Abstract
Upstream of the Streptomyces coelicolor A3(2) chitinase G gene, a small gene (named chb3) is located whose deduced product shares 37% identical amino acids with the previously described CHB1 protein from Streptomyces olivaceoviridis. The chb3 gene and its upstream region were cloned in a multicopy vector and transformed into the plasmid-free Streptomyces lividans TK21 strain. The CHB3 protein (14.9 kDa) was secreted by the S. lividans TK21 transformant during growth in the presence of glucose, N-acetylglucosamine, yeast extract, and chitin. The protein was purified to homogeneity using anionic exchange, hydrophobic interaction chromatographies, and gel filtration. In contrast to CHB1, CHB3 targets α-chitin, β-chitin, and chitosan at pH 6.0 but does so relatively loosely. The ecological implications of the divergence of substrate specificity of various types of chitin-binding proteins are described.
Streptomyces spp. are soil bacteria and main decomposers of chitin, the second most abundant polysaccharide in nature, in soil. Since streptomycetes use chitin as carbon and nitrogen sources, they are assumed to play a major part in the turnover of chitin in natural ecosystems. A number of chitinases have been characterized from various Streptomyces spp. (1, 2, 7, 10, 20, 27, 29). Several genes encoding chitinases (EC 3.2.1.14) that hydrolyze chitin have been cloned from Streptomyces spp. (2, 3, 6, 15, 16, 19–21, 28). The deduced proteins belong to family 18 or 19 of glycosyl hydrolases (3, 19).
The chitin-binding proteins CHB1 and CHB2 lacking hydrolytic activity have been found in Streptomyces olivaceoviridis and Streptomyces reticuli, respectively (11, 25). These proteins specifically bind to α-chitin but not to β-chitin, chitosan, or cellulose. The CBP21 protein produced by a gram-negative, chitin-degrading bacterium, Serratia marcescens (26), shares 45% identical amino acids with CHB1.
We previously found one open reading frame upstream of the Streptomyces coelicolor A3(2) chitinase gene chiG. Its deduced product (21) shares 37% identical amino acids with the previously described chitin-binding protein CHB1 (25). In this report we describe the biochemical characteristics of the protein and give some insights into the regulation of the corresponding gene.
MATERIALS AND METHODS
Bacterial strains, plasmids, and selection of transformants.
Escherichia coli DH5α (23) was used as host for constructs derived from the vector pUC18 (31) or the bifunctional Escherichia coli-Streptomyces vector pWHM3 (30). E. coli DH5α transformants carrying pUC18 or pWHM3 constructs were selected on Luria-Bertani medium agar plates containing ampicillin (50 μg/ml).
TK21, a plasmid-less derivative from Streptomyces lividans 66 (9), served as the host for the pWHM3-based construct (named pWF17) carrying the chb3 gene. Protoplasts from S. lividans TK21 were transformed as described elsewhere (9). Regenerated transformants containing pWHM3-based constructs were selected on complete medium containing thiostrepton (25 μg/ml).
Recombinant DNA techniques and transformations.
Digestions by restriction enzymes were performed as described earlier (23) and according to the suppliers' instructions. Dephosphorylation, blunting, or ligation of DNA fragments was done using bacterial alkaline phosphatase, a DNA blunting kit, or a DNA ligation kit, respectively, according to the manufacturers' instructions. Transformations of E. coli and S. lividans were performed as described previously (9, 23). Bacterial alkaline phosphatase for DNA manipulation was bought from Toyobo, Tokyo, Japan. A DNA blunting kit and a DNA ligation kit were obtained from Takara, Tokyo, Japan. Restriction enzymes were supplied by Gibco-BRL and Roche Diagnostics.
Culture conditions.
