during a co-infection? Indeed, a lot more research is required for in-depth study of co-infection and the role of miRNAs in it.
Footnotes
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Nicky Joshi: Data curation; Formal analysis; Writing – original draft.
Madhuri Chandane Tak: Data curation; Investigation; Writing – original draft.
Anupam Mukherjee: Conceptualization; Funding acquisition; Investigation; Project administration; Supervision; Writing – review & editing.
Disclaimer: All the authors have contributed equally to this work.
ORCID iD: Anupam Mukherjee  https://orcid.org/0000-0002-0612-2258
https://orcid.org/0000-0002-0612-2258
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This review work has been supported by the Indian Council of Medical Research and ICMR-National AIDS Research Institute, Pune.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Availability of data and materials: Not applicable.
Contributor Information
Nicky Joshi, Division of Virology, ICMR-National AIDS Research Institute, Pune, India.
Madhuri Chandane Tak, Division of Virology, ICMR-National AIDS Research Institute, Pune, India.
Anupam Mukherjee, Scientist D & RAMANUJAN Fellow, Division of Virology, ICMR-National AIDS Research Institute, Plot No. 73, ‘G’ Block, MIDC, Bhosari, Pune 411026, Maharashtra, India.
References
- 1. Barré-Sinoussi F, Chermann JC, Rey F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983; 220: 868–871. [DOI] [PubMed] [Google Scholar]
- 2. Gallo RC, Salahuddin SZ, Popovic M, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 1984; 224: 500–503. [DOI] [PubMed] [Google Scholar]
- 3. Levy JA, Hoffman AD, Kramer SM, et al. Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science 1984; 225: 840–842. [DOI] [PubMed] [Google Scholar]
- 4. Bennasser Y, Le S-Y, Yeung ML, et al. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 2004; 1: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shrivastava S, Steele R, Ray R, et al. MicroRNAs: role in hepatitis C virus pathogenesis. Genes Dis 2015; 2: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol 2007; 5: 453–463. [DOI] [PubMed] [Google Scholar]
- 7. Roberts APE, Lewis AP, Jopling CL. The role of microRNAs in viral infection. Prog Mol Biol Transl Sci 2011; 102: 101–139. [DOI] [PubMed] [Google Scholar]
- 8. Simmonds P. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol 2004; 85: 3173–3188. [DOI] [PubMed] [Google Scholar]
- 9. Di Bisceglie AM. Natural history of hepatitis C: its impact on clinical management. Hepatology 2000; 31: 1014–1018. [DOI] [PubMed] [Google Scholar]
- 10. Salmon-Ceron D, Arends J, Leoni C, et al. HIV/HCV coinfection: current challenges. Viral hepatitis: chronic hepatitis C, pp. 141–57, https://link.springer.com/chapter/10.1007/978-3-030-03757-4_7
- 11. Cooper CL. Natural history of HIV and HCV coinfection. J Int Assoc Physicians AIDS Care (Chic) 2003; 2: 147–151. [PubMed] [Google Scholar]
- 12. Dwivedi S, Purohit P, Sharma P. MicroRNAs and diseases: promising biomarkers for diagnosis and therapeutics. Indian J Clin Biochem 2019; 34: 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11: 228–234. [DOI] [PubMed] [Google Scholar]
- 15. Murakami Y, Toyoda H, Tanaka M, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 2011; 6: e16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harwig A, Das AT, Berkhout B. Retroviral microRNAs. Curr Opin Virol 2014; 7: 47–54. [DOI] [PubMed] [Google Scholar]
- 17. Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 2004; 78: 12868–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swaminathan G, Martin-Garcia J, Navas-Martin S. RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiol Genomics 2013; 45: 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bondanese VP, Francisco-Garcia A, Bedke N, et al. Identification of host miRNAs that may limit human rhinovirus replication. World J Biol Chem 2014; 5: 437–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maranini B, Ciancio G, Ferracin M, et al. microRNAs and inflammatory immune response in SARS-CoV-2 infection: a narrative review. Life (Basel) 2022; 12: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mirzaei R, Mahdavi F, Badrzadeh F, et al. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int Immunopharmacol 2021; 90: 107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol 2010; 64: 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhan S, Wang Y, Chen X. RNA virus-encoded microRNAs: biogenesis, functions and perspectives on application. Exrna 2020; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swaminathan G, Navas-Martín S, Martín-García J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol 2014; 426: 1178–1197. [DOI] [PubMed] [Google Scholar]
