Abstract
Purpose:
Genetic disorders often present in the neonatal intensive care unit (NICU), and detecting or confirming these diagnoses has been shown to impact care. However, the availability and usage of genetic testing, particularly exome or genome sequencing, among NICUs varies widely. We therefore sought to investigate practice patterns related to genetic testing in NICUs around the country in order to identify and quantify potential discrepancies.
Methods:
We designed a survey that was distributed to neonatologists via email. The survey contained questions related to test availability and desirability, the process of test ordering in the NICU, and general comfort with ordering and interpreting genetic testing. Demographic data related to the survey participant and characteristics of their NICU were also obtained.
Results:
162 neonatologists completed the survey, representing 40 states and 112 distinct NICUs. While nearly all (93.2%) attributed a high level of importance to identifying a genetic diagnosis for their patients, genetic consultations were only available at 78% of NICUs and exome or genome sequencing was not available on a regular basis (69% of NICUs).
Conclusions:
Among U.S. neonatologists surveyed, although most feel that genetic tests are indicated for their patients, they are not always clinically available. Further research into implementation barriers is warranted.
Introduction
Many genetic conditions present in the neonatal intensive care unit (NICU), where they contribute substantially to admissions, morbidity, and mortality (1–3). Identifying a genetic diagnosis has many potential benefits, such as medical management changes, assistance with end-of-life decision making, and counseling for the family regarding recurrence risk and prognosis (1, 4–6). Indeed, it has been suggested that the NICU may be one of the areas where diagnostic genetic testing has its greatest impact (7, 8).
Multiple tests are currently utilized to identify these diagnoses, from chromosomal testing such as karyotype and chromosomal microarray (CMA), to sequencing tests including gene panels focusing on a particular indication. More recently, broader genomic evaluation with gene panels comprising thousands of genes, or exome or genome sequencing (ES/GS) has become available and has been shown to be of high yield and clinically impactful (9–13).
However, genome-wide sequencing tests such as ES/GS are more costly compared to more traditional testing approaches (2). They are also optimally administered with pre- and post-test counseling by a medical genetics professional, not available in all institutions. Thus, although the clinical utility and cost effectiveness of genetic testing in the NICU is being increasingly recognized (9–13), many NICUs continue to defer it to the outpatient setting.
Though the availability of diagnostic genetic testing in the NICU varies widely, it is continually evolving and has not been previously quantified. Identifying current practice patterns and variability regarding diagnostic genetic testing approaches in NICUs would assist in developing evidence-based practice guidelines for the genetic evaluation of critically-ill infants and identify potential areas for improvement. We therefore surveyed practicing neonatologists across the United States in order to gain insight into these issues.
Materials and Methods
We designed a survey addressing multiple domains regarding to genetic test desirability and availability in the NICU (Figure 1). As no suitable validated instruments were available to capture this information, we developed the survey questions related to commonly used genetic tests in the NICU and common ordering practices using input from practicing clinical geneticists and neonatologists. After creation of the survey instrument, cognitive interviewing with both neonatologists and genetic counselors was performed in order to improve the face validity of our survey items. The full survey is available in the Supplement.
Figure 1.
