Abstract
Motivation
The development of algorithms dedicated to RNA three-dimensional (3D) structures contributes to the demand for training, testing and benchmarking data. A reliable source of such data derived from computational prediction is the RNA-Puzzles repository. In contrast, the largest resource with experimentally determined structures is the Protein Data Bank. However, files in this archive often contain other molecular data in addition to the RNA structure itself, which—to be used by RNA processing algorithms—should be removed.
Results
RNAsolo is a self-updating database dedicated to RNA bioinformatics. It systematically collects experimentally determined RNA 3D structures stored in the PDB, cleans them from non-RNA chains, and groups them into equivalence classes. It allows users to download various subsets of data—clustered by resolution, source, data format, etc.—for further processing and analysis with a single click.
Availability and implementation
The repository is publicly available at https://rnasolo.cs.put.poznan.pl.
1 Introduction
RNA molecules constitute a rich, heterogeneous universe—both at a structural and functional level. They are a fascinating object of basic and applied research in many scientific disciplines. A significant fraction of this research focuses on the tertiary [three-dimensional (3D)] structure. Scientists look for its relationship to intermolecular interactions and, in the longer term, to the molecule’s role in the organism at the molecular and cellular levels. They also try to predict the 3D structure and design molecules with predefined properties.
Computer algorithms specialized for processing structural data are a great help in such studies. Their correctness and precision depend highly on reliable training and test sets (Carrascoza et al., 2022; Popenda et al., 2021). Well-structured datasets also help to perform comparative analyses of different computational methods. Such data collections should be sufficiently numerous and contain non-redundant, representative information selected appropriately for the problem solved. The primary source of reliable structural data is the Protein Data Bank (Berman et al., 2000) that collects molecular structures determined by various experimental methods. Here, researchers interested in ribonucleic acid molecules can find naked (solo) RNAs, protein–RNA complexes and DNA–RNA hybrids. In turn, the repository created by the RNA-Puzzles initiative makes available RNA 3D structures predicted by various state-of-the-art computational methods (Magnus et al., 2020). Searching one of these archives often starts the process of creating a training set (to train an ML algorithm), a test- or a benchmark set (to verify the quality and accuracy of a new algorithm or compare it with the state-of-the-art ones). Found data are usually clustered, stripped of redundancy, cleaned of metadata and non-RNA data, and then supplemented according to the assignment of a collection. Such processing is relatively easy for in silico models from the RNA-Puzzles repository since they are standardized and grouped by challenge and computational method. In contrast, organizing a set of PDB structures requires additional operations and resources—for example, searching the BGSU RNA site (Leontis and Zirbel, 2012) that provides a list of non-redundant RNA 3D structures. Examples of archives created using a similar procedure include RNABase, no longer maintained (Murthy and Rose, 2003) or the recently published RNANet, which collects sequences and structures of RNA homologs (Becquey et al., 2021).
In this work, we respond to the necessity for fast and easy, automatic creation of sets of RNA 3D structures to train, test and benchmark bioinformatics algorithms. We present RNAsolo, designed to systematically collect experimentally determined RNA 3D structures stored in the Protein Data Bank (Berman et al., 2000), clean them from non-RNA data, annotate and assign them to equivalence classes according to Leontis and Zirbel (2012). Its primary advantage is the ability to select RNA structures of interest and download them as a dataset ready for further processing—all with a single click. The RNAsolo database has been freely accessible online since July 2021. It automatically updates every Thursday. As part of each update, 192 benchmark sets are prepared as ZIP archives, so the users have them ready at a glance.
