Abstract
Tissue repair is largely regulated by diverse Mϕ populations whose functions are timing- and context-dependent. The early phase of healing is dominated by proinflammatory Mϕs, also known as M1, followed by the emergence of a distinct and diverse population that is collectively referred to as M2. The extent of the diversity of the M2 population is unknown. M2 Mϕs may originate directly from circulating monocytes or from phenotypic switching of pre-existing M1 Mϕs within the site of injury. The differences between these groups are poorly understood, but have major implications for understanding and treating pathologies characterized by deficient M2 activation, such as chronic wounds, which also exhibit diminished M1 Mϕ behavior. This study investigated the influence of prior M1 activation on human Mϕ polarization to an M2 phenotype in response to IL-4 treatment in vitro. Compared to unactivated (M0) Mϕs, M1 Mϕs up-regulated several receptors that promote the M2 phenotype, including the primary receptor for IL-4. M1 Mϕs also up-regulated M2 markers in response to lower doses of IL-4, including doses as low as 10 pg/mL, and accelerated STAT6 phosphorylation. However, M1 activation appeared to also change the Mϕ response to treatment with IL-4, generating an M2-like phenotype with a distinct gene and protein expression signature compared to M2 Mϕs prepared directly from M0 Mϕs. Functionally, compared to M0-derived M2 Mϕs, M1-derived M2 Mϕs demonstrated increased migratory response to SDF-1α, and conditioned media from these Mϕs promoted increased migration of endothelial cells in transwell assays, although other common Mϕ-associated functions such as phagocytosis were not affected by prior polarization state. In summary, M1 polarization appears to prime Mϕs to transition into a distinct M2 phenotype in response to IL4, which leads to increased expression of some genes and proteins and decreased expression of others, as well as functional differences. Together, these findings indicate the importance of prior M1 activation in regulating subsequent M2 behavior, and suggest that correcting M1 behavior may be a therapeutic target in dysfunctional M2 activation.
Keywords: inflammation, Mϕ polarization, tissue repair
1 ∣. INTRODUCTION
Mϕs are key players in the tissue repair process that regulate the growth of new blood vessels, or angiogenesis. Due to their highly plastic nature, Mϕ phenotype varies with the shifting phases of healing by changing in response to external cues. The early inflammatory response to injury is dominated by a pro-inflammatory population commonly referred to as “M1,” which then gives way to a very different population known as “M2” in later reparative stages. This phenotype-switching behavior is characteristic of normal tissue repair, and may be particularly important for angiogenesis.1-3 The late-stage, reparative Mϕs seen in vivo can emerge in several ways: the recruitment and differentiation of newly arriving non-classical monocytes, the phenotypic switching of pre-existing pro-inflammatory Mϕs, and the proliferation of these populations.1,4-9 It is yet unknown to what extent each mechanism is responsible for generating the collective “M2” population, and it is probable that all are required for normal healing. Nonetheless, studies have shown that when Mϕs are depleted in the early inflammatory stage, late-stage M2 Mϕs are also reduced and healing is inhibited.6,10 Furthermore, it is known that in several diseases in which healing is stalled, such as in chronic wounds, pro-inflammatory M1-like Mϕs are insufficiently activated and also fail to switch to an M2 phenotype.11-16 Thus, M1-to-M2 phenotypic switching is likely a key mechanism in proper healing, and M1 activation may be an important regulator of subsequent M2 polarization.
A few studies have investigated the effects of M1 activation on subsequent M2 Mϕs in vitro, with conflicting results. Based on the reduced expression of 7 prototypical M2 markers, one study concluded that “mouse and human M1 Mϕs fail to convert to M2 cells upon IL-4 exposure”.17 In contrast, another study that used whole transcriptomic analysis following a similar protocol of exposing murine M0 or M1 Mϕs to IL-4 concluded that M1 Mϕs efficiently switch to M2, with “little to no memory of their polarization history”.18 Clearly, there is a need to define the similarities and differences between M2 populations deriving from M0 or M1 Mϕs, especially given the importance of the M1-to-M2 transition in normal tissue repair. Furthermore, the specific functions of this M1-derived M2 population have not yet been reported.
It is important to note that Mϕ phenotypes can be categorized using a number of nomenclatures, none of which are universally employed or accepted. One approach is to describe phenotypes based on function, such as “pro-inflammatory,” “pro-angiogenic,” “anti-inflammatory,” “pro-healing,” and so on. However, this nomenclature can be problematic due to the influence of timing and context on Mϕ behavior; for example, M1 Mϕs have been described as both anti- and pro-angiogenic19,20 and M2 Mϕs, which are commonly called anti-inflammatory, propagate type 2 inflammation.21,22 Alternatively, the dichotomous system of classically activated “M1” and alternatively activated “M2” was originally coined in reference to the polarizing cytokines secreted by Th1 and Th2 cells.23 This M1/M2 system has since expanded to recognize distinct M2 subtypes, including “M2a” (IL-4-activated Mϕs), and is especially useful for describing Mϕs activated in vitro, though it falls short in describing the true complexity of Mϕ phenotypes. Mϕs in vivo can rarely be described as entirely M1 or M2, instead existing somewhere on a wide spectrum of polarization, but the expression of previously-defined M1 and M2 markers determined from in vitro studies can help to identify “M1-like” or “M2-like” features of more complex phenotypes. In this study, we activated Mϕs in vitro using defined chemical stimuli, and will therefore use the “M1” and “M2” terminologies for these experiments. For more extensive reviews of the pros and cons of various Mϕ nomenclature systems, see Murray et al.24 and Spiller and Koh.25
We hypothesized that M1-to-M2 switching generates a unique phenotype that is important for late-stage healing. We first investigated the potential for unactivated (M0) and M1 Mϕs to switch to the M2 phenotype in response to IL-4 in vitro. Then, to understand the impact of M1 activation on subsequent M2 behavior, we compared the gene expression profiles of IL-4-induced M2 Mϕs derived from M0 or M1 Mϕs (called M0→M2 and M1→M2), then conducted several functional assays of Mϕ behavior. The results identify M1→M2 Mϕs as a distinct phenotype that upregulates select M2 markers and several genes and proteins related to angiogenesis, and exhibits some enhanced prohealing functions.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Monocyte culture and differentiation into polarized Mϕs
Primary human monocytes were purchased from the University of Pennsylvania Human Immunology Core, or isolated as previously described26,27 from peripheral blood purchased from the New York Blood Center. Monocytes were cultured on ultra-low attachment plates in RPMI-1640 supplemented with 10% human serum and 1% penicillin/streptomycin. Media was additionally supplemented with 20 ng/mL of MCSF (Peprotech, Rocky Hill, NJ, U.S.) to induce differentiation into Mϕs. On day 5, Mϕs were either maintained in an unactivated M0 state or they were M1-activated with 100 ng/mL of IFNg (Peprotech) and 100 ng/mL of LPS (MilliporeSigma, Burlington, MA, U.S.) for 24–48 hours (Fig. 1A). For IL-4Rα flow cytometry, an additional M2 group, activated with 40 ng/mL of IL-4 (Peprotech) and 20 ng/mL of IL-13 (Peprotech) was included. For dose-response experiments, M0 and M1 Mϕs were treated with four doses of IL-4 (10 ng/mL, 1 ng/mL, 0.1 ng/mL, and 0.01 ng/mL) for 24 hours. A control group in which M1 stimuli were removed but no IL-4 was added was also included. To repolarize the M0→M2 and M1→M2 phenotypes, M0 and M1 Mϕs were each treated with 10 ng/mL of IL-4 on day 6 for an additional 24–72 hours.
