Summary
Routine transrectal ultrasound-guided systematic prostate biopsy only samples a small volume of the prostate and tumors between biopsy cores can be missed, leading to low sensitivity to detect clinically relevant prostate cancers (PCa). Deep learning may enable detection of PCa despite benign biopsies. We included 14,354 hematoxylin-eosin stained benign prostate biopsies from 1,508 men in two groups: men without established PCa diagnosis and men with at least one core biopsy diagnosed with PCa. A 10-Convolutional Neural Network ensemble was optimized to distinguish benign biopsies from benign men or patients with PCa. Area under the receiver operating characteristic curve was estimated at 0.739 (bootstrap 95% CI:0.682–0.796) on man level in the held-out test set. At the specificity of 0.90, the model sensitivity was 0.348. The proposed model can detect men with risk of missed PCa and has the potential to reduce false negatives and to indicate men who could benefit from rebiopsies.
Subject areas: Endocrinology, Biopsy sample, Artificial intelligence
Graphical abstract
Highlights
-
•
Tumor lesions may be missed during a systematic TRUS-guided prostate biopsy
-
•
Improvement of prostate cancer (PCa) detection and reduction of rebiopsies are needed
-
•
The trained deep learning model predicts PCa risk from benign prostate biopsies only
-
•
The model has the potential to reduce false negatives especially in high-grade PCa
Endocrinology; Biopsy sample; Artificial intelligence
Introduction
Prostate cancer (PCa) is the second most common cancer in men worldwide (Rawla, 2019). Transrectal ultrasound (TRUS)-guided 10–12 core needle prostate biopsy is the routine diagnostic tool for men who have elevated prostate-specific antigen (PSA) and/or abnormal digital rectal examination (DRE) and/or other suspicious indications (EAU Guidelines: Prostate Cancer, n.d.). A major limitation of the systematic TRUS biopsy is undersampling, causing cancer lesions between biopsy cores to be missed (Shah and Zhou, 2012). Consequently, the sensitivity of TRUS biopsy has been reported as low as 32%–58% (Delongchamps et al., 2009; Haas et al., 2007) and increasing the number of biopsy cores only marginally improves the sensitivity and mainly detects indolent cancers (Babayan and Katz, 2016). Although magnetic resonance (MRI)-guided targeted biopsy is recommended for biopsy naive patients and patients with indications for repeated biopsy, it is not widely available to patients on a broad scale. Improvements in the sensitivity of TRUS biopsy, especially for more aggressive PCa, are therefore needed to improve the detection of clinically relevant cancer and to reduce unnecessary rebiopsies.
PCa diagnosis based on histopathological inspection of prostate biopsy (hematoxylin-eosin (H&E) stained) slides is challenging and prone to inter-assessor variability (Shah and Zhou, 2012; Sooriakumaran et al., 2005). Currently, there are three features (perineural invasion, glomerulations, and mucinous fibroplasia) not observed in benign prostate glands and therefore are considered to be diagnostic for PCa. However, most PCa are identified based on a combination of non-specific major and minor cancer architectural and cytological features (Shah and Zhou, 2012). The major features (eg. infiltrative growth pattern, absence of basal cells, and nuclear atypia) are strongly linked to cancer, while the minor features (eg. cytoplasmic amphophilia, intraluminal contents, mitosis, and apoptosis) have a weaker link to cancer, and can also be seen in non-cancer lesions (Shah and Zhou, 2012). In addition, premalignant changes of high-grade prostatic intraepithelial neoplasia (HGPIN) and atypical small acinar proliferation (ASAP) are associated with a later PCa diagnosis (Shah and Zhou, 2012). It is well recognized that the development of cancer is a continual process in which cells gradually become malignant as a result of the accumulation of mutations and selection (Hanahan and Weinberg, 2011). Moreover, epigenetic changes have been reported in the early development of cancer and distinct epigenetic signatures may be present in neighboring prostatic tissues adjacent to the PCa foci (Partin et al., 2014). Therefore, subtle cancer-related morphological structures with clinical relevance, which are either non-specific or not detectable by human eyes, could be present in benign cores sampled in the vicinity of the cancer areas.
Deep learning (DL) (LeCun et al., 2015) in the form of convolutional neural networks currently offers the state-of-the-art performance for image classification. DL has recently demonstrated human-level performance in routine pathology tasks (Campanella et al., 2019), including cancer detection and grading (Bulten et al., 2020; Ström et al., 2020). DL has also demonstrated the ability to predict additional factors from H&E-stained whole-slide images (WSIs), which cannot be determined in routine pathology by a human assessor, including status of molecular markers (Coudray et al., 2018; Schmauch et al., 2020) and microsatellite instability (Kather et al., 2019). Consequently, we hypothesize that there might be subtle morphological patterns present in prostate H&E WSIs that can be modeled by DL to predict clinically relevant PCa. In this study, we investigate the potential to apply DL to recognize cancer-associated morphological changes in benign prostate biopsies, and assess to what extent such models could improve the sensitivity to detect clinically relevant PCa in TRUS-guided systematic prostate biopsies.