A total of 100 ml of complete medium (2.0% [wt/vol] tryptic soy broth, 0.5% [wt/vol] yeast extract, and 50 mM MgCl2; pH 7.6) was inoculated with spores of Streptomyces strains and cultivated at 30°C with shaking at 130 rpm for 20 h. The mycelia were then harvested by centrifugation, washed with minimal medium {MM; 1.5 g of KH2PO4, 2.0 g of K2HPO4, 1.4 g of (NH4)2SO4, 0.01 g of CaCl2, 0.1 g of MgSO4, and 1 ml of trace element solution [40 mg of ZnCl2, 200 mg of FeCl3 · 6H2O, 100 mg of CuSO4 · 2H2O, 100 mg of MnCl2 · 4H2O, 100 mg of Na2B4O7 · 10H2O, and 100 mg of (NH4)6Mo7O24 · 4H2O] per liter; pH 7.2}, resuspended in MM, and divided into several aliquots. Then, a 0.75% (wt/vol) solution of each desired soluble carbon source was added to the suspension, and cultivation was continued for up to 24 h. Prewashed mycelia were grown in the presence of chitin (1% crab shells [Sigma], ground to powder) for up to 72 h. All cultivations were done in the presence of thiostrepton (25 μg/ml).
Protein purification.
The spores of S. lividans TK21 harboring pWF17 were inoculated into 300 ml of complete medium (see above) supplemented with 25 μg of thiostrepton per ml in a 1-liter flask with indentations and cultivated at 30°C with shaking at 130 rpm. After 20 h, three portions of 100 ml were diluted with the same medium to 1 liter, and cultivation (in 2-liter flasks) was continued for another 15 h. The mycelia were harvested by centrifugation, washed five times with MM (without supplements), resuspended in the same volume of the MM supplemented with 0.75% (wt/vol) glucose and 25 μg of of thiostrepton per ml, and cultivated for 20 h. The proteins in the culture supernatant were thereafter precipitated [(NH4)2SO4, 90% saturation] and loaded onto a DEAE-Sepharose column (Amersham-Pharmacia Biotech). The CHB3 protein was eluted in 20 mM Tris-HCl (pH 8.0) containing 0.15 M NaCl. Fractions cross-reacting with anti-CHB1 antibodies (25) were applied onto a Phenyl-Sepharose column (Amersham-Pharmacia Biotech), and elution of proteins was done in 20 mM Tris-HCl (pH 7.0) with a continuously decreasing concentration of (NH4)2SO4 from 1.2 to 0 M. The fractions containing the desired protein were immunodetected and rechromatographed using gel filtration.
Analyses of proteins.
Proteins were separated with 15% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (SDS) (12). Western blot hybridization was performed using anti-CHB1 antibodies (11, 25). To determine the amino-terminal amino acids, the protein was blotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore), as described elsewhere (24). The amino-terminal amino acids of the protein were determined by Edman degradation using an LF3000 automated protein sequencer (Beckman).
Binding assay.
Binding of the CHB3 protein to various polysaccharides was performed as outlined previously (32) with some modifications. For binding tests, highly purified cellulose (Avicel; Serva), glucan (from yeast [Sigma]), α-chitin (from crab shells [Sigma]), and chitosan (derived from crab shells, deacetylation of ca. 85%; Sigma) were used. β-Chitin from squid pen and α-chitin from hen lobster tendon were kindly provided by H. Chanzy, Grenoble, France. Chitin from crab shells (practical grade; Sigma) was used for the preparation of colloidal chitin (13). Then, 1 μg of the apparently homogeneous protein in 70 μl of the indicated buffer was added to 3 mg of α-chitin, β-chitin, colloidal chitin, chitosan, Avicel, or glucan; equilibrated with 2.0 ml of the corresponding buffer; and kept overnight at room temperature. We used 100 mM Tris-HCl (pH 7.0 and 8.0), bis-Tris-HCl (pH 6.0), or 100 mM acetic acid (adjusted with NaOH to pH 5.0) as reaction buffer. After centrifugation, the pellets were washed twice with 1 ml of the corresponding buffer, suspended in 70 μl of the buffer containing 1 M NaCl, and shaken for 20 min at room temperature. The pellets were washed twice with 1 ml of the indicated buffer and resuspended in 70 μl of 20 mM Tris-HCl (pH 7.0). To each sample (30 μl), 10 μl of buffer (40% [vol/vol] glycerol, 20% [vol/vol] β-mercaptoethanol, 250 mM Tris-HCl [pH 6.8], 8% [wt/vol] SDS, and 0.1% [wt/vol] bromophenol blue) was added; the mixtures were boiled for 10 min and then subjected to SDS–15% polyacrylamide gel electrophoresis (PAGE). After transfer to a Fluorotrans Transfer membrane (Pall), the ratio of bound and unbound CHB3 protein was immunologically detected using anti-CHB1 antibodies (25).
