Abstract
The marine oligotrophic ultramicrobacterium Sphingomonas alaskensis RB2256 has a physiology that is distinctly different from that of typical copiotrophic marine bacteria, such as Vibrio angustum S14. This includes a high level of inherent stress resistance and the absence of starvation-induced stress resistance to hydrogen peroxide. In addition to periods of starvation in the ocean, slow, nutrient-limited growth is likely to be encountered by oligotrophic bacteria for substantial periods of time. In this study we examined the effects of growth rate on the resistance of S. alaskensis RB2256 to hydrogen peroxide under carbon or nitrogen limitation conditions in nutrient-limited chemostats. Glucose-limited cultures of S. alaskensis RB2256 at a specific growth rate of 0.02 to 0.13 h−1 exhibited 10,000-fold-greater viability following 60 min of exposure to 25 mM hydrogen peroxide than cells growing at a rate of 0.14 h−1 or higher. Growth rate control of stress resistance was found to be specific to carbon and energy limitation in this organism. In contrast, V. angustum S14 did not exhibit growth rate-dependent stress resistance. The dramatic switch in stress resistance that was observed under carbon and energy limitation conditions has not been described previously in bacteria and thus may be a characteristic of the oligotrophic ultramicrobacterium. Catalase activity varied marginally and did not correlate with the growth rate, indicating that hydrogen peroxide breakdown was not the primary mechanism of resistance. More than 1,000 spots were resolved on silver-stained protein gels for cultures growing at rates of 0.026, 0.076, and 0.18 h−1. Twelve protein spots had intensities that varied by more than twofold between growth rates and hence are likely to be important for growth rate-dependent stress resistance. These studies demonstrated the crucial role that nutrient limitation plays in the physiology of S. alaskensis RB2256, especially under oxidative stress conditions.
Ultramicrobacteria are major contributors to the world's biosphere in terms of biological cycling of carbon, nitrogen, and phosphorus (50). As reservoirs of nutrients in oligotrophic marine ecosystems, they interact with all trophic levels and control nutrient fluxes via mineralization, thus having an impact on the productivity of all marine life from microbial primary producers and plankton to whales. Because of predictions of increasing ocean oligotrophy as a consequence of global warming (32, 59), it is clearly important to understand the physiology of this class of bacteria in order to determine the impact that they have on life on earth.
The growth of virtually all microbial cells in nature is limited by the availability of one or more essential growth nutrients (22, 37, 50), and in many regions of the ocean carbon is the primary limiting substrate (1, 4, 5, 27). In the oligotrophic marine environment, bacteria generally adopt one of two different survival strategies; they are either copiotrophic organisms which form resting stage cells with spasmodic bursts of growth (e.g., Vibrio angustum S14) or oligotrophic organisms which grow slowly with intermittent periods of starvation or faster growth (e.g., Sphingomonas sp. strain RB2256) (10).
Despite our relatively good understanding of the physiology and genetics of marine copiotrophic bacteria, oligotrophic bacteria and the roles that they play in environmental processes are poorly understood. Knowledge about the physiology of oligotrophs is limited by the availability of environmental isolates. To date, most insight into the physiology of this class of marine bacteria has been obtained from Sphingomonas sp. strain RB2256 (8, 10, 11, 48, 49), which was isolated as a numerically dominant bacterium from Resurrection Bay, Alaska (3, 47). This strain has been formally described as Sphingomonas alaskensis RB2256 (57).
One of the characteristics that distinguish S. alaskensis RB2256 from V. angustum S14 is its high level of resistance to a variety of stress-inducing agents, including hydrogen peroxide (8). The ability to resist the damaging effects of hydrogen peroxide is an ecologically relevant characteristic because endogenous and exogenous oxidative stress is a common challenge for microorganisms in aquatic environments (6, 17, 42, 50). Reactive oxygen species, such as hydrogen peroxide, damage DNA, RNA, proteins, and lipids, and as a consequence, cells have evolved a broad range of mechanisms to cope with this type of stress (reviewed in reference 52).
Previous studies showed that S. alaskensis RB2256 grown in glucose-limited chemostats at a dilution rate of 0.027 h−1 was more resistant to hydrogen peroxide than logarithmic-phase or starved cells from batch cultures were (8). In view of the fact that slow, nutrient-limited growth is likely to be the type of growth most often exhibited by oligotrophic bacteria, we reasoned that the high degree of resistance observed with chemostat-grown cells may be triggered by nutrient-limited growth and that the precise level of resistance is controlled by the actual specific rate of growth under these conditions.
In order to determine the types of mechanisms and regulatory processes that S. alaskensis RB2256 has evolved to cope with hydrogen peroxide stress, in this study we examined the physiological and molecular responses of cells grown in nutrient-limited chemostats at different growth rates. Growth in chemostats permitted continued growth at a fixed rate, while it also permitted the use of different limiting nutrients (e.g., carbon or nitrogen).
While the growth rate still has been poorly studied, there is evidence that growth rate affects the physiology of Escherichia coli and Vibrio spp. The cell size, cellular composition, and starvation survival of Vibrio sp. strain ANT-300 are affected by growth rate (37, 38), and slow growth induces rpoS-dependent gene expression in E. coli (13, 39). In this study we extended assessment of the growth rate control of hydrogen peroxide resistance to include a marine copiotrophic bacterium (V. angustum S14). We found that growth rate played a critical role in the hydrogen peroxide resistance of S. alaskensis RB2256 and that slowly growing cells were more resistant than fast-growing cells. In contrast, in V. angustum nutrient availability caused an all-or-none type of response, and starved cells were more resistant than growing cells, regardless of the growth rate. These results indicate that the extent of nutrient limitation has fundamentally different consequences for the physiology of oligotrophic ultramicrobacteria and the physiology of copiotrophic bacteria.
MATERIALS AND METHODS
Bacteria, media, and culture conditions.