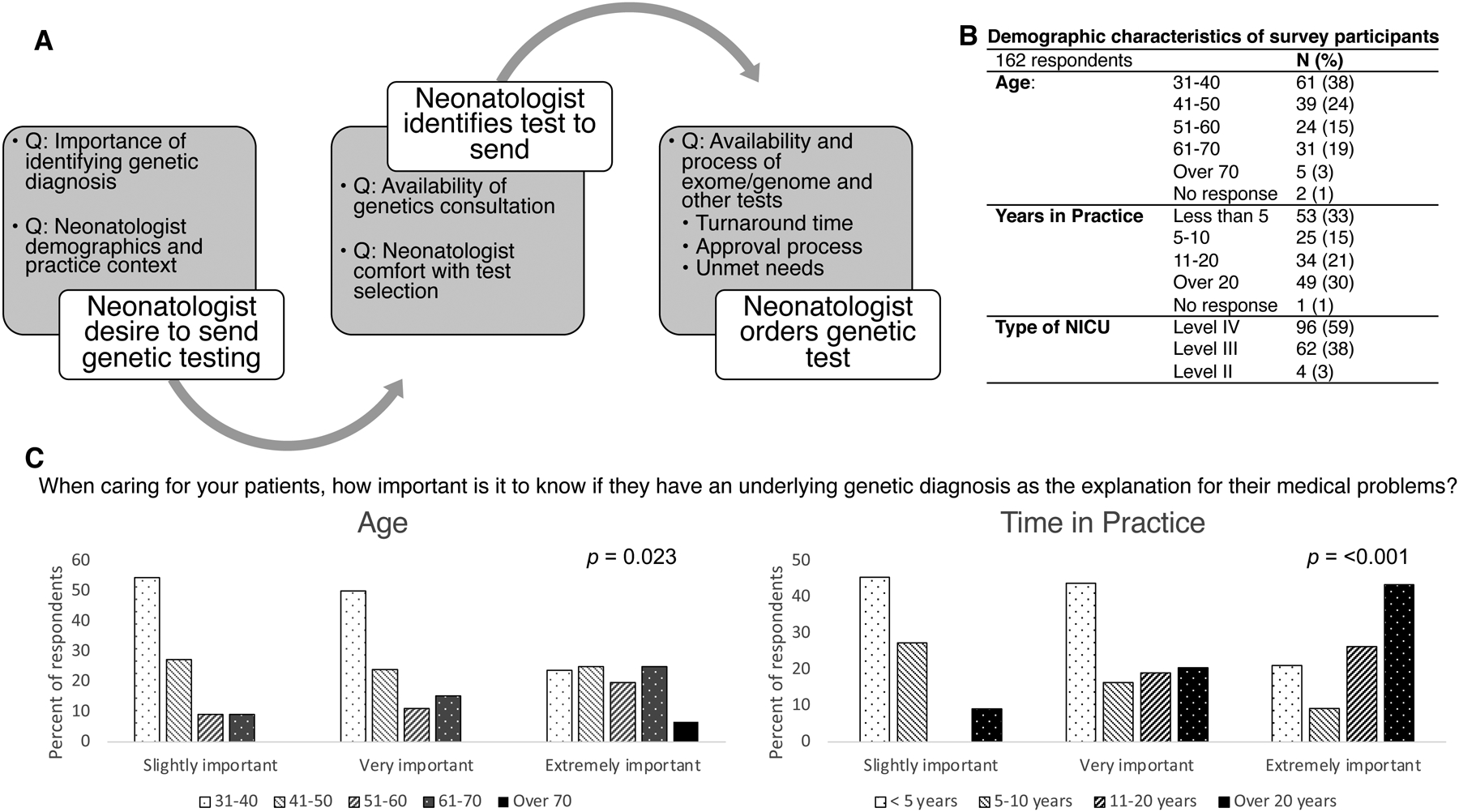
A, Schematic of genetic test ordering process in the neonatal intensive care unit, corresponding to question item domains. B. Demographic characteristics of 162 survey respondents. C. Relationship between age and time in practice to neonatologists’ perceived importance of a genetic diagnosis. Comparisons made by Chi-square analysis across all groups.
The survey was sent via email using the Section on Neonatal and Perinatal Medicine listserv, which comprises over 4,000 specialists (14). Survey responses were collected via REDCap hosted at our institution (15). Participants completed the survey anonymously, though were asked to identify the NICU in which they practice. For the analysis of practice patterns related to a NICU (rather than an individual neonatologist), we restricted responses to distinct NICUs only when two or more responses were obtained from the same site. For completed surveys where the participant did not disclose the NICU name, the survey responses reporting data at the level of the NICU were included if other features allowed us to identify it as a distinct NICU. Responses were aggregated for our NICU-level analyses so that each NICU was represented only once. For questions regarding test availability (in which a single response was requested), if respondents from the same NICU had divergent answers, the response indicating that the test is available was used. Otherwise, if two responses from the same NICU were divergent and unable to be aggregated (i.e. responses were mutually exclusive), the question was left unanswered for that NICU. Questions regarding turnaround time were aggregated choosing the most common response if multiple options were chosen from the same NICU, or an average of divergent responses if there was no majority. Overall, 21 NICUs had more than one respondent for the same NICU and, of these, the responses varied the most for questions pertaining to the process of sending sequencing-based tests, though multiple responses were allowed for these questions (Supplement 2).
