Summary
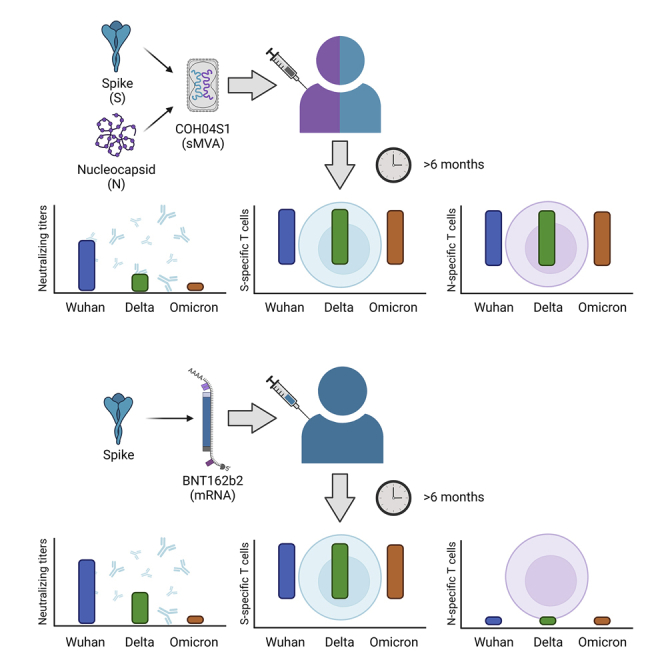
Cell-mediated immunity may contribute to providing protection against SARS-CoV-2 and its variants of concern (VOC). We developed COH04S1, a synthetic multiantigen modified vaccinia Ankara (MVA)-based COVID-19 vaccine that stimulated potent spike (S) and nucleocapsid (N) antigen-specific humoral and cellular immunity in a phase 1 clinical trial in healthy adults. Here, we show that individuals vaccinated with COH04S1 or mRNA vaccine BNT162b2 maintain robust cross-reactive cellular immunity for six or more months post-vaccination. Although neutralizing antibodies induced in COH04S1- and BNT162b2-vaccinees showed reduced activity against Delta and Omicron variants compared to ancestral SARS-CoV-2, S-specific T cells elicited in both COH04S1- and BNT162b2-vaccinees and N-specific T cells elicited in COH04S1-vaccinees demonstrated potent and equivalent cross-reactivity against ancestral SARS-CoV-2 and the major VOC. These results suggest that vaccine-induced T cells to S and N antigens may constitute a critical second line of defense to provide long-term protection against SARS-CoV-2 VOC.
Subject areas: Health sciences, Immunology, Immune response, Virology
Graphical abstract
Highlights
-
•
COH04S1 and BNT162b2 vaccine-elicited cellular immunity sustained for six months
-
•
Neutralizing antibodies with reduced activity against Delta and Omicron variants
-
•
Spike- and nucleocapsid-specific T cells maintain cross-reactivity to variants
-
•
Vaccine-induced T cells may provide long-term protection against SARS-CoV-2
Health sciences; Immunology; Immune response; Virology
Introduction
The rapid genetic evolution of SARS-CoV-2 since its emergence in 2019 resulted in the prompt appearance of several variants of concern (VOC) with defining antigenic sequences and altered infectivity, transmissibility, and pathogenicity. VOC such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) have modified the observed COVID-19 vaccine efficacy in clinical trials taking place during the spread of these variant viruses (Hayawi et al., 2021; Lopez Bernal et al., 2021). More recently, the appearance of the highly contagious Omicron variant (B.1.1.529) and its two sub-lineages BA.1 and BA.2 resulted in observational studies suggesting a significant decrease in vaccine efficacy against COVID-19-associated hospitalizations, especially before the introduction of booster doses (Accorsi et al., 2022; Andrews et al., 2022; Thompson et al., 2022). Omicron contains a remarkable number of mutations of which the majority map to the Spike (S) protein, and almost half of these mutations localize to the receptor-binding domain (RBD), the main target of neutralizing antibody (NAb) responses. Considering the vast number of mutations in the Omicron S protein, it is not surprising that the majority of anti-S monoclonal antibodies for clinical use completely lost their neutralizing activity against the Omicron VOC (VanBlargan et al., 2022; Zhou et al., 2022).
While NAb induced through natural infection or vaccination have been shown to possess significantly reduced neutralizing activity against SARS-CoV-2 VOC, S-specific cellular responses induced after vaccination or natural infection have been proven to be durable and to display a high degree of cross-recognition of different S variants, including Omicron S (Liu et al., 2022; Tarke et al., 2022). These findings suggest that T cell responses could provide long-term protective immunity against emerging SARS-CoV-2 VOC. Approved COVID-19 vaccines are intrinsically limited in their capacity to elicit protective T cell responses owing to their single antigen design based solely on the S protein of the ancestral Wuhan-Hu-1 SARS-CoV-2 reference strain. The inclusion of additional immunodominant T cell targets, such as the nucleocapsid (N) protein, into a COVID-19 vaccine formulation may, therefore, contribute to stimulate durable protective immunity against SARS-CoV-2 and its emerging VOC by broadening the coverage of vaccine-induced T cell responses.
We previously developed clinical vaccine candidate COH04S1, a synthetic multi-antigenic modified vaccinia Ankara (sMVA) vector co-expressing full-length S and N antigen sequences based on the Wuhan-Hu-1 reference strain (Chiuppesi et al., 2020). COH04S1 was highly immunogenic in different animal models and protected against ancestral SARS-CoV-2 and VOC in Syrian hamsters and non-human primates (Chiuppesi et al., 2020, 2022a; Wussow, 2022). COH04S1 has been tested in a combined open label and randomized phase 1 clinical trial in healthy adults, demonstrating high tolerability and robust induction of humoral and cellular responses to both S and N antigens (Chiuppesi et al., 2022b).
Here we show that individuals vaccinated with COH04S1 or the FDA-approved mRNA vaccine BNT162b2 (Comirnaty, Pfizer) maintain potent cellular immune responses for at least six months post-vaccination. Both COH04S1- and BNT162b2-vaccinated subjects showed NAb responses with reduced activity against Delta (B.1.617.2) and Omicron (BA.1) variants compared to ancestral SARS-CoV-2. In contrast, S-specific T cells elicited by COH04S1 and BNT162b2 as well as N-specific T cells elicited by COH04S1 were present at high levels and were similarly activated in the presence of ancestral, Delta-, and Omicron-specific peptide libraries. These results support that vaccine-induced T cell responses maintain potent cross-reactivity against SARS-CoV-2 and its emerging VOC and could be a source of protective immunity in the face of declining NAb responses.
