Abstract
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease globally. miRNAs (miRs) regulate various cellular events that lead to NAFLD. In this study we tested the hypothesis that miR-155 is an important regulator of steatohepatitis and fibrosis pathways. Wild type (WT) or miR-155 deficient (KO) mice received a high fat-high cholesterol-high sugar-diet (HF-HC-HS) for 34 weeks and liver tissues were analyzed. In patients with non-alcoholic steatohepatitis and in the mouse model of HF-HC-HS diet we found increased miR-155 levels in the liver compared to normal livers. Upon HF-HC-HS diet feeding, miR-155 KO mice displayed less liver injury, decreased steatosis, and attenuation in fibrosis compared to WT mice. ALT, triglyceride levels and genes involved in fatty acid metabolic pathway were increased in WT mice whereas miR-155 KO mice showed attenuation in these parameters. HF-HC-HS diet induced significant increase in the expression of NLRP3 inflammasome components in the livers of WT mice compared to chow fed diet. Compared to WT mice, miR-155 KO showed attenuated induction in the NLRP3, ASC and caspase1 inflammasome expression on HF-HC-HS diet. Fibrosis markers such as collagen content and deposition, αSMA, Zeb2, and vimentin were all increased in WT mice and miR-155 KO mice showed attenuated fibrosis marker expression. Overall, our findings highlight a role for miR-155 in HF-HC-HS diet-induced steatosis and liver fibrosis.
Keywords: miRNA-155, macrophages, collagen, NLRP3, ASC, ZEB2, Vimentin, NAFLD, liver injury, TGFbeta
Summary
Non-alcoholic fatty liver disease is the most common cause of chronic liver disease globally and a risk factor for hepatocellular carcinoma. The study highlights the role of miR-155 in non-alcoholic steatohepatitis. miR-155-deficient mice displayed overall protection from diet-induced steatohepatitis and fibrosis. Therapeutic inhibition of miR-155 might be effective approach for treatment of non-alcoholic steatohepatitis.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease globally. The global prevalence of NAFLD is estimated to ~25–30% worldwide 1. NAFLD is a multifactorial disease and comprises a wide array of pathological variations ranging from non-alcoholic fatty liver (NAFL), non-alcoholic steatohepatitis (NASH), fibrosis, NASH cirrhosis, and NASH-related hepatocellular carcinoma (HCC) 1. NAFLD is associated with several metabolic co-morbidities, including obesity, type II diabetes, hyperlipidemia, hypertension, cardiovascular disease and metabolic syndrome 1. Increase in the prevalence and severity of NAFLD is directly correlated with obesity 1. In general the prevalence of NAFLD and NASH is higher in men than women 1. Due to the lack of effective therapies for NAFLD/NASH, care cost and management of associated diseases is a significant economic burden 2.
microRNAs (miRNAs) belong to a class of non-coding small endogenous RNAs that regulate expression of genes either by mRNA degradation or by translational repression in both normal physiological and disease milieus 3. Several studies have revealed the pleiotropic roles of miRNAs in various cellular processes including metabolism, lipogenesis, inflammation, proliferation, differentiation, apoptosis, oncogenesis and metastasis 3,4. Studies have demonstrated the involvement of miRNAs in liver diseases including in the pathogenesis of NAFLD, NASH, and HCC 3–6. miRNA-155 is a highly studied inflammation related miRNA that plays pivotal roles in chronic diseases 3,7. Hepatic macrophages and various immune cells play a significant role in hepatic inflammation, lipid accumulation and fibrosis 8. Previously we demonstrated the role of miR-155 in alcoholic liver disease (ALD) 6 and in methionine/choline-deficient (MCD) diet induced NASH 5.
Several models of NASH/NAFLD have been used by investigators and each model has its own advantages and disadvantages 5. MCD diet results in steatohepatitis in a short period of time, however loss of body weight and lack of insulin resistance are the limitations of this model 5. High-fat/high cholesterol diet is the most dietary method to induce NASH in mice as it mimics Western fast food dietary composition 8. Previously, we reported that HF–HC–HS diet results in the progression of NAFLD (8 weeks) to NASH (27 weeks), fibrosis (27 weeks) and early liver tumor formation (49 weeks) in mice 8. This dietary model causes an increase in body weight, obesity, hepatic steatosis, and insulin resistance, which closely mimic humans NAFLD/NASH 8. Since this model represents the pathology and disease progression similar to humans, we sought to investigate the role of miR-155 in this dietary model using miR-155 KO mouse model system. Using HF–HC–HS diet mouse model in this study we demonstrated that miR-155 KO mice displayed attenuation in liver damage, steatosis, NLRP3 inflammasome complex upregulation and fibrosis after 34 weeks feeding.
Methods
Animal studies
Eight to ten-weeks-old male C57Bl/6 wild type (WT) or miR-155 deficient (knock out/KO, total body) mice were used for the study. This study was approved by the University of Massachusetts Medical School (UMMS, Worcester, MA) and Beth Israel Deaconess Medical Center (BIDMC, Boston, MA) Institutional Animal Use and Care Committee (Worcester, MA). The breeding colonies of miR-155 KO mice were maintained in the animal facility of UMMS and BIDMC as described 6. To induce liver pathology, WT or miR-155 male KO mice (n=8–10) were fed either on a chow diet (control) or a Western diet (treated) equivalent high-fat, high-carbohydrate Surwit diet (58% kcal and 35g% fat) supplemented with 10% cholesterol (Research Diets, NJ, USA) as described previously 8. Drinking water was supplemented with a high-fructose corn syrup equivalent consisting of a total of 42g/L of carbohydrates at a ratio of 55% fructose (Acros Organics, NJ, USA) and 45% sucrose (Sigma-Aldrich, MO, USA) by weight as described 8. Detail diet composition is provided in supplementary figure 2. The mice were given ad libitum access to the food and water and were sacrificed after 34 weeks of feeding. At the end of treatment, blood was collected, and mice were sacrificed. Liver tissues were harvested, washed in PBS and snap frozen in liquid nitrogen for protein extraction and in RNA later (Qiagen, Germany) for RNA extraction. All samples were stored at −80°C.