- 25. Piedade D, Azevedo-Pereira JM. MicroRNAs, HIV and HCV: a complex relation towards pathology. Rev Med Virol 2016; 26: 197–215. [DOI] [PubMed] [Google Scholar]
- 26. Ouellet DL, Plante I, Landry P, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 2008; 36: 2353–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ouellet DL, Vigneault-Edwards J, Létourneau K, et al. Regulation of host gene expression by HIV-1 TAR microRNAs. Retrovirology 2013; 10: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L, Feng H, Da Q, et al. Expression of HIV-encoded microRNA-TAR and its inhibitory effect on viral replication in human primary macrophages. Arch Virol 2016; 161: 1115–1123. [DOI] [PubMed] [Google Scholar]
- 29. Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007; 13: 1241–1247. [DOI] [PubMed] [Google Scholar]
- 30. Witwer KW, Watson AK, Blankson JN, et al. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 2012; 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reynoso R, Laufer N, Hackl M, et al. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Sci Rep 2014; 4: 5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang P, Qu X, Zhou X, et al. Two cellular microRNAs, miR-196b and miR-1290, contribute to HIV-1 latency. Virology 2015; 486: 228–238. [DOI] [PubMed] [Google Scholar]
- 33. Heinson AI, Woo J, Mukim A, et al. Micro RNA targets in HIV latency: insights into novel layers of latency control. AIDS Res Hum Retroviruses 2021; 37: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bukhari MMM, Mir I, Idrees M, et al. Role of MicroRNAs in establishing latency of human immunodeficiency virus. Crit Rev Eukaryot Gene Expr 2020; 30: 337–348. [DOI] [PubMed] [Google Scholar]
- 35. Omoto S, Ito M, Tsutsumi Y, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 2004; 1: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guenzel CA, Hérate C, Benichou S. HIV-1 Vpr-a still ‘enigmatic multitasker’. Front Microbiol 2014; 5: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen AK, Sengupta P, Waki K, et al. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc Natl Acad Sci U S A 2014; 111: E2676–E2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Fan M, Geng G, et al. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 2014; 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barichievy S, Naidoo J, Mhlanga MM. Non-coding RNAs and HIV: viral manipulation of host dark matter to shape the cellular environment. Front Genet 2015; 6: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zucko D, Hayir A, Grinde K, et al. Circular RNA profiles in viremia and ART suppression predict competing circRNA-miRNA-mRNA networks exclusive to HIV-1 viremic patients. Viruses 2022; 14: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu C, Ding Q, Kong X. Integrated analysis of the miRNA-mRNA regulatory network involved in HIV-associated neurocognitive disorder. Pathogens 2022; 11: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Modai S, Farberov L, Herzig E, et al. HIV-1 infection increases microRNAs that inhibit Dicer1, HRB and HIV-EP2, thereby reducing viral replication. PLoS ONE 2019; 14: e0211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shahbaz S, Okoye I, Blevins G, et al. Elevated ATP via enhanced miRNA-30b, 30c, and 30e downregulates the expression of CD73 in CD8+ T cells of HIV-infected individuals. PLoS Pathog 2022; 18: e1010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seddiki N, Brezar V, Ruffin N, et al. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 2014; 142: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agudo J, Ruzo A, Tung N, et al. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol 2014; 15: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Triboulet R, Mari B, Lin Y-L, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007; 315: 1579–1582. [DOI] [PubMed] [Google Scholar]