Primer extension.
The mycelia of S. lividans TK21 transformants frozen at −80°C were disrupted by grinding with alumina Type A-5 (Sigma) and a pellet mixer (Treff Lab). Subsequently, total RNAs were prepared using an RNeasy Mini Preparation Kit (Qiagen) according to the manufacturer's instructions. The oligonucleotide 5′-ATCAGGGCAGAGGTCTTCCGACGTG-3′, the 5′ end of which was labeled with the fluorescent dye fluorescein isothiocyanate (FITC), was utilized as a primer. Then, 12 μg of total RNA and 1 pmol of the primer were denatured at 90°C for 1 min and annealed at 60°C for 2 min in 26 μl of the buffer (50 mM Tris-HCl, pH 8.0; 100 mM KCl). Reverse transcription was done at 42°C for 1 h with ReverTra Ace RNaseH− Reverse Transcriptase (Toyobo) and with dATP, dCTP, dTTP, and 7-deaza-dGTP (Roche Diagnostics). Size ladders were produced by a dideoxy sequencing reaction of the plasmid pCG101 (21) with the primer. The sequencing reaction was performed using a Thermo-Sequenase fluorescence-labeled primer cycling kit with 7-deaza-dGTP (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The primer extension products and the size ladders were separated on a 4.0% (wt/vol) polyacrylamide gel containing 10% (vol/vol) formamide at 40°C by an automated DSQ-2000L laser fluorescent sequencer (Shimadzu).
RESULTS
Physiological and transcriptional studies.
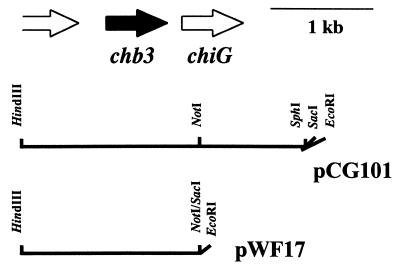
pCG101 (21) was digested with NotI and SacI, blunted, and self-ligated in order to remove the fragment containing the major part of chiG (Fig. 1). The EcoRI-HindIII fragment of the resulting plasmid was inserted into the corresponding sites of pWHM3 to obtain pWF17 (Fig. 1). S. lividans TK21 was transformed with pWF17 or the control vector pWHM3. Several transformants carried the correct constructs, as confirmed by restriction enzyme analysis.
FIG. 1.
Characteristics of constructs. The relative orientations of the chb3 gene, the chitinase (chiG) gene, and a portion of an open reading frame encoding a so-far-unknown protein are shown (top). The relevant restriction sites are indicated for the construct pCG101 (using the vector pUC18) and for the construct pWF17 (using the vector pWHM3).
Precultivated mycelia from S. lividans TK21 carrying pWF17 or the control vector pWHM3 were grown in MM supplemented with glucose, N-acetylglucosamine, yeast extract, or chitin. After concentration, the proteins with the supernatants were tested as to their cross-reactivity with anti-CHB1 antibodies (see Materials and Methods) raised against the previously described chitin-binding protein CHB1 from S. olivaceoviridis (25). About equal amounts of a protein with an apparent molecular mass of ca. 15 to 16 kDa were secreted by S. lividans pWF17 in the presence of each of the carbon sources indicated above. The control strain S. lividans TK21(pWHM3) containing only the vector did not produce the 15- to 16-kDa protein.