Analysis was performed using SPSS version (IBM Corporation, Armonk, NY), with a Fisher’s exact or Chi square test used to compare categorical variables. This study was approved by the Boston Children’s Hospital Institutional Review Board with completion of the survey constituting informed consent.
Results
162 neonatologists completed the study survey from April to July, 2021 for a response rate of approximately 4%. A total of 40 states plus Washington DC were represented (missing: Alaska, Idaho, Maine, New Hampshire, North Dakota, Rhode Island, South Dakota, Vermont, West Virginia, Wyoming) with a range of 1–11 responses from distinct NICUs per state. Two respondents were from outside the US and one was from Puerto Rico; three respondents did not provide state or NICU name. Other demographic features are presented in Figure 1B.
Interest and Comfort with Genetic Testing
Almost half of the responding neonatologists (76/161 (47.2%)), felt that it is “extremely important” to identify a genetic diagnosis while an additional 74/161 (46.0%) responded “very important”. The remaining 11/161 (6.8%) responded “slightly important” and none responded “not at all important”. There was a significant relationship between age category (p = 0.023) and length of time in practice (p < 0.001) and perceived importance of genetic testing, with neonatologists in the older age categories and with a longer period of time in practice feeling that the testing is more important (Figure 2). There was no significant association with NICU level (level II, III, or IV, p = 0.60) or in comparing NICUs where genetics consultants were available or not available (p = 0.81).
Figure 2.
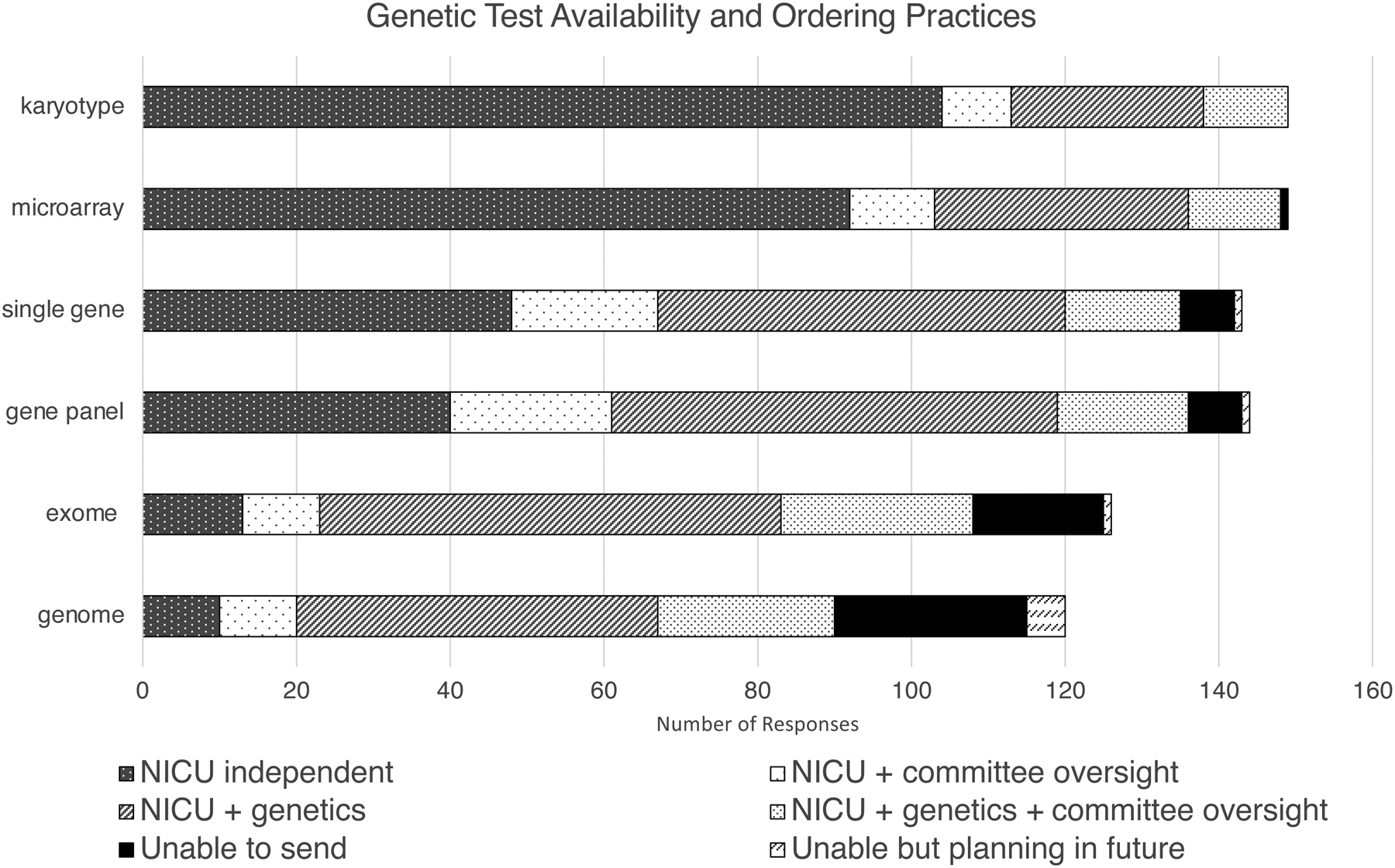
Survey respondents were asked to choose the options best describing how genetic tests are ordered in their NICU. ”NICU independent” = NICU team can send the test independently without genetics consultation or committee oversight. “NICU + committee oversight” = NICU team can send the test after getting permission from an oversight committee. “NICU + genetics” = NICU team can send the test in consultation with the genetics team. “NICU + genetics + committee oversight” = the NICU team must involve the genetics service and get permission from an oversight committee to send the test. Finally, participants could indicate if the test is not currently available or not currently available (“Unable to send”) but with plans in place to increase availability (“Unable but planning in future”). Participants were able to select more than one option.