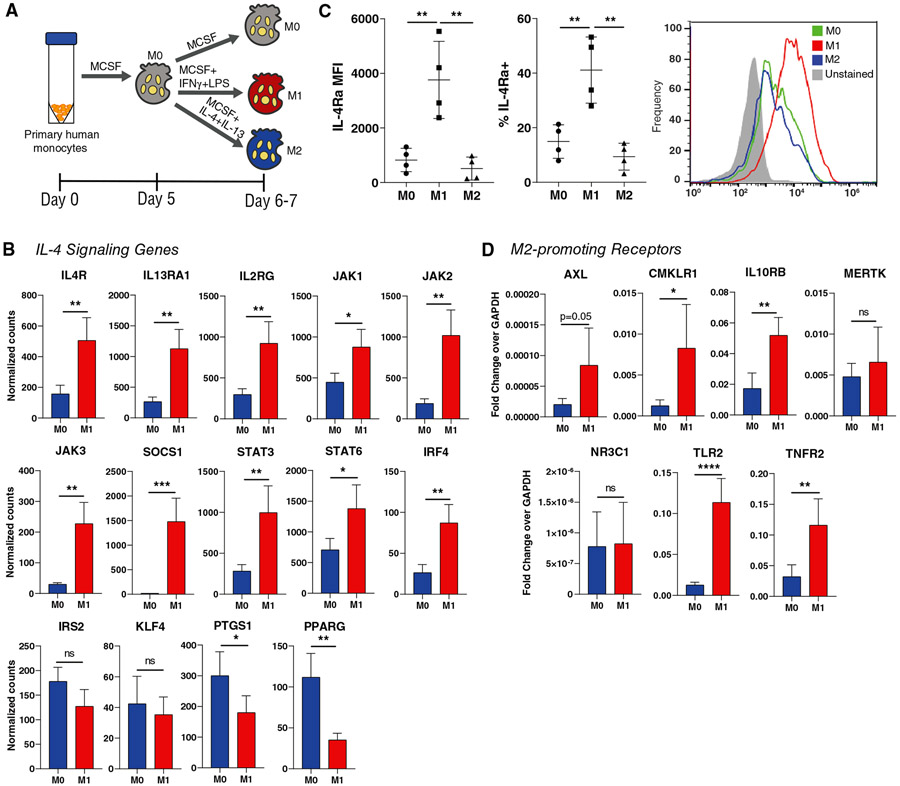
FIGURE 1. M0 and M1 Mϕ expression of M2-promoting receptors.
(A) Polarization of unactivated (M0) Mϕs to the M1 or M2 phenotypes. (B) NanoString counts of genes associated with the IL-4 signaling pathway, using cells collected 24 hours after the addition of polarizing cytokines (day 6). Data are represented as mean ± SD. Unpaired t-test, n = 4 donors, *P < 0.05, **P < 0.01, ***P < 0.001. (C) Flow cytometric analysis of IL-4Rα expression, using cells collected at day 7. Data are represented as mean ± SD. One-way ANOVA with Tukey’s post hoc, n = 4 donors, **P < 0.01. (D) qRT-PCR analysis of receptors associated with promoting an M2-like Mϕ phenotype, using cells collected at day 7. Data are represented as mean ± SD. Unpaired t-test, n = 3-4 donors, *P < 0.05, **P < 0.01, ****P < 0.000
2.2 ∣. NanoString gene expression assays
RNA was extracted from cells using the RNAqueous-Micro Total RNA Isolation Kit (ThermoFisher, Waltham, MA, USA) and RNA concentration was measured using the Nanodrop ND1000. Gene expression was measured with custom-designed gene panels (NanoString Technologies, Seattle, WA, USA), using 100 ng of RNA per sample, according to NanoString’s protocol. Quality control analysis was conducted and raw data were normalized to in-house positive controls using nSolver 4.0 software (NanoString Technologies, Seattle, WA, U.S.) as recommended by the manufacturer.
2.3 ∣. Flow cytometry
Mϕs were incubated with Fc block (BD Biosciences, Franklin Lakes, NJ, U.S.) for 10 min at 4°C, then incubated with primary-conjugated Abs (Biolegend, San Diego, CA, U.S.) and viability stain (Invitrogen, Carlsbad, CA, USA) for 15 min at 4°C. Abs used to stain for IL-4Rα (n = 4 donors) were APC anti-human CD124 (Biolegend, clone G077F6) and Live/Dead Fixable Green (Invitrogen). To amplify IL-4Rα signal, APC FASER kit (Miltenyi Biotec, Bergisch Gladbach, Germany) was used for 2 cycles, according to kit instructions. Antibodies used to stain for CXCR4 (n = 7 donors) were APC anti-human CD184 (Biolegend, clone12G5) and Live/Dead Fixable Green (Invitrogen). The viability stain used to stain Mϕs for phagocytosis was Live/Dead Fixable Aqua (Invitrogen). Samples were fixed and permeabilized using the Foxp3/Transcription Factor Staining Buffer Kit (Tonbo Biosciences, San Diego, CA, USA), then incubated with primary-conjugated Abs for 45 min at 4°C. PE anti-human CD68 (Biolegend, clone Y1/82A) was used as a pan- Mϕ stain.
To stain for phosphorylated STAT6 (n = 3 donors), True-Phos Perm Buffer (Biolegend), and APC anti-STAT6 Phospho (Tyr641) Ab (Biolegend, clone A15137E) were used according to the manufacturer’s instructions.
Data were measured on the Accuri C6 (BD) or LSR II (BD) flow cytometer and analyzed using FlowJo v10 software (BD).
2.4 ∣. Quantitative reverse transcription-polymerase chain reaction
RNA was extracted using the RNAqueous-Micro Total RNA Isolation Kit and RNA concentration was measured using the Tecan Infinite M200 microplate reader. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Invitrogen). qRT-PCR was conducted using custom oligonucleotides (ThermoFisher) and SYBR Green Master Mix (ThermoFisher). Statistical testing was performed using log-transformed data.
2.5 ∣. Enzyme-linked immunosorbent assays
Cell-conditioned media was collected 24–48 h after the addition of polarizing cytokines and frozen at −80°C until ELISAs were run. ELISAs were conducted using kits from Peprotech (PDGF-BB, CCL5, and VEGFA) and R&D Systems (Minneapolis, MN, U.S.) (CCL17). Absorbance was measured on the Tecan Infinite M200 microplate reader. N = 4-11 donors.