Results
COH04S1- and BNT162b2-vaccinees show reduced NAb responses against SARS-CoV-2 Delta and Omicron variants at six-month post-vaccination
We evaluated SARS-CoV-2-specific humoral and cellular immune responses six-to-eight months after vaccination with COH04S1 in 30 volunteers enrolled in phase 1 clinical trial aimed at testing the safety and immunogenicity of COH04S1 at different dose levels (DL) (NCT04639466) (Chiuppesi et al., 2022b). As a comparator, we evaluated SARS-CoV-2-specific humoral and cellular responses measured in 30 healthcare workers (HCW) six-to-fourteen months after vaccination with Pfizer BNT162b2 (Tables S1–S3). COH04S1 subjects were prime-boost vaccinated with low-dose (DL1, 1 × 107 plaque-forming units [pfu]), medium-dose (DL2, 1 × 108 pfu), or high-dose (DL3, 2.5 × 108 pfu) of vaccine. Of the 30 subjects vaccinated with COH04S1, 18 received two DL1 vaccinations 30-90 days apart, six received two DL2 vaccinations 30 days apart, and six received two DL3 vaccinations 30 days apart. All HCW vaccinated with BNT162b2 received two 30 μg doses approximately three weeks apart. Subjects were SARS-CoV-2 naive by history, and subjects vaccinated with COH04S1 had additional negative SARS-CoV-2 IgG and negative SARS-CoV-2 PCR tests at enrollment.
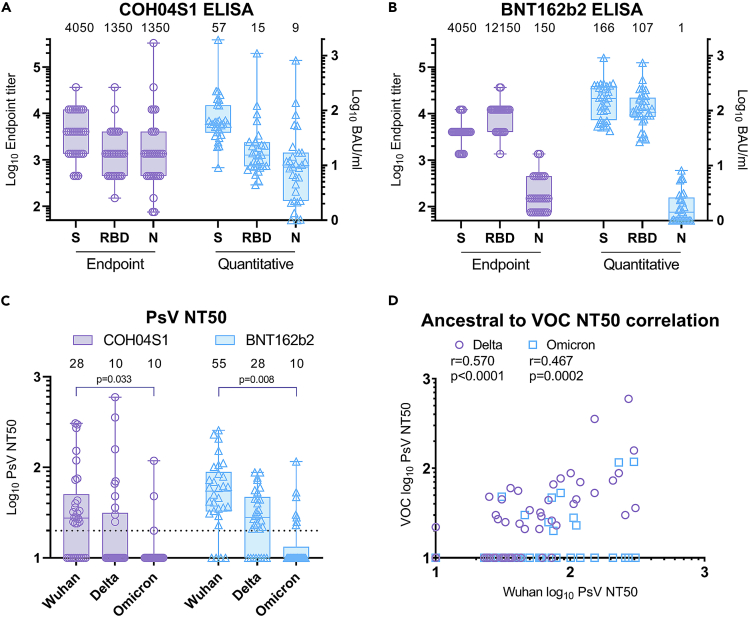
Binding antibodies to S, RBD, and N were measured using Wuhan-Hu-1-specific antigens by endpoint and quantitative ELISA. Using these assays, COH04S1-vaccinees showed robust binding antibody responses to S, RBD, and N antigens, with median S, RBD, and N endpoint IgG titers of 4,050, 1,350, and 1,350 and median WHO IgG international units (BAU/ml) of 57, 15, and nine for S, RBD, and N antigens, respectively. Consistent with the single S antigen design of BNT162b2, robust binding antibody responses in BNT162b2-vaccinated HCW were only measured against S and RBD antigens, with median IgG endpoint of 4,050 and 12,150, respectively, while N-specific IgG endpoint titers were only very low or at the limit of detection. Similarly, BNT162b2-vaccinated HCW showed median WHO IgG international units of 166, 107, and one BAU/ml for S, RBD, and N antigens, respectively (Figures 1A, 1B and S1). Readouts of the two assays were highly correlated independent of the antigen specificity (Figure S2).
Figure 1.
SARS-CoV-2 ancestral and VOC-specific humoral responses in COH04S1- and BNT162b2-vaccinees six months or more after primary vaccination
(A and B). Binding antibodies. Binding antibody titers to Wuhan-Hu-1-specific spike (S), receptor-binding domain (RBD), and nucleocapsid (N) antigens were measured by endpoint and quantitative ELISA in serum samples of COH04S1- (A) and BNT162b2- (B) vaccinated subjects six months or more after primary vaccination. For endpoint, ELISA samples with titers below the limit of quantification were indicated with an endpoint titer of 75. BAU = binding antibody units.
(C). NAb responses. 50% neutralizing antibody titers (NT50) against ancestral SARS-CoV-2 (Wuhan-Hu-1), Delta, and Omicron variants were measured using a pseudovirus (PsV) assay in serum samples of COH04S1- and BNT162b2-vaccinated subjects six months or more after primary vaccination. Samples with titers below the limit of quantification were indicated with an NT50 titer of 10. Dotted line represents the lower limit of quantification. Boxplots in a-c extend from the 25th to the 75th percentiles, medians are shown as a line and median values indicated above each boxplot, whiskers extend from minimum to maximum values. two-way ANOVA followed by Tukey’s multiple comparison test was used. Only p < 0.05 are shown.
(D) Correlation analysis. Correlation between levels of Wuhan-specific neutralizing antibody titers (NT50) and VOC-specific NT50 in subjects vaccinated with COH04S1 and BNT162B2 was assessed. Shown are Pearson’s correlation coefficients (r) and their two-sided significance (p).
NAb responses were measured using ancestral (Wuhan-Hu-1) and VOC-specific pseudovirus (PsV). Both COH04S1- and BNT162b2-vaccinated individuals showed reduced neutralizing activity against Delta and Omicron VOC compared to ancestral SARS-CoV-2. Although a median 50% neutralizing titer (NT50) of 28 was measured against ancestral virus in sera of COH04S1 vaccinated subjects, median NT50 titers measured with Delta and Omicron in COH04S1-vaccinees were below the detection limit (Figures 1C and S1). BNT162b2-vaccinated HCW showed median NT50 titers of 55 and 28 against ancestral and Delta SARS-CoV-2, respectively, while median NT50 titers measured against Omicron in BNT162b2-vaccinated HCW were below the detection limit (Figures 1C and S1). A significant moderately positive correlation between ancestral-specific and VOC-specific NT50 was found (Figure 1D). These results show that ancestral-specific binding antibody and NAb responses in both COH04S1- and BNT162b2-vaccinees were present over six months post-vaccination, whereas VOC-specific NAb responses were low-to-undetectable in most individuals independently of which vaccine was used.
