Abstract
Summary
Diversity of the T-cell receptor (TCR) repertoire is central to adaptive immunity. The TCR is composed of α and β chains, encoded by the TRA and TRB genes, of which the variable regions determine antigen specificity. To generate novel biological insights into the complex functioning of immune cells, combined capture of variable regions and single-cell transcriptomes provides a compelling approach. Recent developments enable the enrichment of TRA and TRB variable regions from widely used technologies for 3′-based single-cell RNA-sequencing (scRNA-seq). However, a comprehensive computational pipeline to process TCR-enriched data from 3′ scRNA-seq is not available. Here, we present an analysis pipeline to process TCR variable regions enriched from 3′ scRNA-seq cDNA. The tool reports TRA and TRB nucleotide and amino acid sequences linked to cell barcodes, enabling the reconstruction of T-cell clonotypes with associated transcriptomes. We demonstrate the software using peripheral blood mononuclear cells from a healthy donor and detect TCR sequences in a high proportion of single T cells. Detection of TCR sequences is low in non-T-cell populations, demonstrating specificity. Finally, we show that TCR clones are larger in CD8 Memory T cells than in other T-cell types, indicating an association between T-cell clonotypes and differentiation states.
Availability and implementation
The Workflow for Association of T-cell receptors from 3′ single-cell RNA-seq (WAT3R), including test data, is available on GitHub (https://github.com/mainciburu/WAT3R), Docker Hub (https://hub.docker.com/r/mainciburu/wat3r) and a workflow on the Terra platform (https://app.terra.bio). The test dataset is available on GEO (accession number GSE195956).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
During T-cell development, a series of recombinations shape the α and β chains that comprise the T-cell receptor (TCR), giving rise to 1015–1021 potential TCRs (La Gruta et al., 2018). The recombinations occur in the variable regions of the TRA and TRB genes that encode the TCR α/β chains. TRA and TRB determine T-cell specificity by shaping TCR recognition of antigens presented by major histocompatibility complex molecules. The recombined sequences can also be used to track T-cell clonotypes. With the advent of single-cell sequencing technologies, simultaneous capture of TRA and TRB sequences combined with transcriptional states provides a powerful approach to studying T-cell biology (Ginhoux et al., 2022).
Several protocols have been developed to combine single-cell RNA-sequencing (scRNA-seq) and TCR sequencing, using cell barcodes to integrate both layers of information. Low-throughput single-cell methods with full-length transcript coverage allow for the recovery of complete TRA and TRB transcripts (Sade-Feldman et al., 2018; Stubbington et al., 2016). Current high-throughput scRNA-seq assays generate sequencing data that are biased toward either the 5′ or the 3′ end of transcripts. The 10× Genomics 5′ scRNA-seq platform provides some coverage of the TCR variable region that can be reconstructed using TRUST4 or, alternatively, further enriched using 10× Genomics TCR amplification (Lowery et al., 2022; Oliveira et al., 2021; Song et al., 2021). In contrast, 3′ scRNA-seq platforms provide poor coverage of the TCR variable region, although this too can be improved with specific TCR amplification (Oh et al., 2020; Singh et al., 2019; Tu et al., 2019). For TCR enrichment from the 10× Genomics 3′ scRNA-seq platform, we recently established the T-cell Receptor Enrichment to linK clonotypes (TREK-seq) protocol (Supplementary Fig. S1; DePasquale et al., 2022; Miller et al., 2022). There is a need for bioinformatics tools that facilitate the analysis of TCR variable regions enriched from 3′ scRNA-seq. Here, we describe WAT3R (pronounced ‘water’), an integrated pipeline that covers TCR-enriched data from preprocessing FASTQ files to alignment and the identification of T-cell clones.
