Abstract
Background:
Telomerase reverse transcriptase (TERT) activation has been shown to be an important cancer hallmark; the activation and expression of TERT has been documented in >90% of tumors and TERT activation has been touted as a prognostic marker in many cancers. However, there is currently no simple testing modality to detect TERT mRNA expression in surgical pathology specimens. In this study we aim to evaluate and validate the utility and reliability of the TERT RNAscope® in-situ hybridization (ISH) assay for the detection of TERT mRNA expression in formalin-fixed, paraffin embedded tissue.
Methods and Materials:
RNAscope® detection for TERT was performed on a Leica Biosystems BOND III research staining robot using the Hs-TERT-O1 (ACD, 481968) probe. Twenty three samples containing 48 tissue types were assessed. TERT genomic alterations were determined by targeted next generation sequencing (NGS), while TERT mRNA expression was determined by both targeted RNA-sequencing and TERT RNAscope® and the results compared. Manual vs automated TERT expression quantification methodologies were evaluated for the ISH assay. The expression levels in normal vs. neoplastic tissues were also compared.
Results:
The RNAscope® assay showed high TERT expression in neoplastic tissues, while most normal tissues have no or very low expression levels (p-value= 0.0001, AUC: 0.99). In addition, there was good correlation of TERT expression between the RNAscope® assay and RNA-sequencing. For RNAscope® quantification, manual calculation of TERT signal/cell ratio based on a count of 100 cells was superior compared to automated signal detection.
Conclusion:
TERT RNAscope® assay is a simple and reliable tool for the evaluation of TERT mRNA expression. TERT signal/cell ratio based on a count of 100 cells is a reproducible and accurate interpretation approach for evaluation of TERT expression.
Keywords: TERT, Telomerase, In situ hybridization, Cancer Diagnosis
1. Introduction
Telomeres are repetitive DNA regions located at the ends of chromosomes that form complexes with proteins and maintain the stability of the genome through multiple divisions. However, with aging and continued cellular division, these segments shorten leading to genomic instability and induction of senescence. Consequently, preserving telomere length is fundamental for tumor survival and immortalization [1]. The maintenance of telomere length is mainly achieved through reactivation of telomerase either through the telomerase reverse transcriptase catalytic subunit (TERT) or through alteration of the template subunit (TERC) [2]. In approximately 10% of tumors, alternative lengthening of telomeres occurs through mechanisms independent of telomerase [3].
Tumorigenesis requires neoplastic cells to acquire cancer hallmarks, one of which is enabling replicative immortality for which TERT activation is needed [4]. However, the role of TERT goes beyond telomere lengthening and enabling immortality. It has been shown that TERT is one of the main regulators of tumor progression as it exerts influence on cellular proliferation rate, resistance to apoptosis, and invasive properties of cancer. Furthermore, TERT appears to alter the metabolic and transcriptional landscape of cells and thus contributes to multiple hallmarks of cancer [5].
In fact, almost all tumors (>90%) have been shown to have increased expression of TERT to maintain telomere length which is essential for tumorigenesis and escape from senescence [6]. There are various mechanisms that lead to increased TERT expression; some tumors acquire TERT promoter mutations [7], while others acquire amplification of TERT gene locus on chromosome 5p15 [8], and yet others acquire TERT activation through TERT gene or promoter fusions [9]. However, alternative pathways can also lead to increased expression of TERT; for example, c-MYC has been shown to induce TERT expression [10], a mechanism that has been implicated in human papillomavirus (HPV) infected cells [11], although various other proteins can also be affected by the viral E6 protein leading to modified methylation of TERT promoter and subsequent increased expression of TERT [12].
Telomerase expression has been touted as a potential diagnostic and prognostic marker in tumors [6]. Various attempts have been made to develop an immunohistochemical assay to evaluate TERT status in formalin-fixed paraffin embedded (FFPE) tissue but to date, attempts using antibodies directed against TERT protein have had limited success and have shown poor correlation with TERT alterations or mRNA expression [13]. In this study we aim to evaluate and validate an mRNA in-situ hybridization brightfield assay (RNAscope assay) for TERT.
2. Methods and materials
2.1. Case selection
Twenty-three neoplastic and normal FFPE samples with known TERT promoter mutation and amplification status were selected; 5 samples contained only benign tissue (benign cervical tissue), 6 samples contained only neoplastic tissue and 12 samples contained a mixture of neoplastic and non-neoplastic tissue. The TERT gene alteration status was determined using a targeted hybrid exon-capture next-generation sequencing assay (MSK-IMPACT) in the neoplastic tissues, as previously described [14]. Neoplastic tissues were chosen to ensure representation of cases harboring TERT promoter mutations, TERT amplification and TERT gene or promoter fusion. TERT amplification was determined using the Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS); copy number gains of more than 5 were considered as gene amplification [15]. A targeted RNA based sequencing assay (MSK-Fusion™) was employed to determine TERT mRNA expression level as well as TERT fusions in a subset of the samples (n = 9) [16]. TERT expression levels were defined using the number of reads of the TERT RNA probe normalized to the average number of reads of the assay’s housekeeping genes CHMP2A, GPI, RAB7A and VCP.
2.2. RNAscope assay
RNAscope detection for TERT was performed on a Leica Biosystems BOND III research staining robot (Leica Biosystems), as follows: four micron thick FFPE sections were mounted on charged slides, which were subsequently placed onto a BOND III staining robot and stained using the ACD Bio BOND RNAscope Detection Reagents – Brown (ACD, 201000), as per the manufacturer’s recommendations. Appropriate positive and negative control probes, UBC (ACD, 200178) and DapB (ACD, 200188), respectively, were used to determine that the optimal pre-treatment conditions were identified, which were found to be 15 min at 40 C with protease while heat-induced epitope retrieval was set at 15 mins at 95 C. The TERT probe used from the vendor was Hs-TERT-O1 (ACD, 481968) – a 40 base pairs long probe targeting the 1487 – 3825 region of the TERT gene – which was incubated for two hours at 40 C. slides were then counterstained with hematoxylin, dehydrated in graded ethanol, cleared in xylene, and coverslipped. Each stained tissue was also interrogated with a positive control probe targeting common housekeeping gene UbC to help qualify the samples and a negative control probe targeting bacterial gene dapB gene to control for background noise.
2.3. Scoring and evaluation
The stained slides were evaluated and scored independently by two reviewers (AMB, EY). For scoring of the TERT mRNA expression in tissues, three approaches were employed. First, five 0.25 mm2 areas with the highest amount of staining (as subjectively and qualitatively determined by the reviewers) for the TERT mRNA probe was selected and the number of brown dots within cells and the total number of cells within the area was counted and the ratio of signal to cell was calculated (Fig. 1); the highest attained number among the five areas was then used for comparisons.
Fig. 1.
