Abstract
Rice straw is a major substrate for the production of methane, a greenhouse gas, in flooded rice fields. The bacterial community degrading rice straw under anoxic conditions was investigated with molecular methods. Rice straw was incubated in paddy soil anaerobically for 71 days. Denaturing gradient gel electrophoresis (DGGE) of the amplified bacterial 16S rRNA genes showed that the composition of the bacterial community changed during the first 15 days but then was stable until the end of incubation. Fifteen DGGE bands with different signal intensities were excised, cloned, and sequenced. In addition, DNA was extracted from straw incubated for 1 and 29 days and the bacterial 16S rRNA genes were amplified and cloned. From these clone libraries 16 clones with different electrophoretic mobilities on a DGGE gel were sequenced. From a total of 31 clones, 20 belonged to different phylogenetic clusters of the clostridia, i.e., clostridial clusters I (14 clones), III (1 clone), IV (1 clone), and XIVa (4 clones). One clone fell also within the clostridia but could not be affiliated to one of the clostridial clusters. Ten clones grouped closely with the genera Bacillus (3 clones), Nitrosospira (1 clone), Fluoribacter (1 clones), and Acidobacterium (2 clones) and with clone sequences previously obtained from rice field soil (3 clones). The relative abundances of various phylogenetic groups in the rice straw-colonizing community were determined by fluorescence in situ hybridization (FISH). Bacteria were detached from the incubated rice straw with an efficiency of about 80 to 90%, as determined by dot blot hybridization of 16S rRNA in extract and residue. The number of active (i.e., a sufficient number of ribosomes) Bacteria detected with a general eubacterial probe (Eub338) after 8 days of incubation was 61% of the total cell counts. This percentage decreased to 17% after 29 days of incubation. Most (55%) of the active cells on day 8 belonged to the genus Clostridium, mainly to clostridial clusters I (24%), III (6%), and XIVa (24%). An additional 5% belonged to the Cytophaga-Flavobacterium cluster of the Cytophaga-Flavobacterium-Bacteroides phylum, 4% belonged to the α, β, and γ Proteobacteria, and 1.3% belonged to the Bacillus subbranch of the gram-positive bacteria with a low G+C content. The results show that the bacterial community colonizing and decomposing rice straw developed during the first 15 days of incubation and was dominated by members of different clostridial clusters, especially clusters I, III, and XIVa.
Wetland rice fields annually release about 60 to 100 million tons of CH4 and thus contribute substantially to the global warming of the atmosphere (50). Methane emission from rice fields starts after the flooding of the fields and stops when the fields are drained for harvest. During this period CH4 emission is driven by the anaerobic degradation of organic matter in the submerged soil (10). One of the main carbon sources is rice straw, commonly incorporated for the fertilization of the fields (53). Previous studies showed that the application of rice straw increases the CH4 emission from rice fields significantly (6, 12, 36, 42). The main components of rice straw are hemicellulose (26 to 35%), cellulose (38 to 41%), lignin (15%), and water-soluble polysaccharides (8%) (16, 52). A complex microbial community is necessary for the degradation of these biopolymers and consists of hydrolytic or cellulolytic, fermenting, homoacetogenic, and syntrophic bacteria and acetate- and H2-utilizing methanogenic archaea (48).
Of all microorganisms existing in nature, approximately 99% have not been detected with cultivation-based techniques (3). Therefore molecular, cultivation-independent methods, such as the cloning and sequencing of the 16S rRNA gene, are often used to get a more complete picture from the community living in a particular habitat (3, 43). Quantitative data can be obtained by fluorescence in situ hybridization (FISH) or by dot blot hybridization using specific probes targeting 16S rRNA (3).
Studies of the microbial community in anoxic rice paddy soil showed a diverse community structure (37). Analysis of phospholipid fatty acids and of group-specific lipids such as diether lipids for methanogens and plasmologen phospolipids for clostridia showed that the total microbial biomass decreased at the flowering stage of flooded rice fields to one-half of the level before flowering. In contrast, individual anaerobic groups such as the methanogens and the fermentative clostridia increased (37). Hengstmann et al. (20) determined the phylogenetic affiliations of 57 clones of 16S rDNA sequences obtained from DNA extracted from rice field soil and showed that 33 of these sequences clustered with the genus Clostridium. Chin et al. (7) isolated three strains (RCel1, RXyl1, and RPec1) from rice field soil by enrichment with defined polysaccharides (cellulose, xylan, and pectin). These three isolates grouped within clostridial clusters I and III (sensu Collins et al. [9]). On the same media, Chin et al. (8) isolated seven strains after dilution of the inoculum to obtain the most abundant culturable phenotypes. These isolates were affiliated with the clostridial clusters I, III, IX, and XIVa. Other abundant 16S rDNA sequences of the domain Bacteria in rice field soil were found to cluster within the division Verrucomicrobia, the Cytophaga-Flavobacterium-Bacteroides (CFB) group, the genus Bacillus, the class Actinobacteria, the family Chlorobiaceae, and the α subdivision of the Proteobacteria (8, 20).
The last step of the anaerobic degradation of biopolymers, the formation of methane, is performed by methanogenic archaea. The responsible microorganisms in rice field soil, detected by sequencing the 16S rDNA, were shown to belong to the genera Methanosaeta, Methanobacterium, and Methanosarcina, to Methanomicrobiales, and to novel groups (17, 18).
So far all information about the microbial community involved in anaerobic degradation processes in rice field soil was obtained either by the investigation of bulk soil without any application of organic matter (20) or by the characterization of isolates obtained from enrichment cultures and most-probable number studies on defined substrates such as cellulose, pectin, and xylan (7, 8). Rice straw, however, is a mixture of many different components in a highly structured composition. Microscopic studies showed a complex colonization pattern of rice straw (24). Glissmann and Conrad (15) recently investigated the fermentation pattern of rice straw and found during the early phase of degradation a qualitative difference in the fermentation pattern of organic matter in the unamended soil. However, nothing is known about the composition of the microbial community colonizing rice straw.
Therefore, we investigated the bacterial community colonizing and degrading rice straw in soil slurries using molecular methods. Community patterns of bacteria via denaturing gradient gel electrophoresis (DGGE) were determined, and clone libraries for the domain Bacteria were created. In addition, quantification of bacterial groups was done by FISH. New 16S rRNA probes for several clusters of the genus Clostridium (sensu Collins et al. [9]) were developed.
MATERIALS AND METHODS
Soil and straw samples.
The soil samples used were obtained from rice fields in Vercelli, Italy. The soil was a sandy loam and has been described before (22). The soil that had been used for growing rice in the greenhouse was drained, air dried, crushed, sieved (1-mm mesh size), and stored at room temperature. The straw originated from rice plants (Oryza sativa var. Roma) and was air dried and stored at room temperature. For the experiments, only the stems of the straw were used; they were cut into pieces approximately 2 cm in length.
Incubation of the straw.
The setup of the experiments was described previously (15): 40 g of dry soil and 0.5 g of straw were mixed with 40 ml of deionized water, and the mixture was poured into 150-ml glass bottles (Müller & Krempel, Bülach, Switzerland) and incubated for up to 10 weeks at 25°C in the dark. The amount of straw corresponds to 37.5 tons ha−1, i.e., about three times higher than normal (42). As controls, straw was incubated in 50 mM phosphate buffer without soil (pH 7.0) and soil was incubated without the application of straw. Methane formation was measured as described by Glissmann and Conrad (15).