When asked about how comfortable neonatologists felt with selecting the appropriate genetic test, 5/162 (3.1%) responded “very uncomfortable”, 46/162 (28.4%) responded “somewhat uncomfortable”, 32/162 (19.8%) responded “neither comfortable nor uncomfortable”, 59/162 (36.4%) responded “somewhat comfortable”, and 20/162 (12.3%) responded “very comfortable”. The level of comfort was not related to age category (p = 0.37), years in practice (p= 0.08), NICU level (p = 0.35), or availability of genetics consultation (p = 0.31).
Respondents were then asked if they felt that their patients were in need of certain tests but unable have them sent from the NICU. Of 154 respondents to this question, the test most commonly identified as needed but not able to send was GS (49/154, 31.8%) followed by ES (40, 26.0%), gene panel (24, 15.6%), single gene (21, 13.6%), chromosomal microarray (11, 7.1%%), and karyotype (2, 1.3%). Respondents from NICUs where genetics consultants were not available (N = 130/162) reported higher rates of inability to send desired tests compared to those NICUs where they were available: karyotype: 0/130 (0%) versus 2/32 (6.7%), p = 0.038; CMA: 3/130 (2.3%) versus 8/32 (25%), p < 0.001; single gene: 8/130 (6.2%) versus 13/32, (41%), p < 0.001; gene panel 11/130 (8.5%) versus 13/32 (41%), p < 0.001; exome: 22/130 (17%) versus 18/32 (56%), p < 0.001; genome: 33/130 (25%) versus 16/32 (50%), p = 0.010.
NICU Genetic Testing Process and Availability
We then performed a NICU-level analysis with responses aggregated by NICU. We found that 87 of these 112 NICUs (78%) had geneticists available for consults. ES was reported to be available clinically at 69/112 NICUs (62%) and 36/112 (32%) had GS available clinically, with 77/112 NICUs (69%) reporting that either modality was available (although 20% responded “I don’t know” to the question regarding GS). Additionally, 4/31 (13%) had ES and 13/49 (27%) had GS available on a research basis. For ES, the genetics team was most commonly responsible for test ordering (56/69, 81%) and consent (57/69, 83%), followed by the NICU team (39/69, 57% order; 26/69, 38% consent) with a small minority reporting that other services also perform these tasks (3/69, 4% order; 2/69, 3% consent). The most common turnaround time for ES was one to three months (36/69, 55%) followed by two weeks to one month (27/69, 39%), with only one NICU (1%) reporting results in less than 2 weeks.