Histopathology
A portion of the liver was fixed in 10% phosphate buffered formalin, embedded in paraffin blocks, and processed for histological analysis. Sections were stained with hematoxylin and eosin (H&E) and Sirius red stain using standard protocols. A portion of liver tissue was embedded in OCT embedding compound, stored at −80°C and processed for sectioning and sections were stained with Oil-Red-O stain 6. Histology slides were observed under light microscopy at 200X, and quantification was performed using Image J software.
Immunohistochemistry
Immunohistochemistry was performed according to the standard protocols. Briefly, formalin-fixed, paraffin-embedded liver tissue sections were deparaffinized and rehydrated. Then tissue sections were heated in a microwave oven in sodium citrate buffer for antigen retrieval. The slides were blocked using goat serum in 1% TBS-T following incubation with CD68 antibody (cell signaling technology, cat no#97778S). Detection was performed using a biotinylated secondary antibody and streptavidin-horseradish peroxidase, followed by colorimetric detection using DAB. Images were captured using light microscope at 20X magnification. CD68 staining was quantified using Image J software from 6 fields per liver section. At least three liver sections were included in each group.
Human samples
Human liver samples were obtained from the National Institutes of Health Liver Tissue Cell Distribution System (Minneapolis, MN; Pittsburgh, PA; Richmond, VA). Liver samples were from 12 control subjects and 16 patients with a clinical diagnosis of NASH. The criteria used to define the patients with NASH were based on alanine aminotransferase levels (>49 U/L) and, in some cases, biopsy.
Biochemical analysis
For the detection of liver injury, serum alanine aminotransferase (ALT) levels were evaluated from serum samples using a kinetic method (D-TEK, PA, USA). Triglyceride levels were determined from whole liver cell lysates using triglyceride kit (Wako Chemicals USA, Inc., VA, USA) according to the manufacturer’s instructions.
Enzyme-linked immunosorbent assay (ELISA)
Protein levels of MCP1 and TNFα were measured from whole liver cell lysates using ELISA as described by manufactures (BioLegend Inc., CA, USA).
Nuclear extraction
Nuclear extraction from the liver tissue samples was performed using nuclear extraction kit as per manufacturer’s protocol (Abcam, cat no# ab113474) with modification. Briefly, 10mg of liver tissue in 1X pre-extraction buffer was lysed using Tissue lyser II (Qiagen) with 2X cycle of 20Hz for 10 secs. The homogenized tissue was incubated on ice for 15 mins, followed by centrifugation at 12,000 rpm for 10 mins. Supernatant (cytoplasmic content) was separated and pellet containing nuclear fraction was washed twice with 1X pre-extraction buffer. The pellet was resuspended in 200 μl of extraction buffer and vortexed after every 3 mins for 10 seconds up to 15 mins. Suspension was centrifuged at 14,000 rpm for 10 mins at 4 °C and the supernatant containing nuclear fraction was aliquoted and stored at −80 °C until further use.
NF-κB p65 transcription factor assay
NF-κB p65 transcription factor assay kit was used for the determination of p65 transcription factor DNA binding activity from the nuclear extracts as described in the manufacturer’s protocol (Abcam, cat no#ab133112, MA, USA). Briefly 20 μg of nuclear extracts was added to the wells pre-coated with NF-κB response elements and incubated overnight at 4°C. Next day, plate was washed three times with wash buffer and incubated with NF-κB p65 primary antibody followed by secondary antibody. Competitor dsDNA and a positive control for p65 subunit were used as controls for the assay. The absorbance was read at 450nm using plate reader.
Western blot analysis
Whole cell lysates were extracted from the frozen liver tissues as described 8 and equal amount of protein was used for Western blotting. The denatured samples were separated in polyacrylamide gel, transferred onto nitrocellulose membrane, and probed with specific primary antibodies followed by HRP-labeled secondary antibodies. Primary antibody for α smooth muscle actin (Abcam, MA, USA, cat no# ab15734), Zeb2 (Santa Cruz Biotechnology, USA, cat no# SC-271984), Vimentin (Cell Signaling Technology, MA, USA, cat no# 5741S), NLRP3 (Adipogen life sciences, cat no# AG-20B-0014-C100), and GAPDH (Cell signaling technology, MA, USA, cat no# 3683S) were used.
RNA extraction and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA from the livers was extracted using Direct-zol™ RNA MiniPrep kit with on column DNA digestion (Zymo Research Corp., CA, USA). For mRNA analysis, cDNA was transcribed from total RNA using iScript Reverse Transcription Supermix for RT-qPCR (Biorad, Hercules, CA). Real-time quantitative polymerase chain reaction was performed using iCycler (Bio-Rad Laboratories Inc., Hercules, CA). Mean Cq of all samples is normalized to 18S mRNA expression and relative expression was calculated by ΔΔCt method. Primer sequences were same as we have described previously 5,6,8. For miRNA quantification, cDNA synthesis and RT-qPCR were performed from total RNA using TaqMan miRNA assays (Ambion, TX, USA). Cq of the target miRNA was normalized either with snoRNA-202 (mouse) or RNU48 (human) Cq as described 5.
Hydroxyproline assay
The collagen contents were determined by quantification of hydroxyproline from the liver tissues. Liver tissue was homogenized in 100 μl dH2O per 10 mg of tissue. 100 μl of 12N HCL was added into the sample homogenate and hydrolyzed in glass tube at 120 °C for 3 hours, followed by measurement according to manufacturer’s instructions (BioVision, cat no# K555–100). Hydroxyproline levels were calculated against standard curves of 4-hydroxy-L-proline and expressed as μg hydroxyproline per gram of the liver tissue.
Statistical analysis
Statistical significance was determined using either the non-parametric Mann-Whitney test or one-way ANOVA in Prism 8 software. Data is presented as mean±standard error and was considered statistically significant at p<0.05.
