Abstract
Current methods for the detection of pathogens in food and water samples generally require a preenrichment step that allows selective enrichment of the test organism. The objective of this research was to eliminate an enrichment step to allow detection of bacteria directly in food and water samples in 30 min. A high-flow-rate, fluidized bed to capture and concentrate large (bacteria and spores) and small (protein) molecules was developed. This format, ImmunoFlow, is volume independent and uses large beads (greater than 3 mm in diameter) when capturing bacteria to prevent sample clogging when testing food samples. Detection of bound targets was done using existing enzyme-linked immunosorbent assay (ELISA) protocols. Four antibodies (anti-Escherichia coli O157:H7, -Bacillus globigii, -bovine serum albumin [BSA], and -ovalbumin [OVA]) were covalently coupled to various glass and ceramic beads. Very small amounts of BSA (<1 ng) and OVA (0.2 to 4.0 μg) were detected. Various industrial and environmental samples were used to observe the effect of the sample composition on the capture of anti-B. globigii and anti-E. coli O157:H7 modified beads. The lower limit of detection for both E. coli O157:H7 and B. globigii was 1 spore/cell independent of the sample size. The activity of anti-B. globigii modified beads declined after 3 days. Anti-E. coli O157:H7 modified beads declined in their capture ability after 2 days in various storage buffers. Storage temperature (4 and 25°C) did not influence the stability. The ImmunoFlow technology is capable of capturing bacteria and spores directly from samples, with subsequent detection in an ELISA format in 30 min.
In 1982, a new pathogen causing hemorrhagic colitis emerged that later became know as Escherichia coli O157:H7. Since then, tremendous efforts have been put forth to identify and characterize this enteropathogen. About 20,000 cases per year of E. coli O157:H7-caused hemorrhagic colitis are found, and this number is increasing. Approximately 5% of the patients develop more serious health problems, such as hemolytic anemia, kidney failure, and thrombocytopenia (27). The route of infection is usually fecal-oral transmission.
E. coli O157:H7 contamination is of concern to the food industry because of the pathogenicity of this organism and the increase in cases (31). It is found in the environment and is prevalent in domestic farm animals, having been isolated from calves, cattle, and sheep (7, 10, 45). Thus, potential cross-contamination at the farm or in commercial meat processing plants can lead to infections from foods (13, 23). Apple juice, apple cider, raw apples, milk, ground beef, radish sprouts, salami, tomatoes, and lettuce have been associated with outbreaks from food sources (2, 5, 9, 11, 19, 28, 35). Other documented outbreaks involve swimming pool and drinking water (30, 41).
Over the past several years, rapid detection methods have been developed for E. coli O157:H7, but all still require at least 6 h of the preenrichment step before the detection phase. Sorbitol MacConkey medium has been the medium of choice in isolating and identifying non-sorbitol-fermenting E. coli O157:H7, followed by additional testing to verify the identification (8, 17, 25). deBoer (12) summarized recent developments in isolation tools for use with solid media. Detection of E. coli is with membrane filtration followed by growth on selective agar containing chromigenic and fluorogenic substrates as an indicator of beta-d-glucuronidase activity; however, this test is not specific for E. coli O157. Further verification after these procedures requires serotyping of the isolates.
Enzyme-linked immunosorbent assays (ELISAs) for detection of E. coli O157:H7 were developed to meet the need for faster detection. ELISAs are performed after the preenrichment step and often require only minutes to visualize the results in a lateral flow device (11, 21, 23, 26, 35). Together with various enrichment methods and ELISA-based detection methods, the analysis time and sensitivity have improved, taking less than 24 h.
Several research groups have developed immunomagnetic separations for the detection of E. coli O157:H7. These methods still require overnight preenrichment followed by capture and concentration of the magnetic beads prior to detection using an ELISA (7, 22, 34, 36, 37, 44). The sensitivity ranges from 10 to 102 CFU/g of ground beef. These tests take more than 8 h to run after preenrichment.
Other research groups have focused on developing filters or other solid supports to capture and concentrate E. coli O157:H7 (11, 23, 29). ELISAs can be performed on these solid supports, and the sensitivity is between 0.1 and 1.3 cells/g of ground beef. However, filtration presents new cell collection problems in complex samples that may clog the filter, thereby limiting the sample volume that can be used for the test.
Numerous comparative studies of detection and identification methods available on the market have also been made (15, 20, 24, 33, 39, 40, 43). They all include preenrichment steps followed by a detection method. The sensitivity and lengths of the tests are similar. Thus, the focus has shifted toward developing methods that omit the preenrichment step to reduce analysis time. To try to meet the collection demand, Tortorello and Stewart (38) developed an antibody-direct epifluorescent filter technique. The sample is homogenized, treated with trypsin and Triton X-100, and concentrated onto a 0.2-μm-pore-size polycarbonate filter. The filter is subsequently stained and analyzed by epifluorescence microscopy. The sensitivity of this test is 16 CFU/g, and the test time was reduced to <1 h. Seo et al. (36) developed a test combining immunomagnetic bead separation with flow cytometry. In <1 h, 103 to 104 CFU/ml of ground beef was detected. Gehring et al. (14) developed an immunoligand assay. The sensitivity of 2.5 × 104 cells/ml was reached in 30 min. Brewster and Mazenko (4) developed a filtration technique, which obtained a sensitivity of 5 × 103 CFU/ml in 25 min. Abdel-Hamid et al. (1) developed the most sensitive method, requiring no preenrichment, by using a flow-through immunofiltration system. This combines flow-through, immunofiltration, and enzyme immunoassay techniques to achieve a sensitivity of 100 cells/ml in 30 min. However, clogging has caused this test to fail with complex food samples.
The need to develop faster and more sensitive methods is apparent. Our research group has focused on developing a biosensor that is more sensitive than 100 cells/ml. This ImmunoFlow system involves the use of existing ELISA protocols together with a new flow-through capture mechanism. We have developed a system which will identify small proteins, i.e., ovalbumin (OVA) and bovine serum albumin (BSA), Bacillus spores, and E. coli O157:H7. The sensitivity for Bacillus and E. coli O157:H7 is <10 total cells independent of the sample size. This new ImmunoFlow (patent awarded) method can easily be adapted for detection and identification of other pathogenic food-borne bacteria, such as Salmonella and Listeria (C. Beer, R. Koka, M. Hill, X. Wang, and B. Weimer, unpublished data).
MATERIALS AND METHODS
Antibody characterization.