The bacterial strains used were the oligotrophic marine ultramicrobacterium strain S. alaskensis RB2256 (46, 47, 57) and V. angustum S14 (31). For cultivation in batch cultures, S. alaskensis RB2256 and V. angustum S14 were grown in an artificial seawater medium (ASW) (8) supplemented with 3 mM d-glucose. For chemostat cultivation, ASW was used with supplements. Glucose-limited feed medium contained d-glucose at a concentration of 3 mM. For ammonium-limited feed medium the concentration of NH4Cl was 94 mM and 5 mM d-glucose was added. For mixed-amino-acid-limited feed medium the concentration of Casamino Acids was 0.5% (wt/vol) and glucose was omitted. The pH values of the media used for batch and chemostat cultures were maintained between 7.5 and 7.8 by addition of morpholinepropanesulfonic acid (MOPS) buffer (1.0 g liter−1) or by continuous automatic adjustment with sterile NaOH (0.25 M). The pH of each medium was adjusted to 7.8 prior to autoclaving. Batch cultures were grown at 30°C with orbital shaking at 120 rpm. Chemostat cultivation was carried out in glass culture vessels (working volume, 450 ml) magnetically stirred at 400 rpm and maintained at a constant temperature of 30°C. Small-scale chemostat cultivation was carried out in 100-ml glass culture vessels constructed from Erlenmyer flasks, which had small headspace volumes, were equipped with two stainless steel baffles, and were stirred magnetically at 400 rpm. The temperature was maintained by a continuous flow of water through a water jacket from a temperature-regulated water bath. Medium entered each vessel through a stainless steel needle (diameter, 0.5 mm) at the bottom of the vessel. Filter-sterilized, prehumidified air was supplied through the same needle. The specific growth rates imposed on chemostat cultures were alternated between high and low values, and hydrogen peroxide resistance was monitored as described below. In chemostat cultures, a steady state was assumed after growth for five to seven generations. The growth of batch and chemostat cultures was monitored by measuring the optical density at 433 nm.
Viability measurements and hydrogen peroxide exposure.
Viable counts of S. alaskensis RB2256 and V. angustum S14 were determined by determining the number of CFU on marine nutrient agar (Bacto Marine Agar 2216) and on ASW-glucose solid medium consisting of ASW, 3 mM d-glucose, and 1.5% agar. A dilution series was prepared with ASW buffered with MOPS (1.0 g liter−1; pH 7.8). Colonies on drop plates (20) were counted with a binocular microscope (magnification, ×25) after 3 and 6 days of incubation at 30°C for S. alaskensis RB2256 and after overnight incubation at 30°C for V. angustum S14. At least five spots from duplicate plates were counted for each experiment. Experiments were performed at least twice. The survival fraction for hydrogen peroxide-treated samples was calculated for each sample separately as a percentage of the survival in untreated control samples. Cells were withdrawn from mid-exponential-phase batch cultures (optical density at 433 nm, 0.4) or steady-state chemostat cultures and exposed to hydrogen peroxide (2 to 75 mM; freshly prepared from a 30% commercial stock solution) at 30°C for up to 60 min. S. alaskensis RB2256 is inherently more resistant to hydrogen peroxide than V. angustum S14. To obtain equivalent levels of survival after 60 min, 25 and 2 mM hydrogen peroxide were used for S. alaskensis RB2256 and V. angustum S14, respectively.
Determination of catalase activity.
Catalase activity was determined with a Clark oxygen electrode by the method of Rørth and Jensen (45). The concentration of oxygen in oxygen-saturated deionized water at 25°C was assumed to be 253 μM (60). The amount of enzyme activity that decomposed 1 μmol of H2O2 to 0.5 μmol of O2 min−1 at 25°C was defined as 1 U of activity. Total catalase activity was determined by subtracting background respiration and oxygen production due to spontaneous decomposition of H2O2. The background activities were less than 10% of the measured oxygen production values. Catalase activities were determined from initial reaction rates from linear regression lines calculated over the first minute. Comparisons of the catalase activities of whole bacteria were made after the addition of H2O2 (final concentration, 1 mM) to 2.0 ml of culture. Enzyme activity was calculated from the initial reaction rate for at least five replicates from at least two independently prepared samples. Protein concentrations were determined by using a bicinchoninic acid kit (Sigma) with bovine serum albumin as the standard.
Two-dimensional polyacrylamide gel electrophoresis.
Triplicate gels of total protein were prepared from at least two independent steady-state cultures of S. alaskensis RB2256 growing at rates of 0.026, 0.076, and 0.18 h−1. Sample preparation, two-dimensional polyacrylamide gel electrophoresis, silver staining, image acquisition, and analysis were carried out as previously described (12).
RESULTS
Effect of growth rate on hydrogen peroxide resistance in S. alaskensis RB2256.
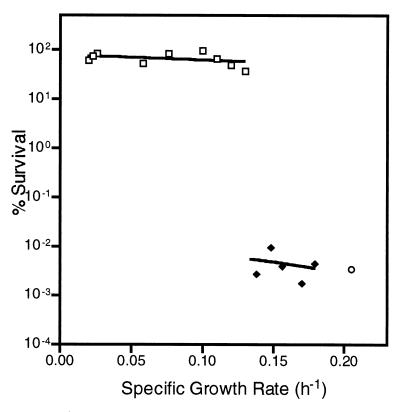
To determine whether growth rate affected the stress resistance of S. alaskensis RB2256, cells were grown at different dilution rates in a glucose-limited chemostat at 30°C, samples were removed, and survival was monitored after exposure to 25 mM hydrogen peroxide for 60 min. The specific growth rates used ranged from 0.02 to 0.18 h−1, which corresponded to approximately 10 to 90% of the maximum specific growth rate (0.21 h−1). In addition, resistance was determined for cells grown at the maximum rate in a batch culture.
Two distinct levels of resistance to hydrogen peroxide stress were observed (Fig. 1). When cultures were grown at a specific growth rate of 0.14 h−1 or higher, the level of stress resistance was equivalent to that of batch-grown cells. The level of resistance was the same for cultures grown at five different rates of growth between 0.14 and 0.18 h−1. Between growth rates of 0.13 and 0.02 h−1, S. alaskensis RB2256 cultures were 10,000 times more resistant to hydrogen peroxide stress than faster-growing cells were, maintaining a level of viability of more than 35% for at least 60 min of exposure to hydrogen peroxide. Results of a series of repeat experiments illustrated the fact that the change from high to low hydrogen peroxide resistance occurred over an extremely narrow range of specific growth rates between 0.13 and 0.14 h−1.
FIG. 1.