Results
Liver damage is decreased in miR-155 KO mice after HF-HC-HS diet
Previously we reported that prolonged high fat-high cholesterol-high sugar (HF–HC–HS) diet resulted in steatohepatitis and early fibrosis in male mice 8. To evaluate the profile of miRNAs in the HF–HC–HS diet-induced steatohepatitis and fibrosis, we checked the expression of miRNAs that are known to be associated with inflammation and various liver diseases 4–6. Among the miRNAs tested, we found a significant induction in miR-155 levels in the livers after 8 and 27 weeks of HF–HC–HS diet (Fig.1A and B), whereas miR-132, miR-146a, and miR-21 showed no changes. To validate the human relevance of this finding, we evaluated human livers with NASH. A significant increase in miR-155 levels was also observed in the livers of NASH patients compared to control subjects (Fig.1C). To assess the role of miR-155 in the development of HF–HC–HS diet induced NAFLD/NASH we employed a miR-155 KO mouse model system. We found that after 34 weeks of HF–HC–HS diet, WT mice has significant liver damage indicated by elevated serum ALT levels, while a significant attenuation in the liver damage was observed in miR-155 KO mice (Fig.1D). The weight gain chart demonstrates that WT mice displayed a progressive increase in weight over the 34 weeks of diet whereas in miR-155 KO mice weight gain reached the plateau after 14 weeks of diet and after 27 weeks a significant decrease in weight gain was detected in KO mice (Fig.1E). The histological evaluations of Hematoxylin and Eosin-stained liver sections revealed an increased accumulation of macro- and micro- vesicular steatosis in WT mice, however attenuation in fat accumulation was found in miR-155 KO mice (Fig.1F). This observation was further supported by the results of Oil- Red-O staining, where decreases in Oil- Red-O-stained fat droplets were found in miR-155 KO mice after HF–HC–HS diet (Fig.1G).
Figure 1. The liver damage and steatosis are decreased in miR-155 KO mice after HF-HC-HS diet.
C57BL/6J wild type (WT) mice (n=6) were either fed with chow or high fat, high cholesterol, high sugar diet (HF-HC-HSD) for 8 and 27 weeks. Total RNA was extracted, and various miRNAs were quantified using TaqMan miRNA RT-qPCR assay and SnoRNA-202 was used to normalize Cq values (A-B). miR-155 levels were determined from total RNA extracted from human NASH patients (n=14) and control subjects (n=11) using TaqMan miRNA RT-qPCR assay and RNU48 was used to normalize Cq values (C). WT or miR-155 KO mice (n=8–10) were either fed with chow or high fat, high cholesterol, high sugar diet (HF-HC-HSD) for 34 weeks. Serum ALT levels were evaluated using biochemical assay (D). Mice were weighed every other week and graph represents weight versus time (E). H&E stained (F) and Oil-Red-O stained (G) liver sections were observed under microscope and representative images are shown. *p<0.05 compared to chow fed WT mice, # p<0.05 compared to HF-HC-HSD fed WT mice, ***p<0.001 compared to control subjects.
miR-155 KO mice exhibit attenuation in fatty acid metabolism genes after HF-HC-HS diet
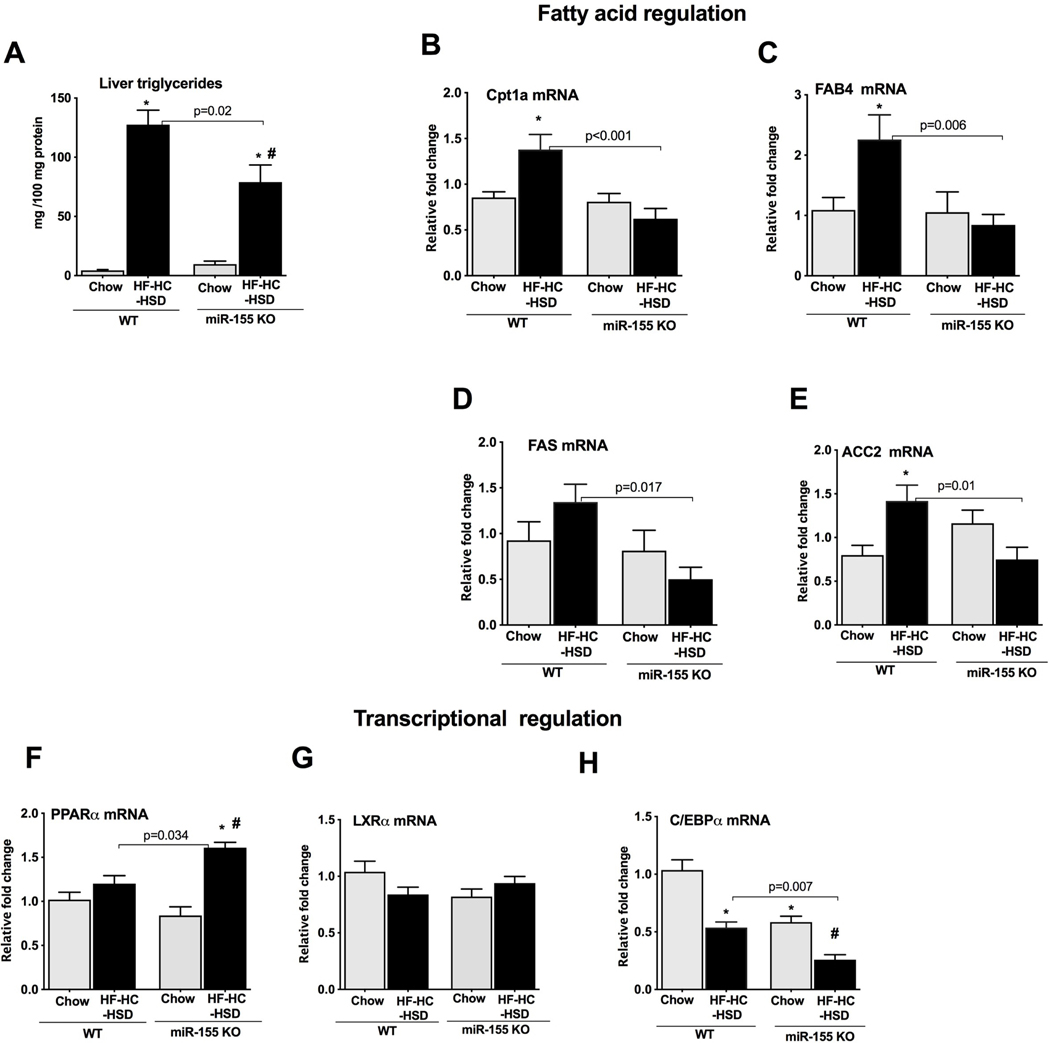
Biochemical evaluations of the liver lysates showed a significant increase in triglyceride levels in WT mice after HF–HC–HS diet (Fig.2A). However, compared to WT mice this increase in triglyceride amount was significantly attenuated in miR-155 KO mice (Fig.2A). Because we found attenuation in the fat accumulation in miR-155 KO mice, we further explored the effect of miR-155 in the fatty acid metabolism pathway. The genes involved in fatty acid metabolism/regulation such as carnitine palmitoyltransferase 1A (Cpt1α) (Fig.2B), fatty acid binding protein 4 (FAB4) (Fig.2C), fatty acid synthase (FAS) (Fig.2D), and acetyl-CoA carboxylase (ACC2) (Fig.2E) were all increased in WT mice whereas no increase in these genes was found in miR-155 KO mice after HF–HC–HS diet (Fig.2B–E). Genes involved in cholesterol regulation such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and lipid binding transport (LDLR) either showed a significant down regulation (HMGCR, Supple. Fig.1A) or no changes (LDLR, (Supple. Fig.1B) in either of genotypes.