COH04S1- and BNT162b2-vaccinees maintain potent antigen-specific and cross-reactive T cells to SARS-CoV-2 Delta and Omicron variants at six-months post-vaccination
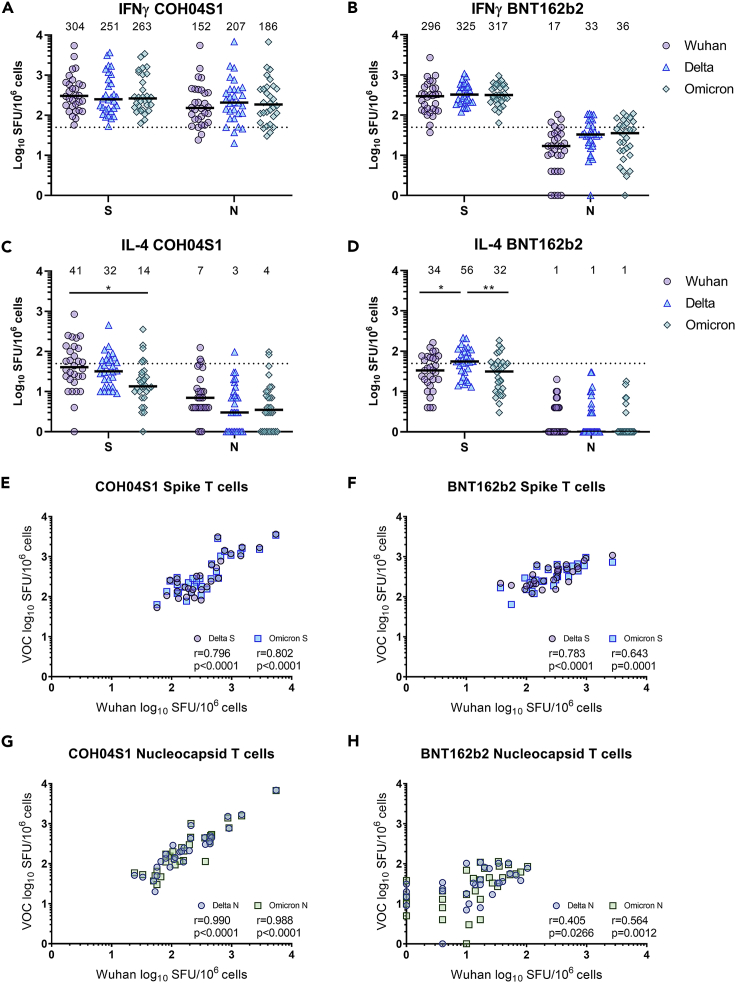
Cellular immune responses to ancestral, Delta, and Omicron S and N antigens were measured by IFNγ/IL-4 ELISPOT using ancestral- and VOC-specific peptide libraries. Robust IFNγ cellular responses to ancestral S were measured in both COH04S1- and BNT162b2-vaccinated subjects, with median levels of 304 or 296 spot forming units (SFU) per 106 cells, respectively (Figures 2A and 2B and S3). Similarly elevated levels of S-specific IFNγ T cell responses were measured against Delta and Omicron S peptides in both vaccine cohorts. COH04S1-vaccinees had median SFU per 106 cells of 251 and 263 against Delta and Omicron S, respectively, while BNT162b2-vaccinated subjects had 325 and 317 median SFU per 106 cells against Delta and Omicron S, respectively. In each cohort, 29 out of 30 (96.7%) volunteers had either an increase or less than a three-fold decrease in the IFNγ cellular response to Delta and Omicron S when compared to the response to ancestral S antigen, indicating preserved S-specific T cell responses across variants (Figure S4). In addition to the elevated S-specific T cell responses, COH04S1-vaccinated subjects showed elevated IFNγ T cell response to ancestral, Delta, and Omicron N antigens. Median SFU per 106 cells in COH04S1-vaccinated subjects were 152, 207, and 186 against ancestral, Delta, and Omicron N, respectively (Figures 2A and S3). All subjects vaccinated with COH04S1 except for one (96.7%) showed a reduction of less than three-fold or an increase of Delta- and Omicron-specific N responses when compared to the ancestral-specific N responses (Figure S4). Consistent with the absence of N in the BNT162b2 vaccine, BNT162b2-vaccinated HCW had low-to-undetectable levels of N-specific T cell responses independently of which peptide library was used (Figures 2B and S3). Median SFU per 106 cells in BNT162b2-vaccinated HCW were 17, 33, and 36 against ancestral, Delta, and Omicron N, respectively. IL-4 cellular responses to ancestral, Delta, and Omicron S in both COH04S1- and BNT162b2-vaccinees were reduced when compared to IFNγ T cell responses and in most cases below the threshold of 50 SFU per 106 cells (Figures 2C, 2D and S5).
Figure 2.
Ancestral- and VOC-specific IFNγ and IL-4 T cell responses in COH04S1-and BNT162b2-vaccinees six months or more after primary vaccination
(A–D). T cell responses. Ancestral- (Wuhan-Hu-1), Delta-, and Omicron-specific spike (S) and nucleocapsid (N) IFNγ (A-B) and IL-4 (C-D) T cell responses were quantified by IFNγ/IL-4 ELISPOT on PBMCs stimulation with S and N peptide libraries in subjects vaccinated with COH04S1 (A, C) or BNT162b2 (B, D) six months or more after primary vaccination. Shown are the IFNγ and IL-4 spot forming units (SFU) measured in 106 PBMCs. two-way ANOVA followed by Tukey’s multiple comparison test was used to compare T cell levels (∗ = p < 0.05, ∗∗ = p < 0.01). Bars represent medians, with median values indicated above each group. Dotted lines represent the arbitrary threshold for a positive response (50 SFU/106 PBMCs).
(C–F). Correlation analysis. Correlation between levels of Wuhan- and VOC-specific T cells recognizing spike (E and F) or nucleocapsid (G and H) in subjects vaccinated with COH04S1 (E, G) or BNT162B2 (F, H) was assessed. Shown in each figure are Pearson’s correlation coefficients (r) and their two-tailed significance (p).
Correlation analysis revealed that COH04S1- and BNT162b2-induced IFNγ T cell responses to ancestral and VOC S antigens were highly correlated (Figures 2E and 2F. 0.643 < r < 0.802, p ≤ 0.0001), while an even stronger correlation was assessed for IFNγ T cell responses to ancestral and VOC N antigens measured following vaccination with COH04S1 (Figure 2G. r > 0.988, p < 0.0001). Although antibody responses to ancestral SARS-CoV-2 showed a marked decline over time independent of the vaccine used, IFNγ T cell responses to ancestral S and N antigens measured six months or more post-vaccination in COH04S1- and BNT162b2-vaccinated subjects were comparable to those measured one month after vaccination (Figure S6), indicating durable S- and N-specific T cell responses. Finally, membrane antigen-specific T cell responses were low-to-undetectable in all subjects, consistent with the absence of intercurrent SARS-CoV-2 infections (Figure S7). These results show that S-specific T cells elicited in COH04S1- and BNT162b2-vaccinees and N-specific T cells elicited in COH04S1-vaccinees maintain potent and equivalent cross-reactivity against ancestral SARS-CoV-2 and Delta and Omicron variants for up to six months post-vaccination.