2.6 ∣. Migration assays
2.6.1 ∣. Mϕ migration in response to SDF-1α
6.5 mm polyester transwell membranes with 8.0 μm pores (Corning, Corning, NY, USA) were coated with 100 μL of 15 μg/mL bovine type I collagen (Gibco, ThermoFisher) for one hour and placed in a 24-well plate. Mϕs (from n = 3 donors, each with 2 experimental replicates) were suspended in serum-free media at a concentration of 500,000 cells/mL and 100 μL were added to each transwell. After allowing cells to settle for 10 min, 600 μL of media supplemented with 10 ng/mL of recombinant human stromal-derived factor-1α (SDF-1α) was added to the bottom chambers of the experimental group. Six hundred microliters of basal media was added to the bottom chambers of the negative control group. Samples were incubated for 24 h, after which media and remaining cells in the upper chamber were removed with sterile cotton swabs, while cells adhered to the lower membrane were fixed with ethanol and stained with 0.2% crystal violet solution (Fisher Chemical, Hampton, NH, U.S.). Images of the transwells were taken on an inverted microscope set to 10x objective and cells were counted using ImageJ software (NIH, Bethesda, MD, U.S.).
2.6.2 ∣. Endothelial cell migration in response to Mϕ-conditioned media
Mϕ-conditioned media without the influence of polarizing cytokines was generated by incubating polarized Mϕs in basal media for 24 h, then frozen at −80°C until assay was conducted. Human umbilical vein endothelial cells (HUVECs) were suspended in serum-free media at a concentration of 500,000 cells/mL and 100 μL were added to each transwell coated with 100 μL of 15 μg/mL bovine type I collagen. After allowing cells to settle for 10 minutes, 600 μL of Mϕ-conditioned media was added to the bottom chambers. Samples were incubated for 24 hours, after which cells on the bottom of the transwell were stained and counted as described above.
2.7 ∣. HUVEC and fibroblast proliferation/metabolism assay
HUVECs were seeded in 96-well plates at 50,000 cells/well and cultured in 50 μL of endothelial cell media (Sciencell) + 50 μL of Mϕ-conditioned media for 24–48 h. Media supplemented with 10 ng/mL of recombinant human VEGF (GenScript, Piscataway, NJ, USA) was used as a positive control.
Fibroblasts were seeded in 96-well plates at 25,000 cells/well and cultured in 50 μL of fibroblast media (DMEM supplemented with fetal bovine serum, l-glutamine, sodium pyruvate, non-essential amino acids, and penicillin/streptomycin) + 50 μL of Mϕ-conditioned media for 24 h.
Ten microliters of MTT reagent from the MTT Cell Proliferation Assay Kit (Cayman Chemical, Ann Arbor, MI, U.S.) was added to each well and incubated for 3–4 h. One hundred microliters of sodium dodecyl sulfate from the same kit was then added to each well and incubated for an additional 18 h. Absorbance at 570 nm was measured on the Tecan Infinite M200 microplate reader.
2.8 ∣. Phagocytosis
Viable HL-60 cells were labeled with CellTrace Far Red (Invitrogen) according to manufacturer instructions, then serum-starved for 2 h. Hydrogen peroxide (Fisher Chemical) was added to the cell suspension at a concentration of 800 μM for 3 h to induce apoptosis. Mϕs (n = 3 donors) were co-cultured with either apoptotic HL-60s or crimson FluoSpheres (Invitrogen) for 3 hours to facilitate phagocytosis. Samples were immediately stained for CD68 and Live/Dead for flow cytometry analysis as described above.
2.9 ∣. Statistics
Results were analyzed using GraphPad Prism 8 software (GraphPad Software, San Diego, CA, U.S.).
3 ∣. RESULTS
3.1 ∣. M1 activation increases expression of IL-4Rα, IL4 signaling genes, and other M2-promoting receptors
In order to test the potential of M0 and M1 Mϕs to switch to the M2 phenotype, we determined their expression of receptors known to promote the M2 phenotype. As IL-4 is the primary cytokine used to polarize M2 Mϕs in vitro and is also strongly implicated in M2 polarization in vivo,28,29 we used NanoString multiplex gene expression analysis to measure the expression of the three components of the IL-4 receptor complexes (IL4R, IL13RA1, IL2RG) as well as their corresponding Janus kinases (JAK1, JAK2, JAK3) and several other genes involved in IL-4 signaling.30 Expression of the receptor complexes, Janus kinases, and other IL-4 signaling genes was significantly up-regulated in M1 Mϕs compared to M0 Mϕs (Fig. 1B). We then verified the surface expression of IL-4 receptor alpha (IL-4Rα) on M1 Mϕs via flow cytometry. Mean fluorescence intensity (MFI) of IL-4Rα was significantly higher on M1 Mϕs than both M0 and M2 Mϕs, as was the percentage of IL-4Rα-positive cells (Fig. 1C).
In order to determine if this upregulation of M2-promoting genes and proteins is exclusive to the IL-4 pathway, we used qRT-PCR to measure gene expression of an additional repertoire of receptors involved in M2 polarization outside of the IL-4 pathway, including AXL, MERTK, CMKLR1, IL10RB, TLR2, TNFR2, and NR3C131-36 (Supplemental Table 1). All receptors except MERTK and NR3C1 were upregulated in M1 Mϕs compared to M0 (Fig. 1D). Together, these data indicate that M1 Mϕs may be primed to switch to the M2 phenotype via upregulation of M2-promoting receptors, particularly IL-4Rα.
3.2 ∣. M1 Mϕs are more sensitive to IL-4 than M0 Mϕs
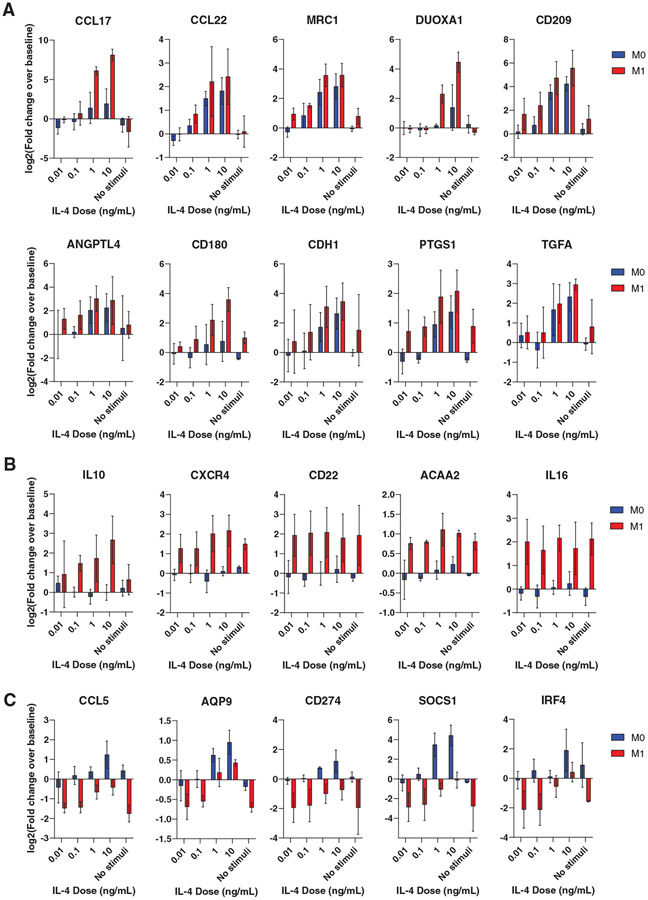
Having confirmed upregulation of IL-4Rα in M1 Mϕs, we hypothesized that this increased expression would translate to increased sensitivity to IL-4. To test this hypothesis, we first compared the response of M0 and M1 Mϕs to low doses of IL-4. After 24 hours of exposure to varying doses (10, 1, 0.1, and 0.01 ng/mL), we used NanoString to measure the expression of a custom panel of 115 genes with mostly M2 markers but also some M1 markers (Supplemental Table 2). We expected the M1 Mϕs to upregulate M2 genes at lower doses compared to the M0 group. Indeed, not only did IL-4-treated M1 Mϕs exhibit greater upregulation of several M2 markers at the lowest doses tested (Fig. 2A), but they also upregulated several M2 genes that were not upregulated in the M0 group (Fig. 2B). Interestingly, some M1-associated genes were downregulated at the lowest IL-4 doses but were upregulated or less downregulated at higher doses, suggesting a unique response to IL-4 (Fig. 2C).
