Abstract
The environmental condition is a critical regulation factor for protein behavior in solution. Several studies have shown that macromolecular crowders can modulate protein structures, interactions, and functions. Recent publications described the regulation of specific interaction by macromolecular crowders. However, the other category of protein–protein interaction, namely, the transient interaction, is rarely investigated, especially from the perspective of protein structure to study transient interactions between proteins. Here, we used nuclear magnetic resonance and small-angle X-ray/neutron scattering methods to structurally investigate the ensemble of the protein complex in dilute buffer and crowded environments. Histidine phosphocarrier protein (HPr) and the N-terminal domain of enzyme I (EIN) are the important components of the bacterial phosphotransfer system. Our results show that the addition of Ficoll-70 promotes HPr molecules to form the encounter complex with EIN maintained by long-range electrostatic interaction. However, when macromolecular crowder BSA is used, the soft interaction between BSA and HPr perturbs the active site of HPr, driving HPr to form an encounter complex with EIN at the weakly charged interface. Our results indicate that different macromolecular crowders could influence transient EIN-HPr interaction through different mechanisms and provide new insights into protein–protein interaction regulation in native environments.
Introduction
The intracellular environment is where proteins interact with their partners to perform biological functions. It is a complex and crowded environment consisting of hundreds of small molecules and thousands of proteins.1 Crowded environments generated by synthetic or natural macromolecules are usually used to mimic the cell interior.2−4 Several protein structures have been studied under such conditions to explore the regulation of the behavior of proteins by the environment.5−8 In a crowded environment, macromolecular crowders occupy a large portion of the available space in the solution. The interaction between the test protein and the background molecules is generally divided into hard-core repulsion and soft interactions.9 The hard-core repulsion between the test protein and macromolecular crowder results in a relatively simple excluded volume effect. The soft interactions refer to the chemical interactions between the test protein and the crowding molecules. Several studies suggest that the presence of the macromolecular crowder would cause various effects on the formation of protein complexes.10−12
The protein–protein interactions are critical to the biological functions, and they are classified into two types: (1) specific interactions and (2) transient interactions.13,14 Specific interactions are the main force for holding specific protein complexes, in which the subunits are uniquely orientated with low interaction energies. The specific complex of proteins is the mode for performing certain biological functions.15 It is evident that the encounter complexes, formed via transient interactions, are essential to forming a specific protein complex in solution. The subunits of an encounter complex are arrayed in a wide range of orientations with multiple interfaces. Although encounter complexes are typically lowly populated due to the transient interprotein interactions, they are believed to be the precursor of the specific complex.16 The encounter complex has been visualized in a simple buffer using the paramagnetic relaxation enhancement (PRE) and chemical cross-linking mass spectrometry (CXMS) methods,13,17−19 but regulation of the transient interaction by different crowding molecules is rarely structurally investigated. Therefore, how the encounter complex behaves in a crowding environment is still not well-answered.
Histidine phosphocarrier protein (HPr) and the N-terminal domain of enzyme I (EIN) are the essential components of the bacterial phosphotransfer system (PTS), which plays a role in bacterial sugar uptake.20 In PTS, EIN interacts with HPr to transfer a phosphoryl group of phosphoenolpyruvate at the initial steps of the total phosphoryl transfer.21 Since the EIN-HPr encounter complex was visualized in 2006 using PRE methods, this protein complex became a classic system for NMR study on protein–protein transient interaction.18,22,23 With the ability of PRE to detect lowly populated protein complexes, it has been shown that the behaviors of EIN-HPr encounter complex are dependent on salt concentration and ligand concentration. This finding emphasizes the solution condition’s regulation of the transient interprotein interaction.
Here, to investigate how the crowding molecules affect the interprotein interactions, we used macromolecules, Ficoll-70 and BSA as the crowding agents, and the protein complex of HPr-EIN under the crowded environment structurally studied using the PRE method and the SANS method.24−26 These two methods can selectively detect target proteins in a complex solution, and their results could be cross-validated with each other. Our findings indicate that although the EIN-HPr specific complex is weakly sensitive to changes in environmental conditions, the solution condition is a critical factor in the behaviors of the encounter complex. Our results show that Ficoll-70 and BSA would selectively influence the transient interactions between EIN and HPr, and different crowding agents differentially modulate the clustering of their encounter complexes. This provides a novel regulation mechanism of interprotein interaction by macromolecular crowders, and it expands the knowledge of biophysical processes in the native environment.
Methods
Sample Preparation
The genes coding HPr, EI, and EIN (N terminal of EI spanning residues from 1 to 249) were cloned into the expression vector pET11a. E25C cysteine mutation of HPr was taken using QuikChange (Stratagene). BL21 star cells were used to express the target proteins, and unlabeled and isotopically labeled proteins were prepared in LB and M9 media. The over-expressed proteins were purified using anion exchange columns and gel-filtration columns. The resultant proteins were checked using SDS-PAGE and ESMS. The purified proteins were buffer-exchanged in the Tris buffer (20 mM Tris, 150 mM NaCl, and pH 7.4).
The phosphorylated EIN (pEIN) was prepared via catalysis of EI. EIN (0.5 mM) was added to the Tris buffer (20 mM Tris, 100 mM NaCl, and pH 7.2) containing 5 mM PEP. EI was then added to the solution with the molar ratio to EIN of 1:50. The reaction was allowed at 37 °C for 4 h. The pEIN was extracted using an anion exchange column, and the successful phosphorylation was confirmed using ESMS. The purified pEIN was buffered and exchanged into the Tris buffer (20 mM Tris, 150 mM NaCl, and pH 7.4).