2 Description
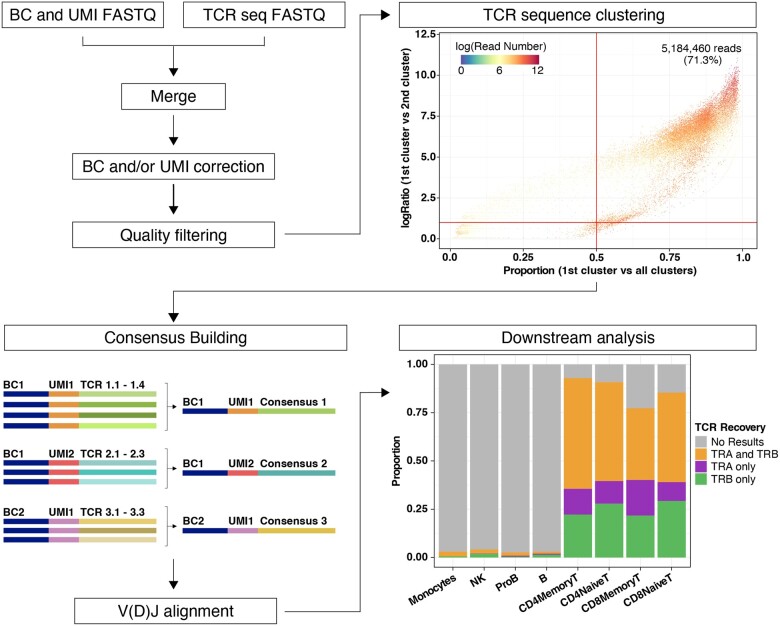
Sequencing data are provided as two compressed FASTQ files, in accordance with the TREK-seq protocol (DePasquale et al., 2022; Miller et al., 2022). One contains the cell barcode and unique molecular identifier (UMI) sequences and the other contains the TCR sequence. First, files are reformatted to join the barcode, UMI and corresponding TCR sequence in a single FASTQ. Next, the user can specify whether barcode and UMI correction should be performed. Barcode correction allows for one mismatch with barcodes in the 10× Single Cell 3′ v3 list (or a custom list provided by the user). UMI correction is performed by clustering together UMIs with one mismatch and considering the most abundant UMI as the correct one. Every barcode and UMI sequence are then added to the corresponding FASTQ read header, to be used as an identifier. Next, we apply a quality filter to remove every read with an average quality score lower than indicated (default is q score <25, Fig. 1, top left). To account for barcode swapping (i.e. incorrect barcode assigning), TCR sequences with an identical barcode and UMI are subjected to clustering based on sequence similarity, using the USEARCH algorithm (Edgar, 2010). The identity threshold to measure similarity can be set by the user (default is 0.9). To reduce the impact of technical artifacts such as barcode swapping (Griffiths et al., 2018), only TCR sequences are kept if the most abundant cluster represents a large proportion of the reads (default is 0.5) and is substantially larger than the second most abundant cluster (default ratio is 2.0; Fig. 1, top right).
Fig. 1.
Overview of WAT3R. The workflow starts by merging two FASTQ files, correction of cell barcodes and UMIs and quality filtering (top left). Clustering of TCR sequences with identical barcode and UMI is then performed. Top right dot plot shows evaluation of cluster quality by comparing the proportion of reads supporting the most abundant cluster (x-axis), the ratio of the most abundant cluster to the second (y-axis) and the number of reads supporting each TCR sequence with a specific barcode and UMI (color). Bottom left: TCR consensus sequences are generated and used for V(D)J alignment. In downstream analysis, results are integrated with a paired scRNA-seq dataset. Bottom right bar plot shows the proportion of cells in the dataset, separated by cell type, for which WAT3R returned information on the TRA gene, TRB gene or both. (A color version of this figure appears in the online version of this article.)
Next, a consensus sequence is built for each of the clusters (Heiden et al., 2014). For a consensus to be constructed, we require a minimum of three reads and allow for a maximum error rate of 0.5 and a gap frequency of 0.5 per position; these parameters can be changed by the user (Fig. 1, bottom left). Consensus sequences are aligned to the V(D)J segments reference provided by IMGT, using IgBLAST with the default parameters (Lefranc et al., 2015; Ye et al., 2013). This task is performed through the interface implemented in the Python package Change-O (Gupta et al., 2015). After selecting TRA and TRB sequences with the highest UMI counts, the CDR3 nucleotide/amino acid sequences and V(D)J calls are assigned to cell barcodes and saved in a results table.
As an optional step, the user can provide a file with cell barcodes and annotations coming from a paired scRNA-seq experiment to integrate with the TRA and TRB calls (Fig. 1, bottom right). Overall, this pipeline returns two tables of results, one at the transcript level and the other at the cell level. In addition, multiple quality control (QC) graphs and metrics are generated. WAT3R, together with the required software, reference data and documentation are available as a docker image. It is also available as a workflow on Terra, which provides access to Google Cloud computing resources through a simple web-based user interface.
3 Results
We analyzed a human peripheral blood sample using 10× Genomics 3′ v3 scRNA-seq and TREK-seq to enrich TRA and TRB variable regions (Miller et al., 2022). The TREK-seq library was sequenced on a MiSeq to a depth of 8 million reads, and WAT3R took 5–8 h to run given moderate resources (Supplementary Table S1). After recovering 4.3% of the cell barcodes using the barcode correction algorithm, 98.3% of reads contained valid barcodes (Supplementary Fig. S2). Likewise, 4.9% of the UMI sequences were corrected. Reads were filtered for an average q score above 25, which retained 92.3% of the original reads. For consensus building and subsequent alignment, 70.4% of the reads were valid. After the removal of TCR sequence clusters below the proportion and ratio thresholds, 65.8% of the original reads were retained. The results were stable with different parameters (Supplementary Table S2). WAT3R generates a results table with TCR nucleotide and amino acid sequences, allele information and quality metrics (Supplementary Table S3).