Preparation of samples for DNA extraction and FISH.
After an incubation times of 1 and 4 weeks, the straw pieces were separated from the soil and washed twice in phosphate-buffered saline (PBS) (0.8% NaCl in 10 mM phosphate, pH 7.0). The straw was then placed together with 5 ml of fresh PBS into a plastic bag and treated with a stomacher (Seward, London, United Kingdom) for 1 min at high speed (13). The PBS was withdrawn into a tube and replaced by 5 ml of fresh PBS, and the procedure was repeated twice. From the collected PBS-cell extract (15 ml), 800 μl was fixed with freshly prepared 4% formaldehyde in PBS for 1 h at room temperature (1). Afterwards the fixed samples were centrifuged (8,765 × g, 5 min), the pellet was resuspended in 1 ml of toluidine blue (45) (Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany; 0.01% [wt/vol] in PBS) and incubated for 1 h at 22°C to reduce the autofluorescence of soil and straw particles. After centrifugation (8,765 × g, 5 min), the pellet was washed three times in PBS and stored in an equal volume of PBS-ethanol at −20°C. The rest of the PBS from the original stomacher extract was concentrated by centrifugation (2,516 × g, 15 min). The pellet and the treated straw were combined and stored at −20°C for later DNA extraction (see below).
DNA extraction.
The procedure used for DNA extraction was similar to a protocol described recently (44), with some modifications. The frozen samples (straw plus concentrated PBS-cell extract) were lyophilized over night (lyophylisator; Christ, Osterode, Germany) and homogenized with a mortar and pestle under liquid nitrogen. About 100 to 200 mg of the sample was dissolved in 1 ml of extraction buffer (0.1 M Tris-HCl [pH 8.0], 50 mM EDTA, 0.5 M NaCl, 1.0 mM dithiothreitol) and shaken vigorously. After three freeze-and-thaw cycles (−196 and +65°C), 5 mg of lysozyme was added and the mixture was incubated for 2 h at 37°C, followed by addition of proteinase K (final concentration: 100 μg ml−1; Promega) and sodium dodecyl sulfate (SDS) (final concentration: 1.7% [wt/vol]) and a further incubation for 1 h at 37°C. The proteins were removed by three chloroform-isoamyl alcohol extractions (24:1 [vol/vol]). The DNA was precipitated with 2 volumes of ethanol at 22°C for 90 min and centrifuged (20,800 × g for 20 min). After being dried (Speed-Vac concentrator; Novodirect GmbH, Kehl, Germany), the DNA was resuspended in 200 μl of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Further purification was done as described elsewhere (17).
PCR amplification.
DNA samples were amplified by PCR as described elsewhere (20). Two bacterial oligonucleotide primer pairs were used: 27f and 1492r (28), with an annealing temperature of 48°C, and F-968 and R-1401/1378 (13), with an annealing temperature of 60°C. The thermal profile for amplification included 30 cycles of denaturation at 92°C for 45 s, primer annealing for 1 min, and primer extension at 72°C for 2 min. For DGGE, primer F-968 was modified at the 5′ end with a 40-bp GC clamp (13). The DNA concentration was 5 to 25 ng for each reaction.
DGGE.
DGGE was done according to previously described protocols (19, 35). DNA was amplified with primer pair F-968-GC and R-1401/1378 (13). The PCR products were separated on a polyacrylamide gel with a denaturing gradient from 20% (6% [wt/vol] acrylamide-bisacrylamide [37.5:1], 8% formamide, 1.4 M urea, 2% glycerol) to 50% (6% [wt/vol] acrylamide-bisacrylamide [37.5:1], 20% formamide, 3.5 M urea, 2% glycerol). The electrophoresis was performed in an electrophoresis cell (Dcode-System; Bio-Rad Laboratories GmbH, Munich, Germany) with 0.5× Tris-acetate-EDTA at 60°C and 150 V for 4.5 h. Visualization of the bands was done as described previously (19). Bands were excised with a 200-μl pipette tip and transferred into 200 μl of Tris-EDTA buffer. The DNA was reamplified and cloned.
Cloning and sequencing.
Cloning and sequencing were done with two different strategies. In the first approach, DNA extracted from DGGE bands was reamplified and cloned. The motilities of the resulting clone fragments were checked by DGGE and compared with the original pattern of the excised band. Clones which had the same motility as the original band, were sequenced with primers F-968 and R-1401/1378 as described by Engelen et al. (13). This cloning step was performed in order to be confident that the clone sequences represented the respective bands in the pattern. In the following, the term RSD is used for these clone libraries. In the second approach, the 16S rDNA of straw samples incubated for 1 day and 4 weeks was amplified with primer pair 27f and 1492r (28) and two clone libraries, RSa (1 day) and RSb (29 days), were created. For each incubation time, the DNA from about 20 randomly selected clones was extracted as described by Rotthauwe et al. (39) and amplified and the mobility of the 16S rDNA was checked by DGGE. Clones with different mobilities were sequenced with primers 9 and 27f (28) and 350f (55).
Cloning was performed by using the original TOPO cloning kit (pCR 2.1 vector for Escherichia coli: TOP 10F′; Invitrogen, Leek, The Netherlands) or the TOPO cloning kit (pCR 2.1 vector for E. coli: TOP 10; Invitrogen) in accordance with the manufacturer's instructions. Extraction of DNA from clones, amplification with primers that target vector sequences, purification of the PCR product, and nonradioactive sequencing were performed as described elsewhere (39).
Phylogenetic analysis.
16S rDNA sequences were added to a database consisting of about 7,000 complete or partial bacterial 16S rRNA sequences which are publicly available. This database is part of the ARB program package (O. Strunk et al., ARB: a software environment for sequence data [http://www.biol.chemie.tu-muenchen.de/pub/ARB/], Department of Microbiology, Technische Universität München, Munich, Germany, 1997) The related sequences XB90, SB90, FCB45, FCB90-1/2 (8), UA3 (AF200699), and OPB54 (AF027087) and that of Clostridium quercicolum (AJ010962), 4C28d-3 (AB034137); and that of a Bacillus sp. (AB030930) obtained by using the Wu-Blast2 tool of the EMBL database were added to the ARB program. In addition sequences from several clones (BSV clones [20]) and from isolates RXy11 and RPec (7) previously obtained from rice field soil were integrated into the database. All 16S rDNA sequences from this study were integrated into the database with the automatic alignment tool of the ARB program package. The resulting alignments were manually checked and corrected if necessary. Distance matrix analyses provided in the ARB program gave pairwise identity values between sequences. Evolutionary distance values between pairs of microorganism were calculated by using the Jukes-Cantor equation (23). Phylogenetic trees were constructed by using the neighbor-joining and maximum-likelihood algorithm provided in the program (40). Various filters for the genus Clostridium with 40 and 50% invariances, respectively, and for clostridial clusters I, III, IV, IX, and XIVa with 50% invariance were used. To exclude obvious chimeric 16S rDNA primary structures prior to the phylogenetic analysis (29), a separate analysis of the terminal 400-nucleotide sequence for the RSa and RSb clones and of the terminal 200 nucleotides for the RSD clones was carried out.
16S rRNA oligonucleotide probes.