2 Materials and methods
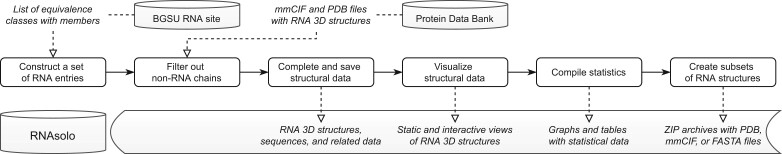
Data processing in the RNAsolo system consists of six steps: primary data collection, non-RNA information removal, structure data completion and populating the database, data visualization, statistics compilation and ZIP archives creation (Fig. 1). At first, RNAsolo connects to the BGSU RNA site (Leontis and Zirbel, 2012). This webpage once a week publishes a list of equivalence classes of PDB-deposited RNA structures. They aim to support building benchmark sets of RNA structures by filtering redundancies that could bias the results while retaining the sequence variation. The classes are defined based on a pairwise analysis of structural redundancy. In general, two RNAs are non-redundant if they come from different species. If associated with the same organism, they may or may not be redundant—this is decided based on sequence comparison and structural superposition focused on geometrics-based similarities and differences. Every class has a representative selected following the three criteria: the number of FR3D-annotated base pairs per nucleotide (Sarver et al., 2008), experimental resolution and release date. High-resolution structures with more recent publication time are preferred (Leontis and Zirbel, 2012). From the BGSU RNA site, we retrieve information about changes in equivalence classes resulting from the recent update of the Protein Data Bank (Berman et al., 2000). Classes are analyzed separately for each resolution cutoff x, where x {1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 20.0}. Additionally, a list of all equivalence classes independent of resolution is retrieved. Based on this information, the RNAsolo procedure downloads from the PDB archive new and modified RNA structures (in mmCIF and PDB format if available) and dereferences entries deleted from the PDB. Assignments to equivalence classes in the RNAsolo system are automatically updated. Then, the cleaning procedure removes non-RNA chains from all downloaded files. In the next step, the system prepares a set of structural data (sequence, 3D structure, number of residues, model and chain identifiers—in the latter case, for consistency between mmCIF and PDB formats, author asym_id is preferred over label asym_id) and metadata (source structure and title) and populates the database. Wherever it is required, the RNAsolo procedure adds symmetry operators. They are extracted from the source mmCIF data, transformed into the PDB format, and saved in the RNAsolo system in the mmCIF and PDB files. If a PDB file does not exist for some RNA in the Protein Data Bank, we produce one for the cleaned tertiary structure if possible (i.e. all restraints of the PDB format are met) and add it to the RNAsolo resources. Note, however, that not all structures can be saved in the PDB format, and therefore the set of PDB files in the RNAsolo database may be less numerous than the corresponding mmCIF set.
Fig. 1.
Data flow in the RNAsolo system
For each RNA molecule, the system creates a Pymol-based, static 3D structure image and visualizes it in Mol* viewer (Sehnal et al., 2021). Basic statistics about equivalence classes and structures in different subgroups are collected and visualized. Finally, RNAsolo creates 192 ZIP archives. They contain different subsets of RNA data grouped by the data format (3D structure in mmCIF or PDB, sequence in FASTA), molecule classification (solo RNA molecules, RNAs from protein–RNA complexes, RNAs from DNA–RNA hybrids, all molecules), redundancy (representatives or all members of equivalence classes) and resolution (1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 20.0, all). The resolution cutoff values have been adopted from the BGSU representative sets.
RNAsolo has a multi-layer architecture. Front-end, implemented in TypeScript with React.js and Ant Design libraries, is served by the Nginx web server. Its responsive UI supports all modern web browsers and platforms, including mobile devices. The back-end layer, written in Python3, uses the Django framework. It integrates a relational database provided by PostgreSQL and Celery—a queueing system for reliable execution of cyclic operations, like database updates. Our Python and BioPython scripts are applied to download data files, preprocess RNA structures and filter non-RNA data.
3 Results
The RNAsolo database is updated every Thursday. All changes are recorded in the Update log accessible from the main menu. The distribution of data is illustrated by graphs and tables on the Database statistics page. The database currently contains 12 914 RNA tertiary structures, including 2101 solo RNAs, 10 694 RNAs from protein–RNA complexes and 119 from DNA–RNA hybrids, determined in different experiments (Table 1). As we rely on representative datasets from the BGSU RNA site (Leontis and Zirbel, 2012), we adopt its understanding of a structure. Thus, the RNA 3D structure files in RNAsolo store individual chains or sometimes multiple chains kept together by the BGSU site. On the other hand, the PDB files may contain more than one structure. It causes that the structure counter in RNAsolo indicates a higher value than in PDB. Structures in RNAsolo are clustered in 3271 equivalence classes counting from 1 to 564 RNAs per class (as of January 5, 2022). A table presented on the RNAsolo Home page lists equivalence classes along with the details of their representatives. It allows downloading sequences or 3D structures of all members of a class (in a ZIP archive) or its representative. Users can sort the table by selected columns, show or hide columns, run a simple search through the table, and save it to the CSV file. On the Search the data page, they can run the database search by PDB identifiers, the minimum and maximum size of a structure (total number of residues), or search for keywords in metadata that describe the entity—such as title and source. Users can save the structures found (all of them or their subset) and all members of the associated equivalence classes in one archive by selecting the checkboxes next to the records of interest and clicking the appropriate download button (FASTA, PDB, CIF) on the results panel. Such a ZIP archive is generated on the fly. Other data collections—192 sets prepared with the database update—can be accessed directly from the Download archive page. Users can get any of them after determining the content of interest based on four criteria: data format, molecule classification, redundancy and experimental resolution. Separately, 64 TXT files listing the instances in every archive are available. Each list contains entries of the form: PDB ID_model id_chain id. Additionally provided web service facilitates data exchange with the RNAsolo database bypassing the GUI. It allows users to download clean RNA 3D structure indicated by PDB id, PDB id + model number or PDB id + model + chain number(s).