For GS, the results were similar with the genetics team most commonly responsible for test ordering (28/36, 78%) and consent (26/36, 72%) followed by the NICU team (20/36 order, 56%; 18/36, 50% consent) and other services (2/36, 6% order; 1/36, 3% consent). GS results were reported to return in less than 2 weeks for 6/36 (1%) NICUs, with 13/36 (53%) reporting 2 weeks to one month and 14/36 (39%) reporting one to three months and 1 NICU (3%) reporting results of GS taking over 3 months to result.
We then asked respondents about the ordering process for various types of testing (Figure 3). Most NICUs reported being able to order and send karyotypes and CMAs independently, though for sequencing tests, particularly gene panels and ES, the majority indicated the need for genetics and/or committee oversight, and test availability decreased as the complexity of the testing increased (from single gene to GS). There was a significant relationship between genetics consultant and ES/GS availability: where genetics consultants were available, 74% (64/87) of NICUs had ES available, 21% (18/86) did not, and 6% (5/86) did not know. Where genetics consultants were not available, 20% had ES available (5/25), 64% did not have ES available (16/25) and 16% (4/25) did not know (p < 0.001). Similar results were found for GS though the results did not reach statistical significance (p = 0.11).
Discussion
We present a description of current practice patterns related to genetic testing in the NICU as described by neonatologists, as well as their views on genetic testing. Overall, we found that most neonatologists perceive importance in a genetic diagnosis, though a substantial proportion identified a discrepancy between desirability and availability, particularly for ES/GS. As we expected, established tests such as karyotype and CMA are easier for the NICU team to order independently while newer, sequencing-based tests typically require oversight to ensure appropriate utilization and aid in test interpretation and counseling.
These results are consistent with prior literature on satisfaction with genomic sequencing, with prior studies of GS for critically-ill infants revealing that NICU providers valued the information gleaned from such testing and used it to direct medical care (16). We also found that a genetic diagnosis is highly valued by neonatologists, however the tests needed to identify a diagnosis are not routinely currently available. Interestingly, neonatologists who were older and had spent a longer time in practice reported higher perceived importance of genetic testing, which may be informed by their prior experience with genetic testing.
Our results suggest a need for increased clinical genetics involvement in the NICU, where test availability has lagged behind current research, as karyotype or CMA are unlikely to identify all infants with rare genetic disorders (2, 4, 17, 18). As the diagnostic and clinical utility of ES/GS in this population is known be high (1, 5, 6, 10), further investigation into incorporation of these diagnostic techniques into the NICU setting seems warranted, particularly where clinical genetics teams are not available.
Limitations to this study include the sample size and potential for non-random self-selection of study participants, where those who are more familiar with genetic testing may have been more likely to complete the survey. While the response rate from 162 neonatologists represents approximately 4% of the entire listserv, which includes not only practicing neonatologists but also trainees such as residents and fellows, prior studies of this cohort report response rates of approximately 400, on the same order of magnitude (19, 20). Furthermore, our sample is geographically diverse, with over 112 NICUs from most states in the country represented. In addition, the reporting of the neonatologist completing the survey may not be accurate, particularly for NICUs in which the genetics team is entirely responsible for test selection and ordering and, in fact, when evaluating responses in aggregate, they were noted to vary within the same NICU (which we accounted for in our analysis).
Additional studies are needed to further characterize practice patterns related to genomic medicine in the NICU, with a particular focus on variation by NICU type and an eye towards implementation where clinical genetics teams are not physically present. These results are critical in order to develop evidence-based recommendations incorporating genomics into NICU care across a variety of practice settings.
Supplementary Material
Acknowledgements:
The authors would like to thank Jill Madden and Casie Genetti for their valuable insights into the survey design.
Footnotes
Conflict of Interest: Dr. Agrawal is a member of the Scientific Advisory Board of Illumina Inc. and GeneDx. The other authors have no relevant conflicts of interest to report.
Ethics Declaration: This study was approved by the Institutional Review Board at Boston Children’s Hospital, and completion of the study survey was taken to constitute consent to participate.
Data availability:
Data are available upon request, contingent upon obtaining a data use agreement.