- 47. Ma L, Shen CJ, Cohen ÉA, et al. miRNA-1236 inhibits HIV-1 infection of monocytes by repressing translation of cellular factor VprBP. PLoS ONE 2014; 9: e99535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chiang K, Sung TL, Rice AP. Regulation of cyclin T1 and HIV-1 Replication by microRNAs in resting CD4+ T lymphocytes. J Virol 2012; 86: 3244–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng Y, Yang Z, Jin C, et al. hsa-miR-191-5p inhibits replication of human immunodeficiency virus type 1 by downregulating the expression of NUP50. Arch Virol 2021; 166: 755–766. [DOI] [PubMed] [Google Scholar]
- 50. Hariharan M, Scaria V, Pillai B, et al. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun 2005; 337: 1214–1218. [DOI] [PubMed] [Google Scholar]
- 51. Bernard MA, Zhao H, Yue SC, et al. Novel HIV-1 miRNAs stimulate TNFα release in human macrophages via TLR8 signaling pathway. PLoS ONE 2014; 9: e106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen CJ, Jia YH, Tian RR, et al. Translation of Pur-α is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J 2012; 26: 4755–4764. [DOI] [PubMed] [Google Scholar]
- 53. Frentzen A, Gürlevik E, Hueging K, et al. Cell entry, efficient RNA replication, and production of infectious hepatitis C virus progeny in mouse liver-derived cells. Hepatology 2014; 59: 78–88. [DOI] [PubMed] [Google Scholar]
- 54. Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 2011; 474: 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin LT, Noyce RS, Pham TN, et al. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol 2010; 84: 9170–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sendi H, Mehrab-Mohseni M, Foureau DM, et al. MiR-122 decreases HCV entry into hepatocytes through binding to the 3′ UTR of OCLN mRNA. Liver Int 2015; 35: 1315–1323. [DOI] [PubMed] [Google Scholar]
- 57. Elhelw DS, Riad SE, Shawer H, et al. Ectopic delivery of miR-200c diminishes hepatitis C virus infectivity through transcriptional and translational repression of Occludin. Arch Virol 2017; 162: 3283–3291. [DOI] [PubMed] [Google Scholar]
- 58. Riad SE, Elhelw DS, Shawer H, et al. Disruption of Claudin-1 expression by miRNA-182 alters the susceptibility to viral infectivity in HCV cell models. Front Genet 2018; 9: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang GJ, Xiao HX, Tian HP, et al. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med 2013; 31: 1375–1380. [DOI] [PubMed] [Google Scholar]
- 60. Mekky RY, El-Ekiaby NM, Hamza MT, et al. Mir-194 is a hepatocyte gate keeper hindering HCV entry through targeting CD81 receptor. J Infect 2015; 70: 78–87. [DOI] [PubMed] [Google Scholar]
- 61. Calland N, Albecka A, Belouzard S, et al. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology 2012; 55: 720–729. [DOI] [PubMed] [Google Scholar]
- 62. Mekky RY, El-Ekiaby N, El Sobky SA, et al. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch Virol 2019; 164: 1587–1595. [DOI] [PubMed] [Google Scholar]
- 63. Bhanja Chowdhury J, Shrivastava S, Steele R, et al. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol 2012; 86: 10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li S, Duan X, Li Y, et al. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat 2014; 21: 121–128. [DOI] [PubMed] [Google Scholar]
- 65. Murakami Y, Aly HH, Tajima A, et al. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol 2009; 50: 453–460. [DOI] [PubMed] [Google Scholar]