Percentages of survival of S. alaskensis RB2256 cells grown at different specific growth rates in glucose-limited chemostats after exposure to 25 mM hydrogen peroxide for 60 min. For the two trendlines the data were grouped. Symbols: □, low specific growth rates (0.020, 0.023, 0.026, 0.058, 0.076, 0.095, 0.11, 0.12, and 0.13 h−1); ⧫, high specific growth rates (0.14, 0.15, 0.16, 0.17, and 0.18 h−1). The level of survival of logarithmic-phase cells in batch cultures (○) is also shown. Duplicate samples were taken from chemostats at every specific growth rate that was tested. Six individual chemostat runs were tested, and at least one sample was taken from a high specific growth rate and a low specific growth rate. The numbers of CFU were determined by using the drop plate method with marine nutrient agar and ASW-glucose solid medium. There was no difference in survival between ASW-glucose solid medium and marine nutrient agar for any time point. The standard deviation for each time point was less than 15%.
For every experiment that was performed at a low dilution rate, the chemostat was switched to a high dilution rate and stress resistance was monitored after a new steady state was obtained (and vice versa). The results demonstrated that the history of growth of the cultures had no bearing on the observed levels of stress resistance. Furthermore, they showed that if mutants arose in the chemostat, since it is unlikely that the same mutant would arise under both regimes (after the growth rate was shifted up and after the growth rated was shifted down), mutants could not account for the observed growth rate dependence of stress resistance.
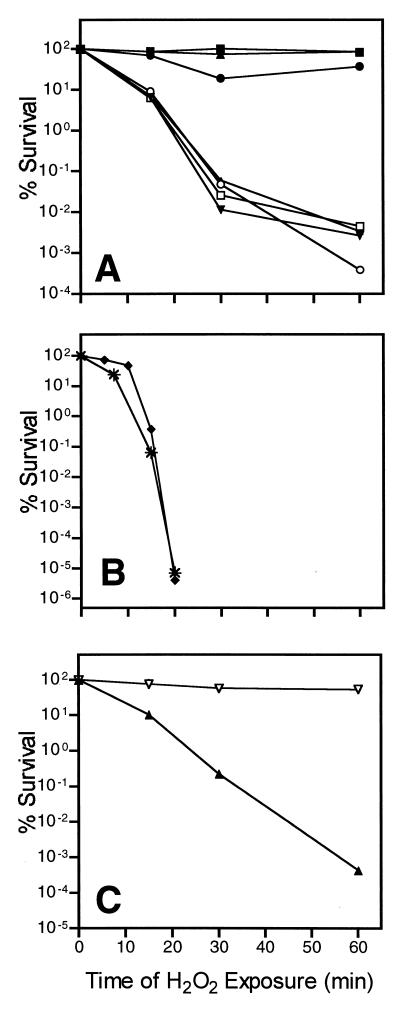
To determine whether the kinetics of survival for cultures with high and low rates of growth were similar, survival in 25 mM hydrogen peroxide was examined throughout a 60-min time course for glucose-limited chemostat cultures grown at a range of specific growth rates from 0.026 to 0.18 h−1 and for a batch culture grown at a rate of 0.21 h−1 (Fig. 2A). The results are consistent with a bimodal response to hydrogen peroxide stress, resulting in two distinct physiological states of S. alaskensis RB2256, in which the highest levels of resistance are achieved when organisms are grown under glucose limitation conditions at a specific growth rate of 0.13 h−1 or lower.
FIG. 2.
Percentages of survival of nutrient-limited chemostat-grown S. alaskensis RB2256 cells following exposure to 25 mM hydrogen peroxide for up to 60 min. Experiments were performed twice, and CFU were counted by using the drop plate method with marine nutrient agar and ASW-glucose solid medium. There was no difference in survival between ASW-glucose solid medium and marine nutrient agar for any time point. The results of representative experiments are shown. The standard deviation for each time point was less than 20%. (A) Samples taken directly from steady-state glucose-limited chemostats at growth rates of 0.026 h−1 (▴), 0.076 h−1 (■), 0.13 h−1 (●), 0.14 h−1 (▾), 0.16 h−1 (○), and 0.18 h−1 (□) and from a batch culture with a maximum specific growth rate of 0.21 h−1 (▵). (B) Samples taken directly from ammonium-limited chemostats at growth rates of 0.15 h−1 (∗) and 0.050 h−1 (⧫). (C) Samples taken directly from mixed-amino-acid-limited steady-state chemostats at growth rates of 0.15 h−1 (▴) and 0.047 h−1 (▿).
A dose-response curve was constructed by using 0 to 75 mM hydrogen peroxide and an exposure time of 60 min (data not shown). After exposure to 0 to 10 mM hydrogen peroxide, cells were 100% viable irrespective of the growth rate. However, the viability of cells grown at a high dilution rate (0.14 h−1 or higher) decreased at least 10,000-fold when the cells were exposed to 25 mM hydrogen peroxide. In contrast, 75 mM hydrogen peroxide was needed to produce a similar decrease in viability for cells grown at a low dilution rate (0.13 h−1 or lower).
Carbon limitation versus nitrogen limitation.
To determine whether the link between growth rate and hydrogen peroxide resistance was affected by the nature of the limiting substrate in the chemostat, the responses of carbon-limited cultures (Fig. 1 and 2A) were compared to the responses of nitrogen-limited cultures (Fig. 2C). Ammonium-limited cultures were grown at low (0.050 h−1) and high (0.15 h−1) rates of growth, and samples taken from cultures after a steady state had been obtained were exposed to 25 mM hydrogen peroxide. These nitrogen-limited cultures were far more sensitive to hydrogen peroxide than any of the glucose-limited cultures were, exhibiting almost undetectable survival after 20 min of exposure. Furthermore, no difference in stress resistance was observed at the two specific growth rates used.
To further test whether the observed bimodal response under glucose limitation conditions was due to carbon or energy limitation per se and was not a specific effect of glucose metabolism, glucose was replaced with mixed amino acids. When mixed amino acids were used as the sole carbon and energy source, the pattern of hydrogen peroxide resistance for low (0.047 h−1) and high (0.15 h−1) rates of growth (Fig. 2C) was essentially the same as the pattern of resistance for glucose-limited cultures (Fig. 2A). These data indicate that the effect of growth rate on hydrogen peroxide resistance is a general phenomenon in S. alaskensis linked to growth under carbon and/or energy limitation conditions.
Effect of growth rate on hydrogen peroxide resistance in V. angustum S14.