COH04S1- and BNT162b2-vaccinees maintain potent and equivalent T cells responses to the S1 and S2 domains of SARS-CoV-2 Delta and Omicron variants
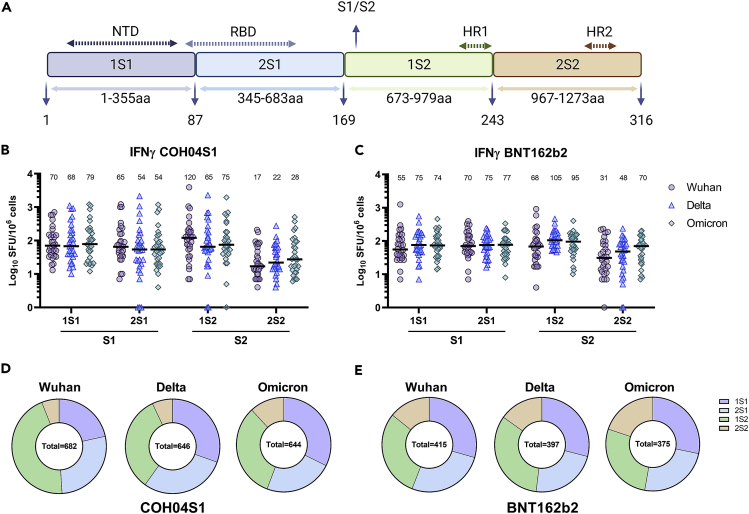
To dissect how differences in ancestral and VOC S sequences affected T cell targeting at the subunit level, we analyzed IFNγ T cell responses to four peptide pools covering N- and C-terminal regions of the S1 (1S1 and 2S1) and S2 domains (2S1 and 2S2) of ancestral, Delta, and Omicron S (Figure 3A). As shown in Figures 3B and 3C, ancestral, Delta, and Omicron S-specific sub-pools were equally recognized in both COH04S1- and BNT162b2-vaccinees. IFNγ responses were evenly distributed between S1 (1S1+2S1 pools) and S2 (1S2+2S2 pools) domains, although COH04S1-vaccinees had more prevalent recognition of the N-terminus of S2 (1S2 pool) than BNT162b2-vaccinees, and in turn, BNT162b2-vaccinees showed better recognition of the C-terminus of S2 (2S2 pool) than COH04S1-vaccinees (Figures 3D and 3E). Interestingly, the 2S1 pools from ancestral, Delta, and Omicron S were evenly recognized in both COH04S1- and BNT162b2-vaccinees, suggesting that the high mutation rate in the RBD does not significantly impact the recognition of T cell epitopes. These results demonstrate that COH04S1 and the FDA-approved BNT162b2 vaccine stimulate robust and equivalent levels of IFNγ T cell responses to the S1 and S2 domains of SARS-CoV-2 ancestral virus and Delta and Omicron variants.
Figure 3.
Subunit-specific IFNγ T cell responses to ancestral and VOC S in COH04S1- and BNT162b2-vaccinees six months or more after primary vaccination
(A). Peptide pools. Shown is the spike (S) peptide library subdivision into four pools of 15mers peptides with 11aa overlaps and their amino acid (aa) composition. 1S1 comprises peptides 1 to 87 (M1-R355); 2S1 comprises peptides 88 to 169 (T345-R683); 1S2 comprises peptides 170 to 243 (S673-D979); 2S2 comprises peptides 244 to 316 (N969-T1273). NTD = N terminal domain, RBD = receptor-binding domain, S1/S2 = S1/S2 cleavage site, HR1 and HR2 = heptad repeat one and 2.
(B and C). Subunit-specific T cells. Ancestral- (Wuhan-Hu-1), Delta-, and Omicron-specific S IFNγ T cell responses were quantified by IFNγ ELISPOT on PBMCs stimulation with S peptide libraries in subjects vaccinated with COH04S1 (B) or BNT162b2 (C) six months or more after the primary vaccination series. Shown are the IFNγ spot forming units (SFU) measured in 106 PBMCs using S-specific sub-libraries 1S1, 2S1, 1S2, and 2S2 as indicated in A. Differences in levels of T cells recognizing Wuhan, Delta, and Omicron S sub-libraries were not significant as evaluated using two-way ANOVA followed by Tukey’s multiple comparison test. Bars represent medians with median values indicated above each group.
(D and E). Percentage distribution of subunit-specific T cells. Mean percentages of Wuhan-, Delta-, Omicron-specific IFNγ T cells reactive to the individual peptide pools in COH04S1- (D) and BNT162b2-vaccinees (E) are shown as pie charts. Sums of mean T cell numbers reactive to each peptide pool are shown inside each pie chart.
Discussion
In this report, we demonstrate that vaccine-induced T cell responses to both S and N antigens preserve potent and long-lasting cross-reactive activity to major SARS-CoV-2 VOC, including the highly mutated Omicron variant. Previous studies of COVID-19 vaccines, including mRNA, adenovirus, and inactivated vaccines, have shown that S-specific T cell responses, unlike NAb responses, remain at elevated levels and confer potent cross-variant reactivity over a prolonged period after vaccination, highlighting the potential of T cells to maintain long-term protective immunity against SARS-CoV-2 and its emerging VOC (Liu et al., 2022; Naranbhai et al., 2022; Tarke et al., 2021, 2022; Zuo et al., 2022). Our findings for COH04S1 and BNT162b2 vaccines extend these prior findings from COVID-19 vaccines and demonstrate that in addition to vaccine-elicited T cell responses to S, vaccine-elicited T cell responses to N are sustained for up to six-to-eight months post-vaccination and confer potent cross-reactivity to major VOC, including Delta and Omicron. These observations are consistent with a recently published study for an inactivated COVID-19 vaccine demonstrating that both S- and N-specific T cells elicited through vaccination maintain potent cross-variant reactivity (Vikkurthi et al., 2022), although Omicron-specific T cell responses induced by the inactivated vaccine were not investigated. These findings support the incorporation of both S and N antigens into a COVID-19 vaccine formulation to elicit durable and cross-reactive T cell responses against continuously emerging SARS-CoV-2 variants.