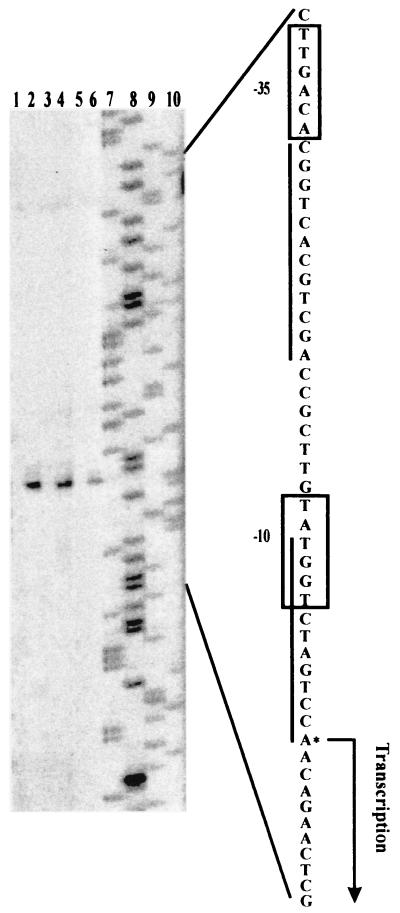
To determine the transcriptional start sites of chb3, primer extension analysis was performed using the total RNAs from S. lividans TK21(pWF17) and the control TK21 strain carrying only the vector pWHM3. The extended products of the same size were detected in S. lividans TK21(pWF17) grown in the presence of chitin, N-acetylglucosamine, or glucose, whereas no product was detected in TK21(pWHM3) (Fig. 2). The dominant extension product corresponded to the A residue at position 188 upstream of the putative translation initiation site. At about 10 and 35 nucleotides upstream of the transcriptional starting site, a sequence matching the consensus of promoters could be recognized by major sigma factors. A motif of 12 bp overlapping the −10 hexamer of the promoter is directly repeated (Fig. 2).
FIG. 2.
Determination of transcriptional initiation sites of the chb3 gene. The S. lividans control strain TK21 containing the vector pWHM3 (lanes 1, 3, and 5) and TK21 carrying the construct pWF17 (lanes 2, 4, and 6) were precultured in complete medium. Cultivation of washed mycelia was then continued in MM supplemented with 0.75% (wt/vol) chitin (lanes 1 and 2), N-acetylglucosamine (lanes 3 and 4), or glucose (lanes 5 and 6). To maintain selection for the plasmid, thiostrepton (25 μg/ml) was added. Primer extension analyses were done as outlined in Materials and Methods. Lanes 7 through 10 show A-, G-, C-, and T-specific dideoxy sequencing reactions, respectively. The corresponding sequences of the coding strands are shown on the right. The nucleotides of transcriptional starts and the −10 and −35 regions of the promoter consensus sequences are indicated. The sequences similar to the 12-bp direct repeat directing the regulation of the S. plicatus chi63 gene (5, 18) are underlined.
Purification of the protein.
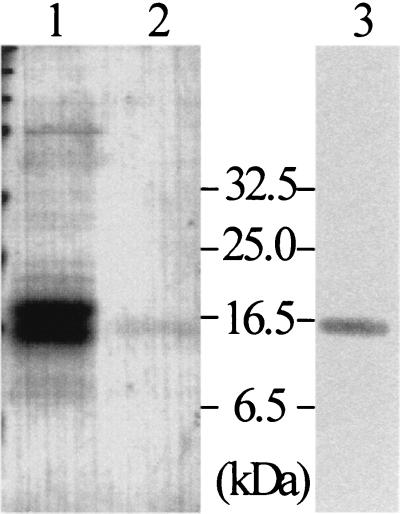
To obtain larger quantities of well-grown mycelia, S. lividans TK21(pWF17) was precultivated in complete medium. After a washing, cultivation was continued in MM supplemented with glucose. The proteins (20 mg/2 liters) from the cleared supernatant were precipitated by (NH4)2SO4. After we tested a number of conditions, we found it best to purify the protein by subsequent chromatographies using DEAE, phenyl-Sepharose, and gel filtration (for details see Materials and Methods). The purified protein (35 μg/2 liters) cross-reacted moderately with anti-CHB1 antibodies (Fig. 3). Its amino-terminal amino acid sequence (HGYISDPPRSQAQ) was determined by Edman degradation, and it corresponded to that of the protein (deduced from DNA sequence data) lacking the signal peptide, the size of which as calculated from the deduced amino acid sequence is 14,964 Da. This protein was designated CHB3.
FIG. 3.
Purification of CHB3. S. lividans TK21(pWF17) was grown in the presence of glucose. The proteins from the culture filtrate (lane 1) and the highly purified CHB3 protein (lane 2) were analyzed by SDS-PAGE. The purified CHB3 protein was also tested for cross-reactions with anti-CHB1 antibodies (lane 3). Size markers are given (in kilodaltons).
Binding specificity of CHB3.