Results
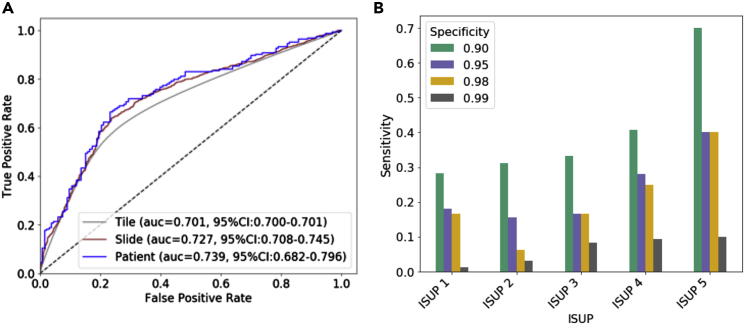
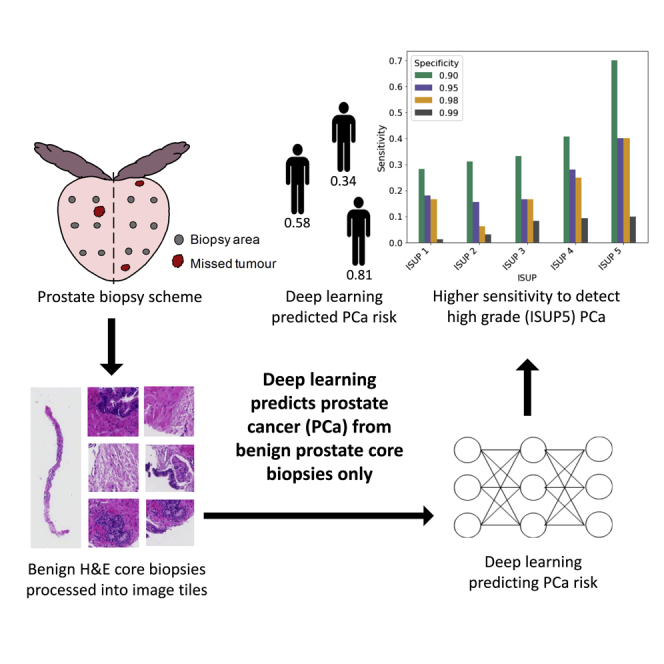
The 10-CNN ensemble model was trained and optimized on 9,192 benign prostate biopsy WSIs (8,780,026 tiles) from 973 men (535 cancer, 438 benign) to distinguish benign biopsies from patients with PCa and benign men (Figure 1). Classification performance was first estimated in the validation set (130 PCa and 108 benign men, 2,311 benign WSIs, 2,192,124 tiles) and subsequently evaluated in the held-out test set (164 PCa and 133 benign men, 2,851 benign WSIs, 2,713,314 tiles) (Figure 2A - ROC curve). We observed a tile-level prediction performance of area under the receiver operating characteristic ROC curve (AUC) = 0.701 (95% CIbootstrap: 0.700–0.701), slide-level AUC = 0.727 (95% CIbootstrap: 0.708–0.745), and a patient-level AUC = 0.739 (95% CIbootstrap: 0.682–0.796) in the held-out test set. Based on patient-level analysis, we assessed sensitivities of the prediction at specificity 0.99, 0.98, 0.95, and 0.90 and reported corresponding cutoffs. (Figure 2B). For overall cancer detection, we observed a sensitivity of 0.043 (95% CIbootstrap: 0.000–0.213) at the specificity of 0.99, a sensitivity of 0.177 (95% CIbootstrap: 0.038–0.240) at the specificity of 0.98, and a sensitivity of 0.224 (95% CIbootstrap: 0.153–0.311) at specificity of 0.95, and a sensitivity of 0.348 (95% CIbootstrap: 0.205–0.468) at the specificity of 0.90. These sensitivities differed by ISUP with higher sensitivity for more aggressive cancers graded as ISUP 4 or 5 (Figure 2B and Table S2). We applied logistic regressions to establish baseline classification performance using only age and PSA as predictors and observed an AUC = 0.562 (95% CIbootstrap: 0.510–0.612) in the held-out test set.
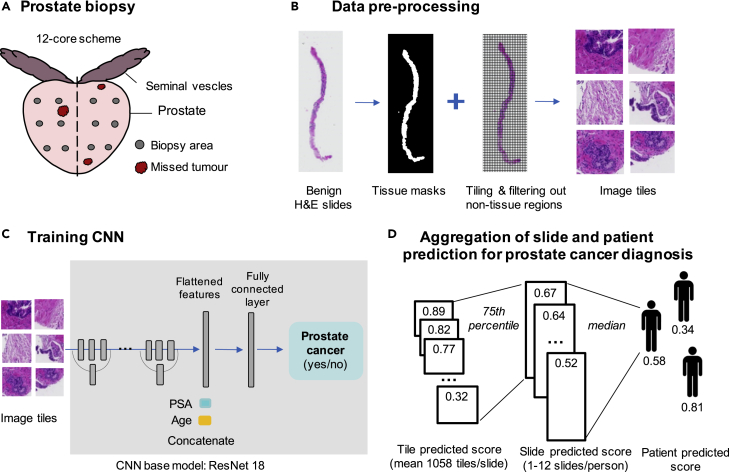
Figure 1.
Overview of image pre-processing and deep learning (DL) modeling for detecting patients with cancer from benign prostate biopsies only
(A) Prostate biopsy scheme showing that tumor may be missed due to sampling error.
(B) Pre-processing of WSIs.
(C) Schematic overview of the image classification by a deep convolutional neural network (CNN). Schematic plot shows that the predicted score for each slide was aggregated using the 75th percentile of the tile-level predicted score and patient-level predicted score was obtained from the median of slide-level predicted score.