For the genus Clostridium, new oligonucleotide probes were designed using the comparative 16S rRNA sequence analysis program PROBE-DESIGN, which is included in the ARB program package (O. Strunk et al., http://www.biol.chemie.tu-muenchen.de/pub/ARB/, 1997). In addition several 16S rDNA sequences of clostridial clones and isolates described by Hengstmann et al. (20) and Chin et al. (8) were added to the database. The specificity of the probes with regard to the target sequences was checked using ARB tool PROBE-MATCH. All oligonucleotide probes were labeled with one of the following reactive dyes: Texas red, Fluorescein isothiocyanate (FITC), CY3 (Amersham, Zürich, Switzerland), or CY5 (MWG-Biotech, Ebersberg, Germany). The probe sequences and the hybridization conditions are given in Table 1. Probes LGC354A, -B, and -C and IRog1 and IRog2 were used together by mixing equivalent amounts. For each of the newly designed probes the optimal formamide concentration was determined by a formamide gradient of 0 to 50% in hybridizations with fixed cells of positive and negative reference strains (Table 2) as described by Manz et al. (32) to achieve optimal stringency.
TABLE 1.
16S rRNA oligonucleotide probes used for hybridization
| Probe | Specificity | Sequence of probe (5′–3′) | Target sitea | FA (%)b | Reference or source |
|---|---|---|---|---|---|
| Univ1392 | All life | ACGGGCGGTGTGTRC | 16S (1392) | NAe | 47 |
| Eub338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S (338) | 0 | 2 |
| Alphalb | α Proteobacteria | CGTTCGYTCTGAGCCAG | 16S (19) | 20 | 32 |
| Beta42a | β Proteobacteria | GCCTTCCCACTTCGTTT | 23S (1027) | 35 | 32 |
| Gam42A | γ Proteobacteria | GCCTTCCCACATCGTTT | 23S (1027) | 35 | 32 |
| CF319A | CFB phylum | TGGTCCGTGTCTCAGTAC | 16S (319) | 35 | 33 |
| HGC69a | Gram-positive bacteria with high G+C content | TATAGTTACCACCGCGT | 23S (1901) | 25 | 38 |
| LGC344A | Gram-positive bacteia with low G+C content | TGGAAGATTCCCTACTGC | 16S (354) | 20 | 34 |
| LGC354B | Same as LGC354A | CGGAAGATTCCCTACTGC | |||
| LGC354C | Same as LGC354A | CCGAAGATTCCCTACTGC | |||
| SRB385 | δ Proteobacteria | CGGCGTCGCTGCGTCAGG | 16S (385) | 35 | 1 |
| VM338c | Verrucomicrobia | TGCTGCCACCCGTAGGTG | 16S (338) | 50 | This study |
| IRog1 | Acidobacterium-Holophaga phylum | AAGGCGGCATCCTGGACC | 16S (353) | 35 | 31 |
| IRog2 | Acidobacterium-Halophaga phylum | GCAGTGGGGAATTGTTC | 16S (728) | ||
| Clost I | Clostridial cluster I | TTCTTCCTAATCTCTACGCA | 16S (696) | 30 | 26 |
| Clost III | 7 sequences of clostridial cluster III; strains FCB90-1 FCB90-2 FCB45, and RCel1 from rice soild | CCTGTGTTATCCCCCTGT | 16S (140) | 30 | This study |
| Clost IV | Clostridial cluster IV, some members of the genera Eubacterium, Rhodothermus, and Chlorobium | GCACCCTTTACACCC | 16S (565) | 30 | This study |
| Clost XIVa | 25 isolates form cluster XIVa of the genus Clustridum | CTGTATGAGGCAGGT | 16S (129) | 30 | This study |
| Clost XIVa-c | 16S rRNA clones related to cluster XIVa but nto detectable with probe clost XIVa; three isolates of cluster XIVa | CAGCTGTTATCCCCCTGTAT | 16S (138) | 40 | This study |
| Clost XIVa- amino | 3 isolates of cluster XIVa | GTACCATGCGGCACT | 16S (183) | 40 | This study |
TABLE 2.
Reference organisms used as positive and negative control for FISH specificity tests
| Strain or species | Origin | 16S rRNA probe(s)a | FISH controlb |
|---|---|---|---|
| FCB90-2c | Chin et al. (8) | Clost III (0) | + |
| Flexibacter ruber | DSM 9560 | Clost III (1) and Clost IV (1) | − |
| C. sporosphaeroides | DSM 1294 | Clost IV (0) | + |
| C. celerecrescens | DSM 5628 | Clost XIVa (0) | + |
| Clost XIVa-amino (0) | + | ||
| Clost XIVa-c (1) | − | ||
| C. butyricum | DSM 6566 | Clost XIVa (1) | − |
| Clost XIVa-amino (1) | − | ||
| C. polysaccharolyticum | DSM 1801 | Clost XIVa-c (0) | + |
Numbers of mismatches are in parentheses.
+, positive control; −, negative control.
Isolate from rice field soil.
Cultivation and preparation of reference organisms.
The reference strains, used as positive and negative controls for the determination of the stringency of 16S rRNA probes, are shown in Table 2. Strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) GmbH, Braunschweig, Germany. Strain FCB 90-2 was kindly provided by Kuk-Jeong Chin. Strain DSM 9560 was cultivated according to the DSMZ description. The other four strains were anaerobically cultivated at 37°C in a medium containing 2% glucose and 1% yeast extract. For FISH, 1 ml of the culture was fixed with formaldehyde as described above.
FISH.
FISH was performed as described elsewhere (4) with some modifications. Fixed samples (1 to 2 μl) were spotted on Teflon-coated eight-field glass slides, air dried overnight, and subsequently dehydrated in 50, 80, and 96% ethanol (3 min each). Hybridizations were carried out in 10 μl of hybridization buffer per field in a water-saturated equilibration chamber at 46°C for 2.5 h. The hybridization buffer contained 0.9 M NaCl, formamide as given in Table 1, 20 mM Tris-HCl (pH 7.4), 7 mM SDS, and 5.3 mM EDTA. In each experiment bacterial probe Eub338 was used together with one or two specific oligonucleotide probes with a final concentration of 8 pmol per 16S rRNA probe. After hybridization the buffer was rinsed with washing buffer and the slides were washed for 20 min at 48°C in a preheated washing buffer containing 20 mM Tris-HCl (pH 7.4), a probe-dependent concentration of NaCl, 7 mM SDS, and 5.3 mM EDTA. The NaCl concentrations in the washing buffers were 980, 220, 150, 100, and 60 mM for 0, 20, 25, 30, and 35% formamide, respectively. The washing buffer was rinsed by deionized water, and the slides were air dried. The cells were counterstained for 10 min at 22°C with 8 μl of 4′, 6-diamidino-2-phenylindole (DAPI; 0.7 mg liter−1), rinsed with deionized water, and air dried. Before examination the slides were covered with antifading agent Citifluor AF1 (in glycerol-PBS; Chemical Laboratory, University of Canterbury, Canterbury, United Kingdom).
Microscopy and quantification.
Fluorescence was detected with a confocal laser scanning microscope, DMR XE, type TCS NT (Leica, Lasertechnique GmbH, Heidelberg, Germany) using a PL-Fluotar objective (×63; 1.32; numerical aperture, oil immersion). Images were analyzed with Leica software package TCS NT, version 1.5.451. For the dyes, the following excitation wavelengths and filter settings were used: Cy5, 647 nm and LP665; CY3 and Texas red, 568 nm and BP 600/30; FITC, 488 nm and BP 530/30; DAPI, 345 nm and BP 440/40.