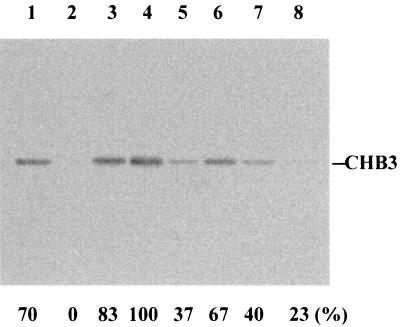
The pI of CHB3 as calculated from its deduced amino acid sequence is 4.58, while those of CHB1 and CHB2 are 8.23 and 8.90, respectively. This suggested that the optimum pH for the binding of CHB3 to chitin is different from those of CHB1 and CHB2. The ability of CHB3 to bind to α-chitin was therefore investigated at pH 5, 6, 7, and 8. At pH 6, all of the added protein bound to α-chitin, part of it at pH 5 and 7 and only a little at pH 8 (Fig. 4), pH 6 thus being the optimum value for efficient targeting of α-chitin. Most of the bound protein could be eluted in the same buffer containing 1 M NaCl (Table 1), indicating that the protein is relatively loosely bound.
FIG. 4.
Binding efficiency of CHB3 in dependence on the pH. As described in Materials and Methods, CHB3 was incubated with ground crab shell powder (α-chitin) at room temperature overnight in buffers adjusted to pH 5 (lanes 1 and 5), pH 6 (lanes 2 and 6), pH 7 (lanes 3 and 7), and pH 8 (lanes 4 and 8). After centrifugation, the supernatants were analyzed by SDS-PAGE (lanes 1 to 4). The pellets were washed with the corresponding buffer, resuspended, boiled in the SDS-PAGE sample buffer for 10 min, and centrifuged. The supernatant was subjected to gel electrophoresis (lanes 5 to 8). CHB3 was detected by immunoblotting with anti-CHB1 antibodies, and the relative quantities were determined by scanning. The highest amount of cross-reacting protein was set as 100%. The sizes of the protein markers are indicated.
TABLE 1.
Binding specificity of CHB3a
| Substrate | % Binding specificity
|
||
|---|---|---|---|
| Unbound | Loosely bound | Bound | |
| Chitin | |||
| Hen lobster tendon (α-chitin) | 0 | 60 | 40 |
| Crab shell (α-chitin) | 0 | 97 | 3 |
| Squid pen (β-chitin) | 0 | 93 | 7 |
| Colloidal chitin | 77 | 23 | 0 |
| Chitosan | 0 | 100 | 0 |
| Avicel | 100 | 0 | 0 |
| Glucan | 100 | 0 | 0 |
CHB3 was incubated with each substrate at pH 6 at room temperature overnight. After centrifugation, the proteins in the supernatant were analyzed by SDS-PAGE and immunodetected (unbound). The proteins which bound to the substrates were then eluted by the buffer containing 1 M NaCl, separated by SDS-PAGE, and immunodetected (loosely bound). Proteins associated with the pellets were released by boiling (95°C, 10 min) and, after removal of the insoluble substrate, analyzed by SDS-PAGE and subsequent immunodetection (bound). After a corresponding photo was scanned, the quantity of cross-reacting protein was related to the total amount of CHB3.
To investigate the binding specificity of CHB3, binding tests were performed at pH 6 using several other polysaccharides (Table 1). The ability to bind to β-chitin was comparable to that of α-chitin. CHB3 bound most strongly to α-chitin from hen lobster tendon, and only about half of it could be removed by 1 M NaCl. CHB3 also bound to chitosan but eluted in the buffer with 1 M NaCl. Its ability to bind to colloidal chitin was lower than that of the four substrates described above. Binding of CHB3 to Avicel and glucan (from yeast) was not detected.
DISCUSSION
Having cloned the corresponding S. coelicolor A3(2) gene, we generated an S. lividans transformant secreting the CHB3 protein (14.9 kDa). We have demonstrated here that the purified CHB3 from S. coelicolor A3(2) binds to both α-chitin and β-chitin, but relatively loosely; furthermore, it binds to chitosan in a low salt concentration. The deduced CHB3 protein shares 37, 37, and 36% identical amino acids with the deduced proteins CHB1 (25), CHB2 (11), and CBP21 (26), respectively.