Figure 2.
Prediction performance in held-out test set
(A) Receiver operating characteristic (ROC) curves and the associated area under the ROC curve (AUC) estimates with bootstrap 95% confidence intervals in the held-out test set for the prediction of PCa versus benign diagnosis on tile, slide, and patient level; (B) corresponding sensitivity by ISUP group at specificities of 0.99 (cutoff = 0.872), 0.98 (cutoff = 0.824), 0.95 (cutoff = 0.812), and 0.90 (cutoff = 0.770).
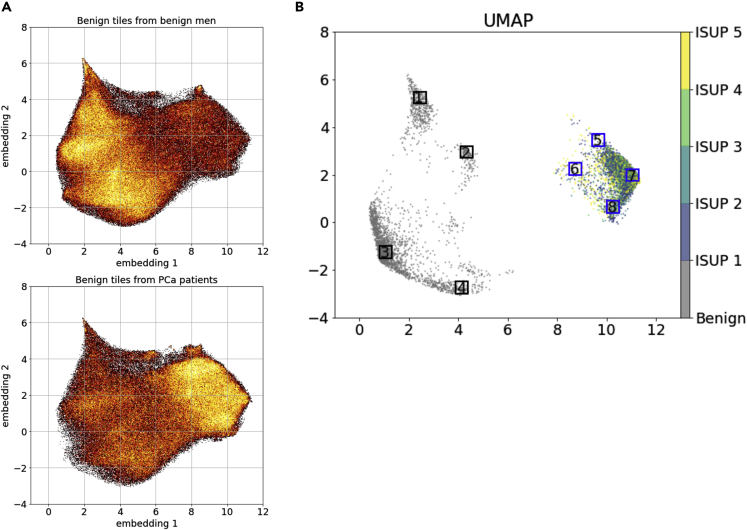
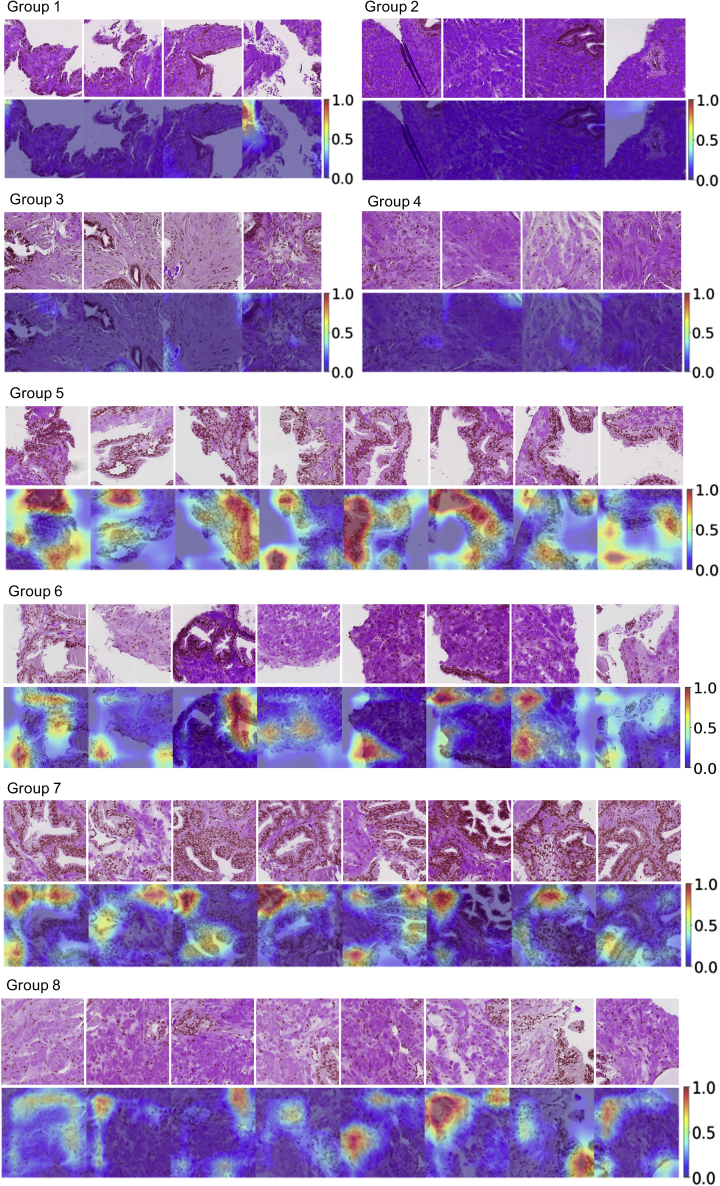
In the UMAP (Figure 3A) based on a random sample of 20% tiles from the entire test dataset, benign tiles from benign men and benign tiles from men with PCa had clear differences in tile density distribution in the UMAP projections. This density difference was more prominent when restricted to cancer-benign tiles with patient-level predicted scores higher than the 90th percentile and benign-benign tiles with the predicted scores below the 10th percentile (Figure 3B). We randomly selected sample tiles from the selected regions in Figure 3B and presented the auxiliary-guided Grad-Cam (Selvaraju et al., 2017; Srivastava et al., 2014) highlighting regions likely linked to PCa. As expected, benign-benign tiles (Figure 4 groups 1–4) generally had fewer hotspots that were linked to PCa diagnosis, while cancer-benign tiles from patients with PCa showed more hotspots (Figure 4 groups 5–8), indicating stronger association with PCa diagnosis. As compared to benign-benign tiles in groups 1–4, cancer-benign tiles in groups 5–8 generally showed more glandular structures which were also frequently highlighted in the Grad-Cam plots. Although unclear, Grad-Cam plots seemingly highlighted regions where luminal cells had irregular shapes of nuclei or nuclei with indistinct boundaries (nucleus membrane) (group 5 and 7).