There is mounting evidence for the important role of T cell immunity to protect against SARS-CoV-2 (Bertoletti et al., 2021; Kingstad-Bakke et al., 2022; McMahan et al., 2021; Moss, 2022; Steiner et al., 2021; Tan et al., 2021). Although the precise contribution of N-specific immune responses to the protection against SARS-CoV-2 is unclear, naturally induced cellular immune responses to N, besides those targeting S, were found to be dominant and durable in patients with COVID-19 (Ferretti et al., 2020; Taus et al., 2022). In addition, vaccine-induced N-specific immune responses have been associated with S-independent protective immunity in animal models (Afkhami et al., 2022; Dangi et al., 2021; Matchett et al., 2021). Recent studies have also shown that N-specific non-NAb promotes antibody-dependent cellular cytotoxicity and other Fc-receptor-mediated functions and can result in the control of SARS-CoV-2 infection (Dangi et al., 2021, 2022; Fielding et al., 2021; Yewdell et al., 2021). These discoveries suggest that the inclusion of N into a COVID-19 vaccine formulation may be important for the induction of T cell responses, and in addition for the stimulation of non-NAb responses. A vaccine combining S and N antigens may utilize an ideal antigen combination to elicit broadly reactive T cell responses as well as broadly functional NAb and non-NAb responses. In addition, our phase 1 clinical study indicates that peak S- and N-specific cellular responses were induced by COH04S1 after the first vaccination (Chiuppesi et al., 2022b), suggesting that a single dose of COH04S1 is sufficient to elicit robust S- and N-specific cellular immunity. A single injection of COH04S1 may elicit or boost S- and N-specific T cell responses in naive or previously infected/vaccinated individuals. Although COH04S1 may have the capacity to broaden vaccine-induced cross-protective immunity through the inclusion of N, inactivated vaccine formulations, such as CoronaVac, Covaxin, or BBIBP-CorV, may have the capacity to stimulate broadly functional immunity that goes beyond the induction of S- and N-specific responses (Deng et al., 2021). Because inactivated vaccines contain a complete set of SARS-CoV-2 antigens they may potentially elicit broadly reactive T cell responses equivalent to those induced during natural infection. Studies directly comparing T cell responses induced by inactivated vaccines or other vaccine approaches are sparse, although a recent study has shown superior induction of S-specific T cells by BNT162b2 compared to BBIBP-CorV or natural infection whereas the response was broader following BBIBP-CorV vaccination or natural infection than in BNT162b2 vaccines (Valyi-Nagy et al., 2021).
In contrast to the robust cross-reactive T cell responses observed in both COH04S1- and BNT162b2-vaccinated subjects, NAb responses observed in these cohorts showed reduced activity against Delta and Omicron VOC when compared to ancestral SARS-CoV-2. NAb responses against the Omicron variant in both COH04S1- and BNT162b2-vaccinated subjects were either very low or undetectable, indicating the exceptional capacity of Omicron to evade NAb. These results are consistent with previous findings for BNT162b2 and other authorized COVID-19 vaccines and confirm that vaccine-induced antibody responses, differently from T cell responses, wane over time and confer reduced neutralizing activity against emerging VOC, suggesting that NAb alone are insufficient to establish long-term protection against COVID-19 (Cheng et al., 2022; Liu et al., 2022; Muik et al., 2022; Rockett et al., 2022). NAb titers induced by COH04S1 were lower than titers induced by BNT162b2. However, it is important to note that COH04S1-vaccinees were in majority (18 out of 30) vaccinated with the lowest COH04S1 dose (DL1), which induced lower NAb titers than the two higher doses (DL2 and DL3) when used as a primary vaccination series in our previously reported phase 1 trial (Chiuppesi et al., 2022b).
NAb are likely the main contributor to the high-level vaccine efficacy against SARS-CoV-2 and its VOC observed shortly after vaccination/booster administration. Although the third dose of BNT162b2 has been shown to significantly improve protection against hospital admission owing to the Delta and Omicron VOC, booster-induced protection rapidly decreased within three months to pre-booster levels of 30-50% (Tartof et al., 2022; Thompson et al., 2022). Cellular responses are thought to contribute to vaccine-induced protection and given their robust maintenance and potent cross-reactivity they may represent an important immune component for the residual 30-50% protection from hospitalization observed in vaccinated individuals several months post-vaccination. Therefore, broadening the coverage of T cell epitopes by using multiantigen vaccines may raise the long-term threshold of protection conferred by T cell immunity beyond the observed 30-50% afforded by vaccines expressing S alone, potentially maintaining protective immunity in the face of waning or reduced cross-reactive neutralizing responses as a result of the continuous emergence of new VOC. Broadly functional vaccine-elicited T cell responses could be particularly important to protect against severe disease in the immunocompromised population, such as transplant recipients, in which mounting of an effective antibody response is impaired owing to B cell aplasia (Jullien et al., 2022). Phase 2 clinical trials are underway testing the use of COH04S1 as a booster vaccine in volunteers previously vaccinated with authorized COVID-19 vaccines (NCT04639466) and COH04S1 as a primary vaccination in patients with hematopoietic cancer following bone marrow transplant or cellular therapy (NCT04977024).
Limitations of the study
This study has several limitations. First, the study did not measure whether the main contributors to the cellular immune responses were CD4+ or CD8+ T cells. However, the analysis of S- and N-specific T cells up to four months post-vaccination with COH04S1 showed that although both CD4+ and CD8+ T cells are present, antigen-specific CD4+ T cells represent the majority of vaccine-induced activated T cells (Chiuppesi et al., 2022b). Similarly, in a study on BNT162b2-vaccinated HCW, the majority IFNγ was produced by CD4+ T cells (Angyal et al., 2022). The preponderance of a CD4+ T cell response is consistent with the T cell phenotype induced after natural SARS-CoV-2 infection (Gao et al., 2022; Tarke et al., 2021). Second, the two cohorts were not precisely balanced in terms of age, sex, and time post-vaccination: the COH04S1 cohort had a more restricted age interval given the limit of 55 years of age imposed by the FDA in the first-in-human phase 1 trial and the BNT162b2 cohort included mostly female healthcare workers. This resulted in a statistically significant difference in age distribution at enrollment between the two cohorts. Finally, while most COH04S1 samples were collected six months post-vaccination, half of BNT162b2 samples were collected at later time points and up to 13 months post-vaccination (Tables S2 and S3).
In conclusion, our study demonstrates that vaccine-induced T cell responses to S and N antigens maintain potent cross-reactive immunity to SARS-CoV-2 and its emerging VOC and warrants the inclusion of N in a vaccine formulation to potentially broaden the reactivity and protective efficacy of vaccine-induced T cell responses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GOAT ANTI HUMAN IgG:HRP antibody | BioRad | Cat# 204,005, RRID:AB_619881 |
| Biological samples | ||
| WHO International reference panel for anti-SARS-CoV-2 immunoglobulin | NIBSC | 20/268 |
| COH04S1 immune serum and PBMCs | This paper | N/A |
| BNT162b2 immune serum and PBMCs | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Spike (S1+S2) | Sino Biological | 40589-V08B1 |
| RBD | Sino Biological | 40592-V08H |
| Nucleocapsid | Sino Biological | 40588-V08B |
| 1 Step TMB-Ultra | Thermo Fisher | 34029 |
| Luciferase Assay Reagent | Promega | E1483 |
| TransIT-Lenti | Mirus | MIR6600 |
| Wuhan Spike and Nucleocapsid peptide library | Genscript | Custom order (Table S4) |
| Delta Spike and Nucleocapsid peptide library | Genscript | Custom order (Table S4) |
| Omicron Spike and Nucleocapsid peptide library | Genscript | Custom order (Table S4) |
| Wuhan Membrane peptide library | Produced in house | N/A |
| Critical commercial assays | ||
| Lenti-XTM p24 Rapid Titer Kit | Takara | 632200 |
| IFNγ/IL-4 FluoroSpot FLEX kit | Mabtech | X-01A16B |
| Experimental models: Cell lines | ||
| HEK293T/17 cells | ATCC | CRL11268 |
| HEK293T-ACE2 cells | Crawford et al., 2020 | N/A |
| Recombinant DNA | ||
| pALD-gag-pol | Aldevron | pALD-gag-pol |
| pALD-rev | Aldevron | pALD-rev |
| pALD-GFP | Aldevron | pALD-GFP |
| pALD-FLuc | This paper | N/A |
| pCMV3-S | Sino Biological | VG40589-UT |
| pTwist-CMV-BetaGlobin-S Delta | Twist Biosciences | Custom order |
| pTwist-CMV-BetaGlobin-S Omicron | Twist Biosciences | Custom order |
| Software and algorithms | ||
| Prism | GraphPad | v8.3.0 |
| SoftMax Pro 7 | Molecular Devices | SMP7 PROF |
| Fluoro-X FluoroSpot | Immunospot | N/A |
| Other | ||
| FilterMax F3 | Molecular Devices | F3 |
| SpectraMax L | Molecular Devices | SpectraMax L |
| Luna-FL cell counter | Logos Biosystems | L20001 |
| CTL-test serum free media | Immunospot | CTLT-010 |
| CTL S6 Fluorocore | Immunospot | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Don J. Diamond (ddiamond@coh.org).