REFERENCES
- 1.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171(12):e173438. doi: 10.1001/jamapediatrics.2017.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swaggart KA, Swarr DT, Tolusso LK, He H, Dawson DB, Suhrie KR. Making a genetic diagnosis in a level IV neonatal intensive care unit population: who, when, how, and at what cost? J Pediatr. 2019;213:211–7 e4. doi: 10.1016/j.jpeds.2019.05.054 [DOI] [PubMed] [Google Scholar]
- 3.Wojcik MH, Schwartz TS, Thiele KE, et al. Infant mortality: the contribution of genetic disorders. J Perinatol. 2019;39(12):1611–1619. doi: 10.1038/s41372-019-0451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malam F, Hartley T, Gillespie MK, et al. Benchmarking outcomes in the neonatal intensive care unit: cytogenetic and molecular diagnostic rates in a retrospective cohort. Am J Med Genet A. 2017;173(7):1839–1847. doi: 10.1002/ajmg.a.38250 [DOI] [PubMed] [Google Scholar]
- 5.Gubbels CS, VanNoy GE, Madden JA, et al. Prospective, phenotype-driven selection of critically ill neonates for rapid exome sequencing is associated with high diagnostic yield. Genet Med. 2020;22:736–744. doi: 10.1038/s41436-019-0708-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18(11):1090–1096. doi: 10.1038/gim.2016.1 [DOI] [PubMed] [Google Scholar]
- 7.Grosse SD, Farnaes L. Genomic sequencing in acutely ill infants: what will it take to demonstrate clinical value? Genet Med. 2019;21(2):269–271. doi: 10.1038/s41436-018-0124-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman JM, Bombard Y, Cornel MC, et al. Genome-wide sequencing in acutely ill infants: genomic medicine’s critical application? Genet Med. 2019;21(2):498–504. doi: 10.1038/s41436-018-0055-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott AM, du Souich C, Lehman A, et al. RAPIDOMICS: rapid genome-wide sequencing in a neonatal intensive care unit-successes and challenges. Eur J Pediatr. 2019;178(8):1207–1218. doi: 10.1007/s00431-019-03399-4 [DOI] [PubMed] [Google Scholar]
- 10.Kingsmore SF, Cakici JA, Clark MM, et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105(4):719–33. doi: 10.1016/j.ajhg.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daoud H, Luco SM, Li R, Bareke E, et al. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. CMAJ. 2016;188(11):254–260. doi: 10.1503/cmaj.150823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maron JL, Kingsmore SF, Wigby K, et al. Novel variant findings and challenges associated with the clinical integration of genomic testing: an interim report of the genomic medicine for ill neonates and infants (GEMINI) study. JAMA Pediatr. 2021;175(5):e205906. doi: 10.1001/jamapediatrics.2020.5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Vandersluis S, Holubowich C, et al. Cost-effectiveness of genome-wide sequencing for unexplained developmental disabilities and multiple congenital anomalies. Genet Med. 2021;23(3):451–60. doi: 10.1038/s41436-020-01012-w. [DOI] [PubMed] [Google Scholar]
- 14.American Academy of Pediatrics. Section on neonatal perinatal medicine. Accessed 25 September. https://www.aap.org/en/community/aap-sections/sonpm/
- 15.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimmock DP, Clark MM, Gaughran M, et al. An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management, and low perceived harm. Am J Hum Genet. 2020;107(5):942–952. doi: 10.1016/j.ajhg.2020.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan NB, Tan TY, Martyn MM, et al. Diagnostic and service impact of genomic testing technologies in a neonatal intensive care unit. J Paediatr Child Health. 2019;55(11):1309–1314. doi: 10.1111/jpc.14398 [DOI] [PubMed] [Google Scholar]
- 18.Marouane A, Keizer R, Frederix GWJ, Vissers L, Boode WP, Zelst-Stams W. Congenital anomalies and genetic disorders in neonates and infants: a single-center observational cohort study. 2021. Eur J Pediatr. (Online ahead of print). doi: 10.1007/s00431-021-04213-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krick JA, Feltman DM. Neonatologists’ preferences regarding guidelines for periviable deliveries: do we really know what we want? J Perinatol. 2019;39(3):445–452. doi: 10.1038/s41372-019-0313-1 [DOI] [PubMed] [Google Scholar]
- 20.Gray MM, Umoren RA, Harris S, Strandjord TP, Sawyer T. Use and perceived safety of stylets for neonatal endotracheal intubation: a national survey. J Perinatol. 2018;38(10):1331–1336. doi: 10.1038/s41372-018-0186-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request, contingent upon obtaining a data use agreement.


