Abstract
SARS-CoV-2 has raised the alarm to search for effective therapy for this virus. To date several vaccines have been approved but few available drugs reported recently still need approval from FDA. Remdesivir was approved for emergency use only. In this report, the SARS-CoV-2 3CLpro was expressed and purified. By using a FRET-based enzymatic assay, we have screened a library consisting of more than 300 different niclosamide derivatives and identified three molecules JMX0286, JMX0301, and JMX0941 as potent allosteric inhibitors against SARS-CoV-2 3CLpro, with IC50 values similar to that of known covalent inhibitor boceprevir. In a cell-based antiviral assay, these inhibitors can inhibit the virus growth with EC50 in the range of 2–3 μM. The mechanism of action of JMX0286, JMX0301, and JMX0941 were characterized by enzyme kinetics, affinity binding and protein-based substrate digestion. Molecular docking, molecular dynamics (MD) simulations and hydration studies suggested that JMX0286, JMX0301, JMX0941 bind specifically to an allosteric pocket of the SARS-CoV-2 3CL protease. This study provides three potent compounds for further studies.
Keywords: SARS-CoV-2, 3CLpro, Protease, Allosteric inhibitor, Niclosamide
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), like SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), belongs to Betacoronavirus genus. The recent outbreak of SARS-CoV-2 becomes a significant concern to public health care worldwide. As of Nov 12, 2021, approximately 5 million people have died due to this deadly virus. In the USA alone, over 46 million people have been infected, and more than 0.75 million people have died. SARS-CoV-2 has shown a higher infection rate and a more extended incubation period than the previous SARS-CoV and MERS-CoV. SARS-CoV-2 binds much tighter than SARS-CoV to the same host receptor, angiotensin-converting enzyme 2 (ACE2) (Lan et al., 2020; Shang et al., 2020; Wang et al., 2020; Wrapp et al., 2020). After entering the host cells, coronavirus translates its genome into two overlapping polyproteins-pp1a and pp1ab, which encode two cysteine proteases, papain-like (PLpro) and 3-chymotrypsin (C)-like (3CLpro) proteases. The two viral proteases of coronavirus are excised from the polyproteins through autocleavage and work together to cleave the polyproteins, leading to 16 functional non-structural proteins (Nsps). The 3CLpro of SARS-CoV-2 specifically cleaves at 11 positions on the large polyprotein 1 ab (790 kDa) (Hilgenfeld, 2014; Zhang et al., 2020).
The cleaved Nsps are essential for assembling the viral replication transcription complex (RTC) to initiate the viral replication. The 3CLpro, also known as the main protease, is one of the most intriguing drug targets due to its unique substrate preference for a glutamine residue at the P1 site and a residue with short sidechain such as Ser, Ala, Gly at the P1’ position. Although Cys-based human proteases exist, none of them have specificity for a P1 Glu (Ma et al., 2020). Therefore, off-target effects are minimized; and the inhibitors of 3CLpro are most likely less toxic to host cells. Although vaccine development is critically important for COVID-19, effective small molecule antiviral drugs are urgently needed.
Recently it was reported that niclosamide, an anthelminthic drug, which is historically used to treat tapeworm infection, could be repurposed for use against SARS-CoV-2 (Garrett et al., 2021). Previously our lab was working on niclosamide and its derivatives as potent inhibitors against flaviviruses (Li et al., 2020b; Samrat et al., 2022; Xu et al., 2020c). It has been shown that niclosamide suppressed the cytopathic effect (CPE) of SARS-CoV at a concentration of as low as 1 μM and inhibited SARS-CoV replication with an EC50 value of less than 0.1 μM in Vero E6 cells (Xu et al., 2020c). In addition, niclosamide can also inhibit MERS-CoV replication by inhibiting autophagosome-lysosome fusion through disrupting autophagy regulator proteins (Gassen et al., 2019). In a recent study, Garrett et al. (2021) found that the niclosamide could reorganize the lipid profile of SARS-CoV-2 infected Vero E6 cells, thereby limiting virus replication. These findings made it an early drug candidate against SARS-CoV-2. Unfortunately, the drug is unlikely to perform well when given in vivo, because of its poor bioavailability. Recently it was reported that niclosamide derivatives could reduce the viral load, modulate cytokine expression, and improve weight loss in mouse model (Blake et al., 2021; Garrett et al., 2021). Interestingly, niclosamide showed no obvious inhibitory activity against SARS-CoV 3CLpro up to 50 μM in our experiments. Mechanistically, it might exert its anti-SARS activity via other modes of actions (Braga et al., 2021; Brunaugh et al., 2021; Kunzelmann, 2021).
In the current study, using a FRET-based enzymatic assay, we have screened a small library of niclosamide derivatives and identified three molecules JMX0286, JMX0301, and JMX0941 as potent inhibitors against SARS-CoV-2 3CLpro, with IC50 values comparable to that of known covalent inhibitor boceprevir (Ma et al., 2020). Two of the derivatives inhibited SARS-CoV-2 in Vero E6 cells with EC50 values in low micromolar range. Kinetics and modeling studies suggest that these inhibitors acted as allosteric inhibitors of 3CLpro.
2. Results
2.1. Antiviral effect of niclosamide derivatives against SARS-CoV-2
Antiviral effect of 30 selected niclosamide derivatives were tested against SARS-CoV-2 in A549-hACE2 cells. SARS-CoV-2-infected cells were treated with a concentration series of each compound (Table 1 , Fig. S1). As shown in Table 1 and Supplementary Fig. S1, except two compounds, 28 compounds showed anti-SARS-CoV-2 efficacy to various degrees. Particularly, compounds JMX308, JMX0325, JMX0320, JMX0887 and JMX0895 showed strong inhibition of SARS-CoV-2 virus with EC50 value in sub-micromolar range.
Table 1.
EC50 of Niclosamide derivatives against SARS-CoV-2.
| Comp Serial# | Comp ID | EC50 (μM) | CC50 (μM) |
|---|---|---|---|
| 1 | JMX0207 | 1.222 | 13.17 |
| 2 | JMX0281 | 2.078 | 8.32 |
| 3 | JMX0285 | 3.031 | 6.13 |
| 4 | JMX0286 | 2.251 | 3.78 |
| 5 | JMX0308 | 0.8065 | 1.80 |
| 6 | JMX0310 | 2.414 | 10.06 |
| 7 | JMX0312 | 1.129 | 1.97 |
| 8 | JMX0317 | 4.025 | 11.70 |
| 9 | JMX0321 | 1.186 | 3.76 |
| 10 | JMX0325 | 0.1228 | 1.81 |
| 11 | JMX0330 | 0.4981 | 10.15 |
| 12 | JMX0510–2 | 4.88 | 11.50 |
| 13 | JMX0670 | 3.202 | 9.17 |
| 14 | JMX0671 | 1.563 | 8.03 |
| 15 | JMX0672 | 1.921 | 8.34 |
| 16 | JMX0673 | 1.743 | 5.20 |
| 17 | JMX0674 | 9.752 | >10 |
| 18 | JMX0679 | 2.514 | 7.14 |
| 19 | JMX0680 | 4.566 | 10.10 |
| 20 | JMX0681 | 9.753 | >10 |
| 21 | JMX0682 | 1.568 | 6.48 |
| 22 | JMX0887 | 0.9203 | 4.18 |
| 23 | JMX0895 | 0.665 | 4.57 |
| 24 | JMX0929 | 2.425 | 5.06 |
| 25 | JMX0936 | 2.214 | 3.74 |
| 26 | JMX0940 | 1.216 | 7.72 |
| 27 | JMX0941 | 1.745 | 4.75 |
| 28 | JMX0942 | 2.891 | 7.20 |
2.2. Fluorescence resonance energy transfer (FRET)-based screening assays
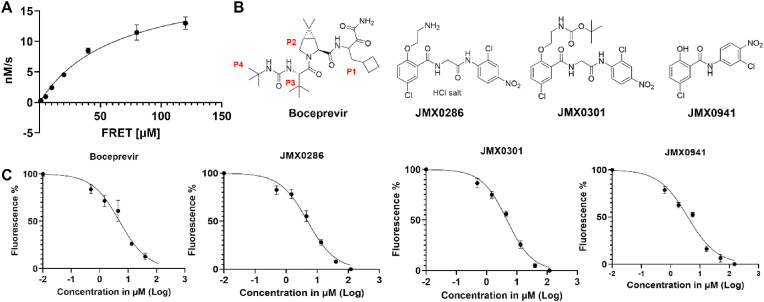
As it was reported that niclosamide was a weak inhibitor of the SARS 3CLpro (Konwar and Sarma, 2021; Wu et al., 2004), we simultaneously tested a library of more than 300 different niclosamide derivatives for 3CLpro inhibition. A FRET-based assay was employed with the FRET peptide substrate for the protease activity measurement. The relative fluorescence units were calculated for the correlation of fluorescence intensity and enzymatic activity. Initial velocity for enzymatic activity was plotted as a function of time which shows the typical fluorescence profile for the hydrolysis of the substrate. Kinetic parameters were determined by fitting experimental curves. The V max of 64.1 nM/s and K m of 68.5 μM were obtained for the SARS-CoV-2 3CLpro (Fig. 1 A).