Figure 3.
Model characterization and interpretation using UMAP
(A) UMAPs of benign tiles from benign men (i.e. benign-benign tiles) and benign tiles from patients with prostate cancer (PCa) (i.e. cancer-benign tiles).
(B) UMAP of benign-benign tiles from benign men with patient level predicted score lower than the 10th percentile and cancer-benign tiles from PCa patients with predicted score higher than the 90th percentile. Eight regions with distinct embeddings were randomly selected to plot example tiles and the corresponding Grad-Cam plots.
Figure 4.
Example tiles and Grad-Cam plots
Tiles were extracted from the eight selected regions in Figure 3B. Groups 1–4 show benign-benign tiles randomly selected in regions 1–4 and groups 5–8 are cancer-benign tiles randomly selected from region 4–8, respectively. The corresponding Grad-Cam plots highlight important structures related to cancer diagnosis.
Discussion
In this study, we developed a DL-based model for classification of WSIs of benign prostate core biopsies from men without a cancer diagnosis and from men with at least one biopsy core with cancer. The model achieved a patient-level AUC of 0.739, and given a specificity of 0.90, the proposed model was able to detect 34.8% of all PCa, and 41% ISUP 4 and 70% of ISUP 5 PCa, from benign biopsy cores only.
There is a plethora of challenges associated with PCa detection by biopsy. PCa is often multifocal and systematic TRUS biopsies only sample a small volume of the prostate; hence, there is an intrinsic and substantial risk that biopsy cores miss a cancer lesion (Djavan et al., 1999). The peripheral zone that harbors 70%–80% of the PCa is most frequently sampled, while the transition zone containing 20% of the cancers is typically not sampled during the initial biopsy (Shah and Zhou, 2012). Sampling templates in clinical practice also vary substantially. A recent study reported that ⅓ of 137 urologists never or seldomly sampled the anterior part of the prostate (Carlsson et al., 2012) despite its high cancer frequency (Chun et al., 2010). The possible false negatives from the initial biopsy require repeated biopsies for patients with abnormal DRE or continuously increased PSA or PSA derivatives. However, due to the nonspecificity of these indicators, cancer detection rate for the second biopsy under the saturation protocol (≥ 20 cores) remains low (22%–24%)(Lane et al., 2008; Pepe and Aragona, 2007). In fact, multiple rebiopsies may be performed before establishing a PCa diagnosis (Babayan and Katz, 2016). The procedure causes patients’ anxiety and potential side effects, including sepsis (Babayan and Katz, 2016).
Our DL model identified 34.8% of PCa cases at a specificity of 0.90 from only benign biopsies, including 70% of ISUP 5 and 41% of ISUP 4 cases. This suggests that there exist distinct, albeit subtle, morphological differences between benign prostate tissue in men with cancer present in the prostate (adjacent, or nearby) and men without any cancer present. Hence, subtle differences can be captured by image analysis using DL, which are typically not captured in routine histopathology assessments by human assessors. A high predicted risk score from a computer model could thus potentially assist pathologists, and indicate either further review of existing biopsies, or guide decision for rebiopsy. Notably, the model showed higher sensitivity for more aggressive PCa (ISUP 4 and 5) compared to low-to-medium risk PCa, although undersampling of cancerous tissue may be less prevalent in poorly differentiated PCa since larger volume tumors are more often seen in more advanced GS patterns (Mcneal et al., 1986). PSA-based screening and subsequent biopsy reduce the prostate-specific mortality; however, it leads to over-treatment of low-risk ISUP 1 cancer (Schröder et al., 2012). In fact, 50% of men diagnosed by systematic biopsy have indolent low-risk PCa (Klotz, 2019), which may only require active surveillance rather than immediate intervention (Sathianathen et al., 2018). The model has desirable properties to contribute to the identification of high-risk PCa cases missed due to sparse sampling of the prostate during biopsy, while it detects low-risk PCa (low ISUP) at lower rate, thus not driving the risk of additional over-treatment. Despite previous success of DL in detection of cancer, cancer grading, and determination of cancer length and prognostic indicators (Arvaniti et al., 2019; Coudray et al., 2018; Lucas et al., 2019; Nir et al., 2018; Ström et al., 2020), there have been few attempts to extract novel information beyond what human assessors can detect in WSIs. We provide the first evidence that deep CNN models can distinguish between benign tissue from men with established cancer, and benign tissue from men with only benign biopsies.