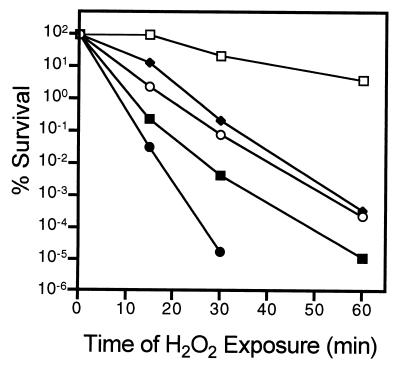
To determine whether the rate of growth affected the ability of V. angustum S14 to survive after exposure to hydrogen peroxide, like S. alaskensis, cells were grown in glucose-limited chemostats at specific growth rates of 0.023, 0.20, and 0.60 h−1 and then exposed to stress in a preparation containing 2 mM hydrogen peroxide for up to 60 min (Fig. 3). In ASW defined minimal medium with 3 mM glucose as the sole carbon and energy source, growth rates of 0.023 and 0.60 h−1 corresponded to approximately 5 and 95% of the maximum specific growth rate (0.62 h−1), respectively. The resistance of the chemostat-grown cells was also compared with the resistance of cells grown at the maximum rate in batch culture and following 24 h of starvation (Fig. 3).
FIG. 3.
Percentages of survival of V. angustum S14 following exposure to 2 mM hydrogen peroxide for up to 60 min after growth at various rates in glucose-limited chemostats. Samples were taken directly from steady-state chemostats at growth rates of 0.023 h−1 (○), 0.20 h−1 (■), and 0.60 h−1 (⧫) and from batch cultures in the logarithmic phase (●) and after 24 h of starvation (□). Experiments were performed twice, and CFU were counted by using the drop plate method with marine nutrient agar and ASW-glucose solid medium for each time point. For each data point the standard deviation was less than 15%.
Starved cells were much more resistant (4.2% survival after 60 min) than cells grown at the maximum specific growth rate (1.7 × 10−5% survival after 30 min), while glucose-limited chemostat-grown cells exhibited an intermediate level of resistance (less than 3 × 10−4% survival after 60 min). In contrast to the bimodal responses exhibited by S. alaskensis RB2256, the resistance of glucose-limited V. angustum S14 varied only about 10-fold, and most interestingly, no correlation with growth rate could be identified. For example, after 60 min of exposure to hydrogen peroxide, the levels of survival were 2.5 × 10−4, 1.2 × 10−5, and 3.4 × 10−4% for growth rates of 0.023, 0.20, and 0.60 h−1, respectively.
Catalase activity and hydrogen peroxide resistance in S. alaskensis RB2256.
When hydrogen peroxide was added to liquid cultures of S. alaskensis RB2256 cells, small bubbles developed after 5 to 30 min. This occurred irrespective of the growth rate and whether the cells were grown in batch or chemostat cultures. The production of bubbles may have resulted from production of O2 due to decomposition of hydrogen peroxide by catalase.
To quantitatively determine whether catalase activity correlated with the degree of hydrogen peroxide resistance observed, catalase activity was measured in whole-cell suspensions of S. alaskensis RB2256 grown at high (0.15 h−1) and low (0.08 h−1) rates of growth in glucose-limited chemostats and at the maximum specific growth rate in batch culture.
The catalase activities of cells grown at low (6.6 U mg of protein−1) and high (4.2 U mg of protein−1) specific growth rates in chemostats and of cells grown in batch culture (1.1 U mg of protein−1) differed marginally (Table 1). In particular, the less-than-twofold difference in catalase activity between cells grown at low and high rates of growth did not correlate with the 10,000-fold difference in stress resistance (Fig. 1). We also examined general peroxidase activity in cell extracts by measuring the disappearance of hydrogen peroxide with a spectrophotometric assay and found essentially no difference in activity between cells grown at low and high dilution rates (data not shown). These data clearly indicate that other cellular factors are responsible for the bimodal response observed in S. alaskensis RB2256.
TABLE 1.
Catalase activities measured in whole cells of S. alaskensis RB2256 grown in glucose-limited chemostats and batch cultures
| Growth conditions | Catalase activitya
|
|
|---|---|---|
| U mg of protein−1 | U 108 cells−1 | |
| Log phase | 1.10 | 0.0117 |
| Growth rate, 0.08 h−1 | 6.63 | 0.0704 |
| Growth rate, 0.15 h−1 | 4.16 | 0.0442 |
One unit of catalase activity was defined as the amount of enzyme required to degrade 1 μmol of H2O2 to 0.5 μmol of O2 in 1 min. The standard deviations were 8 to 12%.
Protein profiles for S. alaskensis RB2256 grown at different rates of growth in glucose-limited chemostat cultures.
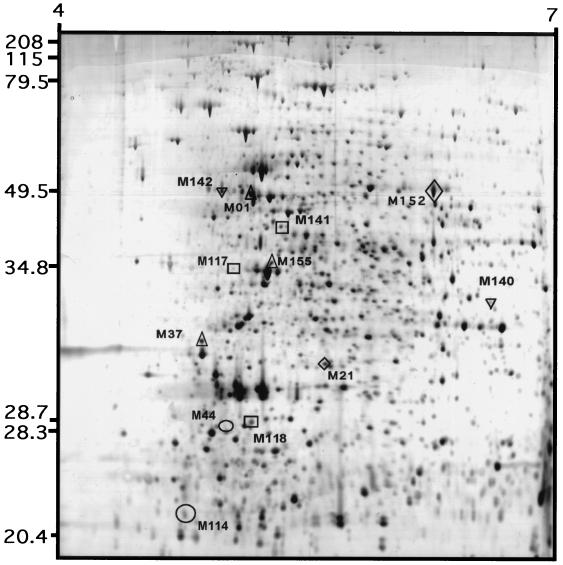
Approximately 1,500 spots can be resolved by high-resolution two-dimensional polyacrylamide gel electrophoresis of proteins from S. alaskensis RB2256 (12). To determine the changes in gene expression that are associated with high and low rates of growth and to identify the genes involved in the increased levels of hydrogen peroxide resistance at low rates of growth, two-dimensional polyacrylamide gel electrophoresis profiles were generated in triplicate for cells grown at rates of 0.026, 0.076, and 0.18 h−1 (Fig. 4). Protein spots for each preparation were separated evenly throughout the resolved pI range (pI 4 to 7) and molecular mass range (10 to 80 kDa). Up to 1,600 protein spots were detected on each silver-stained gel, and 1,049 spots conserved from triplicate gels obtained for each growth rate were analyzed. Relative spot intensity was calculated as the percentage of optical density by comparison with the total optical density of each gel. The intensities of spots ranged from 0.216 to 4.5%.
FIG. 4.