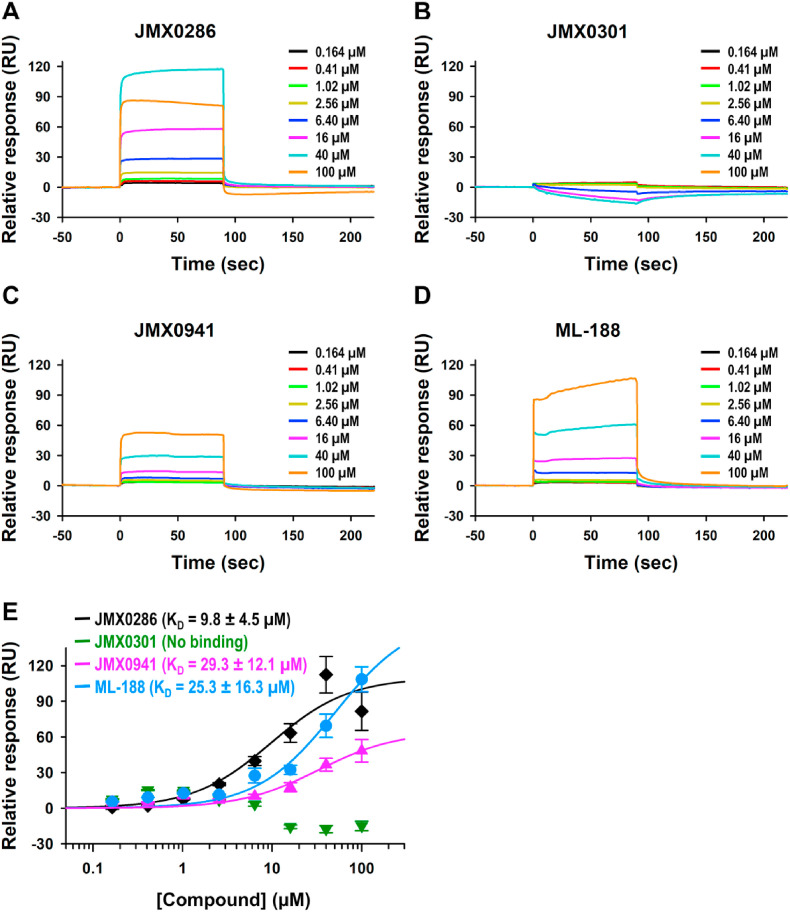
Fig. 1.
Boceprevir, JMX0286, JMX0301 and JMX0941 are potent inhibitors of SARS-CoV-2 3CLpro. A. Michaelis–Menten plot of 100 nM authentic SARS-CoV-2 3CLpro with various concentrations of FRET substrate in Tris buffer. The best-fit Vmax = 64.11 nM/s; Km = 68.48 μM. B. Chemical structure of boceprevir, JMX0286, JMX0301 and JMX0941. C. Dose dependent inhibition of FRET-based substrate digestion by boceprevir, JMX0286, JMX0301 and JMX0941.
Assay was validated using boceprevir, a known covalent inhibitor for the SARS-CoV-2 3CLpro (Fu et al., 2020; Ma et al., 2020). Using the same experimental condition, we screened the niclosamide derivative library at compound concentration of 100 μM. Among them, JMX0286, JMX0301 and JMX0941 were found to have prominent inhibitory activity (Fig. 1B). The three compounds JMX0286, JMX0301 and JMX0941 were subjected to dose-response experiment to determine IC50 values of 4.8, 4.5 and 3.9 μM, respectively (Fig. 1C). These values are similar to that of the positive control boceprevir (IC50: 4.9 μM) (Table 2 ).
Table 2.
IC50, EC50 and CC50 of Boceprevir, JMX0286, JMX0301 and JMX0941 and predicted binding affinities of 3CLpro JMX-inhibitors at the allosteric sites. Docking scores were generated by Glide XP or IFD, as indicated in parenthesis.
| Compound name | SARS-CoV-2 3CLpro |
CatB |
CatL |
Docking Score (kcal/mol) |
||
|---|---|---|---|---|---|---|
| IC50 (μM) | EC50 (μM) | CC50 (μM) | IC50 (μM) | IC50 (μM) | ||
| Boceprevir | 4.9 | 15.6 | >200 | 3.04 | 0.5 | |
| JMX0286 | 4.8 | 2.3 | 53.1 | 46.4 | 110.8 | −1082.28 (IFD) |
| JMX0301 | 4.5 | No Dose response | 342.4 | >200 | 134.6 | −6.02 (XP) |
| JMX0941 | 3.9 | 1.7 | 30 | 82.2 | 100.3 | −5.24 (XP) |
2.3. Cytotoxicity
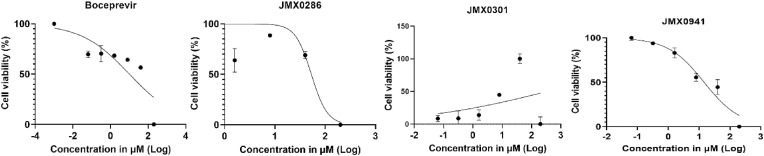
We evaluated the cytotoxicity of these compounds in A549 and Vero E6 cells. A549 and Vero E6 cells were treated with different concentrations of JMX0286, JMX0301 and JMX0941, and boceprevir up to 120 μM, and cell viability was determined using the 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt (WST-8) assay. We found that JMX0286, JMX0301 and JMX0941 show cytotoxicity well above the EC50 in Vero E6 and A549 cells with CC50 values of 53 μM, 342 μM and 30 μM, respectively (Fig. 2 ; Table 2).
Fig. 2.
Cytotoxicity activity of JMX0286, JMX0301 and JMX0941. Vero E6 cells were incubated with various concentrations of boceprevir, JMX0286, JMX0301 and JMX0941 and then viability was assayed after 48 h of incubation.
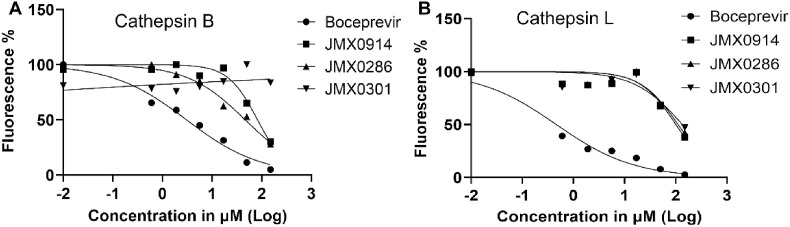
2.4. Inhibitor specificity
It has been shown previously that many compounds showing inhibitory effect against 3CLpro are also active against cathepsin B (CatB) and L (CatL) enzymes (Steuten et al., 2021). We therefore tested these compounds against CatB and CatL. As expected, the positive control boceprevir also strongly inhibited CatB and CatL with IC50 of 3.0 μM and 0.5 μM, respectively. In contrast, our result suggests that JMX0286, JMX0301 and JMX0941 do not inhibit CatB up until very high concentrations. The IC50 values for these compounds against CatB are 46.4 μM, >200 μM and 82.2 μM, respectively. Similarly, JMX0286, JMX0301 and JMX0941 only inhibit CatL with much higher IC50 values of 110.8 μM, 134.6 μM and 100.3 μM, respectively. These results indicate that these three inhibitors are specific for the SARS-CoV-2 3CLpro (Fig. 3 A and B; Table 2).
Fig. 3.
Calculation of IC50for boceprevir, JMX0286, JMX0301 and JMX0941 against Cathepsin B and L. The Cathepsin B and L protease assays were carried in buffer containing 20 mM Hepes 7.4, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT. Protein at concentration 50 μM were mix with different concentration of inhibitors and incubated at room temperature for 30 min. The enzymatic reaction was initiated by adding 10 μM of FRET substrate. The reaction was monitored in a BioTek synergy HI microplate reader with filters for excitation at 360/40 nm and emission at 460/40 nm at 30 °C for 5 h. The IC50 values were calculated by plotting the initial velocity against various concentrations of testing compounds with a dose-response function in Prism 9 software of Cathepsin B (A) and L protease (B).
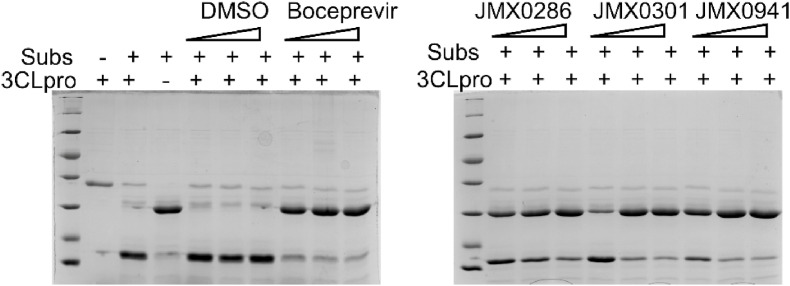
2.5. Inhibition of protein substrate cleavage by JMX0286, JMX0301 and JMX0941
To visualize inhibition of 3CLpro by these compounds, we developed a nanoluciferase-based multipurpose TriPro substrate containing recognition sequences for 3Clpro, PLpro and flavivirus NS2B-NS3 proteases (Li et al., 2017). In the absence of inhibitors, the TriPro substrate was efficiently cleaved by 3CLpro (Fig. 4 ). In contrast, JMX0286, JMX0301 and JMX0941 inhibited the cleavage of the TriPro substrate protein in a dose dependent manner (Fig. 4).
Fig. 4.
Confirmation of inhibition of substrate cleavage by SARS-CoV-2 3CL protease inhibitor. 0.8 μM of 3 C protein was incubated for 1 h with increasing concentration of each inhibitor separately in Tris buffer. After 1 h incubation, substrate protein was added and further incubated for 1 h, followed by SDS-PAGE analysis.
2.6. Analysis of bimolecular interactions between 3CLpro and JMX0286, JMX0301, JMX0941
To determine whether three selected inhibitors, JMX0286, JMX0301, JMX0941 could directly bind to 3CLpro, we performed binding studies using surface plasmon resonance (SPR). The SPR is a very sensitive method that can detect direct binding interaction in real-time, providing three parameters, association rate (k a), dissociation rate (k d), and binding affinity (K D) at equilibrium. We monitored these three selected compound binding behaviors to the immobilized 3CLpro at a series of increasing concentrations to observe their dose-responses. A known 3CLpro non-covalent inhibitor, ML-188, was also tested along with these three compounds as a control. We observed a typical binding pattern of compounds with very fast association and fast dissociation rates, producing square shapes of sensorgrams (Fig. 5 A – D). This made it hard to determine accurate rate constants, and hence we used steady-state affinity model to determine binding affinity (K D) values at equilibrium (Fig. 5E). JMX0286 and JMX0941 showed dose-response bindings with K D values at 9.8 μM and 29.3 μM, respectively, whereas JMX0301 did not show binding. JMX0301 nonspecifically bound higher level to the reference channel than 3CLpro immobilized active channel, resulting in negative response after subtracting reference channel responses. We were expecting tighter binding of the control ML-188 considering its reported IC50 value at ∼2 μM, but its K D value was determined to be 25.3 μM with some nonspecific binding pattern at two high concentrations tested (40 μM and 100 μM). This might have been caused by solubility issue at high concentrations, producing determined K D value higher than the actual K D value. Overall, the binding affinity of JMX0286 and JMX0941 are better or equivalent to that of ML-188, the control inhibitor. Since JMX0301 binding was not detected with SPR, we use microscale thermophoresis (MST) to determine direct binding of JMX0301 with 3CLpro. MST result showed binding of JMX0301 to 3CLpro with a KD value of 34 μM (Fig. S2).