Crowding Environment Preparation
The crowding environment was prepared by mixing the dilute protein solution with the crowding agents of a certain weight. The dilute protein sample was prepared as described above, and the dilute protein sample was mixed with 10% weight/volume (w/v) of Ficoll-70 (purchased from Sigma-Aldrich) or BSA (≥98%, purchased from Sigma-Aldrich) to make the crowding environment.
NMR Titration Assays
A series of 1H-15N HSQC spectra were recorded to monitor the chemical shift changes upon HPr titration at 313 K. The NMR sample of 0.2 mM [2H, 15N] labeled EIN was prepared in the Tris buffer (20 mM Tris, 150 mM NaCl, 10% D2O, and pH 7.4), and an HPr concentration gradient of 0, 20, 40, 80, 120, 160, 220, 300, and 400 μM was added into the NMR sample. This titration was repeated in 10% Ficoll-70 and 10% BSA buffer. The obtained HSQC spectra were processed using NMRPipe,27 and the chemical shift perturbation was calculated with the equation (0.5 × (ΔH)2 + 0.1 × (ΔN)2)0.5, where ΔH and ΔN are the chemical shift changes in the proton and nitrogen dimensions, respectively.
Phosphoryl Transfer Measurement
15N labeled HPr (0.3 mM) was prepared in the Tris buffer (20 mM Tris, 0.15 mM NaCl, 10% D2O, and pH 7.4). This sample was used to optimize the NMR data acquisition parameters, including the spectrum center, the spectrum width, and the 1H pulse width. The stock solution of unlabeled pEIN was equimolar added into the 15N labeled HPr sample. A series of 1H-15N so-fast HMQC spectra were taken at 313 K to monitor the phosphoryl transfer sequentially. The data were collected using a Bruker 600 MHz spectrometer equipped with a cryogenic probe. Peak picking and volume extraction of the NMR data were finished using NMRPipe software.27
PRE Data Acquisition
The 15N labeled E25C HPr was ligated using the EDTA-Mn2+ probe (cat#P996250, Toronto Research Chemicals). The E25C HPr was initially incubated in 2 mM DTT at room temperature for 1 h and then desalted into the HEPES buffer (20 mM HEPES, 0.1 M NaCl, and pH 7.2). The probe in DMSO was mixed with a twofold excess of MnCl2, and the mixture was added to the protein solution with a molar ratio of 1:4. The reaction was taken at room temperature for 4 h. The ligated HPr was extracted using an anion exchange column, and the successful ligation was confirmed using ESMS. The resultant HPr was stored in the Tris buffer (20 mM Tris, 150 mM NaCl, and pH 7.4).
The paramagnetically labeled HPr (300 μM) was mixed with unlabeled EIN (300 μM) in Tris buffer (20 mM Tris, 150 mM NaCl, and pH 7.4). The 1H-15N HSQC spectrum of this sample was recorded as the paramagnetic data. The diamagnetic sample was prepared under the same condition, but 15N labeled E25C HPr was used instead of its paramagnetically labeled form. The 1H-15N HSQC spectrum of the diamagnetic sample was recorded, and the amide signal intensity of the diamagnetic and paramagnetic samples was compared to obtain PRE data.
Generation of EIN/HPr Complex Conformer Library
The HPr structure (PDB code: 1POH) was selected as the model to generate paramagnetically labeled HPr. Three conformations of each paramagnetic probe were patched onto each attachment site (E5C, E25C, and E66C of HPr) using Xplor-NIH.28 The dihedral angles for the rotatable bonds between the paramagnetic center and the protein backbone were randomized. The six structures of resultant HPr with the lowest energy were selected, and each of them was used to generate 3600 EIN/HPr complex structures using ZDOCK29 (version 3.0.2) with Fast Fourier transform (FFT)-based sampling, finally making a library of 21,600 EIN/HPr complex structures. The averaged theoretical intermolecular amide proton PREs caused by the conformers of the paramagnetic center were calculated for each structure, which was used as the target values to retrain the ensemble structure calculation.
Identification of the Protein Structural Ensembles
The experimental PRE input for ensemble calculation was obtained from the published data. A Monte Carlo (MC) simulated annealing method was used to identify an ensemble structure with N members in different environments.30
During the identification of the encounter complex, the PREs caused by E5C paraHPr, E25C paraHPr, and E66C paraHPr were simultaneously used as intermolecular distance restraints. The ensemble member N of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, and 30 are taken to measure agreement between the experimental and calculated values of PRE assessed using Pearson correlation coefficients. The 11 ensembles of encounter complex with the best Pearson correlation coefficients are selected for structure inspection and representation within each ensemble size of the calculation. The structure figures were illustrated with PyMOL (Version 2.2, Schrödinger).
SAXS/SANS Analysis
The small-angle X-ray scattering (SAXS) data for the EIN-HPr in dilute buffer was collected at the National Center for Protein Science Shanghai using the BL19U2 beamline at 25 °C. A total of 20 consecutive frames of 1 s exposure time were recorded and averaged, providing no difference between the frames. On the other hand, SANS measurements were performed on the small-angle neutron scattering (SANS) instrument at the China Spallation Neutron Source (CSNS) to investigate the dynamic conformation of EIN-HPr in crowded agents.24 The sample was prepared at 0.3 mM [2H,15N] EIN and 0.3 mM 2H/15N HPr in Tris buffer (20 mM Tris, 150 mM NaCl, and pH 7.4). After that, 10% Ficoll-70 or 10% BSA was added to the dilute sample. The incident neutrons with a wavelength of 1–10 Å were defined by a double-disc bandwidth chopper, collimated by a pair of apertures to the sample. The experiment used a sample to detector distance of 4 m and a sample aperture of 6 mm. The detector consists of 3He tubes, 8 mm in diameter and 1 m in length. This allows the detector to achieve a resolution of 8 mm and cover a wide Q range from 0.005 to 0.70 Å–1. We collected approximately 120 min scattering information for each sample, including the empty sample holder, sample cell, and solvent. The SANS experiment was also performed at room temperature at about 25 °C. The scattering data were set to the absolute unit after normalization, transmission correction, and standard sample calibration. The scattering profile for the matching buffer was recorded and was substrated. CRYSOL31 in the ATSAS32 software package was used to calculate the theoretical scattering profile and fit a single structure to the experimental scattering profile.