Our DL-based model trained from H&E slides, age, and psa significantly outperformed the logistic regression model only including age and psa (AUC of 0.739 vs 0.562). We attempted to interpret the model by projecting the 516 dimensional feature vector learned from the H&E slides to a 2D UMAP. Although the distribution of the 2D embeddings from benign-benign and cancer-benign tiles weas largely overlapped, clear differences in the density of the distribution were noted. This may indicate that the model prediction of PCa is based on the frequency of shared structures between benign-benign and cancer-benign tiles rather than distinct features presented in each group. We further inspected image tiles with high prediction confidence, which may highlight specific features presented more frequently in the cancer-benign group. Overall, we observed more glandular structures in the cancer-benign tiles, which were also often highlighted by the Grad-Cam plots. However, we did not systematically observe any well-established PCa precancerous lesions that were previously reported in the literature. It is not surprising because the nature of the study is to explore patterns related to PCa in the benign cores, which are very likely non-detectable by human eyes. Albeit very vague, with the guidance of Grad-Cam, we speculate that frequent presence of luminal cells with irregular nucleus shapes (e.g. extremely elongated) or indistinct nucleus edges may be relevant to PCa. A study reported that metastatic cancerous luminal/basal cells demonstrated nuclear plasticity with the loss of nuclear membranes and boundary between nucleus and cytoplasm (Sinha, 2020). However, the results were reported under electron microscopy inspection of prostate biopsy samples at much higher magnification levels. It is difficult to draw any conclusion also because of the intrinsic inaccuracy of the Grad-Cam generated by the non-perfect DL model.
The study has several strengths. The study is based on a large sample of 14,354 WSIs (from 1,508 men) digitized on a single scanner model, precluding potential bias due to use of multiple scanning devices. The sample was part of the well-controlled STHLM3 population-based study. The biopsy procedure was standardized and the pathology report for PCa diagnosis was centralized and blinded for both urologists and the pathologist regarding clinical characteristics.
Limitations of the study
The study has some limitations. Benign biopsies with cancer were taken from men that were not actually missed, since they had at least one positive biopsy. Ideally, we would investigate biopsies from men with initial negative biopsies that developed cancer later on. However, due to lack of suitable datasets at this point in time, we were not able to test the model performance in an independent cohort of initially benign men who were later diagnosed with PCa in rebiopsies. While the well-controlled study design and scanning protocol provides high-quality data and opportunity to demonstrate the feasibility of the approach on a conceptual level, it does not demonstrate generalizability to other populations. Future studies would have to be focused on determining to what extent the model generalizes to other settings. Furthermore, the study was enriched with higher ISUP PCa, which could contribute to the observed higher sensitivity to detect men with high ISUP PCa.
Conclusions
Subtle tumor-associated morphological changes in benign prostate tissue that cannot be distinguished by human eye can be captured by deep CNN models trained on large numbers of benign prostate biopsy histopathology images. The developed model has the ability to detect men with risk of missed PCa after biopsy, caused by undersampling of the prostate. The proposed model has the potential to reduce the number of false negatives caused by sparse sampling of the prostate volume in routine systematic prostate biopsies and to indicate men that could benefit from MRI-guided rebiopsy.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| OpenSlide (v. 1.1.1) | (Goode et al., 2013) | https://openslide.org/api/python/ |
| Scikit-image (v.0.14.2) | (van der Walt et al., 2014) | https://scikit-image.org/docs/0.14.x/api/skimage.morphology.html |
| OpenCV (v.3.4.2) | (Bradski, 2000) | https://docs.opencv.org/3.4.2/ |
| ResNet18 | (He et al., 2016; Yakubovskiy, n.d.) | https://github.com/qubvel/classification_models |
| Keras framework (2.2.4), keras-applications (1.0.7) | Keras | https://keras.io/ |
| Tensorflow (1.12.0) | Google Brain Team | https://www.tensorflow.org |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Mattias Rantalainen (mattias.rantalainen@ki.se).
Materials availability
This study did not generate new unique reagents.
Method details
Study population
The study was based on a subset of men from the STHLM3 study(Grönberg et al., 2015), a prospective population-based diagnostic study for PCa. The study enrolled a random sample of men aged between 50 and 69 years who were free of promoted PCa from the Swedish population register. Out of those, 59,159 men accepted to participate in the study(Grönberg et al., 2015) and 7,406 underwent TRUS prostate biopsies (10-12 cores). The study protocol was reviewed and approved by local and governmental ethics committees. All participants provided written informed consent. The biopsies were reviewed and graded according to the International Society of Urological Pathology (ISUP)(Epstein et al., 2016) system by a single pathologist (Lars Egevad). The study included 606 men with detected cancer (ISUP≥1) randomly sampled together with 134 men with only benign biopsies, for details see(Ström et al., 2020), all benign biopsies from these 740 men were included in the present study. To further increase the sample size, all benign biopsies from participants enrolled during 2015 in STHLM3 (N = 877 men, 585 benign, 292 cancer) were also scanned and included.
To reduce the risk for labeling error, we excluded participants or biopsies if there were suspected errors in gleason score (GS) (i.e. duplicates with inconsistent patient level GS), cancer diagnosis, cancer length. Patients with missing in PSA and age were excluded in the analysis (Figure S1). Only benign biopsy cores were included. In total, 14,354 benign biopsies from 829 men (6523 biopsies) with detected cancer (at least one core biopsy had cancer) (cancer-benign biopsies), and 679 men (7831 biopsies) with no detected cancer (all cores classified as benign) (benign-benign biopsies) were included. All WSIs were scanned at 20X magnification (Hamamatsu Nanozoomer-XR, pixel size = 0.4536μm). The dataset was randomly split on patient level into training (973 men), validation (238 men), and held-out test (297 men) sets. The split (i.e. training and validation versus held-out test) was balanced on age, PSA levels, and cancer diagnosis. For cancer patients, the train-test split was further balanced on ISUP grade and the length of the cancer in the biopsy (Table S1 Sample description). The study was approved by the Stockholm regional ethics board.