Fig. 5.
Analysis ofJMX0286, JMX0301,JMX0941and ML188binding to recombinant SARS-CoV-2 3CLpro: Surface plasmon resonance (SPR). Dose response curve upon titrating 100 μM–0.164 μM for JMX0286 (A), JMX0301 (B) and JMX0941 (C) and ML188 (D) against immobilized 3CLpro. Steady-state affinity model to determine binding affinity (KD) values at equilibrium (E).
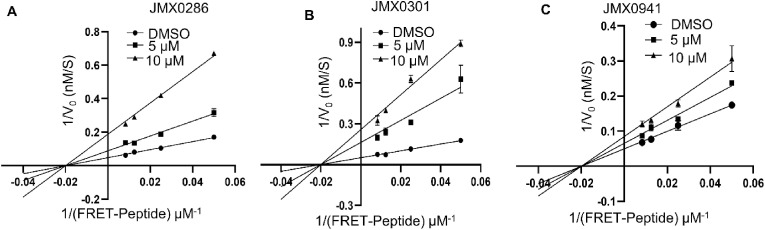
2.7. Allosteric inhibition of the SARS-CoV-2 3CLpro
To determine the mode of action of inhibitors JMX0941, JMX0286 and JMX0301, we performed an enzyme kinetic experiment on 3CLpro in presence or absence of JMX0286 JMX0301 and JMX0941 (Fig. 6 A-C). Compared to the DMSO control, all three inhibitors, JMX0286, JMX0301 and JMX0941 reduced the Vmax values for the 3CLpro whereas the Km values remains similar. The results, showing a lowered Vmax but similar Km values for the inhibitors treated samples in comparison to those of the DMSO control, suggest a non-competitive inhibition mechanism according to classical Michaelis-Menten enzyme kinetics.
Fig. 6.
Inhibition of SARS-CoV-2 3CLpro by a non-competitive mechanism. Lineweaver–Burk plots of kinetics experimental data for inhibition of the SARS-CoV-2 3CLpro by JMX0286, JMX0301 and JMX0941 (A–C). The SARS-CoV-2 3CLpro at 200 nM was mixed with DMSO, JMX0286, JMX0301, or JMX0941 (5 μM or 10 μM) for 30 min. Substrate Edan was added at various concentrations (120 μM–1 μM).
2.8. Molecular modeling
2.8.1. Binding modes of JMX-inhibitors to 3CLpro allosteric site
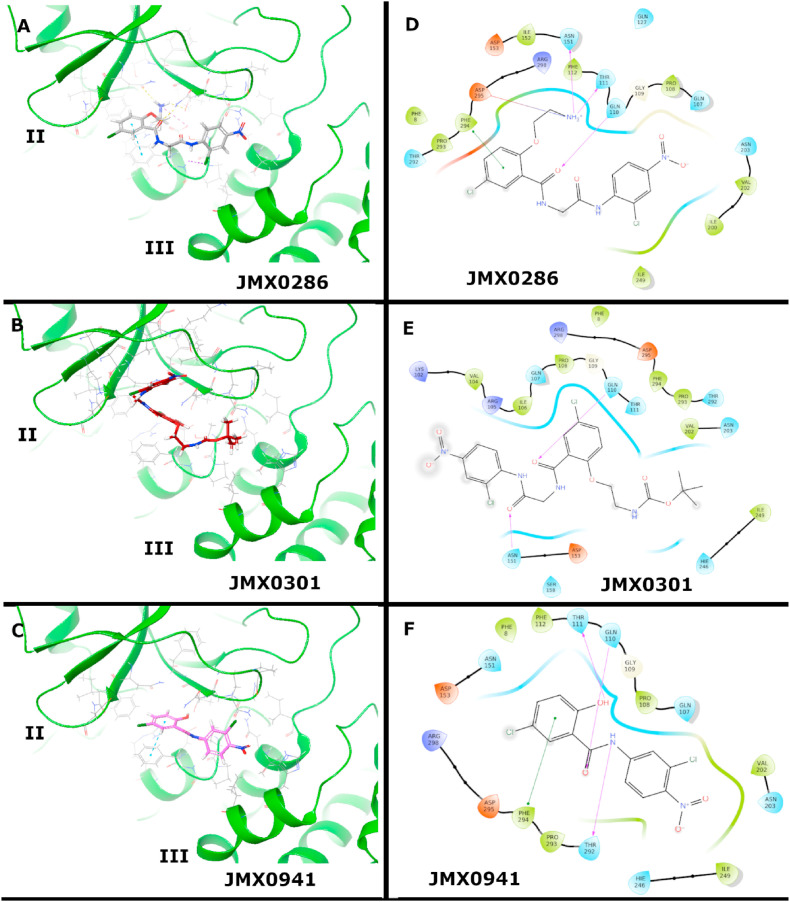
The structure of SARS-CoV-2 3CLpro is made out of three domains, namely a chymotrypsin-like domain (I), a picornavirus 3C protease-like domain (II) and a dimerization domain (III). Furthermore, to achieve optimal catalytic activity, 3CLpro monomers arrange to assemble homodimers (Shi and Song, 2006). Dimerization is guided by interactions established by residue Glu166 of one monomer with the NH2 terminus (N-finger) of the other (Shi and Song, 2006). In addition to the active site, the experimental structure of 3CLpro in complex with inhibitor AT7519 reveals a large allosteric pocket (Günther et al., 2021). Compound AT7519 binds to the cleft between domain II and III facing away from the other protomer. This pocket is comprised of polar (Asn151, Gln107, Asn203, Gln110, and Thr292), hydrophobic (Ile200, Val202, Pro108, Ile249, Pro293, and Phe294) and charged (Arg298) residues.
In this work, the binding modes of novel, non-covalent, allosteric 3CLpro inhibitors JMX0286, JMX0301 and JMX0941 (JMX series), were predicted and evaluated by molecular docking, molecular dynamics (MD) simulations and hydration studies (Table 2, Table 3 ). All three compounds in the JMX series were predicted, via molecular docking, to bind to the allosteric site experimentally observed for the AT7519 ligand (Günther et al., 2021). The binding modes of the three JMX compounds nicely overlap with the experimental pose of AT7519.
Table 3.
Summary of MD simulation studies of 3CLpro JMX-inhibitors. Total number of atoms and number of water atoms are reported per 3CLpro monomer.
| Compound ID | Inhibition Type | Simulation Time | Total Number of Atoms | Number of Water Atoms |
|---|---|---|---|---|
| JMX0286 | Non-competitive | 100 ns | 36181 | 10516 |
| JMX0301 | Non-competitive | 100 ns | 36171 | 10510 |
| JMX0941 | Non-competitive | 100 ns | 36175 | 10517 |
Binding mode of JMX0286. The ethylamine moiety of JMX0286 is deeply buried in the AT7519 allosteric pocket (Fig. 7 A&D). The amine moiety interacts with Asp295, Asn151, and Thr111. Phe294 has a - stacking interaction with the benzene ring in the chlorophenoxy-ethylamine moiety. The chlorine in this ring is solvent exposed. Moving to the right of JMX0286, Gln110 is hydrogen bonding with a carboxyl in the linker. The nitro group is facing the polar Gln107 and Asn203 residues.
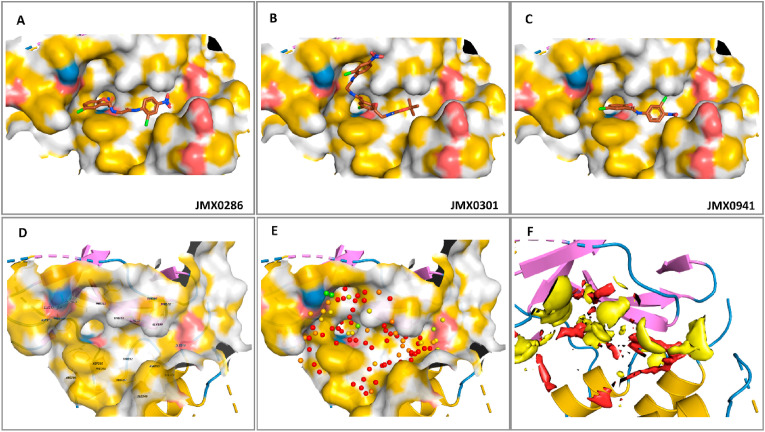
Fig. 7.
Binding modes of JMX0286, JMX0301 and JMX0941 to 3CLpro in the allosteric site. Best docking poses of JMX-inhibitors to the 3CLpro allosteric site. Poses were generated by induced fit docking (A) and molecular docking with Glide at the XP level (B–C), for JMX0286, and JMX0301 and JMX0941, respectively. 2D-Interaction maps are also shown (D–F).
Binding mode of JMX0301. The structure of JMX0301 is quite similar to that of JMX0286, with the protonated amine moiety of the latter substituted by the tert-butyloxycarbonyl protecting group (BOC) (Fig. 7 B&E). The docking pose of JMX0301 is flipped in the binding pocket compared to JMX0286 (Fig. 7 B&E). The chlorine atom in the chlorophenoxy-ethylamine moiety is next to Pro108. The BOC group is in the pocket occupied by the nitro group in the binding mode of JMX0286, near Val202. The nitrobenzene is solvent exposed.
Binding mode of JMX0941. Among the investigated JMX compounds, JMX0941 has the smallest scaffold. The nitro functional group of JMX0941 is in a similar orientation as in JMX0286, interacting with the nearby Val202 (Fig. 7 C&F). The chlorine atom on the nitro-benzene ring is facing into the pocket, as opposed to out of the pocket, a configuration adopted in the binding mode of JMX0286. The linker carboxyl is hydrogen bonding with Gln110 and the linker nitrogen is hydrogen bonding the Thr292. The other benzene ring has a - stacking interaction with Phe294 and the chlorine is solvent exposed, like in JMX0286. The hydroxyl is hydrogen bonding with Thr111.