FIGURE 2. Dose response of M0 and M1 Mϕs to IL-4.
Data are represented as log-transformed fold change over baseline expression. N = 3 donors. (A) Shared M2 genes up-regulated in M1 Mϕs at lower doses of IL-4. (B) M2 genes upregulated in M1→M2 Mϕs, with little to no up-regulation in M0→M2 Mϕs. (C) Genes that are upregulated or less down-regulated in M1→M2 Mϕs with increasing doses of IL-4
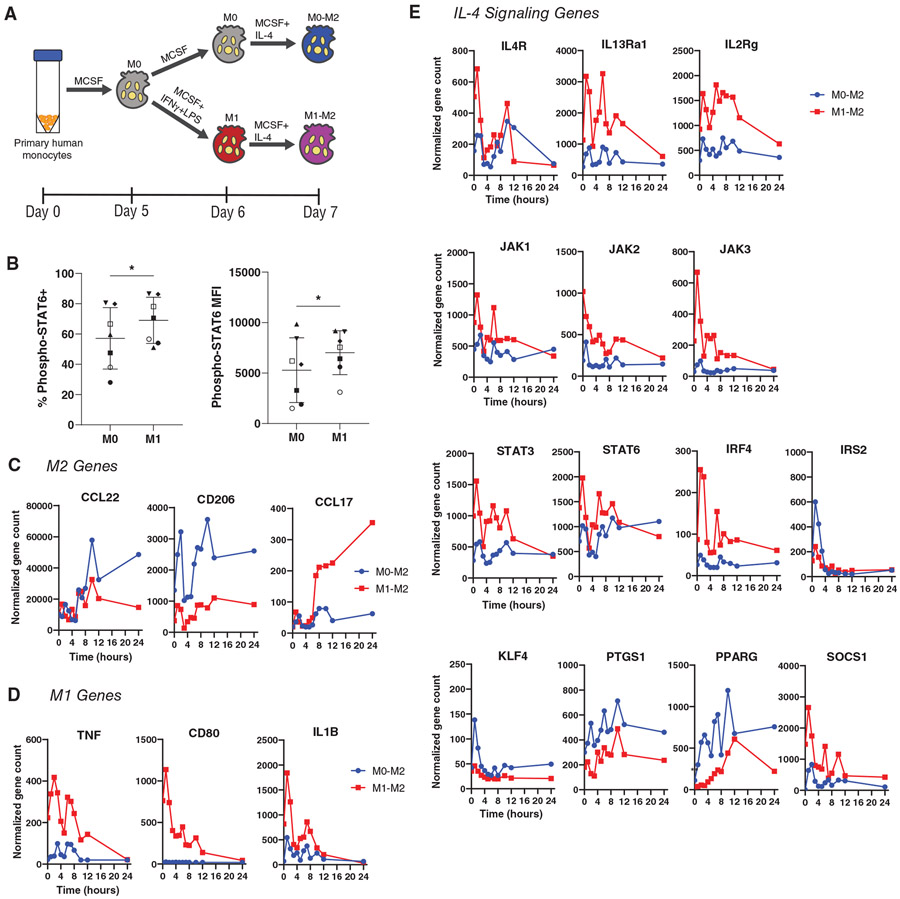
We next tested whether M1 Mϕs would respond more rapidly than M0 Mϕs to IL-4 treatment. Due to the increased IL-4Rα expression in M1 Mϕs, we hypothesized that this would lead to more rapid phosphorylation of STAT6, the transcription factor stimulated by IL-4. After treating M0 and M1 Mϕs with IL-4 for 15 min, we used flow cytometry to determine the extent of STAT6 phosphorylation (Fig. 3B). The M1 group exhibited increased expression of phospho-STAT6, both in terms of proportion of the population and mean fluorescence intensity. Next, using a gene panel of M1, M2, and IL-4 signaling markers, we treated M0 and M1 Mϕs with 10 ng/mL of IL-4 and tracked the expression of these genes over the course of 24 h, expecting M1 Mϕs to upregulate M2 marker genes at earlier time points. We instead found that the two groups upregulated a different set of M2 markers within 24 h of IL-4 exposure, in agreement with the findings from the dose-response experiment. While IL-4-treated M0 Mϕs exhibited increased expression of CD206 and CCL22 immediately after stimulation, IL-4-treated M1 Mϕs up-regulated CCL17, which increased for the duration of the study (Fig. 3C). Additionally, M1 genes were generally downregulated in IL-4-treated M1 Mϕs within 24 hours, confirming a switch in phenotype (Fig. 3D). All but two of the fourteen IL-4 signaling genes evaluated were expressed at higher levels by M1-derived M2 Mϕs for the 24 hour period, although they generally decreased during this time frame (Fig. 3E).
FIGURE 3. Kinetic response of M0 and M1 Mϕs to IL-4.