Results and Discussion
Minor Effect on Phosphoryl Transfer by a Change in the Environment
Initially, the rates of phosphoryl transfer between EIN and HPr in dilute buffer, 10% (w/v) Ficoll-70, and 10% (w/v) BSA were tested to interpret how the environment affects the interprotein interaction between EIN and HPr. Unless indicated, all the experiments in crowded environments were performed in 10% crowding agents. Concentrated Ficoll-70 and BSA are commonly employed to create a crowded environment, mimicking the intracellular environment.11,33−35 Although Ficoll-70 and BSA share very similar molecular weights, the surface properties of these two molecules are different from each other. Ficoll-70 possesses an inert surface, whereas the amino acids make BSA maintain a charged surface. Based on the features of Ficoll-70 and BSA, these two macromolecular crowders can generate crowded environments with different solution properties.
The phosphorylated EIN (pEIN) was synthesized in vitro. A series of 15N HSQC spectra of 15N isotopically labeled HPr under different conditions were recorded to monitor the number of phosphoryl groups transferred from pEIN. His15 is the phosphorylation site of HPr, so the well-resolved signals of T9, G13, L14, V61, and T62, which are spatially proximal to His15, are analyzed to determine the rate of the phosphoryl transfer36 (Figure S1). The comparison indicates that the phosphoryl transfer rates in different environments are very similar (Figure 1). Therefore, the crowding environments bring minor effects on the phosphoryl transfer between EIN and HPr.
Figure 1.
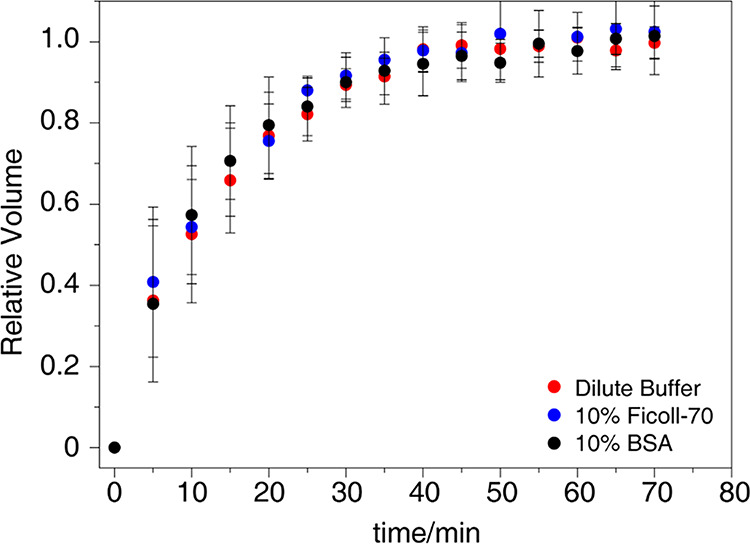
The averaged relative volume ratios of the amide cross-peaks of T9, G13, L14, V61, and T62 are plotted against the duration of the phosphoryl transfer in the dilute buffer (red dots), 10% Ficoll-70 (blue dots), and 10% BSA (black dots). The error bars show the standard deviations.
Phosphoryl transfer is completed only upon specific interaction between EIN and HPr.21 Similar phosphoryl transfer rates in different environments suggest that the specific interaction between EIN and HPr is relatively inert to change environmental conditions. These two proteins may adopt an identical complex structure in the tested environments. Thus, at this stage, our results show that adding the macromolecular crowder possibly causes a weak effect on the specific interaction between EIN and HPr. More detailed structural characterization is needed to explore the regulation of the EIN-HPr interaction in crowded environments.
Weak Influence to EIN-HPR Specific Interaction by the Macromolecular Crowder
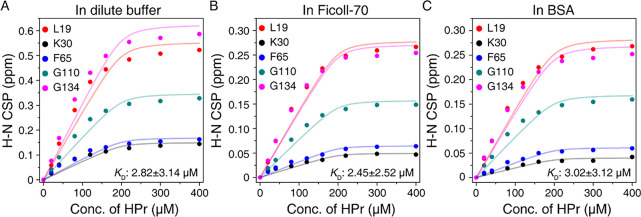
To investigate the regulation of the crowding molecules on EIN-HPr exchange, we carried out the titration experiments monitored by NMR spectroscopy. The superimposed 15N HSQC spectra show that the amide signals of the same group of EIN residues are shifted upon the addition of HPr in the dilute buffer, in Ficoll-70, and BSA (Figure S2). This suggests that EIN uses the same residues to interact with HPr under all conditions. The chemical shift perturbations (CSPs) upon HPr titration were used to fit the dissociation constants (KD) according to a function for a single-site binding model. The resultant KD values between EIN and HPr in the dilute, Ficoll-70, and BSA are 2.82 ± 3.14, 2.45 ± 2.53, and 3.03 ± 3.12 μM, respectively (Figure 2). The KD values are almost identical in different solution environments, suggesting that the crowding effect on the specific interaction is very weak.