Four clones of E. coli O157:H7 monoclonal antibodies (Ab) and one clone of E. coli K-12 monoclonal Ab were screened for cross-reactivity. All E. coli Ab were screened for cross-reactivity and ability to capture E. coli O157:H7 (ATCC 35150, 43888, 43889, 43894, and 43895) using sandwich ELISA procedures (16) (Table 1). Four anti-Bacillus spore monoclonal Ab were screened for cross-reactivity and ability to capture Bacillus globigii spores. All Ab used were immunoglobulin G (IgG) type affinity purified using an A/G column (Pierce Chemical, Rockford, Ill.) either by the manufacturer or by our laboratory according to the manufacturer's instructions.
TABLE 1.
Antibodies investigated for cross-reactivity
| Antibody toa: | Antigen | Host | Clone no. | Lot no. | Source |
|---|---|---|---|---|---|
| B. globigii | Spores | Goat | 56456B | SLH513 | Dugway Proving Grounds |
| B. circulans | Spores | Mouse | BCIRS | BCIRS | Utah State University |
| B. cereus | Spores | Mouse | BCER | BCER | Utah State University |
| B. stearothermophilus | Spores | Mouse | BSTS | BSTS | Utah State University |
| E. coli O157 | Cells | Mouse | 3011 | 2D1048 | Biodesign Internationalb |
| E. coli O157 | Somatic | Mouse | 8-9H.2D | 7A0298 | Biodesign International |
| E. coli O157:H7 | Flagella | Mouse | 4E12.F3.E11 | 2E1225 | Biodesign International |
| E. coli O157:H7 | Cells | Goat | Polyclonal | UG 065 | Kirkegaard & Perry Laboratoriesc |
| E. coli K12 | Cells | Mouse | 1103/77 | E950831 | Biogenesis Inc.d |
E. coli Ab were purchased affinity purified. B. circulans, B. cereus, and B. stearothermophilus antibodies were purified using an A/G column purchased from Pierce and used according to the manufacturer's instructions after production by the method of Blake and Weimer (3).
Kennebunk, Maine.
Gaithersburg, Md.
Sandown, N.H.
Each E. coli Ab (1 μg and 100 μl/well) was bound in 96-well ELISA plates (Costar, VWR, Brisbane, Calif.) overnight at 4°C in 50 mM carbonate buffer (pH 9.5). The plates were washed four times with phosphate-buffered saline (PBS) containing 0.02% (vol/vol) Tween 20 (pH 7.2) (PBST) and blocked overnight with 200 μl of 3% (wt/vol in PBS) BSA/well at 4°C. Subsequently, the plates were washed four times with PBST containing 30 mM NaCl (pH 5.8).
The cells were grown overnight in tryptic soy broth at 37°C, subsequently stabbed into motility agar slants to induce expression of the H7 gene, and incubated for another 16 h (12). The cells were diluted with PBST so that 100 μl of this solution contained 103 CFU of the various E. coli strains. The plates were incubated for 1 h at 37°C with 100 μl of the E. coli-containing buffer and washed four times with PBST-NaCl (pH 7.2). Each E. coli Ab was used as a secondary (2°) Ab in the screening at a concentration of 0.05 μg/well diluted in PBST-NaCl. The complex was incubated for 1 h at 37°C.
Bacillus cereus, Bacillus circulans, Bacillus stearothermophilus (all at 105 spores/well; Difco Laboratories, Detroit, Mich.) and B. globigii (103 spores/well; Dugway Proving Grounds, Tooele, Utah) spore suspensions were bound to the plate in 50 mM carbonate buffer (pH 9.5) and incubated overnight at 4°C (2). All strains were cross-reacted against the various Ab (1 μg/well of 2° anti-Bacillus Ab) mentioned above.
The ELISAs were designed so that each combination of E. coli strain or B. globigii spore was tested against all the respective mono- and polyclonal Ab (Table 1). Following incubation with 2° Ab, the plates were washed four times with PBST (pH 7.2), and 100 μl of a 1:10,000 (from 0.8 mg/ml stock) dilution of anti-IgG-horseradish peroxidase (HRP) (Pierce) in PBST/well (pH 7.2) was added. The plates were incubated for 30 min at 25°C. After incubation, the plates were washed four times with PBST (pH 7.2), and 100 μl of tetramethyl benzidine (1-Step Turbo TMB-ELISA substrate; Pierce)/well was added. The plates were incubated in the dark for 20 min at 37°C. Color generation was stopped with the addition of 50 μl of 2 M H2SO4/well, and signal was determined in a 96-well plate reader at 405 nm (Perkin-Elmer, Norwalk, Conn.). The cross-reactivities were determined in triplicate. Negative controls contained no B. globigii spores or E. coli cells and did not produce color. Positive controls contained the cell or spore that the antibody was specific against.
Other antibodies used in this study included monoclonal mouse anti-egg albumin (clone OVA-14; Sigma, St. Louis, Mo.) and monoclonal goat anti-BSA (clone BSA-33; Sigma).
Surface modification.
The capture ability of antibodies attached to different spacers was investigated. Glass and ceramic surfaces were modified by the methods of Blake and Weimer (3) and Weimer et al. (42). Dextran (molecular weight, 37,500) (Sigma), polyethylene glycol-dicarboxylmethyl (PEG; molecular weight, 3,400) (Shearwater Polymers, Inc., Huntsville, Ala.), and polythreonine (molecular weight, 12,100) (Sigma) were used as spacers (Table 2).
TABLE 2.
List of antibodies and their modifications used to capture BSA, OVA, B. globigii spores, and E. coli O157:H7
| Antibody | Bead | Size | Spacer | Matrices tested |
|---|---|---|---|---|
| Anti-BSA | Polystyrene | 2.8 μm | Polythreonine | PBS |
| Anti-OVA | Glass | 3 mm | PEG | PBS |
| Anti-B. globigii spores | Glass | 3 mm | PEG and dextran | Environmental and industrial water samples, 0.25 M sodium phosphate buffer, pH 7.2 |
| Ceramic | 7 mm | Polythreonine | ||
| Anti-E. coli O157:H7 | Glass | 3 mm | PEG | PBS, meat |
Anti-BSA Ab were bound to 2.8-μm-diameter tosyl-activated polystyrene Dynalbeads (1 mg). Polythreonine (molecular weight, 12,100) (Sigma) was used as a spacer and attached to the beads by the method of Blake and Weimer (3). A total of 100 μl (108 total beads) of modified polystyrene beads was used for each sample. Anti-OVA Ab at a concentration of 1016 molecules/m2 were bound to 3-mm-diameter glass beads by the method of Weimer et al. (42). PEG was used as a spacer and attached using the EDC-facilitated reaction (18). Anti-B. globigii spore Ab were bound to 3-mm-diameter glass and 7-mm-diameter ceramic beads. Polythreonine was used as the spacer for the ceramic beads, whereas PEG and dextran were used as spacers with glass beads (18). The antibody concentration was 1016 molecules/m2 for all anti-B. globigii spore beads. Anti-E. coli O157:H7 (Kirkegaard & Perry Laboratories [KPL], Gaithersburg, Md.) was attached to 3-mm-diameter glass beads using PEG as the spacer (18) at a concentration of 1013 molecules/m2. Hybridization slides (surface area, 2.4 cm2) were also modified with the same concentration of anti-E. coli O157:H7 antibodies using PEG as the spacer.