- 66. Cheng JC, Yeh YJ, Tseng CP, et al. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci 2012; 69: 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. World J Hepatol 2013; 5: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mukherjee A, Shrivastava S, Bhanja Chowdhury J, et al. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol 2014; 88: 7929–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006; 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 70. Roberts APE, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res 2011; 39: 7716–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kunden RD, Khan JQ, Ghezelbash S, et al. The role of the liver-specific microRNA, miRNA-122 in the HCV replication cycle. Int J Mol Sci 2020; 21: E5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wilson JA, Zhang C, Huys A, et al. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J Virol 2011; 85: 2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chang J, Guo JT, Jiang D, et al. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol 2008; 82: 8215–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fukuhara T, Kambara H, Shiokawa M, et al. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J Virol 2012; 86: 7918–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Choi Y, Dienes HP, Krawczynski K. Kinetics of miR-122 expression in the liver during acute HCV infection. PLoS ONE 2013; 8: e76501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jabłonowska E, Wójcik K, Szymańska B, et al. Hepatic HMOX1 expression positively correlates with Bach-1 and miR-122 in patients with HCV mono and HIV/HCV coinfection. PLoS ONE 2014; 9: e95564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van der Meer AJ, Farid WR, Sonneveld MJ, et al. Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122. J Viral Hepat 2013; 20: 158–166. [DOI] [PubMed] [Google Scholar]
- 78. Ishida H, Tatsumi T, Hosui A, et al. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun 2011; 412: 92–97. [DOI] [PubMed] [Google Scholar]
- 79. Yoshikawa T, Takata A, Otsuka M, et al. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep 2012; 2: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang Q, Fu S, Wang J. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Arch Virol 2014; 159: 2997–3003. [DOI] [PubMed] [Google Scholar]
- 81. Xu G, Yang F, Ding CL, et al. MiR-221 accentuates IFN’s anti-HCV effect by downregulating SOCS1 and SOCS3. Virology 2014; 462-463: 343–350. [DOI] [PubMed] [Google Scholar]
- 82. Zhang X, Daucher M, Armistead D, et al. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS ONE 2013; 8: e55733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mukherjee A, Di Bisceglie AM, Ray RB. Hepatitis C virus-mediated enhancement of microRNA miR-373 impairs the JAK/STAT signaling pathway. J Virol 2015; 89: 3356–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zheng F, Liao Y-J, Cai M-Y, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012; 61: 278–289. [DOI] [PubMed] [Google Scholar]
- 85. Bala S, Tilahun Y, Taha O, et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med 2012; 10: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 2010; 107: 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sarma NJ, Tiriveedhi V, Crippin JS, et al. Hepatitis C virus-induced changes in microRNA 107 (miRNA-107) and miRNA-449a modulate CCL2 by targeting the interleukin-6 receptor complex in hepatitis. J Virol 2014; 88: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marquez RT, Bandyopadhyay S, Wendlandt EB, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest 2010; 90: 1727–1736. [DOI] [PubMed] [Google Scholar]
- 89. Ramachandran S, Ilias Basha H, Sarma NJ, et al. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS ONE 2013; 8: e70744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hernandez MD, Sherman KE. HIV/hepatitis C coinfection natural history and disease progression. Curr Opin HIV AIDS 2011; 6: 478–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gadalla SM, Preiss LR, Eyster ME, et al. Correlates of high hepatitis C virus RNA load in a cohort of HIV-negative and HIV-positive individuals with haemophilia. J Viral Hepat 2011; 18: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim AY, Chung RT. Coinfection with HIV-1 and HCV–a one-two punch. Gastroenterology 2009; 137: 795–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Koziel MJ, Peters MG. Viral hepatitis in HIV infection. N Engl J Med 2007; 356: 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Swaminathan G, Pascual D, Rival G, et al. Hepatitis C virus core protein enhances HIV-1 replication in human macrophages through TLR2, JNK, and MEK1/2-dependent upregulation of TNF-α and IL-6. FEBS Lett 2014; 588: 3501–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gupta P. Hepatitis C virus and HIV type 1 co-infection. Infect Dis Rep 2013; 5: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bräu N, Salvatore M, Ríos-Bedoya CF, et al. Slower fibrosis progression in HIV/HCV-coinfected patients with successful HIV suppression using antiretroviral therapy. J Hepatol 2006; 44: 47–55. [DOI] [PubMed] [Google Scholar]