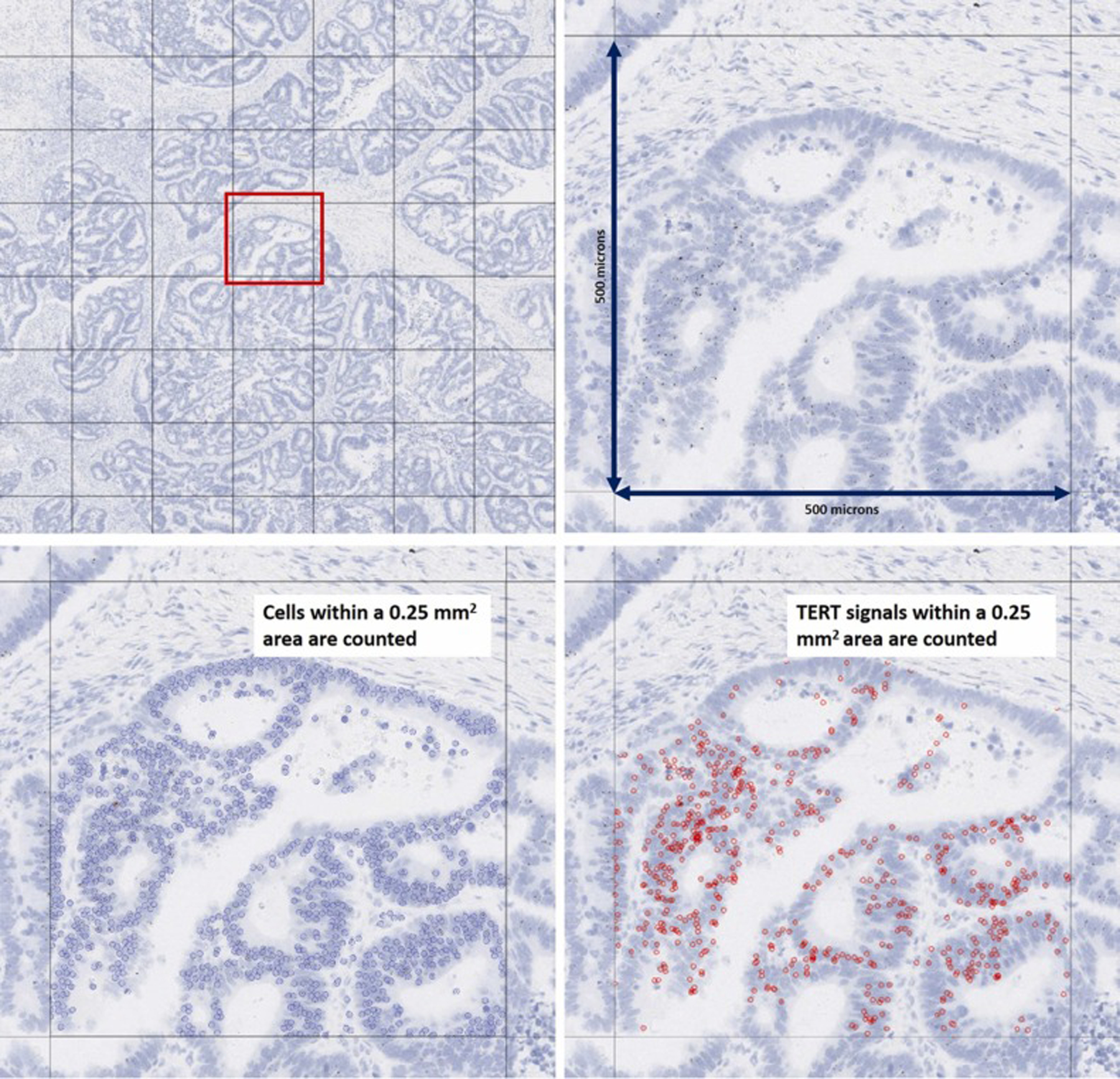
Approach 1 tested for the quantification of TERT ISH expression. In this first approach for quantifying TERT ISH expression, a TERT signal to cell ratio in a 0.25 mm2 area was calculated.
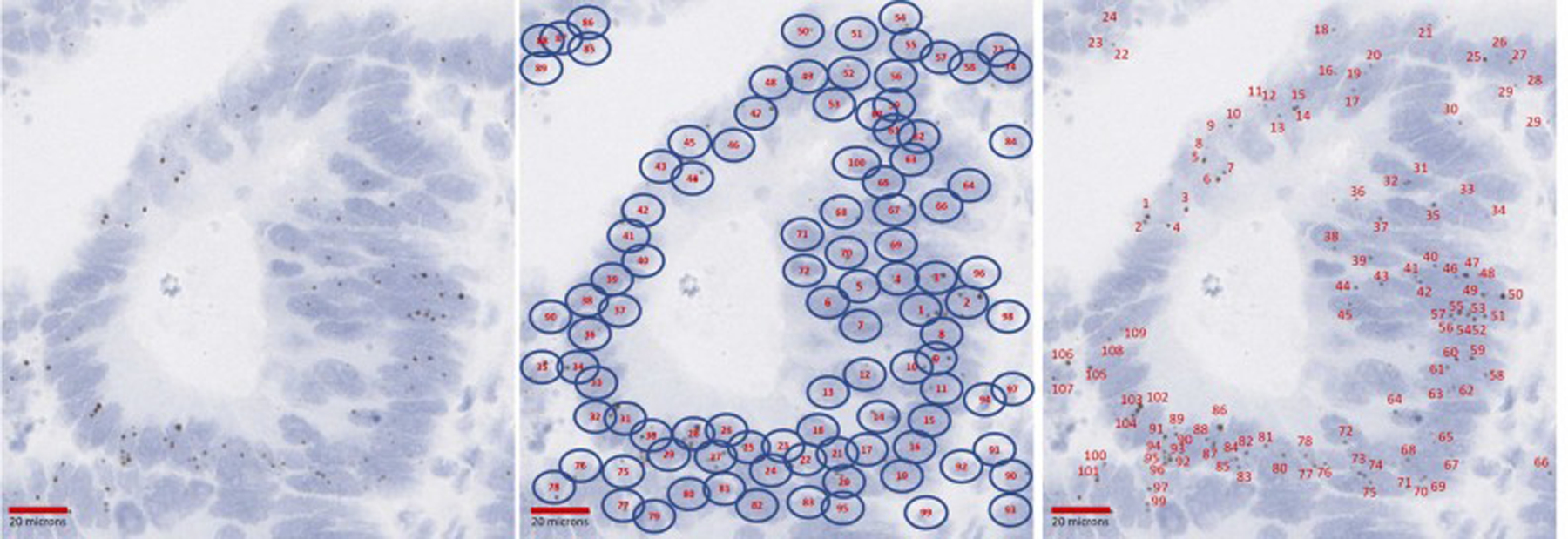
Second, the areas of the tissue with highest TERT expression were selected and 100, 200, 300, 400 and 500 cells with the most TERT signals per nucleus were counted and the total number of TERT signal dots in these cells was also counted by the reviewers. Subsequently the average TERT signal per cell was calculated (Fig. 2).
Fig. 2.
Approach 1 tested for the quantification of TERT ISH expression. In this second approach for quantifying TERT ISH expression, the number of TERT signals in a preset (e.g. 100, 200, 300, …) number of cells with highest TERT expression (here quantification is showed based on a 100-cell count) was counted and a signal to cell ratio calculated.