(A) Treatment of M0 or M1 Mϕs with IL-4 to generate M0-derived M2 (M0→M2) or M1-derived M2 (M1→M2). (B) Phosphorylation of STAT6 after 15 minutes of IL-4 treatment. Data represented as mean ± SD, n = 7 donors. Paired t-test, *P < 0.05. (C) Nanostring counts of genes associated with M2 phenotype, (D) M1 phenotype, and (E) IL-4 signaling pathway, over the course of 24 h of exposure to 10 ng/mL of IL-4. Data are represented as mean only, n = 4 experimental replicates of 1 donor
3.3 ∣. M0-derived and M1-derived M2 Mϕs are distinct phenotypes
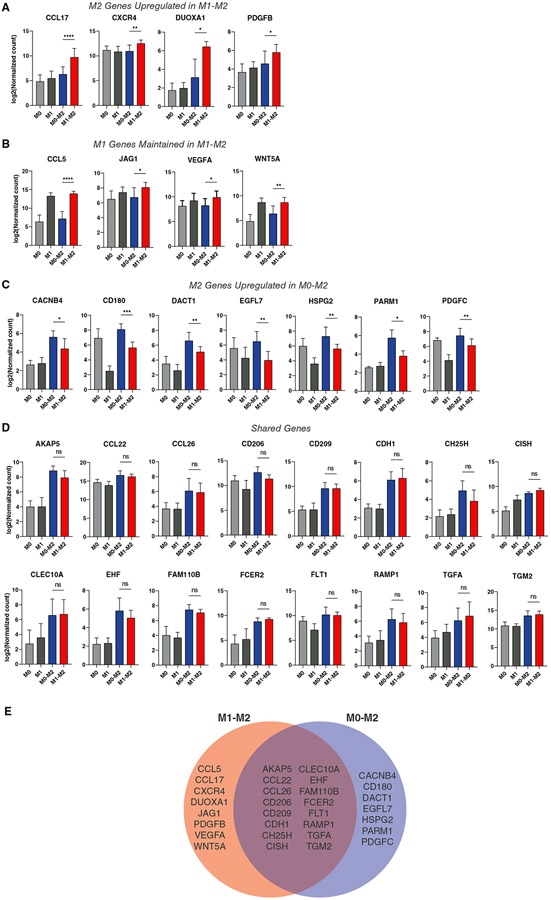
The results of the IL-4 sensitivity experiments led to an updated hypothesis that M1 activation changes the subsequent response to IL-4, as measured by differences in expression of M2 markers and possibly other genes. Using a NanoString panel of genes including Mϕ phenotype markers, genes associated with angiogenesis, and genes associated with tissue deposition or fibrosis (Supplemental Table 3), we evaluated the gene expression profiles of M0 and M1 Mϕs treated with 10 ng/mL IL-4 for 24 hours (M0→M2 vs. M1→M2, respectively). M1→M2 Mϕs expressed higher levels of several typically M2-associated genes compared to M0, M1, and M0→M2 Mϕs, including CCL17, CXCR4, DUOXA1, and PDGFB (Fig. 4A). In addition, while most M1-associated genes were downregulated (Supplemental Figure 1), a few key genes remain elevated, including CCL5, JAG1, VEGFA, and WNT5A (Figure 4B). M0→M2 Mϕs, on the other hand, expressed higher levels of a separate set of genes including PARM1, CD180, DACT1, and HSPG2 (Figure 4C). Both the M0→M2 and M1→M2 groups upregulated a shared set of genes including CD206, CCL22, CD209, CDH1, and CISH, compared to the M0 and M1 controls, but were not significantly different from each other (Figure 4D). Overall, the significant downregulation of M1 markers like CD80, IL1B, and TNF, combined with differential upregulation of CCL17 and other M2 markers, indicates that M1→M2 Mϕs are a distinct M2-like phenotype (Figure 4E).
FIGURE 4. Differential gene expression in M1→M2 Mϕs.
Log-transformed NanoString gene counts of (A) M1→M2 differentially expressed genes (DEGs), (B) M1 markers that remain upregulated in M1→M2, (C) M0→M2 DEGs, and (D) shared M1→M2 and M0→M2 markers. Data are represented as mean ± SD. One-way ANOVA with Tukey’s post hoc, n = 6-10 donors, *P < 0.05, **P < 0.01, ****P < 0.0001. Only significant differences between M0→M2 and M1→M2 are depicted, although differences were detected between other groups. (D) Venn diagram representing shared or differential gene expression among M1→M2 and M0→M2 phenotypes. Markers were selected based on one-way ANOVA of log-transformed gene counts with Tukey’s post hoc, where P < 0.05 indicated significant difference
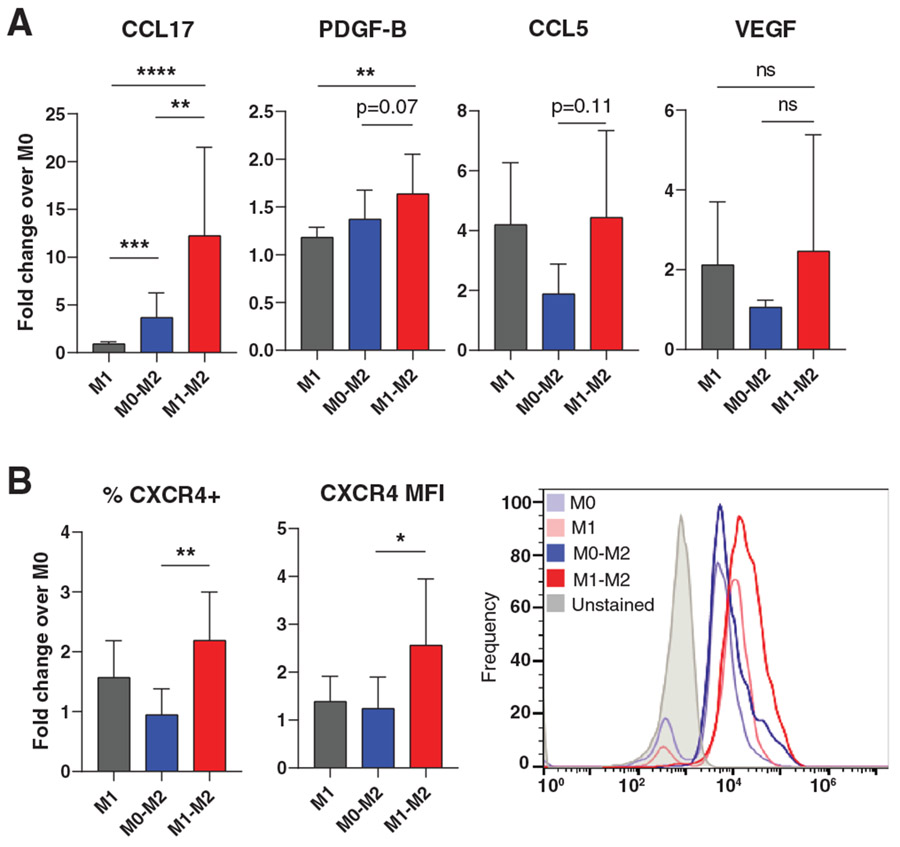
In order to determine whether increased gene expression of CCL17, CCL5, VEGFA, and PDGFB translated to increased protein secretion, we conducted ELISAs to measure concentration of these proteins in the culture media after polarization. As expected, M1→M2 Mϕs secreted higher levels of CCL17, CCL5, and PDGF-BB compared to M0→M2 (Figure 5A). Results of the VEGFA ELISAs, on the other hand, exhibited high variability and no significant differences were found. Flow cytometry was used to confirm protein-level expression of CXCR4 in M1→M2 Mϕs. Both in terms of the percentage of CXCR4-positive cells within the population, as well as mean fluorescence intensity, M1→M2 surface expression of CXCR4 was significantly higher than that of M0→M2 Mϕs (Figure 5B).
FIGURE 5. Differential protein expression in M1→M2 Mϕs.
(A) Protein secretion of selected highly-expressed M1→M2 genes. ELISA analysis of Mϕ-conditioned media. Data are represented as mean ± SD of fold change over M0 control. One-way ANOVA with Tukey’s post hoc conducted on log-transformed data, n = 4-11 donors, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) Flow cytometry analysis of CXCR4 surface expression. Data are represented as mean ± SD of fold change over M0 control. One-way ANOVA with Tukey’s post hoc conducted on log-transformed data, n = 7 donors, *P < 0.05, **P < 0.01. Representative histogram depicts one donor
Collectively, these results indicate that M2 Mϕs that were previously M0 or M1 exhibit distinct phenotypes.