For the determination of active bacterial cells (i.e., cells containing a sufficient number of ribosomes [3]), cells that hybridized with the eubacterial probe Eub338 were counted and correlated to the total number of cells, counted by using DAPI. Quantification of cells hybridized with specific probes relative to the number of Eub338-hybridized cells was done. For each probe at least 1,000 DAPI-stained cells were counted, corresponding to at least 600 Eub338-hybridized cells.
RNA extraction and dot blot hybridization.
RNA was extracted from straw and soil samples, and the 16S rRNA content was quantified to determine the efficiency of the stomacher treatment and to investigate the influence of the straw on the 16S rRNA content in the soil when straw and soil were incubated together. Therefore soil and straw were incubated together in soil slurries and in addition soil was incubated without straw for the same time period. After 8 and 29 days of incubation, RNA was extracted from the following five fractions: straw incubated with soil, washed twice in PBS, and treated with a stomacher as described above (total RNA after stomacher treatment) (I); straw incubated with soil and washed twice in PBS but left untreated (total RNA before stomacher treatment) (II); straw incubated with soil and treated with a stomacher, with the PBS containing the detached microbial cells collected as described above, concentrated, and used for RNA extraction (PBS-cell extract) (III); soil used for straw incubation (IV); and the soil incubated without straw (V). The RNA extraction was carried out as described previously (30).
Dot blot hybridization was carried out as described by Manz et al. (33) with modifications. RNA was denatured by adding 1 volume of 20× SSC (3 M NaCl plus 0.3 M sodium-citrate, pH 7.0)–formaldehyde (37%) at a ratio of 3:2. After 15 min at 65°C, samples were transferred to a positively charged nylon membrane (Amersham Pharmacia Biotech Europe GmbH, Buckinghamshire, United Kingdom) using a Bio-Dot SF blotter (Bio-Rad Laboratories GmbH). Nucleic acids were immobilized by using twice the “auto-cross-link” function (120 mJ) of a UV Stratalinker 2400 (Stratagene, La Jolla, Calif.). Membranes were prehybridized for 1 h at 38°C with 10 ml of a solution containing 5× SSC, 1% blocking solution (Roche Diagnostics GmbH, Mannheim, Germany), 0.1% N-lauroylsarcosine and 0.02% SDS. Hybridizations were performed at 38°C overnight by adding to a 4-ml prehybridization solution 120 pmol of digoxigenin (DIG)-labeled oligonucleotide probe Univ 1392 (Table 1). After hybridization the membranes were washed four times (5 min, 38°C) with 20 ml of washing solution (5× SSC, 0.1% SDS). Nonspecific binding was blocked by incubation of the membranes with 10 ml of 1% blocking solution in a buffer containing 0.1 M maleic acid and 0.15 M NaCl (pH 7.5). The DIG-labeled probes were detected by using anti-DIG antibodies (diluted 1:1,000 [vol/vol] in 1% blocking solution) coupled with an alkaline phosphatase (Roche Diagnostics GmbH) as described by the manufacturer. The signals were detected after addition of substrate ECF (Amersham Pharmacia Biotech Europe GmbH) by a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Quantification of the hybridization signal was done with the program ImageQuant, version 5.0 (Molecular Dynamics) with standard curves of dilution series of rRNA from E. coli (Roche Diagnostics GmbH) on each membrane.
Nucleotide sequence accession numbers.
The sequences were submitted to the EMBL database under the following accession numbers: RSa17, -20, -31, -32, -35, -40, and -48, AJ289201 to AJ289207, respectively; RSb3, -6, -8, -12, -16, -24, -33, -40, and -47, AJ289208 to AJ289216, respectively; RSD1 to RSD15, AJ289217 to AJ289231, respectively.
RESULTS
Community pattern of rice straw after different incubation times.
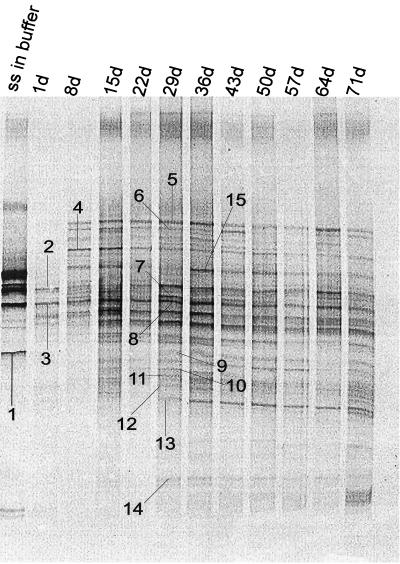
Rice straw was incubated anaerobically in soil slurries for up to 71 days. Methane production started after 8 days and reached quasisteady state after 20 days of incubation (data not shown), similar to what was reported by Glissmann and Conrad (15). Samples of rice straw for molecular investigations were taken after 1 day of incubation and then again every week. As an additional sample, straw was anaerobically incubated for 2 months in phosphate buffer without soil. These samples were used for DNA extraction, followed by amplification of 16S rDNA and DGGE. The DGGE patterns of these samples are shown in Fig. 1. The DGGE pattern of the straw in buffer showed only few bands. The DGGE of straw incubated in soil for only 1 day showed a different pattern but also only a few bands. During longer incubation of straw in soil the number of DGGE bands increased. After 15 days of incubation, the DGGE patterns became similar with respect to the number, migration, and intensity of single bands, and only few differences were observed. Band 6, for example, showed a relatively low intensity and disappeared at 50 and 57 days. Band 13 appeared after 29 days and then increased in intensity (Fig. 1).
FIG. 1.
DGGE patterns of the 16S rDNA fragments of straw samples incubated in soil slurries for different times (1 to 71 days; lanes 2 to 12 [starting from the left]) and from one straw sample (ss) incubated in phosphate buffer for 2 months (lane 1). DGGE bands 1 to 15 were excised from the gels, and the DNA was extracted, amplified, and cloned. Clones with the same electrophoretic mobility as that of the original band were sequenced.
Phylogenetic placement of 16S rDNA sequences retrieved from DGGE bands.
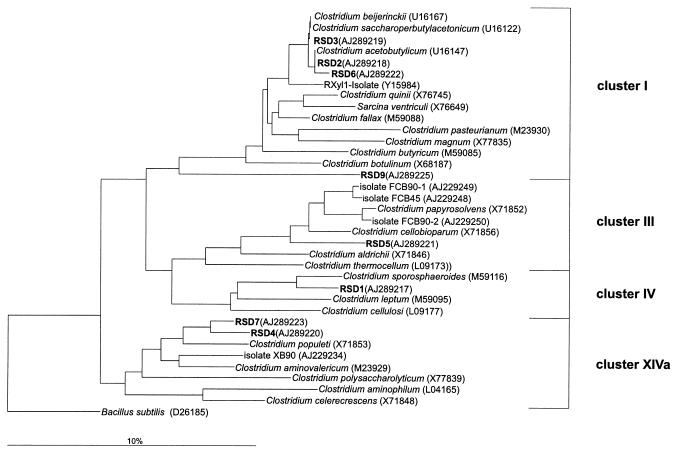
From the DGGE gel (Fig. 1) 15 bands with different signal intensities and from different parts of the gel were arbitrarily selected, excised, amplified, and cloned. After cloning, the mobilities of randomly selected clones from each clone library were checked via DGGE and compared with the original DGGE pattern. Clones that matched the positions of bands of the original DGGE pattern were sequenced (RSD clones). Eight of the 15 sequences clustered with the genus Clostridium, three grouped with clone sequences previously obtained from rice field soil (20), and four clone sequences were distantly related to other bacteria. The dendrogram of Fig. 2 shows the relationship of the RSD clones with the genus Clostridium.