Figure 2. The lipid metabolism genes show attenuation in miR-155 KO mice after HF-HC-HS diet.
The liver triglyceride levels were quantified biochemically from WT and miR-155 KO mice (n=5–8) (A). Expression of genes involved in fatty acid regulation such as Cpt1a (B), FAB4 (C), FAS (D), and ACC2 (E) was evaluated by qRT-PCR from the total RNA extracted from the livers (n=8–10). Expression of transcriptional factors involved in the regulation of fatty metabolic gene cluster, PPARα (F), LXRα (G) and C/EBPα (H) were checked by qRT-PCR. 18S mRNA was used to normalize the cq values. Data is presented as mean ± SEM. *p<0.05 compared to chow fed WT mice, # p<0.05 compared to chow fed miR-155 KO mice.
Transcriptional factors such as peroxisome proliferator-activated receptor alpha (PPARα), liver X receptor alpha (LXRα) and CCAAT/enhancer-binding protein alpha (C/EBPα) regulate lipid and glucose homeostasis and are the validated targets of miRNA-155 6, therefore we checked the expression of these genes. We found no changes in the hepatic PPARα mRNA levels in WT mice whereas a significant increase in its expression was found in miR-155 KO mice after HF–HC–HS diet (Fig.2F). No changes in LXRα expression were observed in either of the genotypes (Fig.2G). WT mice showed a significant decrease in C/EBPα expression and miR-155 KO mice displayed a further reduction in its expression after HF–HC–HS diet (Fig.2H). miR-155 KO mice on chow fed diet also exhibited a decrease in C/EBPα (Fig.2H).
Attenuation of inflammasome upregulation in miR-155 KO mice after HF-HC-HS diet
Previously we demonstrated that prolonged HF-HC-HS diet caused increased liver inflammation and inflammasome activation 8. Hence, we evaluated the inflammation markers and found an increase in TNFα mRNA levels in WT mice after HF-HC-HS diet (Fig.3A) that was significantly attenuated in miR-155 KO mice. No significant changes in hepatic TNFα protein levels were found in either of the genotypes (Supple.Fig.1C). The chemokine MCP1 plays roles in inflammation and steatosis and a significant increase in its levels was observed in the livers of WT mice after HF-HC-HS diet (Fig.3B). Compared to WT mice, a significant attenuation in MCP1 mRNA levels was found in miR-155 KO mice (Fig.3B). The hepatic protein levels of MCP1 were significantly increased in WT mice; however, there was no significant increase in MCP1 in miR-155 KO mice (Supple.Fig.1D). Next, we evaluated NF-κB p65 transcription factor DNA binding activity in the liver nuclear extracts and found a significant increase in p65 DNA binding activity in both WT and miR-155 KO mice (Fig.3C). Inflammasome activation has been shown to play an important role in the development and progression of NAFLD 8,9. We found increased transcription of NLR family pyrin domain containing 3 (NLRP3) (upstream activator gene) in WT mice and a significant attenuation in its expression was observed in miR-155 KO mice after HF-HC-HS diet (Fig.3D). The protein levels of NLRP3 were not significantly increased in WT mice (Fig.3E), however miR-155 KO mice displayed a significant reduction in NLRP3 protein levels after HF-HC-HS diet (Fig.3E). The baseline expression of NLRP3 (mRNA and protein) was also decreased in miR-155 KO chow fed mice (Fig.3D and E). Downstream genes in the inflammasome cascade, apoptosis-associated speck-like protein containing a CARD (ASC) (Fig.3F) and pro-caspase-1 (Fig.3G) were increased in WT mice and a significantly attenuation in all of these genes was found in miR-155 KO mice after HF-HC-HS feeding (Fig.3F–G).
Figure 3. Inflammasome upregulation is decreased in miR-155 KO mice after HF-HC-HS diet.
The expression of inflammation markers TNFα (A) and MCP1 (B) was determined by qRT-PCR from the total RNA extracted from the liver samples (n=8–10). NF-κB p65 transcription factor assay was used for the determination of p65 transcription factor DNA binding activity from the liver nuclear extracts (C). The expression of NLRP3 inflammasome at the mRNA (D) and protein level (E, left panel) was determined by qRT-PCR and Western blot analysis. Densitometery analysis of NLRP3 band intensity compared to chow fed WT mice is presented as a bar graph (E, right panel, n=8–10). mRNA levels of ASC (F) and pro-caspase1 (G) was evaluated by qRT-PCR (n=8–10). 18S mRNA was used to normalize the cq values. GAPDH was used as a loading control for Western blot analysis. Data is presented as mean ± SEM. p<0.05, ** p<0.01 compared to chow fed WT mice, # p<0.05 compared to chow fed miR-155 KO mice. **** p<0.0001.
Macrophage inflammatory markers are decreased in miRNA-155 KO mice after HF-HC-HS diet
Macrophage activation plays a central role in NAFLD/NASH pathogenesis (8). Macrophage activation and changes in macrophage phenotype have been reported after long-term HF-HC-HS diet 8. We found a significant induction in the hepatic expression of macrophage inflammatory markers F4/80 (Fig.4A), CD68 (Fig.4B) and CD11b (Fig.4C) in WT mice whereas miR-155 KO mice showed a decrease in these markers after HF-HC-HS diet (Fig.4A–C). Even chow fed miR-155 KO mice displayed a decreased expression in these markers compared to WT control mice (Fig.4A–C). Next, we checked the number of hepatic macrophages by CD68 immunohistochemistry (Fig.4D, left panel). Quantification of CD68 stained cells by Image J software displayed comparable increase in CD68 macrophages in both genotypes (Fig.4D, right panel), suggesting number of CD68 specific liver macrophages was not affected in miR-155 KO mice (Fig.4D). CD163, a M2 macrophage marker, was significantly decreased in both genotypes after HF-HC-HS diet (Fig.4E). WT mice showed no changes in Arginase1, M2 macrophage marker, whereas it was significantly decreased in miR-155 KO mice after HF-HC-HS diet (Fig.4F). This data suggests that miR-155 KO mice exhibited a decrease in macrophage inflammatory markers.