3.4 ∣. M0→M2 and M1→M2 Mϕs exhibit both distinct and similar functions
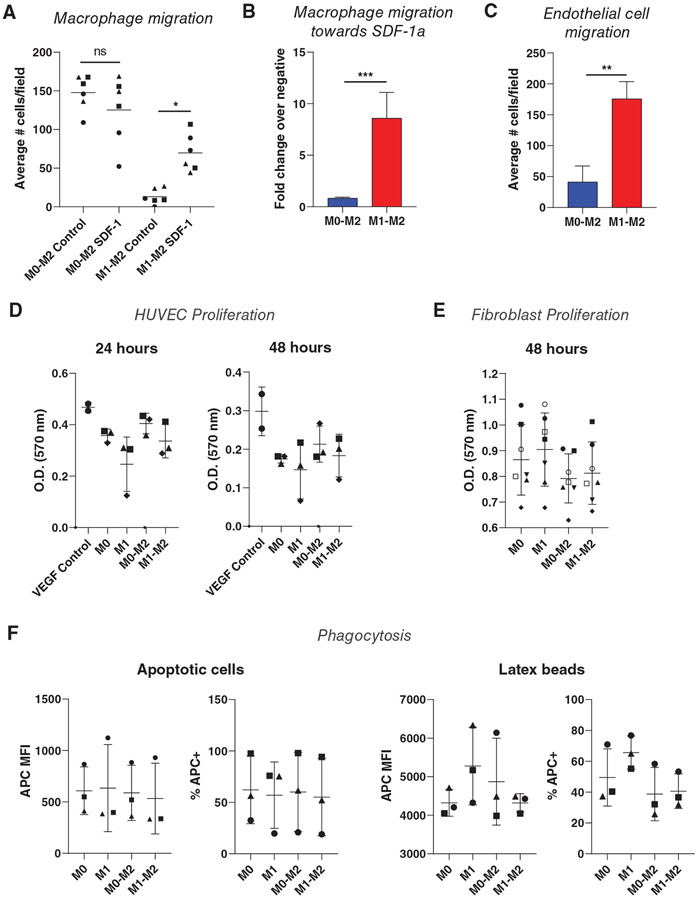
Finally, we set out to evaluate potential differences in functional behavior between M1→M2 and M0→M2 Mϕs. Gene ontology analysis of M1→M2 differentially expressed genes indicated roles in positive chemotaxis of numerous cell types (Supplementary Table 4). Because a critical process in the early inflammatory response is the recruitment of Mϕs to injured cells in large part through SDF-1α, the CXCR4 ligand, and because M1→M2 Mϕs upregulated surface expression of CXCR4 compared to M0→M2 Mϕs, we first tested the ability of these cells to migrate in response to SDF-1α. Using a transwell migration assay, we measured the migratory response of Mϕs to basal media or to media containing SDF-1α. M0→M2 Mϕs were more motile than M1→M2 Mϕs, migrating more in basal media (Figure 6A). However, the inclusion of SDF-1α in the media increased transwell migration of M1→M2 Mϕs, while it had no effect on M0→M2 Mϕs (Figure 6B).
FIGURE 6. Functional assays in M0→M2 and M1→M2 Mϕs.
(A) Migratory response of M0→M2 and M1→M2 Mϕs towards media ± SDF-1. Data are represented as single replicates with mean only. One-way ANOVA with Tukey’s post hoc, n = 6 replicates from 3 donors depicted with different symbols, *P < 0.05. (B) Migratory response of Mϕs towards SDF-1, shown as fold change of experimental cell counts over blank negative control cell counts. Data are represented as mean ± SD. Unpaired t-test conducted on log-transformed data, n = 6 replicates from n = 3 donors, ***P < 0.001. (C) Endothelial cell migration toward Mϕ-conditioned media. Data are represented as mean ± SD. Unpaired t-test, n = 6 replicates from n = 2 donors, **P < 0.01. (D) MTT activity of human umbilical vein endothelial cells after 24 or 48 h of incubation in Mϕ-conditioned media. Data are represented as mean ± SD. One-way ANOVA, n = 3 donors. (E) MTT activity of human fibroblasts after 48 h of incubation in Mϕ-conditioned media. Data are represented as mean ± SD. One-way ANOVA, n = 7 donors. (F) Phagocytosis of apoptotic HL-60 cells or latex beads measured via flow cytometry. Data are represented as mean ± SD. One-way ANOVA, n = 3 donors
Because M1→M2 Mϕs secreted higher levels of proteins known to promote endothelial cell migration, including PDGFBB and CCL5,37-39 we next evaluated HUVEC migration towards conditioned media from the Mϕs, again using a transwell migration assay. Conditioned media from M1→M2 Mϕs induced more HUVEC migration compared to M0→M2 Mϕ-conditioned media (Figure 6C).
Since many of the differentially expressed genes and proteins are also involved in endothelial cell and/or fibroblast proliferation, we also measured proliferation of HUVECs and fibroblasts over 24–48 hours of incubation in Mϕ-conditioned media, but no significant differences were observed between the cells incubated in media from M0→M2 or M1→M2 Mϕs (Figure 6D-E).
Finally, Mϕs are well-known for their role as phagocytes, and some studies have shown increased phagocytosis among M2-activated Mϕs.40,41 In order to assess differences in phagocytic activity between M0→M2 and M1→M2 Mϕs, each group was cultured for 3 h with either fluorescently labeled latex beads or apoptotic HL-60 cells. Using flow cytometry to identify target-containing Mϕs, no significant differences were observed between the groups in percentage of target-positive cells or MFI of target (Figure 6F).
The combined functional similarities and differences observed here suggest that the M0→M2 and M1→M2 phenotypes diverge to perform specialized duties, such as the recruitment of endothelial cells in the case of M1→M2 Mϕs.
4 ∣. DISCUSSION
We have established here that M1 activation primes Mϕs to polarize to an M2-like phenotype that exhibits unique gene and protein signatures compared to M2 Mϕs derived from an unactivated state. M1→M2 Mϕs were more migratory towards SDF-1α, suggesting an enhanced ability to traffic towards sites of injury, and induced more HUVEC migration than did M0→M2 Mϕs, suggesting that they may play an important role in angiogenesis. The characterization of this unique phenotype points to M1 activation as a major influence on subsequent M2 Mϕ behavior. These results have critical implications for understanding how Mϕ phenotype is regulated during tissue repair, and suggest that correcting M1 activation may be a therapeutic target for impaired healing situations characterized by deficient M2 activation.