FIG. 2.
Phylogenetic dendrogram constructed with partial (about 400 bp) 16S rDNA sequences showing the relationship of RSD clones from DGGE gels to members of the genus Clostridium (sensu Collins et al. [9]). The tree was constructed with the neighbor-joining method of the ARB program package and the Jukes-Cantor correction. To omit highly variable regions within the 16S rRNA, a filter with 50% invariance for the genus Clostridium was used. Scale bar, 10% estimated difference in nucleotide sequence position. As the outgroup, Bacillus subtilis was used.
The 16S rDNA sequences of clones RSD2, -3, -6, and -9 (representing DGGE bands 2, 3, 6, and 9, respectively) belonged to clostridial cluster I (sensu Collins et al. [9]). Clones RSD2, -3, and -6 were nearly identical (pairwise identity levels were >99%) and grouped very closely with Clostridium acetobutylicum. The 16S rDNA sequences of clones RSD1 and RSD5 grouped within clostridial clusters IV and III, respectively. Both sequences had a rather low identity (93%) to Clostridium sporosphaeroides and Clostridium papyrosolvens of these clusters. Noteworthy is the comparatively close relation of clone RSD5 to isolates FCB90-1 and FCB90-2, originating from rice field soil (8, 20). Clones RSD4 and RSD7 grouped within clostridial cluster XIVa. Both clones were distantly related to rice field isolate XB90 (8, 20). The two clones showed an identity of 96% to each other.
Clones RSD10, RSD12, and RSD14 grouped together (95% identity) with clones that had previously been obtained from rice field soil, i.e., clone RSD10 grouped with BSV40 (AJ229196), and clones RSD12 and RSD14 grouped with BSV81 (AJ229225), but did not show a clear phylogenetic affiliation. Clones RSD8 and RSD15 also showed only a distant relation to a known species, i.e., a Bacillus sp. (AB030930) and Fluoribacter bozemanii (M36031), with 80 and 77% identities, respectively. The same was true for clones RSD11 and -13, which were distantly related to Acidobacterium capsulatum (D26171).
Phylogenetic placement of 16S rDNA sequences retrieved from clone libraries.
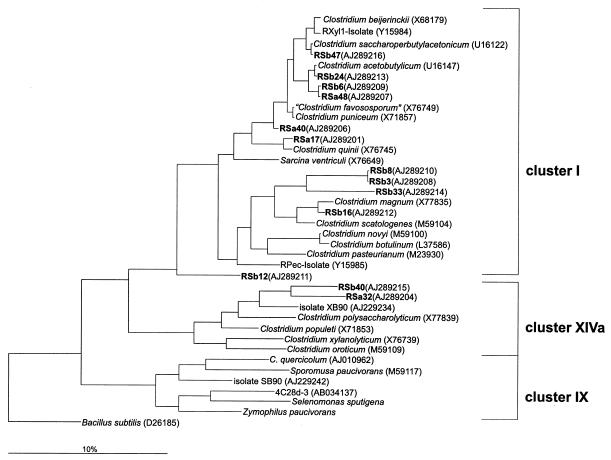
In a parallel approach two clone libraries were generated from DNA extracted from the incubated straw samples to survey a broader spectrum of organisms involved in the degradation of rice straw. One library was from rice straw incubated for 1 day (RSa clones) in anoxic rice soil to investigate the situation at the beginning of the degradation process. The other gene library was generated from rice straw that was incubated for 29 days (RSb clones) to see possible changes in community structure after prolonged incubation. From each library the motilities of 20 randomly chosen clones were compared by DGGE. Sixteen of these clones exhibited different electrophoretic mobilities and were subsequently sequenced. The phylogenetic dendrogram of Fig. 3 shows the relationship of the RSa and RSb clones with the genus Clostridium. The phylogenetic placement of the different 16S rRNA gene fragments again showed that Clostridium was the most common genus present in the clone libraries. Thirteen of the 16 sequenced clones grouped within one of the different clostridial clusters.
FIG. 3.
Phylogenetic dendrogram constructed with 16S rDNA sequences (about 750 bp) showing the relationship of RSa and RSb clones to members of the genus Clostridium (sensu Collins et al. [9]). The tree was constructed with the neighbor-joining method of the ARB program package and the Jukes-Cantor correction. To omit highly variable regions within the 16S rRNA, a filter with 50% invariance for the genus Clostridium was used. Scale bar, 10% estimated difference in nucleotide sequence position. As the outgroup, Bacillus subtilis was used.
Ten of these clones (RSa17, RSa40, and RSa48 and RSb3, RSb6, RSb8, RSb16, RSb24, RSb33, and RSb47) were related to clostridial cluster I. Seven of the ten clones (RSa17, RSa40, RSa48, RSb6, RSb16, RSb24, and RSb47) had pairwise similarities of over 97% to known clostridial species (Fig. 3) and thus may represent members of them (46). Almost-identical clones RSb3 and RSb8 (>99.8% similarity) and clone RSb33 formed a distinct lineage within clostridial cluster I. The next relative to RSb3 and RSb8 was Clostridium quinii, and the next relative to RSb33 was Clostridium scatologenes. Two of the clones (RSa32 and RSb40) were distantly related (91 to 94%) to members of clostridial cluster XIVa, including rice field isolate XB90 (8). One clone (RSb12) belonged to the clostridia but could not be affiliated definitely to one of the clostridial clusters. The two most similar bacteria were isolate SB90, which had been obtained from rice field soil and placed into clostridial cluster IX (8), and C. acetobutylicum, belonging to the clostridial cluster I.
Three of the clones (RSa20, RSa31, and RSa35) did not belong to any of the clostridial clusters and were related to “Bacillus pseudomegaterium” (X77791; 97% identity) and a Nitrosospira sp. (X90820; 88% identity).
Quantification of the bacterial community on straw and in soil.
For the quantification of various bacterial groups degrading rice straw FISH was performed with straw samples incubated for 8 and 29 days. These samples were treated with a stomacher to detach the microbial cells from the straw, resulting in a concentrated PBS-cell extract. The efficiency of the cell extraction by stomacher treatment was tested by dot blot hybridization (Table 3). The total amounts of 16S rRNA in treated and untreated straw samples were determined by using probe Univ 1392 targeting all life (Table 1). The rRNA content of straw was much lower in the stomacher-treated (I) than in the untreated samples (II), amounting to a residual rRNA content of only 6.8 (day-8 sample) and 18.9% (day-29 sample) of the control. This corresponds to extraction efficiencies of 93.2% for the day-8 sample and 81.1% for the day-29 sample. The rRNA content of the concentrated PBS-cell extract (III) was fairly comparable to that in the untreated sample (II) minus that in the stomacher-treated straw (I), with a recovery of 50 to 150% (Table 3). In addition, the influence of straw on the rRNA content in the soil was investigated. The rRNA content in soil (V) was lower than that in untreated straw (II) and was also lower than the rRNA content of soil that had been incubated with straw (IV) (Table 3).