Figure 4. Macrophage inflammatory markers show attenuation in miR-155 KO mice after HF-HC-HS diet.
Expression of M1 macrophage markers F4/80 (A), CD68 (B) and CD11b (C) was determined by qRT-PCR assay (n=8–10). CD68 immunohistochemistry was performed on the paraffin embedded liver sections and representative pictures are shown (D, left panel). The CD68 stained cells were quantified using Image J software and mean intensity was presented as graph (D, right panel). M2 macrophage markers, CD163 (E) and Arg-1 (F) was determined by qRT-PCR assay (n=8–10). 18S mRNA was used to normalize the cq values and data is presented as mean ± SEM. *p<0.05 compared to chow fed WT mice, # p<0.05, ** p<0.01 compared to chow fed miR-155 KO mice, ns: non-significant.
miRNA-155 KO mice display attenuation in liver fibrosis after HF-HC-HS diet
Advanced human NASH/NAFLD is accompanied by fibrosis that over time develops to cirrhosis and increases the risk of hepatocellular carcinoma 8. Consistent with our previous findings of increased fibrosis after long term HF-HC-HS diet 8, we found increased Sirius red staining in the livers of WT mice whereas miR-155 KO mice showed significant attenuation in the stained area as observed under microscope (Fig. 5A, left panel) and quantified by Image J software (Fig. 5A, right panel). Both mRNA and protein levels of alpha smooth muscle actin (αSMA) were increased in WT mice after HF-HC-HS diet (Fig.5B and C). In contrast, miR-155 KO mice showed a significant attenuation in the αSMA levels than WT mice (Fig.5B and C). Consistent with these findings, we found increased hydroxyproline levels in WT mice and a significant attenuation in collagen content was observed in miR-155 KO mice after HF-HC-HS diet (Fig.5D). Evaluation of the mRNA levels of pro-collagen 1α also revealed attenuation in its expression in miR-155 KO mice compared to WT mice after HF-HC-HS diet (Fig.5E). TGFβ and TIMP1 genes were induced in WT mice whereas miR-155 KO mice showed significant decrease in their levels after HF-HC-HS diet (Fig.5F and G). Next, we checked the MMPs levels and found induction in MMP2 expression in WT mice and a significant augmentation in its levels in miR-155 KO mice (Fig.5H). No significant changes were observed in MMP9 levels in either genotype after HF-HC-HS diet (Fig.5I). However, a baseline decrease in MMP9 expression was found in chow fed miR-155 KO mice and this decrease was prevented in KO mice after HF-HC-HS diet (Fig.5I). Zeb1 and Zeb2 belong to the zinc finger-homeodomain transcription factor family and play pivotal role in the epithelial-mesenchymal transition 10 and we found both as predicted targets of miR-155 (www.microrna.org). No significant changes were observed in Zeb1 levels in either of genotype (Fig.5J). However, Zeb2 expression was increased in WT mice at the mRNA (Fig.5K) and protein (Fig.5L) levels and this increase in Zeb2 expression was prevented in miR-155 KO mice after HF-HC-HS diet (Fig.5K and L). Vimentin, a Zeb2 target gene, involved in fibrogenesis was found to be increased in WT mice whereas a significant attenuation in its protein levels was observed in miR-155 KO mice after HF-HC-HS diet (Fig.5M).
Figure 5. miR-155 KO mice show less fibrotic changes after HF-HC-HS diet.
Sirius red stained liver sections were observed under microscope and representative pictures are shown (A, left panel) and stained area was quantified using Image J software (A, right panel) (n=8–10). αSMA levels were evaluated by qRT-PCR (B), and by western blot analysis (C, left panel). Densitometry units are presented as fold change compared to WT chow fed mice after normalization with GAPDH signal (C, right panel) (n=6–7). Total collagen amount was determined from the liver samples by measuring hydroxyproline content biochemically (D) (n=4–6). Expression of genes involved in the fibrotic changes was assessed by qRT-PCR, pro-collagen1α (E), TGFβ (F), TIMP1 (G), MMP2 (H), MMP9 (I), Zeb1 (J) and Zeb2 (K). 18S mRNA was used to normalize the cq values. Zeb2 and vimentin protein levels were determined by western blot analysis (L, and M left panel). Densitometry units are presented as fold change compared to WT chow fed mice after normalization with GAPDH signal (L, and M right panel) (n=6–8). Data is presented as mean ± SEM. *p<0.05 compared to chow fed WT mice, **p<0.01, ***p<0.001, ****p<0.0001, # p<0.05 compared to chow fed miR-155 KO mice.
Discussion
The increasing prevalence of NAFLD is a public health concern and a global economic burden. The pathophysiology of NAFLD is complex and its spectrum ranges from steatosis to steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma if left untreated (11). Previously, we characterized a HFD mouse model supplemented with cholesterol and high fructose corn syrup (HF-HC-HSD) that resulted in liver steatosis at 8 weeks, progressed to steatohepatitis and early fibrosis at 27 weeks, and by 49 weeks, NASH progressed to fibrosis and liver tumor development (8). Multiple parallel factors are suggested to act simultaneously and synergistically to induce NAFLD/NASH 11 and these “hits” of progression of NAFLD-NASH are well represented in our HF-HC-HS diet mouse model (8). NASH-related HCC is currently the fast growing indication for liver transplant in HCC patients 12. Various cellular events are involved in the pathogenesis of NAFLD/NASH, which are regulated at the multiple levels. miRNAs have emerged as fine tuners of various cellular processes. In this study, we show that miR-155 is the one of the regulators of liver injury, steatosis, and fibrotic events caused by high fat-high cholesterol-high sugar (HF-HC-HS) diet. We found induction of miR-155 levels in NASH patients and in the livers of HF-HC-HS diet fed mice. The mechanistic role of miR-155 in NASH is supported by our observations that miR-155 KO mice exhibited attenuation in steatohepatitis and fibrosis in the HF-HC-HS diet model of NASH.