Data pre-processing
Overall, tissue areas in WSIs were first segmented against background, and second, were downsampled by a factor of 2 (10X magnification level, pixel size approximate 0.90 um) and split into tiles of size 299x299 pixels, with 50% overlap. In total, we obtained 8,780,026 tiles for training, 2,192,124 for validation, and 2,713,314 for the test set.
Tissue segmentation
We first generated tissue masks separating the prostate tissue from the background. The images were read at full resolution (0.4536 um/pixel) and downsampled by a factor of five from the 20X images using the resolution pyramid from OpenSlide (v. 1.1.1)(Goode et al., 2013). Next, we converted the RGB images to HSV encoded images using the color module in scikit-image (v.0.14.2) (van der Walt et al., 2014). Tissue areas were defined as regions above the Ostu’s threshold in saturation and above 0.75 threshold in hue. Background areas were discarded from further analysis. In addition, pen marks were present in a small number of benign slides, which marked suspicious regions for cancer or prostatic intraepithelial neoplasia. The pen marks were filtered out from the tissue areas using the following functions from OpenCV (v.3.4.2) (Bradski, 2000): first, the RGB channel was converted to grayscale using COLOR_BGR2GRAY(); second, a “2D” Laplacian operator was applied using Laplacian () with the Sobel kernel size of three and depth of the output image size set as CV_16S; last, the resulting response was converted into absolute value using convertScaleAbs () and the pen marks were subsequently filtered if below the threshold 20 of the absolute value. We further smoothed tissue masks (removing peppers and salts) by applying a morphological opening and losing, with a disk with a radius of six using functions of disk (), opening (), and closing () in scikit-image morphology module (“Module: morphology — skimage v0.14.3 docs,” n.d.).
Image tiling and quality control
To facilitate model training, WSIs were tiled into small image patches. We used a window size of 598 × 598 pixels from the 20X full resolution (0.4536 um/pixel) and a stride of 299 pixels. The tiles were subsequently downsampled by a factor of 2, the resulting image patches were of size 299 × 299 pixels and with 50% overlap between the adjacent tiles. Tiles containing less than 20% of tissue were excluded. To ensure the quality of the images, we excluded unsharp tiles due to poor focusing during scanning. We calculated the variance after the Laplacian filter using variance_to_laplacian () function in OpenCV(Bradski, 2000) and excluded tiles with the variance lower than 200.
Image classification and model development
Model overview
We applied a deep convolutional neural network using the ResNet18 as the base architecture (He et al., 2016; Yakubovskiy, n.d.) for binary classification of benign biopsy cores from benign men versus benign biopsy cores from men with detected cancer. The model took the preprocessed image tiles together with age and PSA as predictors (Figure 1). We randomly split the dataset on the man level into training (973 men), validation (238 men), and held-out test (297 men) sets. Training and validation sets combined, and the held-out test, were balanced on PCa diagnosis, age, PSA, and further balanced on ISUP for PCa patients. The validation set was used only once to validate final model performance, and the held-out test was only used once to evaluate final model performance. The model was optimized using the training set as a weakly supervised binary classifier, with a patient-level label assigned to each tile. The model weights were initialized from weights pre-trained on ImageNet (Russakovsky et al., 2015). This output layer was subsequently concatenated with categorical covariates of age (<= 55, 55- 60, 60-65, 65-70, 70-100 years) and PSA level at the biopsy (1-3, 3-5, 5-10, >=10 ng/ml). We additionally included a fully connected layer to 256 hidden units allowing potential interactions among age, PSA and the learned image features (Figure 1). We applied 50% drop-out after global-average-pooling of the final convolutional layer as well as 50% drop-out and max-norm regularization (Srivastava et al., 2014) in the last fully connected layer of the model to mitigate potential over-fitting. We applied binary cross-entropy loss and the Adam optimizer(Kingma and Ba, 2014). Hyper-parameters were optimized using cross-validation in the training set, including tile size, magnification level, and learning rate. An ensemble of 10 CNN base-models was used to further reduce variance in predictions. Deep learning model was trained using Keras framework (2.2.4) and keras-applications (1.0.7) (Keras: the Python deep learning API, n.d.) with Tensorflow (1.12.0) backend (TensorFlow, n.d.).
Model prediction
The final model prediction performance was only validated once in the validation set (Figure S2 and Table S3) and only tested once in the held-out test set using Receiver Operating Characteristics (ROC) and Area Under the ROC curve (AUC), and 95% confidence intervals for AUC were obtained using bootstrap (2,000 bootstrap samples). Tile level predictions in the validation and test sets were obtained by averaging of the prediction scores from the ten-model ensemble. The slide level predictions were aggregated using the 75th percentile of the predicted class probabilities across all tiles from each WSI. Patient level predictions were defined based on the median across all WSI level predictions of a patient individual. Percentiles for tile-to-slide and slide-to-patient aggregations were optimized using five-fold cross validation. Sensitivity for detecting cancer on patient level was evaluated at specificity levels of 0.99, 0.98, 0.95, and 0.90. The cutoffs corresponding to different specificities were selected based on the ROC curves.