TABLE 3.
Quantification of the 16S rRNA content of straw and soil samples incubated for 8 and 29 days by dot blot hybridization with probe Univ1392
| Incubation time (days) | 16S rRNA content (molecules/g of wet straw or soil) in samplea:
|
||||
|---|---|---|---|---|---|
| I | II | III | IV | V | |
| 8 | 5.3 × 1010 | 7.8 × 1011 | 1.2 × 1012 | 2.4 × 1011 | 1.3 × 1011 |
| 29 | 3.6 × 1011 | 1.9 × 1012 | 7.8 × 1011 | 1.5 × 1011 | 7.4 × 1010 |
Samples are as defined in Materials and Methods.
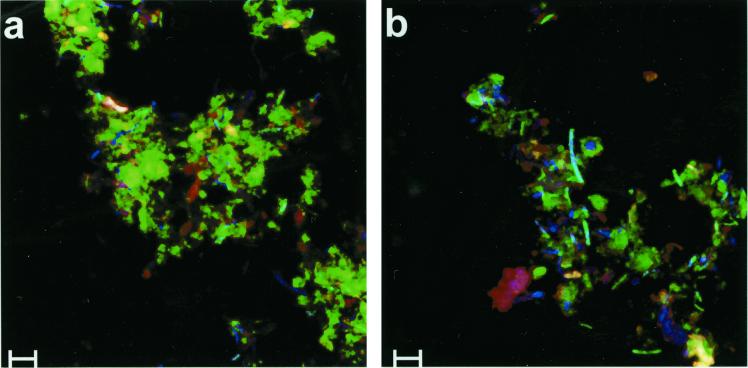
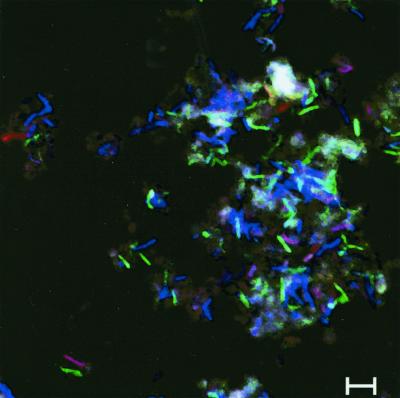
The abundances of different phylotypes of the bacterial populations colonizing the incubated straw were quantified by FISH. The PBS-cell extract was fixed and hybridized with different fluorescence-labeled 16S rRNA probes. In this cell extract the amount of straw particles was relatively low, and therefore the autofluorescence of the straw was not so disturbing. Nevertheless, the remaining straw in the suspension had a high autofluorescence, concealing cells with a relatively low hybridization signal. Therefore, the PBS-cell extract was treated with toluidine blue to reduce this autofluorescence. The result of this treatment is shown in Fig 4. Without toluidine blue treatment the autofluorescence, especially in the green channel, was rather high (Fig. 4a) but was clearly reduced after treatment (Fig. 4b). This improvement made quantification of bacterial cells easier and therefore improved the method significantly.
FIG. 4.
FISH of fixed cells detached mechanically from straw incubated in soil slurries without (a) and with (b) prior treatment by toluidine blue. Two 16S rDNA probes were used simultaneously: CY5-labeled bacterial-domain probe Eub338 (blue) and FITC-labeled probe Clost I (green). One sample (b) was additionally treated with toluidine blue to reduce autofluorescence, especially in the green channel. Scale bars, 5 μm.
The total numbers of microbial cells (Bacteria and Archaea) were determined by DAPI staining. The percentage of active bacterial cells was determined by using probe Eub338, which detects almost all Bacteria (Table 1). In the day-8 sample, 61% ± 16% (mean ± standard deviation) of the DAPI-stained bacteria could be detected with Eub338, whereas only 17% ± 7% were active in the day-29 sample. Treatment of the samples with lysozyme, as suggested by Amann et al. (3), showed no increase in the percentage of Eub338-stained cells (result not shown).
The different phylogenetic bacterial groups were quantified as percentages of the Eub338-stained cells by using different 16S rRNA probes, which were specific for particular phylotypes (Table 1). For the genus Clostridium, a total of five different 16S rRNA probes were used. Some of the probes were newly designed according to the obtained sequences from the clone libraries. Probe Clost I, which had been designed for clostridial cluster I (26), showed no mismatch to 106 sequences belonging to clostridial cluster I. Two clostridia of cluster II available in the ARB database (version April 97) also showed no mismatch with probe Clost I. Probe Clost III was designed for rice field isolates FCB 90-1, FCB 90-2, FCB 45 (8), and RCel1 (7), which belong to clostridial cluster III. Probe Clost III also had no mismatch to seven further clostridial cluster III sequences from the ARB database. Probe Clost IV was designed to detect all clostridia in cluster IV but also showed sequence identity to some bacteria within the genera Eubacterium, Chlorobium, and Rhodothermus. For cluster XIVa three probes were designed. Probe Clost XIVa showed no mismatch to 25 sequences of clostridial cluster XIVa available in the ARB database, not including clones RSa32 and RSb40. These two clones, which were distinctly related to rice field isolate XB90, were detected with probe Clost XIVa-c, which also detect Clostridium polysaccharolyticum, Clostridium herbivorans, and Clostridium aminovalericum. Species C. polysaccharolyticum and C. herbivorans were also detectable by probe Clost XIVa. Probe Clost XIVa-amino was designed for C. aminovalericum but also detected Clostridium sphenoides and Clostridium celerecrescens.
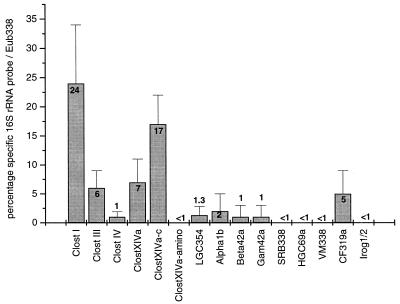
In the straw sample that was incubated for 8 days, the following phylogenetic groups of the domain Bacteria were detected (Fig. 5): the gram-positive bacteria with low G+C-content, including the genera Clostridium and Bacillus, the α, β, and γ subdivisions of the division Proteobacteria, and the CFB group. The gram-positive bacteria with high G+C-content, the δ Proteobacteria, the division Verrucomicrobia, and the Holophaga-Acidobacterium group were not found. A total of 61% of the Eub338-stained cells were detected by phylogenetically more-specific 16S rRNA probes (Fig. 5). Most (about 55%) of these cells belonged to the genus Clostridium, mainly clostridial cluster I and clostridial cluster XIVa (24% each). Double hybridization of single cells with probes Clost XIVa plus Clost XIVa-c should result in the detection of clostridia related to C. polysaccharolyticum and C. herbivorans. However, these species made up only approximately 1% of all Eub338-hybridized cells. In Fig. 6 these two probes together with probe Eub338 were used, but no white cells (overlay of Eub338, Clost XIVa, and Clost XIVa-c) were visible. Next to clostridia, the Cytophaga-Flavobacterium cluster of the CFB phylum, detected with probe CF319a, made up 5% of the active bacteria (Fig. 5).
FIG. 5.