Table 1.
RNAsolo contents according to the experimental method (January 5, 2022)
| Solo RNAs | RNAs from protein–RNA complexes | RNAs from DNA–RNA hybrids | All RNAs | |
|---|---|---|---|---|
| X-ray | 1454 | 6439 | 91 | 7984 |
| NMR | 573 | 146 | 28 | 747 |
| Electron microscopy | 73 | 4104 | 0 | 4177 |
| Multi-method | 1 | 5 | 0 | 6 |
| Total | 2101 | 10 694 | 119 | 12 914 |
4 Conclusions
RNA applications in biomedicine and biotechnology have raised the need to learn this molecule structure and explore its properties. The Protein Data Bank (Berman et al., 2000) currently stores 1586 solo RNAs, 4070 protein–RNA complexes and 96 DNA–RNA hybrids ready to explore (data as of January 5, 2022). Including the two latter subsets, one gets quite a satisfying amount of structural data—5752 PDB structures. However, to process them with a focus on RNA, it is necessary to clean up the structures of complexes and hybrids from non-RNA chains. So far, no online tool existed that could automatically extract naked RNAs from multi-molecule PDB files and make them available to the users. When needed, we used homemade scripts for data cleaning to enable their processing by the RNApolis tools (Szachniuk, 2019). Based on this experience, we have developed RNAsolo, a system that collects cleaned RNA structures, clusters them into equivalence classes, makes them searchable and allows users to create diverse datasets for further study. Although its current functionality is quite broad, the extensions are possible. They include expanding the database scheme to allow storage of the secondary structure, adding new filtering criteria to the RNAsolo search engine and developing additional functions to collect data statistics. We also plan to broaden the scope of web services to facilitate the work of users automatically processing a wide variety of structural data.
Acknowledgements
The authors thank the anonymous reviewers for the comments, which contributed to a significant improvement of the RNAsolo webserver.
Funding
We acknowledge support from the Poznan University of Technology (statutory funds), Institute of Bioorganic Chemistry PAS and the National Science Centre, Poland [2019/35/B/ST6/03074].
Conflict of Interest: none declared.
Contributor Information
Bartosz Adamczyk, Institute of Computing Science, Poznan University of Technology, 60-965 Poznan, Poland.
Maciej Antczak, Institute of Computing Science, Poznan University of Technology, 60-965 Poznan, Poland; Institute of Bioorganic Chemistry, Polish Academy of Sciences, 61-704 Poznan, Poland.
Marta Szachniuk, Institute of Computing Science, Poznan University of Technology, 60-965 Poznan, Poland; Institute of Bioorganic Chemistry, Polish Academy of Sciences, 61-704 Poznan, Poland.
References
- Becquey L. et al. (2021) RNANet: an automatically built dual-source dataset integrating homologous sequences and RNA structures. Bioinformatics, 37, 1218–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. et al. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascoza F. et al. (2022) Evaluation of the stereochemical quality of predicted RNA 3D models in the RNA-Puzzles submissions. RNA, 28, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N., Zirbel C. (2012) Nonredundant 3D Structure Datasets for RNA Knowledge Extraction and Benchmarking. In: Leontis,N and Westhof,E. (eds.) Nucleic Acids and Molecular Biology. Springer, Berlin Heidelberg, pp. 281–298. [Google Scholar]
- Magnus M. et al. (2020) RNA-Puzzles toolkit: a computational resource of RNA 3D structure benchmark datasets, structure manipulation, and evaluation tools. Nucleic Acids Res., 48, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V., Rose G. (2003) RNABase: an annotated database of RNA structures. Nucleic Acids Res., 31, 502–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M. et al. (2021) Entanglements of structure elements revealed in RNA 3D models. Nucleic Acids Res., 49, 9625–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver M. et al. (2008) FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol., 56, 215–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D. et al. (2021) Mol Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res., 49, W431–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szachniuk M. (2019) RNApolis: computational platform for RNA structure analysis. FCDS, 44, 241–257. [Google Scholar]