Model interpretation
Uniform manifold approximation and projection
For model interpretation, we selected the model with the highest tile level AUC obtained in the testset from the ten ensemble models. We randomly selected 20% benign tiles stratified by PCa diagnosis from the test set and applied Uniform Manifold Approximation and Projection (UMAP)(McInnes et al., 2018) to generate a two-dimensional representation of the 512-dimensional feature vector obtained from the DL model. To further explore subtle morphological structures, if exist, that distinguish PCa patients from benign men, we transformed the obtained UMAP to a subset of cancer-benign tiles above the 90th percentile and benign-benign tiles below the 10th percentile of patient level prediction. The selection was performed in a stepwise manner based on patient, slide, and tile level predicted scores. First, we selected PCa patients above 90th percentile and benign men below 10th percentile of patient level predicted score. Next, we selected two slides for each PCa patient and benign man according to the highest and lowest slide level prediction, respectively. Last, among those slides, we chose tiles with top quartile from PCa patients and tiles with bottom quartile from benign men according to the tile level prediction. This selecting procedure was restricted to tiles containing at least 70% of the tissue to facilitate interpretation. We selected four distinct regions for cancer-benign and benign-benign tiles respectively based on UMAP embeddings. For cancer-benign tiles, we randomly sampled 8 image tiles per region and for benign-benign tiles we randomly selected 4 image tiles per region for visualisation. Key patterns in the sample tiles were also plotted using guided gradient-weighted class activation mapping (Guided Grad-CAM)(Selvaraju et al., 2017; Srivastava et al., 2014).
Acknowledgments
This project was supported by funding from the Swedish Research Council (MR, BL), Swedish Cancer Society (Cancerfonden) (MR, ME), Swedish e-science Research Center (SeRC) - eCPC (MR).
Author contributions
Conception and design (MR), acquisition of data (MR, JH, HG, LE), data analysis and interpretation of data (BL, YW, PW, MR), drafting of the manuscript (BL), statistical analysis (BL), obtaining funding (MR), administrative, technical, or material support (MR), supervision (MR), critical revision of the manuscript for important intellectual content (all authors).
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104663.
Supplemental information
Data and code availability
The data reported in this study cannot be deposited in a public repository because they are part of the STHLM3 study and cannot be distributed without access control due to local privacy laws. Requests for data access can be submitted to the STHLM3 study for consideration after ethical permission has been obtained. All data analyses are based upon publicly available software packages (see Method details). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arvaniti E., Fricker K.S., Moret M., Rupp N., Hermanns T., Fankhauser C., Wey N., Wild P.J., Rüschoff J.H., Claassen M. Author Correction: automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 2019;9:7668. doi: 10.1038/s41598-019-43989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayan R.K., Katz M.H. Biopsy prophylaxis, technique, complications, and repeat biopsies. Prostate cancer. 2016. [DOI]
- Bradski G. The OpenCV library. Dr. Dobb’s J. Softw. Tools. 2000;25:120–125. [Google Scholar]
- Bulten W., Pinckaers H., van Boven H., Vink R., de Bel T., van Ginneken B., van der Laak J., Hulsbergen-van de Kaa C., Litjens G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21:233–241. doi: 10.1016/s1470-2045(19)30739-9. [DOI] [PubMed] [Google Scholar]
- Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Werneck Krauss Silva V., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J., et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S., Bratt O., Stattin P., Egevad L. Current routines for transrectal ultrasound-guided prostate biopsy: a web-based survey by the Swedish Urology Network. Scand. J. Urol. Nephrol. 2012;46:405–410. doi: 10.3109/00365599.2012.691111. [DOI] [PubMed] [Google Scholar]
- Chun F.K.-H., Epstein J.I., Ficarra V., Freedland S.J., Montironi R., Montorsi F., Shariat S.F., Schröder F.H., Scattoni V. Optimizing performance and interpretation of prostate biopsy: a critical analysis of the literature. Eur. Urol. 2010;58:851–864. doi: 10.1016/j.eururo.2010.08.041. [DOI] [PubMed] [Google Scholar]
- Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delongchamps N.B., de la Roza G., Jones R., Jumbelic M., Haas G.P. Saturation biopsies on autopsied prostates for detecting and characterizing prostate cancer. BJU Int. 2009;103:49–54. doi: 10.1111/j.1464-410x.2008.07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavan B., Susani M., Bursa B., Basharkhah A., Simak R., Marberger M., BAshArkhAh A. Predictability and significance of multifocal prostate cancer in the radical prostatectomy specimen. Tech. Urol. 1999;5:139–142. [PubMed] [Google Scholar]
- EAU Guidelines: Prostate Cancer [WWW Document], n.d. URL https://uroweb.org/guideline/prostate-cancer/#5 (accessed 3.24.21)
- Epstein J.I., Egevad L., Amin M.B., Delahunt B., Srigley J.R., Humphrey P.A., Committee G. The 2014 international society of urological pathology (ISUP) consensus conference on gleason grading of prostatic carcinoma: Definition of grading patterns and proposal for a new grading system. Am. J. Surg. Pathol. 2016;40:244–252. doi: 10.1097/pas.0000000000000530. [DOI] [PubMed] [Google Scholar]
- Goode A., Gilbert B., Harkes J., Jukic D., Satyanarayanan M. OpenSlide: a vendor-neutral software foundation for digital pathology. J. Pathol. Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönberg H., Adolfsson J., Aly M., Nordström T., Wiklund P., Brandberg Y., Thompson J., Wiklund F., Lindberg J., Clements M., et al. Prostate cancer screening in men aged 50-69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 2015;16:1667–1676. doi: 10.1016/s1470-2045(15)00361-7. [DOI] [PubMed] [Google Scholar]
- Haas G.P., Delongchamps N.B., Jones R.F., Chandan V., Serio A.M., Vickers A.J., Jumbelic M., Threatte G., Korets R., Lilja H., de la Roza G. Needle biopsies on autopsy prostates: sensitivity of cancer detection based on true prevalence. J. Natl. Cancer Inst. 2007;99:1484–1489. doi: 10.1093/jnci/djm153. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011. [DOI] [PubMed]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H., Marx A., Boor P., Tacke F., Neumann U.P., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keras: the Python deep learning API [WWW Document], n.d. URL http://keras.io (accessed 5.25.22)
- Kingma, D.P., Ba, J., 2014. Adam: A method for stochastic optimization. Preprint at arXiv [cs.LG].