Many studies have been conducted to elucidate the roles of M1-like and M2-like Mϕs during angiogenesis and healing in vivo. In the early stages of healing, pro-inflammatory circulating monocytes are recruited to the site of injury where they differentiate to M1-like Mϕs.1,42,43 These early-arriving Mϕs have been shown to be crucial for wound vascularization and complete healing.4,10,44 After this stage, M2-like Mϕs begin to dominate the milieu, further promoting tissue regeneration.10,45-47 These M2 Mϕs are often modeled in vitro via activation with IL-4,31,48-51 although other relevant M2-promoting stimuli include IL-10 and apoptotic cells.20,52 Although the specific roles of each phenotype are still being investigated, the current literature suggests that multiple phenotypes work together in a complementary manner to orchestrate normal healing. Indeed, drug delivery systems designed to sequentially release M1-promoting followed by M2-promoting signals have shown promise in promoting angiogenesis and healing.53 The present study adds to this body of literature by demonstrating the importance of M1 polarization on subsequent M2 polarization.
Several investigations have suggested that M1 polarization may regulate M2 polarization in vivo.3,4,10 For example, M1 Mϕs not only have the capacity to switch to the M2 phenotype,1,5,6,8,9 but stimulation of M1 activity has also been shown to increase subsequent M2 marker expression.54-56 Likewise, studies in which M1 Mϕs were inhibited during early stages of healing resulted in diminished M2 polarization later on.6,10 Correspondent to these findings, several pathologies in which the M2 response is inhibited, such as diabetic wounds, also initially lack robust M1 activity at early stages.11-16 Exactly how M1 activation influences M2 behavior, though, remains poorly understood. M2-like Mϕs that were previously pro-inflammatory and those that were derived from an unactivated state are both present in the site of injury during the later stages of healing.8 Furthermore, a recent study from Liu et al. determined through RNA-sequencing that over 2000 genes are differentially expressed between murine M1- and M0-derived M2 Mϕs.18 Though this would seemingly conflict with work from van den Bossche et al.,17 which indicated M1 Mϕs fail to repolarize to M2 upon IL-4 stimulation, our investigation sheds light on this discrepancy by revealing that M1→M2 Mϕs are a unique phenotype, with higher expression of some M2 genes and lower expression of others. These findings show that M1 activation changes the Mϕ response to IL-4, generating a phenotype that shares some markers and functions with M0→M2 Mϕs, but ultimately differs in important ways.
M1 Mϕs upregulated M2-promoting receptors, and correspondingly exhibited increased responsiveness to IL-4 in terms of sensitivity to lower doses as well as more rapid phosphorylation of STAT6. Moreover, M1 activation seems to change the response to IL-4 by generating a distinct M2-like phenotype with a unique gene signature and set of behaviors. This altered response may stem from a change in the IL-4 signaling pathway upon M1 activation – for example, both STAT3 and STAT6 are more highly expressed in M1 Mϕs compared to M0 Mϕs, while M0 Mϕs exhibit higher expression of PPARγ (Figure 1B). Deeper investigation into IL-4 pathway changes are required to understand the underlying mechanisms of the M1 Mϕ response to IL-4.
This study establishes an initial characterization of human primary M1→M2 Mϕs, and shows that they are phenotypically distinct from M0→M2 Mϕs. In line with their emergence during the later stages of tissue repair and angiogenesis, M1→M2 Mϕs upregulate PDGF-BB, essential for the stabilization of nascent vasculature, and CCL17, which may promote the resolution of inflammation via regulatory T cell recruitment. Though promising, these findings must be investigated further, potentially via transcriptome sequencing to identify all similarities and differences between M0→M2 and M1→M2 Mϕs, additional functional assays such as T cell recruitment, and of course studies to confirm this phenomenon in vivo.
In order to design therapies that promote angiogenesis and healing via Mϕ modulation, a thorough understanding of regulatory mechanisms during normal tissue repair is critical. We have shown here that M1 polarization primes Mϕs to take on a unique M2-like phenotype in response to IL-4, and that this M1→M2 phenotype appears to display important functions for wound healing and angiogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jessica Eager for technical assistance with data analysis, and Jennifer Connors and Dr. Elias El Haddad for helpful discussions. This study was supported by research funding from NHLBI (grant number R01 HL130037).
Abbreviations:
- MTT
3-(4,5-dimethylthiazol-2-yl)–2,5-diphenyltetrazolium bromide
- PDGF-BB
platelet-derived growth factor BB
- SDF-1α
stromal-derived factor-1 alpha
- VEGFA
vascular endothelial growth factor A
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- 1.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. Journal of Experimental Medicine. 2007;204:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troidl CJG, Troidl K, Hoffmann J, et al. The temporal and spatial distribution of macrophage subpopulations during arteriogenesis. Current Vascular Pharmacology. 2013;11:5–12. [PubMed] [Google Scholar]
- 3.Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, et al. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials. 2014;35:4477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurevich DB, Severn CE, Twomey C, Greenhough A, Cash J, Toye AM, et al. Live imaging of wound angiogenesis reveals macrophage orchestrated vessel sprouting and regression. The EMBO Journal. 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bencze M, Negroni E, Vallese D,Yacoub-Youssef H, Chaouch S, Wolff A, et al. Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Molecular Therapy. 2012;20:2168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dal-Secco D, Wang J, Zeng Z, Kolaczkowska E, Wong CHY, Petri B, et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. Journal of Experimental Medicine. 2015;212:447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga T, Mounier R, Horvath A, Cuvellier S, Dumont F, Poliska S, et al. Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. The Journal of Immunology. 2016;196:4771–82. [DOI] [PubMed] [Google Scholar]
- 9.Patsalos A, Pap A, Varga T, Trencsenyi G, Contreras GA, Garai I, et al. In situ macrophage phenotypic transition is affected by altered cellular composition prior to acute sterile muscle injury. The Journal of Physiology. 2017;595:5815–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Muller W, et al. Differential roles of macrophages in diverse phases of skin repair. The Journal of Immunology. 2010;184:3964–77. [DOI] [PubMed] [Google Scholar]
- 11.Sun C, Sun L, Ma H, Peng J, Zhen Y, Duan K, et al. The phenotype and functional alterations of macrophages in mice with hyperglycemia for long term. Journal of Cellular Physiology. 2011;227. [DOI] [PubMed] [Google Scholar]
- 12.Pradhan L, Cai X, Wu S, Andersen ND, Martin M, Malek J, et al. Gene expression of pro-inflammatory cytokines and neuropeptides in diabetic wound healing. Journal of Surgical Research. 2011;167:336–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan R, Geng S, Chen K, Diao N, Chu HW, Li L. Low-grade inflammatory polarization of monocytes impairs wound healing. The Journal of Pathology. 2015;238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon P, Keylock KT, Hartman ME, Freund GG, Woods JA. Macrophage hypo-responsiveness to interferon-γ in aged mice is associated with impaired signaling through Jak-STAT. Mechanisms of Ageing and Development. 2004;125:137–43. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson HN, Roberts ER, Stafford AR, Banyard KL, Matteucci P, Mace KA, et al. Tissue iron promotes wound repair via M2 macrophage polarization and the chemokine (C-C Motif) ligands 17 and 22. The American Journal of Pathology. 2019;189:2196–208. [DOI] [PubMed] [Google Scholar]