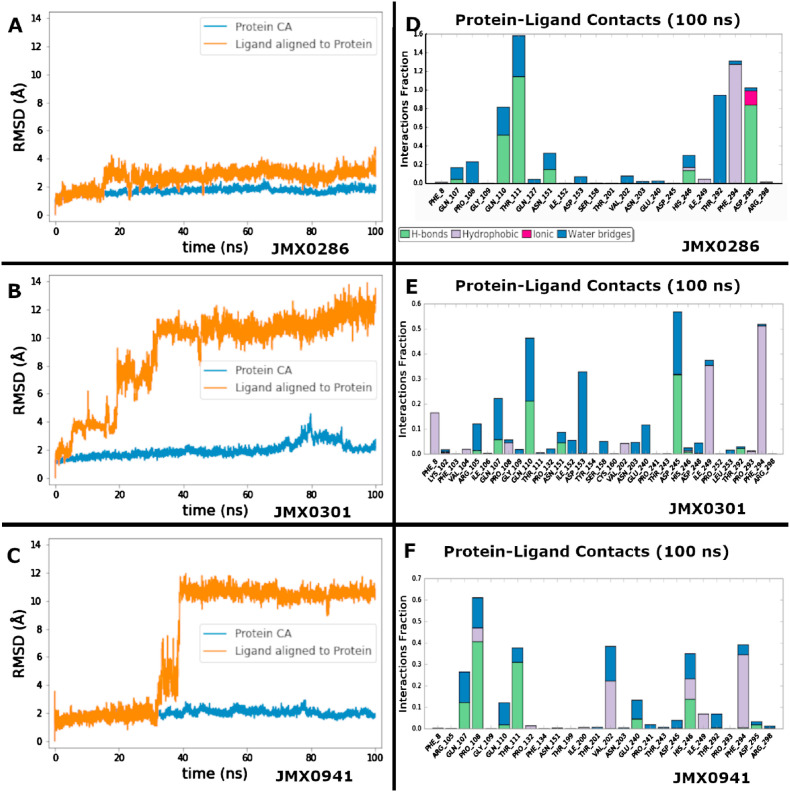
2.8.2. Molecular dynamics (MD) studies
Molecular dynamics (MD) studies were performed to investigate the binding dynamics of non-covalent 3CLpro inhibitors in the JMX series. Hence, three MD simulations were performed, where each system was simulated for 100 ns (Table 3).
Binding dynamics of JMX0286. During the 100 ns simulation, JMX0286 is very stable in the allosteric site. The protein does not have any conformational changes with an average root-mean-square deviation (RMSD) of 2 Å and the ligand does not change its binding pose with an average RMSD, after alignment to the protein, of 3 Å. The strongest interactions of JMX0286 are established with Thr111, where it is hydrogen bonded the full 100 ns, the hydrophobic contact with Phe294 the full 100 ns, the hydrogen bond with Asp295 for 83% of the simulation, and the water bridge with Thr292 for 95% of the simulation. Gln110 also interacts 50% of the time as a hydrogen bond and 30% of the time as a water bridge (Fig. 8 A&D). These strong interactions are maintained over the course of the simulation. An interaction with Asn151 starts strong but becomes much weaker after 17 ns (Fig. S3).
Fig. 8.
Analysis of protein ligand dynamics and interactions of 3CLpro inhibitors JMX0286, JMX0301 and JMX0941 to the allosteric site. Protein-ligand root-mean-square-deviation (RMSD) and protein-ligand interactions (or ‘contacts') for JMX0286, JMX0301 and JMX0941, are shown in (A–C) and (D–F), respectively. Contacts are categorized into four types: Hydrogen Bonds, Hydrophobic, Ionic and Water Bridges. The stacked bar charts are normalized over the course of the trajectory: for example, a value of 0.7 suggests that 70% of the simulation time the specific interaction is maintained. Values over 1.0 are possible as some protein residue may make multiple contacts of same subtype with the ligand.
Binding dynamics of JMX0301. After only 6 ns of simulation, JMX0301 shifts away from its binding pose and moves quite far with a final RMSD of 13 Å right after. The protein remains stable with an average RMSD of 2 Å, reaching a peak of 4 Å at about 80 ns, before coming back down. The nitro-benzene ligand moiety leaves the binding site within 6 ns of simulation and starts to explore the space outside the pocket. The BOC group turns to face outside the pocket becoming very solvent exposed. JMX0301 is anchored by the chloro-benzene ring located at the center of the scaffold. These have moderately strong interactions with Gln110 for 20% in a hydrogen bond and an additional 30% in a water bridge, hydrophobic interactions with Phe294 and Ile249, and a hydrogen bond with Asp245 (Fig. 8 B&E). Gln110 and Gln107 are the most consistent contacts. Several contacts are lost at about 25 ns into the simulation including Phe8, Arg105, Asn151, Ile152, Asp153, and Ser158. A new contact, Asp245 is formed towards the end of the simulation (Fig. S4).
Binding dynamics of JMX0941. The smallest of the JMX ligands, JMX0941, remains stable for 33 ns with a RMSD of 2 Å. After this time, it flips 180° rotating around the nitro group with the p-chloro-phenol ring dipping into a pocket to the right of the binding pose, increasing its RMSD to 11.5 Å. The protein remains stable with a RMSD of 2 Å. Pro108 is the only moderately strong contact which was around for 60% of the simulation (Fig. 8 C&F). For the first 33 ns strong contacts were maintained with Gln107, Pro108, Gln110, Thr111, His246, and Phe294 (Fig. S5).
2.8.3. Allosteric site hydration studies
Solvation and desolvation govern protein-ligand binding. Therefore, SZMAP (SZMAP 1.6.2.1) was used to map and compute the binding free energy of water molecules in regions of the protein surface constituting cavities suitable for ligand binding (i.e., ligand binding pockets). In 3CLpro, the cavities that originate the allosteric site are hydrophobic in nature (Fig. 9 ) as depicted by the yellow surfaces. The positively and negatively chargeable regions of the binding cavities are colored blue and red, respectively. The ligands are mapped favorably with the chemical environment of the binding site. The polarity throughout the binding cavity was mapped by computing the net free energy difference between the water probe and the uncharged (hypothetical) water probe. While the regions occupied by red spheres represent the hydrophobic sites in which polar substituents or water molecules will decrease ligand binding affinity, while areas occupied by yellowish, greenish and bluish spheres (Fig. 9E) are the polar regions that can accommodate water molecules and ligands with variable degrees of substituent polarities. For simplicity, the allosteric region is displayed as red (nonpolar) and yellow (polar) surfaces in the binding cavity (Fig. 9F). The results indicated that the allosteric site (Fig. 9) seems to be quite hydrophobic.
Fig. 9.
Analysis of 3CLpro allosteric site hydration. Yellow/grey surfaces indicate hydrophobic, non-polar regions. Red to blue indicates polar regions (A–C). Binding site with important residues labeled (D). Calculated location of water molecules color coded from red (most hydrophobic) to blue (most hydrophilic) (E). Thermodynamics surface calculated for the apo 3CLpro structure in the allosteric site. Red surface indicates hydrophobic, non-polar regions. Yellow indicates polar regions.
3. Discussion
Coronaviruses with a crown like appearance in electron microscope contain single-stranded RNA of about 30 Kb in length, the largest among the RNA viruses (Masters, 2006; Samrat et al., 2020). The coronavirus genome encodes four structural proteins called spike (S), envelope (E), membrane (M), and nucleocapsid (N), and 16 Nsps (Nsp 1–16) along with 8 accessory proteins (Jiang et al., 2020; Samrat et al., 2020; Siu et al., 2008). Among these Nsps, Nsp 5 represents the 3CLpro. The 3CLpro of coronavirus is a potential drug target since it is responsible for the maturation of itself and the viral polyproteins (Ziebuhr et al., 2000).
Currently approved or authorized COVID-19 vaccines are highly effective against several strains like (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529) of SARS-CoV-2 (National Center for Immunization and Respiratory Diseases NCIRD, Division of Viral Diseases. CDC COVID-19 Science Briefs Internet. Atlanta GA: Centers for Disease Control and Prevention US;, 2020). The current knowledge of SARS-CoV-2 has highlighted the pivotal role of 3CLpro in viral replication, transcription, and it values in developing antivirus drugs (Mengist et al., 2021; Rathnayake et al., 2020).
At present, therapeutic options for SARS-CoV-2 are limited. Recently remdesivir, hydroxychloroquine, molnupiravir, and anti influenzas drug favipiravir has been clinically tested and being used in the treatment of SARS-CoV-2 infected patients (Fischer et al., 2021; Hall et al., 2021; Hassanipour et al., 2021). However, more clinical data regarding doses and safety need to be evaluated. In addition, Pfizer announce a discovery of Paxlovid (PF-07321332), an inhibitor for the SARS-CoV-2 3CL protease (www.Pfizer.com). This drug reduced the risk of hospitalization or death by 89% (within 3 days of symptom onset) and 88% (within 5 days of symptom onset) compared to placebo. Recent study suggests that remdesivir and hydroxychloroquine still need to be evaluated rigorously before generalizing it as a treatment option (Colson et al., 2020; Gautret et al., 2020). It has been shown previously that 3CLpro cleaves critical modulator of inflammatory pathways like TAB1 (TGF-beta-activated Kinase 1 and MAP3K7-binding protein 1) and NLRP12 (NLR Family Pyrin Domain Containing 12), which is a probable cause for cytokines and inflammatory response in Covid-19 positive patients (Moustaqil et al., 2021). The 3CLpro also antagonizes IFN production by retaining phosphorylated IRF3 in the cytoplasm (Fung et al., 2021). In addition, it has been shown previously that virus infection affects the interferon (IFN)-mediated antiviral response which can be rescued by effective protease inhibitor (Hung et al., 2011). These reports suggest that effective inhibition of the SARS-CoV-2 3CLpro not only inhibits the virus replication and propagation but also affects the interruption of antiviral IFN regulatory pathway.
Niclosamide and derivatives were reported as potent protease inhibitors of several viruses, including SARS-CoV (Li et al., 2017, 2020a; Shie et al., 2005; Xu et al., 2020d). Although niclosamide itself does not inhibit the 3CLpros of SARS-CoV and CoV-2 even at 50 μM concentration in our experiment, its derivatives were potent inhibitors against SARS-CoV (Shie et al., 2005). In this project, we synthesized different derivatives of niclosamide and checked their inhibitory potential using FRET and cell-based assays.