Quantification by FISH of the composition of the bacterial community on rice straw incubated for 8 days in rice field soil. The bacterial populations were detached from the straw, fixed with formaldehyde, and hybridized with different group-specific 16S rRNA probes (Table 1). For each group-specific probe about 600 Eub338-marked cells were counted. The percentage of the specific signal was calculated in relation to the number of Eub338-stained cells (mean ± standard deviation; n ≥ 10).
FIG. 6.
FISH of fixed cells detached mechanically from straw incubated for 8 days in soil slurries. Three 16S rDNA probes were used simultaneously. CY5-labeled bacterial-domain probe Eub338 (blue) was used together with CY3-labeled probe Clost XIVa (red), specific for parts of clostridial cluster XIVa, and FITC-labeled probe Clost XIVa-c (green), designed from 16S rDNA sequence data (clones RSa32 and RSb40). The overlay of probes Eub338 and Clost XIVa resulted in pink cells; the overlay of Eub338 and Clost XIVa-c resulted in green-to-turquoise cells. The number of cells hybridized simultaneously with the group-specific probes Clost XIVa and Clost XIVa-c (resulting in white cells) was less then 1% of all Eub338-detected cells. Scale bars, 5 μm.
Because of the low percentage of active cells in the straw samples incubated for 29 days, a quantification of different phylogenetic bacterial groups by FISH was not possible. Therefore, the presence of different phylogenetic bacterial groups was only qualitatively assessed. Values for most of the phylogenetic groups appeared to be in the same range as for the 8-day incubation but were lower (<2%) for the CFB phylum, clostridial cluster I, and the bacteria detected with probe Clost XIVa-c.
DISCUSSION
In parallel to a process-oriented study (15) we have determined the community structure of Bacteria that colonize decomposing rice straw by extraction, amplification, separation, and sequencing of bacterial 16S rDNA by DGGE of the 16S rDNA and by direct hybridization of the bacteria detached from rice straw. Cloning and sequencing as well as DGGE were based on PCR and thus may be biased, e.g., by the preferred amplification of special sequences (51). Therefore, quantitative interpretation of these data should be done with care. However, the direct hybridization of bacteria avoids possible PCR biases and allows quantitative interpretation. Our study presents the first quantification of the dominant bacterial phylotypes in such an environment.
All the approaches (DGGE, cloning, and FISH) consistently showed that the bacterial community on decomposing rice straw mainly consisted of the genus Clostridium. This genus is a physiologically heterogeneous group of anaerobic, gram-positive, endospore-forming bacteria. Many steps of the complex decomposition pathway of straw should be achieved by one of the various species of clostridia. It is known that clostridial cluster III consists of eight cellulolytic species possessing enzymes for hydrolysis of straw biopolymers such as cellulose and hemicellulose. For Clostridium thermocellum, for example, it was shown that a multiprotein complex, the cellulosome, is secreted into the medium (14). Sequence data and hybridization experiments showed that members of clostridial cluster III were indeed involved in the degradation process of complete rice straw. However, FISH detected only 6% of the active bacterial cells with probe Clost III, indicating a limited contribution of members of cluster III to the decomposition of rice straw.
Cellulolytic clostridial species are also found in other clostridial clusters. Cluster XIVa largely consists of species utilizing carbohydrates, some of which are also able to ferment polysaccharides. Clostridium populeti, for example, has the ability to ferment cellulose, hemicellulose, and pectin (21). Strain XB90, which was isolated from rice field soil, is able to utilize xylan and pectin as well as other carbohydrates (8). Three of our clones were related to C. populeti and strain XB90 (8). Hybridization experiments with three differently specific probes for cluster XIVa showed that a high percentage (24%) of the active bacteria belonged to this cluster. Therefore, we assume that cluster XIVa and especially the two strains represented by clones RSa32 and RSb40 were actively involved in the degradation of rice straw carbohydrates.
After the hydrolysis of polysaccharides, the monosaccharides are fermented. During the first 15 days of incubation, rice straw was found to be mainly degraded to H2, CO2, acetate, propionate, butyrate, and caproate. Later on, acetate and propionate were the major fermentation products (15). The DGGE patterns of straw samples (Fig. 1) also showed the most-pronounced changes during the first 15 days of incubation. From 15 to 71 days, on the other hand, the bacterial community on the degrading rice straw exhibited a more stabile DGGE pattern. Possible candidates for the fermentation of straw hydrolysis products are clostridia from cluster I. We obtained 14 clones that were placed into clostridial cluster I, 10 of which had similarities of over 98% to described cluster I species. Because of the high similarity the bacteria represented by the clone sequences may operate similar metabolic pathways. C. quinii (49), C. acetobutylicum (25), and Clostridium magnum (41), for example, ferment carbohydrates to various fatty acids. C. magnum and C. scatologenes (27) are able to perform homoacetogenic fermentation. The quantification with FISH showed that clostridia from cluster I, accounting for 24% of all eubacterial cells, were very active during the rice straw-degrading process.
Only a few clone sequences belonged to phylogenetic groups other than clostridia, i.e., to the genera Bacillus, Nitrosospira (β Proteobacteria), Fluoribacter (γ Proteobacteria), and Acidobacterium (Holophaga-Acidobacterium phylum). FISH also detected only a few (10%) bacteria besides clostridia. These results suggest that all these phylotypes were of minor importance for the degradation of rice straw. Many phylogenetic groups, such as the division Verrucomicrobia, the class Actinobacteria, and the family Chlorobiaceae, were not detected at all, despite their abundance in bulk rice field soil (8, 20, 30). Therefore, the bacterial community colonizing and decomposing rice straw seems to be different from the community found in the bulk soil. This conclusion is in agreement with the process studies of Glissmann and Conrad (15), who reported that the fermentation pattern during the first 15 days of straw incubation was qualitatively different from the fermentation pattern of the organic matter in nonamended rice field soil.
With FISH on 8-day-old straw incubations, 61% of the total microorganisms (stained with DAPI) were also detectable with bacterial probe Eub338. This value decreased to 17% after an incubation time of 29 days. FISH is known to detect only cells with a sufficient number of ribosomes (3). The percentage of active cells apparently decreased during incubation of rice straw. This conclusion does not imply, however, that the total number of bacteria inhabiting the rice straw also decreased. The opposite is probably true, as scanning electron microscopy of degrading rice straw shows that the colonization of decomposing rice straw increases with time (24). In addition, dot blot hybridization experiments showed an increasing amount of 16S rRNA on rice straw after longer incubation. Results obtained by DGGE indicate that the composition of the bacterial community on rice straw was relatively stable after 15 days of incubation. Therefore, we assume that the colonization process is mainly driven by multiplication of the initial colonizers and that cells lose activity with age so that they are no longer detectable by FISH.
About 65% of the active cells detected with probe Eub338 were also recovered with group-specific probes. Only 35% of the active bacteria could not been detected and thus could not be affiliated to a particular phylotype. It should be noted that the 16S rRNA probes used do not cover all possible phylotypes within the domain Bacteria. Probe CF319a, for example, detects only the Cytophaga-Flavobacterium cluster of the CFB phylum, not the Bacteroides. Furthermore, the genus Clostridium is not completely covered by the probes used. Only 4 of the 19 clostridial clusters are partially detectable with the probes presently available. Therefore, it is possible that many of the active bacteria (i.e., those stained by Eub388) were related to other clusters of clostridia but were not detected.