We integrated these results with the paired 3′ scRNA-seq dataset with cell-type annotations based on canonical marker genes (Supplementary Fig. S3; Hao et al., 2021). We detected TRA or TRB sequences in 90% of all T cells, which is similar to alternative protocols (Supplementary Fig. S4; DePasquale et al., 2022; Lowery et al., 2022; Oh et al., 2020; Oliveira et al., 2021; Tu et al., 2019). In contrast, TRA or TRB sequences are present in only 3% of non-T cells, which was partially explained by artifacts in the cDNA library (Supplementary Fig. S5). Specificity was further confirmed by running WAT3R on OT-I mouse T cells, which express transgenic Tcra and Tcrb genes (Supplementary Fig. S6; Blüthmann et al., 1988; Gu et al., 2014; Tu et al., 2019). The TRB variable region is most efficiently enriched: in the peripheral blood mononuclear cells, TRA sequences are detected in 65% of single T cells, TRB in 77% and TRA+TRB in 52% (Fig. 1, bottom right). As expected in a healthy individual, we did not observe any expanded T-cell clones dominating the sample. Nonetheless, the largest detected clones belong mainly to CD8 Memory T-cell subsets, in accordance with previous findings (Supplementary Fig. S7; DePasquale et al., 2022; Penter et al., 2021).
4 Conclusions
WAT3R is a comprehensive pipeline for the analysis of TRA and TRB variable regions enriched from the cDNA of widely used 3′ scRNA-seq protocols. In combination with the TREK-seq protocol, this enables TCR recovery that rivals the 10× 5′ immune profiling platform (Supplementary Fig. S4). From sequencing error correction, alignment and QCs to intersection with cell-type annotations from scRNA-seq, this tool applies state-of-the-art algorithms to reliably detect T-cell clonotypes and initiate new discoveries in immunology.
Supplementary Material
Acknowledgements
We thank the healthy donor for donating peripheral blood cells. We thank Julia Verga, Tyler Miller, Martin Villanueva, Charles Couturier, Daniel Ssozi, Jonathan Good, Jenny Noel and Alex Shalek for help with the TREK-seq protocol development and Yoke Seng Lee and Antonia Kreso for helpful feedback.
Funding
P.v.G. is supported by the Ludwig Center at Harvard, the NIH (R00CA218832), Gilead Sciences, the Bertarelli Rare Cancers Fund, the William Guy Forbeck Research Foundation, and is an awardee of the Glenn Foundation for Medical Research and American Federation for Aging Research (AFAR) Grant for Junior Faculty. M.A. is supported by a PhD fellowship (FPU18/05488) and a mobility scholarship from the Government of Spain.
Conflict of interest: J.C.L. and the Massachusetts Institute of Technology have filed patents related to TREK-seq.
Contributor Information
Marina Ainciburu, Program of Hemato-Oncology, University of Navarra, Pamplona 31008, Spain; Division of Hematology, Brigham and Women's Hospital, Boston, MA 02115, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Duncan M Morgan, Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Erica A K DePasquale, Division of Hematology, Brigham and Women's Hospital, Boston, MA 02115, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
J Christopher Love, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Felipe Prósper, Program of Hemato-Oncology, University of Navarra, Pamplona 31008, Spain.
Peter van Galen, Division of Hematology, Brigham and Women's Hospital, Boston, MA 02115, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Ludwig Center at Harvard, Harvard Medical School, Boston, MA 02115, USA.
References
- Blüthmann H. et al. (1988) T-cell-specific deletion of T-cell receptor transgenes allows functional rearrangement of endogenous alpha- and beta-genes. Nature, 334, 156–159. [DOI] [PubMed] [Google Scholar]
- DePasquale E.A.K. et al. (2022) Single-cell multiomics reveals clonal T-cell expansions and exhaustion in blastic plasmacytoid dendritic cell neoplasm. Front. Immunol., 13, 809414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Ginhoux F. et al. (2022) Single-cell immunology: past, present, and future. Immunity, 55, 393–404. [DOI] [PubMed] [Google Scholar]
- Griffiths J.A. et al. (2018) Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun., 9, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. et al. (2014) Circlize implements and enhances circular visualization in R. Bioinformatics, 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- Gupta N.T. et al. (2015) Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics, 31, 3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y. et al. (2021) Integrated analysis of multimodal single-cell data. Cell, 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden J.A.V. et al. (2014) pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics, 30, 1930–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Gruta N.L. et al. (2018) Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol., 18, 467–478. [DOI] [PubMed] [Google Scholar]
- Lefranc M.-P. et al. (2015) IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res., 43, D413–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery F.J. et al. (2022) Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science, 375, 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T.E. et al. (2022) Mitochondrial variant enrichment from high-throughput single-cell RNA-seq resolves clonal populations. Nat. Biotechnol. https://www.ncbi.nlm.nih.gov/pubmed/35210612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.Y. et al. (2020) Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell, 181, 1612–1625.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G. et al. (2021) Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature, 596, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penter L. et al. (2021) Coevolving JAK2V617F+ relapsed AML and donor T cells with PD-1 blockade after stem cell transplantation: an index case. Blood Adv., 5, 4701–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M. et al. (2018) Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell, 175, 998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. et al. (2019) High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun., 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. et al. (2021) TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods, 18, 627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington M.J.T. et al. (2016) T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods, 13, 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu A.A. et al. (2019) TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures. Nat. Immunol., 20, 1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. et al. (2013) IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res., 41, W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.