Figure 2.
Global fitting of amide CSPs of EIN against a series of HPr concentrations. The 15N HSQC spectra of 200 μM 2H/15N EIN were recorded in dilute buffer (A), in 10% (w/v) Ficoll-70 (B), and in 10% (w/v) BSA (C), respectively. The KD values calculated using the CSPs of L19, K30, F65, G110, and G134 are 2.82 ± 3.14 μM in a dilute environment, 2.45 ± 2.52 μM in a Ficoll-70 environment, and 3.02 ± 3.12 μM in a BSA environment.
Notably, the large deviation between experimental data and the theoretical curve of KD fitting implies that the interaction between EIN and HPr may involve more than a single-site binding. Previously, the existence of the transient interaction between these two proteins besides their specific interaction has been approved.13 In addition, the CSP magnitudes observed in crowded environments are half of those in the dilute buffer. This implies that EIN probably interacts with HPr differently in dilute buffer and crowded environments. Taken together, it is possible to deduce that the crowded environment may influence the transient interaction between EIN and HPr. Below we will discuss this probability in detail.
The Solution Environment Regulates the EIN-HPr Transient Interaction
The previous PRE data shows that the binding between HPr and EIN is mainly composed of specific and nonspecific interactions,13,18 which is responsible for forming the specific EIN-HPr complex, whereas the latter is the driving force for encounter complex formation. Recently, the intermolecular PRE data of the EIN-HPr complex in dilute, Ficoll-70, and BSA environments have been reported.10 It has been found that the specific and transient interactions between EIN and HPr overall remain unchanged in the tested environments. However, a close inspection of the published data shows that the PRE profile is dependent on the solution conditions. To test whether the PRE differences arise from the change in protein–protein interaction upon adding macromolecular crowders, we recorded the intramolecular PREs of HPr complexed with EIN in dilute buffer, Ficoll-70, and BSA environments (Figure 3). There is no noticeable variation in the intramolecular PRE profiles of HPr in different environments. This indicates that the paramagnetic probe behaves identically under different solution conditions. Thus, it can be deduced that the intermolecular PRE variation of the EIN-HPr complexes in different environments is not caused by the crowding effect on the paramagnetic probe, and it arises from subtle differences in protein–protein interaction in different environments.
Figure 3.
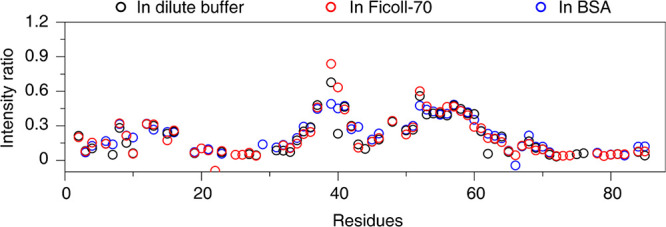
Intramolecular PREs of E25C paramagnetically labeled HPr. The black, red, and blue cycles show the intensity ratio of the amide signals between the diamagnetic and the paramagnetic samples recorded in dilute buffer, Ficoll-70, and BSA environments, respectively.
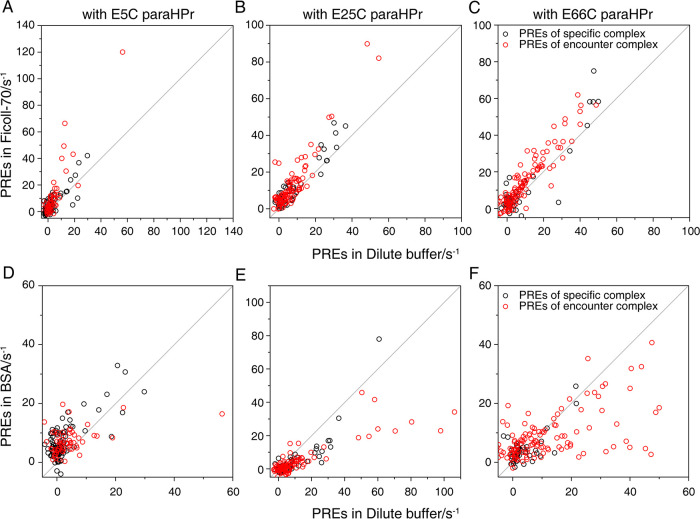
This study used these PRE data (derived from E5C, E25C, or E66C paramagnetically labeled HPr) to investigate how the environmental factor regulates the transient interaction. After the addition of Ficoll-70, the PREs arising from the specific complex almost remain the same, but those derived from the encounter complex are increased (Figure 4). Moreover, the opposite finding is observed for the EIN-HPr sample in the BSA environment (Figure 4). In the presence of BSA, the PREs that originated from the encounter complex decrease more than those from the specific complex. This could be explained as that the specific complex is relatively inert to the change in solution condition, but the encounter complex is sensitive to the presence of crowding agents. Hence, the variation of PRE data in different environments is consistent with the results of our NMR titration. More detailed structural characterization is needed to interpret the PRE differences of the EIN-HPr system in different solution environments.
Figure 4.
Comparison between the PRE data in crowded environments (Y axis) and the PRE data in the dilute environment (X axis). The PRE data acquired with E5C, E25C, and E66C paramagnetically labeled HPr are shown as A/D, B/E, and C/F, respectively. The PREs caused by the specific interaction between EIN and HPr are shown using black cycles, and the PREs arising from the encounter complex are shown using red cycles.