Detection in static environment.
Eight grams of Ab-modified beads was placed into a 50-ml centrifuge tube, and 10 ml of sample was added to the beads. Samples were incubated on a rocker for 1 h at 25°C. The samples were washed six times each with 50 ml of PBST (pH 5.8). Secondary Ab was added (total, 1012 molecules of anti-E. coli O157:H7, 1013 molecules of anti-OVA, 1013 molecules of anti-BSA, and 1012 molecules of anti-B. globigii) in 10 ml of PBST, and beads were again incubated for 1 h. Samples were washed six times with 50 ml of PBST (pH 5.8) and incubated with 10 ml of anti-IgG conjugated to HRP (Pierce) (IgG-HRP, 1 μg per 10 ml of PBST [pH 5.8]). After the last wash step, beads were added to 5 ml of 1-Step Turbo TMB-ELISA substrate (Pierce) and incubated in the dark for 20 min before an A370 reading was taken using a Cary 100-Bio UV/visible light spectrophotometer (Varian, Sugar Land, Tex.). Water blanks were used as the control for the double-beam instrument.
Detection using ImmunoFlow.
ImmunoFlow used a fluidized bed of beads, 8 g for the small unit and 250 g for the large unit, with Ab covalently bound. To generate flow, a vacuum pump was used. The reagents were evacuated from the bead cartridge through the top of the reactor at a constant rate of 0.4 liters/min (or 5 in. of Hg). As soon as all the liquid passed over the beads, the next reagent was allowed to flow through the reactor. This continued until all the reagents flowed across the beads. Just before the substrate (TMB) was added to the bead cartridge, the vacuum was turned off and the TMB was pulled into the reactor with a syringe. Once the TMB solution covered the beads, the cartridge was sealed and placed in the dark for 20 min. To measure the color development at A370, 1 ml of the substrate was placed in a cuvette. Water blanks were used as a control for the double-beam spectrophotometer.
Four liters of 0.25 M sodium phosphate buffer (pH 7.0) or river water was spiked with 106 total B. globigii spores. A stainless steel module was filled with 250 g of modified anti-B. globigii spore ceramic beads. The B. globigii spore solution was recycled over the 7-mm-diameter modified ceramic beads for 60 min at flow rates of 1, 2, and 4 liters/min. Five beads were taken out every 15 min and replaced with five nonmodified ceramic beads, and the capture ability of the beads was investigated using the static method. At the same time, spore counts were determined on plate count agar.
The ability of the detection system to recover B. globigii spores from various environmental and industrial water samples was also investigated. Samples were collected from various environmental and industrial locations in Cache Valley: Logan river water (pH 8.4), Gossner's Cheese Plant tank water (pH 9.2), PBST (pH 7.2), and USU Dairy Plant slush tank water (pH 7.2). Samples were tested in flow using 8 g of Ab-modified beads. Standard curves were generated in these samples with pure cultures in buffer. The ability of the detection system to recover E. coli O157:H7 from meat extract and PBST samples was also investigated with 104 total cells and anti-E. coli O157:H7 Ab attached to 3-mm-diameter glass beads via PEG.
Stability of modified beads.
Stability of anti-B. globigii spore modified beads was tested over a 5-week period. Two spacers, dextran and PEG, were used to attach the Ab to the surface of 3-mm-diameter glass beads at 1016 molecules/m2. Two identical sets of experiments were run. One set of beads was stored in PBST (pH 7.2) containing 0.02% thimerosal at 8°C. The other set of beads was transferred into a stainless-steel module and placed in the river. The flow of the water ran straight through the module. River temperature and pH were recorded during the 5 weeks. Bead samples were taken once a week and tested for the ability to bind 105 total B. globigii spores.
Three-millimeter-diameter glass beads modified with anti-E. coli O157:H7, using PEG as a spacer, were stored in 12 different storage buffers (Table 2) at 8 and 25°C for 25 days. Every 2 days, beads were tested for E. coli O157:H7-capturing ability using 105 total cells.
Confocal microscopy.
B. globigii spores were labeled with Fura-indole C18 using a labeling kit from Molecular Probes (Eugene, Oreg.) according to the manufacturer's manual. Labeled spores showed a blue color under fluorescence (Ex, 481 nm; Em, 694 nm). E. coli O157:H7 cells were stained with the LIVE/DEAD BacLight kit (Molecular Probes). Live bacteria showed a green color (Ex, 480 nm; Em, 520 nm), and dead bacteria showed a red color (Ex, 550 nm; Em, 580 nm) under fluorescence.
Hybridization slides were modified with 1016 molecules of anti-B. globigii spore Ab/m2 or 1016 molecules of anti-E. coli O157:H7 Ab/m2 using PEG as a spacer. Labeled B. globigii spores (104 total spores) or labeled E. coli O157:H7 cells (104 total cells) were added to the slides and incubated in the dark for 30 min. Slides were then washed with distilled water, and the captured cells or spores were visualized using a confocal microscope (MRC 1024; Bio-Rad, Hercules, Calif.).
RESULTS
Cross-reactivity of antibodies.
All of the E. coli O157 Ab tested reacted to all strains of E. coli O157:H7. Escherichia coli K-12 cells did not cross-react with any of the E. coli O157 Ab but reacted to their own Ab. Because of this, the monoclonal mouse anti-E. coli K-12 Ab was used as a negative control in the experiments.
Anti-B. globigii spore Ab showed no cross-reactivity with B. circulans, B. cereus, and B. stearothermophilus spores but reacted with the B. globigii spores used to produce the Ab. Thus, the Ab was very specific for B. globigii, not for the major Bacillus strains commonly found in the environment, and was well suited for development of the assay described.