Two-dimensional gel showing proteins from S. alaskensis RB2256 grown in glucose-limited chemostats. Total protein was extracted from steady-state cultures growing at rates of 0.026, 0.076, and 0.18 h−1. Protein profiles were visualized by silver staining. The protein profile shown is the profile from a culture growing at a rate of 0.18 h−1, and the highlighted spots have relative intensities that were at least two-fold less or greater in at least one growth condition. Symbols: ○, spots unique to a growth rate of 0.026 h−1; ▿, spots unique to a growth rate of 0.076 h−1; ▵, spots unique to a growth rate of 0.18 h−1; □, spots found at growth rates of 0.026 and 0.076 h−1; ◊, spots found at growth rates of 0.076 and 0.18 h−1. The molecular weight and pI values for specific spots were assigned by using Melanie II software (Bio-Rad) and are shown in Table 1.
Spots whose intensities differed by at least twofold when two growth rates were compared are indicated in Fig. 4. This figure shows that only 12 spots (∼1%) were significantly different in different gels; 7 of these spots were specific for low rates of growth (0.026 and 0.076 h−1), and 3 were specific for a high rate of growth (0.18 h−1). Each spot indicated in Fig. 4. was characterized to determine its approximate molecular weight, isoelectric point, and difference in relative spot intensity (Table 2).
TABLE 2.
Characteristics of protein spots on two-dimensional protein gels that have spot intensities that vary twofold or more for different glucose-limited specific rates of growth
| Spota | Relative intensity at a growth rate of b:
|
pI | Molecular mass (kDa) | ||
|---|---|---|---|---|---|
| 0.026 h−1 | 0.076 h−1 | 0.18 h−1 | |||
| M114 | 2.2 | 1.0 | 1.0 | 4.49 | 20.5 |
| M44 | 3.3 | 0.9 | 1.0 | 4.67 | 28.8 |
| M118 | 4.0 | 4.5 | 1.0 | 4.84 | 37.0 |
| M117 | 2.4 | 3.1 | 1.0 | 4.67 | 29.5 |
| M141 | 2.2 | 3.4 | 1.0 | 4.97 | 40.5 |
| M140 | 2.6 | 5.9 | 1.0 | 6.37 | 31.5 |
| M142 | NDc | 2.1 | 1.0 | 4.59 | 42.5 |
| M21 | ND | 1.0 | 1.0 | 5.28 | 30.0 |
| M152 | 0.3 | 1.2 | 1.0 | 5.98 | 63.5 |
| M37 | 0.5 | 0.5 | 1.0 | 4.26 | 30.3 |
| M155 | ND | ND | 1.0 | 4.92 | 36.5 |
| M01 | ND | ND | 1.0 | 4.79 | 52.0 |
See Fig. 4.
The intensities of protein spots from cells grown at the high dilution rate (0.18 h−1) were defined as 1.0, and the relative intensities of spots from cells grown at low rates of growth (0.026 and 0.076 h−1) were determined by comparison with these intensities.
ND, not detected.
The predicted molecular masses of the differentially expressed proteins varied from 20.5 to 63.5 kDa, and the predicted isoelectric points varied from 4.26 to 6.37. The largest difference in intensity for a spot that was present in all three gels was the difference for spot M140, which was 5.9-fold more intense at a growth rate of 0.076 h−1 than at a growth rate of 0.18 h−1. Spots M142, M21, M155, and M01 were not detectable under one or two growth conditions. Based on the detection limit for the 1,049 spots examined (0.216% optical density), spot M01 had the highest level of intensity (2.373% optical density), indicating that the level of spot intensity in gels for a growth rate of 0.18 h−1 was at least 11-fold higher than the level of spot intensity in gels for the lower growth rates.
DISCUSSION
Growth rate control of hydrogen peroxide resistance.
We previously found that cultures of S. alaskensis RB2256 grown at 25°C in a glucose-limited chemostat (growth rate, 0.027 h−1) were more resistant to hydrogen peroxide than cells grown in a batch culture (growth rate, 0.16 h−1) (8). In the present study we examined whether there was a link between the actual specific growth rate of a culture and hydrogen peroxide resistance and whether the resistance of cells was significantly affected by the mode of growth (e.g., carbon- or nitrogen-limited growth in chemostats and growth under nutrient-excess conditions in batch cultures). It is indeed apparent that hydrogen peroxide resistance in S. alaskensis RB2256 is strongly influenced by the specific growth rates of cultures when glucose or mixed amino acids are the limiting substrates (Fig. 1 and 2A and C); however, growth rate control of hydrogen peroxide stress resistance was not apparent under nitrogen limitation conditions (Fig. 2B). Thus, a comparison of stress survival of cells grown under carbon or energy limitation conditions with stress survival of ammonium-limited cells grown at comparable dilution rates in chemostats (approximately 0.03 to 0.05 and 0.14 to 0.16 h−1, respectively) clearly showed that the specific rate of growth is not the only major determinant of the level of stress resistance in S. alaskensis RB2256. The results indicate that hydrogen peroxide stress resistance in S. alaskensis RB2256 is influenced directly or indirectly both by the nature of the limiting substrate (carbon or energy versus nitrogen) and by the extent of nutrient limitation in carbon- and/or energy-limited chemostat cultures. In contrast, exponentially growing or starved cells from batch cultures had levels of resistance equivalent to those of carbon-limited, chemostat-grown cells at high dilution rates.
In order to establish whether the physiological responses of S. alaskensis RB2256 were unique, we also examined the hydrogen peroxide resistance of V. angustum S14 in response to specific growth rates in glucose-limited chemostats. The rate of growth had some influence on peroxide stress survival; however, starved and exponentially growing cultures represented the extremes of peroxide stress resistance and sensitivity, respectively. These observations are in line with several earlier observations which all indicated that starvation evokes strong cross-protection against heat or peroxide stress in V. angustum S14 and other phylogenetically unrelated organisms (14, 15, 23, 24, 40, 41, 44). In support of this, E. coli grown in batch cultures and glucose-limited chemostats had a response to hydrogen peroxide stress similar to that observed for V. angustum S14 (M. Ostrowski, J. C. Gottschal, and R. Cavicchioli, unpublished results; T. Ferenci, personal communication). This emphasizes the uniqueness of the response observed in S. alaskensis RB2256, in which starvation does not elicit any cross-protection against hydrogen peroxide or heat stress (8), whereas substrate-limited growth rates that are less than approximately 75% of the maximum specific growth rate result in large increases in hydrogen peroxide resistance.