In conclusion, the anaerobic degradation process of rice straw in paddy soil was dominated by the genus Clostridium, especially by clostridial clusters I, III, and XIVa within the domain Bacteria. Other groups such as the Proteobacteria, the genus Bacillus, and the CFB phylum were present but not very abundant. The bacterial community colonizing and degrading rice straw was established during the first 15 days of incubation and was moderately different from the community found in bulk rice field soil without the application of rice straw.
ACKNOWLEDGMENT
This study was part of the Sonderforschungsbereich 395 of the Deutsche Forschungsgemeinschaft “Interaction, adaptation and catalytic capacity of terrestrial microorganisms.”
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chrisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence J R, Hartmann A. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Chidthaisong A, Inubushi K, Muramatsu Y, Watanabe I. Production potential and emission of methane in flooded rice soil microcosms after continuous application of straw. Microbes Environ. 1996;11:73–78. [Google Scholar]
- 7.Chin K J, Rainey F A, Janssen P H, Conrad R. Methanogenic degradation of polysaccharides and the characterization of polysaccharolytic clostridia from anoxic rice field soil. Syst Appl Microbiol. 1998;21:185–200. [Google Scholar]
- 8.Chin K-J, Hahn D, Hengstmann U, Liesack W, Janssen P H. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl Environ Microbiol. 1999;65:5042–5049. doi: 10.1128/aem.65.11.5042-5049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 10.Conrad R. Mechanism controlling methane emission from wetland rice fields. In: Oremland R S, editor. The biogeochemistry of global change: radiative trace gases. New York, N.Y: Chapman & Hall; 1993. pp. 317–335. [Google Scholar]
- 11.Daims H, Brühl A, Amann R, Schleifer K H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 12.Denier van der Gon H, Neue H U. Influence of organic matter incorporation on the methane emission from a wetland rice field. Global Biogeochem Cycles. 1995;9:11–22. [Google Scholar]
- 13.Engelen B, Meinken K, von Wintzingerode F, Heuer H, Malkomes H-P, Backhaus H. Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl Environ Microbiol. 1998;64:2814–2821. doi: 10.1128/aem.64.8.2814-2821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felix C, Ljungdahl L G. The cellulosome: the exocellular organelle of Clostridium thermocellum. Annu Rev Microbiol. 1993;47:791–819. doi: 10.1146/annurev.mi.47.100193.004043. [DOI] [PubMed] [Google Scholar]
- 15.Glissmann K, Conrad R. Fermentation pattern of methanogenic degradation of rice straw in anoxic paddy soil. FEMS Microbiol Ecol. 2000;31:117–126. doi: 10.1111/j.1574-6941.2000.tb00677.x. [DOI] [PubMed] [Google Scholar]
- 16.Grant R F. Simulation of methanogenesis in the mathematical model ECOSYS. Soil Biol Biochem. 1998;30:883–896. [Google Scholar]
- 17.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Großkopf R, Stubner S, Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1998;64:4983–4989. doi: 10.1128/aem.64.12.4983-4989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengstmann U, Chin K J, Janssen P J, Liesack W. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundent culturable bacteria from an anoxic rice paddy soil. Appl Environ Microbiol. 1999;65:5050–5058. doi: 10.1128/aem.65.11.5050-5058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hippe H, Andreesen J R, Gottschalk G. The genus Clostridium—nonmedical. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer; 1992. pp. 1800–1866. [Google Scholar]
- 22.Holzapfel-Pschorn A, Conrad R, Seiler W. Effects of vegetation on the emission of methane from submerged paddy soil. Plant Soil. 1986;92:223–233. [Google Scholar]
- 23.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 24.Kimura M, Tun C C. Microscopic observation of the decomposition process of leaf sheath of rice straw and colonizing microorganisms during the cultivation period of paddy rice. Soil Sci Plant Nutr. 1999;45:427–437. [Google Scholar]
- 25.Kreis S, Bennett C F, Ward V K, Jones D T. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int J Syst Bacteriol. 1995;45:693–705. doi: 10.1099/00207713-45-4-693. [DOI] [PubMed] [Google Scholar]
- 26.Küsel K, Pinkart H C, Drake H L, Devereux R. Acetogenic and sulfate-reducing-bacteria inhabiting the rhizoplane and deep cortex cells of the sea grass Halodule wrightii. Appl Environ Microbiol. 1999;65:5117–5123. doi: 10.1128/aem.65.11.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Küsel K, Dorsch T, Acker G, Stackebrandt E, Drake H L. Clostridium scatologenes strain SL1 isolated as an acetogenic bacterium from acidic sediments. Int J Syst Environ Microbiol. 2000;50:537–546. doi: 10.1099/00207713-50-2-537. [DOI] [PubMed] [Google Scholar]
- 28.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 29.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 30.Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl Environ Microbiol. 2000;66:754–762. doi: 10.1128/aem.66.2.754-762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 32.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 33.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. System Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 34.Meier H, Amann R, Ludwig W, Schleifer K H. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 35.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rath A K, Mohanty S R, Mishra S, Kumaraswamy S, Ramakrishnan B, Sethunathan N. Methane production in unamended and rice-straw-amended soil at different moisture levels. Biol Fertil Soils. 1999;28:145–149. [Google Scholar]
- 37.Reichardt W, Mascarina G, Padre B, Doll J. Microbial communities of continuously cropped, irrigated rice fields. Appl Environ Microbiol. 1997;63:233–238. doi: 10.1128/aem.63.1.233-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K H. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA targeted oligonucleotides. Microbiology & (Reading) 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 39.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Schink B. Clostridium magnum sp. non., a non-autotrophic homoacetogenic bacterium. Arch Microbiol. 1984;137:250–255. [Google Scholar]
- 42.Schütz H, Holzapfel-Pschorn A, Conrad R, Rennenberg H, Seiler W. A 3-year continuous record on the influence of daytime, season, and fertilizer treatment on methane emission rates from an Italian rice paddy. J Geophys Res. 1989;94:16405–16416. [Google Scholar]
- 43.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 45.Smith M M, McCully M E. Enhancing aniline blue fluorescent staining of cell wall structures. Stain Technol. 1978;53:79–85. doi: 10.3109/10520297809111446. [DOI] [PubMed] [Google Scholar]
- 46.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 47.Stahl D A, Fletsher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stams A J M. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Leeuwenhoek. 1994;66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 49.Svensson B H, Dubourguier H-C, Prensier G, Zehnder A J B. Clostridium quinii sp. nov., a new saccharolytic anaerobic bacterium isolated from granular sludge. Arch Microbiol. 1992;157:97–103. [Google Scholar]
- 50.van Breemen N, Feijtel T C J. Soil processes and properties involved in the production of greenhouse gases, with special relevance to soil taxonomic systems. In: Bouwman A F, editor. Soils and the greenhouse effect. New York, N.Y: John Wiley & Sons; 1990. pp. 195–224. [Google Scholar]
- 51.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe A, Katoh K, Kimura M. Effect of rice straw application of CH4-emission from paddy fields. 2. Contribution of organic constituents in rice straw. Soil Sci Plant Nutr. 1993;39:707–712. [Google Scholar]
- 53.Watanabe A, Takeda T, Kimura M. Evaluation of origins of CH4 carbon emitted from rice paddies. J Geophys Res. 1999;104:23623–23629. [Google Scholar]
- 54.Weisburg W G, Oyaizu Y, Oyaizu H, Woese C R. Natural relationship between Bacteroides and Flavobacteria. J Bacteriol. 1985;164:230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]