Static capture ability of modified beads.
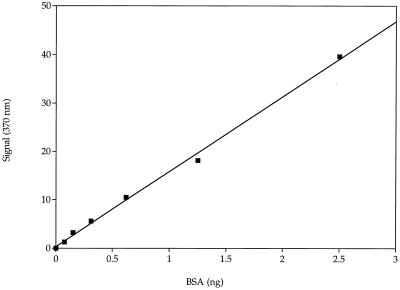
Figure 1 shows the standard curve obtained with anti-BSA-modified immunomagnetic beads and static detection. Ab-modified polystyrene beads, 108 total beads, successfully captured BSA. Very small amounts (<1 ng) of BSA can be detected with these beads. The linear response of the signal to the increase in BSA was 99.7%, which makes this test very sensitive.
FIG. 1.
Standard curve of BSA capture with tosyl-activated poly-Thr-modified immunomagnetic beads. Standard errors of the means are masked by the symbols. The y axis indicates absorbance.
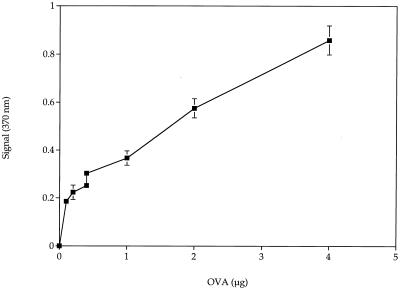
Figure 2 shows the standard curve obtained for 3-mm PEG–anti-OVA-modified beads tested without flow. The lower limit of detection is 0.2 μg. There was a linear response of increased signal to the increase in OVA from 0.2 to 4.0 μg. We did not test beyond 4 μg, because our objective was to develop a sensitive test.
FIG. 2.
Standard curve of OVA capture using 3-mm-diameter PEG-modified glass beads. Error bars represent standard errors of the means.
Flow capture ability of modified beads.
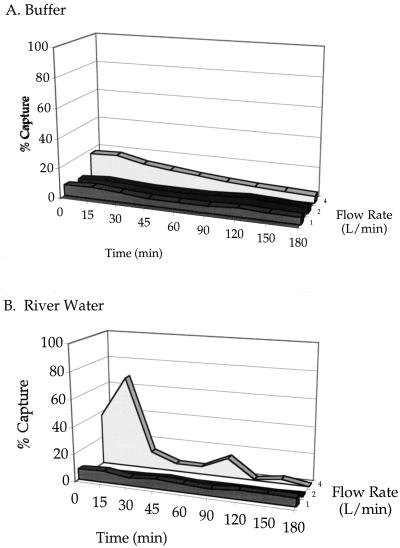
Figure 3 shows the capture of B. globigii spores at various flow rates in a continuous cycle. Figure 3A shows the capture efficiency in 0.25 M sodium phosphate buffer, pH 7.2, and Fig. 3B shows the efficiency in river water. Unmodified beads served as controls and were tested at the same time a sample was collected. Values for controls were subtracted from the sample signal, and the percent capture was calculated. Spores suspended in sodium phosphate buffer were not captured efficiently at the flow rates tested. Only 13% capture was seen within the first 45 min at 4 liters/min, and <10% was observed at lower flow rates. However, the spore count showed that most of the spores were captured at the end of the test period (data not shown). Spores spiked into river water were captured more efficiently. After 15 min, 70% of the spores were captured by the beads, but the capture efficiency dropped to <10% after 15 min.
FIG. 3.
Capture ability of anti-B. globigii-modified polythreonine ceramic beads in sodium phosphate buffer (pH 7.2) (A) and river water (B).
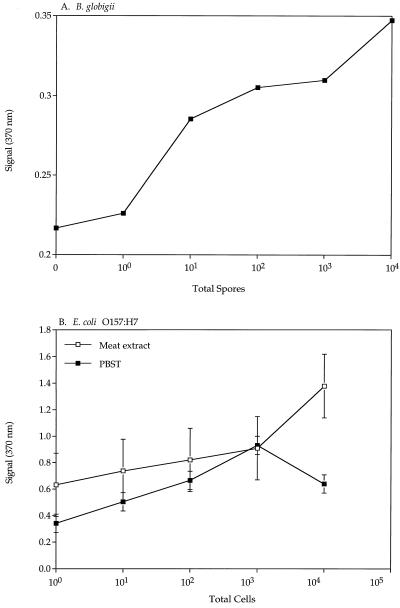
Figure 4A shows the standard curves obtained for B. globigii spore capture at a constant sample flow of 0.4 liters/min using 8 g of 3-mm-diameter PEG-modified glass beads. A linear increase in the signal was observed as the spore concentration increased. Figure 4B shows the ImmunoFlow capture of E. coli O157:H7 at a constant sample flow rate of 0.4 liters/min with 8 g of 3-mm-diameter PEG-modified glass beads. To observe the influence of sample composition, PBST and sterile meat extract were used. Meat extract spiked with cells consistently showed a higher signal than cells spiked into buffer.
FIG. 4.
Standard curve of capture of B. globigii spores in PBST (A) and E. coli O157:H7 in meat extract and PBST (B) using 3-mm-diameter PEG-modified glass beads. Standard errors of the means are masked by the symbols. The y axis indicates absorbance.
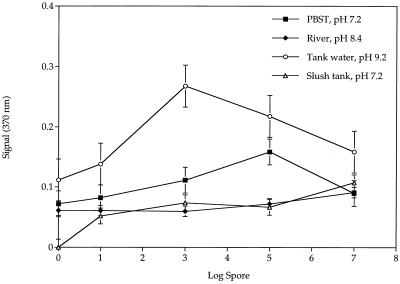
Figure 5 shows the standard curve obtained for B. globigii spores spiked into the various environmental and industrial water samples. All samples were sterilized prior to the addition of B. globigii spores. No linear response to the increased spore concentration was observed using river or slush tank water. This resulted in a constant flat line, as seen in Fig. 5. A linear response was observed using tank water from Gossner's Cheese Plant and PBST. The lower limit of detection for both tank water and PBST was 1 spore/sample. However, the upper limit with PBST was 105 total spores, and for tank water, 103 total spores. All environmental and industrial samples had pHs ranging from 7.2 to 9.2. The beads were active and captured spores over this range.
FIG. 5.