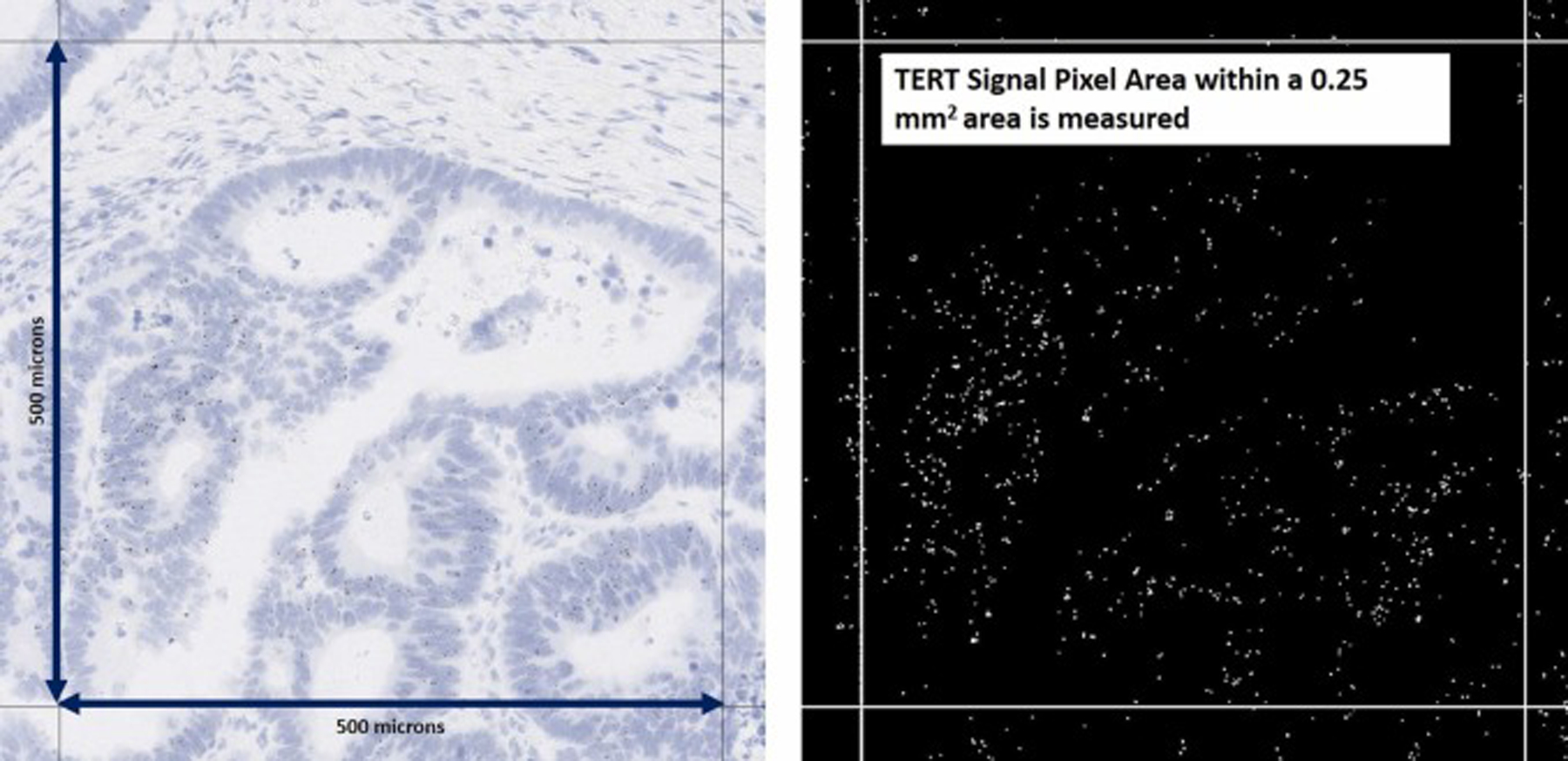
Third, using scanned whole slides of the stained slides, after selection of five 0.25 mm2 areas with highest TERT expression by reviewers, the ratio of DAPI (brown) colored pixels to the overall pixels within that area was calculated using the QuPath software [17] (Fig. 3).
Fig. 3.
Approach 3 tested for the quantification of TERT ISH expression. In this third approach for quantifying TERT ISH expression computational tools of the QuPath software were employed to measure the TERT DAPI signal area within a 0.25 mm2 area.
For tissue sections containing various tissue components (e.g. carcinoma, benign epithelium, benign stroma, …), each tissue component on the slide was scored separately.
The scoring approaches were evaluated using ANOVA with post-hoc Tukey test comparing scores among malignant, benign and intraepithelial neoplasia. Multinomial regression analysis was also used to evaluate the discrimination capacity of TERT scoring approaches to differentiate between malignant, benign and dysplastic tissue. TERT scores were also compared between cases based on TERT gene alteration status (i.e. promoter mutation vs amplification vs fusion vs unknown vs normal). Interobserver reproducibility was determined by calculating the Pearson’s R coefficient. Linear regression analysis was used to evaluate the correlation of the TERT scores with mRNA expression level as determined by the RNA sequencing assay for 9 cases for which TERT RNA expression data was available.
3. Results
In this study, 23 slides were stained using the TERT RNAscope® assay, which corresponded to 48 different tissue types. In total, 29 benign tissue samples, 3 dysplastic tissue samples and 16 malignant tissue samples were evaluated. The breakdown of the tissue sections evaluated is provided in Table 1.
Table 1.
Tissue types evaluated for TERT gene status and TERT RNAscope expression.
| Tissue section type | Tissue section type detailed | Number of tissue sections | Somatic TERT alteration status | Median TERT/cell ratio based on 100 cells |
|---|---|---|---|---|
| Benign (N = 29) | Colon Deep Crypts | 2 | NA | 0.675 |
| Colon Surface Crypts | 2 | NA | 0.000 | |
| Endocervical Glands | 4 | NA | 0.000 | |
| Squamous epithelium | 6 | NA | 0.000 | |
| Anal glandular epithelium | 1 | NA | 0.000 | |
| Stroma from benign and neoplastic samples | 14 | NA | 0.000 | |
| Intraepithelial neoplasia (N = 3) | Endocervical adenocarcinoma in situ | 1 | None | 4.000 |
| High-grade Squamous Intraepithelial Lesion (CIN3) | 1 | None | 1.000 | |
| Differentiated Vulvar Intraepithelial Neoplasia | 1 | Unknown | 0.250 | |
| Malignant (N = 16) | Melanoma | 1 | TERT Amplification | 4.048 |
| Dedifferentiated Liposarcoma | 1 | TERT Gene Fusion | 5.350 | |
| Desmoplastic Small Round Cell Tumor | 1 | TERT Amplification | 13.550 | |
| Osteosarcoma | 1 | TERT Amplification | 7.091 | |
| Myxoid Liposarcoma | 1 | TERT Promoter Mutation | 0.750 | |
| Uterine Serous Carcinoma | 1 | TERT Amplification | 0.350 | |
| Colon Adenocarcinoma | 3 | TERT Amplification [1], None [2] | 1.65 | |
| Vulvar Squamous Cell Carcinoma | 5 | TERT Promoter Mutation [5] | 1.952 | |
| Cervical Squamous Cell Carcinoma | 1 | TERT Amplification | 6.423 | |
| Hepatocellular Carcinoma | 1 | TERT Amplification | 5.700 | |
| Total | 48 |
All neoplastic tissue sections underwent NGS using the MSK-IMPACT assay to determine the somatic TERT status. As expected, the 3 intraepithelial neoplasias did not have any TERT alterations, whereas 14 of 16 malignant tissues harbored alterations in the TERT gene with TERT amplification being the most common (n = 7, 44%), followed by TERT promoter hotspot mutations (n = 6, 38%) and TERT gene fusion (n = 1, 6%). Two colonic adenocarcinomas did not show any genomic TERT alterations based on the targeted DNA or RNA sequencing assays.
Of the 16 malignant cases, 9 underwent testing using the Archer fusion assay and the quantitative TERT RNA expression was extracted, normalized against the housekeeping genes and converted to log-scale. The tested cases had variably increased TERT expression with the lowest expression seen in cancers lacking TERT genomic alteration, however, the numbers were too small for evaluation of statistically significant difference (Fig. 4-A).
Fig. 4.