Visualization of the EIN-HPr Encounter Complex in Crowding Environments
To visualize the EIN-HPr complex in different environments, we used the PRE data as the experimental restraints to calculate the ensemble structure of this protein system. A structure pool of 3600 EIN-HPr complexes was generated using molecular docking,29 and the theoretical PRE profiles were calculated for each member of the structure pool. During the calculation of the ensemble structure, the theoretical PREs of the N membered ensemble were compared with the experimental PRE restraints.30 The 10 sets of the ensemble possessing the best correlation with the experimental data were used to visualize the EIN-HPr complex. N (from 1 to 30) membered ensembles were tested to obtain the best correlation and avoid over-fitting the experimental data.30 The correlation R rapidly grows as the number of the structure members increases, and after an ensemble of 20 structures, the increase in R almost stops (Figure S3). Thus, the ensemble of 20 structures is used as the model to study the encounter complex of EIN and HPr in different environments.
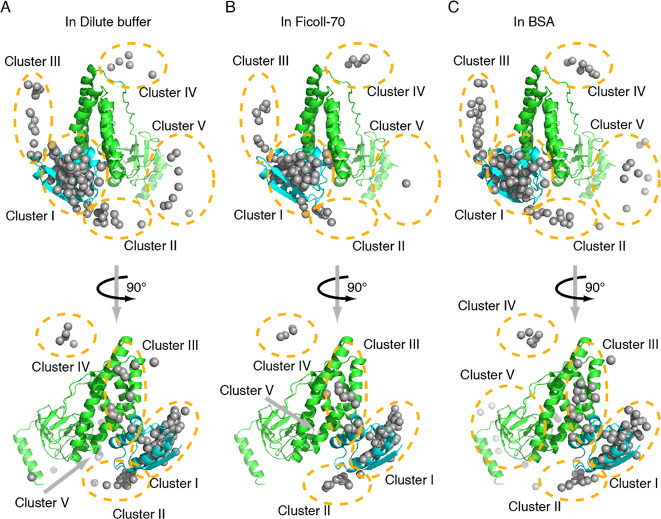
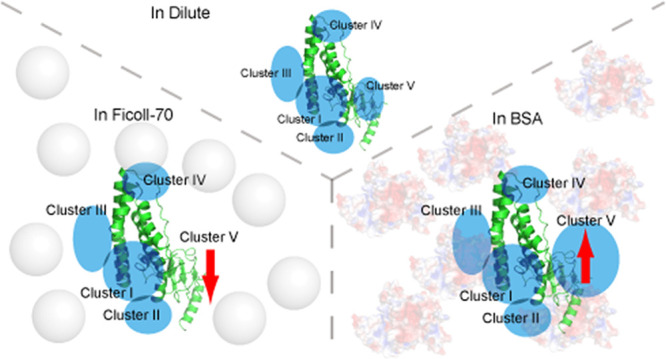
Our calculated EIN-HPr encounter complex in dilute buffer indicates that HPr molecules are located surrounding the active site of EIN (Figure 5). The polar projection of the calculated encounter complexes against the EIN-HPr specific complex was plotted. It indicates that the polar projection of some HPr molecules in the ensemble is away from the origin, and they could be spatially divided into five clusters (Figure S4). A similar clustering pattern of HPr molecules on the EIN surface has been observed previously using PRE13,18 and CXMS.17 Cluster I is located on the specific interface between EIN and HPr, and HPr molecules in this cluster represent HPr in the specific complex and HPr in a similar orientation to that in the specific complex. HPr molecules in Clusters II and III stay closer to the active site of EIN, and HPr molecules in Clusters I–III are believed to undergo rapid rotation and translation, helping the specific binding and alignment of their active sites. In addition to these three clusters, Clusters IV and V are located relatively distant from the active site of EIN. HPr molecules in Clusters IV and V are futile to the specific interaction between EIN and HPr, but they could co-exist with the EIN-HPr specific complex.
Figure 5.
Calculated ensemble structures of the EIN-HPr complex in dilute buffer (A), Ficoll-70 (B), and BSA (C). The specific complex of EIN (green) and HPr (cyan) are shown as cartoon, and the mass center of HPr molecules of the calculated EIN-HPr complex is shown as gray dots. According to the location on the EIN surface, HPr molecules are divided into five clusters marked using dashed cycles or noted as arrows.
Similar to the encounter complex of EIN and HPr in dilute buffer, those in the Ficoll-70 environment and BSA environment are also formed by the HPr molecules clustered in these five regions on the EIN surface. Hence, EIN and HPr interact via similar binding modes in different environments. However, the in-depth inspection of the encounter complexes in different environments reveals that the populations of HPr molecules in each cluster vary with the solution environment (Table 1). It confirms our hypothesis that the crowding environment can regulate the transient interaction between EIN and HPr.
Table 1. Statistics of the HPr Populations (%) in Different Clusters on the EIN Surfacea.
| Cluster I–III | Cluster IV | Cluster V | |
|---|---|---|---|
| in dilute buffer | 93.2 ± 3.4 | 2.3 ± 3.4 | 4.6 ± 2.7 |
| in Ficoll-70 | 94.6 ± 1.5 | 5.0 ± 0.0 | 0.5 ± 1.5 |
| in BSA | 87.7 ± 7.2 | 3.6 ± 2.3 | 8.6 ± 6.0 |
The HPr population is calculated using the averaged data of 10 sets of the calculation ensemble (N = 20) structures.
Table 1 shows that, in the three tested environments, most HPr molecules on the EIN surface stay in Clusters I–III, and the addition of Ficoll-70 or BSA mildly influences the HPr population in this region. The Cluster I–III region on the EIN surface is highly charged, which helps yield long-range electrostatic interaction between EIN and HPr (Figure S5A), benefiting the formation of numerous stable encounter complexes. Thus, most EIN and HPr molecules are expected to interact with each other in Clusters I–III.