Our results suggest that niclosamide derivatives JMX0286, JMX0301 and JMX0941 inhibit enzymatic activity of the SARS-CoV-2 3CLpro with IC50 of 4.8, 4.5 and 3.9 μM respectively (Fig. 1C). Niclosamide itself has IC50 more than 50 μM against the SARS-CoV-2 3CLpro, suggesting that it has no inhibitory activity. We used boceprevir as a positive control in our experiment which showed IC50 as of 5.9 μM in the in vitro enzymatic assay (Hu et al., 2021; Ma et al., 2020). Our results indicated that these compounds were better than boceprevir in inhibition of the viral 3CLpro.
Since above results were based on inhibition of fluorescence of the FRET substrate, we performed a protein-based inhibition assay, in which protease cleavage can be visualized on SDS page. We found that all three compounds JMX0286, JMX0301 and JMX0941 inhibited the digestion of protein substrate in dose dependent manner (Fig. 4 A and B). It has been reported that many SARS-CoV-2 3CLpro inhibitors also inhibit cathepsin proteases, CatB and CatL (Steuten et al., 2021). To test this possibility, we evaluated our compounds against CatB and CatL proteins. We found that these compounds did not inhibit the cathepsins’ protease activity unless given high concentrations (Fig. 3 A and B) (Ma et al., 2020). These results indicated that these niclosamide derivatives were inhibitors specific for the 3CLpro, which is in contrast to boceprevir that could inhibit cathepsins more potently than did 3CLpro (Liu et al., 2021).
Moreover, we tested the efficacy of JMX0286, JMX0301 and JMX0941 in cell-based antiviral assay. We found that JMX0286 and JMX0941 inhibited the SARS-CoV-2 viral replication with EC50 of 2.2 and 1.7 μM, respectively (Table 2).
Although virus inhibition in cell-based assay for JMX0286 and JMX0941 are in 1–2 μM range, our SPR experiment shows that JMX0286 and JMX0941 showed dose-response bindings with K D values at 9.8 μM and 29.3 μM, respectively, whereas JMX0301 did not show binding with 3CLpro (Fig. 5). However, our MST result suggested binding of JMX0301 with 3CLpro with a K D value of 34 μM Fig. S2). Our molecular docking, along with MD simulation and hydration site thermodynamics studies suggested that all three compounds bind well to an allosteric site, experimentally known to accommodate the binding of AT7519 ligand (Fig. 7, Fig. 8, Fig. 9; Fig. S6). Our enzyme kinetic assay also indicated that all three compounds showed a non-competitive inhibition mechanism (Fig. 6). In addition, at higher concentrations, the compounds displayed mixed inhibition mechanism (data not shown). Docking and simulation studies suggested that the compounds could also bind to the active site as competitive inhibitors (data not shown). Nevertheless, these data provide new niclosamide derivatives with both in vitro as well as cellular antiviral activity against SARS-CoV-2. Further, determining crystal structures of the SARS-CoV-2 3CLpro in complex with these inhibitors along with testing in animal models will be helpful for COVID-19 antiviral drug development. They can be used alone or in combination with other known antiviral drugs.
4. Material and methods
4.1. Synthesis of niclosamide derivatives
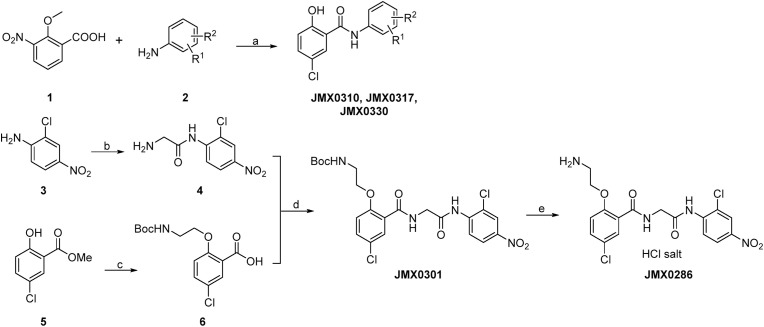
The synthetic routes of compounds JMX0286, JMX0301, JMX0310, JMX0317 and JMX0330 were described as below (Scheme 1 ), and the synthesis of other niclosamide derivatives was reported in our previous publications (Li et al., 2020b; Xu et al., 2020a, 2020b). The structures and purity of all synthesized compounds were confirmed by 1H and 13C NMR, HRMS, and HPLC analysis, and all biologically evaluated compounds are >95% pure.
Scheme 1.
Synthesis of Compounds JMX0286, JMX0301, JMX0310, JMX0317 and JMX0330.
aReagents and conditions: (a) i. PCl3, toluene, 100 °C; ii. BBr3, CH2Cl2, -78 °C–0 °C, 95% in two steps. (b) I. Fmoc-Gly-OH, PCl3, toluene, 100 °C; ii. Piperidine, CH3CN, r. t., 69% in two steps (c) i. BocNHCH2CH2OH, Ph3P, DIAD, THF, r. t.; ii NaOH, H2O, MeOH, r. t., 97% in two steps (d) EDCI, DMAP, CH2Cl2, r. t., 41% (e) HCl (aq.), MeOH, 70 °C, 38%.
General procedure for synthesizing compounds JMX0310, JMX0317 and JMX0330.
To a solution of acid 1 (1 eq) and amine 2 (1 eq) in toluene (15 mL/mmol) was added PCl3 (1.5 eq). The resulting mixture was stirred at 100 °C for 6 h and concentrated. Then MeOH (15 mL/mmol) was added, and the mixture was stirred at r. t. For 20 min. The amide product was isolated by filtration. The amide intermediate was dissolved in DCM (20 mL/mmol) and then BBr3 (3 eq, 1 M in DCM) was added at 0 °C. The mixture was stirred at r. t for 2 h. The mixture was diluted with DCM, washed with H2O and brine, dried (Na2SO4) and concentrated. The residue was purified by column chromatography or recrystallization (MeOH) to give the final product.
N-(2-Chloro-4-(trifluoromethyl)phenyl)-2-hydroxy-3-nitrobenzamide (JMX0310): 100 mg, 54% in two steps. Yellow solid. HPLC purity 99.9% (t R = 21.19 min). 1H NMR (300 MHz, CDCl3) δ 10.37 (s, 1H), 8.83 (d, J = 8.7 Hz, 1H), 8.45 (dd, J = 7.8, 1.8 Hz, 1H), 8.04 (dd, J = 8.1, 1.8 Hz, 1H), 7.72 (d, J = 1.8 Hz, 1H), 7.60 (dd, J = 8.7, 2.1 Hz, 1H), 7.44 (t, J = 8.1 Hz, 1H), 4.11 (s, 3H). 13C NMR (75 MHz, CDCl3) δ 161.8, 151.6, 144.4, 138.1, 136.9, 129.5, 128.7, 127.1 (q, J = 33.4 Hz), 126.6 (q, J = 3.8 Hz), 125.2 (q, J = 3.7 Hz), 125.2, 123.4 (q, J = 267.3 Hz), 123.2, 121.7, 64.8. HRMS (ESI) calcd for C14H9ClF3N2O4 361.0203 (M + H)+, found 361.0196.
N-(3-Fluoro-5-(trifluoromethyl)phenyl)-2-hydroxy-3-nitrobenzamide (JMX0317): 100 mg, 47% in two steps. Yellow solid. HPLC purity 99.5% (t R = 19.91 min). 1H NMR (300 MHz, DMSO‑d 6) δ 13.21 (s, 1H), 8.06 (dd, J = 7.5, 1.8 Hz, 1H), 7.98–7.86 (m, 3H), 7.36 (d, J = 8.4 Hz, 1H), 6.67 (t, J = 7.8 Hz, 1H). 13C NMR (75 MHz, DMSO‑d 6) δ 165.9, 162.1 (d, J = 242.6 Hz), 159.2, 141.8 (d, J = 11.3 Hz), 139.6, 134.8, 131.1 (qd, J = 32.6, 9.9 Hz), 129.1, 123.3 (qd, J = 271.0, 3.6 Hz), 123.0, 112.8, 112.4 (quint, J = 3.5 Hz), 110.3 (d, J = 25.9 Hz), 106.9 (dq, J = 25.4, 3.8 Hz). HRMS (ESI) calcd for C14H9F4N2O4 345.0498 (M + H)+, found 345.0494.
N-(3,5-Bis(trifluoromethyl)phenyl)-2-hydroxy-3-nitrobenzamide (JMX0330): 155 mg, 57% in two steps. Yellow solid. HPLC purity 99.7% (t R = 20.72 min). 1H NMR (300 MHz, CDCl3) δ 12.49 (s, 1H), 10.05 (s, 1H), 8.67 (dd, J = 7.8, 1.8 Hz, 1H), 8.40 (dd, J = 8.4, 1.8 Hz, 1H), 8.22 (s, 2H), 7.67 (s, 1H), 7.26 (t, J = 8.1 Hz, 1H). 13C NMR (75 MHz, CDCl3) δ 161.5, 152.9, 141.2, 139.3, 134.5, 132.7 (q, J = 33.4 Hz, 2C), 129.8, 123.2 (q, J = 271.1 Hz, 2C), 122.9, 121.0, 120.5 (q, J = 3.0 Hz, 2C), 118.2 (hept, J = 3.8 Hz). HRMS (ESI) calcd for C15H9F6N2O4 395.0467 (M + H)+, found 395.0462.
Tert-Butyl (2-(4-chloro-2-((2-((2-chloro-4-nitrophenyl)amino)-2-oxoethyl)carbamoyl)phenoxy)ethyl)carbamate (JMX0301). To a solution of 2-chloro-4-nitroaniline (1.0 g, 5.8 mmol) and Fmoc-Gly-OH (2.2 g, 7.5 mmol) in toluene (100 mL) was added PCl3 (1.0 g, 7.5 mmol). The mixture was stirred at 80 °C for 1 h. The mixture was diluted with AcOEt, washed with water, dried (Na2SO4) and concentrated to give the intermediate. The intermediate was dissolved in CH3CN (200 mL) and piperidine (1.3 g, 15.1 mmol) was added. The mixture was stirred at r. t. Overnight and then concentrated. The residue was purified by column chromatography to give 2-amino-N-(2-chloro-4-nitrophenyl)acetamide (4) (800 mg, 69% in two steps) as a yellow solid. 1H NMR (300 MHz, DMSO‑d 6) δ 8.67 (d, J = 9.3 Hz, 1H), 8.42 (d, J = 2.7 Hz, 1H), 8.27 (dd, J = 9.0, 2.7 Hz, 1H), 5.33 (s, 2H), 3.37 (s, 2H).