Lipid metabolism plays an essential role in NAFLD pathogenesis and miR-155 regulates various transcriptional factors involved in lipid metabolism such as PPARα, PPARγ 6, LXRα 13, SREBP-1c 14, C/EBPα (15), C/EBPβ 15, and SIRT1 16. PPARα and PPARγ have opposing functions in the regulation of fat metabolism, PPARα promotes utilization, while activation of PPARγ promotes storage 17. miR-155 regulates both PPARα and PPARγ 6 and we found an increase in PPARα transcripts in miR-155 KO mice. Though PPARα transcripts were increased in miR-155 KO mice, we can not rule out the possibility of Cpt1α regulation by other miR-155 target transcriptional factors involved in lipid metabolism. Fatty acid synthesis and regulatory genes such as FAS, ACC2, and FAB4 were all attenuated in miR-155 KO mice after HF-HC-HC diet, suggesting a regulatory role of miR-155 in fatty acid homeostasis in our NASH model. LXRα is another regulator of lipid metabolism and miR-155 was shown to target LXRα in NAFLD 13. We didn’t find any changes in LXRα expression in WT or miR-155 KO mice after HF-HC-HS diet. This could be due to differences in the duration of feeding, diet composition and phase of the liver injury.
The C/EBPα transcription factor regulates various functions of the liver including hepatic glucose and lipid metabolism and proliferation 18. miR-155 was shown to regulate C/EBPα levels in adipocytes 18 and mice overexpressing C/EBPα were found to be more susceptible to hepatic steatosis 19. Consistent with this, in our study, we found decreased expression of C/EBPα in WT mice and miR-155 KO exhibited a further reduction in its levels. Our results support the studies suggesting miR-155 an important direct or indirect regulator of genes involved in lipid metabolism 13. Since miR-155 targets various transcriptional factors involved in lipid metabolism it is likely that the phenotype we observed in miR-155 KO mice is due to cumulative effects of miR-155 on several genes rather than on a single gene and miR-155 could also be affecting lipogenesis at multiple levels.
Though we found improved steatosis in miR-155 KO mice in our HF- HF-HC-HS diet mouse model, others have reported enhanced steatosis in miR-155 KO-HFD mice and vice versa (14). The contradictory findings could be due to differences in the diet composition (cholesterol, 0.2% −2% versus 10%, and high sugar), duration of diet (8–12 weeks versus 34 weeks), and stage of the disease (fatty liver and early hepatitis versus steatohepatitis with fibrosis) between previous and our study.
It has been reported in the literature that differences in diet composition can result in distinctions in metabolic and liver pathobiology. One study compared two dietary mouse models of NAFLD and showed that modest differences in diet can significantly uncouple glucose homeostasis and liver damage 20. The differential effects of miR-155 deficiency on steatohepatitis in different diets suggest that miR-155 might have distinctive roles at the different phases of the liver disease [early (fatty liver/NASH), mid (NASH with fibrosis) or chronic (cirrhosis/HCC)]. In accordance with this notion, we found higher ALT levels in miR-155 KO mice after 14 weeks of HF-HC-HS diet. WT mice exhibited a progressive increase in ALT levels over course of diet whereas a decrease in ALT levels was found in miR-155 KO mice after 14 weeks (Supplementary Fig.1).
miR-155 is known to promote inflammation via targeting various genes involved in inflammatory pathway 21. MCP-1, a macrophage regulatory chemokine was increased in the livers of WT, but not in miR-155 KO mice after HF-HC-HS diet that correlated with decreased F4/80, CD68, and CD11b levels in miR-155 KO mice. Our results further indicated that number of CD68 specific hepatic macrophages was not affected in miR155 KO mice. Though the number of CD68 macrophages/KC was not affected in miR155 KO mice, it is possible they might be less active as miR-155 was shown to regulate macrophage/ monocyte activation 21,22. Also, it is possible that a number of other macrophage types such as F4/80 macrophages, and monocytes/neutrophils might have affected in miR-155 KO mice. Previously, we demonstrated that although the number of inflammatory monocytes was not attenuated in miR-155 KO mice in ALD mouse model, the alcohol induced neutrophil infiltration was prevented and inflammation was improved in miR-155 KO mice 6. In MCD diet induced NASH mouse model, miR-155 KO mice still showed increased inflammation, but the steatosis and fibrosis were improved 5, suggesting miR-155 may have a context-dependent function.
Though we found reduction in TNFα and MCP1 transcripts in miR-155 KO mice, the NF-κB p65 nuclear binding activity was induced in both genotypes to a similar extent. It is possible that miR-155 is regulating TNFα and MCP1 directly as previously we showed that miR-155 regulates TNFα mRNA stability 21 and others have shown MCP1 regulation via miR-155 23. NF-κB regulates distinct functions in a cell type dependent manner 24. In parenchymal cells, activation of NF-κB is crucial for survival, whereas in Kupffer cells, it is known to promote inflammation 24.
In recent years, inflammasomes have emerged as new players in the pathogenesis of NAFLD. The NLRP3 inflammasome is comprised of NLRP3, ASC, and caspase1 genes and has been shown to play various roles in NASH/NAFLD 25. Increased hepatic NLRP3 levels have been reported in NALFD/NASH (mouse and humans) and pharmacological inhibition of NLRP3 or NLRP3 deficient mice showed protection from steaohepatitis and liver fibrosis 25,26. Consistent with previous studies, we found induction of NLRP3, ASC and caspase 1 inflammasome in WT mice and upregulation of inflammasome complex was either prevented or reduced in miR-155 KO mice. NLRP3 was shown to induce miR-155 expression and miR-155 regulated NLRP3 mediated inflammation and fibrosis 27,28. It was revealed that miR-155 regulates inflammatory responses in psoriasis via NLRP3 pathway and miR-155 silencing significantly suppressed NLRP3 and caspase-1 in keratinocyte cell lines 27. In systemic sclerosis fibroblasts miR-155 was shown to be required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis 28. In our study the observed protection from liver fibrosis in miR-155 KO mice could be in part due to miR-155’s effect on NLRP3. Our results show a potent attenuation in liver fibrosis in miR-155 KO mice, however inflammatory markers showed minimal reduction. Similar observation was reported in NLRP3 activated mice where fibrotic changes were independent of hepatic inflammatory macrophage infiltrates 26.