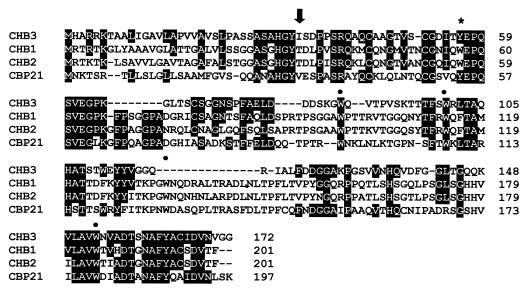
The Streptomyces proteins CHB1 (S. olivaceoviridis) and CHB2 (S. reticuli) share five tryptophan residues in corresponding positions. Substitution of tryptophan Trp-57 by tyrosine (Tyr) or leucine (Leu) in CHB1 resulted in a mutant protein whose binding to crab shell chitin (α-chitin) was reduced to ca. 10% in the presence of 1 M NaCl compared to the wild-type protein (33). Under the same conditions, interaction of CHB3 with the same substrate was about threefold less pronounced than the binding of the W57L CHB1 mutant protein (exchange of the tryptophan residue [Trp, W] by a lysine residue). CHB3 contains three Trp residues in positions corresponding to those in CHB1 and CHB2; however, it lacks a Trp residue corresponding to the one in position 57. Considering, in addition, the data gathered about the W57L mutant CHB1 (33), the findings indicate that to some extent the differences are due to the lack of this Trp residue. A region that corresponds to the one including Trp-134 in CHB1 is also absent in CHB3. This region of CHB1 contributes to its interaction with α-chitin (33).
Unlike CHB1 and CHB2, CHB3 bound to β-chitin (Table 1), a feature shared with CBP21 from S. marcescens, which predominantly interacts with β-chitin (26). CHB3 shares 10 identical amino acids with CBP21 that are not present in CHB1 and CHB2, and 5 of them are gathered around position 130 in CHB3 (Fig. 5). It will thus be interesting to determine whether this region is essential for the specific interaction with β-chitin.
FIG. 5.
Alignment of deduced amino acid sequences of CHB3 from S. coelicolor A3(2), CHB1 from S. olivaceoviridis (25), CHB2 from S. reticuli (11), and CBP21 from S. marcescens (26). Amino acids which are identical between CHB3 and any of the other three proteins are given in white against a black background. The black arrow indicates the predicted cleavage site of a signal peptidase. The cleavage site could be predicted because the N termini of the mature CHB3 (see Results) and of CHB1, CHB2, and CBP21 had been determined previously (11, 25, 26). Tryptophan residues (W) which are shared with CHB1 and CHB2 are indicated by a heavy dot; those which are exchanged by tyrosine in CHB3 are marked by an asterisk.
Several Streptomyces chitinase genes, i.e., chi63 of S. plicatus (5); chiA and chiC of S. lividans (6, 14); exo-chiO1 of S. olivaceoviridis (3); chiA, chiB, chiC, chiD, and chiF of S. coelicolor A3(2) (22); and chb1 of S. olivaceoviridis (25), are transcribed in the presence of chitin but seem to be repressed in the presence of glucose. The upstream regions of all these chitinase genes and of the Streptomyces chitin-binding genes chb1, chb2, and chb3 contain one (exo-chiO1) or two repeats of a 12-bp repeat, which vary from one another to different degrees. In this context, it is interesting that the first nucleotide T of the direct repeat is substituted by C only within the chb3 gene (Fig. 6). Whether this exchange leads to cessation of binding of a repressor protein implicated to interact with the 12-bp repeat upstream of the S. plicatus chi63 gene (18) remains to be elucidated.
FIG. 6.
Alignment of upstream regions of chitinase genes and the genes for chitin-binding proteins. The experimentally determined transcriptional start sites are wavy underlined. The predicted −35 and −10 sequences for the RNA polymerase binding sites are double and single underlined, respectively. The sequences corresponding to the 12-bp direct repeat of the S. plicatus chi63 gene (5) are given in capitals; those identical with the direct repeat of the chi63 gene are shown in white on a black background. The strains have been abbreviated as follows: (c), S. coelicolor A3(2); (p), S. plicatus; (l), S. lividans; (o), S. olivaceoviridis.