To a solution of methyl 5-chloro-2-hydroxybenzoate (2.0 g, 10.7 mmol), N-Boc-ethanolamine (3.46 g, 21.4 mmol) and PPh3 (5.6 g, 21.4 mmol) in 100 mL of THF was added DIAD (4.2 g, 21.4 mmol) dropwise at 0 °C. The mixture was stirred at 0 °C for 2 h and then concentrated. The residue was purified by column chromatography (Hex/AcOEt = 6/1 to 4/1) to give the crude intermediate methyl 2-(2-((tert-butoxycarbonyl)amino)ethoxy)-5-chlorobenzoate as light-yellow oil. The intermediate was dissolved in 20 mL of MeOH and then NaOH (1.2 g, in 8 mL of H2O) was added. The mixture was stirred at r. t. For 2 h. The pH of the mixture was adjusted to 3–4 with 1 M HCl (aq.) at 0 °C and then extracted with AcOEt (2 × 150 mL), washed with brine (60 mL), dried (Na2SO4) and concentrated. The residue was purified by column chromatography (Hex/AcOEt/Et3N = 3/1/0.03 to DCM/MeOH/AcOH = 10/1/0.1) to give 2-(2-((tert-butoxycarbonyl)amino)ethoxy)-5-chlorobenzoic acid (5) (3.3 g, 97% in two steps) as a white solid. 1H NMR (300 MHz, DMSO‑d 6) δ 7.60 (d, J = 2.7 Hz, 1H), 7.52 (dd, J = 8.7, 2.7 Hz, 1H), 7.17 (d, J = 8.7 Hz, 1H), 6.85 (t, J = 5.1 Hz, 1H), 4.04 (t, J = 6.0 Hz, 2H), 3.28 (q, J = 6.0 Hz, 2H), 1.37 (s, 9H). 13C NMR (75 MHz, DMSO‑d 6) δ 166.1, 156.0, 155.6, 132.2, 129.8, 124.2, 124.0, 116.2, 77.9, 68.0, 39.1, 28.2 (3C).
To a solution of amine 4 (150 mg, 0.65 mmol), acid 6 (248 mg, 0.78 mmol) and DMAP (8 mg, 0.07 mmol) in 20 mL of DCM was added EDCI (250 mg, 1.31 mmol). The mixture was stirred at r. t. Overnight and then concentrated. The residue was purified by column chromatography (Hex/AcOEt = 2/1 to 1/1) to give compound JMX0301 (140 mg, 41%) as a white solid. HPLC purity 98.8% (t R = 19.03 min). 1H NMR (300 MHz, CDCl3) δ 9.00 (s, 2H), 8.70 (d, J = 9.0 Hz, 1H), 8.29 (d, J = 2.7 Hz, 1H), 8.25 (d, J = 2.7 Hz, 1H), 8.16 (dd, J = 9.1, 2.4 Hz, 1H), 7.43 (dd, J = 8.7, 2.7 Hz, 1H), 6.91 (d, J = 9.0 Hz, 1H), 5.31 (s, 1H), 4.46 (d, J = 5.4 Hz, 2H), 4.16 (t, J = 4.5 Hz, 2H), 3.71–3.62 (m, 2H), 1.41 (s, 9H). 13C NMR (75 MHz, CDCl3) δ 168.9, 165.1, 156.80, 155.8, 143.3, 140.4, 133.3, 132.5, 127.2, 125.0, 123.6, 122.8, 121.8, 120.5, 113.5, 80.3, 70.3, 45.9, 40.4, 28.5 (3C). HRMS (ESI) calcd for C22H25Cl2N4O7 527.1100 (M + H)+, found 527.1093.
2-(2-Aminoethoxy)-5-chloro-N-(2-((2-chloro-4-nitrophenyl)amino)-2-oxoethyl)benzamide hydrochloride (JMX0286). To a solution of compound JMX0301 (90 mg, 0.17 mmol) in 5 mL of MeOH was added 4 mL of concentrated HCl. The resulting mixture was stirred at 70 °C for 8 h, and then cooled to r. t. The white solid was isolated by filtration to afford compound JMX0286 (30 mg, 38%). HPLC purity 95.0% (t R = 14.94 min). 1H NMR (300 MHz, DMSO‑d 6) δ 10.14 (s, 1H), 8.71 (t, J = 5.4 Hz, 1H), 8.39 (d, J = 2.7 Hz, 1H), 8.30 (d, J = 9.3 Hz, 1H), 8.26–8.08 (m, 4H), 7.74 (d, J = 3.0 Hz, 1H), 7.59 (dd, J = 9.0, 3.0 Hz, 1H), 7.27 (d, J = 9.0 Hz, 1H), 4.40–4.31 (m, 4H), 3.40–3.34 (m, 2H). 13C NMR (75 MHz, DMSO‑d 6) δ 169.2, 164.1, 154.4, 143.4, 140.7, 132.1, 130.0, 125.2, 125.0, 124.8, 124.5, 123.8, 123.2, 115.7, 66.2, 43.7, 38.3. HRMS (ESI) calcd for C17H17Cl2N4O5 427.0576 (M - Cl)+, found 427.0572.
4.2. Cell lines and viruses
HEK293T and Vero E6 cells were maintained in Dulbecco's modified Eagle's medium (DMEM); A549 cell were maintained in MEM medium. Each medium was supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin antibiotics. Cells were incubated at 37 °C in a 5% CO2 atmosphere. The A549-hACE2 cell line was maintained in a high-glucose DMEM supplemented with 10% fetal bovine serum, 1% P/S and 1% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES); ThermoFisher Scientific), 10 μg/mL Blasticidin S.
The recombinant SARS-CoV-2 (strain 2019-nCoV/USA_WA1/2020) with a Nanoluciferase reporter gene (SARS-CoV-2-Nluc) (Xie et al., 2020) was used in this study.
4.3. SARS-CoV-2-nluc antiviral assay
The antiviral activities were evaluated in A549-hACE2 using a protocol described previously (Xie et al., 2020). In brief, 12,000 cells per well in phenol-red free medium containing 2% FBS were plated into a white opaque 96-well plate (Corning). On the next day, 2-fold serial dilutions of compounds were prepared in dimethyl sulfoxide (DMSO). The compounds were further diluted 100-fold in the phenol-red free culture medium containing 2% FBS. Cell culture fluids were removed and incubated with 50 μL of diluted compound solutions and 50 μL of SARS-CoV2-Nluc viruses (MOI 0.01). At 48 h post-infection, 50 μL Nano luciferase substrates (Promega) were added to each well. Luciferase signals were measured using a Synergy™ Neo 2 microplate reader. The relative luciferase signals were calculated by normalizing the luciferase signals of the compound-treated groups to that of the DMSO-treated groups (set as 100%). The relative luciferase signal (Y-axis) versus the log 10 values of compound concentration (X-axis) was plotted in software Prism 8. The EC50 (compound concentration for reducing 50% of luciferase signal) were calculated using a nonlinear regression model (four parameters). Two experiments were performed with technical duplicates.
4.4. Construction, expression, and purification of SARS-CoV-2 3CLpro
Codon-optimized gene sequence of the SARS-CoV-2 3CLpro was synthesized and replaced the SARS-CoV 3CLpro sequence in the Addgene plasmid 61692 through seamless cloning technology by Gene Universal. The construct contained a modified pGEX-6P-1 backbone to generate authentic N-terminus of the 3CLpro through autocleavage, and a C-terminal His-tag GPHHHHHH to facilitate purification. The His-tag could be cleaved by the HRV 3C protease to generate authentic 3CLpro C-terminus.
The SARS-CoV-2 3CLpro protein was purified as describe previously (Jin et al., 2020). In brief, the expression plasmid 3CLpro SARS-CoV-2 pGEX-6P-1 was transformed into E. coli Rosetta (DE3) cells and then cultured in Sper broth medium containing 100 μg/mL ampicillin at 37 °C till OD reached to 0.6 at 600 nm. Then the cells were induced with 0.5 mM IPTG and further incubated with shaking at 16 °C. After 16 h, the cells were collected by centrifugation at 7000 rpm for 15 min. The cell pellets were resuspended in lysis buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl), lysed by sonication, and then centrifuged at 20,000 rpm for 30 min. The supernatant was loaded onto Ni-NTA affinity column (Qiagen) and washed in the resuspension buffer containing 20 mM imidazole. The His-tagged 3CLpro was eluted by 300 mM of imidazole in lysis buffer. Human rhinovirus 3C protease was added to remove the C-terminal His tag. The 3CLpro was further purified by size-exclusion chromatography using 75 Superdex column. Peak fractions were collected and pooled together. The purified 3CLpro was stored in a buffer containing 20 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM DTT.
4.5. Expression and purification of 3CLpro-PLpro-nanoluc substrate protein
A 3CLpro-PLpro-NS3 (TriPro) nanoluciferase (nanoluc) substate was constructed. Codon-optimized nanoluciferase gene sequence encoding nanoluciferase with GGGGG [ERELNGGAPIKS]GGGG (KTSAVLQSGFRKME)GGGGRRRRSAGGGSGGG sequence inserted between nanoluciferase residues 51 and 52 was synthesized and inserted between the Nco 1 and Xho 1 sites of the pET28a vector. The inserted extra sequence contains recognition sequences for PLpro (square brackets), 3CLpro (parenthesis), and flavivirus NS2B-NS3 protease (italicized). Purification of the TriPro nanoluc substrate was carried out similarly as described above for 3CLpro, with the following modifications. Upon elution from the Ni-NTA column, the protein was dialyzed in a buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl and 1 mM DTT. Dialysed proteins were stored in −80 °C.
4.6. Enzymatic assays
The SARS-CoV-2 3CLpro FRET peptide substrate Dabcyl-KTSAVLQ/SGFRKME (Edans) were custom synthesized by Genescript. Edans standard curve was generated as described below: 200 nM SARS-CoV-2 3CLpro was incubated with different concentrations of the FRET substrate (1–100 μM). The reaction progress was monitored until the fluorescence signals reached plateau, at that point we assumed that all the FRET substrate was digested by the SARS-CoV-2 3CLpro. The endpoint fluorescence signal was plotted against FRET substrate concentration with a linear regression function in Prism 8.