Materials availability
All requests for resources and reagents should be directed to the lead contact author. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Human subjects
COH04S1 immunogenicity was investigated at City of Hope (COH) as part of a clinical protocol (IRB#20447) approved by an external Institutional Review Board (Advarra IRB). This open-label and randomized, placebo controlled, phase 1 clinical study is registered (NCT04639466). Among others, exclusion criteria included age<18 or >55, previous SARS-CoV-2 infection, BMI<18 or >35, and underlying health conditions. Out of the 51 subjects who received one or two doses of COH04S1, 30 subjects were selected based on 2 doses regimen, absence of SARS-CoV-2 intercurrent infection and lack of additional vaccination with EUA SARS-CoV-2 vaccines. Immunogenicity of BNT162b2 was investigated as part of an observational study conducted at COH and approved by City of Hope IRB (IRB#20720). Participants were ≥18 years old and no exclusion criteria based on BMI or underlying health conditions were applied. This study first enrolled 19 subjects who were followed up to 1-year post-vaccination and additional 34 subjects with a single sample six months or more post primary series vaccination. Out of 53 subjects who were enrolled, 30 subjects were selected based on the availability of frozen PBMC samples and absence of N-specific binding antibodies or M-specific IFNγ T cell responses. All subjects gave informed consent at enrollment. Study populations are described on Tables S1–S3.
Method details
IgG ELISA
SARS-CoV-2-specific binding antibodies were detected by endpoint and quantitative ELISAs utilizing purified S, RBD, and N proteins (Sino Biological 40589-V08B1, 40592-V08H, 40588-V08B). Briefly, 96-well plates (Costar 3361) were coated with 100 μL/well of S, RBD, or N proteins at a concentration of 1 μg/mL in PBS pH 7.4 and incubated overnight at 4°C. Plates were washed 5X with wash buffer (0.1% Tween-20/PBS), then blocked with 250 μl/well of assay buffer (0.5% casein/154mM NaCl/10mM Tris-HCl/0.1% Tween-20 [pH 7.6]/8% Normal goat serum) for 2 h 37°C. After washing, 3-fold diluted heat-inactivated serum in blocking buffer was added to the plates starting from a dilution of 1:150. Plates were wrapped in foil and incubated 2 h at 37°C. Plates were washed and 1:3,000 dilution of anti-human IgG HRP secondary antibody (BioRad, 204005) in assay buffer was added for 1 h at room temperature. Plates were washed and developed with 1 Step TMB-Ultra (Thermo Fisher 34029). After 2-4 min the reaction was stopped with 1M H2SO4 and 450nm absorbance was immediately quantified on FilterMax F3 (Molecular Devices). Endpoint titers were calculated as the highest dilution to have an absorbance >0.100 nm. Positive and negative controls were included in each plate and consisted of serum pools of SARS-CoV-2 seropositive (S, RBD, and N endpoint titer 36,450) and seronegative individuals (S, RBD, and N endpoint titer <150). For quantitative ELISA the same protocol was used with the difference that in each plate each member of the WHO International reference panel for anti-SARS-CoV-2 immunoglobulin (UK National Institute for Biological Standards and Control code, 20/268) was added diluted 1:1,350 and the WHO assigned values used to create a standard curve. Samples were six-fold diluted in duplicates starting from a 1:150 dilution. For each sample, the first absorbance value to fall within the standard curve range was used to calculate the IgG titer and expressed as BAU/ml. Samples with IgG<1 BAU/ml were indicated as 1 BAU/ml.
SARS-CoV-2 PsV production and neutralization assay
SARS-CoV-2 PsV was produced using a plasmid lentiviral system based on pALD-gag-pol, pALD-rev, and pALD-GFP (Aldevron). Plasmid pALD-GFP was modified to express Firefly luciferase (pALD-Fluc). Plasmid pCMV3-S (Sino Biological VG40589-UT) was utilized and modified to express SARS-CoV-2 Wuhan-Hu-1 S with D614G modification. Customized gene sequences cloned into pTwist-CMV-BetaGlobin (Twist Biosciences) were used to express SARS-CoV-2 VOC-specific S variants. All S antigens were expressed with C-terminal 19 aa deletion. The S sequence of the Delta (B.1.617.2)-specific PsV contained the following mutations: T19R, G142D, Δ156-157, R158G, A222V, L452R, T478K, D614G, P681R, D950N. The S sequence of the BA.1 Omicron-specific PsV contained the following mutations: A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, Ins214(EPE), G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F. A transfection mixture was prepared 1 ml OptiMEM that contained 30 μl of TransIT-Lenti transfection reagent (Mirus MIR6600) and 6 μg pALD-Fluc, 6 μg pALD-gag-pol, 2.4 μg pALD-rev, and 6.6 μg S expression plasmid. The transfection mix was added to 5 × 106 HEK293T/17 cells (ATCC CRL11268) seeded the day before in 10 cm dishes and the cells were incubated for 72 h at 37°C. Supernatant containing the PsV was harvested and frozen in aliquots at 80°C. Lentivirus was titrated using the Lenti-XTM p24 Rapid Titer Kit (Takara) according to the manufacturer’s instructions.