An interesting finding is that HPr molecules in Cluster V of the sample in Ficoll-70 almost disappear. In contrast, more HPr molecules are observed in Cluster V when the solution condition is changed to the BSA-crowded solution. Ficoll-70 is an inert synthetic crowding agent, and it only interacts with the EIN-HPr system via hard-core repulsion.10 With concentrated Ficoll-70, a small fraction of space in the solution is left for the test protein, leading to the preference for the compact and stable protein complexes under this condition.33,37,38 Thus, it is possible that the hard-core repulsion could protect relatively stable encounter complexes in Clusters I–III. It probably pushes HPr molecules to the Cluster I–III region, which stay in Cluster V in a dilute buffer. Consequently, when comparing encounter complexes in the tested environments, the Cluster V region of the sample in Ficoll-70 possesses the least number of HPr molecules. The encounter complexes in Clusters I–III exhibit the highest population in Ficoll-70.
It has been proven that there exists soft interaction between BSA and HPr.10 BSA can neutralize the charged active interface of HPr. This would weaken the ability of HPr to electrostatically interact with EIN, disturbing the formation of encounter complexes, especially those formed via long-range electrostatic interaction. As a result, HPr molecules in the Cluster I–III region have decreased population in the BSA environment. Since the bound form of the protein is favorable in a crowded environment, HPr molecules tend to form a complex with EIN even though it is neutralized. The Cluster V region is weakly charged (Figure S5B), so the transient interaction around this area would be poorly dependent on electrostatic interaction. This makes Cluster V an ideal location for neutralized HPr to form encounter complexes. Consequently, the number of HPr molecules in the Cluster V region is increased when the environment condition is shifted from diluted to BSA-crowded solution. Overall, the HPr distributions in different environments suggest that hard-core repulsion and soft interaction can affect the clustering of the EIN-HPr encounter complexes through different mechanisms.
Cross-Validation between NMR Data and SANS/SAXS Data
To further confirm our NMR findings, we examined the EIN-HPr complex using small-angle X-ray scattering (SAXS)39 and small-angle neutron scattering (SANS).24,25 SAXS and SANS are twin techniques of small angle scattering, and both of them are powerful methods to resolve protein structures. Moreover, with contrast variation (proton and deuterium labeling), SANS could be used to selectively capture the structure data of the target protein in a complex system.40 Taking advantage of this feature of SANS, we recorded the scattering data of the fully deuterated protein complex of EIN and HPr in Ficoll-70 and BSA environments. These data were compared with the SAXS data of the EIN-HPr complex in the dilute buffer.
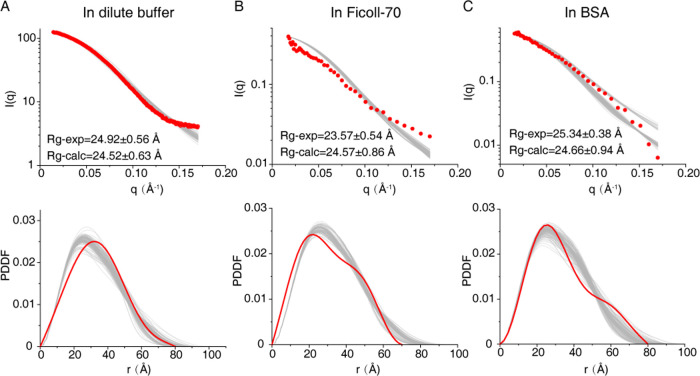
The experimental scattering data in the three tested environments correlate well with the theoretical scattering profiles calculated from the corresponding ensemble structures derived from our NMR data (Figure 6). These data show that the radius of gyration (Rg) of the sample in Ficoll-70 (23.6 Å) is smaller than those of the samples in dilute and in BSA (24.9 and 25.3 Å, respectively). The experimental paired-distance distribution function (PDDF) plots also agree with the observation of Rg in different environments. It shows that the sample in Ficoll-70 possesses the smallest value of Dmax (∼70 Å), and those in dilute buffer and BSA exhibit a larger Dmax value (∼80 Å for both conditions).
Figure 6.
SAXS analysis of the EIN-HPr complex in dilute buffer (A) and SANS analysis of this protein complex in the Ficoll-70 environment (B) and BSA environment (C). The top panels show the experimental scattering profiles (red dots) and the theoretical scattering profiles of the calculated EIN-HPr complexes in different environments (gray lines). The lower panels display the experimental paired-distance distribution function (PDDF) plots (red lines) and the theoretical PDDF plots of the calculated ensemble EIN-HPr structures in different environments (gray lines).
The apparent difference between the calculated EIN-HPr ensemble structures in different environments is the variation of the HPr populations in Cluster V. With the spatial shift of the whole cluster of HPr molecules, the shape of the EIN-HPr complex would be correspondingly changed. In Ficoll-70, the HPr molecules in Cluster V nearly totally re-distributed to Clusters I–III, and the overall size of the EIN-HPr complex shall be decreased. This could explain why the Rg value of the EIN-HPr complex is lower in Ficoll-70. Although the BSA environment enriches the HPr molecules in Cluster V, the clustering pattern of HPr molecules remains the same as that in the dilute buffer. Thus, the size of the EIN-HPr complexes in the dilute buffer and the BSA environment would be approximately close to each other, supported by our SAXS and SANS data. Taken together, our NMR and SAXS/SANS data all support that the crowding environment can regulate the interaction between EIN and HPr, especially the transient interaction between these two proteins.