Standard curves of capture of B. globigii spores in environmental and industrial water samples using PEG-modified glass beads. Error bars represent standard errors of the means. The y axis indicates absorbance.
Stability of modified beads.
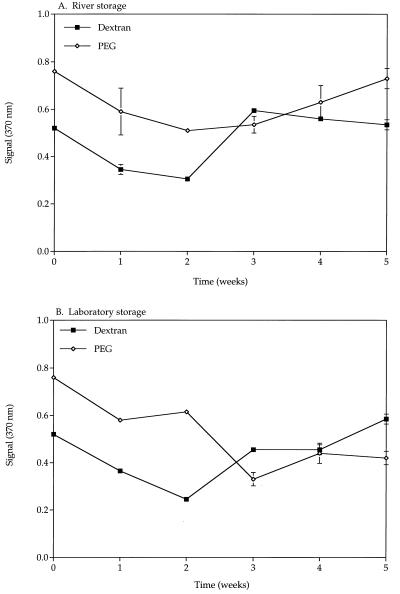
Both dextran- and PEG-modified beads stored in the river and the laboratory lost activity within the first 2 weeks (Fig. 6). The pH and temperature of the river stayed between 8.23 and 8.38 and between 3 and 7°C, respectively. These results led us to repeat the experiment, taking a sample every day for 14 days. After 1 day, the ability of both river and laboratory beads to capture 105 B. globigii spores declined. No further decrease was observed after day 3, and the beads showed similar capturing capabilities through 14 days. Differences in storage temperature (lab, 8°C; river, 1 to 4°C) and pHs (lab, 7.2; river, 8.1 to 8.5) did not seem to influence the stability of the beads.
FIG. 6.
Stability of anti-B. globigii modified glass beads using both dextran and PEG as spacers in river water and (A) the laboratory (B). Error bars represent standard errors of the means. The y axis indicates absorbance.
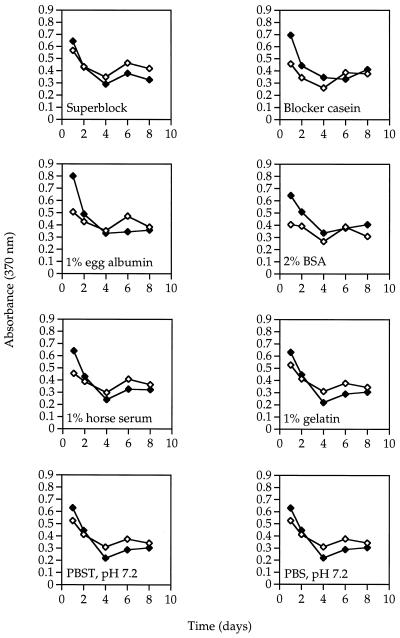
The same decrease in capture ability was observed with anti-E. coli O157:H7 PEG-modified beads. These results led us to investigate the use of storage and blocking buffers (Table 3). All buffers contained 0.02% of an antimicrobial agent, either provided by the company or added by us in the laboratory. Beads were stored at 7 and 25°C. Samples were taken every 2 days for 14 days (Fig. 7). The activity of all beads declined after 2 days, followed by constant activity throughout the test period. The type of blocker and the storage temperature did not influence the capture ability.
TABLE 3.
Storage and blocking buffers used to test 3-mm-diameter anti-E. coli O157:H7 PEG-modified glass beads
| Storage buffera | pH | Company or institution |
|---|---|---|
| Blocker casein | 7.4 | Pierce |
| Superblock Blocking Buffer | 7.4 | Pierce |
| 1% Egg albumin | 7.2 | Sigma Chemicals |
| 2% BSA | 7.2 | Utah State University |
| 1% Horse serum | 7.2 | HyCloneb |
| 1% Gelatin | 7.2 | Sigma Chemicals |
| PBST | 7.2 | Utah State University |
| PBS | 7.2 | Utah State University |
All buffers contained 0.02% thimerosal or an antimicrobial agent supplied by the manufacturer. Beads were stored at 7 and 25°C. All buffers were diluted in PBS unless otherwise stated.
Logan, Utah.
FIG. 7.
Effect of blocking buffers on signal generation using 103 total E. coli O157:H7 cells over time stored at 25°C (⧫) and 7°C (◊). Standard errors of the means are masked by the symbols.
Confocal microscopy.
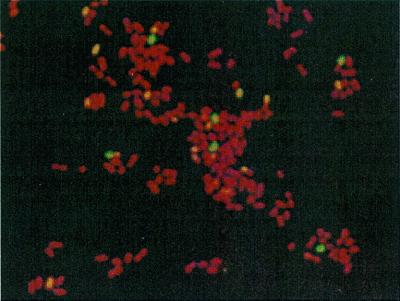
Hybridization slides modified with anti-B. globigii spore Ab and anti-E. coli O157:H7 Ab using PEG were examined with confocal microscopy. B. globigii spores were seen as bright blue oval shapes (Fig. 8), and live E. coli O157:H7 cells were seen as bright green spheres (Fig. 9). The slides captured both spores and cells. Control slides with no Ab attached did not show any capture. We used this technique to verify that the spores and cells were captured by the Ab-modified glass slides and not just physically adsorbed onto them.
FIG. 8.
Confocal image of captured B. globigii spores. Spores are seen as small blue spheres. Slides were coated with PEG and anti-B. globigii Ab. The inset graphically shows the capture of the spore (blue) by an immobilized antibody.
FIG. 9.
Confocal image of captured E. coli O157:H7 cells. Live cells are stained green, and dead cells are stained red. Slides were coated with PEG and anti-E. coli O157:H7 Ab.
DISCUSSION
The rapid detection of microbes in samples such as food is becoming more critical. Our research group has focused on developing a biosensor that is more rapid than and at least as sensitive as currently commercially available microbial tests kits. Currently commercially available kits for detecting bacteria in food rely on a preenrichment step that allows selective enrichment of the test organism, followed by rapid lateral flow ELISA or a PCR test (20, 21, 24). These tests vary in sensitivity but generally require ∼105 to 108 CFU to generate a signal (unpublished data). This level is obtained during the preenrichment step, depending on the cellular state during incubation. Therefore, a need exists to develop a more sensitive test that produces results in a shorter time frame. The objective of this work was to eliminate the enrichment step to allow detection of bacteria directly in samples. Direct comparison of the ImmunoFlow test and commercial tests is currently being studied.