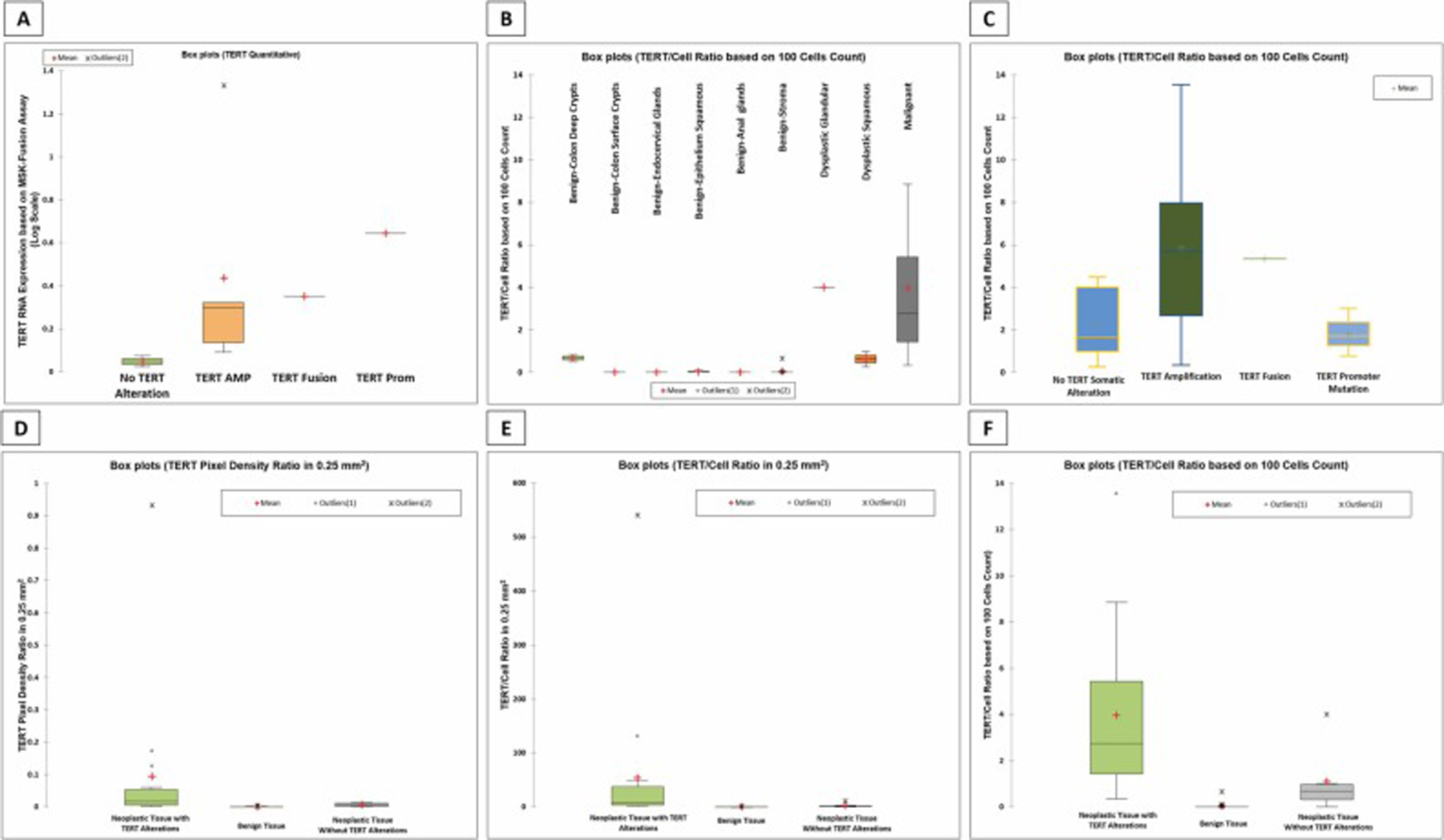
Boxplot showing TERT RNA expression levels in the 9 samples tested using the MSK-Fusion™ assay. The subcategories represent the somatic TERT alterations identified in the samples. (B) Boxplot showing TERT RNAscope® expression as quantified TERT/Cell ratio based on 100 cells count for different tissue types. (C) Boxplots showing TERT RNAscope® expression as quantified TERT/Cell ratio based on 100 cells count in malignant tissue and the corresponding TERT somatic alteration. (D – F) Boxplots depicting the TERT expression quantification using the TERT RNAscope® assay. Panel (D) shows quantification based on TERT pixel density ratio, panel (E) shows TERT/cell ratio in 0.25 mm2 and panel (F) shows TERT/cell ratio based on 100 cells count. Note that the differences between the three tissue categories is best highlighted in the panel on the right.
Three different approaches were used to quantify TERT expression based on the TERT RNAscope® assay, including DAPI color pixel density ratio in a 0.25 mm2 area of tissue (automated using QuPath software), signal to cell ratio in a 0.25 mm2 area (manual counting by two observers), and finally signal to cell ratio based on a set number of cells (manual counting by two observers). For the latter approach, the ratio was obtained in increments of 100 cells in all cases (i.e. signal to cell ratio based on 100, 200, 300, 400 and 500 counted cells) to establish the minimum number of cells that needs to be counted for determining TERT expression status. An ANOVA test followed by post-hoc pairwise comparison failed to show any statistically significant difference in the signal to cell ratio when counting different number of cells (p value: 0.187) and as a result hereafter we will only discuss the signal to cell ratio based on 100 cells along with the two other previously mentioned quantification approaches.
Using regression analysis, we tested to see whether TERT RNAscope® expression correlates with TERT expression as determined by the MSK-Fusion assay for the 9 samples that had the latter assay performed. Our results showed that a linear correlation exists between the two measures (p value = 0.039, R2 = 0.478), further confirming the ability of the TERT RNAscope® assay to determine the TERT mRNA status in FFPE samples.
Comparison of the TERT expression based on TERT RNAscope® assay was made between neoplastic tissue with genomic TERT alterations identified by either the MSK-IMPACT or MSK-Fusion assays, benign tissue and neoplastic tissue without identifiable TERT genomic alterations. While all three TERT quantification approaches showed variations in TERT expression amongst the three tissue groups (Fig. 4-D, E, F), ANOVA analysis revealed that these differences were not statistically significant from each other, with the exception when quantification was performed using the signal to cell ratio based on 100 cells (p value: 0.0001). The average TERT signal per cell ratio based on 100 cells was 3.95 in the neoplastic tissues with TERT genomic alteration, 1.1 in neoplastic tissue without TERT genomic alteration, and 0.03 in benign tissue.
A breakdown of the various tissue types (Fig. 4-B) showed that benign tissue is generally negative for TERT expression, with the only exception being deep crypts of colonic mucosa where cells have low level TERT expression; these cells possibly represent colonic epithelial stem cells which have been shown to overexpress TERT [18].
Among intraepithelial lesions, the HPV associated lesions, including a case of endocervical HPV associated adenocarcinoma in situ and a separate case with high grade squamous intraepithelial lesion, showed relatively high TERT expression (4 and 1 TERT/Cell ratio based on 100 cells count respectively). In contrast, a case of differentiated vulvar intraepithelial neoplasia (dVIN) showed low level expression (0.25 TERT/Cell ratio based on 100 cells count) which was nonetheless significantly higher than benign squamous epithelium which only rarely had a cell expressing TERT (an average of 0.026 TERT/Cell ratio based on 100 cells count). Malignant tissue sections had invariably increased TERT expression levels, which was significantly higher than benign tissue (p value <0.0001). In fact, using the TERT RNAscope® expression alone, there is excellent delineation of non-neoplastic versus neoplastic tissue with a 0.99 area under curve (AUC) on ROC curve analysis and a TERT/cell ratio based on 100 cell-count of 0.448 corresponding to a sensitivity of 89.5% and specificity of 93.1%. Examples of TERT RNAscope® expression are shown in Figs. 5, 6 and 7.
Fig. 5.