A striking characteristic of the response of S. alaskensis RB2256 was that the increased stress survival was not gradual; lowering the growth rate below 0.13 h−1 seemed to act like a switch. This is in contrast to the general picture which emerged from most earlier observations on the influence of changes in the specific growth rates of cells in carbon- or energy-limited chemostats, all of which indicated that the changes in enzyme levels (18, 33–35, 39, 53), viability (16, 37, 43, 54), cell size (28, 30, 36–38, 54), concentrations of soluble and structural cell components (18, 21, 26, 29, 30, 56), and abundance and influence of stationary-phase sigma factors (39, 51) are gradual.
Possible mechanisms of growth rate control of hydrogen peroxide resistance.
The phenotypic responses of S. alaskensis RB2256 are consistent with a regulatory cascade in which very small changes in the concentration of effector molecules and/or proteins become amplified through the cascade. Such regulatory events may be mediated by global regulators, such as rpoS, oxyR, and soxRS, and other mechanisms, such as DNA methylation and stringent control, which are important in oxidative stress responses in E. coli (9, 52).
The fact that the growth rate-dependent hydrogen peroxide resistance is linked to carbon limitation (Fig. 1 and 2A and C) but not to nitrogen limitation (Fig. 2B) indicates that the sensing mechanism involves a response to the flux through a metabolic pathway related to carbon uptake or carbon and energy metabolism. When uptake of glucose is restricted in a glucose-limited chemostat, acetate excretion by E. coli decreases until it becomes zero at a growth rate of 0.72 h−1 (21). While a reduction in growth rate leads to a gradual change in flux through central metabolic pathways, there is a point when acetate is no longer produced. It is possible that a similar change in flux through carbon metabolism in S. alaskensis RB2256 provides the signal that leads to the sudden change in stress resistance.
The mechanism that leads to dramatically enhanced hydrogen peroxide resistance in S. alaskensis RB2256 has not been determined. However, catalase activity does not appear to be linked to the resistance state of the cells; total catalase activity varied about sixfold (Table 1) and did not correlate with stress resistance. In his review of the regulation of enzyme synthesis in bacteria grown in chemostats, Matin (33) reported five types of changes in enzyme activity in response to growth rate. Interestingly, while about 50% of the enzymes exhibited increased activity with a decreasing growth rate (33), in E. coli superoxide dismutase activity increased with growth rate, peroxidase activity decreased with growth rate, and catalase activity did not vary until the growth rate exceeded 0.4 h−1, and it then declined (19).
While these studies on E. coli and our studies on S. alaskensis RB2256 indicate that regulation of catalase expression appears to be largely independent of growth rate, the involvement of the katC locus in E. coli illustrates the complexity of the mechanism by which catalase gene expression may be regulated. Volkert et al. (58) have reported that the katC locus is responsible for the sensitivity of wild-type strains to hydrogen peroxide. Deletion of this locus in an argF-lacZ strain or interruption of the locus with Tn9 apparently leads to a dramatic increase in hydrogen peroxide resistance (∼103-fold-greater survival after 25 min of exposure to 150 mM hydrogen peroxide compared to the wild type). Interestingly, while the increased resistance is starvation dependent and requires functional katE and katF genes, catalase activity and katE expression are not elevated in katC mutant strains compared to the wild type. Volkert et al. speculated that the product of the katC gene (IS1B-IS30B fusion) may affect a function of KatF that involves an activity other than catalase activity. While the function of katC is unknown, it provides a precedent for a genetic element that affects the hydrogen peroxide resistance of isogenic strains by at least 3 orders of magnitude.
Hydrogen peroxide and other reactive oxygen species are capable of causing damage to DNA, RNA, proteins, and lipids (52). Catalase activity is only one means of reducing the damaging effects of hydrogen peroxide. The factors that may be involved in the increased resistance of slowly growing cells include increased levels of detoxifying enzymes, such as proteins that reduce disulfide bridges caused by oxidative stress (e.g., glutathione reductase), organic hydroperoxidases other than catalase (e.g., Ahp), or nucleic acid binding proteins (e.g., Dps). In addition, the cell wall or cytoplasmic membrane may be modified to reduce hydrogen peroxide penetration into the cell. It is noteworthy that S. alaskensis RB2256 produces a pigment that appears to be the carotenoid nostoxanthin (A. Nouwens and R. Cavicchioli, unpublished results), and carotenoids are known to be effective scavengers of singlet oxygen (2, 55). Resistance also may be afforded by improved DNA protection or repair mechanisms. Joux et al. (25) have shown that S. alaskensis RB2256 does not accumulate cyclopyrimidine dimers during UV-B irradiation and have suggested that this may be due to a constitutive photoprotective mechanism. While UV-B causes DNA strand breakage, the protective mechanism may also repair damage caused by reactive oxygen species.
A rapid means of extensively surveying the gene products required for differential stress resistance is analysis of two-dimensional protein profiles (Fig. 4). Interestingly, the number of differences between the profiles of fast-growing and slowly growing cells is relatively small (Table 2). This may indicate that the alterations in gene expression are associated with a narrow range of physiological responses, including resistance to hydrogen peroxide. At present, we are examining the link between growth rate and resistance to other stresses. In addition, we are determining the identities of differentially expressed proteins. As the hydrogen peroxide resistance of cells with a growth rate of 0.026 and the hydrogen peroxide resistance of cells with a growth rate of 0.076 h−1 are equivalent and greater than the hydrogen peroxide resistance of cells grown at a rate of 0.18 h−1, the candidate spots that represent proteins of particular importance for stress resistance of the cells are the spots whose intensities are decreased or increased only at a growth rate of 0.18 h−1; that is, proteins that are specifically upregulated or repressed at low rates of growth may either activate or derepress the associated stress resistance mechanism(s). Such candidate spots are M118, M117, M141, M140, M37, M155, and M01 (Fig. 4 and Table 2).
Starvation versus low-growth-rate induction of peroxide stress resistance.
In almost all cases starvation survival and in some cases starvation-induced cross-protection are dependent on the substrate for which the culture is starved. V. angustum starvation-induced survival, miniaturization, and stress resistance are specific for carbon starvation and not for nitrogen or phosphorus starvation (41). E. coli viability is more sensitive to phosphate starvation than to carbon or nitrogen starvation (7), while porin regulation in response to glucose limitation is not observed in nitrogen-limited cultures (29). An explanation based on the ecology of marine bacteria may be related to the fact that most pelagic heterotrophs are limited by the availability of carbon (1, 4, 5, 27) and adaptive responses are geared for carbon starvation.