It is interesting that the Bacillus circulans chitinase A1 comprises a small domain (45 amino acids) targeting various forms of insoluble chitin at a wide pH range (8). The 12-kDa N-terminal domain of the exoChiO1 chitinase from S. olivaceoviridis recognizes α- and β-chitin (3). The AKWWTQG of the binding domains of the B. circulans chitinase A1 and some other deduced chitinases is assumed to be required for the recognition of chitin. A similar motif is absent in the biochemically characterized binding domain of the exoChiO1 chitinase and in the CHB1, CHB2, and CHB3 proteins from streptomycetes. CHB3 also differs from the recently identified small (9.8-kDa) AFP1 protein from Streptomyces tendae interacting with chitin and chitosan (4).
Native chitin varies as to the length and arrangement of its chains and as to its accessory organic and inorganic compounds. Chitosan shows different degrees of deacetylation (17). Chitin and chitosan can be encountered separately or together (i.e., in certain fungal cell walls), as well in association with other polysaccharides (e.g., glucan). The studies performed up to now have shown that the recognition properties of the individual chitin-binding proteins and of the binding domains of chitinases considerably differ.
Representing a large population in soil, several Streptomyces strains, depending on the type of binding protein, have a selective advantage in colonizing different types of chitin-containing substrates which they then degrade, due to their large repertoire of chitinases.
ACKNOWLEDGMENTS
A.S. thanks the members of the study group of Applied Genetics of Microorganisms at the University of Osnabrück, D. Müller for support in subcloning, and A. Bećirević and D. Ortiz de Orué Lucana for advice on the fast-performance liquid chromatography system, the performance of binding tests, and transformations. We are grateful to M. Lemme for her help with the writing of the manuscript.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (Schr. 203/6-3) to H.S., by a short-term fellowship awarded to A.S. by the DAAD (Deutscher Akademischer Auslandsdienst), and by the Grant-in-Aid Design Program from the MAFF (Ministry of Agriculture, Forestry, and Fisheries of Japan) (BDP-00-VI-2-6) to K.M.
REFERENCES
- 1.Beyer M, Diekmann H. The chitinase system of Streptomyces sp. ATCC 11238 and its significance for fungal cell wall degradation. Appl Microbiol Biotechnol. 1985;23:140–146. [Google Scholar]
- 2.Blaak H, Schnellmann J, Walter S, Henrissat B, Schrempf H. Characteristics of an exochitinase from Streptomyces olivaceoviridis, its corresponding gene, putative protein domains and relationship to other chitinases. Eur J Biochem. 1993;214:659–669. doi: 10.1111/j.1432-1033.1993.tb17966.x. [DOI] [PubMed] [Google Scholar]
- 3.Blaak H, Schrempf H. Binding and substrate specificities of a Streptomyces olivaceoviridis chitinase in comparison with its proteolytically processed form. Eur J Biochem. 1995;229:132–139. doi: 10.1111/j.1432-1033.1995.tb20447.x. [DOI] [PubMed] [Google Scholar]
- 4.Bormann C, Baier D, Hörr I, Raps C, Berger J, Jung G, Schwarz H. Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tü901 that interferes with growth polarity. J Bacteriol. 1999;181:7421–7429. doi: 10.1128/jb.181.24.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delic I, Robbins P, Westpheling J. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci USA. 1992;89:1885–1889. doi: 10.1073/pnas.89.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii T, Miyashita K. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J Gen Microbiol. 1993;139:677–686. doi: 10.1099/00221287-139-4-677. [DOI] [PubMed] [Google Scholar]
- 7.Hara S, Yamamura Y, Fujii Y, Mega T, Ikenada T. Purification and characterization of chitinase produced by Streptomyces erythraeus. J Biochem. 1989;105:484–489. doi: 10.1093/oxfordjournals.jbchem.a122691. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto M, Ikegami T, Seino S, Ohuchi N, Fukada H, Sugiyama J, Shirakawa M, Watanabe T. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J Bacteriol. 2000;182:3045–3054. doi: 10.1128/jb.182.11.3045-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 10.Jeuniaux C. Chitinases. Methods Enzymol. 1966;8:644–650. [Google Scholar]