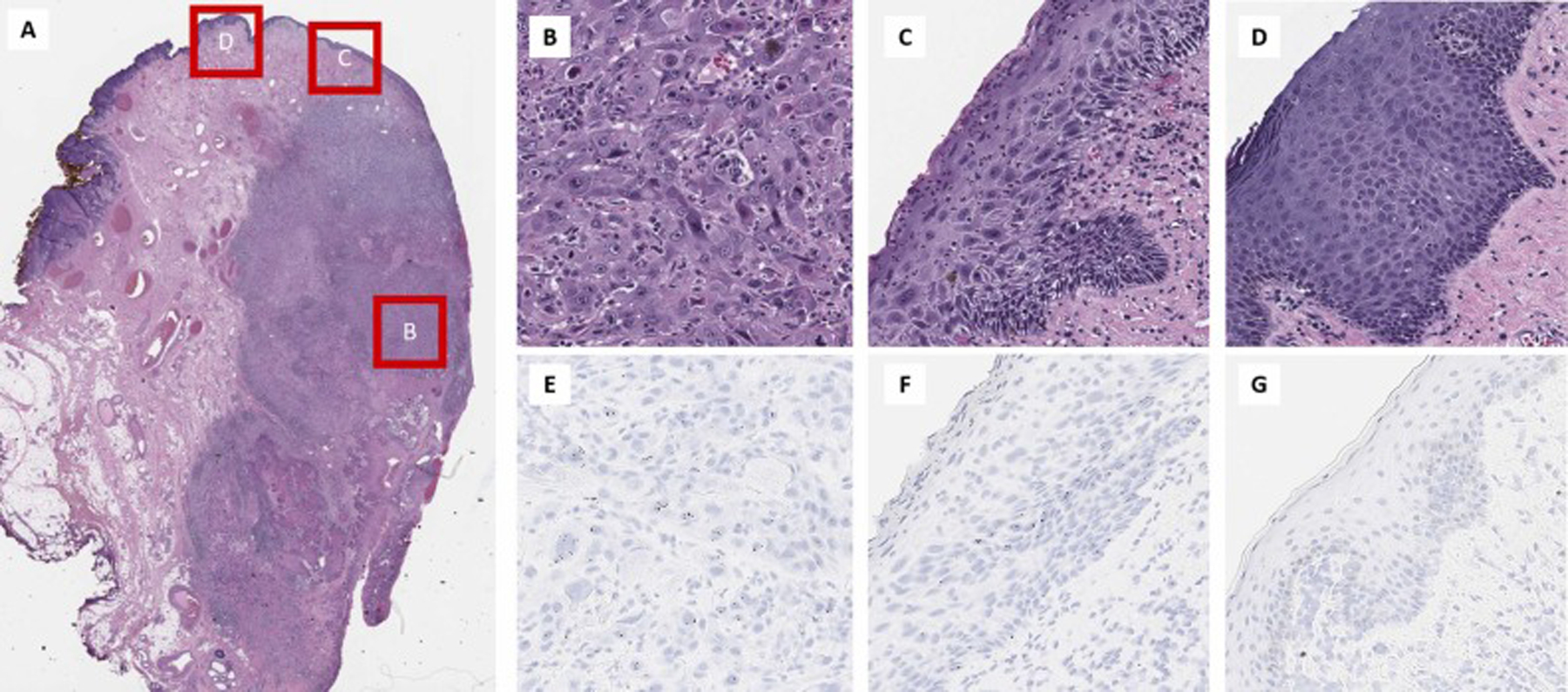
TERT RNAscope® expression in a case vulvar squamous cell carcinoma. (A) Low power magnification of the vulvar squamous carcinoma. The invasive squamous cell carcinoma component (B) has high TERT expression in the tumor cells (E), while the differentiated vulvar intraepithelial neoplasm component (C) has increased expression limited to the basal and parabasal cells (F). Normal squamous epithelium (D) however shows no TERT expression (G).
Fig. 6.
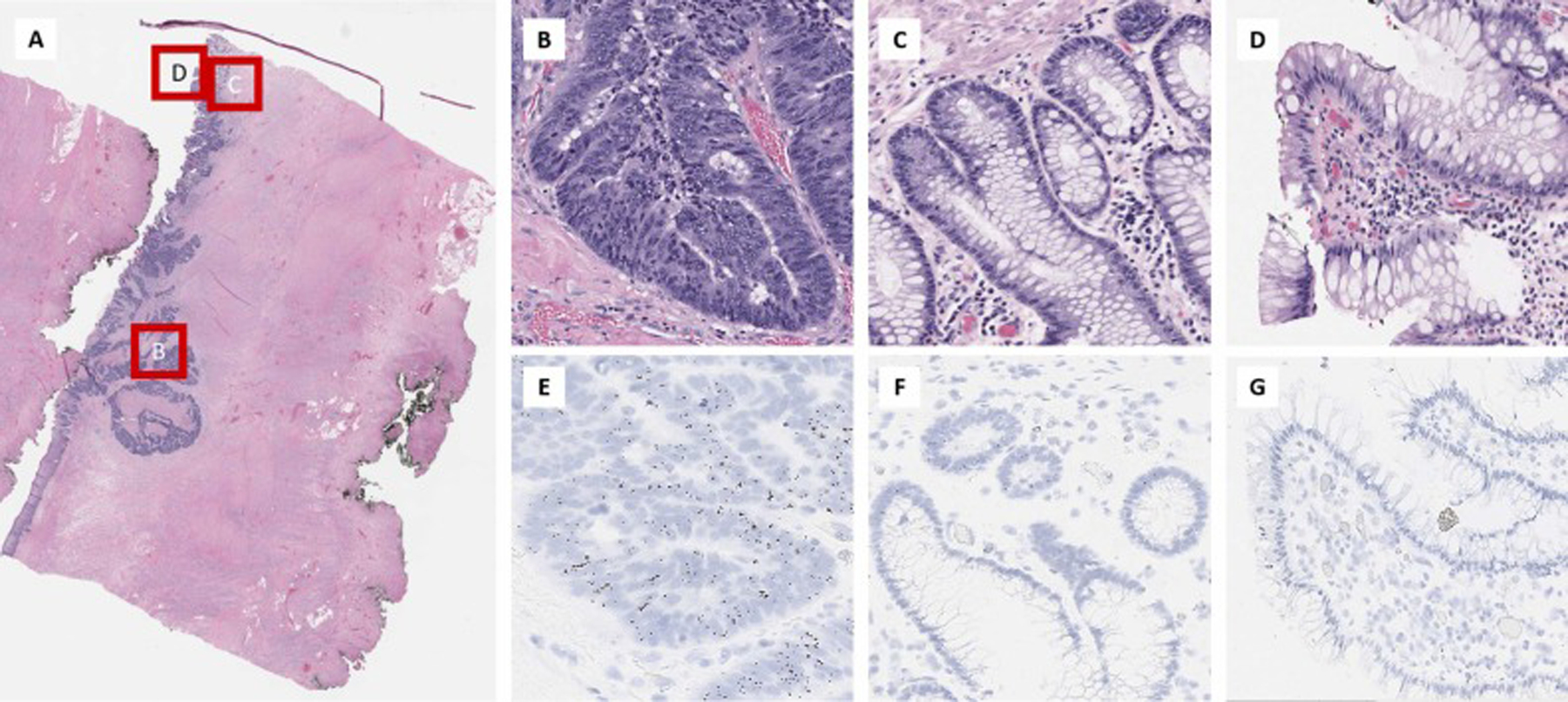
TERT RNAscope® expression in a case of colon adenocarcinoma. (A). Low power magnification of the colon adenocarcinoma. The invasive colon adenocarcinoma (B) has high TERT expression in the tumor cells (E) which is significantly different from normal colonic deep crypts in the same tissue section (C) which show slightly increased TERT expression (F) and superficial colonic epithelium (D) which shows no TERT expression (G).
Fig. 7.