In this regard it is interesting that even though S. alaskensis RB2256 is physiologically suited to growth in oligotrophic conditions, it is genetically geared to respond to carbon starvation (10). It is likely that in terms of the total available oceanic carbon and gradients of nutrients present in microzones, starvation in the marine environment is potentially significant for all members of the microbiota. Clearly, however, S. alaskensis RB2256 evolved a genotype that enabled it to be a numerically dominant bacterium at the time of its isolation, compared to the significantly lower number (<1%) of copiotrophic bacteria (3). It is therefore not surprising that in contrast to copiotrophic bacteria, which are likely to survive in oligotrophic water by producing stress-resistant, resting stage cells, S. alaskensis RB2256 is likely to have developed a strategy of maintaining optimal stress resistance for survival during slow growth.
ACKNOWLEDGMENTS
We thank Tom Ferenci for providing unpublished results, Staffan Kjelleberg and Mitsuru Eguchi for valuable discussions, and Tassia Kolesnikow, Scott Rice, and Fitri Fegatella for critical reviews of the manuscript.
The research performed by R.C. and M.O. was supported by the Australian Research Council. M.O. was supported by an Australian Postgraduate Award.
REFERENCES
- 1.Børsheim K Y. Bacterial production rates and concentrations of organic carbon at the end of the growing season in the Greenland Sea. Aquat Microb Ecol. 2000;21:115–123. [Google Scholar]
- 2.Bridges B A, Timms A. Effect of endogenous carotenoids and defective RpoS sigma factor on spontaneous mutation under starvation conditions in Escherichia coli—evidence for the possible involvement of singlet oxygen. Mutat Res. 1998;403:21–28. doi: 10.1016/s0027-5107(98)00013-x. [DOI] [PubMed] [Google Scholar]
- 3.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson C A, Ducklow H W. Growth of bacterioplankton and consumption of dissolved organic carbon in the Sargasso Sea. Aquat Microb Ecol. 1996;10:69–85. [Google Scholar]
- 5.Church M J, Hutchins D A, Ducklow H W. Limitation of bacterial growth by dissolved organic matter and iron in the southern ocean. Appl Environ Microbiol. 2000;66:455–466. doi: 10.1128/aem.66.2.455-466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper W J, Zika R G. Photochemical formation of hydrogen peroxide in surface and ground waters exposed to sunlight. Science. 1983;220:711–712. doi: 10.1126/science.220.4598.711. [DOI] [PubMed] [Google Scholar]
- 7.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eguchi M, Nishikawa T, MacDonald K, Cavicchioli R, Gottschal J C, Kjelleberg S. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1996;62:1287–1294. doi: 10.1128/aem.62.4.1287-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fegatella F, Cavicchioli R. Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 2000;66:2037–2044. doi: 10.1128/aem.66.5.2037-2044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegatella F, Ostrowski M, Cavicchioli R. An assessment of protein profiles from the marine oligotrophic ultramicrobacterium, Sphingomonas sp. strain RB2256. Electrophoresis. 1999;20:2094–2098. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2094::AID-ELPS2094>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Ferenci T. Regulation by nutrient limitation. Curr Opin Microbiol. 1999;2:208–213. doi: 10.1016/S1369-5274(99)80036-8. [DOI] [PubMed] [Google Scholar]
- 14.Givskov M, Eberl L, Møller S, Poulsen L K, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovlev E L. An introduction to the biology of the stationary-phase of bacteria: the mechanism of the common response to stresses. Microbiology (Engl Transl Mikrobiologiya) 1999;68:543–550. [Google Scholar]
- 16.Gottschal J C. Phenotypic responses to environmental changes. FEMS Microbiol Ecol. 1990;74:93–102. [Google Scholar]
- 17.Gourmelon M, Cillard J, Pommepuy M. Visible light damage to Escherichia coli in seawater: oxidative stress hypothesis. J Appl Bacteriol. 1994;77:105–112. doi: 10.1111/j.1365-2672.1994.tb03051.x. [DOI] [PubMed] [Google Scholar]
- 18.Harder W, Dijkhuizen L. Physiological responses to nutrient limitation. Annu Rev Microbiol. 1983;37:1–23. doi: 10.1146/annurev.mi.37.100183.000245. [DOI] [PubMed] [Google Scholar]
- 19.Hassan H M, Fridovich I. Physiological function of superoxide dismutase in glucose-limited chemostat cultures of Escherichia coli. J Bacteriol. 1977;130:805–811. doi: 10.1128/jb.130.2.805-811.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoben H S P. Comparison of the pour, spread, and drop plate methods for the enumeration of Rhizobium spp. in inoculants made from presterilized peats. Appl Environ Microbiol. 1982;44:1246–1247. doi: 10.1128/aem.44.5.1246-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holms H. Flux analysis and control of the central metabolic pathways in Escherichi coli. FEMS Microbiol Rev. 1996;19:85–116. doi: 10.1111/j.1574-6976.1996.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 22.Janssen P H, Schuhmann A, Mörschel E, Rainey F A. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol. 1997;63:1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1998;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jouper-Jaan A, Goodman A, Kjelleberg S. Bacteria starved for prolonged periods develop increased protection against lethal temperatures. FEMS Microbiol Ecol. 1992;101:229–236. [Google Scholar]
- 25.Joux F, Jeffrey W H, Lebaron P, Mitchell D L. Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol. 1999;65:3820–3827. doi: 10.1128/aem.65.9.3820-3827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp P F, Lee S, Laroche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchman D L. Limitation of bacterial growth by dissolved organic matter in the subarctic Pacific. Mar Ecol Prog Ser. 1990;62:47–54. [Google Scholar]
- 28.Koch A L. Microbial growth in low concentrations of nutrients. In: Shilo M, editor. Strategies in microbial life in extreme environments, Dahlem Konferenzen—1978. Weinheim, Germany: Verlag Chemie; 1979. pp. 261–279. [Google Scholar]