Conclusions
In summary, the native cellular environment is where proteins fulfill their physiological functions, and this environment is composed of thousands of molecules of various sizes. The interactions between proteins are sensitive to environmental factors. Several pieces of evidence show that changes in environmental conditions could modulate the structure, interaction, dynamics, and functions of proteins.4,37,41,42 Thus, studies on protein in crowded environments are essential to help us understand the mechanism of protein interactions and functions under native conditions. Our current study indicates that, as far as in Ficoll-70 and BSA, the transient interaction between EIN and HPr is a preferable regulating target for macromolecular crowders. The repulsive interaction induced by Ficoll-70 allows HPr shifting between different regions on the EIN surface for transient interaction. However, the soft interaction between BSA and HPr would downgrade the local transient interaction between EIN and HPr, leading to the re-distribution of HPr molecules on the EIN surface. The tested crowded environments could cause a more substantial regulation effect on EIN-HPr transient interaction than their specific interaction, which highlights a novel way for the environmental condition to modulate interprotein interactions. Furthermore, the NMR experiment can be performed in near physiological or even in cells, and SANS can also be effective for selective analysis of research objects. The methods used here can be applied to other protein–protein interaction studies, especially those interactions under complex environmental conditions.
Acknowledgments
We thank the staff of BL19U2 beamlines at the National Facility for Protein Science Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility, Shanghai, People’s Republic of China, for assistance during SAXS data collection. We acknowledge the use of the resources of the China Spallation Neutron Source in Dongguan of Guangdong Province of P. R. China. We are grateful to Prof. Maili Liu, Prof. Conggang Li, and Prof Yunfei Hu for their helpful comments on the manuscript. The work has been supported by the National Key R&D Program of China (2017YFA0505400 to X.D., 2018YFA0507700 to Z.G. and C.T.), the National Natural Science Foundation of China (31971155 to Z.G.), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (under grant nos. 2020329 and 2020010 for Z.G. and Y.K., respectively).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.2c02713.
Figures showing rate measurement of phosphoryl transfer, NMR titration of EIN with HPr in tested environments, correlation of the ensemble calculation, and inspection of the ensemble of the EIN-HPr encounter complex (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Zimmerman S. B.; Trach S. O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991, 222, 599–620. 10.1016/0022-2836(91)90499-V. [DOI] [PubMed] [Google Scholar]
- Zhou Y. L.; Liao J. M.; Chen J.; Liang Y. Macromolecular crowding enhances the binding of superoxide dismutase to xanthine oxidase: implications for protein-protein interactions in intracellular environments. Int. J. Biochem. Cell Biol. 2006, 38, 1986–1994. 10.1016/j.biocel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Zhou B. R.; Zhou Z.; Hu Q. L.; Chen J.; Liang Y. Mixed macromolecular crowding inhibits amyloid formation of hen egg white lysozyme. Biochim. Biophys. Acta 2008, 1784, 472–480. 10.1016/j.bbapap.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Stagg L.; Zhang S. Q.; Cheung M. S.; Wittung-Stafshede P. Molecular crowding enhances native structure and stability of alpha/beta protein flavodoxin. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 18976–18981. 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.; Cheng K.; Wu Q.; Liu M.; Li C. Confinement Alters the Structure and Function of Calmodulin. Angew. Chem., Int. Ed. 2017, 56, 530–534. 10.1002/anie.201609639. [DOI] [PubMed] [Google Scholar]
- Pan B. B.; Yang F.; Ye Y.; Wu Q.; Li C.; Huber T.; Su X. C. 3D structure determination of a protein in living cells using paramagnetic NMR spectroscopy. Chem. Commun. 2016, 52, 10237–10240. 10.1039/C6CC05490K. [DOI] [PubMed] [Google Scholar]
- Ye Y.; Liu X.; Xu G.; Liu M.; Li C. Direct observation of Ca(2+) -induced calmodulin conformational transitions in intact Xenopus laevis oocytes by (19) F NMR spectroscopy. Angew. Chem., Int. Ed. 2015, 54, 5328–5330. 10.1002/anie.201500261. [DOI] [PubMed] [Google Scholar]
- Homouz D.; Perham M.; Samiotakis A.; Cheung M. S.; Wittung-Stafshede P. Crowded, cell-like environment induces shape changes in aspherical protein. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 11754–11759. 10.1073/pnas.0803672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M.; Li C.; Pielak G. J. Soft interactions and crowding. Biophys. Rev. 2013, 5, 187–194. 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.; Qin L. Y.; Gong Z.; Qin S.; Zhou H. X.; Tang C. Preferential Interactions of a Crowder Protein with the Specific Binding Site of a Native Protein Complex. J. Phys. Chem. Lett. 2022, 13, 792–800. 10.1021/acs.jpclett.1c03794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman A. J.; Pielak G. J. Cosolute and Crowding Effects on a Side-By-Side Protein Dimer. Biochemistry 2017, 56, 971–976. 10.1021/acs.biochem.6b01251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. X.; Qin S. Simulation and Modeling of Crowding Effects on the Thermodynamic and Kinetic Properties of Proteins with Atomic Details. Biophys. Rev. 2013, 5, 207–215. 10.1007/s12551-013-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.; Iwahara J.; Clore G. M. Visualization of transient encounter complexes in protein-protein association. Nature 2006, 444, 383–386. 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- Iwahara J.; Clore G. M. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 2006, 440, 1227–1230. 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Nooren I. M.; Thornton J. M. Diversity of protein-protein interactions. EMBO J. 2003, 22, 3486–3492. 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R.Structure and Mechanism in Protein Science: a Guide to Enzyme Catalysis and Protein Folding; Freeman & Co, 1999.