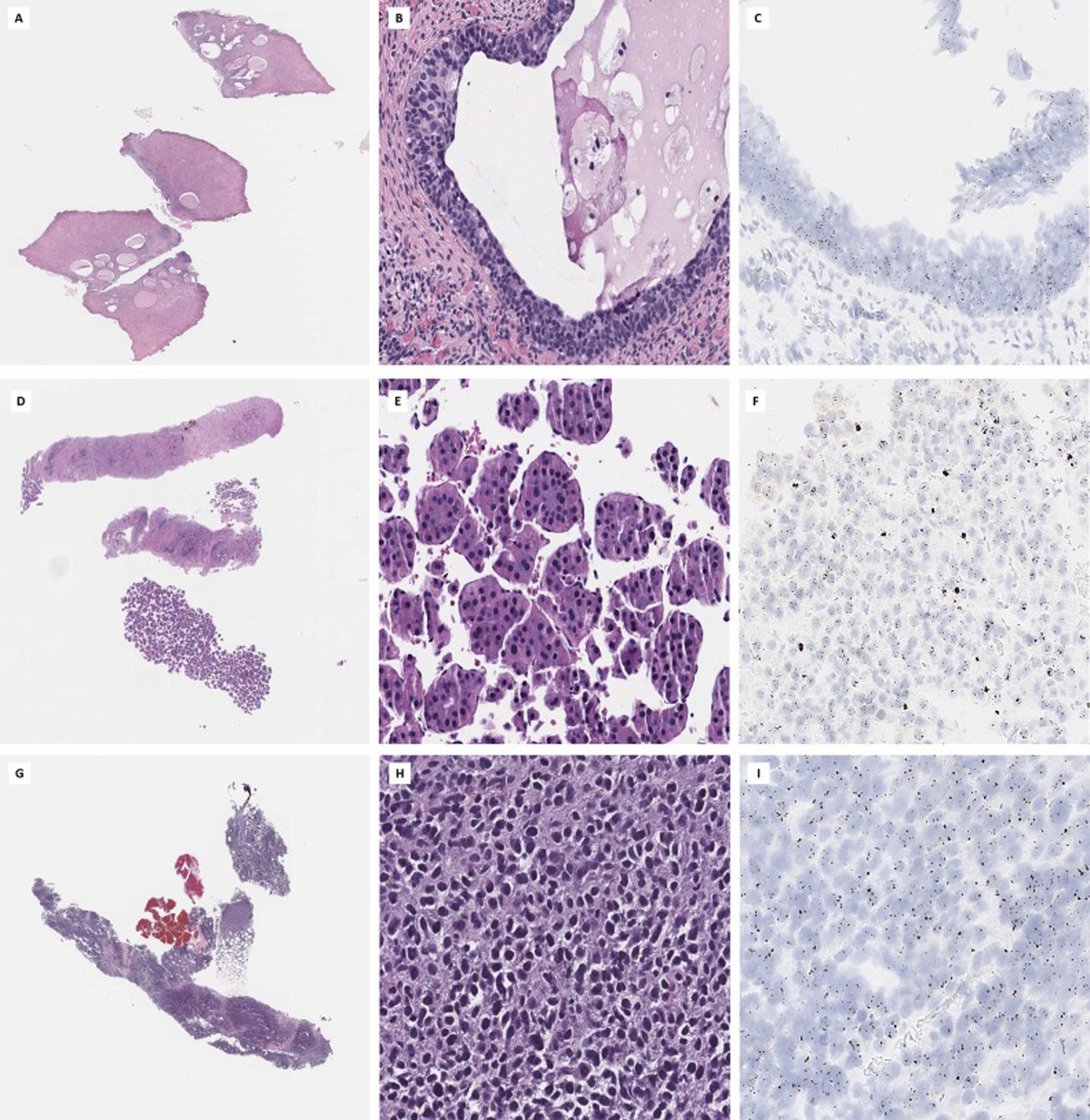
Neoplastic tissues show high levels of TERT RNAscope® expression. Examples of cervical adenocarcinoma in situ (A, B, C), hepatocellular carcinoma (D, E, F) and malignant melanoma (G, H, I) are shown, all of which have high levels of TERT expression by TERT ISH (right).
Consistent with the NGS-based gene expression analysis (Fig. 4A), evaluation of TERT RNAscope® expression in malignant tissue showed that TERT expression is increased regardless of the TERT genomic alteration status, with no significant differences in TERT expression levels amongst the various TERT genomic alterations (p value=0.115) (Fig. 7-C). TERT gene amplification shows the highest degree of variability in TERT expression but the level of TERT mRNA expression does not appear to correlate with the total copy number of the TERT gene as determined by the FACETS algorithm (p value=0.430), however the lack of statistical significance may be due to small number of samples in the cohort.
Interobserver agreement was measured using Pearson’s r correlation measure and showed good interobserver agreement for scoring using the TERT/cell ratio based on 100 cells count approach (Pearson’s r = 0.81).
4. Discussion
TERT reactivation and expression is an essential milestone in many tumors; the neoplastic transformation of cells requires TERT to be activated either through genomic or epigenomic mechanisms. In fact, for many neoplasms, activation of TERT marks malignant transformation and/or acquisition of aggressive behavior and poor prognosis.
Among genomic mechanisms of TERT upregulation and reactivation, TERT promoter mutation is the most common; these alterations often involve G>A substitutions leading to formation of novel binding sites for E-twenty-six (ETS) transcription factor which causes upregulation of TERT transcription [19]. Alternatively, tumor cells can acquire TERT gene amplification. In some tumors, the transcriptional upregulation occurs through TERT gene or promoter fusion with other genes or promoter sequences; these fusion events often involve approximation of enhancer motifs of highly expressed genes to upstream of TERT gene leading to a robust increase in TERT expression and activity [9]. These alterations are often mutually exclusive, and tumors only need one of these alterations for upregulation and reactivation of TERT to occur [20].
TERT upregulation can also occur through epigenomic mechanisms including alteration in the methylation landscape of the upstream regulatory sequences of TERT; hypomethylation at specific CpG islands upstream of the transcriptional start site as well as hypermethylation within the promoter sequence have been correlated with increased TERT gene expression [12,21].
Irrespective of the underlying reactivation, the result is increased TERT mRNA expression, a neoplastic hallmark that is present in the majority of cancers and neoplastic processes. Thus, being able to document and quantify TERT expression is of high value in diagnostic pathology; such a tool can help in distinguishing non-neoplastic versus neoplastic processes (with exception of perhaps stem cell niches which have intrinsic TERT expression), serve as a marker of malignant transformation in some tumors and/or be a surrogate for prognosis in other tumors. We describe some examples for each of these applications below.
Hepatocellular carcinoma invariably has high TERT expression, and genomic TERT alterations are often present in these tumors. TERT promoter mutation has been shown to be an early carcinogenetic event in liver and these alterations are often present in hepatocellular carcinoma and preneoplastic liver lesions including cirrhotic preneoplastic macronodules, and low-grade and high-grade dysplastic nodules. In addition, acquisition of TERT promoter mutation has been linked to malignant transformation of hepatocellular adenoma with TERT mutation status being touted as a possible distinguishing factor between adenoma and carcinoma in liver [22,23].
Thus, evaluation and quantification of TERT expression using the ISH probe can potentially serve as a surrogate of malignant transformation especially when dealing with small and difficult to interpret liver biopsies. In our cohort, we stained one case of hepatocellular carcinoma with the TERT ISH probe (Fig. 7-D, E, F) and subjected it to targeted RNA capture assay. As expected, high level of TERT mRNA expression was shown by both assays. TERT quantification may also serve as a prognostic marker in hepatocellular carcinoma [24].
TERT can also serve as a prognostic marker in tumors as well. For example, in thyroid carcinoma, TERT alterations are common and the frequency of TERT alterations in tumors is directly related to their aggressive behavior, i.e., TERT promoter mutations are shown in ~40% of poorly differentiated and anaplastic thyroid carcinomas versus 11% in papillary thyroid carcinoma and 17% in follicular thyroid carcinoma. TERT status has been proposed as a prognostic marker in thyroid carcinoma and this is a field in which TERT expression measurement using the ISH probe can potentially serve as a prognostic marker [25].