SARS-CoV-2 PsV were titrated in vitro to calculate the virus stock amount that equals 200,000- 500,000 relative luciferase units. Flat-bottom 96-well plates were coated with 100 μl poly-L-lysine (0.01%). Serial 2-fold serum dilutions starting from 1:20 were prepared in 50 μl media and added to the plates in triplicates, followed by 50 μl of PsV. Plates were incubated overnight at 4°C. The following day, 10,000 HEK293T-ACE2 cells (Crawford et al., 2020) were added to each well in the presence of 3 μg/ml polybrene and plates were incubated at 37°C. After 48h of incubation, luciferase lysis buffer (Promega E1531) was added and luminescence was quantified using SpectraMax L (Molecular Devices) after adding Luciferase Assay Reagent (Promega E1483, 100 μl/well). For each plate, positive (PsV only) and negative (cells only) controls were added. The neutralization titer for each dilution was calculated as follows: NT = [1−(mean luminescence with immune sera/mean luminescence without immune sera)] × 100. The titers that gave 50% neutralization (NT50) were calculated by determining the linear slope of the graph plotting NT versus serum dilution by using the next higher and lower NT using Office Excel (v2019).
IFNγ/IL-4 ELISPOT
Peripheral blood mononuclear cells (PBMC) were isolated from fresh blood using Ficoll and counted using Luna-FL cell counter (Logos Biosystems). Frozen PBMCs were thawed and IFNγ/IL-4 secretion evaluated using human IFNγ/IL-4 FluoroSpot FLEX kit (Mabtech, X-01A16B) following manufacturer instructions. Briefly, 150,000 cells/well in CTL-test serum free media (Immunospot CTLT-010) were added to duplicate wells and stimulated with peptide pools (15-mers, 11 aa overlap, >70% purity). Ancestral, Delta (B.1.617.2) and Omicron (BA.1) S peptide libraries (GenScript) were divided into 4 sub-pools spanning the S1 and S2 domains (Table S4. 1S1=1-86; 1S287-168; 2S1 = 169-242; 2S2 = 243-316; peptides 173 and 304-309 were not successfully synthesized therefore excluded from the pools). Ancestral, Delta and Omicron N peptide libraries consisted of 102 peptides (Table S4. GenScript). Membrane (in house synthesized) library based on the Wuhan sequence included 53 peptides. Each peptide pool (2 μg/mL) and αCD28 (0.1 μg/ml, Mabtech) were added to the cells and plates were incubated for 48 h at 37°C. Control cells (50,000/well) were stimulated with PHA (10 μg/mL). After incubation, plates were washed with PBS and primary and secondary antibodies were added according to manufacturer’s protocol. Fluorescent spots forming units (SFU) were acquired using CTL S6 Fluorocore (Immunospot). For each sample, spots in unstimulated DMSO-only control wells were subtracted from spots in stimulated wells. For plotting zero spots were indicated as one. Total spike response was calculated as the sum of the response to each spike sub-pool. Fifty spots/106 cells were chosen as an arbitrary threshold for positivity.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 8.3.0. Groups were compared using 2-way ANOVA followed by Tukey’s multiple comparison test. Prism was used to calculate Pearson correlation coefficients and their p values.
Additional resources
COH04S1 phase I clinical trial has been registered on ClinicalTrials.gov (Identifier: NCT04639466, URL: https://clinicaltrials.gov/ct2/show/NCT04639466): A Synthetic MVA-based SARS-CoV-2 Vaccine, COH04S1, for the Prevention of COVID-19 Infection.
Acknowledgments
The authors would like to thank all the participants who volunteered in the study and all the investigators and study site personnel who assisted in the clinical trial completion. Funding was provided by the Carol Moss Foundation, donors Julie and Roger Baskes, Judd Malkin, Michael Sweig, and the City of Hope Integrated Drug Development Venture program. We acknowledge and thank Christoph Pittius and Yuriy Shostak (Research Business Development, City of Hope) for excellent project management. We thank Duygu Ercan Laguna for her help with sample processing and immunological analysis. We thank Karen Gutierrez and Christina Ulloa (Department of Hematology & HCT, City of Hope) for the excellent support of investigators and meeting coordination. Figure 3A and the Graphical Abstract were created with BioRender.com.
Author contributions
Study conceptualization: FC, FW, SD, DJD. Study design: FC, KF. Immunological analysis: KF, DJ, VK, ML, TK. Sample processing: QZ, YP, SOF, KF, VK. Clinical PIs: JAZ, RAT, SD, SR. Article writing: FC, FW, SD, JAZ, DJD. Statistical analysis: PHF. Study data collection and participant’s oversight: CLR, JD, JM, AMA, SD All authors contributed to and approved the final version of this article.
Declaration of interests
While unknown whether the publication of this report will aid in receiving grants and contracts, it is possible that this publication will be of benefit to the City of Hope (COH). COH had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the article. DJD and FW are co-inventors on a patent application covering the design and construction of the synthetic MVA platform (PCT/US2021/016,247). DJD, FW, and FC are co-inventors on a patent application covering the development of a COVID-19 vaccine (PCT/US2021/032,821). DJD is a consultant for GeoVax. All other authors declare no competing interests. GeoVax Labs Inc. has taken a worldwide exclusive license for COH04S1 under the name of GEO-CM04S1.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104745.
Supplemental information
Data and code availability
All data supporting the findings of this study are available within the paper and are available from the lead contact upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Accorsi E.K., Britton A., Shang N., Fleming-Dutra K.E., Link-Gelles R., Smith Z.R., Derado G., Miller J., Schrag S.J., Verani J.R. Effectiveness of homologous and heterologous covid-19 boosters against omicron. N. Engl. J. Med. 2022;386:2433–2435. doi: 10.1056/NEJMc2203165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afkhami S., D'Agostino M.R., Zhang A., Stacey H.D., Marzok A., Kang A., Singh R., Bavananthasivam J., Ye G., Luo X., et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell. 2022;185:896–915.e19. doi: 10.1016/j.cell.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angyal A., Longet S., Moore S.C., Payne R.P., Harding A., Tipton T., Rongkard P., Ali M., Hering L.M., Meardon N., et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2-naive UK health-care workers: a multicentre prospective cohort study. Lancet. Microbe. 2022;3:e21–e31. doi: 10.1016/S2666-5247(21)00275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A., Le Bert N., Qui M., Tan A.T. SARS-CoV-2-specific T cells in infection and vaccination. Cell. Mol. Immunol. 2021;18:2307–2312. doi: 10.1038/s41423-021-00743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., Ng S.S., Chan K.C.K., Ko F.W., Chen C., Yiu K., Lam B.H.S., Lau E.H.Y., et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022;28:486–489. doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F., Nguyen V.H., Park Y., Contreras H., Karpinski V., Faircloth K., Nguyen J., Kha M., Johnson D., Martinez J., et al. Synthetic multiantigen MVA vaccine COH04S1 protects against SARS-CoV-2 in Syrian hamsters and non-human primates. NPJ Vaccines. 