- 97. Qurishi N, Kreuzberg C, Lüchters G, et al. Effect of antiretroviral therapy on liver-related mortality in patients with HIV and hepatitis C virus coinfection. Lancet 2003; 362: 1708–1713. [DOI] [PubMed] [Google Scholar]
- 98. Valle-Millares D, Brochado-Kith Ó, Gómez-Sanz A, et al. HCV eradication with DAAs differently affects HIV males and females: a whole miRNA sequencing characterization. Biomed Pharmacother 2022; 145: 112405. [DOI] [PubMed] [Google Scholar]
- 99. Li Y, Masaki T, Yamane D, et al. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci U S A 2013; 110: 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yousefpouran S, Mostafaei S, Manesh PV, et al. The assessment of selected MiRNAs profile in HIV, HBV, HCV, HIV/HCV, HIV/HBV Co-infection and elite controllers for determination of biomarker. Microb Pathog 2020; 147: 104355. [DOI] [PubMed] [Google Scholar]
- 101. Gupta A, Swaminathan G, Martin-Garcia J, et al. MicroRNAs, hepatitis C virus, and HCV/HIV-1 co-infection: new insights in pathogenesis and therapy. Viruses 2012; 4: 2485–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Su C, Hou Z, Zhang C, et al. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J 2011; 8: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nathans R, Chu C-Y, Serquina AK, et al. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 2009; 34: 696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pilakka-Kanthikeel S, Nair MP. Interaction of drugs of abuse and microRNA with HIV: a brief review. Front Microbiol 2015; 6: 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Martínez-González E, Brochado-Kith Gómez-Sanz ÓA, Martín-Carbonero L, et al. Comparison of methods and characterization of small RNAs from plasma extracellular vesicles of HIV/HCV coinfected patients. Sci Rep 2020; 10: 11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu X, Wang T, Wakita T, et al. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology 2010; 398: 57–67. [DOI] [PubMed] [Google Scholar]
- 107. Hayes AM, Qian S, Yu L, et al. Tat RNA silencing suppressor activity contributes to perturbation of lymphocyte miRNA by HIV-1. Retrovirology 2011; 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Anadol E, Schierwagen R, Elfimova N, et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology 2015; 61: 46–55. [DOI] [PubMed] [Google Scholar]
- 109. Franco S, Buccione D, Tural C, et al. Circulating microRNA signatures that predict liver fibrosis progression in patients with HIV-1/hepatitis C virus coinfections. AIDS 2021; 35: 1355–1363. [DOI] [PubMed] [Google Scholar]
- 110. Moghoofei M, Najafipour S, Mostafaei S, et al. MicroRNAs profiling in HIV, HCV, and HIV/HCV co-infected patients. Curr HIV Res 2021; 19: 27–34. [DOI] [PubMed] [Google Scholar]
- 111. Bulfoni M, Pravisani R, Dalla E, et al. miRNA expression profiles in liver grafts of HCV and HIV/HCV-infected recipients, 6 months after liver transplantation. J Med Virol 2021; 93: 4992–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Dalla E, Bulfoni M, Cesselli D, et al. Reinfection of transplanted livers in HCV- and HCV/HIV-infected patients is characterized by a different MicroRNA expression profile. Cells 2022; 11: 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sun B, Abadjian L, Monto A, et al. Hepatitis C virus cure in human immunodeficiency virus coinfection dampens inflammation and improves cognition through multiple mechanisms. J Infect Dis 2020; 222: 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tu X, Zhang H, Zhang J, et al. MicroRNA-101 suppresses liver fibrosis by targeting the TGFβ signalling pathway. J Pathol 2014; 234: 46–59. [DOI] [PubMed] [Google Scholar]