This new method described here, ImmunoFlow, involves the use of specific Ab immobilized onto glass beads (typically 3 mm in diameter) housed in a cartridge to allow flow in a fluidized bed format. The use of a fluidized bed allows real-time testing of samples that contain particulates commonly encountered in food and environmental samples and is independent of sample size. Detection of bound microbes involves the use of existing ELISA protocols.
Since this is an Ab-based method, it is directly dependent on the specificity of the immobilized Ab. Cross-reactivity of commercially available Ab was investigated to select the most species-specific Ab for immobilization. Using two different Ab as the 1° and 2° Ab in an ELISA format generally gives a greater signal than using the same antibody as both the 1° and 2° Ab (16). Therefore, we used each of the E. coli Ab mentioned in Table 1 as 1° Ab and tested them against the remaining three Ab. The combination of the polyclonal KPL Ab as 1° Ab with the monoclonal 3011 Ab as 2° Ab was very specific but gave a low signal. It was found that using KPL as both 1° and 2° Ab produced the strongest signal. Therefore, this Ab combination was used in the development of the ImmunoFlow system for the capture and detection of E. coli O157:H7. One of the problems encountered was the commercial availability of E. coli O157:H7 antibodies. A handful of Ab are available, most of which are monoclonal and thus very specific. The other antibody tested for cross-reactivity, anti-B. globigii Ab, showed no cross-reactivity to the spores tested; therefore, this antibody was used as both the 1° and 2° Ab. The specific capture of B. globigii and E. coli O157:H7 molecules was demonstrated with confocal microscopy (Fig. 8).
The capture of small molecules without flow was demonstrated with BSA and OVA (BSA molecular weight, 66,000; OVA molecular weight, 43,000) with PEG- or polyThr-modified beads (Fig. 1 and 2). BSA and OVA were used as the toxin simulants. Some of the toxins of concern to the food industry are staph and botulinum toxins and aflatoxins. Very small amounts of these toxins will cause the onset of disease; thus, more-sensitive detection methods need to be available. The kits available today will recognize small amounts of toxins, but the test time is 4 h or less. The test described in this paper detects <1 ng of small proteins using Dynalbeads, whereas 3-mm-diameter modified glass beads will detect up to 4 μg. These levels are sensitive enough to detect and quantify the toxin of interest. However, the test becomes inadequate at higher concentrations because BSA will bind nonspecifically to the beads in addition to specific antibody capture (32).
The capture of B. globigii spores from both buffer and river water was demonstrated using 7-mm-diameter ceramic beads by recycling solutions spiked with 106 total spores at various flow rates (Fig. 3). There was variability in the spore capture efficiency with respect to the sample medium and flow rate. Some of this variation can be explained by the size of the sample that was collected. The amount of Ab per surface area is sufficient to capture 106 spores. However, the randomly selected beads taken out as samples and tested for spore capture may or may not have spores attached to them. The optimum sample size would have included testing all the beads in the module and then returning them to the module for continued capture. However, this was not done for practical reasons, because once the test was performed on the beads they could no longer be used for capture.
A comparison of bead size and linker type in relation to the capture of B. globigii was demonstrated with ceramic beads with a polythreonine linker and 3-mm-diameter glass beads with a PEG linker (Fig. 3, 4A, and 5). Both bead types were capable of capturing B. globigii spores, with a linear relationship between the spore count and the signal generated (Fig. 4A). The lower limit of detection was 10 spores, and this limit can potentially be increased by adding more antibody-modified beads for capture. However, the upper limit with PBST was 105 total spores, and for tank water it was 103 total spores. All environmental and industrial samples had pHs ranging from 7.2 to 9.2. The beads were active and captured spores over this range. An unexpected result was found with tank water, because the pH was high, 9.2. At this pH, antibodies begin to denature but the bound antibodies were stable.
The signal drop-offs and plateaus observed within the standard curves (Fig. 4B and 5) can be explained. Overloading Ab-modified beads with cells or spores can create a “hook” effect commonly observed between two competing antibodies (35). Another explanation is the “falling off” (31) of spores or cells that were interacting with the antibodies on the beads. If too many targets are available for potential interaction, they compete for binding sites and thus interfere with one another. This may lead to dissociation between antibody and target. It may be necessary to use several dilutions of a sample to bring it within the detection range when using this test and a sample with an unknown number of cells or spores.
There was no increase in the signal when using slush tank or river water as samples. This may be due to the already high level of other, interfering, spores. This may also help to explain the high background level seen in the control. The protein present in the slush tank, which contained whey and other cheese production byproducts, can coat the surface of the modified beads and thus prohibit the interactions between the capture molecule and the target (32).
The capture of E. coli O157:H7 in flow with 3-mm-diameter glass beads containing a PEG spacer in two sample types was demonstrated (Fig. 4B). In both sample types, there was an increase in the signal with an increase in the number of cells, with a maximum signal generated at 104 cells in meat extract and 103 cells in PBST. These results are similar to what we observed with the capture of B. globigii, in that a true unknown sample may have to be tested at several dilutions to fit within the detection range of this test. There was a higher signal and background level observed in the meat extract sample. This phenomenon may be due to the interaction of the fat and proteins of the meat with the test components. Secondary and tertiary Ab are difficult to wash away completely, and thus nonspecific bindings of these antibodies contribute to the stronger signal generated.
The stability of the Ab beads and the influence of the spacer type were investigated with anti-B. globigii beads and PEG and dextran (Fig. 6.). The PEG beads consistently showed a higher signal and background level than the dextran beads, which is probably due to nonspecific interaction of the 2° and/or 3° Ab with the surface of the beads. Both bead types showed a decrease in activity beginning at day 1. This decrease continued to a plateau at 2 to 3 weeks, at which time the signal indicated that spore capture and detections were still possible under the conditions tested. Immobilized E. coli beads showed a similar decrease in activity from time zero to approximately 4 days (Fig. 7), independent of the blocker type and temperature of storage. The bead activity was then fairly constant from 4 to 8 days.
We then investigated the use of several blocking agents that can be used to reduce nonspecific adsorption of 2° and 3° Ab and maintain activity of the immobilized Ab beads (Fig. 7). We found that the stability of the beads was independent of the blocker type and temperature of storage. Guire (16) investigated the use of several blocking agents to reduce nonspecific binding. He blocked mono- and polyclonal Ab bound to 96-well plates with various blocking agents, dried the plates, and tested for activity over 18 months. Three blocking agents, Stabilguard, Stabilcoat, and Superblock, together with two controls, BSA and PBS, were used. All three blocking agents maintained 70 to 80% activity after 3 months, whereas PBS- and BSA-stored plates only maintained about 10% activity. In our lab the beads were stored in a BSA blocking liquid, and we observed an average 70% loss of activity after 4 days.