2022;7:7. doi: 10.1038/s41541-022-00436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F., Salazar M.D., Contreras H., Nguyen V.H., Martinez J., Park Y., Nguyen J., Kha M., Iniguez A., Zhou Q., et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020;11:6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F., Zaia J.A., Frankel P.H., Stan R., Drake J., Williams B., Acosta A.M., Francis K., Taplitz R.A., Dickter J.K., et al. Safety and immunogenicity of a synthetic multiantigen modified vaccinia virus Ankara-based COVID-19 vaccine (COH04S1): an open-label and randomised, phase 1 trial. Lancet. Microbe. 2022;3:e252–e264. doi: 10.1016/S2666-5247(22)00027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12 doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi T., Class J., Palacio N., Richner J.M., Penaloza MacMaster P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 2021;36:109664. doi: 10.1016/j.celrep.2021.109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi T., Sanchez S., Park M., Class J., Richner M., Richner J.M., Penaloza-MacMaster P. Nucleocapsid-specific humoral responses improve the control of SARS-CoV-2. bioRxiv. 2022 doi: 10.1101/2022.03.09.483635. Preprint at. [DOI] [Google Scholar]
- Deng Y., Li Y., Yang R., Tan W. SARS-CoV-2-specific T cell immunity to structural proteins in inactivated COVID-19 vaccine recipients. Cell. Mol. Immunol. 2021;18:2040–2041. doi: 10.1038/s41423-021-00730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding C., Sabberwal P., Williamson J., Greenwood E., Crozier T., Zelek W., Seow J., Graham C., Huettner I., Edgeworth J., et al. ADNKA overcomes SARS-CoV2-mediated NK cell inhibition through non-spike antibodies. bioRxiv. 2021 doi: 10.1101/2021.04.06.438630. Preprint at. [DOI] [Google Scholar]
- Gao Y., Cai C., Grifoni A., Müller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Österborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayawi K., Shahriar S., Serhani M.A., Alashwal H., Masud M.M. Vaccine versus variants (3Vs): are the COVID-19 vaccines effective against the variants? A systematic review. Vaccines. 2021;9:1305. doi: 10.3390/vaccines9111305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien M., Le Bourgeois A., Coste-Burel M., Peterlin P., Garnier A., Rimbert M., Imbert B.M., Le Gouill S., Moreau P., Mahe B., et al. B cell aplasia is the most powerful predictive marker for poor humoral response after BNT162b2 mRNA SARS-CoV-2 vaccination in recipients of allogeneic hematopoietic stem cell transplantation. Transplant. Cell. Ther. 2022;28:279.e1–279.e4. doi: 10.1016/j.jtct.2022.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingstad-Bakke B., Lee W., Chandrasekar S.S., Gasper D.J., Salas-Quinchucua C., Cleven T., Sullivan J.A., Talaat A., Osorio J.E., Suresh M. Vaccine-induced systemic and mucosal T cell immunity to SARS-CoV-2 viral variants. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2118312119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matchett W.E., Joag V., Stolley J.M., Shepherd F.K., Quarnstrom C.F., Mickelson C.K., Wijeyesinghe S., Soerens A.G., Becker S., Thiede J.M., et al. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J. Immunol. 2021;207:376–379. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- Muik A., Lui B.G., Wallisch A.K., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., Güimil Garcia R.d.l.C., Poran A., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranbhai V., Nathan A., Kaseke C., Berrios C., Khatri A., Choi S., Getz M.A., Tano-Menka R., Ofoman O., Gayton A., et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041–1051.e6. doi: 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett R., Basile K., Maddocks S., Fong W., Agius J.E., Johnson-Mackinnon J., Arnott A., Chandra S., Gall M., Draper J., et al. Resistance mutations in SARS-CoV-2 delta variant after sotrovimab use. N. Engl. J. Med. 2022;386:1477–1479. doi: 10.1056/NEJMc2120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S., Schwarz T., Corman V.M., Sotzny F., Bauer S., Drosten C., Volk H.D., Scheibenbogen C., Hanitsch L.G. Reactive T cells in convalescent COVID-19 patients with negative SARS-CoV-2 antibody serology. Front. Immunol. 2021;12:687449. doi: 10.3389/fimmu.2021.687449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof S.Y., Slezak J.M., Puzniak L., Hong V., Xie F., Ackerson B.K., Valluri S.R., Jodar L., McLaughlin J.M. Durability of BNT162b2 vaccine against hospital and emergency department admissions due to the omicron and delta variants in a large health system in the USA: a test-negative case-control study. Lancet Respir. Med. 2022;10:689–699. doi: 10.1016/S2213-2600(22)00101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus E., Hofmann C., Ibarrondo F.J., Hausner M.A., Fulcher J.A., Krogstad P., Ferbas K.G., Tobin N.H., Rimoin A.W., Aldrovandi G.M., Yang O.O. Dominant CD8(+) T cell nucleocapsid targeting in SARS-CoV-2 infection and broad spike targeting from vaccination. Front. Immunol. 2022;13:835830. doi: 10.3389/fimmu.2022.835830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.G., Natarajan K., Irving S.A., Rowley E.A., Griggs E.P., Gaglani M., Klein N.P., Grannis S.J., DeSilva M.B., Stenehjem E., et al. Effectiveness of a third dose of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of delta and omicron variant predominance - VISION network, 10 states, august 2021-January 2022. Morb. Mortal. Wkly. Rep. 2022;71:139–145. doi: 10.15585/mmwr.mm7104e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vályi-Nagy I., Matula Z., Gönczi M., Tasnády S., Bekő G., Réti M., Ajzner É., Uher F. Comparison of antibody and T cell responses elicited by BBIBP-CorV (Sinopharm) and BNT162b2 (Pfizer-BioNTech) vaccines against SARS-CoV-2 in healthy adult humans. Geroscience. 2021;43:2321–2331. doi: 10.1007/s11357-021-00471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikkurthi R., Ansari A., Pai A.R., Jha S.N., Sachan S., Pandit S., Nikam B., Kalia A., Jit B.P., Parray H.A., et al. Inactivated whole-virion vaccine BBV152/Covaxin elicits robust cellular immune memory to SARS-CoV-2 and variants of concern. Nat. Microbiol. 2022;7:974–985. doi: 10.1038/s41564-022-01161-5. [DOI] [PubMed] [Google Scholar]
- López-Muñoz A.D., Kosik I., Holly J., Yewdell J.W. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. bioRxiv. 2021 doi: 10.1101/2F2021.12.10.472169. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wussow Felix, et al. COH04S1 and beta sequence-modified vaccine protect hamsters from SARS-CoV-2 variants. iScience. 2022;25(6) doi: 10.1016/j.isci.2022.104457. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Tada T., Dcosta B.M., Landau N.R. Neutralization of SARS-CoV-2 Omicron BA.2 by therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1016/S1473-3099(22)00365-6. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo F., Abolhassani H., Du L., Piralla A., Bertoglio F., de Campos-Mata L., Wan H., Schubert M., Cassaniti I., Wang Y., et al. Heterologous immunization with inactivated vaccine followed by mRNA-booster elicits strong immunity against SARS-CoV-2 Omicron variant. Nat. Commun. 2022;13:2670. doi: 10.1038/s41467-022-30340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the lead contact upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.