- Klotz L. Contemporary approach to active surveillance for favorable risk prostate cancer. Asian J. Urol. 2019;6:146–152. doi: 10.1016/j.ajur.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane B.R., Zippe C.D., Abouassaly R., Schoenfield L., Magi-Galluzzi C., Jones J.S. Saturation technique does not decrease cancer detection during followup after initial prostate biopsy. J. Urol. 2008;179:1746–1750. doi: 10.1016/j.juro.2008.01.049. discussion 1750. [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lucas M., Jansen I., Savci-Heijink C.D., Meijer S.L., de Boer O.J., van Leeuwen T.G., de Bruin D.M., Marquering H.A. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475:77–83. doi: 10.1007/s00428-019-02577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. UMAP: Uniform Manifold approximation and projection for dimension reduction. arXiv. 2018 doi: 10.48550/arXiv.1802.03426. stat.ML Preprint at. [DOI] [Google Scholar]
- Mcneal J., Kindrachuk R., Freiha F., Bostwick D., Redwine E., Stamey T. Patterns of progression in prostate cancer. Lancet. 1986;327:60–63. doi: 10.1016/s0140-6736(86)90715-4. [DOI] [PubMed] [Google Scholar]
- Module: morphology — skimage v0.14.3 docs [WWW Document], n.d. URL https://scikit-image.org/docs/0.14.x/api/skimage.morphology.html (accessed 6.22.21)
- Nir G., Hor S., Karimi D., Fazli L., Skinnider B.F., Tavassoli P., Turbin D., Villamil C.F., Wang G., Wilson R.S., et al. Automatic grading of prostate cancer in digitized histopathology images: learning from multiple experts. Med. Image Anal. 2018;50:167–180. doi: 10.1016/j.media.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Partin A.W., Van Neste L., Klein E.A., Marks L.S., Gee J.R., Troyer D.A., Rieger-Christ K., Jones J.S., Magi-Galluzzi C., Mangold L.A., et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J. Urol. 2014;192:1081–1087. doi: 10.1016/j.juro.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe P., Aragona F. Saturation prostate needle biopsy and prostate cancer detection at initial and repeat evaluation. Urology. 2007;70:1131–1135. doi: 10.1016/j.urology.2007.07.068. [DOI] [PubMed] [Google Scholar]
- Rawla P. Epidemiology of prostate cancer. Oncol. 2019;10:63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- Sathianathen N.J., Konety B.R., Crook J., Saad F., Lawrentschuk N. Landmarks in prostate cancer. Nat. Rev. Urol. 2018;15:627–642. doi: 10.1038/s41585-018-0060-7. [DOI] [PubMed] [Google Scholar]
- Schmauch B., Romagnoni A., Pronier E., Saillard C., Maillé P., Calderaro J., Kamoun A., Sefta M., Toldo S., Zaslavskiy M., et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Commun. Now. 2020;11:3877. doi: 10.1038/s41467-020-17678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder F.H., Hugosson J., Roobol M.J., Tammela T.L.J., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., et al. Prostate-cancer mortality at 11 years of follow-up. J. Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- Shah R.B., Zhou M. Prostate biopsy interpretation: an illustrated guide. 2012. [DOI]
- Sinha A.A. Identification of metastatic cell nucleus in human prostate cancer by electron microscopy. Future Sci. OA. 2020;6:FSO609. doi: 10.2144/fsoa-2019-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooriakumaran P., Lovell D.P., Henderson A., Denham P., Langley S.E.M., Laing R.W. Gleason scoring varies among pathologists and this affects clinical risk in patients with prostate cancer. Clin. Oncol. 2005;17:655–658. doi: 10.1016/j.clon.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- Ström P., Kartasalo K., Olsson H., Solorzano L., Delahunt B., Berney D.M., Bostwick D.G., Evans A.J., Grignon D.J., Humphrey P.A., et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 2020;21:222–232. doi: 10.1016/s1470-2045(19)30738-7. [DOI] [PubMed] [Google Scholar]
- TensorFlow [WWW Document], n.d. TensorFlow. URL https://www.tensorflow.org (accessed 5.25.22)
- van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., scikit-image contributors scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskiy, P., n.d. classification_models [WWW Document]. URL https://github.com/qubvel/classification_models (accessed 79.21)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this study cannot be deposited in a public repository because they are part of the STHLM3 study and cannot be distributed without access control due to local privacy laws. Requests for data access can be submitted to the STHLM3 study for consideration after ethical permission has been obtained. All data analyses are based upon publicly available software packages (see Method details). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.