For reaction condition optimization, proteins at concentrations of 0.1 μM, 0.2 μM and 0.4 μM were added with 10 mM and 20 mM of FRET substrate respectively in assay buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT and 1 mM of EDTA. Reaction progress was monitored for 2 h. Based on the linear graph, 0.2 μM of protein and 10 μM of substrate was used for all future experiment in the buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT and 1 mM of EDTA. For the measurements of K m/V max, screening of the protease inhibitor library, as well as IC50 measurements, proteolytic reaction with 200 nM 3CLpro in 100 μL of Tris buffer was carried out at 30 °C in a BioTek synergy HI microplate reader with filters for excitation at 360/40 nm and emission at 460/40 nm. Reactions were monitored every 10 min.
The initial velocity of the proteolytic activity was calculated by linear regression for the first 10 min of the kinetic progress curves. The initial velocity was plotted against the FRET substrate concentration with the classic Michaelis–Menten equation in Prism 8 software.
To calculate he enzyme kinetics and Lineweaver–Burk plots of JMX0286, JMX0301, JMX0941, the assay was carried out at 200 nM protein concentration in buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT and 1 mM of EDTA in final volume of 30 μL in 384 well plates. Various concentration of FRET based peptide-Edans substrate and inhibitors were added to initiate the enzyme reaction. The initial velocity of the enzymatic reaction with different inhibitors and DMSO were calculated by linear regression for the first 6 min of the kinetic progress curve, and then plotted against substrate concentrations in Prism 9 with the Michaelis–Menten equation and linear regression of double reciprocal plot.
4.7. Inhibition of cleavage of 3CL substrate protein with inhibitors
0.8 μM 3CLpro was incubated for 1 h with different concentration of each inhibitor separately in Tris buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT and 1 mM EDTA). Then, the TriPro substrate protein was added at 5 μM concentration and further incubated for 1 h, followed by SDS-PAGE analysis.
4.8. Cytotoxicity measurement
A549 and Vero E6 cells were used for cell viability and cytotoxicity measurement using cell counting kit-8 (CCK-8) (GLPBIO) as per manufacturer protocol. In brief, 100 μL of cells at concentration 2 × 105 cells/well were seeded and grown overnight at 37 °C in a 5% CO2 atmosphere to ∼90% confluence on the next day. Cells were then treated with various concentrations of protease inhibitors. After 48 h of treatment 10 μL of CCK8 solution was added to each well of the plate using a repeating pipettor and incubate the plate for 1–4 h in the incubator. Absorbance was taken at 460 nm using a BioTek synergy HI microplate reader. The CC50 values were calculated by fitting dose-response curves using the GraphPad Prism 8 software.
4.9. Surface plasmon resonance
The purified native SARS-CoV-2 3CLpro was buffer exchanged with immobilization buffer containing 10 mM phosphate, pH 7.4, 2.7 mM KCl, 137 mM NaCl, and 0.05% Tween-20) to remove Tris from the storage buffer. Then it was diluted to 25 μg/mL with 10 mM sodium acetate at pH 4.5 and immobilized to flow channels 2–4 on a CM5 sensor surface after being first activated by a 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-hydroxy succinimide (NHS) mixture using a Biacore T200 (Cytiva, former GE Healthcare). Ethanolamine blocking was performed next to deactivate the unoccupied surface area of the sensor chip. Unmodified flow channel 1 was used as a reference. Three compounds were prepared at a series of increasing concentrations (0.164–100 μM at 2.5-fold dilution) in binding buffer containing 25 mM Tris, pH 7.4, 2.7 mM KCl, 137 mM NaCl, 0.05% Tween-20, and 4% DMSO and were applied to all four channels at a 30 μL/min flow rate with 90 s and 120 s of association and dissociation times, respectively, at 25 °C. The data were double referenced with a reference channel and zero concentration (DMSO control) responses, and reference subtracted sensorgrams were fitted with 1 to 1 Langmuir kinetic model using a Biacore Insight evaluation software, producing two rate constants (k a and k d). The equilibrium dissociation constants (K D) were determined by fitting the data with steady-state affinity model. For steady-state affinity fittings, response units at each concentration were measured during the equilibration phase, and the K D values were determined by fitting the data to a single rectangular hyperbolic curve equation (1), where y is the response, ymax is the maximum response and x is the compound concentration.
| (1) |
SPR binding analyses were done in triplicates, and standard deviations were calculated from three repeats (n = 3).
4.10. Ligand binding by microscale thermophoresis
Monolith NT.115 Microscale Thermophoresis (MST) instrument (NanoTemper Technologies) was used for this assay. Monolith protein labeling kit RED-NHS was purchased from NanoTemper Technologies. Briefly, 3CLpro protein was labeled using RED-NHS labelling kit (NanoTemper) following manufacturer's instructions. A serial dilution of ligand JMX0301 (0.6 mM–9.1 nM) was prepared and titrated against 10 nM labeled 3CLpro. The assay was read in 20% excitation power and medium MST power.
4.11. Computational modeling
A multi-step computational procedure was implemented, similar to the one reported previously to study the binding modes and dynamics of niclosamide derivatives JMX0941, JMX0286 and JMX0301 to 3CLpro (Manandhar et al., 2021).
Ligand preparation. Ligprep was used to prepare ligand molecules, including JMX0941, JMX0286 and JMX0301 (Sastry et al., 2013). Tautomers and stereoisomers were assigned by using Epik at pH 7.0 (Greenwood et al., 2010; Shelley et al., 2007).
Protein preparation. The Protein Preparation Wizard (PrepWizard) (Sastry et al., 2013), available in the Schrödinger suite, was used to prepare the crystal structures of 3CLpro as in complex with the allosteric inhibitor AT7519 (RCSB-PDB ID 7AGA) (Günther et al., 2021). Prime was used to model missing residues and protein loops (Jacobson et al., 2002, 2004). Protonation and tautomerization states were assigned for the pH range of 7.0 ± 2.0. During the initial stage of structure preparation, original hydrogen atoms were replaced by hydrogen atoms; no water molecules were retained. Then, hydrogen bond networks were optimized at pH 7.0 and only water molecules with at least three hydrogen bonds to non-water molecules were retained. Last, the OPLS3e force field was used to energy minimize the obtained structures (RMSD <0.30 Å heavy atoms cut-off) (Harder et al., 2016).
Binding site mapping. The program SiteMap (Halgren, 2007), available in the Schrödinger suite, was used (default settings) to explore binding sites on the surface of 3CLpro structure in complex with the allosteric inhibitor AT7519 (RCSB-PDB ID 7AGA) (Günther et al., 2021).
Molecular Docking. The program Glide (Friesner et al., 2004, 2006) was used for molecular docking, with the extra precision (XP) scoring function. In the 3CLpro/AT7519 complex, the Glide docking protocol was used with default settings. Coordinates of the ligands in the obtained systems were used as centroid of the docking grids. Prime was used to refine docking poses by allowing flexibility of protein residues within 10 Å of the ligand. For the AT7519 pocket (allosteric site), the best XP-binding pose (selected by taking into account docking score, visual inspection and match between ligand substructure moieties and SiteMap pockets) of JMX0286 was used as input for induced fit docking studies before simulating the best induced-fit system.
Induced fit docking. Induced fit docking (Sherman et al., 2006) was preformed using the Schrödinger suite to optimize the binding mode of JMX0286 to the AT7519 pocket (allosteric site). The best pose from XP docking was used to define the centroid of the workspace and default settings were used.
Solvent Mapping. The semi-continuum solvation approach, SZMAP, was used to compute the stabilizing and destabilizing effects of water molecules on 3CLpro-ligand complex. Explicit probe molecules, water and hypothetical water, were used to analyze the binding site and evaluate the solvent thermodynamics at different sites on the protein surface (Truchon et al., 2010). The crystal structure of 3CLpro in complex with the allosteric inhibitor AT7519 (RCSB-PDB ID 7AGA) (Günther et al., 2021) was processed by Spruce (Spruce 1.3.0.1) to enumerate possible alternate amino acid locations, build missing partial sidechains, cap chain breaks, model missing loops, and optimize hydrogen atoms. AmberFF94 charges (Cornell et al., 1995) were assigned for the protein. The grids for the complex, apo and ligand were calculated by SZMAP around the ligand, and the structural waters were excluded from the stabilization calculation. The energy values were calculated at each grid point. The results were then analyzed by VIDA (Vida 5.0.0.1) and PyMol.
Molecular Dynamics (MD) Simulations. The Desmond (Bowers, 2006) program, as distributed in the Schrödinger suite, was used to perform molecular dynamics (MD) studies of 3CLpro in the docking complexes with non-covalent inhibitors JMX0941, JMX0286 and JMX0301. The OPLS3e force field was used to model ligand, protein and Na+ atoms. The TIP3P model (Jorgensen et al., 1983) was used for water molecules. Systems were simulated in an NPT ensemble; constant pressure was set to 1 bar, constant temperature to 300 K, by applying the Nose−́ Hoover chain and Martyna−Tobias−Klein coupling schemes, respectively (Martyna et al., 1992; Martyna et al., 1994). Numerical integration was implemented by the RESPA integrator, by updating short-range/bonded and long-range/nonbonded interactions every 2 and 6 ps, respectively (Tuckerman et al., 1993). While a 9.0 Å cutoff was set for short-range Coloumb, long-range interactions were calculated using the particle mesh Ewald method (1 × 10−9 tolerance).
Structure visualization & MD analyses. Image rendering was obtained by using the visualization tools PyMOL (Schrödinger and DeLano, 2020) and VMD (Humphrey et al., 1996). Simulation interaction diagrams were obtained for each simulated protein/ligand complex; analyses include Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF; C-α for protein residues and ligand heavy atoms) plots; Protein Secondary Structural Element (PSSE) composition plots; and protein ligand contacts (i.e., H-bonds, hydrophobic contacts, ionic interactions and water bridges). The latter are represented as stacked bar charts, normalized over the course of the trajectory. (As protein residues may engage in multiple interactions with the ligand, values greater than 1.0 are possible.) RMSD plots were plotted using the Matplotlib python package (Hunter, 2007).
4.12. Statistical analysis
All statistical analysis was done using GraphPad version 8.