Though we didn’t detect cleaved form of IL-1β in our model we can not rule out the possibility of direct involvement of NLRP3 alone in fibrosis independent of inflammasome activation. Previously, we found earlier induction in NLRP3 (8 and 27 weeks) but IL-1β was detectable only after 49 weeks of HF-HC-HSD. Recent studies suggest a direct role for NLRP3 in fibrosis independent of inflammasome activation 29. It is suggested that the EMT process might happen in two ways: one is dependent on the inflammasome activation via IL-1β -TGFβ pathway; another is independent of the inflammasome activation 29. It was revealed that NLRP3 promotes TGFβ signaling and R-Smad activation in epithelial cells independent of the inflammasome 30. Another study demonstrated that the NLRP3 expression but not NLRP3 inflammasome complex activation was required for EMT in colorectal cancer cells 31.
Matrix metalloproteinases (MMPs) play a crucial role in homeostatic regulation of the extracellular environment and degradation of the liver matrix 32. Depending upon the cell injury milieu, MMP2 has been reported to play dual roles, either in promoting or resolving fibrosis 32,33. In chronic CCl4 administration mouse model, MMP2 deficient mice displayed enhanced liver fibrosis and TIMP1 upregulation 32. Another study reported that MMP2 inhibits type I collagen synthesis in activated hepatic stellate cells and plays a protective rather than pathologic role 33. Consistent with these reports we found enhanced MMP2 levels, decreased TIMP1 and procoallgen1α levels and reduction in hydroxyproline content in miR-155 KO mice, suggesting that absence of miR-155 protects from fibrosis. miR-155 is shown to modulate MMP2 and MMP9 levels either directly or indirectly 34.
Epithelial to mesenchymal transition (EMT) is a biological transition of polarized epithelial cells into mobile mesenchymal cells 10. ZEB protein family of transcription factors (Zeb1 and 2) promotes EMT via repression of epithelial genes 10. In our study we found an increases in Zeb2 and its target gene, vimentin, in WT mice whereas miR-155 KO mice showed attenuation in these genes. Smads transcriptionally activate ZEB1/2 35 and miR-155 is a known regulator of Smad2/3 36. It is possible that in our model miR-155 is regulating fibrotic events in part via modulating Zeb2-vimentin axis directly or indirectly via SMAD2/3.
Though we found improved fibrosis in miR-155 KO mice in our HF-HC-HSD model, Dai et al reported inhibition of miR-155 in rat hepatic stellate cell line promoted the accumulation of myofibroblastic stellate cells via EMT and Erk signaling. Our unpublished results suggest a reduction in miR-155 in LX2 cells (a human HSC cell line) after TGFβ treatment. Considering the complexity of in vivo system, it is likely that phenotype we have observed for fibrosis in miR-155 KO mice may not be due to miR155 role in one cell type, but complex interactions of various cell types. Since miR-155 is expressed in various liver cells (parenchymal and non-parenchymal cells) and regulates multiple genes, dissecting its role in vivo at the cellular level will be able to define precise role.
Previously, we showed that miR-155 KO mice exhibited reduced fibrosis phenotype in alcoholic liver disease and CCl4-induced liver fibrosis 6, and MCD diet induced NASH 5 and our current study suggests improvement in HF-HC-HSD-induced liver fibrosis in miR-155 KO mice. Since miR-155 targets various transcriptional factors involves in inflammation, lipid metabolism and fibrogenesis, it is possible that depending upon the stimuli, and stage of the disease, alternation in the transcriptional factors in different cell types might result in differential outcome. Similar opposing effects of miR-155, anti-fibrotic and pro-fibrotic have been reported in lung fibrosis 37–39. It was suggested that contradictory findings on miR-155 in lung fibrosis may have been the results of different study design and use of different cells, cytokines and pathways and at different stage of the disease 37.
Our study further raises questions whether miR-155 plays differential roles in healthy, and NALFD/NASH livers via its ability to simultaneously fine tune the expression of various transcriptional factors. Depending upon the diet composition, and stimuli, miR-155 may be able to execute distinct functions via modulating the abundance of target genes. Based on studies conducted on different HFD models it is clearer that miR-155 has a complex role in liver homeostasis and is subjective to the diet composition, duration, cell type and phase/stage of the liver injury. Our study highlights the hierarchy of miR-155 regulation and function in a context-dependent fashion. These criteria should be taken into consideration for miR-155 based therapeutic approaches.
In summary, our findings suggest a role for miR-155 in regulating key cellular events in NAFLD/NASH including liver injury and steatosis and to a greater extent fibrosis in the mouse model of HF-HC-HS diet. Genes such as NLRP3, Zeb2, and vimentin that are known to modulate fibrotic events are attenuated in miR-155 KO mice. Based on our findings, we speculate that miR-155 exerts its effect on fibrosis via targeting various genes involved in the fibrogenesis.
Supplementary Material
Acknowledgments
The authors thank Karen Kodys and Donna Catalano for helping in the animal feeding.
Funding
Research reported in this publication was supported by National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under award number R01AA0207440 to Dr. Szabo.
Footnotes
Conflict of Interest
G.S. reports being a paid consult for Allergan, Alnylam, Arrow, Durcect Corporation, Generon, Glympse Bio, Terra Firma, Quest Diagnostics, Pandion Therapeutics, Surrozen, and Zomagen. G.S. has received grants from Gilead, Genfit, Intercept, Novartis, SignaBlok, and Shire; she also holds intellectual property rights with Up to Date. Other authors declare no conflicts of interest to the research conducted.