- 29.Liu X, Ferenci T. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J Bacteriol. 1998;180:3917–3922. doi: 10.1128/jb.180.15.3917-3922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maalöe O, Kjeldgaard N O. Control of macromolecular synthesis. W. A. New York, N.Y: Benjamin; 1966. [Google Scholar]
- 31.Mården P, Nyström T, Kjelleberg S. Uptake of leucine by a marine gram negative bacterium during exposure to starvation conditions. FEMS Microbiol Ecol. 1987;45:233–241. [Google Scholar]
- 32.Matear R J, Hirst A C. Climate change feedback on the future oceanic CO2 uptake. Tellus. 1999;51:722–733. [Google Scholar]
- 33.Matin A. Regulation of enzyme synthesis as studied in continuous culture. In: Calcott P H, editor. Continuous culture of cells. Vol. 12. Boca Raton, Fla: CRC Press; 1981. pp. 69–97. [Google Scholar]
- 34.Matin A, Matin M K. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coli grown in continuous culture. J Bacteriol. 1982;149:801–807. doi: 10.1128/jb.149.3.801-807.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matin A, Grootjans A, Hogenhuis H. Influence of dilution rate on enzymes of intermediary metabolism. J Gen Microbiol. 1976;94:323–332. doi: 10.1099/00221287-94-2-323. [DOI] [PubMed] [Google Scholar]
- 36.Matin A A, Veldkamp H. Physiological basis of the selective advantage of a Spirillum sp. in a carbon-limited environment. J Gen Microbiol. 1978;105:187–197. doi: 10.1099/00221287-105-2-187. [DOI] [PubMed] [Google Scholar]
- 37.Moyer C L, Morita R Y. Effect of growth rate and starvation survival on the viability and stability of a psychrophilic marine bacterium. Appl Environ Microbiol. 1989;55:1122–1127. doi: 10.1128/aem.55.5.1122-1127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyer C L, Morita R Y. Effect of growth rate and starvation survival on cellular DNA, RNA, and protein of a psychrophilic marine bacterium. Appl Environ Microbiol. 1989;55:2710–2716. doi: 10.1128/aem.55.10.2710-2716.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyström T. Starvation, cessation of growth and bacterial aging. Curr Opin Microbiol. 1999;2:214–219. doi: 10.1016/S1369-5274(99)80037-X. [DOI] [PubMed] [Google Scholar]
- 41.Nyström T, Olsson R M, Kjelleberg S. Survival, stress resistance and alterations in protein expression in the marine Vibrio sp. strain S14 during starvation for different individual nutrients. Appl Environ Microbiol. 1992;58:55–65. doi: 10.1128/aem.58.1.55-65.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oda T, Atsushi N, Midori S, Ienobu K, Atsushi I, Tsuyoshi M. Generation of reactive oxygen species by raphidophycean phytoplankton. Biosci Biotechnol Biochem. 1997;61:1658–1662. doi: 10.1271/bbb.61.1658. [DOI] [PubMed] [Google Scholar]
- 43.Poolman B, Smid E J, Veldkamp H, Konings W N. Bioenergetic consequences of lactose starvation in continuously cultured Streptococcus cremoris. J Bacteriol. 1987;169:1460–1468. doi: 10.1128/jb.169.4.1460-1468.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preyer J M, Oliver J D. Starvation-induced thermal tolerance as a survival mechanism in a psychrophilic marine bacterium. Appl Environ Microbiol. 1993;59:2653–2656. doi: 10.1128/aem.59.8.2653-2656.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rørth M, Jensen P K. Determination of catalase activity by means of the Clark oxygen electrode. Biochim Biophys Acta. 1967;139:173–176. doi: 10.1016/0005-2744(67)90124-6. [DOI] [PubMed] [Google Scholar]
- 46.Schut F. Ph. D. thesis. Groningen, The Netherlands: University of Groningen; 1993. [Google Scholar]
- 47.Schut F, de Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schut F, Gottschal J C, Prins R A. Isolation and characterisation of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol Rev. 1997;20:363–369. [Google Scholar]
- 49.Schut F, Jansen M, Pedro Gomes T M, Gottschal J C, Harder W, Prins R A. Substrate uptake and utilization by a marine ultramicrobacterium. Microbiology. 1995;141:351–361. doi: 10.1099/13500872-141-2-351. [DOI] [PubMed] [Google Scholar]
- 50.Schut F, Prins R A, Gottschal J C. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat Microb Ecol. 1997;12:177–202. [Google Scholar]
- 51.Schweder T, Kolyschkow A, Völker U, Hecker M. Analysis of the expression and function of the ςB-dependent general stress regulon of Bacillus subtilus during slow growth. Arch Microbiol. 1999;171:439–434. doi: 10.1007/s002030050731. [DOI] [PubMed] [Google Scholar]
- 52.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 53.Tempest D W, Neijssel O M. Eco-physiological aspects of microbial growth in aerobic nutrient-limited environments. Adv Microb Ecol. 1978;2:105–153. [Google Scholar]
- 54.Tempest D W, Herbert D, Phipps P J. Studies on the growth of Aerobacter aerogenes at low dilution rates in a chemostat. In: Powell E O, Evans C G T, Strange R E, Tempest D W, editors. Microbial physiology and continuous culture. London, United Kingdom: Her Majesty's Stationery Office; 1967. pp. 240–253. [Google Scholar]
- 55.Tuveson R W, Sandmann G. Protection by cloned carotenoid genes expressed in Escherichia coli against phototoxic molecules activated by near-ultraviolet light. Methods Enzymol. 1993;214:323–330. doi: 10.1016/0076-6879(93)14075-t. [DOI] [PubMed] [Google Scholar]
- 56.Tweeddale H, Notley-McRobb L, Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J Bacteriol. 1998;180:5109–5116. doi: 10.1128/jb.180.19.5109-5116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vancanneyt M, Schut F, Snauwaert C, Goris J, Swings J, Gottschal J C. Sphingomonas alaskensis sp. nov., a dominant organism from a marine oligotrophic environment. Int J Syst Evol Microbiol. 2001;51:73–79. doi: 10.1099/00207713-51-1-73. [DOI] [PubMed] [Google Scholar]
- 58.Volkert M R, Loewen P C, Switala J, Crowley D, Conley M. The Δ(argF-lacZ)205(U169) deletion greatly enhances resistance to hydrogen peroxide in stationary-phase Escherichia coli. J Bacteriol. 1994;176:1297–1302. doi: 10.1128/jb.176.5.1297-1302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wignall P B, Twitchett R J. Oceanic anoxia and the end Permian mass extinction. Science. 1996;272:1155–1158. doi: 10.1126/science.272.5265.1155. [DOI] [PubMed] [Google Scholar]
- 60.Wilhelm E R, Battino R, Wilcock R J. Low pressure solubility of gases in liquid water. Chem Rev. 1977;77:219–262. [Google Scholar]