- Gong Z.; Ding Y. H.; Dong X.; Liu N.; Zhang E. E.; Dong M. Q.; Tang C. Visualizing the Ensemble Structures of Protein Complexes Using Chemical Cross-Linking Coupled with Mass Spectrometry. Biophys. Rep. 2015, 1, 127–138. 10.1007/s41048-015-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzi N. L.; Doucleff M.; Suh J. Y.; Clore G. M. Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1379–1384. 10.1073/pnas.0909370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. H.; Gong Z.; Dong X.; Liu K.; Liu Z.; Liu C.; He S. M.; Dong M. Q.; Tang C. Modeling Protein Excited-state Structures from ″Over-length″ Chemical Cross-links. J. Biol. Chem. 2017, 292, 1187–1196. 10.1074/jbc.M116.761841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J.; Francke C.; Postma P. W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett D. S.; Seok Y. J.; Peterkofsky A.; Gronenborn A. M.; Clore G. M. Solution structure of the 40,000 M-r phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat. Struct. Biol. 1999, 6, 166–173. 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- Suh J. Y.; Tang C.; Clore G. M. Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2007, 129, 12954–12955. 10.1021/ja0760978. [DOI] [PubMed] [Google Scholar]
- Kozakov D.; Li K.; Hall D. R.; Beglov D.; Zheng J.; Vakili P.; Schueler-Furman O.; Paschalidis I.; Clore G. M.; Vajda S. Encounter complexes and dimensionality reduction in protein-protein association. Elife 2014, 3, e01370 10.7554/eLife.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.; He C.; Zheng H.; Geng Y.; Fu J.; Zhang S.; Hu H.; Wang S.; Zhou B.; Wang F.; et al. The time-of-flight Small-Angle Neutron Spectrometer at China Spallation Neutron Source. Neutron News 2018, 29, 14–17. 10.1080/10448632.2018.1514197. [DOI] [Google Scholar]
- Sonntag M.; Jagtap P. K. A.; Simon B.; Appavou M. S.; Geerlof A.; Stehle R.; Gabel F.; Hennig J.; Sattler M. Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins. Angew. Chem., Int. Ed. 2017, 56, 9322–9325. 10.1002/anie.201702904. [DOI] [PubMed] [Google Scholar]
- Zhu W.; Guseman A. J.; Bhinderwala F.; Lu M.; Su X.-C.; Gronenborn A. M. Visualizing Proteins in Mammalian Cells by 19F NMR Spectroscopy. Angew. Chem., Int. Ed. 2022, e202201097 10.1002/anie.202201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Clore G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Chen R.; Li L.; Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 2003, 52, 80–87. 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- Gong Z.; Schwieters C. D.; Tang C. Theory and practice of using solvent paramagnetic relaxation enhancement to characterize protein conformational dynamics. Methods 2018, 148, 48–56. 10.1016/j.ymeth.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D.; Barberato C.; Koch M. H. J. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. 10.1107/S0021889895007047. [DOI] [Google Scholar]
- Franke D.; Petoukhov M. V.; Konarev P. V.; Panjkovich A.; Tuukkanen A.; Mertens H. D. T.; Kikhney A. G.; Hajizadeh N. R.; Franklin J. M.; Jeffries C. M.; Svergun D. I. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova I.; Turoverov K.; Uversky V. What Macromolecular Crowding Can Do to a Protein. Int. J. Mol. Sci. 2014, 15, 23090–23140. 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozer N.; Kuttner Y. Y.; Haran G.; Schreiber G. Protein-protein association in polymer solutions: From dilute to semidilute to concentrated. Biophys. J. 2007, 92, 2139–2149. 10.1529/biophysj.106.097717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas G.; Fernández J. A.; Minton A. P. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: Indefinite linear self-association of bacterial cell division protein FtsZ. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 3150–3155. 10.1073/pnas.051634398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland N. A. J.; Boelens R.; Scheek R. M.; Robillard G. T. High-resolution structure of the phosphorylated form of the histidine-containing phosphocarrier protein HPr from Escherichia coli determined by restrained molecular dynamics from NMR-NOE data. J. Mol. Biol. 1995, 246, 180–193. 10.1006/jmbi.1994.0075. [DOI] [PubMed] [Google Scholar]
- Guseman A. J.; Perez Goncalves G. M.; Speer S. L.; Young G. B.; Pielak G. J. Protein shape modulates crowding effects. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 10965–10970. 10.1073/pnas.1810054115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X.; Rivas G.; Minton A. P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37, 375–397. 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar A.; Herranz-Trillo F.; Langkilde A. E.; Vestergaard B.; Bernadó P. Structure and thermodynamics of transient protein-protein complexes by chemometric decomposition of SAXS datasets. Structure 2021, 29, 1074–1090. 10.1016/j.str.2021.03.017. [DOI] [PubMed] [Google Scholar]
- Johansen D.; Jeffries C. M. J.; Hammouda B.; Trewhella J.; Goldenberg D. P. Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys. J. 2011, 100, 1120–1128. 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. A.; Buell A. K.; Knowles T. P. J.; Welland M. E.; Dobson C. M. Protein aggregation in crowded environments. J. Am. Chem. Soc. 2010, 132, 5170–5175. 10.1021/ja909997e. [DOI] [PubMed] [Google Scholar]
- Wirth A. J.; Platkov M.; Gruebele M. Temporal variation of a protein folding energy landscape in the cell. J. Am. Chem. Soc. 2013, 135, 19215–19221. 10.1021/ja4087165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.