Fig. 4 |. Nonsynonymous differences between B and D Mutyh alleles segregate in both wild and inbred mouse strains and appear to be ancestral in DBA/2J.
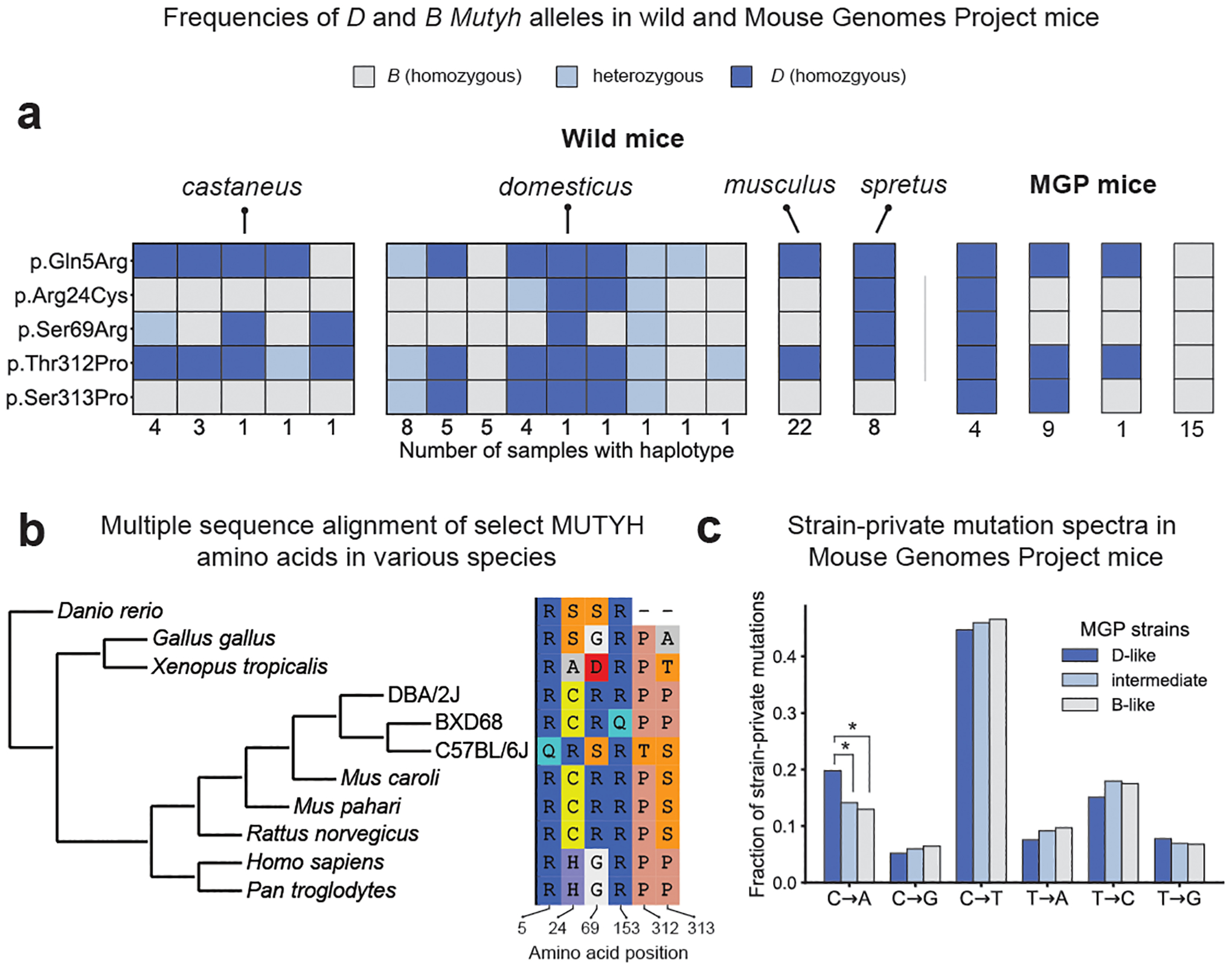
a, Presence of D or B Mutyh alleles in 67 wild mice35 and in 29 Sanger Mouse Genomes Project (MGP) strains that have associated strain-private singleton data17. Unique combinations of Mutyh alleles are represented using columns of coloured boxes. Wild mice are grouped by species or subspecies, and the number of mice with each unique combination of Mutyh alleles is listed below each column. MGP mice are grouped by Mutyh genotype, and each genotype is labelled with the number of lines where this allele combination is found. b, Multiple sequence alignment of MUTYH amino acids is subsetted to only show the six amino acids affected by moderate- or high-impact mutations in the BXD. Positions of amino acids in the mouse MUTYH peptide sequence (ENSMUST00000102699.7) are shown below each column. c, Mutation spectra of MGP strains17 with D-like, intermediate, and B-like Mutyh genotypes. D-like strains have significantly higher C>A fractions than B-like (P = 1.4 × 10−10) and intermediate strains (P = 3.3 × 10−7; C>A fractions of intermediate and B-like strains are not significantly different (P = 0.12; Chi-square test).