Declaration of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
HL is supported by the NIH grants: AI161845, AI131669 and AI140406, and by University of Arizona College of Pharmacy faculty Startup fund, and by R. Ken and Donna Coit Endowed Chair fund in Drug Discovery. Jia Zhou is partly supported by the John D. Stobo, M.D. Distinguished Chair Endowment Fund. P.-Y.S. was supported by NIH grants U19AI171413 and U01AI151801, and awards from the Sealy & Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation. The research conducted at Temple University includes calculations carried out on HPC resources supported in part by the National Science Foundation through major research instrumentation grant number 1625061 and by the US Army Research Laboratory under contract number W911NF-16-2-0189. This project is funded, in part, under a Grant with the Pennsylvania Department of Health (PA CURE). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. We acknowledge OpenEye Scientific Software for providing the academic license.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105381.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Blake S., Shaabani N., Eubanks L.M., Maruyama J., Manning J.T., Beutler N., Paessler S., Ji H., Teijaro J.R., Janda K.D. Salicylanilides reduce SARS-CoV-2 replication and suppress induction of inflammatory cytokines in a rodent model. ACS Infect. Dis. 2021;7:2229–2237. doi: 10.1021/acsinfecdis.1c00253. [DOI] [PubMed] [Google Scholar]
- Bowers J.K.e.a. 2006. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters, SC '06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing; p. 84. [Google Scholar]
- Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeño J.M., Ortega-Prieto A.M., Bussani R., Cannatà A., Rizzari G., Collesi C., Schneider E., Arosio D., Shah A.M., Barclay W.S., Malim M.H., Burrone J., Giacca M. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunaugh A.D., Seo H., Warnken Z., Ding L., Seo S.H., Smyth H.D.C. Development and evaluation of inhalable composite niclosamide-lysozyme particles: a broad-spectrum, patient-adaptable treatment for coronavirus infections and sequalae. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.M., Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., Duke E.R., Azizad M.M., Borroto-Esoda K., Wohl D.A., Loftis A.J., Alabanza P., Lipansky F., Painter W.P. Cold Spring Harbor Laboratory; 2021. Molnupiravir, an Oral Antiviral Treatment for COVID-19. [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S.Y., Siu K.L., Lin H., Yeung M.L., Jin D.Y. SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int. J. Biol. Sci. 2021;17:1547–1554. doi: 10.7150/ijbs.59943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett T.J., Coatsworth H., Mahmud I., Hamerly T., Stephenson C.J., Yazd H.S., Ayers J., Miller M.R., Lednicky J.A., Dinglasan R.R. Cold Spring Harbor Laboratory; 2021. Niclosamide Reverses SARS-CoV-2 Control of Lipophagy. [DOI] [Google Scholar]
- Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mosbauer K., Zellner A., Zannas A.S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Muller-Myhsok B., Drosten C., Muller M.A., Rein T. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Tissot Dupont H., Honore S., Colson P., Chabriere E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Greenwood J.R., Calkins D., Sullivan A.P., Shelley J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Peña-Murillo G.E., Mashhour A.R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. New method for fast and accurate binding‐site identification and analysis. Chem. Biol. Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- Hall K., Mfone F., Shallcross M., Pathak V. Review of pharmacotherapy trialed for management of the coronavirus disease-19. Eurasian J Med. 2021;53:137–143. doi: 10.5152/eurasianjmed.2021.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., Kaus J.W., Cerutti D.S., Krilov G., Jorgensen W.L., Abel R., Friesner R.A. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theor. Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Hassanipour S., Arab-Zozani M., Amani B., Heidarzad F., Fathalipour M., Martinez-De-Hoyo R. The efficacy and safety of Favipiravir in treatment of COVID-19: a systematic review and meta-analysis of clinical trials. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90551-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Ma C., Szeto T., Hurst B., Tarbet B., Wang J. Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses. ACS Infect. Dis. 2021;7:586–597. doi: 10.1021/acsinfecdis.0c00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hung H.C., Wang H.C., Shih S.R., Teng I.F., Tseng C.P., Hsu J.T. Synergistic inhibition of enterovirus 71 replication by interferon and rupintrivir. J. Infect. Dis. 2011;203:1784–1790. doi: 10.1093/infdis/jir174. [DOI] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- Jacobson M.P., Friesner R.A., Xiang Z., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J., Honig B., Shaw D.E., Friesner R.A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Konwar M., Sarma D. Advances in developing small molecule SARS 3CL(pro) inhibitors as potential remedy for corona virus infection. Tetrahedron. 2021;77 doi: 10.1016/j.tet.2020.131761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann K. Getting hands on a drug for covid-19: inhaled and intranasal niclosamide. The Lancet Regional Health - Europe. 2021;4 doi: 10.1016/j.lanepe.2021.100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li Z., Brecher M., Deng Y.-Q., Zhang J., Sakamuru S., Liu B., Huang R., Koetzner C.A., Allen C.A., Jones S.A., Chen H., Zhang N.-N., Tian M., Gao F., Lin Q., Banavali N., Zhou J., Boles N., Xia M., Kramer L.D., Qin C.-F., Li H. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017;27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Xu J., Lang Y., Fan X., Kuo L., D'Brant L., Hu S., Samrat S.K., Trudeau N., Tharappel A.M., Rugenstein N., Koetzner C.A., Zhang J., Chen H., Kramer L.D., Butler D., Zhang Q.Y., Zhou J., Li H. JMX0207, a niclosamide derivative with improved pharmacokinetics, suppresses zika virus infection both in vitro and in vivo. ACS Infect. Dis. 2020;6:2616–2628. doi: 10.1021/acsinfecdis.0c00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Xu J., Lang Y., Fan X., Kuo L., D'Brant L., Hu S., Samrat S.K., Trudeau N., Tharappel A.M., Rugenstein N., Koetzner C.A., Zhang J., Chen H., Kramer L.D., Butler D., Zhang Q.-Y., Zhou J., Li H. JMX0207, a niclosamide derivative with improved pharmacokinetics, suppresses zika virus infection both in vitro and in vivo. ACS Infect. Dis. 2020;6:2616–2628. doi: 10.1021/acsinfecdis.0c00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Boland S., Scholle M.D., Bardiot D., Marchand A., Chaltin P., Blatt L.M., Beigelman L., Symons J.A., Raboisson P., Gurard-Levin Z.A., Vandyck K., Deval J. Dual inhibition of SARS-CoV-2 and human rhinovirus with protease inhibitors in clinical development. Antivir. Res. 2021;187 doi: 10.1016/j.antiviral.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar A., Blass B.E., Colussi D.J., Almi I., Abou-Gharbia M., Klein M.L., Elokely K.M. Targeting SARS-CoV-2 M3CLpro by HCV NS3/4a inhibitors: in silico modeling and in vitro screening. J. Chem. Inf. Model. 2021;61:1020–1032. doi: 10.1021/acs.jcim.0c01457. [DOI] [PubMed] [Google Scholar]
- Martyna G.J., Klein M.L., Tuckerman M. Nosé–Hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys. 1992;97:2635–2643. [Google Scholar]
- Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994:4177–4189. [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.P., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J.B., Freiberg A.N., Jacques D., Lee B., Sierecki E., Gambin Y. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microb. Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfizer Announces Additional Phase 2/3 Study Results Confirming Robust Efficacy of Novel COVID-19 Oral Antiviral Treatment Candidate in Reducing Risk of Hospitalization or Death.
- Science Brief: COVID-19 Vaccines and Vaccination. 2021 Sep 15. PMID: 34009769.; 2020-.. National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. CDC COVID-19 Science Briefs [Internet]. Atlanta (GA): Centers for Disease Control and Prevention (US); [PubMed] [Google Scholar]
- Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., Kashipathy M.M., Battaile K.P., Lovell S., Perlman S., Groutas W.C., Chang K.O. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samrat S.K., Xu J., Li Z., Zhou J., Li H. Antiviral agents against flavivirus protease: prospect and future direction. Pathogens. 2022;11 doi: 10.3390/pathogens11030293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006. Novel;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie J.J., Fang J.M., Kuo C.J., Kuo T.H., Liang P.H., Huang H.J., Yang W.B., Lin C.H., Chen J.L., Wu Y.T., Wong C.H. Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease. J. Med. Chem. 2005;48:4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce 1.3.0.1: OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com.
- Steuten K., Kim H., Widen J.C., Babin B.M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C.J., Cortese M., Muir R.K., Bennett J.M., Geiss-Friedlander R., Peters C., Bartenschlager R., Bogyo M. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect. Dis. 2021;7:1457–1468. doi: 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZMAP 1.6.2.1. OpenEye Scientific Software; Santa Fe, NM: 2013. http://www.eyesopen.com [Google Scholar]
- Truchon J.F., Nicholl's A., Grant J.A., Iftimie R.I., Roux B., Bayly C.I. Using electronic polarization from the internal continuum (EPIC) for intermolecular interactions. J. Comput. Chem. 2010;31:811–824. doi: 10.1002/jcc.21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman M.E., Berne B.J., Martyna G.J., Klein M.L. Efficient molecular dynamics and hybrid Monte Carlo algorithms for path integrals. J. Chem. Phys. 1993;99:2796–2808. [Google Scholar]
- Schrödinger, L., & DeLano, W., 2020. PyMOL. Retrieved from http://www.pymol.org/pymol.
- Vida 5.0.0.1: OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com.
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Jan J.T., Chen C.M., Hsieh H.P., Hwang D.R., Liu H.W., Liu C.Y., Huang H.W., Chen S.C., Hong C.F., Lin R.K., Chao Y.S., Hsu J.T. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., Tseng C.K., Makino S., Menachery V.D., Bilello J.P., Shi P.Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Berastegui-Cabrera J., Chen H., Pachón J., Zhou J., Sánchez-Céspedes J. Structure-activity relationship studies on diversified salicylamide derivatives as potent inhibitors of human adenovirus infection. J. Med. Chem. 2020;63:3142–3160. doi: 10.1021/acs.jmedchem.9b01950. [DOI] [PubMed] [Google Scholar]
- Xu J., Berastegui-Cabrera J., Ye N., Carretero-Ledesma M., Pachón-Díaz J., Chen H., Pachón-Ibáñez M.E., Sánchez-Céspedes J., Zhou J. Discovery of novel substituted N-(4-Amino-2-chlorophenyl)-5-chloro-2-hydroxybenzamide analogues as potent human adenovirus inhibitors. J. Med. Chem. 2020;63:12830–12852. doi: 10.1021/acs.jmedchem.0c01226. [DOI] [PubMed] [Google Scholar]
- Xu J., Shi P.-Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Shi P.Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.