Squamous cell carcinoma often has TERT reactivation [26]; however, depending on the underlying etiology for development of the squamous cell carcinoma, different mechanisms will be utilized by the tumor to activate TERT. For example, in HPV independent squamous cell carcinoma of head and neck and vulva, TERT promoter mutations are the most common mechanism of TERT activation. Conversely, in HPV driven squamous cell carcinoma of cervix, vulva and head and neck, genomic alterations of TERT are uncommon; rather TERT is activated through epigenomic alterations as a direct result of HPV infection. Yet, regardless of the TERT activation mechanism, all squamous cell carcinomas and their neoplastic precursors have increased expression of TERT. As we have shown in this study, squamous cell carcinoma of the cervix (Fig. 8) and vulva have high expression of TERT (Fig. 5). Furthermore, both cervical high-grade squamous intraepithelial lesion and differentiated vulvar intraepithelial neoplasia (dVIN) have increased TERT expression (Fig. 4). In contrast, normal non-dysplastic squamous epithelium shows only very rare cells expressing TERT. As a result, TERT expression can be used as a surrogate for squamous precursor lesions; this would be especially helpful in diagnosis of dVIN, which is a notoriously difficult lesion to diagnose [27–30].
Fig. 8.
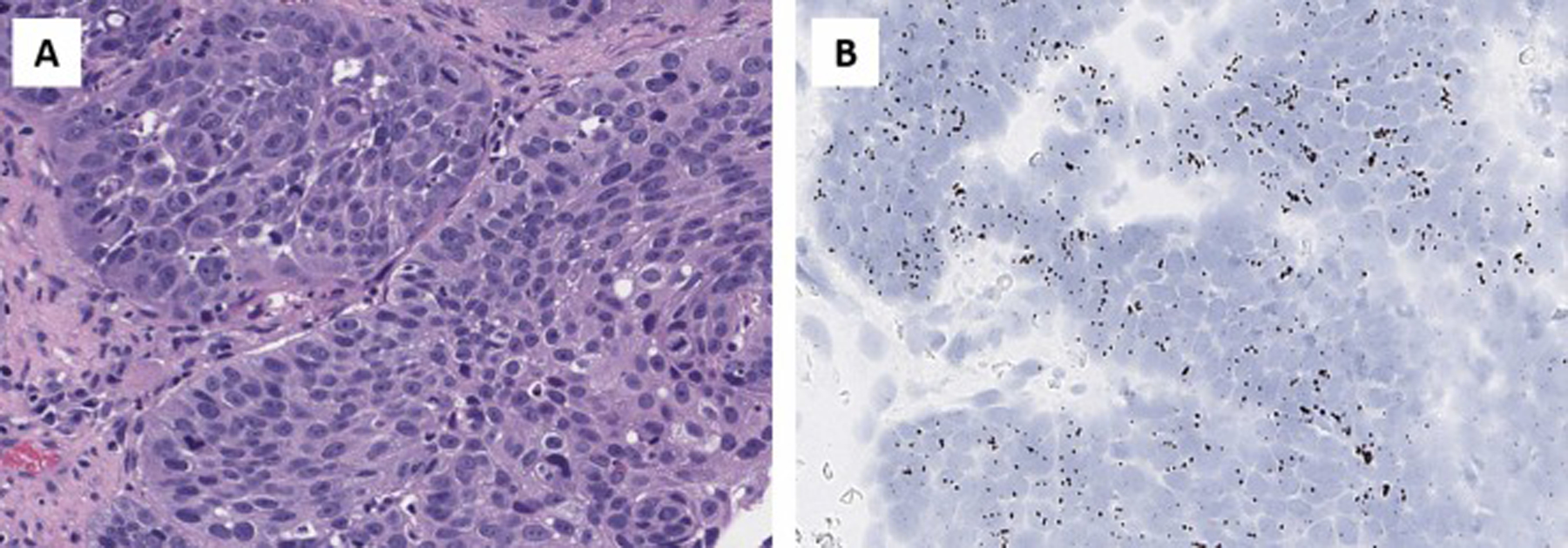
TERT ISH expression in HPV-associated cervical squamous cell carcinoma.
In addition, there have been studies that have suggested that TERT expression levels differ between low-grade and high-grade squamous intraepithelial lesions. Consequently, TERT expression may also be used in differentiating these lesions in difficult cases where other ancillary tests are not helpful [31,32].
Cutaneous melanoma is another malignant entity that has been shown to require TERT expression. In fact, in xenograft models it was shown that cutaneous melanoma requires TERT activation alongside a mitogenic driver (e.g. NRAS mutation), senescence evasion (e.g. CDK4 mutation) and antiapoptotic alterations (e.g., TP53 mutation) to acquire invasive properties; consequently, TERT alterations are categorized as immortalizing mutations that occur later in tumor evolution and allow for invasion and metastasis [33].
TERT activation is an important evolutionary landmark in cutaneous melanomas [34]; studies have shown that the majority (70%) of invasive melanomas have TERT mutations [35], while nevi or radial growth--phase melanomas rarely have TERT alterations [36]. On the other hand, recent studies have shown that TERT promoter mutations may be present at subclonal levels in precursor melanocytic lesions [37]. As such, a quantifiable biomarker such as the TERT RNAscope® may be better suited for difficult to classify melanocytic lesions compared to high-sensitivity sequencing assays, as it allows for the visualization of tissue level expression, as well as correlation with histomorphology. Among our samples we had a single case of invasive melanoma which showed remarkably high TERT expression levels (Fig. 7 – G, H, I) in all tumor cells, highlighting the possible utility of this assay. Furthermore, TERT alterations have been shown to be a significant predictor of adverse outcome in melanoma suggesting TERT expression may also serve as a prognostic tool in these tumors [38].
Our results show that the TERT RNAscope® assay is a reliable tool for the evaluation of TERT expression in formalin fixed paraffin embedded tissue as it correlates with TERT expression as determined by next generation sequencing assay. Interestingly, the RNAscope® assay shows that TERT expression can be variable throughout neoplastic tissue which may lead to underestimation of TERT expression by sequencing methodologies. Furthermore, the RNAscope® assay allows for morphology/expression correlation and can reliably distinguish neoplastic from non-neoplastic tissues, making this a valuable tool in diagnostic histopathology.
We also presented a relatively simple yet reliable method for documenting and quantifying TERT expression. In some instances, TERT mRNA expression by RNAscope® can be qualitatively interpreted since neoplastic tissue often has robust expression in contrast to the almost entirely negative expression in non-neoplastic tissue. However, quantification may be necessary in differentiating non-neoplastic from neoplastic tissue in some cases and we have shown that counting TERT signals in 100 cells with the highest TERT expression (as determined by a visual inspection and screening of the stained slide) is a reliable and reproducible approach in quantification of TERT expression: TERT/cell ratio based on 100 cells of more than 0.448 reliably separates neoplastic tissues from non-neoplastic tissues (AUC: 0.99). Cutoffs, however, would have to be adjusted based on specific use intended for TERT and it remains to be seen whether quantification of TERT expression is correlated with prognosis in various tumors. Additional studies are warranted to define these applications.
The main limitation of our study was the small size of the cohort; however, the main aim of this study was to serve as a proof of concept to show that TERT expression can be adequately measured using the RNAscope® assay. Previous attempts at TERT RNA in-situ hybridization were made using custom designed probes with similar results to our cohort [39], however, the current methodology is promising in that it shows excellent sensitivity and specificity combined with standardized methodology and ease of utilization.