- 16.Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, et al. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLOS One. 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Reports. 2016;17:684–96. [DOI] [PubMed] [Google Scholar]
- 18.Liu SX, Gustafson HH, Jackson DL, Pun SH, Trapnell C. Trajectory analysis quantifies transcriptional plasticity during macrophage polarization. Scientific Reports. 2020:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MPJ, MMPC Donners. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis. 2014;17:109–18. [DOI] [PubMed] [Google Scholar]
- 20.Graney PL, Ben-Shaul S, Landau S, Bajpai A, Singh B, Eager J, et al. Macrophages of diverse phenotypes drive vascularization of engineered tissues. Science Advances. 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siracusa MC, Reece JJ, Urban JF, Scott AL. Dynamics of lung macrophage activation in response to helminth infection. Journal of Leukocyte Biology. 2008;84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anthony RM, Urban JF, Alem F, Hamed HA, Rozo CT, Boucher JL, et al. Memory TH2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nature Medicine. 2006;12:955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. Journal of Experimental Medicine. 1992;176:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiller KL, Koh TJ. Macrophage-based therapeutic strategies in regenerative medicine. Advanced Drug Delivery Reviews. 2017;122:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danciger JS, Lutz M, Hama S, Cruz D, Castrillo A, Lazaro J, et al. Method for large scale isolation, culture and cryopreservation of human monocytes suitable for chemotaxis, cellular adhesion assays, macrophage and dendritic cell differentiation. Journal of Immunological Methods. 2004;288:123–34. [DOI] [PubMed] [Google Scholar]
- 27.Spiller KL, Nassiri S, Witherel CE, Anfang RR, Ng J, Nakazawa KR, et al. Sequential delivery of immunomodulatory cytokines to facilitate the M1-to-M2 transition of macrophages and enhance vascularization of bone scaffolds. Biomaterials. 2015;37:194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipper JA, Willenborg S, Brinckmann J, Bloch W, Maab T, Wagener R, et al. Interleukin-4 receptor α signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity. 2015;43:803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gocheva V, Wang H-W, Gadea BB, Shree T, Hunter KE, Garfall AL, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes & Development. 2010;24:241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormickSM Heller NM. Commentary: iL-4 and IL-13 receptors and signaling. Cytokine. 2015;75:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Y, et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. 2017;356:1072–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani F, Roncucci L. Chemerin/chemR23 axis in inflammation onset and resolution. Inflammation Research. 2015;64:85–95. [DOI] [PubMed] [Google Scholar]
- 33.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, et al. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. Journal of Experimental Medicine. 1998;187: 571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang HZ, Cui B, Liu HZ, Chen ZR, Yan HM, Hua F, et al. Targeting TLR2 attenuates pulmonary inflammation and fibrosis by reversion of suppressive immune microenvironment. The Journal of Immunology. 2009;182:692–702. [DOI] [PubMed] [Google Scholar]
- 35.Candel S, de Oliveira S, Lopez-Munoz A, Garcia-Moreno D, Espin-Palazon R, Tyrkalska SD. Tnfa signaling through Tnfr2 protects skin against oxidative stress-induced inflammation. PLoS Biology. 2014;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nature Reviews Immunology. 2010;10:365–76. [DOI] [PubMed] [Google Scholar]
- 37.Asahara T, Takahashi T, Masuda H, Kalka C, Chen D, Iwaguro H, et al. VEGF contributes to postnatal neovascularization by mobilizing bone marrow-derived endothelial progenitor cells. The EMBO Journal. 1999;18:3964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thommen R, Humar R, Misevic G, Pepper MS, Hahn AWA, John M, et al. PDGF-BB increases endothelial migration and cord movements during angiogenesis in vitro. Journal of Cellular Biochemistry. 1998;64:403–13. [PubMed] [Google Scholar]
- 39.Suffee N, Hlawaty H, Meddahi-Pelle A, Maillard L, Louedec L, Haddad O, et al. RANTES/CCL5-induced pro-angiogenic effects depend on CCR1, CCR5 and glycosaminoglycans. Angiogenesis. 2012;15:727–44. [DOI] [PubMed] [Google Scholar]
- 40.Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Phenotypic FantinoE. Functional, and plasticity features of classical and alternatively activated human macrophages. American Journal of Respiratory Cell and Molecular Biology. 2015;53:676–88. [DOI] [PubMed] [Google Scholar]
- 41.Gratchev A, Kzhyshkowska J, Utikal J, Goerdt S. Interleukin-4 and dexamethasone counterregulate extracellular matrix remodelling and phagocytosis in type-2 macrophages. Scandinavian Journal of Immunology. 2005;61:10–7. [DOI] [PubMed] [Google Scholar]
- 42.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai J, J Feng, K Liu, S Zhou, F Lu. Early macrophage infiltration improves fat graft survival by inducing angiogenesis and hematopoietic stem cell recruitment. Plastic and Reconstructive Surgery. 2018;141:376–86. [DOI] [PubMed] [Google Scholar]
- 45.Mirza R, Koh TJ. Dysregulation of monocyte/macrophage phenotype in wounds of diabetic mice. Cytokine. 2011;56:256–64. [DOI] [PubMed] [Google Scholar]
- 46.Saclier M, Yacoub-Youssef H, Mackey AL, Arnold L, Ardjoune H, Magnan M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells. 2013;31:384–96. [DOI] [PubMed] [Google Scholar]
- 47.Perdiguero E, Sousa-Victor P, Ruiz-Bonilla V, Jardi M, Caelles C, Serrano AL, et al. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. Journal of Cell Biology. 2011;195:307–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Liu J, Zhao S, Zhang H, Cai W, Cai M, et al. Interleukin-4 is essential for microglia/macrophage m2 polarization and long-term recovery after cerebral ischemia. Stroke. 2016;47:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balce DR, Li B, Allan ERO, Rybicka JM, Krohn RM, Yates RM. Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood. 2011;118:4199–208. [DOI] [PubMed] [Google Scholar]
- 50.Salmon-Ehr V, Ramont L, Godeau G, Birembaut P, Guenounou M, Bernard P, et al. Implication of interleukin-4 in wound healing. Laboratory Investigation. 2000;80:1337–43. [DOI] [PubMed] [Google Scholar]
- 51.Francos-Quijorna IA-AJ, Martinez-Muriana A, Lopez-Vales R. IL-4 drives microglia and macrophages toward a phenotype conducive for tissue repair and functional recovery after spinal cord injury. Glia. 2016;64:2079–92. [DOI] [PubMed] [Google Scholar]
- 52.Lurier EB, Dalton D, Dampier W, Raman P, Nassiri S, Ferraro NM, et al. Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology. 2017;222:847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Brien EM, Risser GE, Spiller KL. Sequential drug delivery to modulate macrophage behavior and enhance implant integration. Adv Drug Deliv Rev. 2019;149-150:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao AJ, Gibon E, Ma T, Yao Z, Smith RL, SB Goodman. Revision joint replacement, wear particles, and macrophage polarization. Acta Biomaterial. 2012;8:2815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leal EC, Carvalho E, Tellechea A, Kafanas A, Tecilazich F, Kearney C, et al. Substance P promotes wound healing in diabetes by modulating inflammation and macrophage phenotype. Am J Pathol. 2015;185:1638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rybalko V, Hsieh PL, Merscham-Banda M, Suggs LJ, Farrar RP. The development of macrophage-mediated cell therapy to improve skeletal muscle function after injury. PLOS One. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.