- 11.Kolbe S, Fischer S, Becirevic A, Hinz P, Schrempf H. The Streptomyces reticuli α-chitin-binding protein CHB2 and its gene. Microbiology. 1998;144:1291–1297. doi: 10.1099/00221287-144-5-1291. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lingappa Y, Lockwood J L. Chitin media for selective isolation and culture of actinomycetes. Phytopathology. 1962;52:317–323. [Google Scholar]
- 14.Miyashita K, Fujii T. Nucleotide sequence and analysis of a gene (chiA) for a chitinase from Streptomyces lividans 66. Biosci Biotechnol Biochem. 1993;57:1691–1698. doi: 10.1271/bbb.57.1691. [DOI] [PubMed] [Google Scholar]
- 15.Miyashita K, Fujii T, Sawada Y. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J Gen Microbiol. 1991;137:2065–2072. [Google Scholar]
- 16.Miyashita K, Fujii T, Watanabe A, Ueno H. Nucleotide sequence and expression of a gene (chiB) for a chitinase from Streptomyces lividans. J Ferment Bioeng. 1997;83:26–31. [Google Scholar]
- 17.Muzzarelli R A A. Chitin. Oxford, England: Pergamon Press; 1977. [Google Scholar]
- 18.Ni X, Westpheling J. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc Natl Acad Sci USA. 1997;94:13116–13121. doi: 10.1073/pnas.94.24.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, Watanabe T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol. 1996;178:5065–5070. doi: 10.1128/jb.178.17.5065-5070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins P W, Albright C, Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988;263:443–447. [PubMed] [Google Scholar]
- 21.Saito A, Fujii T, Yoneyama T, Redenbach M, Ohno T, Watanabe T, Miyashita K. High-multiplicity of chitinase genes in Streptomyces coelicolor A3(2) Biosci Biotechnol Biochem. 1999;63:710–718. doi: 10.1271/bbb.63.710. [DOI] [PubMed] [Google Scholar]
- 22.Saito A, Ishizaka M, Francisco P B, Jr, Fujii T, Miyashita K. Transcriptional co-regulation of the five chitinase genes scattered on the Streptomyces coelicolor A3(2) chromosome. Microbiology. 2000;146:2937–2946. doi: 10.1099/00221287-146-11-2937. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schlochtermeier A, Walter S, Schröder J, Moormann M, Schrempf H. The gene encoding the cellulase (Avicelase) Cel1 from Streptomyces reticuli and analysis of protein domains. Mol Microbiol. 1992;6:3611–3621. doi: 10.1111/j.1365-2958.1992.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 25.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to crystalline α-chitin of fungi and other organisms. Mol Microbiol. 1994;13:807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K, Suzuki M, Taiyoji M, Nikaidou N, Watanabe T. Chitin binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci Biotechnol Biochem. 1998;62:128–135. doi: 10.1271/bbb.62.128. [DOI] [PubMed] [Google Scholar]
- 27.Tarentino A L, Maley F. Purification and properties of an endo-β-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249:811–817. [PubMed] [Google Scholar]
- 28.Tsujibo H, Hatano N, Endo H, Miyamoto K, Inamori Y. Purification and characterization of a thermostable chitinase from Streptomyces thermoviolaceus OPC-520 and cloning of the encoding gene. Biosci Biotechnol Biochem. 2000;64:96–102. doi: 10.1271/bbb.64.96. [DOI] [PubMed] [Google Scholar]
- 29.Ueno H, Miyashita K, Sawada Y, Oba Y. Purification and some properties of extracellular chitinases from Streptomyces sp. S-84. J Gen Appl Microbiol. 1990;36:377–392. [Google Scholar]
- 30.Vara J, Lewandowska-Skarbek M, Wang Y-G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 32.Zeltins A, Schrempf H. Visualization of α-chitin with a specific chitin-binding protein (CHB1) from Streptomyces olivaceoviridis. Anal Biochem. 1995;231:287–294. doi: 10.1006/abio.1995.0053. [DOI] [PubMed] [Google Scholar]
- 33.Zeltins A, Schrempf H. Specific interaction of the Streptomyces chitin-binding protein CHB1 with α-chitin: the role of individual tryptophan residues. Eur J Biochem. 1997;246:557–564. doi: 10.1111/j.1432-1033.1997.t01-1-00557.x. [DOI] [PubMed] [Google Scholar]