References
- 1.Polyzos SA, Kountouras J. & Mantzoros CS Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metabolism 92, 82–97 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y. et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64, 1577–1586 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Szabo G. & Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 10, 542–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabo G. & Csak T. Role of MicroRNAs in NAFLD/NASH. Dig Dis Sci 61, 1314–1324 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Csak T, Bala S, Lippai D, Kodys K, Catalano D, Iracheta-Vellve A. et al. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS One 10, e0129251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D. et al. The proinflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol 64, 1378–1387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahesh G. & Biswas R. MicroRNA-155: A Master Regulator of Inflammation. J Interferon Cytokine Res 39, 321–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz M, Bukong TN, Csak T, Saha B, Park JK, Ambade A. et al. Progression of non-alcoholic steatosis to steatohepatitis and fibrosis parallels cumulative accumulation of danger signals that promote inflammation and liver tumors in a high fat-cholesterol-sugar diet model in mice. J Transl Med 13, 193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Lee HE & Lee JY A pharmacological inhibitor of NLRP3 inflammasome prevents non-alcoholic fatty liver disease in a mouse model induced by high fat diet. Sci Rep 6, 24399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott CL & Omilusik KD ZEBs: Novel Players in Immune Cell Development and Function. Trends Immunol 40, 431–446 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Buzzetti E, Pinzani M. & Tsochatzis EA The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 65, 1038–1048 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Wong RJ, Cheung R. & Ahmed A. Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology 59, 2188–2195 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Miller AM, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One 8, e72324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Zhang N, Wang Z, Ai D. m., Cao Z. y. & Pan H -p. Decreased MiR155 level in the peripheral blood of non-alcoholic fatty liver disease patients may serve as a biomarker and may influence LXR activity. Cellular Physiology and Biochemistry 39, 2239–2248 (2016). [DOI] [PubMed] [Google Scholar]
- 15.He M, Xu Z, Ding T, Kuang D-M & Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cellular & molecular immunology 6, 343–352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zheng Z. j., Jia Y. j., Yang Y. l. & Xue Y -m. Role of p53/miR-1555p/sirt1 loop in renal tubular injury of diabetic kidney disease. Journal of translational medicine 16, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berlanga A, Guiu-Jurado E, Porras JA & Auguet T. Molecular pathways in non-alcoholic fatty liver disease. Clin Exp Gastroenterol 7, 221–239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karkeni E, Astier J, Tourniaire F, El Abed M, Romier B, Gouranton E. et al. Obesity-associated Inflammation Induces microRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function. J Clin Endocrinol Metab 101, 1615–1626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin J, Iakova P, Breaux M, Sullivan E, Jawanmardi N, Chen D. et al. Increased expression of enzymes of triglyceride synthesis is essential for the development of hepatic steatosis. Cell Rep 3, 831–843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Léveillé M, Courty E, Gunes A, Nguyen NB & Estall JL Differences in metabolic and liver pathobiology induced by two dietary mouse models of nonalcoholic fatty liver disease. American Journal of PhysiologyEndocrinology and Metabolism 319, E863–E876 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P. et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. Journal of Biological Chemistry 286, 1436–1444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alivernini S, Gremese E, McSharry C, Tolusso B, Ferraccioli G, McInnes IB et al. MicroRNA-155—at the critical interface of innate and adaptive immunity in arthritis. Frontiers in immunology 8, 1932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elmesmari A, Fraser AR, Wood C, Gilchrist D, Vaughan D, Stewart L. et al. MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in Rheumatoid Arthritis. Rheumatology 55, 2056–2065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czauderna C, Castven D, Mahn FL & Marquardt JU Context-Dependent Role of NF-kappaB Signaling in Primary Liver Cancer-from Tumor Development to Therapeutic Implications. Cancers (Basel) 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mridha AR, Wree A, Robertson AAB, Yeh MM, Johnson CD, Van Rooyen DM et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol 66, 1037–1046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inzaugarat ME, Johnson CD, Holtmann TM, McGeough MD, Trautwein C, Papouchado BG et al. NLR Family Pyrin Domain-Containing 3 Inflammasome Activation in Hepatic Stellate Cells Induces Liver Fibrosis in Mice. Hepatology 69, 845–859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Q, Zeng J, Li W, Lin L, Zhou X, Tian X. et al. Silencing of miR155 suppresses inflammatory responses in psoriasis through inflammasome NLRP3 regulation. Int J Mol Med 42, 1086–1095 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Artlett CM, Sassi-Gaha S, Hope JL, Feghali-Bostwick CA & Katsikis PD Mir-155 is overexpressed in systemic sclerosis fibroblasts and is required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis. Arthritis Res Ther 19, 144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alyaseer AAA, Lima MHS & Braga TT The role of NLRP3 inflammasome activation in the epithelial to mesenchymal transition process during the fibrosis. Frontiers in Immunology 11, 883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Wang X, Chun J, Vilaysane A, Clark S, French G. et al. Inflammasome-independent NLRP3 augments TGF-β signaling in kidney epithelium. The Journal of Immunology 190, 1239–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Wang Y, Du Q, Lu P, Fan H, Lu J. et al. Inflammasome-independent NLRP3 is required for epithelial-mesenchymal transition in colon cancer cells. Experimental cell research 342, 184–192 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Onozuka I, Kakinuma S, Kamiya A, Miyoshi M, Sakamoto N, Kiyohashi K. et al. Cholestatic liver fibrosis and toxin-induced fibrosis are exacerbated in matrix metalloproteinase-2 deficient mice. Biochem Biophys Res Commun 406, 134–140 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Radbill BD, Gupta R, Ramirez MC, DiFeo A, Martignetti JA, Alvarez CE et al. Loss of matrix metalloproteinase-2 amplifies murine toxin-induced liver fibrosis by upregulating collagen I expression. Dig Dis Sci 56, 406–416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, Zheng Z, Zhou Q, Bai X, Fan L, Yang C. et al. miR-155 promotes cutaneous wound healing through enhanced keratinocytes migration by MMP-2. J Mol Histol 48, 147–155 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Chen Y, Liu L, Shen A. & Zheng W. MicroRNA-155–5p suppresses the migration and invasion of lung adenocarcinoma A549 cells by targeting Smad2. Oncol Lett 16, 2444–2452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louafi F, Martinez-Nunez RT & Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta}. J Biol Chem 285, 41328–41336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Mao J, Ye X, Zhang F, Kerr WG, Zheng T. et al. SHIP-1, a target of miR-155, regulates endothelial cell responses in lung fibrosis. The FASEB Journal 34, 2011–2023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurowska-Stolarska M, Hasoo MK, Welsh DJ, Stewart L, McIntyre D, Morton BE et al. The role of microRNA-155/liver X receptor pathway in experimental and idiopathic pulmonary fibrosis. Journal of Allergy and Clinical Immunology 139, 1946–1956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christmann RB, Wooten A, Sampaio-Barros P, Borges CL, Carvalho CR, Kairalla RA et al. miR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis research & therapy 18, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.