Conclusion.
Various surfaces can successfully be modified with antibodies to capture small proteins, spores, and bacterial cells. This capture mechanism can easily be adapted for the capture of other targets, such as small toxins and other food-borne bacteria.
ACKNOWLEDGMENTS
We thank Cultor Food Science for funding this project.
We thank Dugway Proving Grounds, Tooele, Utah, for providing us with B. globigii antibodies and spores.
Footnotes
Approved as journal paper 7244 of the Utah Agricultural Experiment Station.
REFERENCES
- 1.Abdel-Hamid I, Ivnitski D, Atanasov P, Wilkins E. Flow-through immunofiltration assay system for rapid detection of E. coli O157:H7. Biosens Bioelectron. 1999;14:309–316. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 2.Beuchat L R, Nail B V, Adler B B, Clavero M R. Efficacy of spray application of chlorinated water in killing pathogenic bacteria on raw apples, tomatoes, and lettuce. J Food Prot. 1998;61:1305–1311. doi: 10.4315/0362-028x-61.10.1305. [DOI] [PubMed] [Google Scholar]
- 3.Blake M, Weimer B C. Immunomagnetic detection of Bacillus stearothermophilus spores in food and environmental samples. Appl Environ Microbiol. 1997;63:1643–1646. doi: 10.1128/aem.63.5.1643-1646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewster J D, Mazenko R S. Filtration capture and immunoelectrochemical detection for rapid assay of Escherichia coli O157:H7. J Immunol Methods. 1998;211:1–8. doi: 10.1016/s0022-1759(97)00161-0. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan R L, Edelson S G, Snipes K, Boyd G. Inactivation of Escherichia coli O157:H7 in apple juice by irradiation. Appl Environ Microbiol. 1998;64:4533–4535. doi: 10.1128/aem.64.11.4533-4535.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burley S K, Petsko G A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 7.Chapman P A, Cerdan Malo A T, Siddons C A, Harkin M. Use of commercial enzyme immunoassays and immunomagnetic separation systems for detecting Escherichia coli O157 in bovine fecal samples. Appl Environ Microbiol. 1997;63:2549–2553. doi: 10.1128/aem.63.7.2549-2553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavero M R S, Beuchat L R. Suitability of selective plating media for recovering heat- or freeze-stressed Escherichia coli O157:H7 from tryptic soy broth and ground beef. Appl Environ Microbiol. 1995;61:3268–3273. doi: 10.1128/aem.61.9.3268-3273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavero M R S, Beuchat L R. Survival of Escherichia coli O157:H7 in broth and processed salami as influenced by pH, water activity, and temperature and suitability of media for its recovery. Appl Environ Microbiol. 1996;62:2735–2740. doi: 10.1128/aem.62.8.2735-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cray W C, Casey T A, Bosworth B T, Rasmussen M A. Effect of dietary stress on fecal shedding of Escherichia coli O157:H7 in calves. Appl Environ Microbiol. 1998;64:1975–1979. doi: 10.1128/aem.64.5.1975-1979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czajka J, Batt C A. A solid phase fluorescent capillary immunoassay for the detection of Escherichia coli O157:H7 in ground beef and apple cider. J Appl Bacteriol. 1996;81:601–607. doi: 10.1111/j.1365-2672.1996.tb03553.x. [DOI] [PubMed] [Google Scholar]
- 12.deBoer E. Update on media for isolation of Enterobacteriaceae from foods. Int J Food Microbiol. 1998;45:43–53. doi: 10.1016/s0168-1605(98)00146-9. [DOI] [PubMed] [Google Scholar]
- 13.Fratamico P M, Strobaugh T P. Evaluation of an enzyme-linked immunosorbent assay, direct immunofluorescent filter technique, and multiplex polymerase chain reaction for detection of Escherichia coli O157:H7 seeded in beef carcass wash water. J Food Prot. 1998;61:934–938. doi: 10.4315/0362-028x-61.8.934. [DOI] [PubMed] [Google Scholar]
- 14.Gehring A G, Patterson D L, Tu S I. Use of a light-addressable potentiometric sensor for the detection of Escherichia coli O157:H7. Anal Biochem. 1998;258:293–298. doi: 10.1006/abio.1998.2597. [DOI] [PubMed] [Google Scholar]
- 15.Grif K, Dierich M P, Allerberger F. Dynabeads plus 3M Petrifilm HEC versus Vitek Immunodiagnostic Kit Assay System for detection of E. coli O157:H7 in minced meat. Lett Appl Microbiol. 1998;26:199–204. doi: 10.1046/j.1472-765x.1998.00318.x. [DOI] [PubMed] [Google Scholar]
- 16.Guire P E. Stability issues for protein-based in vitro diagnostic products. IVD Technol. 1999;March/April:50–56. [Google Scholar]
- 17.Hammack T S, Feng P, Amaguana R M, June G A, Sherrod P S, Andrews W H. Comparison of sorbitol MacConkey and hemorrhagic coli agars for recovery of Escherichia coli O157:H7 from brie, ice cream, and whole milk. J AOAC Int. 1997;80:335–340. [PubMed] [Google Scholar]
- 18.Hermanson G T. Bioconjugate techniques. New York, N.Y: Academic Press; 1996. Zero length cross-linkers; p. 170. [Google Scholar]
- 19.Janisiewicz W J, Conway W S, Brown M W, Sapers C M, Fratamico P, Buchanan R L. Fate of Escherichia coli O157:H7 on fresh-cut apple tissue and its potential for transmission by fruit flies. Appl Environ Microbiol. 1999;65:1–5. doi: 10.1128/aem.65.1.1-5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson J L, Brooke C L, Fritschel S J. Comparison of the BAX for screening/E. coli O157:H7 method with conventional methods for detection of extremely low levels of Escherichia coli O157:H7 in ground beef. Appl Environ Microbiol. 1998;64:4390–4395. doi: 10.1128/aem.64.11.4390-4395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R P, Durham R J, Johnson S T, MacDonald L A, Jeffrey S R, Butman B T. Detection of Escherichia coli O157:H7 in meat by an enzyme-linked immunosorbent assay, EHEC-Tek. Appl Environ Microbiol. 1995;61:386–388. doi: 10.1128/aem.61.1.386-388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karch H, Janetzki-Mittmann C, Aleksic S, Datz M. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J Clin Microbiol. 1996;34:516–519. doi: 10.1128/jcm.34.3.516-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M S, Doyle M P. Dipstick immunoassay to detect enterohemorrhagic Escherichia coli O157:H7 in retail ground beef. Appl Environ Microbiol. 1992;58:1764–1767. doi: 10.1128/aem.58.5.1764-1767.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie A M R, Lebel P, Orrbine E, Rowe P C, Hyde L, Chan F, Johnson W, McLaine P N the Synsorb Pk Study Investigators. Sensitivities and specificities of Premier E. coli O157:H7 and Premier EHEC enzyme immunoassays for diagnosis of infection with verotoxin (shiga-like toxin)-producing Escherichia coli. J Clin Microbiol. 1998;36:1608–1611. doi: 10.1128/jcm.36.6.1608-1611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.March S B, Ratnam S. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1986;23:869–872. doi: 10.1128/jcm.23.5.869-872.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milley D G, Sekla L H. An enzyme-linked immunosorbent assay-based isolation procedure for verotoxigenic Escherichia coli. Appl Environ Microbiol. 1993;59:4223–4229. doi: 10.1128/aem.59.12.4223-4229.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nauschuetz W. Emerging foodborne pathogens: enterohemorrhagic Escherichia coli. Clin Lab Sci. 1998;11:298–304. [PubMed] [Google Scholar]
- 28.Onoue Y, Konuma H, Nakagawa H, Hara-Kudo Y, Fujita T, Kumagai S. Collaborative evaluation of detection methods for Escherichia coli O157:H7 from radish sprouts and ground beef. Int J Food Microbiol. 1999;46:27–36. doi: 10.1016/s0168-1605(98)00174-3. [DOI] [PubMed] [Google Scholar]
- 29.Padhye N V, Doyle M P. Rapid procedure for detecting enterohemorrhagic Escherichia coli O157:H7 in food. Appl Environ Microbiol. 1991;57:2693–2698. doi: 10.1128/aem.57.9.2693-2698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paunio M, Pebody R, Keskimaki M, Kokki M, Ruutu P, Oinonen S, Vuotari V, Siitonen A, Lahti E, Leinikki P. Swimming-associated outbreak of Escherichia coli O157:H7. Epidemiol Infect. 1999;122:1–5. doi: 10.1017/s0950268898001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawelzik M. Pathogenic Escherichia coli O157:H7 and their detection. Acta Microbiol Hung. 1991;38:315–320. [PubMed] [Google Scholar]
- 32.Pesce A J, Michael J G. Artifacts and limitations of enzyme immunoassay. J Immunol Methods. 1992;150:111–119. doi: 10.1016/0022-1759(92)90070-a. [DOI] [PubMed] [Google Scholar]
- 33.Power C A, McEwen S A, Johnson R P, Shonkri M M, Rahu K, Griffiths M W, De Grandis S A. Repeatability of the Petrifilm HEC test and agreement with hydrophobic grid membrane filtration method for the enumeration of Escherichia coli O157:H7 on beef carcasses. J Food Prot. 1998;61:402–408. doi: 10.4315/0362-028x-61.4.402. [DOI] [PubMed] [Google Scholar]
- 34.Pyle B H, Broadaway S C, McFeters G A. Sensitive detection of Escherichia coli O157:H7 in food and water by immunomagnetic separation and solid-phase laser cytometry. Appl Environ Microbiol. 1999;65:1966–1972. doi: 10.1128/aem.65.5.1966-1972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo K H, Brackett R E, Frank J F. Rapid detection of Escherichia coli O157:H7 using immunomagnetic flow cytometry in ground beef, apple juice, and milk. Int J Food Microbiol. 1998;44:115–123. doi: 10.1016/s0168-1605(98)00129-9. [DOI] [PubMed] [Google Scholar]
- 36.Seo K H, Brackett R E, Frank J F, Hillard S. Immunomagnetic separation and flow cytometry for rapid detection of Escherichia coli O157. J Food Prot. 1998;61:812–816. doi: 10.4315/0362-028x-61.7.812. [DOI] [PubMed] [Google Scholar]
- 37.Tomoyasu T. Improvement of the immunomagnetic separation method selective for Escherichia coli O157 strains. Appl Environ Microbiol. 1998;64:376–382. doi: 10.1128/aem.64.1.376-382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tortorello M L, Stewart D S. Antibody-directed epifluorescent filter technique for rapid, direct enumeration of Escherichia coli O157:H7 in beef. Appl Environ Microbiol. 1994;60:3553–3559. doi: 10.1128/aem.60.10.3553-3559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortorello M L, Reineke K F, Stewart D S, Raybourne R B. Comparison of methods for determining the presence of Escherichia coli O157:H7 in apple juice. J Food Prot. 1998;61:1425–1430. doi: 10.4315/0362-028x-61.11.1425. [DOI] [PubMed] [Google Scholar]
- 40.Tsukamoto T, Kawai T, Takeda Y. Evaluation of commercial kits for a rapid detection of Escherichia coli O157 in stool. Kanenshogaky Zasshi. 1998;72:40–44. doi: 10.11150/kansenshogakuzasshi1970.72.40. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Doyle M P. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J Food Prot. 1998;61:662–667. doi: 10.4315/0362-028x-61.6.662. [DOI] [PubMed] [Google Scholar]
- 42.Weimer B C, Walsh M K, Wang X. Influence of polyethylene glycol spacer on antigen capture by immobilized antibodies. Biochem Biophys Methods. 2000;45:211–219. doi: 10.1016/s0165-022x(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 43.Woody J M, Stevensson J A, Wilson R A, Knabel S J. Comparison of the DifcoEZ Coli rapid detection system and Petrifilm test kit-HEC for detection of Escherichia coli O157:H7 in fresh and frozen ground beef. J Food Prot. 1998;61:110–112. doi: 10.4315/0362-028x-61.1.110. [DOI] [PubMed] [Google Scholar]
- 44.Wright D J, Chapman P A, Siddons C A. Immunomagnetic separation as a sensitive method for isolating Escherichia coli O157:H7 from food samples. Epidemiol Infect. 1994;113:31–39. doi: 10.1017/s0950268800051438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao T, Doyle M P, Harmon B G, Brown C A, Mueller P O E, Parks A H. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J Clin Microbiol. 1998;36:641–647. doi: 10.1128/jcm.36.3.641-647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]