In summary, we have shown the utility of TERT mRNA expression analysis using the RNAscope® assay and we have also suggested a reproducible and simple quantification and interpretation approach for evaluation of TERT expression. The assay has potential to become a useful diagnostic tool in the arsenal of histopathologists; however, further work, including validation studies focusing on specific pathologic entities, is needed in order to establish the scope of the clinical utility of TERT expression analysis.
Acknowledgements
This study was funded in part through the NIH/NCI Support Grant P30 CA008748 for Memorial Sloan Kettering Cancer Center.
References
- [1].Jafri MA, Ansari SA, Alqahtani MH, Shay JW, Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies, Genome Med. 8 (1) (2016) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Akincilar SC, Unal B, Tergaonkar V, Reactivation of telomerase in cancer, Cell. Mol. Life Sci 73 (8) (2016) 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cesare AJ, Reddel RR, Alternative lengthening of telomeres: models, mechanisms and implications, Nat. Rev. Genet 11 (5) (2010) 319–330. [DOI] [PubMed] [Google Scholar]
- [4].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (5) (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [5].Low KC, Tergaonkar V, Telomerase: central regulator of all of the hallmarks of cancer, Trends Biochem. Sci 38 (9) (2013) 426–434. [DOI] [PubMed] [Google Scholar]
- [6].Mathon NF, Lloyd AC, Cell senescence and cancer, Nat. Rev. Cancer 1 (3) (2001) 203–213. [DOI] [PubMed] [Google Scholar]
- [7].Heidenreich B, Rachakonda PS, Hemminki K, Kumar R, TERT promoter mutations in cancer development, Curr. Opin. Genet. Dev 24 (2014) 30–37. [DOI] [PubMed] [Google Scholar]
- [8].Cao Y, Bryan TM, Reddel RR, Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells, Cancer Sci. 99 (6) (2008) 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Karlsson J, Lilljebjorn H, Holmquist Mengelbier L, Valind A, Rissler M, Ora I, et al. , Activation of human telomerase reverse transcriptase through gene fusion in clear cell sarcoma of the kidney, Cancer Lett. 357 (2) (2015) 498–501. [DOI] [PubMed] [Google Scholar]
- [10].Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. , Direct activation of TERT transcription by c-MYC, Nat. Genet 21 (2) (1999) 220–224. [DOI] [PubMed] [Google Scholar]
- [11].McMurray HR, McCance DJ, Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression, J. Virol 77 (18) (2003) 9852–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jiang J, Zhao LJ, Zhao C, Zhang G, Zhao Y, Li JR, et al. , Hypomethylated CpG around the transcription start site enables TERT expression and HPV16 E6 regulates TERT methylation in cervical cancer cells, Gynecol. Oncol 124 (3) (2012) 534–541. [DOI] [PubMed] [Google Scholar]
- [13].Paulsson JO, Olander A, Haglund F, Zedenius J, Juhlin CC, TERT immunohistochemistry is a poor predictor of TERT promoter mutations and gene expression in follicular thyroid carcinoma, Endocr. Pathol 29 (4) (2018) 380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. , Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology, J. Mol. Diagn 17 (3) (2015) 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shen R, Seshan VE, FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing, Nucleic Acids Res. 44 (16) (2016), e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, et al. , High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden, Clin. Cancer Res 25 (15) (2019) 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. , QuPath: open source software for digital pathology image analysis, Sci. Rep 7 (1) (2017) 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Flores I, Cayuela ML, Blasco MA, Effects of telomerase and telomere length on epidermal stem cell behavior, Science 309 (5738) (2005) 1253–1256. [DOI] [PubMed] [Google Scholar]
- [19].Lorbeer FK, Hockemeyer D, TERT promoter mutations and telomeres during tumorigenesis, Curr. Opin. Genet. Dev 60 (2020) 56–62. [DOI] [PubMed] [Google Scholar]
- [20].Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, et al. , Systematic analysis of telomere length and somatic alterations in 31 cancer types, Nat. Genet 49 (3) (2017) 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee DD, Leao R, Komosa M, Gallo M, Zhang CH, Lipman T, et al. , DNA hypermethylation within TERT promoter upregulates TERT expression in cancer, J. Clin. Investig 129 (1) (2019) 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. , High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions, Nat. Commun 4 (2013) 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nault JC, Zucman-Rossi J, TERT promoter mutations in primary liver tumors, Clin. Res. Hepatol. Gastroenterol 40 (1) (2016) 9–14. [DOI] [PubMed] [Google Scholar]
- [24].Yu JI, Choi C, Ha SY, Park CK, Kang SY, Joh JW, et al. , Clinical importance of TERT overexpression in hepatocellular carcinoma treated with curative surgical resection in HBV endemic area, Sci. Rep 7 (1) (2017) 12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu R, Xing M, TERT promoter mutations in thyroid cancer, Endocr. Relat. Cancer 23 (3) (2016) R143–R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Griewank KG, Murali R, Schilling B, Schimming T, Moller I, Moll I, et al. , TERT promoter mutations are frequent in cutaneous basal cell carcinoma and squamous cell carcinoma, PLOS One 8 (11) (2013), e80354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, et al. , Genomic, pathway network, and immunologic features distinguishing squamous carcinomas, Cell Rep. 23 (1) (2018) 194–212, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yuan X, Larsson C, Xu D, Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players, Oncogene 38 (34) (2019) 6172–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Morris LGT, Chandramohan R, West L, Zehir A, Chakravarty D, Pfister DG, et al. , The molecular landscape of recurrent and metastatic head and neck cancers: insights from a precision oncology sequencing platform, JAMA Oncol. 3 (2) (2017) 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jin C, Liang S, Differentiated vulvar intraepithelial neoplasia: a brief review of clinicopathologic features, Arch. Pathol. Lab. Med 143 (6) (2019) 768–771. [DOI] [PubMed] [Google Scholar]
- [31].Wisman GB, De Jong S, Meersma GJ, Helder MN, Hollema H, de Vries EG, et al. , Telomerase in (pre)neoplastic cervical disease, Hum. Pathol 31 (10) (2000) 1304–1312. [DOI] [PubMed] [Google Scholar]
- [32].Ravindranathan A, Cimini B, Diolaiti ME, Stohr BA, Preliminary development of an assay for detection of TERT expression, telomere length, and telomere elongation in single cells, PLOS One 13 (12) (2018), e0206525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bennett DC, Genetics of melanoma progression: the rise and fall of cell senescence, Pigment Cell Melanoma Res. 29 (2) (2016) 122–140. [DOI] [PubMed] [Google Scholar]
- [34].Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. , The genetic evolution of melanoma from precursor lesions, N. Engl. J. Med 373 (20) (2015) 1926–1936. [DOI] [PubMed] [Google Scholar]
- [35].Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA, Highly recurrent TERT promoter mutations in human melanoma, Science 339 (6122) (2013) 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, et al. , TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma, J. Natl. Cancer Inst 106 (2014) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Colebatch AJ, Ferguson P, Newell F, Kazakoff SH, Witkowski T, Dobrovic A, et al. , Molecular genomic profiling of melanocytic nevi, J. Investig. Dermatol 139 (8) (2019) 1762–1768. [DOI] [PubMed] [Google Scholar]
- [38].Nagore E, Heidenreich B, Rachakonda S, Garcia-Casado Z, Requena C, Soriano V, et al. , TERT promoter mutations in melanoma survival, Int. J. Cancer 139 (1) (2016) 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kolquist KA, Ellisen LW, Counter CM, Meyerson MM, Tan LK, Weinberg RA, Haber DA, Gerald WL, Expression of TERT in early premalignant lesions and a subset of cells in normal tissues, Nat. Genet 19 (2) (1998) 182–186. [DOI] [PubMed] [Google Scholar]


